Abstract
Background
The effective detection and monitoring of potentially malignant oral lesions (PMOL) are critical to identifying early‐stage cancer and improving outcomes. In the current study, the authors described cytopathology tools, including machine learning algorithms, clinical algorithms, and test reports developed to assist pathologists and clinicians with PMOL evaluation.
Methods
Data were acquired from a multisite clinical validation study of 999 subjects with PMOLs and oral squamous cell carcinoma (OSCC) using a cytology‐on‐a‐chip approach. A machine learning model was trained to recognize and quantify the distributions of 4 cell phenotypes. A least absolute shrinkage and selection operator (lasso) logistic regression model was trained to distinguish PMOLs and cancer across a spectrum of histopathologic diagnoses ranging from benign, to increasing grades of oral epithelial dysplasia (OED), to OSCC using demographics, lesion characteristics, and cell phenotypes. Cytopathology software was developed to assist pathologists in reviewing brush cytology test results, including high‐content cell analyses, data visualization tools, and results reporting.
Results
Cell phenotypes were determined accurately through an automated cytological assay and machine learning approach (99.3% accuracy). Significant differences in cell phenotype distributions across diagnostic categories were found in 3 phenotypes (type 1 [“mature squamous”], type 2 [“small round”], and type 3 [“leukocytes”]). The clinical algorithms resulted in acceptable performance characteristics (area under the curve of 0.81 for benign vs mild dysplasia and 0.95 for benign vs malignancy).
Conclusions
These new cytopathology tools represent a practical solution for rapid PMOL assessment, with the potential to facilitate screening and longitudinal monitoring in primary, secondary, and tertiary clinical care settings.
Keywords: artificial intelligence, biomarkers, cytology, oral epithelial dysplasia, point‐of‐care testing, single‐cell analysis, squamous cell carcinoma
Short abstract
A point‐of‐care oral cytology tool has been developed for the noninvasive detection and monitoring of potentially malignant oral lesions. The distribution of cell phenotypes identified by machine learning and a cytology‐on‐a‐chip approach provides useful information as part of the assessment of oral lesions, with improved interpretability, calibration, and generalizability compared with conventional methods.
Introduction
Cancers of the lip, oral cavity, and pharyngeal subsites are estimated to affect >500,000 individuals globally each year.1 The National Cancer Institute's Surveillance, Epidemiology, and End Results (SEER) program has estimated 53,000 new cases and 10,860 deaths attributed to oral and pharyngeal cancer (OPC) in 2019 in the United States alone, of which approximately 50% will involve oral cavity subsites. Collectively, OPCs represent approximately 3% of all cancers.2 Approximately two‐thirds of OPCs are diagnosed at stage III or stage IV when the 5‐year survival rates are just 45% and 32%, respectively.3 For the remaining one‐third of OPCs detected at early stages (localized),4 survival increases to 84%.2 Despite steady improvements in overall survival rates for patients with OPC over the last 40 years, identifying OPCs at an early stage remains a challenge for oral health care providers.5 The current diagnostic paradigm of procuring a biopsy is based on remote laboratory services, which can take days or weeks to provide results, and this further prolongs anxiety for patients. A point‐of‐care (POC) solution could provide immediate feedback within the same visit. Thus, there is a strong need for technology‐driven solutions that can precisely and rapidly diagnose the entire spectrum of oral epithelial dysplasia (OED) and oral squamous cell carcinoma (OSCC) using minimally invasive sampling at the POC.
A successful diagnostic adjunctive test for primary care settings should be able to discriminate potentially malignant oral lesions (PMOLs) that are at “risk” (ie, malignant lesions or those with an elevated risk of undergoing malignant transformation) from more common benign lesions with no malignant potential, thus improving the referral efficiency to secondary or tertiary care (eg, reducing overreferral of patients with benign lesions and improving the early identification and prompt referral of those with malignant or high‐grade dysplastic PMOLs for oncologic care). Numerous adjunctive tests are available to assist in the diagnosis of PMOLs. In a meta‐analysis of oral cancer adjuncts, vital staining and visualization adjuncts (eg, autofluorescence and tissue reflectance) demonstrated insufficient accuracy to be recommended for use as lesion triage tools by general dentists.6 However, cytology has demonstrated greater sensitivity and specificity compared with the other adjuncts, suggesting its potential as a surrogate for gold‐standard histopathology. This evidence to support the accuracy of cytology is based largely on accuracy studies performed in secondary and tertiary care settings. Although cytology is unable to replace histopathologic diagnosis based on tissue architecture, this relatively inexpensive, easy to perform, and minimally invasive method may be useful for triaging lesions in any setting: primary care settings such as a dental office, low‐resource/remote settings, and secondary and/or tertiary settings. Incisional biopsy followed by histopathologic examination represents the current standard of care for diagnosing PMOLs. However, incisional biopsy of PMOLs, particularly in those that are large, nonhomogeneous leukoplakias, leads to an underestimation of the severity of OED up to 30% of the time because the biopsy sample (typically measuring 5 mm in diameter) may not be representative of the variable pathology across the field of the entire PMOL.7 Brush cytology could enable a wider sampling of PMOLs that encompass larger areas or are multifocal with the potential to reduce sampling errors encountered with incisional biopsies.
Previously, we have demonstrated the conceptual basis and the efficacy of chip‐based cell capture, multispectral fluorescence measurements, and single‐cell analysis approaches yielding high‐content diagnostic information related to oral lesions.8, 9, 10 This compact and integrated lesion diagnostic adjunct approach has been studied previously through a multisite clinical validation effort that has led to the development of what to our knowledge is one of the largest oral cytology databases ever assembled for PMOLs.11, 12 These efforts included the development of an “enhanced gold‐standard” adjudication process12 that was used to correlate brush cytology measurements with 6 levels of histopathological diagnosis, ranging from benign, to OED, to OSCC. The same approach demonstrated strong promise for OSCC surveillance in patients with Fanconi anemia13 and for the development of a cytology‐based numerical risk index for cancer progression.14 Overall, these past efforts have demonstrated that microfluidic‐based cell capture systems with integrated imaging and embedded diagnostic algorithms can yield diagnostic accuracies that rival and exceed the capabilities of previously developed adjunct devices. These tools were developed previously to serve as adjunctive aids capable of distinguishing between high‐risk and low‐risk oral lesions with the goal of improving the pipeline of referrals from primary care settings to secondary and tertiary treatment centers. Thus, these models were intended to assist primary care providers in making binary referral decisions and considered hundreds of complicated image‐based cytomorphometric features with minimal clinical interpretability (ie, “black box”).
