ABSTRACT
This review focusses on the functions of intracellular and extracellular calmodulin, its target proteins and their binding proteins during the asexual life cycle of Dictyostelium discoideum. Calmodulin is a primary regulatory protein of calcium signal transduction that functions throughout all stages. During growth, it mediates autophagy, the cell cycle, folic acid chemotaxis, phagocytosis, and other functions. During mitosis, specific calmodulin‐binding proteins translocate to alternative locations. Translocation of at least one cell adhesion protein is calmodulin dependent. When starved, cells undergo calmodulin‐dependent chemotaxis to cyclic AMP generating a multicellular pseudoplasmodium. Calmodulin‐dependent signalling within the slug sets up a defined pattern and polarity that sets the stage for the final events of morphogenesis and cell differentiation. Transected slugs undergo calmodulin‐dependent transdifferentiation to re‐establish the disrupted pattern and polarity. Calmodulin function is critical for stalk cell differentiation but also functions in spore formation, events that begin in the pseudoplasmodium. The asexual life cycle restarts with the calmodulin‐dependent germination of spores. Specific calmodulin‐binding proteins as well as some of their binding partners have been linked to each of these events. The functions of extracellular calmodulin during growth and development are also discussed. This overview brings to the forefront the central role of calmodulin, working through its numerous binding proteins, as a primary downstream regulator of the critical calcium signalling pathways that have been well established in this model eukaryote. This is the first time the function of calmodulin and its target proteins have been documented through the complete life cycle of any eukaryote.
Keywords: calcium signalling, calmodulin, asexual development, Dictyostelium discoideum, mitosis, osmoregulation, chemotaxis, cell differentiation, morphogenesis, extracellular matrix
I. INTRODUCTION
The eukaryotic social amoebozoan Dictyostelium discoideum has been a popular research organism for more than 80 years. Its original appeal was based on its lifestyle transition from single cell growth to multicellular asexual development characterized by chemotaxis, morphogenesis and cell differentiation. There are multiple reasons for this popularity: a well‐defined, rapid 24‐h multicellular life cycle; a haploid, sequenced genome; the ease of generating conditional and knockout mutants via diverse techniques; and the availability of multiple curated resources (e.g. strains, antibodies, data analysis programs) available through http://dictybase.org. Initially recognized for studies on fundamental biological processes, more recently it has been used a biomedical model for research into autophagy, cell stress, extracellular vesicles, osmoregulation, wound healing, neurodegenerative diseases, and mitochondrial diseases, among others (e.g. Annesley & Fisher, Annesley & Fisher, 2001; Huber, 2016; Mathavarajah, O'Day, & Huber, 2018b; Dunn et al., 2018; Myre, Huber, & O'Day, 2018; Tatischeff, 2019).
The asexual life cycle of this eukaryotic microbe is summarized in Fig. 1. After feeding on bacteria, where folic acid‐mediated chemotaxis aids in food detection and ingestion, starved cells switch to cyclic AMP (cAMP)‐mediated chemotaxis that guides their pulsatile migration towards aggregation centres. The aggregates grow upwards until they topple over as multicellular pseudoplasmodia that, depending on environmental conditions, can either culminate directly or migrate for extended periods in response to heat, light and other factors. The cellular nature of the amoebozoan pseudoplasmodia differ from the multinucleate plasmodia of mycomycetes (true slime molds). Within these pseudoplasmodia, also commonly referred to as ‘slugs’, cell differentiation begins with a well‐defined pattern and polarity. The pre‐spore cells reside in the hind two‐thirds to three‐quarters of the slug while the anterior one‐third to one‐quarter of the slug contains cells destined to differentiate into stalk cells. The pre‐stalk cohort is heterogeneous consisting of A, AB, and O cells, each with distinct fates and patterns of gene expression. Anterior‐like cells (ALCs) distribute throughout the pre‐spore region where they serve as a source of renewal for the anterior pre‐stalk cells. Pseudoplasmodia can regenerate if disrupted or transected, as detailed below.
Figure 1.
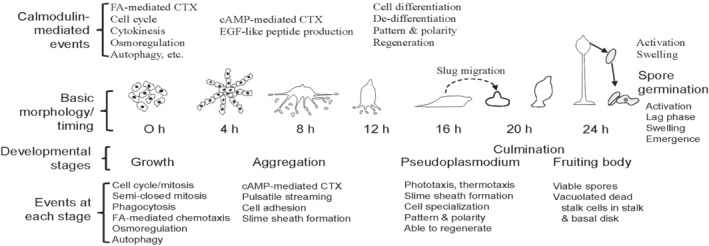
Calmodulin functions during asexual development in Dictyostelium discoideum. cAMP, cyclic AMP; CTX, chemotaxis; EGF, epidermal growth factor; FA, folic acid.
During culmination, cells in the pre‐stalk region flow like a reverse fountain through the pre‐spore region where cells from three groups of pre‐stalk cells (i.e. AB, B, ALCs) will form the basal disc that supports the growing stalk. Differentiation of other prestalk cells (i.e. A, AB, O) causes the stalk to elongate lifting the differentiating spore mass off the substratum ultimately to reside at the tip of the elongated stalk. During these events, the stalk cells die and become the supporting structure for the viable spores that will start the next generation. In the spore mass, the spores of Dictyostelium float freely in an aqueous, protein‐rich extracellular fluid. Since there is no surrounding wall around this spore droplet, it cannot be considered a sorus or sporangium, terms well established for plants and fungi.
