Abstract
HIV‐1 Tat is essential for HIV‐1 replication and appears to play an important role in the pathogenesis of HIV‐associated neurological complications. Secreted from infected or transfected cells, Tat has the extraordinary ability to cross the plasma membrane. In the brain, Tat can be taken up by CNS cells via receptor‐mediated endocytosis. Following endocytosis and its internalization into endolysosomes, Tat must be released in order for it to activate the HIV‐1 LTR promoter and facilitate HIV‐1 viral replication in the nucleus. However, the underlying mechanisms whereby Tat escapes endolysosomes remain unclear. Because Tat disrupts intracellular calcium homeostasis, we investigated the involvement of calcium in Tat endolysosome escape and subsequent LTR transactivation. We demonstrated that chelating endolysosome calcium with high‐affinity rhodamine‐dextran or chelating cytosolic calcium with BAPTA‐AM attenuated Tat endolysosome escape and LTR transactivation. Significantly, we demonstrated that pharmacologically blocking and knocking down the endolysosome‐resident two‐pore channels (TPCs) attenuated Tat endolysosome escape and LTR transactivation. This calcium‐mediated effect appears to be selective for TPCs because knocking down TRPML1 calcium channels was without effect. Our findings suggest that calcium released from TPCs is involved in Tat endolysosome escape and subsequent LTR transactivation. TPCs might represent a novel therapeutic target against HIV‐1 infection and HIV‐associated neurological complications.
Keywords: HIV‐1 LTR transactivation, HIV‐1 Tat, Tat endolysosome escape, two‐pore channels
Abbreviations
- BAPTA‐AM
1,2‐bis(2‐aminophenoxy)ethane‐N,N,N′,N′‐tetraacetic acid tetrakis(acetoxymethyl ester)
- CD26
cluster of differentiation 4
- CD4+
cluster of differentiation 4
- CNS
central nervous system
- CRISPR
clustered regularly interspaced short palindromic repeats
- CSF
cerebrospinal fluid
- CXCR4
C‐X‐C chemokine receptor type 4
- DMEM
Dulbecco's Modified Eagle Medium
- FBS
fetal bovine serum
- FITC
fluorescein isothiocyanate
- GAPDH
glyceraldehyde 3‐phosphate dehydrogenase
- GFP
green fluorescent protein
- gp120
envelope glycoprotein 120
- HCl
hydrogen chloride
- HIV‐1
human immunodeficiency virus‐1
- kDa
kilodalton
- LRP1
low density lipoprotein receptor‐related protein 1
- LTR
long terminal repeat
- MERS‐CoV
middle east respiratory syndrome coronavirus
- MW
molecular weight
- NAADP
nicotinic acid adenine dinucleotide phosphate
- P2X4
purinergic P2X4 receptors
- PAGE
polyacrylamide gel electrophoresis
- PBS
phosphate‐buffered saline
- PI(3,5)P2
phosphatidylinositol 3,5‐bisphosphate
- PVDF
polyvinylidene difluoride
- RIPA
radioimmunoprecipitation assay buffer
- SD
standard deviation
- SDS
sodium dodecyl sulfate
- shRNA
short hairpin RNA
- Tat
transcriptional activator
- TFEB
transcription factor EB
- TPCs
two‐pore channels
- Trans‐Ned19
(1R,3S)‐1‐[3‐[[4‐(2‐Fluorophenyl)piperazin‐1‐yl]methyl]‐4‐methoxyphenyl]‐2,3,4,9‐tetrahydro‐1H‐pyrido[3,4‐b]indole‐3‐carboxylic acid
- TRPM2
transient receptor potential melastatin 2
- TRPML1
transient receptor potential mucolipin 1
1. INTRODUCTION
Infecting 36.9 million people globally, HIV‐1 virus can enter the brain early after infection1, 2 and it can harbor in CSF, perivascular macrophages, microglia, and astrocytes.3 Although combined antiretroviral therapy (ART) effectively suppresses HIV‐1 replication, it does not eliminate the virus. Resting memory CD4+ T cells in the blood are the best characterized cellular HIV‐1 reservoirs.4 However, there are other sites of HIV persistence in the body. The brain is unique in terms of its “immune privileged” status and evidence does suggest that the brain is a viral reservoir for HIV‐1.5, 6 In brain, macrophages, microglia, and astrocytes can be infected by HIV‐1; HIV‐1 infection of macrophages and microglia is more productive than astrocytes.7 Although endocytosis is involved in HIV‐1 entry in both macrophages7, 8 and astrocytes,7 different receptors are used for HIV‐1 entry in macrophages or astrocytes and this could help explain the restrictive nature of HIV‐1 infection in astrocytes. It has been reported that up to 19% astrocytes carry HIV‐1 DNA in HIV‐1 infected brain9, 10 and because of their abundance, astrocytes could be an important viral reservoir in the brain.
The existence of HIV‐1 reservoirs in peripheral cells and brain cells5, 6 and the persistence of chronic low levels of neuroinflammation might be responsible for the 30%‐50% prevalence rates of HIV‐associated neurocognitive disorders in this ART era.11, 12 The existence of such viral reservoirs that act as sanctuaries for HIV‐1 has made the complete eradication of HIV‐1 extremely challenging.10, 13, 14 Therefore, additional potential strategies are needed to block viral reactivation to maintain HIV‐1 in a state of prolonged silencing and to prevent disease progression and the development of HIV‐induced CNS dysfunction.
The HIV‐1 viral protein Tat represents an attractive target for the eradication of HIV‐1 because Tat is essential for viral transcription15, 16, 17 and because current ART does not block the secretion of Tat in HIV‐1 infected individuals.18, 19 Further, brain levels of Tat remain elevated even though HIV‐1 virus is below detectable levels.19 Actively secreted from HIV‐1 infected cells,20, 21, 22, 23 Tat can bind to cell surface receptors including LRP1, heparin sulfate proteoglycan, CD26, and CXCR4, and enters cells via receptor‐mediated endocytosis.24, 25, 26, 27 The arginine‐rich basic domain of Tat48‐60 is responsible for cellular entry,28, 29, 30, 31 and, as such, Tat48‐60 is widely used for transporting a variety of macromolecules into cells.32, 33 In brain, secreted Tat is taken up rapidly by CNS cells via endocytosis by interacting with specific cell surface proteins and receptors.24, 25, 26, 27, 34, 35 Following its endocytosis and internalization into endolysosomes, Tat must undergo endolysosome escape into the cytosol, where it can transit to the nucleus and activate the HIV‐1 LTR promoter.36, 37, 38 However, it is not clear how Tat escapes endolysosomes.
Others and we have shown that exogenous Tat leads to calcium dyshomeostasis in neuronal and non‐neuronal cells.39, 40, 41, 42, 43, 44, 45 Accordingly, we investigated the involvement of calcium in Tat endolysosome escape and subsequent Tat‐mediated LTR transactivation using split‐GFP Tat endolysosome escape and Tat‐mediated LTR transactivation assays. We demonstrated blocking endolysosome‐resident two‐pore channels (TPCs) attenuated Tat endolysosome escape and subsequent Tat‐mediated LTR transactivation.
2. MATERIAL AND METHODS
2.1. Cell culture
U87MG glioblastoma cells were cultured in DMEM (Invitrogen) supplemented with 10% fetal calf serum and penicillin/streptomycin (Invitrogen). U87MG cells were stably transfected with luciferase reporter gene under the control of HIV‐1 LTR promoter following neomycin (Sigma‐Aldrich) selection pressure. These cells were provided generously by Dr Lena Al‐Harthi (Rush University, Chicago). H1299 cells with stable GFPβ1‐10 expression under hygromycin selection were maintained in DMEM supplemented with 10% FBS and penicillin/streptomycin (Invitrogen).
2.2. Luciferase reporter assay for Tat‐mediated LTR transactivation
U87MG cells with luciferase reporter gene under the control of HIV‐1 LTR promoter were incubated with 2 μg/ml HIV‐1 Tat (ABL Inc and NIH AIDS program) in the presence of 100 μM chloroquine. Forty‐eight hours post‐incubation, luciferase activity assays (Promega) were performed and relative luminescence units were quantified using a fluorometer/luminometer plate reader (Spectra MAX GEMINI EM).
2.3. Split GFP assay for Tat endolysosome escape
H1299 cells with stable GFPβ1‐10 expression under hygromycin selection were treated with various concentrations of GFPβ11‐Tat for up to 4 hours with GFPβ11 plus Tat as controls. GFP fluorescence was analyzed with flow cytometry (Attune NxT). The peptide sequences were as follows; GFPβ11‐Tat RDHMVLHEYVNAAGITGRKKRRQRRRPPQ, GFPβ11 RDHMVLHEYVNAAGIT, and Tat GRKKRRQRRRPPQ.
2.4. Tat internalization assay
Quantitative analysis of Tat internalization in H1299 cells was performed using a method as described previously,46 but with minor modifications. Cells plated on glass‐bottom 35‐mm2 tissue culture dishes were treated with 5 μg/mL Tat‐FITC (ImmunoDX, 1002‐F) for 4 hours at 37°C. Following treatment, 100 μL of cell media was used to determine the remaining levels of Tat‐FITC and relative fluorescence intensity was quantified using a fluorometer plate reader (Spectra MAX GEMINI EM). Cells were washed with an acid wash solution (0.2 M acetic acid, 0.5 M NaCl, pH 2.8) at 4°C for 10 minutes and then washed with ice‐cold PBS for 5 minutes to remove surface‐bound Tat‐FITC. Cells were fixed in 4% paraformaldehyde and images were taken with a confocal laser‐scanning microscope (Zeiss LSM800). The average integrated intensity of the Tat‐FITC signal per cell was quantified using ImageJ software.