The current study targeted the development of a Point‐of‐Care Oral Cytology Tool (POCOCT), which to our knowledge is the first precision oncology technology capable of high‐content cell analysis for near patient testing. The POCOCT platform comprises a minimally invasive brush cytology test kit, disposable assay cartridge, instrument, clinical algorithms, and cloud‐based software services that automate the quantification and analysis of cellular and molecular signatures of dysplasia, with results available in a matter of minutes compared with days for traditional, labor‐intensive, laboratory‐based pathology methods. This article features the development of new diagnostic models using the same database described above with the goal of greatly simplifying the diagnostic algorithms and their interpretation through the classification and quantification of cellular phenotypes, resulting in more informative and transparent models for cytopathologists. Likewise, this work has explored the usefulness of cell phenotype identification through machine learning, their implementation in diagnostic models with interpretable predictors and responses, and the practical application of these software tools in a cytopathology service.
Materials and Methods
Oral Cytology Data
Data used in the current study originated from the 999‐patient, multisite, prospective, noninterventional study evaluating the cytology‐on‐a‐chip system for the measurement of cytological parameters on brush cytology samples to assist in the diagnosis of PMOL.11, 12 Briefly, both histopathological and brush cytological samples for 714 subjects from 3 patient groups were measured: 1) subjects with PMOL who underwent scalpel biopsy as part of the standard of care for microscopic diagnosis; 2) subjects with recently diagnosed malignant lesions; and 3) healthy volunteers without lesions. Histopathological assessment of scalpel biopsy specimens classified lesions into 6 categories (benign; mild, moderate, or severe dysplasia; carcinoma in situ; and OSCC), including healthy controls without lesions. Although the grading of OED traditionally has been considered subjective and lacking intraobserver and interobserver reproducibility,15, 16 this new study implemented an “enhanced gold standard” adjudication.12 Here, 2 adjacent serial histologic sections were scored independently by 2 pathologists. In the event that the pathologists disagreed, a third independent adjudicating pathologist reviewed both sections. If the adjudicator did not agree with either of the initial 2 pathologists, a third‐stage consensus review was conducted to attain a final diagnosis. This “enhanced gold‐standard” process was able to achieve 100% consensus agreement compared with an initial preadjudication agreement rate of 69.9%.
Brush cytology specimens were collected and processed using protocols published previously.11, 12 Cytopathological assessment of brush cytology specimens implemented a cytology‐on‐a‐chip approach, which measured morphological and intensity‐based cell metrics as well as the expression of 6 molecular biomarkers (αvβ6, EGFR, CD147, McM2, geminin, and Ki‐67), resulting in a total of 13 million cells analyzed using >150 image‐based parameters. The molecular biomarkers were selected based on their capacity to distinguish benign, dysplastic, and malignant oral epithelial cells through prior immunohistochemistry studies.9, 17, 18 Specific details regarding the molecular biomarker selection, patient characteristics, sample collection and processing, cytology assay, and cytological parameters were published previously,11 and are summarized in the Supporting Methods.
Cell Identification Model Training and Validation
A cell phenotype classification model was explored for its ability to discriminate and quantitate the frequency and distributions of 4 cell phenotypes: type 1, in which cells present as polygonal in shape with a low nuclear‐cytoplasmic ratio (NC ratio) representing mature squamous epithelial cells; type 2, in which cells present as small round cells representing immature parabasal cells; type 3 in which cells present as mononuclear leukocytes; and type 4, in which cells are represented by lone (naked) nuclei without a cell membrane and cytoplasm. To recognize these cell types, a machine learning algorithm was trained on 144 cellular/nuclear features from single‐cell analyses, including morphological and intensity‐based measurements. Prior to model development, principal component analysis (PCA) was performed on the training set. The PCA method is an unsupervised statistical learning technique for exploratory data analysis that improves data visualization by reducing the dimensionality of complex data sets19 and has been used for phenotypic identification in flow cytometric data.20 Detailed methods for the training and validation of the cell identification model are provided in the Supporting Methods.
Numerical Index and Diagnostic Models for Assessing PMOL
A numerical index was developed for the purpose of discriminating benign from dysplasia/malignant lesions (OED spectrum model 2|3). Detailed methods for the training and validation of the numerical index and a detailed definition of predictors are provided in the Supporting Methods. Briefly, subjects were dichotomized into “case” and “noncase” outcomes according to their lesion determination (noncase for benign lesions and case for [mild, moderate, or severe] dysplasia and malignant lesions). Due to the relatively few numbers of patients with moderate and severe dysplasia (total of 21 patients), these lesion determinations were combined. Least absolute shrinkage and selection operator (lasso) logistic regression was selected for its ability to reduce the number of predictors in high‐dimensional data sets to improve prediction performance and generalizability.21, 22, 23, 24 Nonzero lasso logistic regression coefficients were retained for the following predictors: percentage of nonmature squamous cells, percentage of small round cells, percentage of leukocytes, age, sex, smoking pack‐years, major axis diameter of the lesion, clinical impression of lichen planus, and lesion color (red, white, or red/white). Diagnostic performance was characterized by the area under the curve (AUC), sensitivity, and specificity. Median numerical indices were compared for each diagnostic classification using a 2‐sided Wilcoxon rank sum test at a significance level of P = .05. Internal calibration was performed by sorting and grouping the predicted responses (ie, numerical index) into deciles and measuring the observed percentages of dysplasia/malignant lesions in each decile. The Hosmer‐Lemeshow goodness of fit statistic was used to assess the model fit.21
Following this same method, diagnostic algorithms for mild versus moderate dysplasia (OED‐spectrum model 3|4), low versus high risk (model 4|4), moderate versus severe dysplasia (model 4|5), healthy control (no lesion) versus malignant (model 0|6), and benign versus malignant (model 2|6) also were developed, and the AUC, sensitivity, and specificity were reported as the mean and 95% CIs for the cross‐validated test set.
Cytopathology Software
Measurements of individual cells, such as morphometric appearance and biomarker staining intensity, were recorded using the open‐source software CellProfiler.25 All model development and data analyses were completed using MATLAB R2017b (MathWorks, Natick, Massachusetts) software. A graphical user interface for visualizing cytopathology results was developed in MATLAB R2017b. The results summary report tool was developed using Python 3.6.3. Figures for the cytopathology software interface and the results summary were compiled from tests performed on the integrated POCOCT instrument.
Level of Integration
Data originating from our 999‐patient National Institutes of Health Grand Opportunity (GO) study and used in the cell identification and diagnostic models were collected using nonintegrated cytology‐on‐a‐chip flow cell prototypes, syringe pumps, research microscope stations, and a collection of commercial and open‐source software packages (see Supporting Methods for more details).11 More recently, we integrated the cytology‐on‐a‐chip technology into a POC device comprising integrated instrument, microfluidic cartridges with onboard blister packs, and dedicated software. Likewise, sample processing steps have been reduced significantly. Cell identification and diagnostic models developed on the nonintegrated platform were translated into the POC instrument, and software screenshots and results reports presented herein were completed using this integrated POC platform.