Calcium (Ca2+) function is universal in eukaryotes and evolutionary research indicates that the first signal transduction systems utilized this ubiquitous divalent cation (e.g. Peterson, Michalak, & Verkhratsky, 2015; Plattner & Verkhratsky, 2015). The transition from single‐celled eukaryotes to metazoans was accompanied by increasing complexity in how Ca2+ is utilized for signal transduction. This includes the evolution of distinct protein machinery in protozoans, such as the emergence of store‐operated Ca2+ entry, prior to the appearance of animals (Collins & Meyer, 2011). Undergoing developmental phase transitions from single cells to a bona fide multicellular tissue during development puts D. discoideum in a unique position to provide critical insight into the evolution of calcium signal transduction. During animal development, Ca2+ functions in multiple events including cell death, differentiation, division and motility. It is involved in biomembrane fusion (e.g. fertilization, secretion, endocytosis, etc.), morphogenesis and the specialization of bone, heart and neurons, to name a few cell types (e.g. Webb & Miller, 2003; Zhou et al., 2011).
II. CALCIUM–CALMODULIN SIGNALLING IN DICTYOSTELIUM DISCOIDEUM
As in all other eukaryotes, Ca2+ signalling is central to the survival and physiological function of D. discoideum. Ca2+‐mediated events are, in turn, dependent on the activation of Ca2+‐binding proteins (CBPs) of which calmodulin (CaM) is the most predominant. D. discoideum CaM [152 amino acids (aas), 17 kDa] is encoded by the calA gene (http://dictybase.org/gene/DDB_G0279407). It is expressed throughout growth, steadily decreasing in expression throughout asexual development (Van Driessche et al., 2002). It contains four Ca2+‐binding EF hands (a helix–loop–helix structural motif) and is over 90% identical to mammalian CaM, being able to fully activate mammalian CaM‐binding proteins (CaMBPs) such as bovine brain cyclic nucleotide phosphodiesterase and erythrocyte Ca2+/Mg2+‐ATPase (Bazari & Clarke, 1981). It differs in the lack of Lys‐118 trimethylation, a mechanism of post‐translational regulation. It is the primary CBP involved in activating a diversity of downstream proteins in its Ca2+‐free (apo‐CaM) or Ca2+‐bound (Ca‐CaM) states as detailed herein. Intracellularly, CaM is found in the nucleus, nucleolus and contractile vacuole (CV) system as well as various regions throughout the cytoplasm (e.g. cytoskeleton). CaM is also constitutively secreted where it functions extracellularly to inhibit cell proliferation and increase cAMP‐mediated chemotaxis. (O'Day, Huber, & Suarez, 2012; see Section V).
The related calB encodes a Ca2+‐binding, CaM‐like protein consisting of 149 aas of 16.8 kDa. While its lowest level of expression occurs during growth, calB is differentially expressed during asexual development (Rosel et al., 2000). During aggregation through to the pseudoplasmodium stage it increases in expression and is expressed at the highest level in spores. CalB shows about 50% similarity to other CaMs but contains only two Ca2+‐binding EF hands. However, the precise function of CalB remains to be studied. Another CaM‐related protein, Myosin‐ID light chain (MlcD; 147 aas, 16.5 kDa) is a low‐affinity Ca2+‐binding protein that operates as the light chain for myosin‐D (MyoD) but not MyoB or MyoC (De La Roche, Lee, & Coté, 2003).
In eukaryotes, CaM is a highly conserved Ca2+‐sensor and effector that mediates a diversity of biological processes through its regulation of hundreds of target CaMBPs (Chin & Means, 2000). In the absence or presence of Ca2+, it regulates the activity of ion channels, enzymes, heterotrimeric G‐proteins, transcription factors, transport systems and other critical proteins. As the primary mediator of intracellular Ca2+ signal transduction, it oversees the operation of a diversity of growth factors, hormones and neurotransmitters. As such, it carries out critical operations in a diversity of biological processes (e.g. apoptosis, autophagy, cell differentiation, cell division, morphogenesis, osmoregulation, etc.) and pathophysiological states (e.g. cancer, neurodegeneration, heart arrythmias, etc.) (Berchtold & Villalobo, 2014; Villalobo, 2018; Urrutia et al., 2019). The importance of D. discoideum as a model system to investigate fundamental biological processes and disease states is gaining increased attention (Annesley & Fisher, 2001; Huber, 2016; Dunn et al., 2018; Myre et al., 2018; Tatischeff, 2019). Here we show the critical role of CaM and its CaMBPs throughout the full developmental cycle of D. discoideum to emphasize its importance for continued study of specific cellular events and disease processes. Considering the diversity and central importance of CaM function, understanding how it can modulate such a diversity of precise events, often in the same cell, in different physiological and developmental events, argues that this area of research has the potential to yield extraordinary insight and pharmaceutical targets.
Just over a decade ago, Catalano & O'Day (2008) catalogued the CaMBPs of D. discoideum, their specific functions and their CaM‐binding domains (CaMBDs). CaMBDs are unique since they do not involve a precise sequence of specific amino acids (Hoeflich & Ikura, 2002; Berchtold & Villalobo, 2014). Calcium‐independent CaM‐binding occurs through IQ [(FILV)Qxxx(RK)Gxxx(RK)xx(FILVWY)] or IQ‐like [(FILV)Qxxx(RK)Gxxxxxxxx] motifs. Calcium‐dependent CaMBDs employ sequences that fall into two categories: canonical and non‐canonical binding motifs. Canonical CaMBDs comprise a diverse collection that are defined by the positions of hydrophobic residues [e.g. 1–5‐10 motif: (FILVW)xxxx(FAILVW)xxxx(FILVW); 1–5–8‐14 motif: (FILVW)xxx(FAILVW)xx(FAILVW)xxxxx(FILVW)]. Non‐canonical binding does not appear to involve definable amino acid patterns.