2.5. shRNA knockdown
Cells were stably transfected with TPC1 and TPC2 targeted shRNA (Santa Cruz) and scrambeled control shRNA following puromycin (Invitrogen) antibiotic selection pressure (25.0 μg/mL). Knockdown efficiency of TPC1 and TPC2 was confirmed by immunoblotting (Ab94731 and Ab119915 from Abcam) and by flow cytometry (Attune NxT) using immunofluorescence antibodies targeting TPC1 (Abcam, Ab94731) and TPC2 (Novus Biologicals, NBP1‐92152).
2.6. TPCs overexpression
Overexpression of TPC1 and TPC2 proteins was conducted using CRISPR‐based gene activation plasmid‐encoded lentivirus particles against TPC1 (Santa‐Cruz, Sc‐404943‐LAC) and TPC2 proteins (Santa‐Cruz, Sc‐402960‐LAC); scrambled lentivirus (Santa‐Cruz, Sc‐437282) was used as a control. Following 48 hours post‐transduction, 10 μg/mL of puromycin (Fisher Scientific), hygromycin (Santa‐Cruz), and blasticidin‐HCl (Fisher Scientific) were added to stable activation of CRISPR‐based gene activation plasmids. After incubation for 3‐4 days, cells were passaged to remove the dead and dying cells, and the remaining living cells were cultured for an additional 48 hours prior to being taken for experimentation. Gene activation or overexpression efficiency was confirmed by immunoblotting using specific antibodies against TPC1 (Abcam, Ab94731) and TPC2 (Abcam, Ab119915).
2.7. Endolysosome leakage assay
The release of fluorescent‐dextran from endolysosomes into the cytosol and the detection of the translocation of galectins to damaged endolysosomes were used to assess endolysosome membrane permeability. For the detection of released fluorescent‐dextran from endolysosomes into the cytosol, cells were seeded on 35 mm2 dishes. The next day, Alex488‐Dextran (MW: 10K) (10 μM, Thermo Fisher) was incubated with the cells for 5 hours. Then, chloroquine (100 μM) or Trans‐Ned19 (50 μM) was added to the cells and incubated for 4 hours. Cells were then washed three times by 1X PBS and fixed with 4% paraformaldehyde for 10 minutes. Cells were examined by confocal microscopy (Zeiss LSM800). Green fluorescence in the cytoplasm and in puncta structure was quantified (Image J) and analyzed. Increased green fluorescence in the cytoplasm and decreased green fluorescence in the puncta pattern indicates the leakage of dextran from the endolysosomes. For the detection of the translocation of galectins to damaged endolysosomes, cells were seeded on 35 mm2 dishes. The next day, cells were treated with chloroquine (100 μM) or Trans‐Ned19 (50 μM) for 24 hours. Cells were then fixed with 1% paraformaldehyde for 10 minutes followed by cold methanol (−20°C) for 10 minutes. The cells were then washed with PBS, blocked with 5% goat serum, and incubated overnight at 4°C with the primary antibody Alexa‐647 conjugated Galectin‐3 (Abcam, Ab207358). Cells were examined by confocal microscopy (Zeiss LSM800). Galectin‐3 positive puncta in each single cell were quantified and analyzed. Increased numbers of galectin‐3 positive puncta indicate the increased leakage of endolysosomes.
2.8. Immunoblotting
Cells were harvested and lysed in 1X RIPA lysis buffer (Thermo Fisher) plus 10 mM NaF, 1 mM Na3VO4, and protease inhibitor cocktail (Sigma‐Aldrich). After centrifugation (14 000 g for 10 minutes at 4°C), supernatants were collected and protein concentrations were determined with a DC protein assay (Bio‐Rad). Proteins (10 μg) were separated by SDS‐PAGE (12% gel) and transferred to PVDF membranes with iBlot 2 (Invitrogen). The membranes were incubated overnight at 4°C with antibodies against GAPDH (Abcam, Ab181603), TPC1 (Abcam, Ab94731), and TPC2 (Abcam, Ab119915). The blots were developed with enhanced chemiluminescence and quantified with our LI‐COR Odyssey Fc Imaging System. Quantification of results was performed by densitometry and the results were analyzed as total integrated densitometric volume values (arbitrary units).
2.9. Statistical analysis
All data were presented as means and standard deviation (SD). Statistical significance between two groups was analyzed by Student's t‐test and the statistical significance among multiple groups was analyzed by one‐way or two‐way ANOVA plus a Tukey post hoc test. P < .05 was accepted to be statistically significant.
3. RESULTS
3.1. Calcium is involved in Tat‐mediated LTR transactivation
Because Tat disrupts intracellular calcium homeostasis,39, 40, 41, 42, 43, 44, 45 we investigated the involvement of calcium in Tat‐mediated LTR transactivation in U87MG cells stably expressing luciferase reporter gene under the control of the HIV‐1 LTR promoter.47, 48 We first determined the extent to which cytosolic calcium is involved in Tat‐mediated LTR transactivation. Here, free cytosolic calcium was decreased using BAPTA‐AM, a plasma membrane permeable calcium chelator. BAPTA‐AM (1‐4 μM) significantly attenuated Tat‐mediated LTR transactivation (Figure 1A). Using a cell‐free assay, we demonstrated that Tat did not affect BAPTA's ability to chelate calcium (Data not shown). Given that endolysosomes have readily releasable stores of intracellular calcium ranging in concentration from 400 to 600 μM,49, 50 we next determined if endolysosome calcium affected Tat‐mediated LTR transactivation. Endolysosome calcium depleting using a high‐affinity rhodamine‐dextran (MW: 10 000) that enters cells via endocytosis and efficiently chelates endolysosome calcium51 significantly inhibited Tat‐mediated LTR transactivation (Figure 1B). These findings indicate that endolysosome calcium plays a role in Tat‐mediated LTR transactivation.
Figure 1.
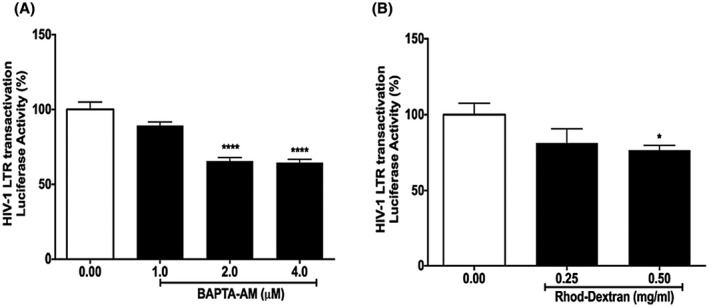
Calcium is involved in Tat‐mediated LTR transactivation. A, Chelating cytosolic calcium with BAPTA‐AM (1‐4 μM) significantly decreased Tat‐mediated LTR transactivation (n = 3; ***P < .001). B, Chelating endolysosome calcium with high‐affinity rhodamine‐dextran (0.5 mg/mL) significantly attenuated Tat‐mediated LTR transactivation (n = 3; *P < .05)
3.2. Calcium is involved in Tat endolysosome escape
To activate LTR transactivation in the nucleus, exogenous Tat must first escape endolysosomes. Here, we used a quantitative split‐GFP fluorescence assay for the direct measurement of Tat endolysosome escape.52 In this assay, H1299 cells stably expressing the GFPβ1‐10 protein fragment were treated with a 29‐amino acid GFPβ11‐Tat peptide. The exogenously added GFPβ11‐Tat peptide, once released from endolysosomes, induced fluorescence complementation with the intracellularly expressed GFPβ1‐10 protein fragment (Figure 2A). Using flow cytometry, we first determined concentration (0‐100 μM)‐ and time (0‐6 hours)‐dependent responses of exogenous GFPβ11‐Tat‐induced GFP fluorescence complementation. We demonstrated that 50 μM of exogenous GFPβ11‐Tat‐induced robust GFP fluorescence complementation that plateaued at 4 hours and that GFPβ11‐Tat treatment (50 μM for 4 hours) did not have cytotoxicity as indicated by LDH assay (Data not shown). We demonstrated that GFPβ11‐Tat‐induced concentration‐dependent increases in GFP fluorescence (Figure 2B) was enhanced in the presence of chloroquine, a lysosomotropic agent that enhances the efficiency for extracellular Tat‐induced LTR transactivation27, 38, 53, 54, 55 and enhances HIV‐1 infectivity in cells that require endocytosis for HIV‐1 virus entry.56, 57 Using confocal microscopy imaging, we confirmed that GFPβ11‐Tat‐induced the fluorescence complementation of the intracellularly expressed GFPβ1‐10 protein fragment (Figure 2C) and that this effect was enhanced by chloroquine. Using this split‐GFP Tat endolysosome escape assay, we determined next the involvement of calcium in Tat endolysosome escape. We demonstrated that chelating cytosolic calcium with BAPTA‐AM (2.5‐10 μM) significantly attenuated Tat endolysosome escape (Figure 2D). Using a cell‐free assay, we demonstrated that BAPTA did not affect GFPβ11‐Tat‐induced GFP fluorescence complementation with GFPβ1‐10 protein fragment (Data not shown). Furthermore, we demonstrated that chelating endolysosome calcium with rhodamine‐dextran significantly attenuated Tat endolysosome escape (Figure 2E). Using a cell‐free assay, we demonstrated that rhodamine‐dextran did not affect GFPβ11‐Tat‐induced GFP fluorescence complementation with GFPβ1‐10 protein fragment (Data not shown). Given that calcium affects vesicular trafficking,58 we then determined the extent to which chelating endolysosome calcium with rhodamine‐dextran affects Tat internalization; rhodamine‐dextran did not affect Tat‐FITC internalization nor did it affect Tat‐FITC levels in the media (Figure 2F). These findings indicate that endolysosome calcium plays a role in Tat endolysosome escape and subsequent Tat‐mediated LTR transactivation.
Figure 2.
