Abstract
Background
Despite the success of immune checkpoint blockade therapy in the treatment of certain cancer types, only a small percentage of patients with solid malignancies achieve a durable response. Consequently, there is a need to develop novel approaches that could overcome mechanisms of tumor resistance to checkpoint inhibition. Emerging evidence has implicated the phenomenon of cancer plasticity or acquisition of mesenchymal features by epithelial tumor cells, as an immune resistance mechanism.
Methods
Two soluble factors that mediate tumor cell plasticity in the context of epithelial-mesenchymal transition are interleukin 8 (IL-8) and transforming growth factor beta (TGF-β). In an attempt to overcome escape mechanisms mediated by these cytokines, here we investigated the use of a small molecule inhibitor of the IL-8 receptors CXCR1/2, and a bifunctional agent that simultaneously blocks programmed death ligand 1 (PD-L1) and traps soluble TGF-β.
Results
We demonstrate that simultaneous inhibition of CXCR1/2, TGF-β, and PD-L1 signaling synergizes to reduce mesenchymal tumor features in murine models of breast and lung cancer, and to markedly increase expression of tumor epithelial E-cadherin while reducing infiltration with suppressive granulocytic myeloid-derived suppressor cells, significantly enhancing T-cell infiltration and activation in tumors, and leading to improved antitumor activity.
Conclusions
This study highlights the potential benefit of combined blockade of CXCR1/2 and TGF-β signaling for modulation of tumor plasticity and potential enhancement of tumor responses to PD-L1 blockade. The data provide rationale for the evaluation of this novel approach in the clinic.
Keywords: immunology, oncology
Background
Breakthrough immunotherapies of antagonist monoclonal antibodies directed against the checkpoint programmed cell death protein 1 (PD-1) or its ligand programmed death ligand 1 (PD-L1) have revolutionized the treatment of many cancer types, yet as monotherapies these drugs result in durable clinical responses in only 20%–30% of patients with solid malignancies.1–5 A novel mechanism that has been implicated in primary resistance to checkpoint inhibition, i.e., in patients who never respond, is the phenomenon of tumor cell plasticity induced in the context of an epithelial-mesenchymal transition (EMT). This process allows epithelial cells to lose cell-to-cell contacts and apicobasal polarity while gaining mesenchymal protein expression, motility and invasiveness.6–8 Overall, carcinomas with molecular features of EMT have been previously associated with worse overall survival9–11 and more recently, a transcriptomic signature enriched in genes involved in EMT was found to be associated with primary resistance to anti-PD-1 therapy in patients with metastatic melanoma.12
Extensive work from our laboratory and others has identified soluble factors secreted by tumor cells as key drivers of cancer cell plasticity in the context of EMT.13–17 We have shown that autocrine interleukin 8 (IL-8) secretion by carcinoma cells induces their acquisition of mesenchymal features. Furthermore, IL-8, together with other chemokines of the CXC family (CXCL1-3, CXCL5–7) that bind to the IL-8 receptors CXCR1 and CXCR2, can drive an immune suppressive tumor microenvironment (TME) by chemoattracting granulocytic myeloid-derived suppressor cells (G-MDSC) to the tumor.18–20 Transforming growth factor beta (TGF)-β is another factor that has been identified as a key driver of tumor plasticity and a recent study has linked a TGF-β-associated mesenchymal gene signature with T-cell exclusion from tumors, with consequential lack of response to atezolizumab in patients with urothelial carcinoma.21 In addition, TGF-β promotes tumor progression through angiogenesis together with T-cell and natural killer (NK)-cell suppression and promotion of Tregs.22–25
Here we hypothesized that reducing tumor plasticity mediated by EMT could lead to enhanced antitumor activity of the PD-1/PD-L1 axis blockade. We postulated that such a goal could be achieved by a combination of IL-8 receptor blockade together with TGF-β sequestration. SX-682, a clinical stage, small molecule inhibitor that allosterically binds to the intracellular domain of CXCR1/2 and irreversibly inhibits downstream signaling, provided IL-8 signal inhibition. TGF-β sequestration in the TME was coupled with PD-L1 blockade via the use of bintrafusp alfa (M7824), a bifunctional anti-PD-L1/TGF-βRII agent that simultaneously blocks PD-L1 while trapping soluble TGF-β via two TGF-βRII molecules. Our data demonstrate that combination of CXCR1/2 blockade, TGF-β trapping and PD-L1 inhibition induces marked changes in the tumor phenotype, promoting a reduction of mesenchymal markers and an increase of epithelial E-cadherin in the tumor cells, and significantly enhances tumor infiltration with CD4+ and CD8+ T cells, leading to improved antitumor activity in murine models of breast and lung cancer. These results provide rationale for future evaluation of this multimodal therapy in the clinic.
Methods
Cell lines
Human cell lines MDA-MB-231, BALB/c-derived 4T1 cells, and C57BL/6-derived Lewis lung carcinoma (LLC) were obtained and cultured as recommended by the American Type Culture Collection (Manassas, VA). Cell lines were determined to be mycoplasma free by using a MycoAlert Mycoplasma Detection Kit (Lonza, Basel, Switzerland) and used at low passage number from the date of acquisition.
Mice
Four to six-week-old female BALB/c, C57BL/6, and NSG mice were obtained from the NCI Frederick Cancer Research Facility. Mice were maintained under pathogen-free conditions in accordance with the Association for Assessment and Accreditation of Laboratory Animal Care guidelines.
Tumor inoculation, treatment, and metastasis assay
BALB/c mice were inoculated in the abdominal mammary fat pad with 3×104 4T1 cells; C57BL/6 mice were inoculated subcutaneously in the flank with 5×105 LLC cells. Control diet feed or SX-682 feed (1428.5 mg/kg, equivalent to a dose of 200 mg/kg body weight/day; Research Diets, New Brunswick, NJ) was administered to mice starting on day 7. SX-682 was obtained under a Cooperative Research and Development Agreement with Syntrix Pharmaceuticals. In some experiments, intraperitoneal injections of bintrafusp alfa (492 µg per mouse; obtained under a Cooperative Research and Development Agreement with EMD Serono) were given on days 9 and 11. NSG mice were inoculated subcutaneously in the flank with 4×106 MDA-MB-231 mammary carcinoma cells. Control diet feed or SX-682 feed (500 mg/kg body weight/day; Research Diets) was administered to mice starting on day 7. In all experiments, tumors were measured with a Vernier caliper every 2–3 days in two perpendicular diameters. Tumor volume = (short diameter2 × long diameter)/2. For metastasis assays, lungs were harvested from 4T1 tumor-bearing mice and digested and plated in 6-thioguanine supplemented medium as previously described.26
Serum cytokine array
Blood was collected from tumor-bearing mice by submandibular blood collection with a lancet (Medipoint, Mineola, NY) and serum was separated in Microtainer SST tubes (Becton Dickinson, Franklin Lakes, NJ). Cytokines were quantified using the G-Series Mouse Cytokine Array 1 (RayBiotech, Norcross, GA) per the manufacturer’s instructions. Slide scanning and quantification were performed on an Axio Scan.Z1 and Zen Blue software (Zeiss, Oberkochen, Germany). Values for each cytokine were normalized to an internal positive control.
Flow cytometry
Prior to flow cytometry analysis, spleens and tumors were processed into single-cell suspensions. Spleens were crushed through a 70 µm filter and red cell lysis was performed with ammonium-chloride-potassium (ACK) buffer (Gibco). Tumors were weighed, mechanically dissociated, incubated at 37°C in a buffer of RPMI, 5% fetal bovine serum (FBS), 5 mg/mL collagenases IV and I (Gibco), and 40 U/mL DNase, and then passed through a 70 µm filter. In 4T1 experiments, the cell suspension was resuspended in a 40% Percoll PLUS (GE Healthcare, Chicago, IL)/Hanks’ balanced salt solution, layered over a 70% Percoll PLUS/HBSS, and centrifuged at 800g for 20 min. Cells in the interface were collected and stained for flow cytometry analysis. In LLC experiments, CD45+ cells were purified from the cell suspension with a CD45 (tumor-infiltrating lymphocyte, TIL) Mouse MicroBeads Kit (Miltenyi Biotec, Bergisch Gladbach, Germany) per the manufacturer’s instructions prior to flow cytometry analysis. All antibodies used for flow cytometry were purchased from Thermo Fisher Scientific (Waltham, MA), BioLegend (San Diego, CA), or BD Biosciences (San Jose, CA). Cells were stained for cell surface expression in flat-bottom 96-well plates on ice in phosphate buffered saline with 2% FBS. Fluorescently conjugated antibodies for CD3 (500A2), CD4 (RM4-5), CD8 (53-6.7), PD-1 (29F.1A12), CD44 (IM7), CD45 (30-F11), CD62L (MEL14), Ly6G (1A8), Ly6C (HK1.4), CD11b (M1/70), F4/80 (BM8), Ki67 (16A8), and GzmB (QA18A28) were used per the manufacturers’ instructions. LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (Thermo Fisher Scientific) was used to gate on live cells; when necessary, cells were enumerated using 123count eBeads (Thermo Fisher Scientific) per the manufacturer’s instructions. Cytometry data were acquired via Attune NxT Flow Cytometer (Thermo Fisher Scientific). Data were analyzed via FlowJo (FlowJo, Ashland, OR). Flow cytometry analysis of immune cell subsets is defined as: CD4=CD3+CD4+; CD8=CD3+CD8+; TCM=CD3+CD44+CD62L+; TEff&EM=CD3+CD44+CD62L−; G-MDSC=CD11b+F4/80−Ly6CloLy6G+; M-MDSC=CD11b+F4/80−Ly6G−Ly6C+; Macrophage=CD11b+F4/80+.