Results
Cell Identification Model
A cell identification tool to assist in the accurate and precise estimation of histopathological endpoints for the entire spectrum of OED and OSCC was developed. Figure 1 shows the diagnostic categories and rates for oral cancer and dysplasia based on the World Health Organization classification26 found during mass screening,27 demonstrating 5‐year malignant transformations28 and 5‐year cancer recurrence rates.29 The literature presents a range of 5‐year transformation and recurrence rates, and we believe the ones listed herein are representative of those reported previously.30
Figure 1.
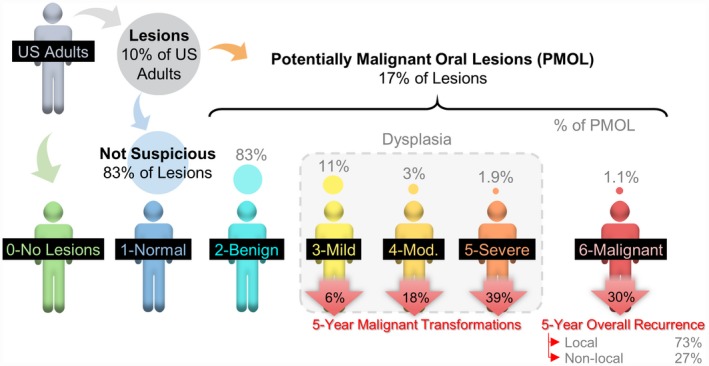
Diagnostic categories for oral cancer and dysplasia based on the World Health Organization classification with 5‐year malignant transformations and 5‐year cancer recurrence rates. Although approximately 10% of US adults may present to their dentist for a routine care visit with an abnormal oral cavity lesion, approximately 83% of these lesions are diagnosed clinically as having no malignant potential, and 17% have unknown significance and meet the clinical criteria for potentially malignant oral lesions (PMOLs). Approximately 17% of patients with PMOLs are histopathologically diagnosed with oral epithelial dysplasia (OED) or oral squamous cell carcinoma (OSCC). OED is approximately 15 times more common than OSCC, yet only a small percentage of patients with dysplastic PMOLs undergo malignant transformation.
The POCOCT platform (Fig. 2) comprises a minimally invasive brush cytology test kit, disposable assay cartridge, instrument, clinical algorithms, and cloud‐based software services to automate the quantification and analysis of cellular and molecular signatures of dysplasia and OSCC. The cell identification tool automatically classified 4 distinct cell phenotypes (Fig. 3A). Type 1 (“mature squamous” or “mature keratinocytes”) were broad/flat cells, measured approximately 50 to 100 µm in diameter, had a low NC ratio, and demonstrated a relatively low cytoplasm staining intensity (Phalloidin‐Alexa Fluor 647; Thermo Fisher Scientific, Waltham, Massachusetts). Type 2 “small round” cells were small (12‐30 µm in diameter), highly circular cells with a high NC ratio and a brightly stained cytoplasm representing immature basaloid keratinocytes. Type 3 “leukocytes” appeared as small, brightly stained pink objects measuring 6 to 23 µm in diameter representing mononuclear leukocytes. Type 4 “lone nuclei” represented by lone or naked nuclei without a cytoplasm appeared as brightly stained blue objects measuring approximately 5 to 12 µm in diameter.
Figure 2.
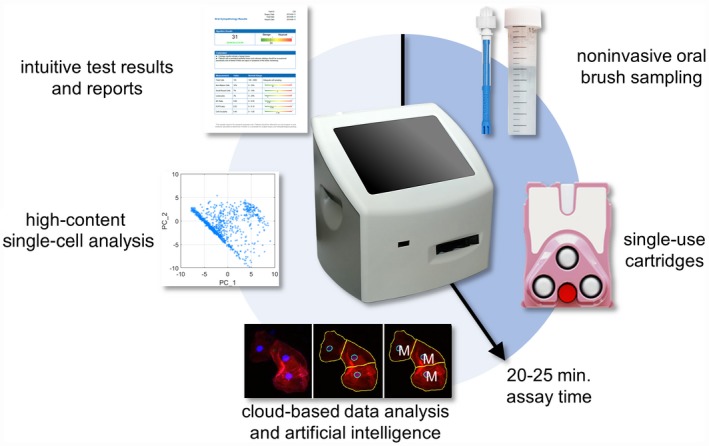
The Point‐of‐Care Oral Cytology Tool (POCOCT) assay platform allows for the analysis of cellular samples obtained from a minimally invasive brush cytology sample. The cell suspension collected in this manner allows for the simultaneous quantification of cell morphometric data and the expression of molecular biomarkers of malignant potential in an automated manner using refined image analysis algorithms based on pattern recognition techniques and advanced statistical methods. This novel approach turns around cytology results in a matter of minutes compared with days for traditional pathology methods, thereby making it amenable to POC settings. The POC testing is expected to have tremendous implications for disease management by enabling dental practitioners and primary care physicians to circumvent the need for multiple referrals and consultations before obtaining assessment of molecular risk of PMOL.
Figure 3.
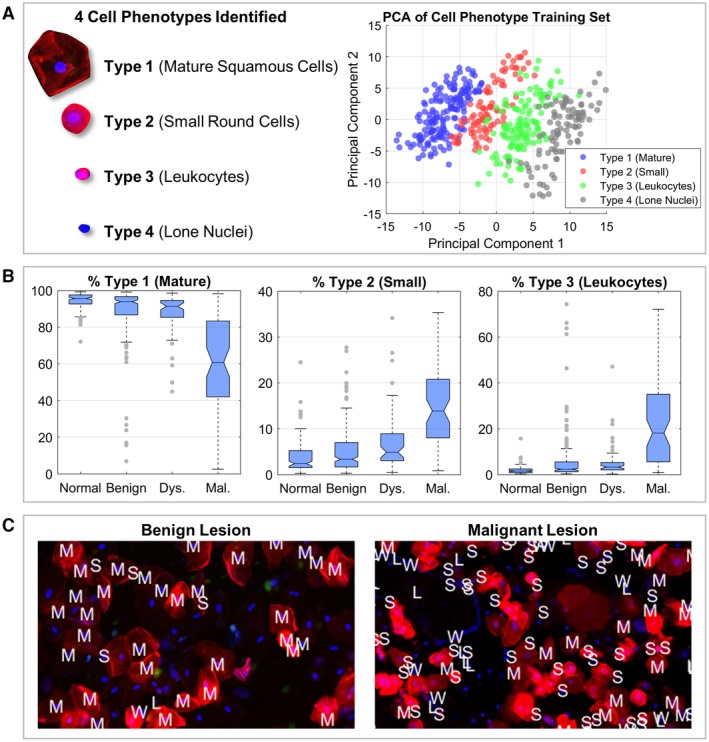
A cell type identification model was developed to automatically classify cell types 1 to 4. Panel A (left) shows the 4 distinct cell phenotypes that were identified: type 1 (“mature squamous cells”), type 2 (“small round cells”), type 3 (“leukocytes”), and type 4 (“lone nuclei”). Principal component analysis (PCA) (right) shows cell phenotypes clustered into distinct groups with substantial separation between cell phenotype labels, demonstrating strong promise for an effective cell phenotype recognition algorithm. Boxplots in panel B show the study population distributions of mature squamous cells (left), small round cells (center), and leukocytes (right), representing the predicted mean cell type percentages across 6 biomarker assays (αvβ6, CD‐147, EGFR, geminin, Ki‐67, and MCM2) within each lesion class: normal (121 cases), benign (241 cases), dysplasia (59 cases), and malignant (65 cases). The results shown include only patients with definitive lesion determinations and patients with evaluable data for all 6 biomarkers. Panel C shows limited field‐of‐view cytology pseudocolor images (fluorescence images acquired with a monochrome camera and digitally assigned to red, green, and blue color channels) of benign (left) and malignant (right) lesions with the cell phenotype model output labels overlaid as follows: “M” indicates mature squamous cells, “S” indicates small round cells, “W” indicates leukocytes, and “L” indicates lone nuclei (unknown type [“U”] not shown). Fluorescent staining showed the cytoplasm (red), nuclei (blue), and Ki‐67 biomarker (green). Dys indicates dysplasia; Mal, malignant.