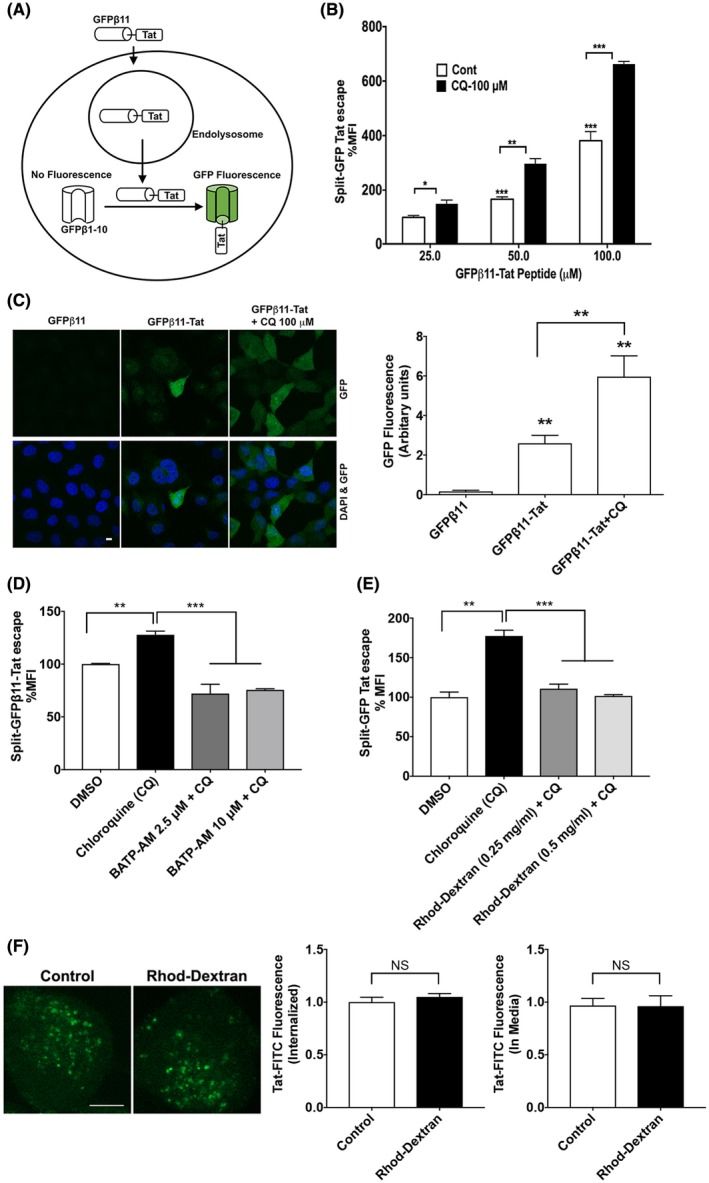
Calcium is involved in Tat endolysosome escape. A, A split‐GFP fluorescence assay was used to assess Tat endolysosome escape in H1299 cells stably expressing fluorescence complementation of intracellularly GFPβ1‐10 protein fragment. Following endolysosome escape, extracellular addition of GFPβ11‐Tat peptide induces the completion of GFP fluorescence in a concentration‐dependent manner. B, Flow cytometry data show that chloroquine (100 μM) dramatically enhanced Tat endolysosome escape (n = 3; *P < .05; **P < .01; ***P < .001). C, Confocal imaging shows that GFPβ11‐Tat (50 μM), but not GFPβ11 (50 μM) significantly induced GFP fluorescence, and chloroquine (100 μM) enhanced GFPβ11‐Tat‐induced GFP fluorescence (scale bar = 10 μm) (n = 5; **P < .01). D, Chelating cytosolic calcium with BAPTA‐AM (2.5‐10 μM) significantly decreased Tat‐endolysosome escape (n = 3; **P < .01; ***P < .001). E, Chelating endolysosome calcium with high‐affinity rhodamine‐dextran (0.5 mg/mL) significantly prevented chloroquine‐induced increases in Tat‐mediated LTR transactivation (n = 3; *P < .05). F, Chelating endolysosome calcium with high‐affinity rhodamine‐dextran (0.5 mg/mL) did not affect the internalization of Tat‐FITC (n = 4, P > .05, bar = 10 μm)
3.3. TPCs are involved in Tat‐mediated LTR transactivation
Next, we determined the involvement of endolysosome‐resident calcium channels in Tat‐mediated LTR transactivation. A variety of endolysosome‐resident calcium channels have been implicated in endolysosome calcium release including TRPML, TPC, TRPM2, P2X4, P‐type, and L‐type.59, 60, 61 Using pharmacological approaches, we determined the extent to which blocking endolysosome calcium channels affected Tat‐mediated LTR‐transactivation. For these studies we blocked TPC with Ned‐19, TRPML with ML‐SI1, TRPM2 with flufenamic acid, P2X4 with 5‐BDBD, P‐type calcium channel with ω‐agatoxin TK, and L‐type calcium channels with verapamil. Ned‐19 significantly inhibited Tat‐mediated LTR transactivation in a concentration‐dependent manner (Figure 3E), but none of the other calcium channel blockers significantly affected Tat‐induced LTR transactivation (Figure 3A‐D). However, verapamil at the high concentration of 50 μM did significantly inhibit Tat‐mediated LTR transactivation (Figure 3F), but it is likely that this was due to its ability to block TPCs at such high concentrations.62 Using the newer TPC blockers tetrandrine and fangchinoline63, 64, 65 at subtoxic concentrations of 0.5 and 1 µM66 we showed that tetrandrine (Figure 3G) and fangchinoline (Figure 3H) both significantly attenuated Tat‐mediated LTR transactivation. Thus, our pharmacological studies suggest strongly that blocking TPC attenuates Tat‐mediated LTR transactivation.
Figure 3.
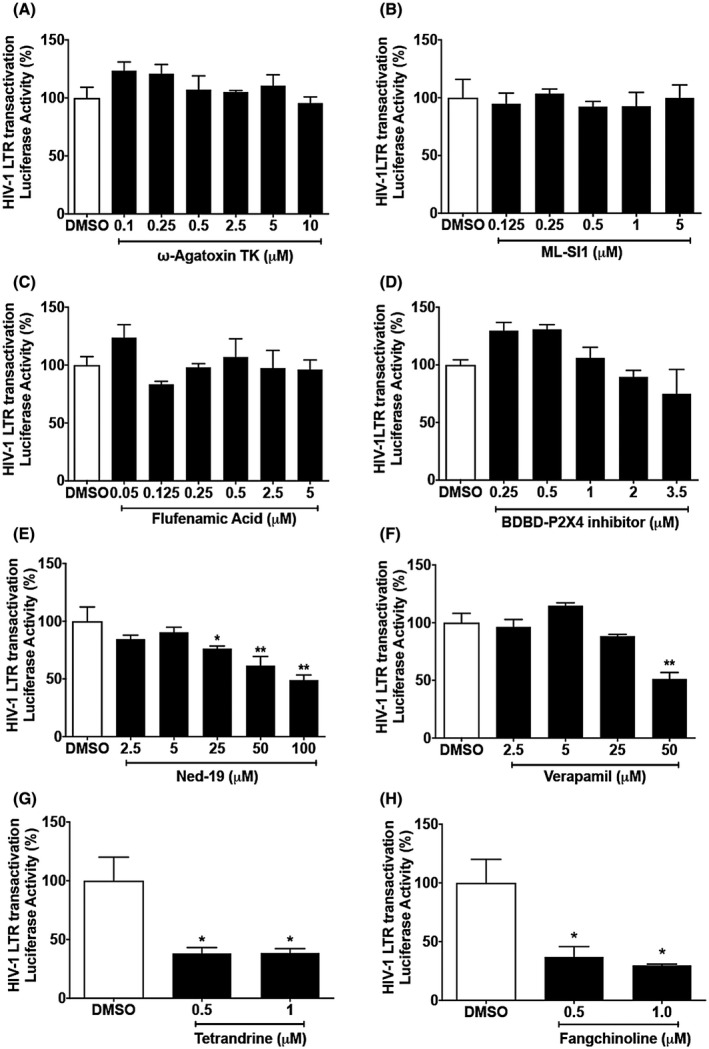
TPCs are involved Tat‐mediated LTR transactivation. A‐D, Blocking P‐type calcium channel with ω‐agatoxin TK, TRPMLs with ML‐SI1, TRPM2 with flufenamic acid, or P2X4 with 5‐BDBD did not affect Tat‐mediated LTR transactivation in U87MG cells. (n = 3, P > .05). E, Blocking TPCs with Ned‐19 significantly inhibited Tat‐induced LTR transactivation in a concentration‐dependent manner (n = 3; **P < .01). F, Blocking L‐type calcium channel with verapamil at high concentration (50 μM) significantly inhibited Tat‐mediated LTR transactivation (n = 3; **P < .01). G, Blocking TPCs with tetrandrine (0.5‐1 μM) significantly inhibited Tat‐induced LTR transactivation (n = 3; *P < .05). H, Blocking TPCs with fangchinoline (0.5‐1 μM) significantly inhibited Tat‐induced LTR transactivation (n = 3; *P < .05)
3.4. TPCs are involved in Tat endolysosome escape
We next determined the involvement of TPCs in Tat endolysosome escape using the split‐GFP fluorescence Tat endolysosome escape assay. Blocking TPCs with Ned‐19 significantly attenuated Tat endolysosome escape in the absence and presence of chloroquine (Figure 4A). To further investigate whether these effects resulted from altered Tat internalization, we then determined the extent to which blocking TPCs with Ned‐19 affected Tat internalization. Ned‐19 did not affect the internalization of Tat‐FITC nor did it affect the remaining levels of Tat‐FITC in the media (Figure 4B). These observations indicate that blocking TPCs prevents Tat endolysosome escape and subsequent LTR transactivation.
Figure 4.