OPAL immunofluorescence
Tumor tissue was fixed in Z-fix (Anatech, Battle Creek, MI) overnight, embedded in paraffin, and sectioned onto glass slides (American HistoLabs, Gaithersburg, MD). Slides were stained using the Opal 4-Color Manual IHC Kit (PerkinElmer, Waltham, MA) per the manufacturer’s instructions. Briefly, slides were deparaffinized and rehydrated with xylene and ethanol gradients, microwaved with pH6, pH9, or Rodent Decloaker (BioCare Medial, Pacheco, CA) antigen retrieval solution, cooled, rinsed with tris-buffered saline, 0.1% tween (TBST), and blocked with BLOXALL Blocking Solution (Vector Laboratories, Burlingame, CA). Staining with primary and secondary antibodies and OPAL fluorophore working solution was conducted following the manufacturer’s instructions. Antibodies used included anti-E-cadherin (3195, Cell Signaling Technology, Danvers, MA), anti-vimentin (GTX100619, GeneTex, Irvine, CA), anti-ZEB1 (NBP1-05987, Novus Biologicals, Centennial, CO), anti-CD4 (4SM95, Invitrogen, Carlsbad, CA), anti-CD8a (4SM16, Invitrogen), anti-FoxP3 (5H10L18, Invitrogen), anti-versican (Vcan; ab177037, Abcam, Cambridge, UK), anti-occludin (OCLN; NBP1-77037, Novus Biologicals), anti-fibronectin (GTX112794, GeneTex), and anti-osteopontin (ab8448, Abcam). Image quantification was performed by random sampling of tumor sections containing a minimum of 300 cells and no obvious signs of necrosis. Slide scanning and quantification were performed on an Axio Scan.Z1 and Zen Blue software (Zeiss).
RNA fluorescent in situ hybridization
CXCL1, TGF-β1, CXCR2, and PD-L1 RNA in situ hybridization was performed on Z-fixed paraffin-embedded tumor tissues using the RNAscope technology (Advanced Cell Diagnostics (ACD), Newark, CA), following the manufacturer’s protocol. In some experiments, slides were stained with anti-wide spectrum cytokeratin (ab9377, Abcam) according to ACD’s recommended protocol. Slide scanning and quantification were performed on an Axio Scan.Z1 and Zen Blue software (Zeiss).
Real-time PCR and NanoString analysis
Total RNA from tissue culture cells or flash-frozen tumor sections was prepared using the RNeasy Mini Kit (Qiagen, Hilden, Germany). For some experiments, RNA was then reverse transcribed using SMARTer PCR cDNA Synthesis Kit (Takara Bio, Mountain View, CA) per the manufacturer’s instructions. cDNA (20 ng) was amplified in triplicate using TaqMan Master Mix in an Applied Biosystems 7500 Real-Time PCR System (Thermo Fisher Scientific). The following Taqman gene expression primers were used (Thermo Fisher Scientific): CDH1 (Mm01247357_m1), Vim (Mm01333430_m1), Snai1 (Mm0041533_g1), Snai2 (Mm00441531_m1), Zeb1 (Mm00495564_m1), Vcan (Mm01283063_m1), Ocln (Hs00170162_m1), Vim (Hs00958116_m1), Cdh2 (Hs00983062_m1), IL-8 (Hs00174103_m1), TGFB1 (Hs00998133_m1), TGFBR1 (Hs00610320_m1), Spp1 (Hs00959010_m1), Fn1 (Hs00415006_m1), Cdh1 (Hs01013959_m1), Zo-1 (Hs01551861_m1), Snai1 (Hs00195591_m1), Snai2 (Hs00161904), Zeb1 (Hs00232783_m1), TGFB3 (Hs01086000_m1), Cxcr1 (Hs04965770_s1), Cxcr2 (Hs01891184), MMP2 (Hs01548727_m1), Col3a1 (Hs00943809_m1), and Col5a2 (Hs00893878_m1). Human GAPDH (4325792, Applied Biosystems, Foster City, CA) and mouse GAPD (4352339E, Applied Biosystems) were also used. Expression of all target genes relative to GAPDH was calculated 2−[Ct(GAPDH)–Ct(target gene)]. Expression of gene ratio was calculated (2−[Ct(GAPDH)–Ct(target gene1)])/(2−[Ct(GAPDH)–Ct(target gene2)]).
NanoString analysis was performed on purified RNA samples from indicated tumors using the PanCancer IO 360 Gene Expression Panel and nSolver analysis software was used for data normalization (NanoString Technologies, Seattle, WA). Further hierarchical clustering of log2-transformed gene expression counts was performed on Partek Genomics Suite analysis software (Partek, St Louis, MO).
Proliferation assay
Tumor cell proliferation was measured using the CellTiter-Glo assay (Promega, Madison, WI) per the manufacturer’s instructions. Cells were plated (300 cells per well) in a white-bottomed 96-well plate (Greiner Bio-One, Monroe, NC) with dimethyl sulfoxide (DMSO) or SX-682 and assayed on day 7.
T-cell proliferation assays were performed by coculturing splenocytes from BALB/c mice depleted of red cells in 96-well plates at 37°C in proliferation medium (RPMI, 10% FBS, 1% HEPES, 1% penicillin, 1% streptomycin, 1% L-Glutamine, and 50 µM 2-ME) at 105 cells per well with irradiated (2500 rad) purified G-MDSCs from spleens of either control or SX-682-treated mice at 2:1, 1:1, 0.5:1 or 0.25:1 ratio of G-MDSC-to-splenocytes with 4 μg/mL of concanavalin A (Sigma-Aldrich, St Louis, MO) for 48 hours in 5% CO2. Cells were pulsed with 1 µCi of [3H ]-Thymidine per well for 24 additional hours. Counts were measured using a 1450 Microbeta liquid scintillation counter (PerkinElmer). Data are shown as cpm mean±SD of triplicate cultures.
Migration and invasion assays
Cell invasion was evaluated by using Cultrex 96 Well BME Cell Invasion Assay (Trevigen, Gaithersburg, MD) per the manufacturer’s instructions. Chemoattractants used were either murine CXCL1 at 100 ng/mL in Iscove's Modified Dulbecco's Media with 10% FBS, or human IL-8 at 200 ng/mL in RPMI with 10% FBS. MDSC migration assays were performed using Neuro Probe Blind Well Chambers with a 5 μM pore filter (Neuro Probe, Gaithersburg, MD) as per the manufacturer’s instructions. MDSCs were purified using a Miltenyi MDSC Isolation Kit; 2×105 Ly6G+ or Ly6C+ cells were plated and chemoattracted towards 100 ng/mL of CXCL1 and CXCL2 in IMDM with 10% FBS. Chambers were incubated for 6 hours before counting.
Cytotoxicity assay
NK cells were isolated from NIH Blood Bank healthy donor peripheral blood mononuclear cells by using a magnetic NK Cell Isolation Kit (Miltenyi Biotech). Target cells were incubated with DMSO, 2.5 µM SX-682, 0.2-25 µg/mL bintrafusp alfa, or 2.5 µM SX-682 plus 0.2-25 µg/mL bintrafusp alfa for 3 days in RPMI media containing 10% FBS. On the day of the assay, cells were harvested, washed, and labeled with 10 µM calcein-AM (Invitrogen) for 20 min at 37°C and subsequently plated at 5×103 cells per well in 384-well flat-bottom culture plates. NK effector cells were added from different donors at a 20:1 ratio of effector:target cells. Following 6-hour culture, viable cells were counted with a Celigo Image Cytometer (Nexcelom Bioscience, Lawrence, MA).
Microscopy
4T1 cells were cultured alone or with 2.5 µM SX-682, 25 µg/mL bintrafusp alfa, or 2.5 µM SX-682 plus 25 µg/mL bintrafusp alfa for 3 days in IMDM media containing 10% FBS. Cells were observed and photographed using a Leica DMI400B microscope with a DFC340FX black and white camera.
