NOVEL ALLELE REPORT
Vel‐negative individuals may become alloimmunized after blood transfusion with Vel‐positive blood or through pregnancy after carrying a Vel‐positive child, potentially leading to transfusion reactions or to hemolytic disease of the newborn in subsequent pregnancies. Previously it was found that the single‐pass transmembrane protein SMIM1 harbors the Vel blood group system, ISBT34.1, 2, 3 Vel‐negative individuals have a homozygous 17‐bp deletion (c.64‐80del) in SMIM1, but specific single‐nucleotide polymorphisms in SMIM1 additionally regulate Vel expression. For instance, weak Vel expression is correlated with single‐nucleotide polymorphism rs1175550 in Intron 2, with the minor G allele associated with higher SMIM1 transcript levels compared to the major A allele,1, 4 and previously described single heterozygous missense mutations in Exon 4 of VEL*01 W.01 (c.152T>A, p.Met51Lys) and of VEL*01 W.02 (c.152T>G, p.Met51Arg) are shown to significantly decrease Vel expression.4 To facilitate appropriately matched transfusions it is important to identify mutations in SMIM1 that lead to very weak Vel expression that are difficult to phenotype when needed.
Nine putative Vel‐negative donors were identified by automated serologic screening using a human anti‐Vel serum (BN60) in a period of 3 weeks (1488 donors tested). Genotyping of the SMIM1 gene confirmed that one donor was homozygote for the c.64_80del in SMIM1 and thus indeed Vel negative. In seven donors heterozygosity was revealed for c64_80del and AA for rs1175550 (see Fig. 1A). In one donor, who was homozygous AA for rs1175550, a novel mutation was detected: SMIM1*c.161T>C (p.Leu54Pro; GenBank Accession Number MK783942; see Fig. 1B). This missense mutation resulted in an amino acid change in the transmembrane region of SMIM1. The Leu54 is a conserved hydrophobic amino acid, and the change into proline is deleterious according to in silico predictive methods PolyPhen‐2, PROVEAN, and mutation tasting (SMIM1 NM_001163724.2 c.161T>C; Fig. 1C). Manual serology analysis of the red blood cells (RBCs) carrying the SMIM1*c.161T>C mutation showed that Vel expression was not detected by human anti‐Vel serum (MB46) by column cards (see Fig. 1D) or tube testing, but was only detected by agglutination with anti‐Vel monoclonal antibody M5F3S (data shown in van der Rijst et al.5). Flow cytometry on the RBCs of this SMIM1 missense mutation donor showed that the Vel antigen could be detected but was very low. The RBCs of this donor had lower Vel antigen expression compared to RBCs from donors with a heterozygous 17‐bp deletion in SMIM1 (see Fig. 1E). Ectopic expression of wild‐type SMIM1 and SMIM1*c.161T>C in HEK293T cells showed that SMIM1*c.161T>C resulted in lower Vel expression compared to wild‐type (Fig. 1F) but did not show the same low Vel expression as was observed in RBCs.
Figure 1.
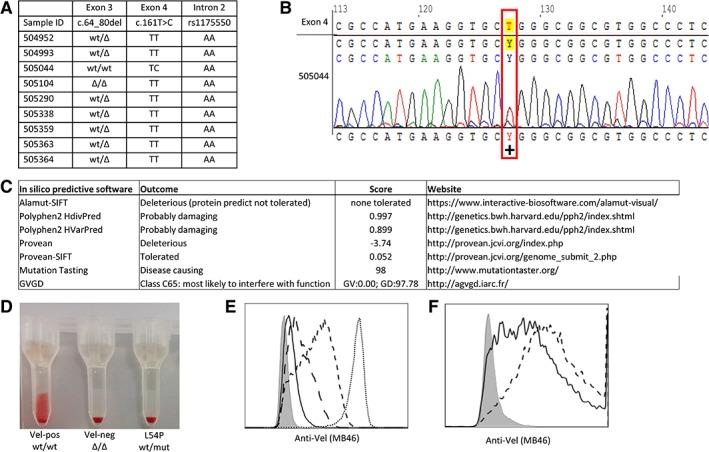
(A) Genotyping analysis of the SMIM1 gene of nine putative Vel‐negative donors. (B) Sequence analysis of a novel heterozygote mutation in exon 4 of the SMIM1 gene: SMIM1*161T>C (p.Leu54Pro). (C) Scores and outcome of the used in silico predictive methods. (D) Serology analysis with serum of MB46 of p.Leu54Pro in column cards with a Vel‐positive donor as positive control (Vel‐pos wt/wt) and a homozygote deletion donor as a negative control (Vel‐neg Δ/Δ). (E) Flow cytometry Vel expression analysis of SMIM1 genotyped RBCs: Vel‐negative (Δ/Δ; filled), SMIM1*161T>C (p.Leu54Pro; solid), heterozygote donor (wt/Δ; long‐dashed), and Vel‐positive (wt/wt + rs1175550 AA; dashed, wt/wt + rs1175550 GG; dotted). (F) Flow cytometry analysis of transfected HEK293T cell line with pcDNA3.1+ empty vector (filled), wild‐type SMIM1 (dashed), and SMIM1*161T>C (p.Leu54Pro); solid. Mut = mutation; wt = wild‐type; Δ = c.del64_80.