D. discoideum CaMBPs with identified binding domains include calcineurin (CanA), cysteine‐rich protein A (CyrA), nucleomorphin (NumA1) and phosphoglycerate kinase (PgkA). CaMBPs where the CaM binding was experimentally verified and putative CaMBDs were identified but not experimentally proven include: calmodulin‐binding protein 46 (CmbB), Ras GTPase‐activating‐like protein (RgaA), histone H1 (H1), ribosomal subunit protein L19 (Rpl19), thymidine kinase (ThyB) and Von Willebrand factor kinase A (VwkA). In addition to covering all the newly identified CaMBPs and their functions, the attributes and roles of proteins previously reviewed by Catalano & O'Day (2008) are updated and linked to specific events during the D. discoideum life cycle. Other CaMBPs that were reviewed by Catalano & O'Day (2008) but which since have not been detailed further are not covered here: PgkA, histone H1, Rpl19 and ThyB.
Studying a group of proteins that are controlled by a common regulator can lead to new insights that may not arise when studying a single protein. The study of CaMBPs in D. discoideum has led to the discovery of many proteins that at first seemed unrelated in localization and function but, with continued work, revealed a series of previously unexpected interplays. By focussing for the first time on CaM during a complete eukaryotic life cycle, this review provides greater insight into the importance of this regulatory protein in this model eukaryotic amoebozoan while simultaneously revealing specific translational insight into various fundamental cellular events.
III. CELL MOTILITY AND CHEMOTAXIS DURING GROWTH
During growth, D. discoideum amoebae feed on bacteria and other microbes. Their ability to locate their food supply is enhanced through their positive chemotactic response to folic acid secreted by bacteria (Van Driel, 1981). Recently, Pan et al. (2016) revealed that folic acid binds to a receptor that not only allows cells to locate bacteria but also to ingest them. Schlatterer & Malchow (1993) demonstrated the importance of Ca2+ signalling in chemotaxis to folic acid. Subsequent research showed that a diversity of CaM‐dependent effectors is involved in the motility of D. discoideum cells: RgaA, myosin light chain kinase (MlkA), myosin heavy chain kinase (MhkA), and many myosin I isoforms (reviewed in Catalano & O'Day, 2008). Thus, it came as no surprise when chemotaxis to folic acid was shown to be CaM‐dependent (Gauthier & O'Day, Gauthier & O'Day, 2001). Profiling revealed the presence of dozens of Ca2+‐dependent CaMBPs in cytosolic and nuclear fractions plus a small population of membrane‐localized proteins in growth‐phase cells (Gauthier & O'Day, Gauthier & O'Day, 2001). Starvation and refeeding experiments linked a handful of these to events including chemotaxis, but most of the proteins remain uncharacterized.
Other experiments revealed the CaM‐dependent serine/threonine phosphorylation of proteins linked to folic acid chemotaxis, thus reinforcing the role of CaM in this process (Gauthier & O'Day, Gauthier & O'Day, 2001). Since CanA, the sole CaM‐dependent protein phosphatase, dephosphorylates both serine and threonine residues, it was suggested that it could function during folic acid‐mediated chemotaxis. CanA was the first CaMBP to be characterized in D. discoideum (Catalano & O'Day, 2008; Damman et al., Dammann et al., 1996; Thewes et al., 2016). However, despite its role in a diversity of chemotactic events in other species, investigations into the role of CanA suggest it is not linked to folic acid‐mediated chemotaxis. Instead, it is critical to numerous other basic cellular processes essential for growth and, as detailed later, asexual development (Kobel‐Höller et al., 2018). However, CanA has been implicated in cAMP‐mediated chemotaxis, fitting with the results of Gauthier & O'Day (Gauthier & O'Day, 2001) who showed significant differences in the CaMBP profiles of cells capable of folic acid‐mediated chemotaxis versus those undergoing cAMP‐mediated chemotaxis.
Various myosins and kinases function in motility and chemotaxis. Myosin light chain C (MlcC), a regulator of myosin‐1C (Myo1C), is one of the best characterized CaMBPs in D. discoideum (e.g. Langelaan et al., 2016). It binds to apo‐CaM via the Ca2+‐independent IQ motif (summarized in Catalano & O'Day, 2008). During cell motility, Myo1C localizes to the cell cortex where it interacts with actin to generate cortical tension as part of the events leading to the extension and retraction of the cell membrane (Langelaan et al., 2016). The CaM‐mediated regulation of myosin‐based contraction involves MhkA, myosin heavy chain kinase B (MhkB) and the myosin I heavy chain kinase P21‐activated protein kinase (PakB), each of which has been shown to contain presumptive CaMBDs (see Table 1 in Catalano & O'Day, 2008). Other functions for cortical Myo1C during the growth phase include pinocytosis (Langelaan et al., 2016).
Until recently, the receptor for folic acid remained elusive. Using a novel quantitative phosphoproteomic technique, Pan et al. (2016) identified fAR1, a G‐protein coupled receptor (GPCR) that is essential for folic acid‐mediated signal transduction. In addition, the receptor is essential for both chemotaxis and bacterial phagocytosis. Because of the importance of CaM in folic acid‐mediated chemotaxis and since many G‐protein‐coupled receptors are CaMBPs, a scan of fAR1 was performed to assess the presence of potential CaMBDs (Yap et al., 2000; Hoeflich & Ikura, 2002). A CaM‐binding region (611CVLIIFGAKFWKIYKPVEDD630) was identified within which six distinct canonical binding motifs were present, strongly supporting fAR1 as a CaMBP (D.H. O'Day, unpublished data).