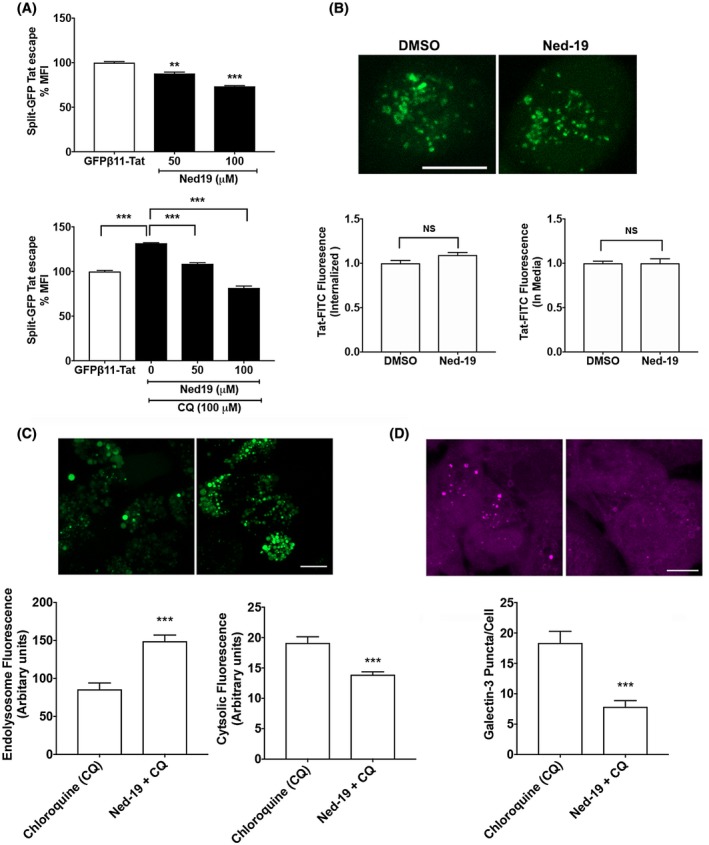
TPCs are involved in Tat endolysosome escape. A, Blocking TPCs with Ned‐19 significantly inhibited Tat endolysosome escape in the absence of chloroquine (n = 3; **P < .01; ***P < .001). Blocking TPCs with Ned‐19 significantly blocked Tat endolysosome escape in H1299 cells in the presence of chloroquine (n = 3; ***P < .001). B, Blocking TPCs with Ned‐19 did not affect the internalization of Tat‐FITC (n = 4, P > .05, bar = 10 μm). C, Blocking TPCs with Ned‐19 significantly attenuated chloroquine (100 μM)‐induced leakage of dextran from endolysosomes into cytosol (n = 15, ***P < .001). D, Blocking TPCs with Ned‐19 significantly attenuated chloroquine (100 μM)‐induced increases in the numbers of galectin‐3 positive puncta (n = 10, ***P < .001)
Given recent findings that blocking TPCs prevents the entry of virus into cells,63, 64, 65 blocking TPCs may generally affect the escape of a variety of macromolecules from endolysosomes. Thus, we determined the extent to which blocking TPC affects the release of fluorescent‐dextran (MW: 10 000) from endolysosome into the cytosol and the translocation of galectin‐3 (26 kDa) to damaged endolysosomes.67, 68 As a gene delivery enhancing agent, chloroquine's ability to promote endolysosome escape has long been recognized.69, 70, 71 Chloroquine can rapidly diffuse into cells where it can be trapped in endolysosomes,72 and upon protonation and subsequent influx of water molecules, chloroquine can induce osmotic swelling of the endolysosome and endolysosome leakage.73, 74 Thus, we used chloroquine as a positive control for endolysosome leakage and determine the extent to which blocking TPCs affects endolysosome leakage. Indeed, we demonstrated that chloroquine induced the endolysosome leakage of fluorescent‐dextran and galectin‐3 (Supplementary Figure S1). Significantly, we demonstrated that blocking TPCs with Ned‐19 significantly attenuated chloroquine‐induced endolysosome leakage of fluorescent‐dextran (Figure 4C); Ned‐19 attenuated chloroquine‐induced increases in cytosolic green dextran fluorescence and decreases in endolysosome green dextran fluorescence. In addition, using the galectin‐3 puncta assay we found that Ned‐19 significantly reduced chloroquine‐induced endolysosome leakage (Figure 4D); Ned‐19 attenuated chloroquine‐induced increases in the number of galectin‐3 puncta. Our finding suggests that blocking TPCs exerts a general protective effect on endolysosome leakage.
3.5. TPC knockdown attenuates Tat‐mediated LTR transactivation
To further explore the involvement of TPCs in Tat‐mediated LTR transactivation, we knocked down the expression levels of TPC1 (Figure 5A) and TPC2 (Figure 5C) using shRNA strategies in U87MG cells, as confirmed by immunoblotting. Both TPC1 and TPC2 antibodies were validated in overexpression studies. The knockdown of TPC1 (Figure 5B) and TPC2 (Figure 5D) both significantly attenuated Tat‐mediated LTR transactivation. Consistent with our pharmacological findings, TRPML1 knockdown did not affect the Tat‐mediated LTR transactivation (Supplementary Figure S2).
Figure 5.
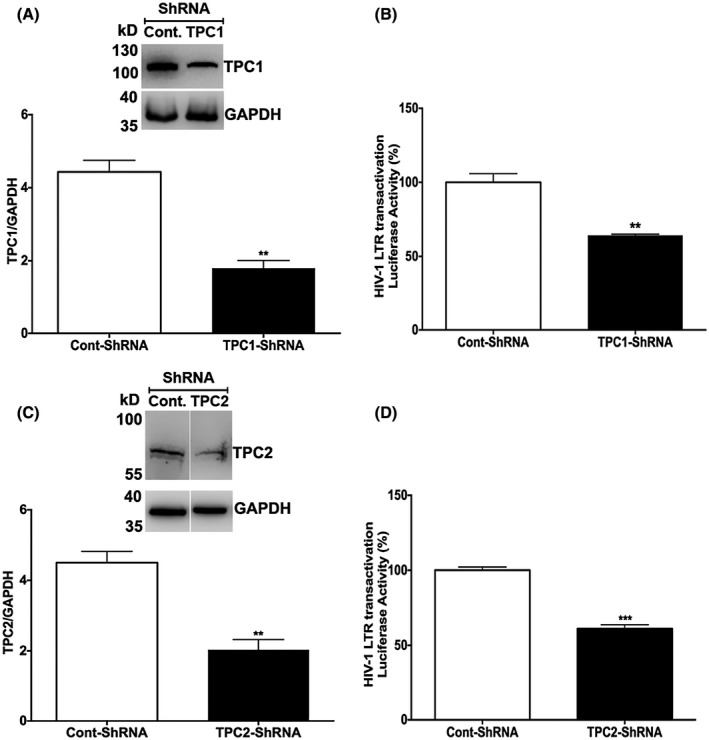
TPCs knockdown attenuates Tat‐mediated LTR transactivation. A, Quantitative immunoblotting data showed that the expression of TPC1 was knocked down with specific shRNAs in U87MG cell (n = 3; **P < .01). B, Knockdown of TPC1 significantly attenuated Tat‐mediated LTR transactivation in U87MG cell (n = 3; **P < .01). C, Quantitative immunoblotting data showed that the expression of TPC2 was knocked down with specific shRNAs in U87MG cell (n = 3; *P < .05; **P < .01). D, Knockdown of TPC2 significantly attenuated Tat‐mediated LTR transactivation in U87MG cell (n = 3; ***P < .001)
3.6. TPCs knockdown attenuates Tat endolysosome escape
To further determine the extent to which TPCs are involved in Tat endolysosome escape, we knocked down the expression of TPC1 (Figure 6A) in H1299 cells using shRNA strategy. The knockdown of TPC1 attenuated Tat endolysosome scape (Figure 6B). Blocking TPCs with Ned‐19 did not further block Tat endolysosome escape in TPC1 knockdown cells (Figure 6B). We then determined the extent to which TPC1 knockdown affects Tat internalization and found that TPC1 knockdown did not affect the internalization of Tat‐FITC nor did it affect the remaining levels of Tat‐FITC in media (Figure 6C). Next, we knocked down the expression of TPC2 (Figure 7A) in H1299 cells using shRNA strategy and found that knockdown TPC2 also attenuated Tat endolysosome scape (Figure 7B). Blocking TPCs with Ned‐19 could not further block Tat endolysosome escape in TPC2 knockdown cells (Figure 7B). We also determined the effect of TPCs knockdown on Tat endolysosome escape in the presence of chloroquine and found that TPC1 knockdown, but not TPC2 knockdown attenuated Tat endolysosome escape in the presence of chloroquine (Supplementary Figure S3). To further investigate whether these effects resulted from altered Tat internalization, we then determined the extent to which blocking TPC2 knockdown affects Tat internalization. We found that TPC2 knockdown significantly increased the internalization of Tat‐FITC and decreased the remaining levels of Tat‐FITC in media (Figure 7C). In addition, we knocked down the expression of TRPML1 using shRNA strategy (Figure 7D) and found that TRPML1 knockdown did not affect Tat endolysosome escape (Figure 7E).
Figure 6.