Statistical methods
Statistical analysis of tumor growth curves was conducted using two-way analysis of variance (ANOVA). Two-tailed Student’s t-test was used to determine statistical difference between two sets of data while one-way ANOVA with Tukey’s post hoc test was used to determine statistical differences among three or more sets of data. Pearson r scores and p values (two tailed) were calculated for correlation data. All statistical analyses were performed using GraphPad Prism V.7 for Windows (GraphPad Software, La Jolla, CA, www.graphpad.com). Error bars represent SD. Asterisks indicate that the experimental p value is statistically significantly different from the associated controls at *p≤0.05; **p≤0.01; ***p≤0.001, ****p≤0.0001.
Results
Signaling through the IL-8 receptors, CXCR1 and CXCR2 regulates plasticity of human triple negative breast cancer cells
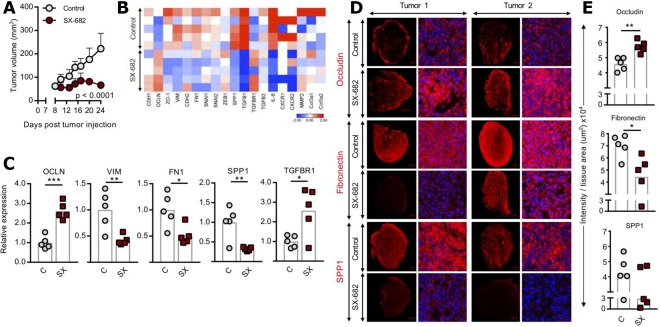
In a previous study we have shown the ability of a neutralizing anti-IL-8 antibody to modulate the phenotype of mesenchymal, triple negative breast cancer cells.17 Since the IL-8 receptors CXCR1 and CXCR2 are also activated via binding of other CXC chemokines of the same family as IL-8, we have now investigated the effect of dual blockade of CXCR1/2 in cancer cell plasticity. For the initial studies, triple negative, mesenchymal MDA-MB-231 cells were injected into the flank of NSG mice, followed by administration of control diet or a diet containing SX-682 starting on day 7. As shown in figure 1A, SX-682 treatment significantly delayed tumor growth as a monotherapy, an effect likely associated with the direct antiproliferative activity of CXCR1/2 blockade in MDA-MB-231 cells (online supplementary figure 1). Treatment with SX-682 also impacted the phenotype of the tumor resulting in increased expression of genes encoding for epithelial OCLN and significantly lower expression of numerous mesenchymal markers including VIM, FN1, and SPP1 (figure 1B, C). The modulation of OCLN, fibronectin and SPP1 proteins was also confirmed by immunofluorescence (figure 1D, E). Interestingly, among the genes upregulated by SX-682 treatment was TGFBR1, potentially indicating a compensatory feedback mechanism involving TGF-β signaling in cells blocked for CXCR1/2 signaling. These data corroborated our hypothesis that SX-682 can drive tumor cells into a less mesenchymal phenotype.
Figure 1.
SX-682 inhibits growth of MDA-MB-231 tumors and reduces tumor cell mesenchymal features in vivo. (A) NSG mice were injected subcutaneously with 4×106 MDA-MB-231 cells in Matrigel. On day 7 mice were started on a control or SX-682 diet. Mice were sacrificed on day 24. Graph shows average tumor volume±SD; n=5 mice/group. Data represent one of three independent experiments. (B, C) qPCR analysis of indicated genes in MDA-MB-231 tumors, shown as a heat map (B) and a bar graph (C), respectively. Expression of each gene was normalized to the average expression in the control. (D) OPAL immunofluorescence images of two representative tumors in the control and SX-682–treated groups, stained for occludin, fibronectin, and SPP1. (E) Quantification of OPAL images in (D). Individual points represent averaged data from one tumor. Error bars indicate mean±SD. *P≤0.05; **p≤0.01; ***p≤0.001; ****p≤0.0001 for two-way analysis of variance (ANOVA) in (A), two-tailed Student’s t-test in (C, E).
jitc-2019-000326supp001.pdf (93.1KB, pdf)
CXCR1/2 blockade synergizes with dual TGF-β/PD-L1 blockade in vitro
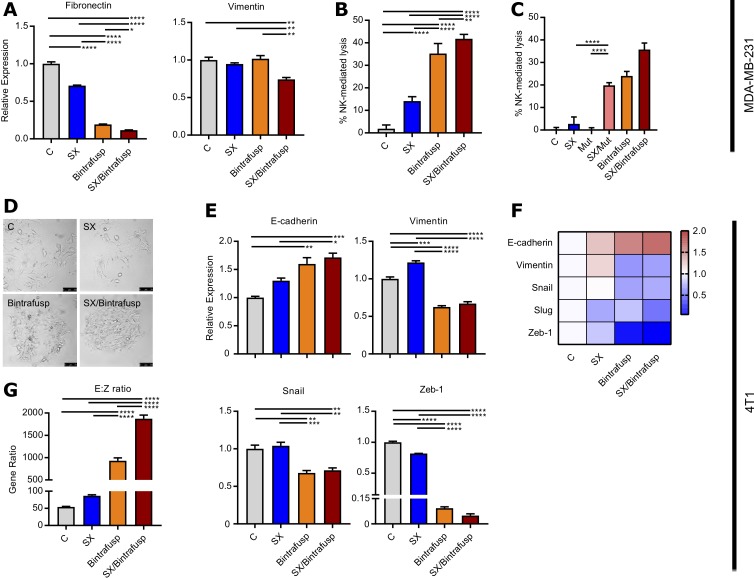
Perhaps the major challenge of any strategy aimed at modulating tumor cell plasticity is that the signaling pathways that control this phenomenon are multiple, making the blockade of one such pathway potentially ineffective when other alternative signaling events that trigger the acquisition of mesenchymal features are activated. In agreement, blockade of CXCR1/2 with SX-682 (SX) or trapping of TGF-β with bintrafusp alfa (Bintrafusp) partially reduced expression of mesenchymal fibronectin but had no impact on vimentin expression, while combined treatment with SX-682 and bintrafusp alfa (SX/Bintrafusp) exhibited the greatest decrease in fibronectin and additionally a reduction in vimentin expression (figure 2A). Moreover, MDA-MB-231 cells treated with the combination SX/Bintrafusp also exhibited the highest increase in susceptibility to NK-mediated lysis (figure 2B). Due to the bifunctional nature of bintrafusp alfa, we also evaluated the effect of blockade of TGF-β in the absence of PD-L1 engagement by using a mutant form of the molecule comprising human TGFβRII bound to a mutated αPD-L1 moiety (Mut, figure 2C). Tumor cells treated with the mutant were not susceptible to NK-mediated cell lysis; however, cells treated with the combination of SX/Mut exhibited an increased susceptibility to NK-mediated lysis in the absence of PD-L1 binding, thus indicating that combined blockade of CXCR1/2 and TGF-β signaling is needed to improve the tumor susceptibility to immune-mediated lysis.
Figure 2.
Blockade of CXCR1/2 and transforming growth factor beta (TGF-β) enhances immune-mediated tumor cell lysis. (A) MDA-MB-231 cells were cultured for 72 hours alone or with SX-682, bintrafusp alpha, or SX-682 plus bintrafusp alfa and assayed via qPCR. Expression of each gene was normalized to the average expression in the control. (B) MDA-MB-231 cells from (A) were used as target cells in a natural killer (NK) cell-based killing assay. Data represent one of two independent experiments with two independent donors. (C) NK-cell-mediated lysis of tumors treated as indicated, including tumors treated with a mutant reagent (Mut) that traps TGF-β in the absence of programmed death ligand 1 (PD-L1) binding. (D) Morphological changes in 4T1 cells that were cultured for 72 hours alone or with indicated agents. (E, F) qPCR analysis of indicated genes in treated 4T1 cells shown as a bar graph (E) and a heat map (F), respectively. Expression of each gene was normalized to the average expression in the control. (G) Gene ratio of E-cadherin and Zeb-1 expression (E:Z ratio) in treated 4T1 cells. Error bars indicate mean±SD of the average of three biological replicates. *P≤0.05; **p≤0.01; ***p≤0.001; ****p≤0.0001 for one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test in (A, B, C, E, G).