IV. GROWTH PHASE AND SEMI‐CLOSED MITOSIS
While researchers have been unable to synchronize the cell cycle of D. discoideum sufficiently, this has not limited its value in the study of this critical process, not only as a biomedical research model but also for elucidation of related evolutionary events. The presence of CaM in the nucleus of this microbe has been experimentally verified (Huber, Catalano, & O'Day, 2013). The cell cycle includes semi‐closed mitosis wherein the nuclear envelope does not break down but allows the regulated nucleocytoplasmic transport of various CaMBPs and some of their binding partners (O'Day & Budniak, 2015). For example, cyclin‐dependent kinase 5 (Cdk5) is a nuclear CaMBP that progressively translocates from the nucleoplasm to the cytoplasm during mitosis, re‐entering the nucleus during cytokinesis (Huber et al., 2013). D. discoideum Cdk5 is involved in multiple events during growth and development including cell differentiation, endocytosis, phagocytosis, proliferation and secretion (Sharma et al., 2002; Huber & O'Day, 2011b, 2012b). During interphase, Cdk5 binds to the metalloproteinase puromycin‐sensitive aminopeptidase A (PsaA) in the nucleus in a Ca2+/CaM‐dependent manner. PsaA also possesses CaMBDs, and immunoprecipitation experiments with nuclear extracts co‐precipitate CaM, Cdk5 and PsaA (Huber et al., 2013).
In keeping with its role in other organisms, the significance of PsaA in the cell cycle was suggested initially by its nuclear co‐localization and co‐immunoprecipitation with NumA1 (Catalano, Poloz, & O'Day, 2011). NumA1 is a nucleolar/nucleoplasmic, BRCA1 C‐terminal (BRCT) domain‐containing CaMBP involved in mitosis in D. discoideum (Myre & O'Day, 2002, 2004a, 2004b, 2005). Showing a predominantly nuclear localization during interphase, PsaA translocates to the cytoplasm during mitosis (Catalano et al., 2011). The study also identified the first nuclear localization sequence (680RKRF683) for D. discoideum. Despite the presence of a putative CaMBD and binding to two CaMBPs, NumA and Cdk5, neither Ca2+ nor CaM are required for the nuclear localization of PsaA. More about the role of this enzyme in multicellular development and differentiation is discussed below (see Section XIV).
In addition to containing experimentally verified CaM‐binding and BRCT domains, as well as nuclear (NLS) and nucleolar localization sequences (NoLS), the nuclear/nucleolar protein NumA also contains a DEED domain, an extensive palindromic stretch of repeating aspartic acid and glutamic acid residues (Myre & O'Day, 2002, 2004a, 2004b). The highly acidic DEED domain shares similarities with a sequence found in nucleoplasmin, a protein that binds to core histones where it is involved in chromatin condensation and DNA transcription (Onikubo et al., 2015). Overexpression of NumA constructs lacking the DEED domain results in multinuclearity while not altering the nucleolar/nuclear localization of the protein (Myre & O'Day, 2002). It should be noted that during mitosis, as in other organisms, the nucleolus disappears during prophase, reappearing during telophase. Fitting with this, NumA localization becomes nucleoplasmic during prophase then it becomes predominantly cytoplasmic until nucleolar reorganization (O'Day, 2019). To understand how the DEED domain might mediate nuclear number, immunoprecipitation and yeast two‐hybrid studies revealed that Ca2+‐binding protein 4a (Cbp4a) bound to NumA via the DEED sequence (Myre & O'Day, 2004a). While these studies so far have failed to reveal the answer, they provide valuable insight into critical aspects of mitosis in this eukaryotic microbe. Unlike NumA, PsaA and Cdk5 that translocate from the nucleus to the cytoplasm during mitosis, Cbp4a localizes to the nucleoplasm after nucleolar dissolution where it organizes into multiple, discrete nucleoplasmic islands (Catalano & O'Day, 2013; O'Day, 2019). These translocations along with those of other proteins led to the realization that D. discoideum has a semi‐open mitosis as opposed to a closed mitosis as previously thought (O'Day & Budniak, 2015). Semi‐open mitosis refers to a form of mitosis in which the nuclear envelope undergoes fenestration to be partially but not completely disrupted. It is observed in certain fungal species such as Ustilago maydis and Schizosaccharomyces japonicas (Boettcher & Barral, 2013).
Less is known about CaMBPs involved in the final event of cell division: cytokinesis. However, the Dictyostelium WW domain‐containing protein A (DwwA) is a calcium‐independent, single IQ‐domain CaMBP that is essential for this event (Catalano & O'Day, 2008).