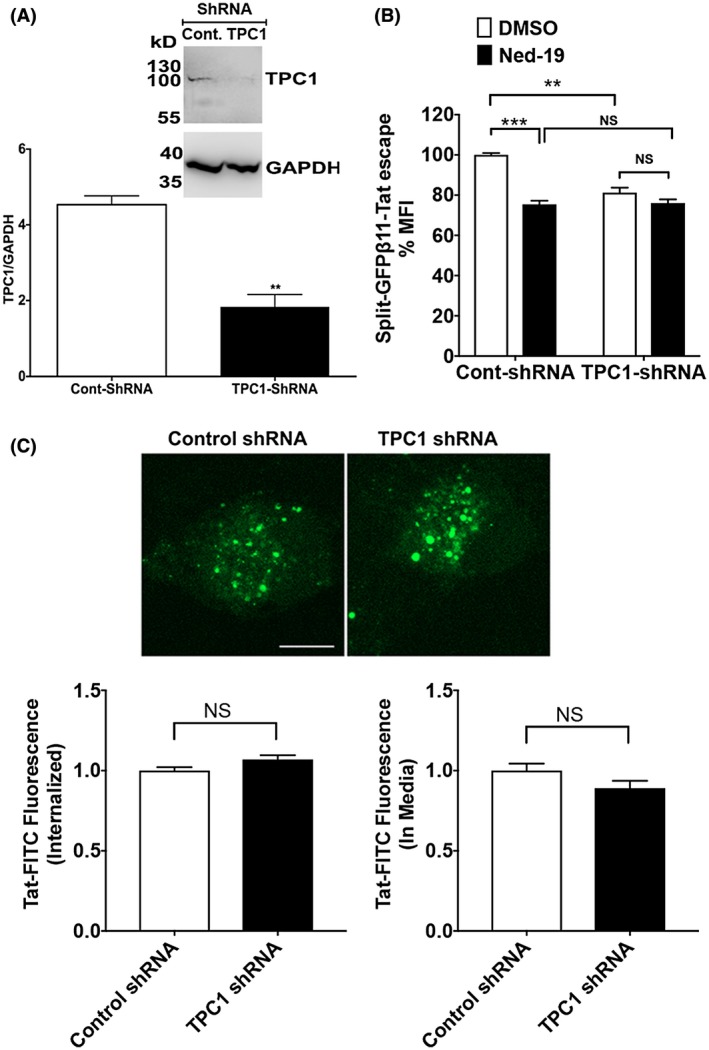
TPC1 knockdown attenuates Tat endolysosome escape. A, Quantitative immunoblotting data showed that the expression of TPC1 was knocked down with specific shRNAs in H1299 cells (n = 3; **P < .01). B, Knockdown of TPC1 significantly attenuated Tat endolysosome escape in the absence of chloroquine (n = 3; **P < .01). Blocking TPCs with Ned‐19 decreased Tat endolysosome escape in control shRNA‐treated cells but did not further decrease Tat endolysosome escape in TPC1 knockdown cells (n = 3; **P < .01; ***P < .001). C, TPC1 knockdown did not affect internalization of Tat‐FITC (n = 4, P > .05, bar = 10 μm)
Figure 7.
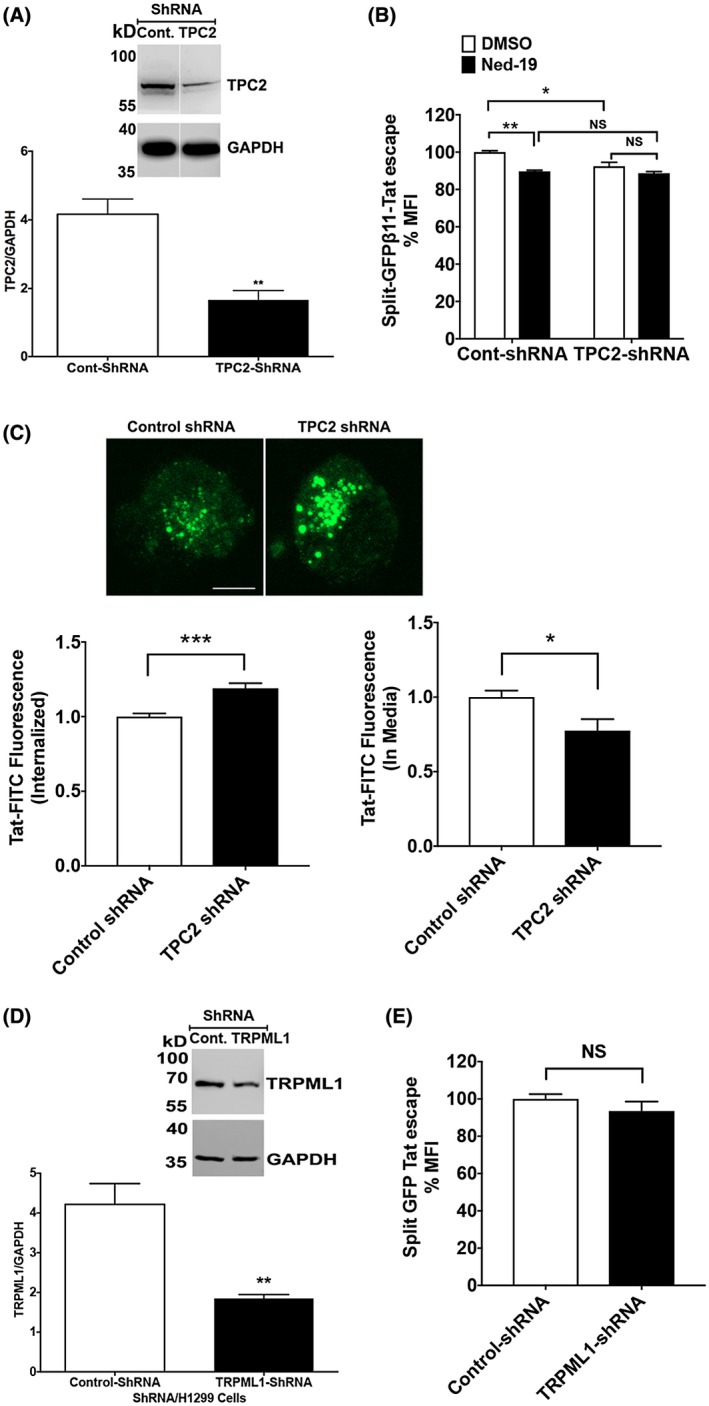
TPC2 knockdown attenuates Tat endolysosome escape. A, Quantitative immunoblotting data showed that the expression of TPC2 was knocked down with specific shRNAs in H1299 cells (n = 3; **P < .01). B, Knockdown of TPC2 significantly attenuated Tat endolysosome escape in the absence of chloroquine (n = 3; *P < .05). Blocking TPCs with Ned‐19 decreased Tat endolysosome escape in control shRNA‐treated cells but did not further decrease Tat endolysosome escape in TPC2 knockdown cells (n = 3; *P < .05). C, TPC2 knockdown significantly enhanced the internalization of Tat‐FITC (n = 4, *P < .05, ***P < .001, bar = 10 μm). D, Quantitative immunoblotting data showed that the expression of TRPML1 was knocked down with specific shRNAs in H1299 cells (n = 3; **P < .01). E, Knockdown of TRPML1 did not affect Tat endolysosome escape (n = 3; P > .05)
4. DISCUSSION
It is well known that extracellular Tat enters cells via endocytosis, but little is known about how Tat escapes endolysosomes before entering the nucleus to activate the HIV‐1 LTR promoter. Here we explored underlying mechanisms whereby Tat escapes endolysosomes using both split‐GFP Tat endolysosome escape and Tat‐mediated LTR transactivation assays. The main findings of these studies are that blocking endolysosome‐resident TPCs attenuated endolysosome leakage, Tat endolysosome escape, and ultimately Tat‐mediated LTR transactivation.
4.1. Tat endolysosome escape
The endocytic delivery of extracellular macromolecules and plasma membrane components for intracellular degradation requires traffic through early to late endosomes followed by fusions between late endosomes and lysosomes.75, 76, 77, 78 Endolysosomes are complex and dynamic organelles that exhibit morphological and functional heterogeneity as well as the ability to physically and functionally interact with other organelles. An important hallmark of endolysosomes is their acidic luminal pH, which is maintained largely by the electrogenic pumping of protons by the v‐ATPase in conjunction with vesicular chloride transporters that shunt the membrane potential and allow for a build‐up of luminal protons.77, 79 The acidic pH of endolysosomes is critical for the activity of up to 60 different pH‐sensitive hydrolytic enzymes including proteases, lipases, and nucleases thus enabling the endolysosomes to break down a wide range of endogenous and exogenous cargos.78
In order to replicate, viruses need to enter host cells, and many viruses including HIV‐1 enter host cells through the endocytic pathway.80, 81, 82, 83 Once endocytosed, viruses can be degraded by pH‐sensitive hydrolytic enzymes in endolysosomes or alternatively they can escape the endolysosomes following which they can transit to the nucleus or recycled back into the extracellular space.84 Similar to viruses, secreted viral proteins from infected cells can also enter cells via endocytosis. For instance, released HIV‐1 viral proteins gp12085, 86 and Tat24, 25, 26, 27 have been shown to enter cells via endocytosis, where they can exert direct cytotoxic effects. Tat protein is of particular importance for HIV‐1 infection, HIV‐1 disease progression, and the development of HIV‐associated neurological complications because HIV‐1 Tat is essential for viral transcription,15, 16, 17 ART does not block the secretion of Tat in HIV infected individuals,18, 19 and because brain levels of Tat remains elevated even though HIV‐1 levels are undetectable.19 Secreted from HIV‐1 infected cells,20, 21, 22, 23 Tat can bind to cell surface receptors including LRP1, heparin sulfate proteoglycan, CD26, and CXCR4, and enters CNS cells via receptor‐mediated endocytosis.24, 25, 26, 27 Following its endocytosis and internalization into endolysosomes, Tat must undergo endolysosome escape and transit to the nucleus where it can activate the HIV‐1 LTR promoter.36, 37, 38 Tat transits through late endosomes to the cytosol,38 but it is not clear how Tat escapes endolysosomes.