The PCA scatter plot of the first 2 principal components revealed a glimpse of the internal data structure and variance (Fig. 3A). Here, populations according to each cell type were clearly observed. Furthermore, >90% of the variance was explained by the first 20 principal components from a total of 144, with 30% and 14% variance, respectively, explained in the first and second principal components. Despite types 2 and 3 having similar cytomorphology, the features with the largest association with the first principal component were NC ratio and mean cytoplasm intensity, suggesting that cell size and cellular actin content and/or distribution play a dominant role in explaining the variance among these cell phenotypes.
The cross‐validated k‐nearest neighbors (k‐NN) algorithm resulted in an overall accuracy of 96.9% and an accuracy of 100%, 90.1%, 96.0%, and 99.0%, respectively, for types 1 (mature), 2 (small), 3 (leukocytes), and 4 (lone nuclei) cells. An additional label (“unknown”) was added for cells that had ≤4 similar neighbors. After accounting for this “unknown” cell type, the overall accuracy was 99.3%. When applied to the study population, cell phenotype distributions demonstrated significant differences across all diagnostic categories (Fig. 3B). The percentage of type 1 (mature) cells decreased with more advanced disease. In contrast, the percentages of type 2 (small) and type 3 (leukocytes) cells increased with disease progression. Median values for type 1 (mature) and type 2 (small) cells were significantly different between all lesion determinations. For type 3 cells (leukocytes), all lesion determinations had significantly different median values except for benign versus dysplasia (P = .0539).
The same cell identification model development process was completed on recently developed integrated instrumentation, cartridges, and cloud‐based analysis tools. Images from 2 samples, one each from benign and malignant lesions, were collected using the POCOCT platform, and cell phenotype labels were overlaid on each recognized cell object (Fig. 3C). Here, the benign lesion sample contained mostly type 1 (mature) cells, whereas the malignant sample contained a mixture of primarily type 2 (small), type 3 (leukocytes), and type 4 (lone nuclei) cells.
Numerical Index and Diagnostic Models for Assessing PMOL
Expanding on this capability, a numerical index for discriminating benign from dysplasia/malignant lesions was developed using the cell phenotypes as predictors. Figure 4A shows the receiver operating characteristic curve representing discrimination performance of the multivariate model. The numerical index is a score between 0 and 100 that can be interpreted literally as the probability of dysplasia/malignancy. The diagnostic accuracy of the model is defined by the cutoff score that maximizes its AUC (benign vs dysplasia/malignant numerical index cutoff value of 36). Predictors for the model were retained as follows: cell phenotype distributions (types 1, 2, and 3), age, sex, smoking pack‐years (ie, packs smoked per day times years of smoking), lesion size (maximum diameter), clinical impression of lesion as lichen planus, and lesion color (white, red, or both) (Fig. 4B). Minimal differences were observed between training and test error (28% and 27% misclassification rate, respectively, on the training and test sets), which suggests no evidence of overfitting. The numerical index demonstrated significant differences between all lesion diagnostic categories studied (P < .01) except for mild versus moderate/severe dysplasia (P = .1519) (Fig. 4C); however, significant differences were observed in a dichotomous model for mild versus moderate dysplasia (ie, model 3|4) (P = .04). Model calibration demonstrated the numerical index compared with the observed percentages of dysplasia/malignant subjects when sorted and grouped into deciles (Fig. 4D). A nonsignificant result of the Hosmer‐Lemeshow goodness of fit test suggested that there was no evidence of a poor fit (P = .6259).
Figure 4.
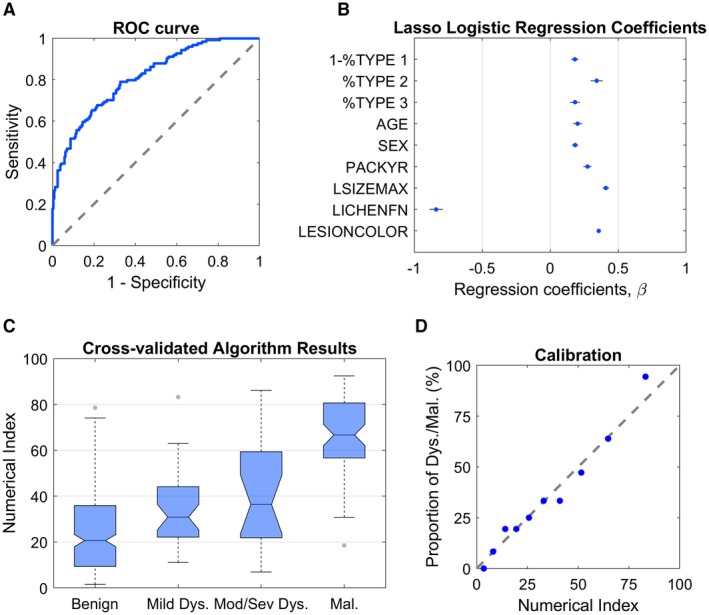
Algorithm results of the dichotomous benign versus dysplasia/malignant lesion model from 241 subjects with benign lesions and 124 subjects with dysplasia and malignant lesions for 6 molecular biomarker assays on the Point‐of‐Care Oral Cytology Tool (POCOCT) system. Panel A shows the receiver operating characteristic (ROC) curve for the model. The least absolute shrinkage and selection operator (lasso) logistic regression coefficients are provided in panel B. The predictors were as follows: “1‐%TYPE 1” (percentage of cells that were nonmature squamous cells), “%TYPE 2” (percentage of cells that were small round cells), “%TYPE 3” (percentage of cells that were leukocytes), “AGE,” “SEX,” “PACKYR” (pack‐years of smoking), “LSIZEMAX” (lesion diameter of the major axis), “LICHENFN” (clinical impression of lichen planus), and “LESIONCOLOR” (red, white, or red/white). The boxplot in panel C shows cross‐validated algorithm response (“numerical index”) for the lasso logistic regression on the test set averaged over all biomarker assays. Distribution of scores are represented for benign (241 lesions), mild dysplasia (38 lesions), moderate/severe dysplasia (21 lesions), and malignant (65 lesions). Panel D shows a model calibration plot of the predicted responses (numerical index) sorted and grouped into deciles versus the observed percentages of dysplasia and malignant lesions. Mod/sev dys indicates moderate/severe dysplasia.