V. EXTRACELLULAR CALMODULIN DURING GROWTH
Before solid experimental evidence was presented for the extracellular presence and function of CaM, several studies had suggested its presence in a diversity of plant and animal species. Although not noted in their publication, a scan of the data from a proteomics‐based analysis of secreted proteins during D. discoideum development indicated the presence of extracellular CaM (eCaM; Bakthavatsalam & Gomer, 2010). Also present were several CaMBPs including Cbp4a, PsaA, CmbB and CmbC. Suarez et al. (2011) subsequently revealed that eCaM was involved in the processing of the matricellular protein CyrA as discussed below (see Section XII). O'Day et al. (2012) followed up with a detailed study on eCaM showing that it is present at high and consistent levels throughout asexual development where it functions during at least three different events. It dose‐dependently inhibits cell proliferation during growth and enhances cAMP‐mediated chemotaxis during development. eCaM localizes to the extracellular matrix (ECM) where it is involved in processing CyrA to release fragments containing bioactive epidermal growth factor (EGF)‐like repeats (see Section XII). While the mechanism of eCaM secretion remains to be clarified, two primary routes seem reasonable. First, it could be translocated in association with one or more extracellular CaMBPs, such as CyrA, or it could be released during the expulsion phase of CV system function. The relationship between CaM and the CV system is detailed below (see Section VIII). Since CaM serves multiple functions intracellularly, it is likely that more extracellular functions will be revealed in future studies.
VI. PHAGOCYTOSIS
D. discoideum is a professional phagocyte that has provided valuable insights into the early evolution of the immune system (Chen, Zhuchenko, & Kuspa, 2007; Dunn et al., 2018). However, while Ca2+ signal transduction mediated by CaM is essential for phagocytosis in both animal, protozoa and plant cells, the importance of Ca2+/CaM signalling in this process in D. discoideum remains unstudied. As discussed above, Pan et al. (2016) identified fAR1, a folic acid G‐protein‐linked receptor that mediates both chemotaxis and phagocytosis which, as indicated above, is a potential CaMBP. They proposed that mammalian phagocytes might use the same system of chase (chemotaxis) and engulf (phagocytosis) that D. discoideum employs during bacterial feeding. The raw data on the genes linked to phagocytosis, based on transcriptional changes, from a study by Sillo et al. (2008) were analysed for the presence of CaMBP‐encoding genes, but only one was identified (CmbB). CmbB is almost entirely comprised of tandem IP22 (FNIP) repeats (O'Day et al., 2006). It binds CaM via a CaMBD (179IPKSLRSLFLGKGYNQPLEF198) that contains canonical 1–10 and 1–15 binding motifs.
The highly conserved, multi‐functional protein nucleotide diphosphate kinase (NDPK) is an enigma. Its primary housekeeping function is to phosphorylate nucleotides, but it also is involved in a diversity of other processes that are typically unrelated to its enzymatic function. D. discoideum has three NDPK genes (mitochondrial ndkM, cytosolic ndkC, and uncharacterized gene DDB_G0292928) of which cytosolic nucleotide diphosphatase kinase C (NdkC) is the most predominant (Annesley et al., 2011). NdkC functions to inhibit phagocytosis and macropinocytosis, and it co‐localizes with CaM at the CV system. It should be noted that the phagocytic data of Sillo et al. (2008) did not reveal NdkC. Citing unpublished data, Annesley et al. (2011) stated that NdkC did not co‐immunoprecipitate with CaM. While this suggests it is not a CaMBP, sequence analysis reveals that cytosolic NdkC contains a CaMBD (23GLVGEIIARYEKKGFVLVGLKQLV46) with six different canonical binding motifs (3 × 1–14, 2 × 1–16, 1 × 1–5‐10, 1 × 1–10; D.H. O'Day, unpublished data). Further to this, the authors did not provide details on the methodology or antibodies that were used to assesss an interaction between CaM and NdkC. This suggests that NdkC remains a potential CaMBP that requires further study.
VII. AUTOPHAGY DURING GROWTH AND DEVELOPMENT
Autophagy is a critical metabolic pathway that is highly conserved in eukaryotes. The term autophagy describes a group of cellular pathways (including macro‐ and microautophagy) that oversee the elimination of intracellular material through lysosomal digestion. Seminal work in yeast established the genetics underlying autophagy (Levine & Klionsky, 2017). Many of the genes in yeast that regulate autophagy also function in D. discoideum (Calvo‐Garrido et al., 2010). While autophagosomes in yeast originate from a concentrated cytoplasmic region, autophagy induction in D. discoideum resembles the stages in mammalian autophagy, where nascent autophagosomes originate from multiple cytoplasmic locations (Otto et al., 2004; Tekinay et al., 2006). The autophagy‐related 1 (Atg1/ULK) complex initiates autophagy in unikonts (which includes yeast, Dictyostelium and animal cells) but alternative strategies have also been suggested for non‐unikont parasites (such as Trypanosoma) (Földvári‐Nagy et al., 2014). In D. discoideum, this complex is composed of Atg1, Atg13, Atg101 and DDB_G0285767 (ortholog of Atg11) (Mesquita et al., 2015, 2017). By utilizing the Calmodulin Target Database, we found that each protein from the Atg1 complex in D. discoideum possesses putative CaMBDs (Yap et al., 2000). One or more Ca2+‐dependent non‐canoncal and canonical CaMBDs were identified for Atg1 (non‐canonical 110EKALYFMKQLAN121), Atg13 (1–10, 1–16, 104FYKNVIILIRTIYAML129), Atg101 (1–14 and 1–16, 72LTSIMKRKAKTAQISI86), and DDB_G0285767 (1–16, 172FFEYRENLIKTANKSF191; 1–14, 1–16, 1–5‐10, 240WYKHLKEEFSKVKLKV255). The potential for CaM to bind the Atg1 complex aligns with recent observations in D. discoideum where rapamycin (a drug which initiates autophagy) treatment increases intracellular Ca2+ levels (Swer, Lohia, & Saran, 2014).