Here we identified a novel SMIM1 mutation, p.Leu54Pro, resulting in very weak Vel expression on RBCs. As the expression levels were lower than in donors with a heterozygous deletion, as was the case for two previously described SMIM1 mutations VEL*01 W.01 and VEL*01 W.02,4 this weak Vel expression suggests a dominant negative effect of the mutated SMIM1 protein. This missense mutation is not present in the gnomAD database containing more than 125,000 exomes (https://gnomad.broadinstitute.org/gene/ENSG00000235169, accessed April 10, 2019).
In conclusion, to date all reported heterozygous SMIM1 missense mutations in donors with very weak Vel expression show to exert the same dominant negative effect on wild‐type SMIM1 resulting in very weak Vel expression. The origin of this dominant effect remains however unknown. From a clinical point of view, it is important to genotype for the c.64_80del mutation to identify Vel‐negative RBC donors, as this case again demonstrates that serology might fail to identify donors carrying a missense mutation as Vel very weak.
STUDY DESIGN AND METHODS
Donor material
Dutch donors were serologically screened for the Vel blood group by automated screening using a very strong human anti‐Vel serum (BN60 titer 1:32) on the Magister Blood Grouping System (Sanquin Reagents). Screened Vel‐negative RBC were cryopreserved.4 Informed consent was in accordance with the Declaration of Helsinki and Dutch national and Sanquin internal ethics boards.
Flow cytometry
Flow cytometry analysis was performed as described previously.5 HEK293T cells were transfected with either empty pcDNA3.1+ vector (Invitrogen), wild‐type SMIM1, or SMIM1*c.161T>C (double‐stranded DNA ordered at gBlocks IDT) as described previously.5 Antibodies used were as follows: immunopurified anti‐Vel (MB46) as primary antibody and Alexa Fluor‐647 anti‐human IgG (Invitrogen) as secondary antibody. Results were acquired on a flow cytometer (BD CantoII/LSRII/Fortessa, BD Bioscience, Breda, the Netherlands) and analyzed using computer software (FloJo, Treestar).
Agglutination assays
Donor blood samples were serologically tested with anti‐Vel serum of immunized patients. Automated screening on the Magister was performed by adding 20 μL anti‐Vel serum (BN60) with 40 μL 0.6% (vol/vol) RBCs in low‐ionic‐strength saline (LISS) in screen column cards (both Cellbind, Sanquin Reagents), for 15 minutes at 37°C; subsequently the cards were centrifuged with increasing speed: 120 sec at 75 g, 60 sec at 200 g, and 420 sec at 1790 g. After centrifugation the reactions in the column cards were photographed and thereupon interpreted by the computer software (Magister). Manual testing was performed as described previously,5 with anti‐Vel serum MB46, using standard LISS/antiglobulin test column cards (BIO‐RAD) or tubes.
Genetic analysis
All donor samples were genotyped for the SMIM1 gene using a Taqman assay specific for the c.64_80del, Exons 3 and 4, and Intron 2 as described previously.4, 5 Sequence reactions were performed on a thermocycler (Veriti, Thermo Fisher Scientific), and sequence products were analyzed on a genetic analyzer (3130, Applied Biosystems) with use of computer software (ContigExpress module of Vector NTI Advance 11.5.4 software, Invitrogen) and Vel‐reference sequence (ENST000004444870.2).
AUTHOR CONTRIBUTIONS
LV and JJ performed the automated screening. MvdR performed the other experiments. MvdR, EvdA, and EvdS designed the experiments, analyzed the data, and wrote the manuscript. The manuscript was critically revised by all authors. The authors declare no competing financial interest.
CONFLICT OF INTEREST
The authors have disclosed no conflicts of interest
ACKNOWLEDGMENTS
The authors thank ImmunoHematology‐Diagnostic Sanquin and Molecular Platform Sanquin for the technical assistance and data acquisition regarding blood samples, DNA samples, and sequencing. We thank the Central Facility of Sanquin for their assistance regarding flow cytometry analysis.
Supported by the Landsteiner Foundation for Blood Transfusion Research (LSBR Grant 1351).
The copyright line for this article was changed on 25 February 2020 after original online publication.
REFERENCES
- 1. Cvejic A, Haer‐Wigman L, Stephens JC, et al. SMIM1 underlies the Vel blood group and influences red blood cell traits. Nat Genet 2013;45:542‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Storry JR, Joud M, Christophersen MK, et al. Homozygosity for a null allele of SMIM1 defines the Vel‐negative blood group phenotype. Nat Genet 2013;45:537‐41. [DOI] [PubMed] [Google Scholar]
- 3. Ballif BA, Helias V, Peyrard T, et al. Disruption of SMIM1 causes the Vel‐ blood type. EMBO Mol Med 2013;5:751‐61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Haer‐Wigman L, Stegmann TC, Solati S, et al. Impact of genetic variation in the SMIM1 gene on Vel expression levels. Transfusion 2015;55(6 Pt 2):1457‐66. [DOI] [PubMed] [Google Scholar]
- 5. van der Rijst MVE, Lissenberg‐Thunnissen SN, Ligthart PC, et al. Development of a recombinant anti‐Vel immunoglobulin M to identify Vel‐negative donors. Transfusion 2019;59:1359‐66. [DOI] [PubMed] [Google Scholar]