Similar experiments were then conducted with murine models of breast cancer. 4T1 cells were cultured alone or in the presence of SX-682, bintrafusp alfa, or a combination of both. As shown in figure 2D, SX-682 or bintrafusp alfa-treated cells exhibited a more clustered morphology in comparison to control-treated cells; however, the most compact cell clusters were observed in the combination treatment group (figure 2D). Further qPCR analysis revealed that 4T1 cells treated with the combination CXCR1/2, TGF-β and PD-L1 blockade exhibited the greatest upregulation of epithelial E-cadherin and downregulation of mesenchymal vimentin and the mesenchymal transcription factors Snail and Zeb-1 (figure 2E, F). The gene ratio of E-cadherin to Zeb-1 was the clearest indicator that 4T1 cells treated with combination SX/Bintrafusp underwent the greatest change in tumor cell plasticity (figure 2G).
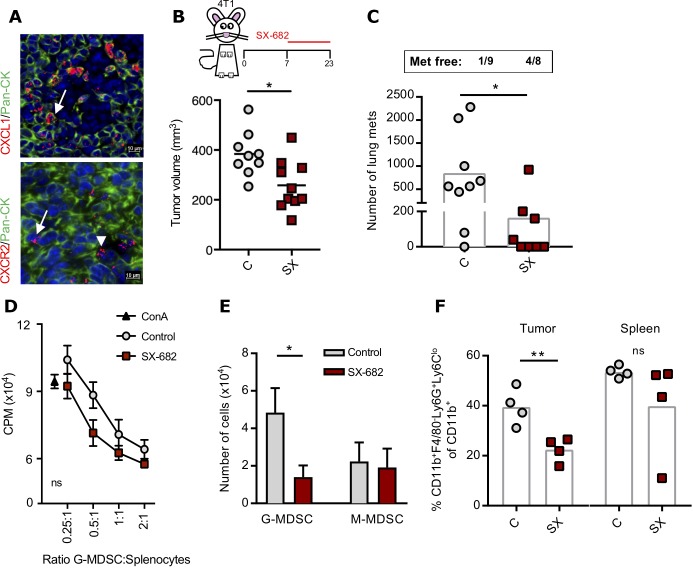
CXCR1/2 blockade inhibits growth of 4T1 tumors and G-MDSC migration
The effect of dual blockade of CXCR1/2 in the murine 4T1 breast cancer model has not been previously investigated. A multiplex fluorescent in situ RNA hybridization assay in 4T1 tumors confirmed robust expression of the chemokine CXCL1 (KC) mostly localized to the tumor cell compartment (labeled by costaining with a pan-cytokeratin antibody), and a modest expression of CXCR2 observed both in the tumor cells and in the surrounding stroma or immune cell infiltrate (figure 3A). To test the monotherapy effect of SX-682, BALB/c mice bearing 4T1 tumors in the mammary fat pad were fed an SX-682-containing diet starting on day 7. SX-682 monotherapy slightly delayed primary tumor growth (figure 3B) and impaired tumor dissemination to the lungs, with 4/8 (50%) mice free of metastases in the SX-682-treated group (figure 3C). Further in vitro studies with low doses of SX-682 demonstrated the ability of CXCR1/2 blockade to impair invasion of 4T1 cells towards a CXCL1 gradient (online supplementary figure 2). Ly6G+Ly6Clo cells purified from the spleens of both control and SX-682-treated mice were immunosuppressive and therefore corresponded to G-MDSC (figure 3D). To determine whether SX-682 reduced the migration of G-MDSC, BALB/c mice bearing 7-day 4T1 subcutaneous tumors were treated with control or SX-682-containing diet for 7 additional days. Splenic G-MDSC and monocytic myeloid-derived suppressor cells (M-MDSC) were purified and assayed ex vivo. SX-682 treatment in vivo markedly impaired the migration of G-MDSC towards CXCL1/CXCL2 (figure 3E), while M-MDSC, which have lower CXCR1/CXCR2 levels,27 did not migrate. In agreement with these and previous results,27 SX-682 treatment resulted in a significantly lower percentage of tumor infiltrating G-MDSC while percentages were equal in the spleens of the same animals (figure 3F).
Figure 3.
SX-682 monotherapy inhibits 4T1 mammary carcinoma growth and G-MDSC migration. (A) Representative images of 4T1 tumors analyzed by in situ hybridization for mRNA expression of CXCL1 and CXCR2, simultaneously stained for pan-cytokeratin (Pan-CK, green). DAPI was used for nuclei staining (blue). Arrows indicate tumor cells and arrowheads indicate stroma/immune cells positive for each marker. (B) BALB/c mice were inoculated subcutaneously with 3×104 4T1 cells in the mammary fat pad; on day 7 mice were started on a control or SX-682 diet. Tumors were measured until day 23; n=9 or 10 mice/group. (C) Lungs were harvested and processed for metastases. (D) Proliferation assay was performed with purified CD11b+Ly6G+ cells from spleens of either control or SX-682–treated mice and splenocytes from untreated mice stimulated with ConA. (E) Purified, splenic G-MDSC from control or SX-682–treated mice was assayed for migration towards medium containing CXCL1 and CXCL2. (F) BALB/c mice were inoculated subcutaneously with 3×104 4T1 cells in the mammary fat pad; on day 7 mice were started on a control or SX-682 diet with flow cytometry analysis performed on tumors and spleens on day 14. Individual points represent data from one tumor. Error bars indicate mean±SD of three biological replicates. *P≤0.05; **p≤0.01; for two-tailed Student’s t-test in (B, C, E, F) and two-way analysis of variance (ANOVA) in (D).; CPM, counts per minute; G-MDSC, granulocytic myeloid-derived suppressor cells; M-MDSC, monocytic myeloid-derived suppressor cells.
CXCR1/2/TGF-β/PD-L1 blockade reduces tumor cellular plasticity and enhances antitumor immunity
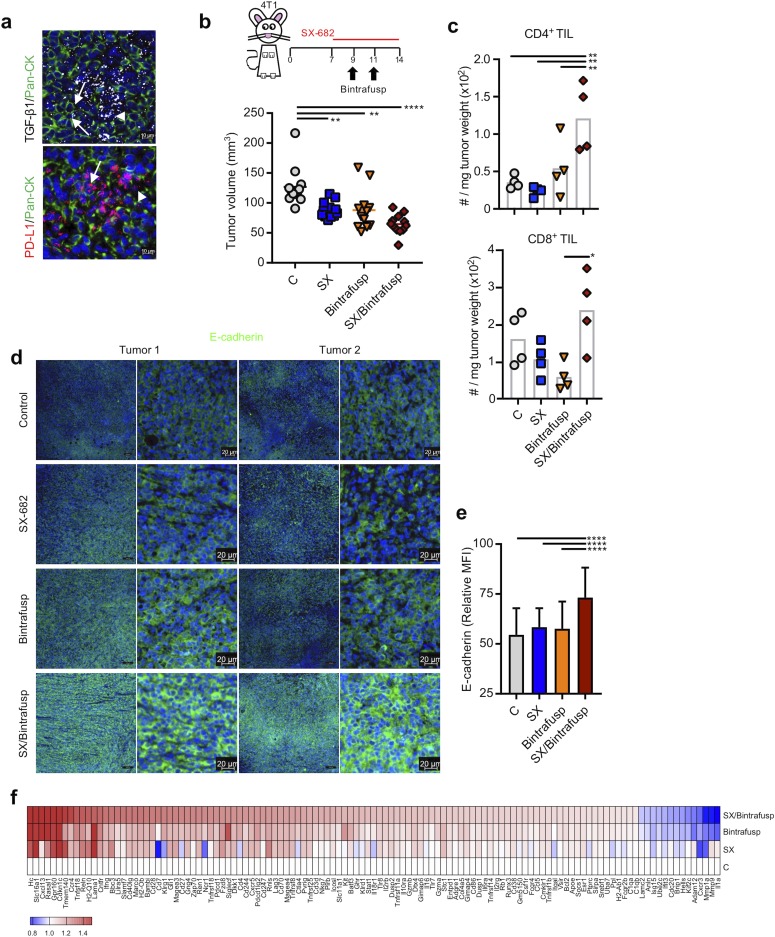
We next sought to determine the effect of CXCR1/2 blockade in combination therapy with TGF-β/PD-L1 blockade in vivo. 4T1 tumors were further characterized for expression of TGF-β1 and PD-L1 through multiplex fluorescent in situ RNA hybridization. Robust expression of TGF-β1 and PD-L1 (figure 4A) was observed both in the tumor cells as well as in the surrounding stroma or immune cell infiltrate. The TME profile of 4T1 suggested that combination therapy blocking CXCR1/2, TGF-β, and PD-L1 could be effective therapeutically.
Figure 4.