During growth in nutrient‐rich media, many of the D. discoideum autophagy genes appear to be dispensable (Otto et al., 2003, 2004). Known exceptions to this are atg8a and atg9, where their loss leads to reduced growth (Tung et al., 2010; Messling et al., 2017). Atg9 is found on distinct Golgi‐derived vesicles that are a source of autophagosomal membrane (they fuse to the autophagosomal outer membrane) and it is consequently involved in forming autophagosomes (Orsi et al., 2012; Yamamoto et al., 2012). It has been shown that the induction of autophagy by thapsigargin occurs through increased intracellular Ca2+ signalling (Luciani et al., 2011; Marcassa et al., 2017). The resulting Ca2+‐activated calpains mobilize Atg9‐positive vesicles for autophagosome formation in human cells. Early research had shown that calpains often target CaMBPs (Wang, Villalobo, & Roufogalis, 1989). A Calmodulin Target Database scan revealed a putative, unusually short CaMBD in D. discoideum Atg9 (221IANRIMRK228) suggesting the ortholog may also bind to CaM but via a non‐canonical motif (S. Mathavarajah, unpublished data). This idea is strengthened by the result that CaM co‐immunoprecipitated during a proteomic screen of Atg9 interactors in human cells (Kakuta et al., 2017). These results link Ca2+ fluxes to the progression of autophagy and suggest a potential role for CaM in regulating autophagosome formation.
During starvation, increased intracellular Ca2+ is thought to be involved in the signalling cascade regulating the transition to multicellular development (Tanaka et al., 1998). Similarly, nutrient depletion in mammalian cells causes a transient increase in intracellular Ca2+ levels leading to the initiation of autophagy (Høyer‐Hansen et al., 2007). This signalling mechanism may also initiate developmental autophagy in D. discoideum (Otto et al., 2003). The independent loss of atg5, atg6, atg7, atg8a, atg9 or atg101 leads to multi‐tipped aggregates (Otto et al., 2004; Calvo‐Garrido et al., 2010; Tung et al., 2010; Messling et al., 2017). Multi‐tipped aggregates are a characteristic phenotype of D. discoideum autophagy mutants, and this has been used to identify other autophagy‐regulating proteins in D. discoideum multiple tip mutations (e.g. TipC, TipD) (Muñoz‐Braceras, Calvo, & Escalante, 2015). The nature of this phenotype also suggests that autophagic machinery is involved in the differentiation of cell types (Mesquita et al., 2017).
As discussed below (see Section XIV), Ca2+/CaM signalling is critical in stalk cell differentiation. It involves a process that resembles autophagic cell death (ACD). Work has shown that the induction of ACD in D. discoideum requires the Atg1 complex (Kosta et al., 2004). Based on the putative CaMBDs of the Atg1 complex proteins and Atg9, we hypothesize that there is potential for CaM regulation of autophagy induction during ACD. In addition to this, ACD is regulated by the inositol 1,4,5‐trisphosphate receptor (encoded by iplA) that governs the efflux of Ca2+ from the endoplasmic reticulum (ER) (Lam et al., 2008). IplA has been studied extensively as a CaMBP: it is inhibited by the actions of CaM at high Ca2+ concentrations but not at low Ca2+ levels (Kasri et al., 2002). IplA also has a predicted CaMBD (1–10, 841VSKGRNYNGI850; 1–14, 850IRLVGQRITHKECL863) based on Calmodulin Target Database scanning and is likely regulated in a similar way (Yap et al., 2000; D.H. O'Day & S. Mathavarajah, unpublished data). This indicates that in addition to stalk cell differentiation, ACD may be regulated by CaM through its interactions with IplA. This idea is taken further in a discussion of IplA and CaM in spore germination (see Section XVI).
VIII. OSMOREGULATION AND VACUOLE FUNCTION
The CV system of D. discoideum and its relationship to those found in other organisms has been reviewed previously (Plattner, 2013). As a soil microbe, the osmoregulatory function of the D. discoideum CV system is critical for survival. Plattner (2013) summarized the importance of CaM in CV function. The CV system is bipartite consisting of a tubular spongiomal network that feeds into a bladder‐like vacuole from which a pore can form to release water from the distended vacuole (Nolta & Steck, 1994). Taking direction from previous work on the protozoan Paramecium, wherein CaM binds to the membranes of its well‐defined CV complex, Zhu & Clarke (1992) first showed that the less‐well‐defined CV system of D. discoideum also contained membrane‐bound CaM along with multiple, unidentified CaMBPs. A CaM‐binding unconventional myosin I isoform was one of these proteins. Subsequent work verified that MyoJ (a type V myosin) localizes with CaM in CV system bladders but it is likely that binding between the two is not essential for CaM localization or CV system function (Jung, Titus, & Hammer, 2009). It was also shown that CaM binds to a modifier of myosin II that localizes to the CV system, VwkA (a kinase that affects myosin II abundance and assembly). The autophosphorylation activity of VwkA is enhanced by CaM binding (Betapudi et al., 2005). Taken together, CaM appears to have a role in regulating the cytoskeletal changes that occur in response to osmotic stress.