4.2. TPCs and Tat endolysosome escape
Besides protons, endolysosomes contain high levels of calcium second only to the endoplasmic reticulum.49, 87 The release of calcium from endolysosomes has been implicated in numerous functions central to cellular well‐being including lysosomal exocytosis,88 endosome‐lysosome fusion,89 the activation of TFEB,90 and the regulation of oxidative stress.91 The release of calcium from endolysosomes is mediated by a variety of endolysosome‐resident calcium channels.60, 61 Because others and we have shown consistently that Tat treatment leads to calcium dyshomeostasis in neuronal and non‐neuronal cells,39, 40, 41, 42, 43, 44, 45 we investigated the involvement of calcium in Tat endolysosome escape and subsequent LTR transactivation using split‐GFP Tat endolysosome escape and Tat‐mediated LTR transactivation assays. We found that chelating endolysosome calcium attenuated Tat endolysosome escape and Tat‐mediated LTR transactivation, and that these affects were not the result of altered Tat internalization.
Endolysosomes contain multiple calcium channels including a family of TPCs; TPC1, TPC2, and TPC3 (not present in mice, rats, and primates).92, 93 TPCs are located in endosomes and lysosomes where they trigger the release of Ca2+, Na+ or other cations from acidic organelles in response to various activators.65, 92, 93 TPC1 is broadly localized to recycling endosomes, early and late endosomes, and lysosomes, while TPC2 is localized more with lysosomes.92, 94, 95, 96 One fundamental difference between TPC1 and TPC2 is their voltage dependence97, 98, 99, 100, 101, 102, 103; TPC1 is a voltage‐gated channel able to be activated by the NAADP, Ca2+, and PI(3,5)P2, whereas TPC2 is voltage‐insensitive.
TPCs participate in the regulation of multiple endolysosome trafficking pathways97, 98, 99, 100, 101 and TPC1 helps control the uptake and processing of proteins in endolysosomes through the release for calcium.104 TPCs are also involved in the pathogenicity of viruses like Ebola and middle east respiratory syndrome coronavirus (MERS‐CoV). In particular, the inhibition of or the absence of TPCs blocks the entry of the Ebola virus into the host cell62, 63 and impairs the progression of MERS‐CoV through the endolysosome system.64 Consistent with these findings, our findings provide evidence that TPCs are involved in Tat endolysosome escape and subsequent Tat‐mediated LTR activation. First, we demonstrated that a set of pharmacological blockers (Ned‐19, verapamil, tetrandrine, and fangchinoline) of TPCs attenuated Tat‐mediated LTR transactivation. Second, we demonstrated that blocking TPCs with Ned‐19 attenuated general endolysosome leakage as well as specific Tat endolysosome escape. Third, the knockdown of TPC1 or TPC2 attenuated Tat endolysosome escape and subsequent Tat‐mediated LTR transactivation. These effects were selective to TPCs because knocking down the endolysosome‐resident TRPML1 did not affect Tat endolysosome escape or subsequent Tat‐mediated LTR transactivation. Our findings suggest that blocking TPCs exert a general protective against the leakage of endolysosomes and especially the escape of larger molecules (MW ≥ 10 000) from endolysosomes because pharmacologically blocking TPCs attenuated the leakage of dextran (10 kDa) and increased the accumulation of galectin‐3 (26 kDa), and because knocking down TPC1 enhanced Tat (14 kDa)‐mediated LTR transactivation. However, knockdown TPC2 had a modest effect on endolysosome escape of the 29‐amino acid GFPβ11‐Tat peptide (MW ~ 3.5 kDa) used in the Tat endolysosome escape assay possibly due to enhanced internalization of Tat‐FITC.
The significance of these findings is threefold. First, because Tat is essential for HIV‐1 replication and important for the activation of latent HIV reservoir, our findings that TPCs are involved in Tat endolysosome escape indicate that TPCs might be involved in latent HIV‐1 infection especially in cells that require endocytosis for HIV‐1 virus entry. Thus, blocking TPCs represent a novel therapeutic strategy against latent HIV‐1 infection. Second, Tat is a neurotoxic protein, and the neurotoxic effect could result from Tat disturbing endolysosome function or Tat disturbing the function of other organelles such as mitochondria following Tat endolysosome escape. Thus, Ned‐19, by inhibiting endolysosome leakage and preventing Tat endolysosome escape, may ameliorate Tat‐induced neurotoxicity and may represent a novel therapeutic strategy against HIV‐associated neurological comorbidities. It is important to note that the advantage of blocking TPCs as a therapeutic strategy is that TPC1 as well as TPC2 knockout mice do not have a reduced life span or an obvious reduction in quality of life.97 Third, it is well known that Tat has the extraordinary ability to cross the plasma membrane and has been used to transport a variety of macromolecules into a plethora of cell types.105, 106, 107, 108 Our findings that TPCs are involved in Tat‐endolysosome escape provide TPCs as a novel target for enhancing the transport of macromolecules into cells. Although we explored the involvement of TPCs in Tat endolysosome escape in the present study, the underlying mechanisms whereby Tat escapes endolysosomes remain unclear. Thus, further mechanistic studies are warranted.
In summary, our finding demonstrated that TPCs are involved in Tat endolysosome escape and subsequent LTR transactivation. Our findings suggest that TPCs might be a therapeutic target against latent HIV‐1 infection and HIV‐associated neurological complications, and our findings represent a step forward toward developing TPC antagonists as a novel therapeutic approach against HIV. In addition, our findings suggest that TPCs might represent a novel target for enhancing the transport of macromolecules into cells.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
AUTHOR CONTRIBUTIONS
X. Chen and N. Khan designed the research; N. Khan, P.W. Halcrow, K.L. Lakpa, Z. Afghah, and N.M. Miller performed the research and analyzed the data; S.F. Dowdy provided new reagents; X. Chen, N. Khan, and J.D. Geiger wrote the paper.
Supporting information
ACKNOWLEDGMENTS
This work was supported by R01MH100972, R01MH105329, R01NS065957, and R01DA032444.
Khan N, Halcrow PW, Lakpa KL, et al. Two‐pore channels regulate Tat endolysosome escape and Tat‐mediated HIV‐1 LTR transactivation. The FASEB Journal. 2020;34:4147–4162. 10.1096/fj.201902534R
REFERENCES
- 1. Spudich S, Gonzalez‐Scarano F. HIV‐1‐related central nervous system disease: current issues in pathogenesis, diagnosis, and treatment. Cold Spring Harb Perspect Med. 2012;2:a007120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Valcour, V. , Chalermchai, T. , Sailasuta, N. , et al. Central nervous system viral invasion and inflammation during acute HIV infection. J Infect Dis. 2012;206:275‐282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Saylor D, Dickens AM, Sacktor N, et al. HIV‐associated neurocognitive disorder—pathogenesis and prospects for treatment. Nat Rev Neurol. 2016;12:234‐248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Eisele E, Siliciano RF. Redefining the viral reservoirs that prevent HIV‐1 eradication. Immunity. 2012;37:377‐388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marban C, Forouzanfar F, Ait‐Ammar A, et al. Targeting the brain reservoirs: toward an HIV cure. Front Immunol. 2016;7:1‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gray LR, Roche M, Flynn JK, Wesselingh SL, Gorry PR, Churchill MJ. Is the central nervous system a reservoir of HIV‐1? Curr Opin HIV AIDS. 2014;9:552‐558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chauhan A, Mehla R, Vijayakumar TS, Handy I. Endocytosis‐mediated HIV‐1 entry and its significance in the elusive behavior of the virus in astrocytes. Virology. 2014;456–457:1‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carter GC, Bernstone L, Baskaran D, James W. HIV‐1 infects macrophages by exploiting an endocytic route dependent on dynamin, Rac1 and Pak1. Virology. 2011;409:234‐250. [DOI] [PubMed] [Google Scholar]
- 9. Trillo‐Pazos G, Diamanturos A, Rislove L, et al. Detection of HIV‐1 DNA in microglia/macrophages, astrocytes and neurons isolated from brain tissue with HIV‐1 encephalitis by laser capture microdissection. Brain Pathol. 2003;13:144‐154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Churchill MJ, Wesselingh SL, Cowley D, et al. Extensive astrocyte infection is prominent in human immunodeficiency virus‐associated dementia. Ann Neurol. 2009;66:253‐258. [DOI] [PubMed] [Google Scholar]
- 11. Ellis RJ, Rosario D, Clifford DB, et al. Continued high prevalence and adverse clinical impact of human immunodeficiency virus‐associated sensory neuropathy in the era of combination antiretroviral therapy: the CHARTER Study. Arch Neurol. 2010;67:552‐558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Heaton RK, Clifford DB, Franklin DR Jr, et al. HIV‐associated neurocognitive disorders persist in the era of potent antiretroviral therapy: CHARTER Study. Neurology. 2010;75:2087‐2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Churchill MJ, Gorry PR, Cowley D, et al. Use of laser capture microdissection to detect integrated HIV‐1 DNA in macrophages and astrocytes from autopsy brain tissues. J Neurovirol. 2006;12:146‐152. [DOI] [PubMed] [Google Scholar]