Models also were developed for dichotomous classification across the OED spectrum, and Figure 5 summarizes the diagnostic performance of these models. The clinical algorithms resulted in AUCs ranging from 0.81 (95% CI, 0.76‐0.86) for benign versus mild dysplasia (model 3|4) to 0.97 (95% CI, 0.94‐1.00) for healthy controls (no lesion) versus malignancy (model 0|6). Although previous work demonstrated AUCs of 0.836 for the binary low‐risk versus high‐risk (model 4|4) split and 0.883 for moderate versus severe dysplasia (model 4|5),11 these new optimized models presented herein resulted in improved AUCs of 0.88 (95% CI, 0.84‐0.93) and 0.92 (95% CI, 0.88‐0.96) for the same diagnostic splits, respectively.
Figure 5.
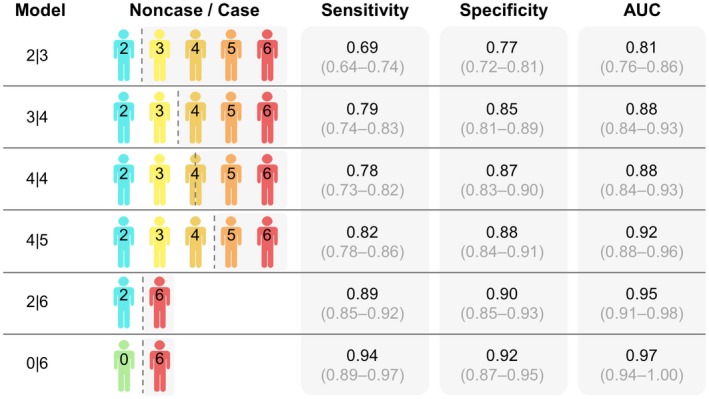
Diagnostic models for the oral epithelial dysplasia (OED) spectrum. Results are shown for the cross‐validated clinical algorithms for benign versus dysplasia (model 2|3), mild versus moderate dysplasia (model 3|4), low versus high risk (model 4|4), moderate versus severe dysplasia (model 4|5), healthy control (no lesion) versus malignant (model 0|6), and benign dysplasia versus malignant (model 2|6) models. Model responses for each subject were averaged over all biomarker assays to inform diagnostic performance. The area under the curve (AUC), sensitivity, and specificity are shown as the mean and 95% CI values for the cross‐validated test set.
Cytopathology Software
A cytopathology interface tool was developed to assist pathologists in reviewing the brush cytology test results, enabling rich content cellular analyses on single‐cell and multicell levels (Fig. 6) (see Supporting Figs. 1‐6). This interface enabled the pathologist users to access data stored and processed on cloud‐based services, view results summaries, explore cytology results through data visualization tools, and generate automated oral cytopathology reports (Fig. 7) which provide the adjunctive referral recommendations and summarize important information from cytology, including total cell count, cell phenotype distributions (types 1, 2, and 3), and mean values for the NC ratio, molecular biomarker fluorescence intensity, and cell circularity. The ability to assess cumulative data on this cloud‐based cytopathology platform may improve pathologist decision making (eg, through learning about their own histopathologic assessment versus the POCOCT and, ultimately, the surgical pathology).
Figure 6.
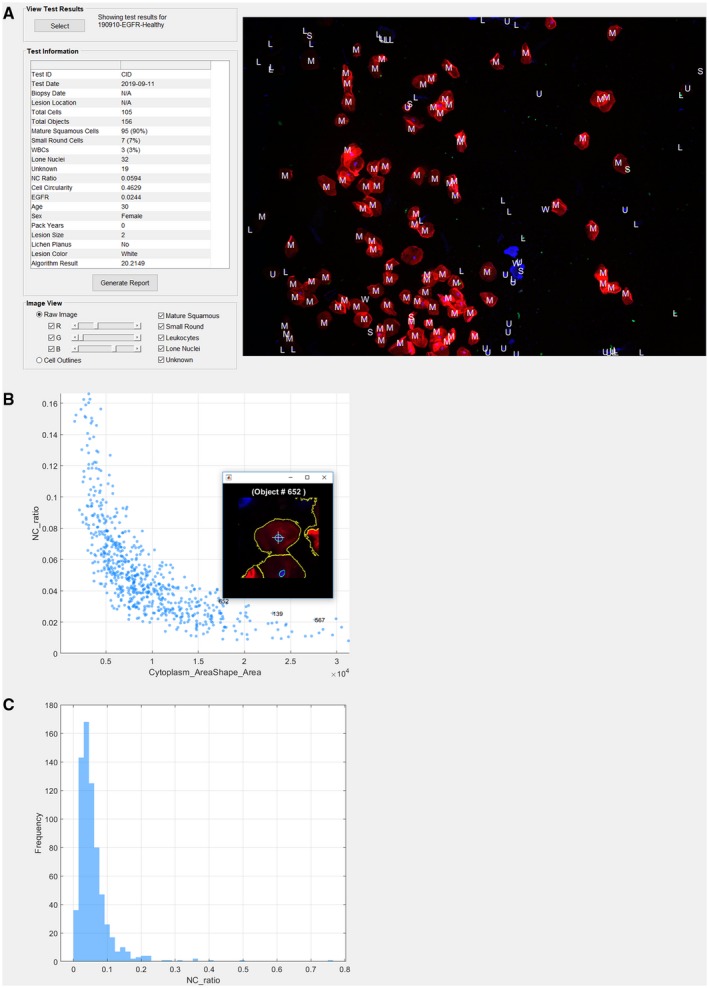
The cytopathology interface tool provides pathologists with cloud access to test results summaries and detailed (A) data visualizations, (B) scatter plots, and (C) histograms for >150 different cytology parameters. With this tool, pathologists can view all cells within the field of view, zoom in for more detail, and isolate individual cells of interest. L indicates lone nuclei; M, mature squamous cells; NC, nuclear‐cytoplasmic ratio; S, small round cells; U, unknown type; W, leukocytes; WBC, white blood cell.