Zhu & Clarke (1992) also offered unpublished support for the presence of an essential vacuolar H+‐ATPase (V‐ATPase), a discovery supported by other research (Liu & Clarke, 1996; Plattner, 2013). Despite their early work on the importance of CaM to CV system function, the CaM binding of the V‐ATPase was not assessed. V‐ATPase is a multi‐subunit mechanical protein consisting of two subcomplexes: a water‐soluble V1 shaft and a membrane‐embedded V0 channel (Tirtom et al., 2013). In D. discoideum, the 100 kDa V0 channel is encoded by the vatM gene. V‐ATPase is critical for the acidification of endosomal compartments and contributes to the membrane potential of the CV system (Clarke et al., 2002; Grønlien et al., 2002). This H + ‐ATPase was recently also shown to be localized to phagosomes, alluding to multiple functions within the cell (Clarke et al., 2002). A Calmodulin Target Database scan of V‐ATPase subunit M (VatM) revealed a non‐canonical Ca2+‐dependent CaMBD motif (291DHKRQTLAGIV301) and one Ca2+‐independent IQ‐like motif (357IQLALRTATTRSGA360) suggesting that VatM may bind to CaM in both the presence and absence of Ca2+ (D.H. O'Day, unpublished data). This interaction is supported by work using CaM antagonists (W‐7 and W‐5) that reduce the ATP‐dependent acidification of acidic compartments (Malchow, Lusche, & Schlatterer, 2004). The highly potent W‐7 [N‐(6‐aminohexyl)‐5‐chloro‐1‐naphthalenesulfonamide] significantly reduced acidification, suggesting that the normal function of V‐ATPase requires CaM binding. Independent of acidification, CaM interaction with an ortholog of VatM in Drosophila melanogaster regulates the assembly of soluble N‐ethylmaleimide‐sensitive factor attachment protein receptors (SNAREs), that have been shown to facilitate the membrane‐to‐membrane contact required for CV system discharge in Paramecium tetraurelia (Schönemann et al., 2013; Wang et al., 2014). This suggests that the interaction of CaM with V‐ATPase subunits influences osmoregulation in a multi‐faceted manner in terms of CV system acidification (or membrane potential) and SNARE‐mediated CV system discharge.
Over 24 different proteins have been found to be associated with the CV system in D. discoideum, a few only transiently (Plattner, 2013). Of these and those subsequently identified, only a limited number have been identified as CaMBPs. The BEACH (Beige and Chediak‐Higashi) domain‐containing protein LvsA, a critical protein for the maintenance of vacuole integrity, requires CaM for its binding to CV system membranes (Malchow et al., 2006). This is a reciprocal relationship between CaM and LvsA, as the loss of LvsA leads to the absence of CaM at CV system membranes (Gerald, Siano, & De Lozanne, 2002).
Cad1, a Ca2+‐dependent, homophilic cell‐to‐cell adhesion molecule that binds to and enters vacuoles, was identified and verified as a CaMBP by Sriskanthadevan et al. (2009). Produced as a soluble protein early in development, Cad1 is inserted into the plasma membrane through its association with the vacuoles of the CV system. Co‐immunoprecipitation analyses revealed that vacuole docking and uptake of Cad1 is Ca2+/CaM dependent and that both events are inhibited by CaM antagonists. Cad1 also has functions in cell sorting and morphogenesis as discussed below (see Section XIV).
The P‐type ATPase (PatA), a plasma membrane Ca2+‐ATPase‐type pump, is a CV system protein but was identified as lacking the conserved inhibitory CaMBD found in other species (Pittman, 2011). However, a binding site scan revealed one CaMBD in PatA (1–10, 1–14, 1064WQIVRQTHKKLVVINALKE1085) that contains multiple canonical binding motifs, a strong indicator of CaM binding. The importance of PatA and other Ca2+ pumps led to the discovery that the CV system is also critical for the regulation of Ca2+ levels during cAMP‐mediated chemotaxis, a subject that is discussed in Section IX (Malchow et al., 2006).
Du et al. (2008) proposed a disgorgin‐based model for the regulation of CV system function that included the sequential functions of dajumin, Ras‐related protein Rab‐1A (Rab11A), Ras‐related protein Rab‐1C (Rab11C), drainin, Ras‐related protein Rab‐8A (Rab‐8A), disgorgin and BEACH domain‐containing protein lvsA (LvsA), but not other critical proteins such as PatA or VatM. More importantly, despite its central role in CV system structure and function, CaM was not included in the model. Various Rab family proteins either bind CaM directly or via their activating proteins (e.g. Zhu et al., 2016). Sequence analysis of D. discoideum Rab11A and Rab11C revealed they both contain canonical Ca2+‐dependent CaMBDs (D.H. O'Day, unpublished data). Rab11A has one CaMBD (69TAGQERYRAITSAYYRGAVGALLV92) that possesses multiple binding motifs (1–8‐14, two 1–5‐10, 1–16, 1–14, 1–10) while Rab11C has a shorter CaMBD (68GQERFRAVTSGYYRGAVGAMI88) within which three motifs (1–10, 1–5‐10, 1–14) are evident. Drainin also contains a short Ca2+‐dependent CaMBD (380RTALSILRYFIS391) but it lacks any canonical motifs. Rab8A, on the other hand, reveals a putative CaMBD that lacks canonical motifs (71TAGQERFRTITTAYYRGAMGI91).
Na, Tunggal, & Eichinger (2007) examined the transcriptional changes during hypertonic stress in D. discoideum and identified 809 genes that were differentially expressed. Utilizing the Gene Ontology (GO) database, we identified an enrichment (binomial test) of genes encoding CaMBPs (GO: 0005516): rgaA, mhcA, myoA, cdk5, thyB, canA, and uncharacterized gene DDB_G0269864 (Fig. 2). Two of the seven genes are linked to the cytoskeleton, encoding the myosin IA heavy chain (myoA) and the myosin‐2 heavy chain (mhcA). Cytoskeletal changes alter cell shape during hypertonic stress (where cells shrink and become rounded) and strengthen the cell cortex (Kuwayama et al., 1996). Therefore, these two cytoskeletal CaMBPs are potentially regulated by CaM during the remodelling of the cytoskeleton during hypertonic stress.