SX-682 plus bintrafusp alfa modulates tumor cell plasticity in vivo. (A) Representative images of 4T1 tumors analyzed by in situ hybridization for mRNA expression of TGF-β1 and PD-L1, simultaneously stained for pan-cytokeratin (Pan-CK, green). DAPI was used for nuclei staining (blue). Arrows indicate tumor cells and arrowheads indicate stroma/immune cells positive for each marker. (B) 4T1 tumor-bearing mice were started on a control or SX-682 diet on day 7. On days 9 and 11 mice received intraperitoneal injections of 492 µg bintrafusp alfa. Four representative tumors were harvested and analyzed on day 14. Graph shows individual tumor volumes and mean per group at day 13; n=12 mice/group. Data represent one of two independent experiments. (C) Flow cytometry analysis of tumors on day 14 for indicated cell subsets. (D) Representative images of OPAL immunofluorescence staining of 4T1 tumors in each group for E-cadherin. (E) Quantification of OPAL images in (D). (F) Heat map of genes differentially expressed in tumors from two (C, SX, Bintrafusp) or three (SX/Bintrafusp) mice on day 14 commonly found among treatment groups or expressed exclusively in the combination SX/Bintrafusp group. Individual points represent data from one tumor. Error bars indicate mean±SD of biological replicates. *P≤0.05; **p≤0.01; ***p≤0.001; ****p≤0.0001 for one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test in (B, C, E). Bintrafusp, bintrafusp alfa monotherapy; C, control diet; MFI, mean fluorescence intensity; PD-L1, programmed death ligand 1; SX, SX-682 diet; SX/Bintrafusp, SX-682 plus bintrafusp alfa combination therapy; TGF-β, transforming growth factor beta; TIL, tumor-infiltrating lymphocyte.
BALB/c mice bearing 7-day 4T1 tumors in the mammary fat pad were initiated on a continuous control (C) or SX-682 (SX) containing diet. Bintrafusp alfa was administered intraperitoneally on days 9 and 11 to mice in each diet. Early during treatment (day 13), SX-682 (SX), bintrafusp alfa (Bintrafusp), and SX-682 plus bintrafusp alfa (SX/Bintrafusp)-treated mice showed a delay in tumor growth. Although not statistically significant, there was a trend towards improved tumor control in the combination SX/Bintrafusp group (figure 4B); however, flow cytometric analysis of tumor immune infiltrates at this early time point revealed higher numbers of CD4+ TIL but few differences in the quantities of CD8+ TIL (figure 4C). Immunofluorescence staining showed a homogeneous distribution of CD8+ and CD4+ TIL across the whole tumor section in the combination group, with only a small fraction of infiltrating CD4+/Foxp3+ Tregs (online supplementary figure 3).
At this early time point, prior to large influxes of cytotoxic CD8+ T cells, tumors were also assayed by immunofluorescence for changes in tumor cellular plasticity. Combination of SX-682 and bintrafusp alfa induced a significant upregulation of epithelial E-cadherin on 4T1 tumor cells as shown by representative images (figure 4D) and mean fluorescence intensity quantification (figure 4E). Single treatment with each agent, however, had no effect on the tumor expression of epithelial E-cadherin, therefore confirming that combination CXCR1/2 and TGF-β blockade was needed for modulation of tumor cell plasticity.
To better understand what SX-682 and bintrafusp alfa were each contributing to the combination therapy, NanoString gene expression RNA analysis was performed on whole tumor tissue of the C, SX, Bintrafusp and SX/Bintrafusp-treated mice (figure 4F). A subset of genes was found to be similarly modulated in tumors treated with either SX-682 or bintrafusp alfa and the combination SX/Bintrafusp. Among those genes were the matrix metalloproteinases Mmp9 and Mmp1a that were two of the three most downregulated genes in the combination group, together with IL1a. Several immune activation genes found in this group were upregulated by bintrafusp alfa monotherapy and also found in the combination group, including CD3d, CD4, CD27, Zap70, Gzma, Gzmb and Ctla-4. Indicative of an additive effect employing SX-682 and bintrafusp alfa, the level of modulation of the majority of genes in figure 4F was the greatest in the SX/Bintrafusp group. Additionally, we observed genes that were exclusively modulated in the combination SX/Bintrafusp group and not in either monotherapy treatment. These genes included interferon gamma (IFN-γ), CD40lg, Icosl, IL2rg and numerous other genes involved in remodeling of the TME. In agreement with an enhanced immune response in tumors receiving bintrafusp alfa, quantification of serum cytokines from these mice revealed high levels of the Th1 polarizing cytokines IL-12 and tumor necrosis factor alpha and the Th2 polarizing cytokines IL-4 and IL-5 in the blood of Bintrafusp and SX/Bintrafusp mice but not the control (C) or SX-682 (SX) groups (online supplementary figure 4). The proinflammatory cytokine IL-1β was reduced in all treatment groups, and the potent Th1 cytokine IFN-γ was highest in the serum of mice in the SX/Bintrafusp combination group, in agreement with the RNA data in tumors.
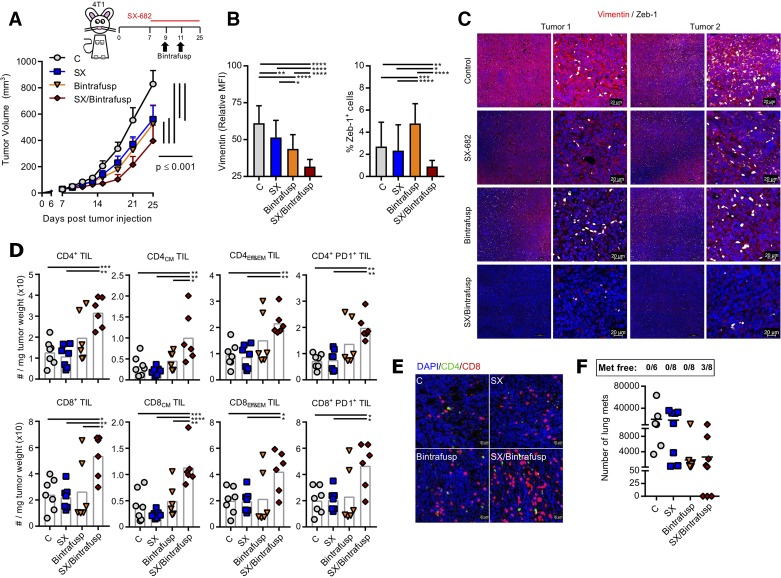
In order to track changes in TIL and tumor cellular plasticity in larger SX/Bintrafusp-treated tumors, the remaining 4T1 tumor-bearing mice were allowed to progress until day 25 with no additional bintrafusp alfa injections (figure 5A). Both SX and bintrafusp alfa monotherapies continued to delay tumor growth while the combination group of SX/Bintrafusp elicited the best antitumor response. Tumors were also assayed by immunofluorescence for changes in tumor cellular plasticity. Combination of SX-682 and bintrafusp alfa induced the greatest reduction of mesenchymal vimentin and significantly decreased the number of Zeb-1-positive tumor cells (figure 5B, C).
Figure 5.
SX-682 plus bintrafusp alfa in the 4T1 mammary carcinoma model enhances immune activation. (A) BALB/c mice were inoculated subcutaneously with 3×104 4T1 in the mammary fat pad. On day 7 mice were started on a control or SX-682 diet. On days 9 and 11 mice received intraperitoneal injections of 492 µg bintrafusp alfa. Tumors and organs were harvested and analyzed on day 25. Graph shows average tumor growth; n=7 (C) or 8 (SX, Bintrafusp, SX/Bintrafusp) mice/group. Data in (A) are representative of one of two independent experiments. (B, C) OPAL immunofluorescence staining of 4T1 tumors in each group was performed for vimentin and Zeb-1 and were quantified (B) with representative images shown (C). (D) Flow cytometry analysis of tumors on day 25. Immune cell subsets are defined as number of cells per tumor weight. (E) Representative images of tumors stained for CD4+ (green) and CD8+ (red) T cells by immunofluorescence. DAPI (blue) was used to stain nuclei. (F) Number of micrometastases quantified in the lungs of 4T1 tumor-bearing mice. Individual points represent data from one tumor. Error bars indicate mean±SD of biological replicates. *P≤0.05; **p≤0.01; ***p≤0.001; ****p≤0.0001 for two-way analysis of variance (ANOVA) in (A), one-way ANOVA followed by Tukey’s post hoc test in (B, D). MFI, mean fluorescence intensity; PD1, programmed death-1 receptor; TIL, tumor-infiltrating lymphocyte.
Flow cytometry analysis of the immune subsets at day 25 showed high numbers of tumor infiltrating CD4+ and CD8+ TCM and TEff&EM cells, many of which were PD-1+ (figure 5D). Immunofluorescence analysis of TIL showed a robust infiltration with CD8+ TIL in the combination-treated tumors mostly organized in cell clusters across the whole tumor area (figure 5E). Although no tumor cures were observed in primary tumors, the SX/Bintrafusp group was the only one with mice free of lung metastases (3/8 mice, figure 5F).