Figure 7.
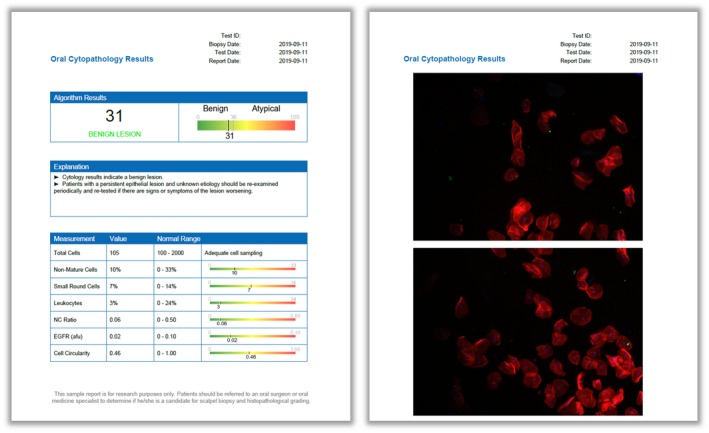
Oral cytopathology test results. The algorithm result is a numerical index between 0 and 100 with a cutoff value of 36 that distinguishes benign and dysplasia/malignant (“atypical”) lesions (left). Other informative cytopathology results are shown on a reference range, including total cell counts, cell phenotype distributions, mean values for the nuclear‐cytoplasmic (NC) ratio, molecular biomarker fluorescence intensity, and cell circularity. Images and outlines of the cells are provided for additional test context (right). afu indicates arbitrary fluorescence units.
Discussion
The results of the current study demonstrated an evolution of the POCOCT technology toward a rapid and simple brush cytology analysis for POC or in a remote laboratory setting. We demonstrated that: 1) cell phenotypes can be determined accurately through the automated cytological assay and machine learning approach; 2) significant differences in cell phenotype distributions across diagnostic categories are found in 3 phenotypes (types 1, 2, and 3); and 3) these cell phenotypes are valuable predictors in distinguishing lesion diagnostic categories in a multivariate lasso logistic regression model. The compilation of these results has suggested that the observed cellular phenotypic variations within cytological samples are equated with disease severity, and therefore may be useful in the evaluation of PMOLs. Although cell phenotyping can be completed by a pathologist by manually identifying cells in a cytological sample, this is a lengthy process that is subject to human errors. Providing a means with which to automate metrics, such as the distributions of cell phenotypes, may increase the adoption of this POCOCT approach through a cytopathology service and allow for pathologists to complete more efficient and more effective recommendations.
The optimized numerical index for evaluating PMOLs developed herein represents a simple, practical, and effective approach that is directly applicable to clinical implementation and interpretation. Although previous models have relied on complicated high‐dimensional cytological parameters, the classification and quantitation of cell phenotypes greatly simplifies the predictive algorithm and its interpretation, substantially improves performance for diagnostic splits compared with these earlier efforts,11, 14 and supports the translation of research methodologies from laboratory‐based microscopy stations into an integrated POC instrument. With a total of 9 predictors, the practical model developed in the current study represents a sparse solution (ie, reduction of >150 variables to 9) with greater potential generalizability without sacrificing any diagnostic performance. Furthermore, excellent model calibration performance and significant differences between the diagnostic endpoints has demonstrated strong potential for the numerical index as a continuous indicator of PMOL risk. Although previous work was focused primarily on delivering binary results for referral decisions,11 this new work involves a cytopathology interface tool, which was developed to assist pathologists in reviewing the brush cytology test results, and a numerical index, thereby enabling rich content cellular analyses on single‐cell and multicell levels. This interface enables the pathologist to access data stored on cloud‐based services, view results summaries, explore cytology data through data visualization tools, and generate a report that provides recommendations. Accurate diagnostic models spanning the entire OED spectrum also have demonstrated the potential for the POCOCT to be used for multiple applications, such as screening PMOLs in primary care and the surveillance of patients with a history of OED and OSCC in secondary or tertiary care settings.
Although light‐based adjuncts offer clinicians a new perspective with which to view a lesion at the POC, to the best of our knowledge their diagnostic usefulness remains unproven.5 Rashid and Warnakulasuriya reviewed the performance of light‐based adjuncts in discriminating between low‐risk and high‐risk lesions (VELscope [Apteryx Imaging; Akron, Ohio] [sensitivity/specificity: 30%‐100%/15%‐100%], ViziLite Plus [Zila Pharmaceuticals, Phoenix, Arizona] [sensitivity/specificity: 0%‐100%/0%‐78%], and Microlux DL [Addent Inc, Danbury, Connecticut] [sensitivity/specificity: 78%/71%]) and concluded that there was insufficient evidence to validate their efficacy as screening adjuncts.31 Despite the numerous adjunctive tests currently available to assist in the diagnosis of PMOLs, to the best of our knowledge only cytology has demonstrated potential as a surrogate for gold‐standard histopathology.32 Several commercial cytopathology services currently exist, including OralCDx (CDx Diagnostics Inc, Suffern, New York), OralCyte (ClearCyte Diagnostics Inc, Bellevue, Washington), Cyt ID (Forward Science, Houston, Texas), and ClearPrep OC (Resolution Biomedical, Tustin, California). OralCDx, for example, provides an oral brush sample collection kit for their BrushTest.33 Despite the ease of collection, samples need to be shipped to a commercial laboratory for analysis, resulting in delays between sample collection and test results. Furthermore, the test often returns an ambiguous “atypical” result for which the positive predictive value for dysplasia or carcinoma has been determined to be only 30% to 40%.34 In addition, prior studies of cytology adjuncts demonstrated methodological gaps by performing only matched gold‐standard histopathology on a subset of lesions with a higher index of suspicion for malignancy, and not for lesions with a lower index of suspicion, which frequently are encountered in primary care settings.35, 36 A clinically validated POC cytology service capable of distinguishing the degree of OED in PMOL and stratifying the risk of malignant progression as a numerical index in near real time would fulfill a significant unmet need, mitigating unnecessary referrals to experts, leading to a more efficient process in surveillance clinics, and reducing the patient distress related to waiting for test results.
One limitation of the current study was that previous studies of the POCOCT, and cytology adjuncts in general, primarily focused on PMOL evaluation in secondary care settings in which the prevalence of dysplastic and malignant lesions may be substantially higher than in the primary care setting. In addition, although expert clinicians in secondary and tertiary care settings have extensive training and experience in the recognition and risk stratification of PMOLs, primary care clinicians may have difficulty distinguishing PMOLs from normal/nonneoplastic lesions. Thus, the POCOCT technology may potentially have a larger impact in primary care settings, in which there is a strong need to accurately interrogate the PMOLs detected there and generate a dichotomous outcome to indicate whether referral of patients to higher care settings for expert evaluation and possible biopsy is required and if such referral should be urgent.