- 14. Thompson KA, Cherry CL, Bell JE, McLean CA. Brain cell reservoirs of latent virus in presymptomatic HIV‐infected individuals. Am J Pathol. 2011;179:1623‐1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Romani B, Engelbrecht S, Glashoff RH. Functions of Tat: the versatile protein of human immunodeficiency virus type 1. J Gen Virol. 2010;91:1‐12. [DOI] [PubMed] [Google Scholar]
- 16. Jeang KT, Xiao H, Rich EA. Multifaceted activities of the HIV‐1 transactivator of transcription, Tat. J Biol Chem. 1999;274:28837–28840. [DOI] [PubMed] [Google Scholar]
- 17. Clark E, Brenda N, Massimo C. Tat is a multifunctional viral protein that modulates cellular gene expression and functions. Oncotarget. 2017;8:27569‐27581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mediouni S, Darque A, Baillat G, et al. Antiretroviral therapy does not block the secretion of the human immunodeficiency virus Tat protein. Infect Disord Drug Targets. 2012;12:81‐86. [DOI] [PubMed] [Google Scholar]
- 19. Johnson TP, Patel K, Johnson KR, et al. Induction of IL‐17 and nonclassical T‐cell activation by HIV‐Tat protein. Proc Natl Acad Sci U S A. 2013;110:13588‐13593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Agostini S, Ali H, Vardabasso C, et al. Inhibition of non canonical HIV‐1 Tat secretion through the cellular Na+, K+‐ATPase Blocks HIV‐1 Infection. EBioMedicine. 2017;21:170–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ensoli B, Barillari G, Salahuddin SZ, Gallo RC, Wong‐Staal F. Tat protein of HIV‐1 stimulates growth of cells derived from Kaposi's sarcoma lesions of AIDS patients. Nature. 1990;345:84‐86. [DOI] [PubMed] [Google Scholar]
- 22. Chang HC, Samaniego F, Nair BC, Buonaguro L, Ensoli B. HIV‐1 Tat protein exits from cells via a leaderless secretory pathway and binds to extracellular matrix‐associated heparan sulfate proteoglycans through its basic region. AIDS. 1997;11:1421‐1431. [DOI] [PubMed] [Google Scholar]
- 23. Rayne F, Debaisieux S, Yezid H, et al. Phosphatidylinositol‐(4,5)‐bisphosphate enables efficient secretion of HIV‐1 Tat by infected T‐cells. EMBO J. 2010;29:1348‐1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Debaisieux S, Rayne F, Yezid H, Beaumelle B. The ins and outs of HIV‐1 Tat. Traffic. 2012;13:355‐363. [DOI] [PubMed] [Google Scholar]
- 25. Tyagi M, Rusnati M, Presta M, Giacca M. Internalization of HIV‐1 Tat requires cell surface heparan sulfate proteoglycans. J Biol Chem. 2001;276:3254‐3261. [DOI] [PubMed] [Google Scholar]
- 26. Liu Y, Jones M, Hingtgen CM, et al. Uptake of HIV‐1 tat protein mediated by low‐density lipoprotein receptor‐related protein disrupts the neuronal metabolic balance of the receptor ligands. Nat Med. 2000;6:1380‐1387. [DOI] [PubMed] [Google Scholar]
- 27. Frankel AD, Pabo CO. Cellular uptake of the tat protein from human immunodeficiency virus. Cell. 1988;55:1189‐1193. [DOI] [PubMed] [Google Scholar]
- 28. Nakase I, Niwa M, Takeuchi T, et al. Cellular uptake of arginine‐rich peptides: roles for macropinocytosis and actin rearrangement. Mol Ther. 2004;10:1011‐1022. [DOI] [PubMed] [Google Scholar]
- 29. Green M, Loewenstein PM. Autonomous functional domains of chemically synthesized human immunodeficiency virus Tat trans‐activator protein. Cell. 1988;55:1179‐1188. [DOI] [PubMed] [Google Scholar]
- 30. Futaki S, Suzuki T, Ohashi W, et al. Arginine‐rich peptides. An abundant source of membrane‐permeable peptides having potential as carriers for intracellular protein delivery. J Biol Chem. 2001;276:5836‐5840. [DOI] [PubMed] [Google Scholar]
- 31. Richard JP, Melikov K, Brooks H, Prevot P, Lebleu B, Chernomordik LV. Cellular uptake of unconjugated TAT peptide involves clathrin‐dependent endocytosis and heparan sulfate receptors. J Biol Chem. 2005;280:15300‐15306. [DOI] [PubMed] [Google Scholar]
- 32. Wadia JS, Stan RV, Dowdy SF. Transducible TAT‐HA fusogenic peptide enhances escape of TAT‐fusion proteins after lipid raft macropinocytosis. Nat Med. 2004;10:310‐315. [DOI] [PubMed] [Google Scholar]
- 33. Kaplan IM, Wadia JS, Dowdy SF. Cationic TAT peptide transduction domain enters cells by macropinocytosis. J Control Release. 2005;102:247‐253. [DOI] [PubMed] [Google Scholar]
- 34. Mann DA, Frankel AD. Endocytosis and targeting of exogenous HIV‐1 Tat protein. EMBO J. 1991;10:1733‐1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gaskill PJ, Miller DR, Gamble‐George J, Yano H, Khoshbouei H. HIV, Tat and dopamine transmission. Neurobiol Dis. 2017;105:51‐73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vives E. Cellular uptake [correction of utake] of the Tat peptide: an endocytosis mechanism following ionic interactions. J Mol Recognit. 2003;16:265‐271. [DOI] [PubMed] [Google Scholar]
- 37. Ensoli B, Buonaguro L, Barillari G, et al. Release, uptake, and effects of extracellular human immunodeficiency virus type 1 Tat protein on cell growth and viral transactivation. J Virol. 1993;67:277‐287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vendeville A, Rayne F, Bonhoure A, Bettache N, Montcourrier P, Beaumelle B. HIV‐1 Tat enters T cells using coated pits before translocating from acidified endosomes and eliciting biological responses. Mol Biol Cell. 2004;15:2347‐2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Norman JP, Perry SW, Reynolds HM, et al. HIV‐1 Tat activates neuronal ryanodine receptors with rapid induction of the unfolded protein response and mitochondrial hyperpolarization. PLoS ONE. 2008;3:e3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Haughey NJ, Holden CP, Nath A, Geiger JD. Involvement of inositol 1,4,5‐trisphosphate‐regulated stores of intracellular calcium in calcium dysregulation and neuron cell death caused by HIV‐1 protein tat. J Neurochem. 1999;73:1363‐1374. [DOI] [PubMed] [Google Scholar]
- 41. Nath A, Haughey NJ, Jones M, Anderson C, Bell JE, Geiger JD. Synergistic neurotoxicity by human immunodeficiency virus proteins Tat and gp120: protection by memantine. Ann Neurol. 2000;47:186‐194. [PubMed] [Google Scholar]
- 42. Haughey NJ, Nath A, Mattson MP, Slevin JT, Geiger JD. HIV‐1 Tat through phosphorylation of NMDA receptors potentiates glutamate excitotoxicity. J Neurochem. 2001;78:457‐467. [DOI] [PubMed] [Google Scholar]
- 43. Peng F, Yao H, Akturk HK, Buch S. Platelet‐derived growth factor CC‐mediated neuroprotection against HIV Tat involves TRPC‐mediated inactivation of GSK 3beta. PLoS One. 2012;7:e47572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hu XT. HIV‐1 Tat‐mediated calcium dysregulation and neuronal dysfunction in vulnerable brain regions. Curr Drug Targets. 2016;17:4‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hui L, Geiger NH, Bloor‐Young D, Churchill GC, Geiger JD, Chen X. Release of calcium from endolysosomes increases calcium influx through N‐type calcium channels: evidence for acidic store‐operated calcium entry in neurons. Cell Calcium. 2015;58:617‐627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li S, Geiger NH, Soliman ML, Hui L, Geiger JD, Chen X. Caffeine, through adenosine A3 receptor‐mediated actions, suppresses amyloid‐beta protein precursor internalization and amyloid‐beta generation. J Alzheimers Dis. 2015;47:73‐83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Khan N, Datta G, Geiger JD, Chen X. Apolipoprotein E isoform dependently affects Tat‐mediated HIV‐1 LTR transactivation. J Neuroinflammation. 2018;15:1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Khan N, Lakpa KL, Halcrow PW, et al. BK channels regulate extracellular Tat‐mediated HIV‐1 LTR transactivation. Sci Rep. 2019;9:1‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Christensen KA, Myers JT, Swanson JA. pH‐dependent regulation of lysosomal calcium in macrophages. J Cell Sci. 2002;115:599‐607. [DOI] [PubMed] [Google Scholar]
- 50. Morgan AJ, Platt FM, Lloyd‐Evans E, Galione A. Molecular mechanisms of endolysosomal Ca2+ signalling in health and disease. Biochem J. 2011;439:349‐374. [DOI] [PubMed] [Google Scholar]
- 51. Lloyd‐Evans E, Morgan AJ, He X, et al. Niemann‐Pick disease type C1 is a sphingosine storage disease that causes deregulation of lysosomal calcium. Nat Med. 2008;14:1247‐1255. [DOI] [PubMed] [Google Scholar]
- 52. Lonn P, Kacsinta AD, Cui XS, et al. Enhancing endosomal escape for intracellular delivery of macromolecular biologic therapeutics. Sci Rep. 2016;6:1‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li GH, Li W, Mumper RJ, Nath A. Molecular mechanisms in the dramatic enhancement of HIV‐1 Tat transduction by cationic liposomes. FASEB J. 2012;26:2824‐2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kolson DL, Collman R, Hrin R, et al. Human immunodeficiency virus type 1 Tat activity in human neuronal cells: uptake and trans‐activation. J Gen Virol. 1994;75(Pt 8):1927‐1934. [DOI] [PubMed] [Google Scholar]
- 55. Slice LW, Codner E, Antelman D, et al. Characterization of recombinant HIV‐1 Tat and its interaction with TAR RNA. Biochemistry. 1992;31:12062‐12068. [DOI] [PubMed] [Google Scholar]
- 56. Vijaykumar TS, Nath A, Chauhan A. Chloroquine mediated molecular tuning of astrocytes for enhanced permissiveness to HIV infection. Virology. 2008;381:1‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fredericksen BL, Wei BL, Yao J, Luo T, Garcia JV. Inhibition of endosomal/lysosomal degradation increases the infectivity of human immunodeficiency virus. J Virol. 2002;76:11440‐11446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wu LG, Hamid E, Shin W, Chiang HC. Exocytosis and endocytosis: modes, functions, and coupling mechanisms. Annu Rev Physiol. 2014;76:301‐331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lloyd‐Evans E. On the move, lysosomal CAX drives Ca2+ transport and motility. J Cell Biol. 2016;212:755‐757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhong XZ, Yang Y, Sun X, Dong XP. Methods for monitoring Ca(2+) and ion channels in the lysosome. Cell Calcium. 2017;64:20‐28. [DOI] [PubMed] [Google Scholar]