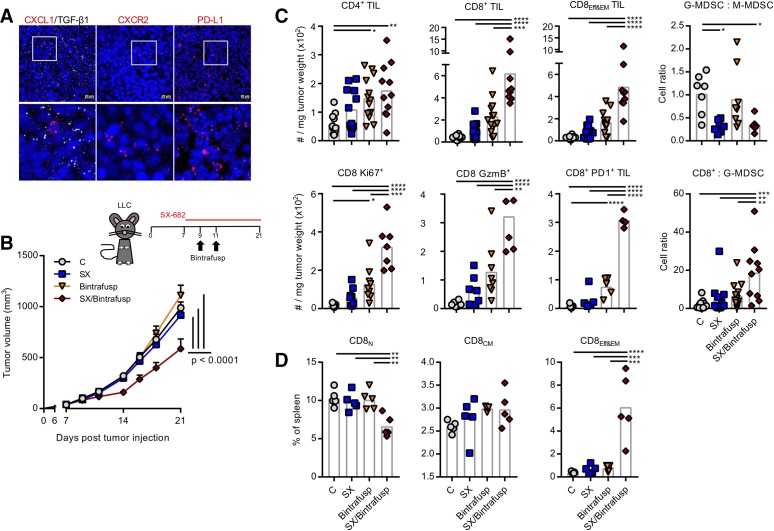
The effect of the combination therapy was also evaluated in the LLC model. Tumors analyzed by in situ RNA hybridization showed significant expression of TGF-β1 and PD-L1 mRNA, with a discreet number of cells positive for CXCL1 or CXCR2 (figure 6A). To test if the combination therapy could also be effective in the LLC model, C57BL/6 mice were injected with LLC cells subcutaneously in the flank; tumors were allowed to grow for 7 days before starting control (C) or SX-682 (SX) diet, with administration of bintrafusp alfa intraperitoneal on days 9 and 11 (figure 6B). Unlike the 4T1 model, LLC tumors were impervious to SX-682 or bintrafusp alfa monotherapy and only the combination SX/Bintrafusp elicited a delay in tumor growth. Flow cytometry analysis of tumors on day 21 revealed a significant infiltrate with both CD4+ and CD8+ TEff&EM present only in the SX/Bintrafusp tumors (figure 6C). It is interesting to note that effector-type T cells were increased only in the SX/Bintrafusp-treated tumors, while CD8+ TCM cell populations were consistent among all treatment groups (data not shown). This influx of highly activated Ki67+/GzmB+/PD-1+ T cells into the tumors resulted in a high CD8-to-G-MDSC ratio. Corresponding to the increase of CD8+ TEff&EM in the tumor, SX/Bintrafusp-treated mice had higher levels of CD8+ TEff&EM and reduced levels of CD8+ TNaive cells in their spleens (figure 6D).
Figure 6.
SX-682 plus bintrafusp alfa inhibits LLC tumor growth. (A) Representative images of LLC tumors analyzed by in situ hybridization for mRNA expression of the target molecules CXCL1 (red), TGF-β1 (white), PD-L1, and CXCR2. DAPI was used for nuclei staining (blue). Detailed areas are shown below each picture at a higher magnification. (B) LLC tumor-bearing mice were administered a control or SX-682 diet on day 7. On days 9 and 11 mice received intraperitoneal injections of 492 µg bintrafusp alfa. Tumors and organs were harvested and analyzed on day 21. Graph shows average tumor growth; n=11 (C) or 13 (SX, Bintrafusp, SX/Bintrafusp) mice/group. Graph represents combined data from two independent experiments. (C, D) Flow cytometry analysis of tumors showing indicated immune cell numbers per tumor weight or immune cell ratios (B), and spleens (D) as percentage of live cells on day 21. Individual points represent data from one tumor. Error bars indicate mean±SD. *P≤0.05; **p≤0.01; ***p≤0.001; ****p≤0.0001 for two-way analysis of variance (ANOVA) in (B), one-way ANOVA followed by Tukey’s post hoc test in (C, D). G-MDSC, granulocytic myeloid-derived suppressor cells; LLC, Lewis lung carcinoma; PD1, programmed death-1 receptor; PD-L1, programmed death ligand 1; TGF-β, transforming growth factor beta; TIL, tumor-infiltrating lymphocyte.
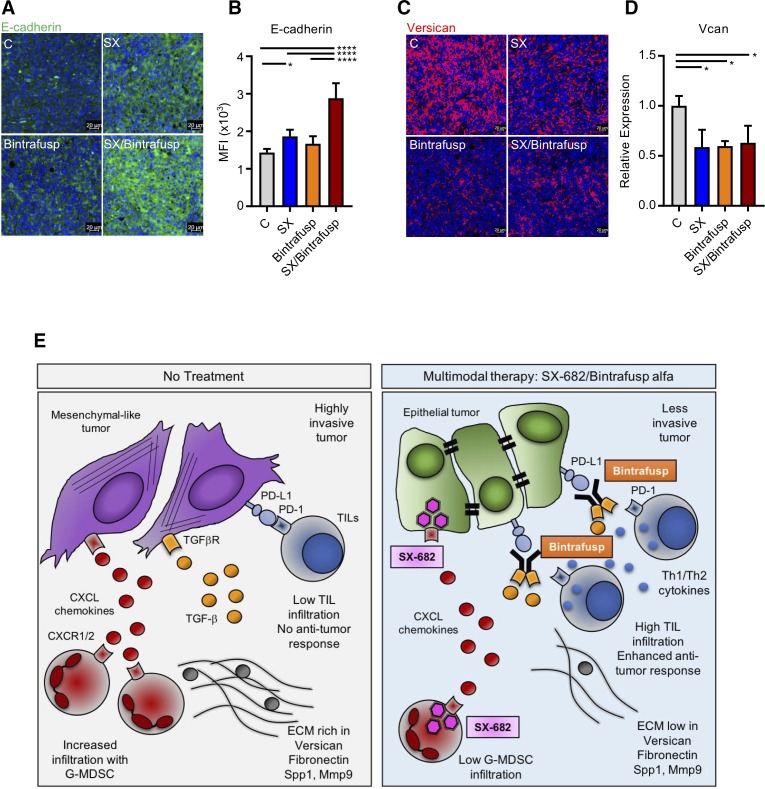
We then focused our attention on elucidating why only the combination of SX-682 and bintrafusp alfa was able to elicit an antitumor response in LLC tumors. We hypothesized that the SX/Bintrafusp combination treatment may have altered the phenotype of these highly mesenchymal tumors, therefore driving TIL infiltration and enhancing immune-mediated tumor cell killing. To test this hypothesis, epithelial and mesenchymal markers were measured on LLC tumors in all treatment groups from figure 6B. We first quantified and confirmed the expression of epithelial E-cadherin on all treatment groups via OPAL immunofluorescence and qPCR, respectively (figure 7A, B). The combined treatment with SX-682 and bintrafusp alfa exclusively upregulated the expression of tumor E-cadherin, thus driving the LLC tumor cells towards a more epithelial state. Additionally, we quantified the expression of Vcan, a potent promoter of angiogenesis that has been shown to reduce CD8+ T-cell recruitment to LLC tumors.28 29 LLC tumors treated with either SX-682 or bintrafusp alfa as monotherapies or with the combination SX/Bintrafusp all significantly downregulated Vcan expression (figure 7C, D).
Figure 7.
SX-682 plus bintrafusp alfa modulates tumor cell plasticity in Lewis lung carcinoma (LLC) tumors. (A, B) E-cadherin (green) expression in a representative section of a tumor in each treatment group (A) and quantification of E-cadherin expression from three individual tumors in each group (B). Shown are the average mean fluorescence intensities (MFI) of total fluorescence. (C, D) Versican (red) expression in a representative section of a tumor in each treatment group (C), and qPCR quantification of versican mRNA in CD45 negative cells isolated from three tumors in each group (D). Error bars indicate mean±SD of biological replicates. *P≤0.05; ****p≤0.0001 for one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test in (B, D). (E) Synopsis of the proposed mechanism of action of the combination SX-682 plus bintrafusp alfa.; ECM, extracellular matrix; G-MDSC, granulocytic myeloid-derived suppressor cell; PD-1, programmed death-1 receptor; PD-L1, programmed death ligand 1; TGF, transforming growth factor; TIL, tumor-infiltrating lymphocyte.
Discussion
In the present work we demonstrated the efficacy and mechanism of action of a multimodal immunotherapy approach combining dual CXCR1/2 inhibition with neutralization of TGF-β and PD-L1 signaling blockade. Our data showed that this immunotherapeutic regime can successfully increase T-cell infiltration and activation in tumors, reduce infiltration with suppressive G-MDSC, and modulate tumor cell plasticity for improved susceptibility to immune lysis and decreased tumor proliferation and invasiveness (figure 7E).