The current study has provided a key step toward the development of new tools that could pave the way for new capabilities in the area of “precision lesion diagnostics.” Helping to push forward this theme, we have demonstrated the usefulness of temporal changes in the numerical index in a pilot study of patients with Fanconi anemia.13 These efforts demonstrated strong potential for patient‐specific temporal changes in the lesion numerical index to track early signs of disease for this high‐risk population. Plans currently are in place to: 1) evaluate the POCOCT's precision lesion diagnostic capabilities through a prospective longitudinal study of malignant transformation and cancer recurrence; and 2) move the POCOCT into a clinical trial to assess its diagnostic performance versus routine care in primary care clinics.
Conclusions
The results of the current study have demonstrated the usefulness of a POC‐amenable cytology platform that has the potential to screen and monitor oral lesions across the entire diagnostic spectrum of OED. Cell phenotype distributions provided additional information in the assessment of PMOL. Furthermore, a practical model comprised of patient information, lesion characteristics, and cell types from cytology demonstrated performance characteristics similar to those of more complicated models that have been developed previously. Cytopathology software may assist expert pathologists and nonexpert care providers in reviewing and understanding the brush cytology test results. We developed data visualization tools to provide high‐content cellular analyses on single‐cell and multicell levels with full transparency of test results data for pathologists. In addition, oral cytopathology results summarized the test's most important predictors through indications of potential lesion progression for care providers and patients. Along with recently developed instrumentation and cartridges, this simple and sensitive system could provide noninvasive triage for PMOLs detected in primary, secondary, and tertiary care settings.
Future work may expand the use of molecular biomarkers and explore the identification of additional rare cell phenotypes to further improve performance. Future clinical studies also may be directed to determine whether brush cytology could enable a wider sampling of large/multifocal lesions compared with incisional biopsies via multiple site‐precise samplings, the effect of inflammation on the cytological analysis, whether the system can identify candida and distinguish clinical leukoplakia from neoplastic versus nonneoplastic conditions, and its placement in existing monitoring algorithms for PMOLs. Clinical trials are needed to assess the POCOCT's ability to identify early‐stage cancer compared with existing protocols and to validate the POCOCT as a substitute for biopsy. Future publications will describe and validate the integrated POC hardware (ie, instrument, cartridge, and assay). To accelerate the translation and expand the adoption of the POCOCT platform, a cytopathology service for secondary and tertiary care oral cytology applications currently is in development. Scaling and distribution of this versatile cytology approach is now underway with the potential to serve diagnostic and surveillance applications in primary, secondary, and tertiary care settings.
Funding Support
Funding for this work was provided by the National Institutes of Health through the National Institute of Dental and Craniofacial Research (awards 1RC2DE020785‐01, 5RC2DE020785‐02, 3RC2DE020785‐02S1, 3RC2DE020785‐02S2, 4R44DE025798‐02, and R01DE024392). The content of this article is solely the responsibility of the authors and does not necessarily represent or reflect the official views of the National Institutes of Health or the US government. Segments of this work were supported by Renaissance Health Service Corporation and Delta Dental of Michigan.
Conflict of Interest Disclosures
Michael P. McRae has served as paid consultant for SensoDx and has a provisional patent pending. Glennon W. Simmons has patents US10060937B2 and US7781226B2 issued. Denise A. Trochesset has received grants from the New York University College of Dentistry for work performed as part of the current study. Martin H. Thornhill has received National Institutes of Health grant 1RC2DE020785‐01 for work performed as part of the current study. Spencer W. Redding has patent US9535068B2 issued. Stella K. Kang has received royalties from Wolters Kluwer for work performed outside of the current study. John T. McDevitt has received grants from the National Institutes of Health for work performed as part of the current study (grants 1RC2DE020785‐01, 4R44DE025798‐02, and R01DE024392) and has a provisional patent pending. In addition, he has an ownership position and an equity interest in SensoDx II LLC and also serves on its Scientific Advisory Board. The other authors made no disclosures.
Author Contributions
Michael P. McRae: Conceptualization, investigation, data curation, formal analysis, methodology, software, validation, visualization, writing–original draft, and writing–review and editing. Sayli S. Modak: Investigation, validation, and writing–review and editing. Glennon W. Simmons: Project administration, investigation, methodology, data collection, and writing–review and editing. Denise A. Trochesset: Conceptualization and writing–review and editing. A. Ross Kerr: Conceptualization and writing–review and editing. Martin H. Thornhill: Conceptualization and writing–review and editing. Spencer W. Redding: Conceptualization and writing–review and editing. Nadarajah Vigneswaran: Conceptualization and writing–review and editing. Stella K. Kang: Conceptualization and writing–review and editing. Nicolaos J. Christodoulides: Conceptualization, writing–original draft, and writing–review and editing. Craig Murdoch: Investigation and writing–review and editing. Steven J. Dietl: Methodology and writing–review and editing. Roger Markham: Methodology and writing–review and editing. John T. McDevitt: Conceptualization, funding acquisition, project administration, resources, supervision, writing–original draft, and writing–review and editing.
Supporting information
We thank the University of Texas Health Science Center at San Antonio (Stephanie Rowan, Chih‐Ko Yeh, Stan McGuff, and Frank Miller), the University of Texas Health Science Center at Houston (Jerry Bouquot, Nagi Demian, Etan Weinstock, and Nancy Bass), New York University/Bluestone Center for Clinical Research (Joan Phelan, Patricia Corby, and Ismael Khouly), and Sheffield Teaching Hospitals NHS Foundation Trust and the University of Sheffield (Paul Speight, Christine Freeman, Anne Hegarty, and Katy D’Apice) for assistance in obtaining clinical samples. We also thank Rho Inc (Julie Vick) for assisting with patient data management and for statistical and data analysis support (Robert James). Finally, we thank Shannon Weigum, Pierre Floriano and Timothy J. Abram for early contributions to the project, including assay development and database organization.