- 61. Xiong J, Zhu MX. Regulation of lysosomal ion homeostasis by channels and transporters. Sci China Life Sci. 2016;59:777‐791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sakurai Y. Ebola virus host cell entry. Uirusu. 2015;65:71‐82. [DOI] [PubMed] [Google Scholar]
- 63. Sakurai Y, Kolokoltsov AA, Chen CC, et al. Ebola virus. Two‐pore channels control Ebola virus host cell entry and are drug targets for disease treatment. Science. 2015;347:995‐998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gunaratne GS, Yang Y, Li F, Walseth TF, Marchant JS. NAADP‐dependent Ca(2+) signaling regulates Middle East respiratory syndrome‐coronavirus pseudovirus translocation through the endolysosomal system. Cell Calcium. 2018;75:30‐41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rahman T, Cai X, Brailoiu GC, Abood ME, Brailoiu E, Patel S. Two‐pore channels provide insight into the evolution of voltage‐gated Ca2+ and Na+ channels. Sci Signal. 2014;7:ra109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wong VKW, Zeng W, Chen J, et al. Tetrandrine, an activator of autophagy, induces autophagic cell death via PKC‐alpha inhibition and mTOR‐dependent mechanisms. Front Pharmacol. 2017;8:1‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Aits S, Kricker J, Liu B, et al. Sensitive detection of lysosomal membrane permeabilization by lysosomal galectin puncta assay. Autophagy. 2015;11:1408‐1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Aits S, Jaattela M, Nylandsted J. Methods for the quantification of lysosomal membrane permeabilization: a hallmark of lysosomal cell death. Methods Cell Biol. 2015;126:261‐285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Cheng J, Zeidan R, Mishra S, et al. Structure‐function correlation of chloroquine and analogues as transgene expression enhancers in nonviral gene delivery. J Med Chem. 2006;49:6522‐6531. [DOI] [PubMed] [Google Scholar]
- 70. Wolfram J, Nizzero S, Liu H, et al. A chloroquine‐induced macrophage‐preconditioning strategy for improved nanodelivery. Sci Rep. 2017;7:1‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zhang X, Sawyer GJ, Dong X, Qiu Y, Collins L, Fabre JW. The in vivo use of chloroquine to promote non‐viral gene delivery to the liver via the portal vein and bile duct. J Gene Med. 2003;5:209‐218. [DOI] [PubMed] [Google Scholar]
- 72. Solomon VR, Lee H. Chloroquine and its analogs: a new promise of an old drug for effective and safe cancer therapies. Eur J Pharmacol. 2009;625:220‐233. [DOI] [PubMed] [Google Scholar]
- 73. Morissette G, Moreau E, C.‐Gaudreault R, Marceau F. Massive cell vacuolization induced by organic amines such as procainamide. J Pharmacol Exp Ther. 2004;310:395‐406. [DOI] [PubMed] [Google Scholar]
- 74. Marceau F, Bawolak MT, Lodge R, et al. Cation trapping by cellular acidic compartments: beyond the concept of lysosomotropic drugs. Toxicol Appl Pharmacol. 2012;259:1‐12. [DOI] [PubMed] [Google Scholar]
- 75. Bright NA, Gratian MJ, Luzio JP. Endocytic delivery to lysosomes mediated by concurrent fusion and kissing events in living cells. Curr Biol. 2005;15:360‐365. [DOI] [PubMed] [Google Scholar]
- 76. Luzio JP, Pryor PR, Bright NA. Lysosomes: fusion and function. Nat Rev Mol Cell Biol. 2007;8:622‐632. [DOI] [PubMed] [Google Scholar]
- 77. Huotari J, Helenius A. Endosome maturation. EMBO J. 2011;30:3481‐3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. de Duve C. The lysosome turns fifty. Nat Cell Biol. 2005;7:847‐849. [DOI] [PubMed] [Google Scholar]
- 79. Mindell JA. Lysosomal acidification mechanisms. Annu Rev Physiol. 2012;74:69‐86. [DOI] [PubMed] [Google Scholar]
- 80. Yamauchi Y, Helenius A. Virus entry at a glance. J Cell Sci. 2013;126:1289‐1295. [DOI] [PubMed] [Google Scholar]
- 81. Cossart P, Helenius A. Endocytosis of viruses and bacteria. Cold Spring Harb Perspect Biol. 2014;6:a016972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Chauhan A, Khandkar M. Endocytosis of human immunodeficiency virus 1 (HIV‐1) in astrocytes: a fiery path to its destination. Microb Pathog. 2015;78:1‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Miyauchi K, Kim Y, Latinovic O, Morozov V, Melikyan GB. HIV enters cells via endocytosis and dynamin‐dependent fusion with endosomes. Cell. 2009;137:433‐444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Staring J, Raaben M, Brummelkamp TR. Viral escape from endosomes and host detection at a glance. J Cell Sci. 2018;131:jcs216259. [DOI] [PubMed] [Google Scholar]
- 85. Wenzel ED, Bachis A, Avdoshina V, Taraballi F, Tasciotti E, Mocchetti I. Endocytic trafficking of HIV gp120 is mediated by dynamin and plays a role in gp120 neurotoxicity. J Neuroimmune Pharmacol. 2017;12:492‐503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Datta G, Miller NM, Afghah Z, Geiger JD, Chen X. HIV‐1 gp120 promotes lysosomal exocytosis in human schwann cells. Front Cell Neurosci. 2019;13:1‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Xu H, Ren D. Lysosomal physiology. Annu Rev Physiol. 2015;77:57‐80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Cheng X, Zhang X, Gao Q, et al. The intracellular Ca(2)(+) channel MCOLN1 is required for sarcolemma repair to prevent muscular dystrophy. Nat Med. 2014;20:1187‐1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Li X, Rydzewski N, Hider A, et al. A molecular mechanism to regulate lysosome motility for lysosome positioning and tubulation. Nat Cell Biol. 2016;18:404‐417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Wang W, Gao Q, Yang M, et al. Up‐regulation of lysosomal TRPML1 channels is essential for lysosomal adaptation to nutrient starvation. Proc Natl Acad Sci U S A. 2015;112:E1373‐E1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zhang X, Cheng X, Yu L, et al. MCOLN1 is a ROS sensor in lysosomes that regulates autophagy. Nat Commun. 2016;7:1‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Calcraft PJ, Ruas M, Pan Z, et al. NAADP mobilizes calcium from acidic organelles through two‐pore channels. Nature. 2009;459:596‐600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Zhu MX, Ma J, Parrington J, Calcraft PJ, Galione A, Evans AM. Calcium signaling via two‐pore channels: local or global, that is the question. Am J Physiol Cell Physiol. 2010;298:C430‐C441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Zong X, Schieder M, Cuny H, et al. The two‐pore channel TPCN2 mediates NAADP‐dependent Ca(2+)‐release from lysosomal stores. Pflugers Arch. 2009;458:891‐899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Brailoiu E, Churamani D, Cai X, et al. Essential requirement for two‐pore channel 1 in NAADP‐mediated calcium signaling. J Cell Biol. 2009;186:201‐209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Ruas M, Rietdorf K, Arredouani A, et al. Purified TPC isoforms form NAADP receptors with distinct roles for Ca(2+) signaling and endolysosomal trafficking. Curr Biol. 2010;20:703‐709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Grimm C, Chen CC, Wahl‐Schott C, Biel M. Two‐pore channels: catalyzers of endolysosomal transport and function. Front Pharmacol. 2017;8:1‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Sterea AM, Almasi S, El Hiani Y. The hidden potential of lysosomal ion channels: a new era of oncogenes. Cell Calcium. 2018;72:91‐103. [DOI] [PubMed] [Google Scholar]
- 99. Marchant JS, Patel S. Two‐pore channels at the intersection of endolysosomal membrane traffic. Biochem Soc Trans. 2015;43:434‐441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Ruas M, Galione A, Parrington J. Two‐pore channels: lessons from mutant mouse models. Messenger (Los Angel). 2015;4:4‐22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Kintzer AF, Stroud RM. On the structure and mechanism of two‐pore channels. FEBS J. 2018;285:233‐243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Lagostena L, Festa M, Pusch M, Carpaneto A. The human two‐pore channel 1 is modulated by cytosolic and luminal calcium. Sci Rep. 2017;7:1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Cang C, Bekele B, Ren D. The voltage‐gated sodium channel TPC1 confers endolysosomal excitability. Nat Chem Biol. 2014;10:463‐469. [DOI] [PubMed] [Google Scholar]
- 104. Castonguay J, Orth JHC, Muller T, et al. The two‐pore channel TPC1 is required for efficient protein processing through early and recycling endosomes. Sci Rep. 2017;7:1‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Lonn P, Dowdy SF. Cationic PTD/CPP‐mediated macromolecular delivery: charging into the cell. Expert Opin Drug Deliv. 2015;12:1627‐1636. [DOI] [PubMed] [Google Scholar]
- 106. Glogau R, Blitzer A, Brandt F, Kane M, Monheit GD, Waugh JM. Results of a randomized, double‐blind, placebo‐controlled study to evaluate the efficacy and safety of a botulinum toxin type A topical gel for the treatment of moderate‐to‐severe lateral canthal lines. J Drugs Dermatol. 2012;11:38‐45. [PubMed] [Google Scholar]
- 107. Nakase I, Tanaka G, Futaki S. Cell‐penetrating peptides (CPPs) as a vector for the delivery of siRNAs into cells. Mol Biosyst. 2013;9:855‐861. [DOI] [PubMed] [Google Scholar]
- 108. Koren E, Torchilin VP. Cell‐penetrating peptides: breaking through to the other side. Trends Mol Med. 2012;18:385‐393. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