In addition to poor prognosis, plasticity induced by the acquisition of mesenchymal features by cancer cells has been proposed as one of the mechanisms of resistance to checkpoint inhibition.12 21 Emerging research has also found higher levels of PD-L1 in tumors with a more mesenchymal phenotype in vivo,30 31 and previous work from our laboratory and others demonstrated a significant upregulation of PD-L1 in tumors undergoing tumor cell plasticity via exposure to TGF-β1.32 33 Bintrafusp alfa is a bifunctional fusion protein that simultaneously blocks the PD-1/PD-L1 interaction and TGF-β signaling in the TME. Extensive in vitro and in vivo evaluations have shown the ability of this molecule to promote NK and T-cell-mediated killing of tumor cells and delay tumor growth in multiple mouse models as a monotherapy and in combination approaches,34 35 demonstrating significantly greater antitumor efficacy than blockade of either PD-L1 or TGF-β alone, or the combination of untethered agents that individually block PD-L1 or TGF-β.35 Bintrafusp alfa was also shown to mediate direct killing of tumor cells by the mechanism of antibody-dependent cell cytotoxicity,36 and to be able to prevent or revert TGF-β-induced tumor cell phenotypic plasticity.32 A phase 1 clinical trial of bintrafusp alfa showed safety and clinical activity in patients with heavily pretreated, advanced cancer,37 and multiple clinical studies are currently ongoing.
A different TME factor that has been associated with tumor progression as well as with decreased responses to checkpoint inhibition is the inflammatory chemokine IL-8.38 Upregulation of this chemokine has been observed in multiple cancer types,39 with IL-8/IL-8R signaling being reported to foster tumor progression by several mechanisms, including by stimulating the recruitment of G-MDSC to the TME,19 promoting the acquisition of mesenchymal features by tumor cells,13 15–17 and increasing the survival of cancer stem-like cells in the tumor.40 41 In the context of immunotherapy, recent studies have also shown that upregulation of IL-8 and enhanced mesenchymal tumor features are enriched in melanoma, renal cell carcinoma, and non-small-cell lung cancer patient populations that do not respond to anti-PD-1 therapy.12 38 42 Pretreatment serum IL-8 levels have also been shown in patients with melanoma to correlate with responses to treatment with the checkpoint inhibitor ipilimumab and chemotherapy, with high levels correlating with failure to treatment.43 Blockade of IL-8 signaling may be achieved by direct neutralization of IL-8 or via inhibition of its receptors. Previously, our group showed that neutralization of IL-8 via a monoclonal anti-IL-8 in vitro and in vivo was able to revert mesenchymal features in claudin-low, triple negative tumor models, resulting in enhanced immune-mediated lysis with NK and antigen-specific T cells in vitro, and reduced tumor infiltration with G-MDSC in vivo.17 The strategy of inhibiting the CXCR1/2 receptors, on the other hand, could achieve blockade of signaling mediated by IL-8 and by all other chemokines that bind to CXCR1/2. In addition, murine cells do not produce IL-8 but rather respond to other CXC chemokines via binding to the murine CXCR1/2 receptors, thus making possible the investigation of the approach in syngeneic murine models. Two previous studies have explored the use of SX-682 as an inhibitor of MDSC migration to the TME resulting in the enhancement of checkpoint blockade with anti-PD-1 antibodies.27 44 To our knowledge, the present work is the first one to attempt to overcome escape mechanisms mediated by redundancy of signaling that controls tumor phenotype by using a combination approach whereby both CXCR1/2 inhibition and TGF-β neutralization act in synergy to enhance the antitumor activity of anti-PD-L1 inhibition. Our data in vivo with syngeneic, murine mesenchymal tumors showed that while mesenchymal markers could be slightly decreased by single treatments with SX-682 or bintrafusp alfa, the combined treatment with SX-682 and bintrafusp alfa was the only one able to significantly upregulate epithelial and downregulate mesenchymal markers in vivo, thus driving the tumor phenotype towards a more epithelial state.
It is important to note that even though SX-682 plus bintrafusp alfa combination therapy resulted in a greater influx of TIL, reduced tumor cell plasticity, and delayed tumor growth, cures were not observed in primary tumors in any combination-treated mice. This could be explained by the rapid tumor growth of these models exceeding the inherent maximum rate of immune-mediated tumor clearance, the limited window of drug delivery due to the use of a human drug (bintrafusp alfa) in fully immune competent mice, or due to the presence of additional mechanisms of immune suppression. One could speculate that additional doses of bintrafusp alfa could neutralize newly emerging PD-L1 and TGF-β at later time points in the TME and result in a more potent antitumor response including cures. Additionally, blockade of other immune suppressive factors could potentially afford an improved antitumor response. NanoString analysis performed on whole tumor tissue of the C, SX, Bintrafusp and SX/Bintrafusp-treated 4T1 tumor-bearing mice showed, for example, upregulation of CTLA-4 in tumors treated with the combination therapy. Antagonist CTLA-4 therapy has been shown to be a potent inducer of antitumor responses against late-stage cancers and is now regularly being combined with PD-1/PD-L1 therapies in some tumor types to induce synergistic efficacy.1 45 Although our data did not show an increased number of Tregs in the combination group tumors, eliminating this suppressive cell population could also potentially enhance antitumor immunity.
Conclusions
This study demonstrated for the first time the efficacy and mechanism of action of the combination of a CXCR1/2 inhibitor, SX-682, with bintrafusp alfa, a PD-L1/TGF-βRII bifunctional fusion protein. These agents were shown to have synergistic effects in two tumor models with no evidence of antagonism. This multimodal immunotherapy successfully increased T-cell infiltration and activation in tumors, reduced infiltration with suppressive G-MDSC, and modulated the phenotypic plasticity of the tumor for improved susceptibility to immune lysis and decreased invasiveness. The approach demonstrated synergy in tumor models with varying immune cell infiltrates, degrees of plasticity, and responses to immunotherapy. Efficacy of the combination was found in models with relatively high versus low quantities of G-MDSC, suggesting that patient efficacy may not require a particular myelosuppressive cell population to be present in the TME. Future clinical trials to evaluate the use of SX-682 plus bintrafusp alfa in triple negative breast cancer and potentially other malignancies are currently being planned.
Acknowledgments
The authors thank Debra Weingarten for her editorial assistance in the preparation of this manuscript.
Footnotes
Funding: This work was supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute (NCI), National Institutes of Health (NIH), as well as through Cooperative Research and Development Agreements (CRADA) between the NCI/NIH and Syntrix Pharmaceuticals and the NCI/NIH and EMD Serono.
Competing interests: The authors from the National Cancer Institute (NCI), NIH do not have any competing interests to disclose. The authors from Syntrix Pharmaceuticals and EMD Serono are employees or officers of the respective companies. The NCI/NIH has ongoing Collaborative Research and Development Agreements (CRADA) with Syntrix Pharmaceuticals and EMD Serono.
Patient consent for publication: Not required.
Ethics approval: Human peripheral blood was obtained from NIH Blood Bank healthy donors. All animal studies were approved by the NIH Intramural Animal Care and Use Committee.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: Data are available upon reasonable request. Data generated in this study are available from the corresponding author.