References
- 1. Shield KD, Ferlay J, Jemal A, et al. The global incidence of lip, oral cavity, and pharyngeal cancers by subsite in 2012. CA Cancer J Clin. 2017;67:51‐64. [DOI] [PubMed] [Google Scholar]
- 2. National Cancer Institute Surveillance, Epidemiology, and End Results Program . Cancer stat facts: oral cancer and pharynx cancer. Accessed May 10, 2019. https://seer.cancer.gov/statfacts/html/oralcav.html
- 3. Neville BW, Damm DD, Allen CM, Chi AC. Epithelial pathology In: Neville BW, Damm DD, Allen CM, Chi AC, eds. Oral Maxillofacial Pathology. 4th ed Elsevier Health Sciences; 2015:331‐421. [Google Scholar]
- 4. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7‐30. [DOI] [PubMed] [Google Scholar]
- 5. Huber MA. Adjunctive diagnostic techniques for oral and oropharyngeal cancer discovery. Dent Clin North Am. 2018;62:59‐75. [DOI] [PubMed] [Google Scholar]
- 6. Lingen MW, Abt E, Agrawal N, et al. Evidence‐based clinical practice guideline for the evaluation of potentially malignant disorders in the oral cavity: a report of the American Dental Association. J Am Dent Assoc. 2017;148:712‐727.e10. [DOI] [PubMed] [Google Scholar]
- 7. Lee JJ, Hung HC, Cheng SJ, et al. Factors associated with underdiagnosis from incisional biopsy of oral leukoplakic lesions. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2007;104:217‐225. [DOI] [PubMed] [Google Scholar]
- 8. Weigum SE, Floriano PN, Christodoulides N, McDevitt JT. Cell‐based sensor for analysis of EGFR biomarker expression in oral cancer. Lab Chip. 2007;7:995‐1003. [DOI] [PubMed] [Google Scholar]
- 9. Weigum SE, Floriano PN, Redding SW, et al. Nano‐bio‐chip sensor platform for examination of oral exfoliative cytology. Cancer Prev Res. 2010;3:518‐528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McDevitt JT, Weigum SE, Floriano PN, et al. A new bio‐nanochip sensor aids oral cancer detection. SPIE Newsroom. March 28, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Abram TJ, Floriano PN, Christodoulides N, et al. ‘Cytology‐on‐a‐chip’ based sensors for monitoring of potentially malignant oral lesions. Oral Oncol. 2016;60:103‐111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Speight PM, Abram TJ, Floriano PN, et al. Interobserver agreement in dysplasia grading: toward an enhanced gold standard for clinical pathology trials. Oral Surg Oral Med Oral Pathol Oral Radiol. 2015;120:474‐482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Abram TJ, Pickering CR, Lang AK, et al. Risk stratification of oral potentially malignant disorders in Fanconi Anemia patients using autofluorescence imaging and cytology‐on‐a chip assay. Transl Oncol. 2018;11:477‐486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Abram TJ, Floriano PN, James R, et al. Development of a cytology‐based multivariate analytical risk index for oral cancer. Oral Oncol. 2019;92:6‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bosman FT. Dysplasia classification: pathology in disgrace? J Pathol. 2001;194:143‐144. [DOI] [PubMed] [Google Scholar]
- 16. Warnakulasuriya S, Reibel J, Bouquot J, Dabelsteen E. Oral epithelial dysplasia classification systems: predictive value, utility, weaknesses and scope for improvement. J Oral Pathol Med. 2008;37:127‐133. [DOI] [PubMed] [Google Scholar]
- 17. Vigneswaran N, Beckers S, Waigel S, et al. Increased EMMPRIN (CD 147) expression during oral carcinogenesis. Exp Mol Pathol. 2006;80:147‐159. [DOI] [PubMed] [Google Scholar]
- 18. Torres‐Rendon A, Roy S, Craig GT, Speight PM. Expression of Mcm2, geminin and Ki67 in normal oral mucosa, oral epithelial dysplasias and their corresponding squamous‐cell carcinomas. Br J Cancer. 2009;100:1128‐1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jolliffe I. Principal Component Analysis. 2nd ed Springer; 2011. [Google Scholar]
- 20. Lugli E, Pinti M, Nasi M, et al. Subject classification obtained by cluster analysis and principal component analysis applied to flow cytometric data. Cytometry A. 2007;71A:334‐344. [DOI] [PubMed] [Google Scholar]
- 21. Hosmer DW, Lemeshow S. Applied Logistic Regression. 2nd ed John Wiley & Sons Inc; 2004. [Google Scholar]
- 22. LaValley MP. Logistic regression. Circulation. 2008;117:2395‐2399. [DOI] [PubMed] [Google Scholar]
- 23. Hastie T, Tibshirani R, Friedman JH. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. 2nd ed Springer; 2009. [Google Scholar]
- 24. Wang D, Zhang W, Bakhai A. Comparison of Bayesian model averaging and stepwise methods for model selection in logistic regression. Stat Med. 2004;23:3451‐3467. [DOI] [PubMed] [Google Scholar]
- 25. Carpenter AE, Jones TR, Lamprecht MR, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. El‐Naggar AK, Chan JK, Grandis JR, Takata T, Slootweg PJ. WHO Classification of Tumours of the Head and Neck. 4th ed IARC Press; 2017. [Google Scholar]
- 27. Bouquot JE. Common oral lesions found during a mass screening examination. J Am Dent Assoc. 1986;112:50‐57. [DOI] [PubMed] [Google Scholar]
- 28. Sperandio M, Brown AL, Lock C, et al. Predictive value of dysplasia grading and DNA ploidy in malignant transformation of oral potentially malignant disorders. Cancer Prev Res. 2013;6:822‐831. [DOI] [PubMed] [Google Scholar]
- 29. Brands MT, Smeekens EAJ, Takes RP, et al. Time patterns of recurrence and second primary tumors in a large cohort of patients treated for oral cavity cancer. Cancer Med. 2019;8:5810‐5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mehanna HM, Rattay T, Smith J, McConkey CC. Treatment and follow‐up of oral dysplasia–a systematic review and meta‐analysis. Head Neck. 2009;31:1600‐1609. [DOI] [PubMed] [Google Scholar]
- 31. Rashid A, Warnakulasuriya S. The use of light‐based (optical) detection systems as adjuncts in the detection of oral cancer and oral potentially malignant disorders: a systematic review. J Oral Pathol Med. 2015;44:307‐328. [DOI] [PubMed] [Google Scholar]
- 32. Lingen MW, Tampi MP, Urquhart O, et al. Adjuncts for the evaluation of potentially malignant disorders in the oral cavity: diagnostic test accuracy systematic review and meta‐analysis–a report of the American Dental Association. J Am Dent Assoc. 2017;148:797‐813.e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. CDx Diagnostics . The painless test for common oral spots. Accessed May 10, 2019. https://www.cdxdiagnostics.com/brushtest/
- 34. Svirsky JA, Burns JC, Carpenter WM, et al. Comparison of computer‐assisted brush biopsy results with follow up scalpel biopsy and histology. Gen Dent. 2002;50:500‐503. [PubMed] [Google Scholar]
- 35. Sciubba JJ. Improving detection of precancerous and cancerous oral lesions: computer‐assisted analysis of the oral brush biopsy. J Am Dent Assoc. 1999;130:1445‐1457. [DOI] [PubMed] [Google Scholar]
- 36. Poate TW, Buchanan JA, Hodgson TA, et al. An audit of the efficacy of the oral brush biopsy technique in a specialist Oral Medicine unit. Oral Oncol. 2004;40:829‐834. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