References
- 1. Hodi FS, O'Day SJ, McDermott DF, et al. . Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 2010;363:711–23. 10.1056/NEJMoa1003466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brahmer J, Reckamp KL, Baas P, et al. . Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med 2015;373:123–35. 10.1056/NEJMoa1504627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Motzer RJ, Escudier B, McDermott DF, et al. . Nivolumab versus everolimus in advanced renal-cell carcinoma. N Engl J Med 2015;373:1803–13. 10.1056/NEJMoa1510665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pennock GK, Chow LQM. The evolving role of immune checkpoint inhibitors in cancer treatment. Oncologist 2015;20:812–22. 10.1634/theoncologist.2014-0422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fares CM, Van Allen EM, Drake CG, et al. . Mechanisms of resistance to immune checkpoint blockade: why does checkpoint inhibitor immunotherapy not work for all patients? Am Soc Clin Oncol Educ Book 2019;39:147–64. 10.1200/EDBK_240837 [DOI] [PubMed] [Google Scholar]
- 6. Shibue T, Weinberg RA. Emt, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol 2017;14:611–29. 10.1038/nrclinonc.2017.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brabletz T, Kalluri R, Nieto MA, et al. . Emt in cancer. Nat Rev Cancer 2018;18:128–34. 10.1038/nrc.2017.118 [DOI] [PubMed] [Google Scholar]
- 8. Santamaría PG, Moreno-Bueno G, Cano A. Contribution of epithelial plasticity to therapy resistance. J Clin Med 2019;8:E676(5:676 10.3390/jcm8050676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cristescu R, Lee J, Nebozhyn M, et al. . Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med 2015;21:449–56. 10.1038/nm.3850 [DOI] [PubMed] [Google Scholar]
- 10. Guinney J, Dienstmann R, Wang X, et al. . The consensus molecular subtypes of colorectal cancer. Nat Med 2015;21:1350–6. 10.1038/nm.3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kardos J, Chai S, Mose LE, et al. . Claudin-Low bladder tumors are immune infiltrated and actively immune suppressed. JCI Insight 2016;1:e85902 10.1172/jci.insight.85902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hugo W, Zaretsky JM, Sun L, et al. . Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell 2016;165:35–44. 10.1016/j.cell.2016.02.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Benoy IH, Salgado R, Van Dam P, et al. . Increased serum interleukin-8 in patients with early and metastatic breast cancer correlates with early dissemination and survival. Clin Cancer Res 2004;10:7157–62. 10.1158/1078-0432.CCR-04-0812 [DOI] [PubMed] [Google Scholar]
- 14. Thiery JP, Acloque H, Huang RYJ, et al. . Epithelial-Mesenchymal transitions in development and disease. Cell 2009;139:871–90. 10.1016/j.cell.2009.11.007 [DOI] [PubMed] [Google Scholar]
- 15. Fernando RI, Castillo MD, Litzinger M, et al. . Il-8 signaling plays a critical role in the epithelial-mesenchymal transition of human carcinoma cells. Cancer Res 2011;71:5296–306. 10.1158/0008-5472.CAN-11-0156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fernando RI, Hamilton DH, Dominguez C, et al. . Il-8 signaling is involved in resistance of lung carcinoma cells to erlotinib. Oncotarget 2016;7:42031–44. 10.18632/oncotarget.9662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dominguez C, McCampbell KK, David JM, et al. . Neutralization of IL-8 decreases tumor PMN-MDSCs and reduces mesenchymalization of claudin-low triple-negative breast cancer. JCI Insight 2017;2 10.1172/jci.insight.94296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Waugh DJJ, Wilson C. The interleukin-8 pathway in cancer. Clin Cancer Res 2008;14:6735–41. 10.1158/1078-0432.CCR-07-4843 [DOI] [PubMed] [Google Scholar]
- 19. Highfill SL, Cui Y, Giles AJ, et al. . Disruption of CXCR2-mediated MDSC tumor trafficking enhances anti-PD1 efficacy. Sci Transl Med 2014;6:237ra67 10.1126/scitranslmed.3007974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Patel SA, Minn AJ. Combination cancer therapy with immune checkpoint blockade: mechanisms and strategies. Immunity 2018;48:417–33. 10.1016/j.immuni.2018.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mariathasan S, Turley SJ, Nickles D, et al. . Tgfβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018;554:544–8. 10.1038/nature25501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moo-Young TA, Larson JW, Belt BA, et al. . Tumor-Derived TGF-beta mediates conversion of CD4+Foxp3+ regulatory T cells in a murine model of pancreas cancer. J Immunother 2009;32:12–21. 10.1097/CJI.0b013e318189f13c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Travis MA, Sheppard D. TGF-β activation and function in immunity. Annu Rev Immunol 2014;32:51–82. 10.1146/annurev-immunol-032713-120257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Batlle E, Massagué J. Transforming growth factor-β signaling in immunity and cancer. Immunity 2019;50:924–40. 10.1016/j.immuni.2019.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Massagué J. Tgfbeta in cancer. Cell 2008;134:215–30. 10.1016/j.cell.2008.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pulaski BA, Ostrand-Rosenberg S. Mouse 4T1 breast tumor model. Curr Protoc Immunol 2001;Chapter 20:Unit 20.2 10.1002/0471142735.im2002s39 [DOI] [PubMed] [Google Scholar]
- 27. Sun L, Clavijo PE, Robbins Y, et al. . Inhibiting myeloid-derived suppressor cell trafficking enhances T cell immunotherapy. JCI Insight 2019;4 10.1172/jci.insight.126853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang Z, Li Z, Wang Y, et al. . Versican silencing improves the antitumor efficacy of endostatin by alleviating its induced inflammatory and immunosuppressive changes in the tumor microenvironment. Oncol Rep 2015;33:2981–91. 10.3892/or.2015.3903 [DOI] [PubMed] [Google Scholar]
- 29. Asano K, Nelson CM, Nandadasa S, et al. . Stromal versican regulates tumor growth by promoting angiogenesis. Sci Rep 2017;7:17225 10.1038/s41598-017-17613-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lou Y, Diao L, Cuentas ERP, et al. . Epithelial-Mesenchymal transition is associated with a distinct tumor microenvironment including elevation of inflammatory signals and multiple immune checkpoints in lung adenocarcinoma. Clin Cancer Res 2016;22:3630–42. 10.1158/1078-0432.CCR-15-1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ock C-Y, Kim S, Keam B, et al. . Pd-L1 expression is associated with epithelial-mesenchymal transition in head and neck squamous cell carcinoma. Oncotarget 2016;7:15901–14. 10.18632/oncotarget.7431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. David JM, Dominguez C, McCampbell KK, et al. . A novel bifunctional anti-PD-L1/TGF-β trap fusion protein (M7824) efficiently reverts mesenchymalization of human lung cancer cells. Oncoimmunology 2017;6:e1349589 10.1080/2162402X.2017.1349589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen L, Gibbons DL, Goswami S, et al. . Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat Commun 2014;5:5241 10.1038/ncomms6241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Knudson KM, Hicks KC, Luo X, et al. . M7824, a novel bifunctional anti-PD-L1/TGFβ trap fusion protein, promotes anti-tumor efficacy as monotherapy and in combination with vaccine. Oncoimmunology 2018;7:e1426519 10.1080/2162402X.2018.1426519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lan Y, Zhang D, Xu C, et al. . Enhanced preclinical antitumor activity of M7824, a bifunctional fusion protein simultaneously targeting PD-L1 and TGF-β. Sci Transl Med 2018;10:eaan5488 10.1126/scitranslmed.aan5488 [DOI] [PubMed] [Google Scholar]
- 36. Jochems C, Tritsch SR, Pellom ST, Wang P, et al. . Analyses of functions of an anti-PD-L1/TGFβR2 bispecific fusion protein (M7824). Oncotarget 2017;8:75217–31. 10.18632/oncotarget.20680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Strauss J, Heery CR, Schlom J, et al. . Phase I trial of M7824 (MSB0011359C), a bifunctional fusion protein targeting PD-L1 and TGFβ, in advanced solid tumors. Clin Cancer Res 2018;24:1287–95. 10.1158/1078-0432.CCR-17-2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Carleton M, Zhou M, De Henau O, et al. . Serum interleukin 8 (IL-8) may serve as a biomarker of response to immuno-oncology (I-O) therapy. JCO 2018;36:3025 10.1200/JCO.2018.36.15_suppl.3025 [DOI] [Google Scholar]
- 39. David J, Dominguez C, Hamilton D, et al. . The IL-8/IL-8R axis: a double agent in tumor immune resistance. Vaccines 2016;4:22 10.3390/vaccines4030022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ginestier C, Liu S, Diebel ME, et al. . Cxcr1 blockade selectively targets human breast cancer stem cells in vitro and in xenografts. J Clin Invest 2010;120:485–97. 10.1172/JCI39397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu S, Ginestier C, Ou SJ, et al. . Breast cancer stem cells are regulated by mesenchymal stem cells through cytokine networks. Cancer Res 2011;71:614–24. 10.1158/0008-5472.CAN-10-0538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sanmamed MF, Perez-Gracia JL, Schalper KA, et al. . Changes in serum interleukin-8 (IL-8) levels reflect and predict response to anti-PD-1 treatment in melanoma and non-small-cell lung cancer patients. Ann Oncol 2017;28:1988–95. 10.1093/annonc/mdx190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jamal R, Lapointe R, Cocolakis E, et al. . Peripheral and local predictive immune signatures identified in a phase II trial of ipilimumab with carboplatin/paclitaxel in unresectable stage III or stage IV melanoma. J Immunother Cancer 2017;5:83 10.1186/s40425-017-0290-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liao W, Overman MJ, Boutin AT, et al. . KRAS-IRF2 axis drives immune suppression and immune therapy resistance in colorectal cancer. Cancer Cell 2019;35:559–72. 10.1016/j.ccell.2019.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gubens MA, Sequist LV, Stevenson JP, et al. . Pembrolizumab in combination with ipilimumab as second-line or later therapy for advanced non-small-cell lung cancer: KEYNOTE-021 cohorts D and H. Lung Cancer 2019;130:59–66. 10.1016/j.lungcan.2018.12.015 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2019-000326supp001.pdf (93.1KB, pdf)