Figure 2.

Calmodulin‐binding proteins are differentially regulated under hypertonic stress. A transcriptomic approach by Na et al. (2007) revealed genes that are differentially expressed at various time points (0–120 min) during hypertonic stress. Gene enrichment analysis of the 809 genes through the Gene Ontology (GO) database revealed an enrichment of calmodulin binding proteins (CaMBPs) (GO: 0005516). A binomial test was used to determine the enriched genes on the GO database. Seven of the identified genes encoded CaMBPs (listed to the right of the heat map); these are shown in the heat map with the associated log‐fold changes at various time points. A dendrogram (left of the heat map) describes the similarity in the transcriptional changes at various time points.
CanA and Cdk5 are two well‐studied CaMBPs in D. discoideum (Dammann et al., 1996; Huber & O'Day, 2011b, 2012b; Huber et al., 2013). They also show time‐dependent co‐expression during hypertonic stress (Fig. 2). These transcriptional changes imply roles for these CaMBPs in osmoregulation. There is experimental evidence for this function for CanA, as reductions in its expression (through RNA interference) causes D. discoideum cells to be sensitive to cation‐induced hypertonic stress (Thewes et al., 2016).
In response to hypertonic stress, D. discoideum cells secrete both cyclic GMP (cGMP) and H+ (Oyama, 1996; Oyama & Kubota, 1997). It is possible that this secretion is regulated by CanA and Cdk5. In human neurons, the combined activities of calcineurin and CDK5 regulate the exocytosis of neurotransmitters (Kim & Ryan, 2013). Interestingly, in D. discoideum, Cdk5 influences protein secretion (Sharma et al., 2002). By contrast, CanA is required for the secretion of Ca2+ via its regulation of PatA (Moniakis, Coukell, & Janiec, 1999). Therefore, the actions of these known and potential CaMBPs may be key in regulating exocytosis during the hypertonic stress response in D. discoideum.
Mercanti et al. (2006) reported on a D. discoideum protein called Rh50 (Rhesus‐like glycoprotein, encoded by the rhgA gene) that is targeted to the CV system by a sequence of acidic residues (DDEEE) in the C‐terminal of the protein. Immunolocalization studies revealed that Rh50 colocalized with CaM at the membranes of the CV system but was not involved in osmoregulation. Uniprot lists two D. discoideum Rhesus‐like transmembrane glycoproteins, A and B, each of which possesses a single CaMBD in the cytoplasmic tail of the protein. The putative CaMBD for RhgA (425VILLQLKKIKGLKSKEY442) has a 1–10 binding motif while the RhgB CaMBD (431ILKALKKVGGLKAKQYY447) has 1–10, 1–12 and 1–5‐10 motifs. If both the CaM binding of these resident transmembrane proteins and their proposed role in ammonium transport can be validated, this would greatly advance our understanding of CV system function in D. discoideum.
IX. CHEMOTAXIS AND AGGREGATION
D. discoideum cells can undergo random motility as well as chemotactic and chemokinetic movement directed either by folic acid (see Section III) or cAMP (developmental phase). There is also evidence for chemotaxis towards gradients of Ca2+ and arachidonic acid (Schaloske et al., 2007; Lusche et al., 2012). Understanding the genetic mechanisms that regulate CaM‐dependent chemotaxis in D. discoideum provides a unique opportunity to define the role of CaM across species due to the conserved chemotactic machinery shared with mammalian neutrophils and other white blood cell types. Granulocytes are a category of white blood cells characterized by the presence of granules in their cytoplasm. The CaM inhibitors trifluoperazine and W‐7 strongly inhibit chemotactic behaviour in rabbit peritoneal granulocytes (Elferlink, Deierkauf, & Riemersma, 1982). Regulation of serotonin‐induced human and mouse eosinophil chemotaxis may also require CaM. Using specific inhibitors, disruption of phosphatidylinositol 3‐kinase (PI3K), protein kinase C (PKC) and CaM, but not G(αi)‐proteins, prevents eosinophil rolling and actin polymerization suggesting that cell migration is dependent on these signalling molecules (Kang et al., 2013).
D. discoideum as a model for chemotaxis continues to allow us to identify conserved genes not previously known to be involved in directed cell motility, and to test mutant cell lines for altered CaM‐binding kinetics, expression and intracellular localization. When starved on non‐nutrient agar plates, D. discoideum cells aggregate by chemotaxis in response to outwardly relayed, pulsatile waves of cAMP that are released from aggregation centres (Konijn et al., 1967; Gerisch et al., 1975). Myre et al. (2011) showed that cells deficient for orthologous huntingtin protein (HTT) were defective in chemotaxis in a spatial gradient of cAMP. Although mutations in the huntingtin gene due to poly‐Q expansion cause Huntington's disease (HD) in humans, the normal function of HTT remains obscure. Sequencing of the D. discoideum genome (Eichinger et al., 2005) and many more invertebrate genomes presents an unprecedented opportunity to understand HTT function and its evolution. Suppression of HTT messenger RNA (mRNA) in Ciona intestinalis resulted in decreased motility; primary microglia from early postnatal HD mice are greatly impaired in their migration to chemotactic stimuli; compared to HTT(7Q/7Q) neuronal stem cells (NS), mutant Htt(140Q/140Q) and HTT knock‐out NS cells also showed significantly impaired motility (Kwan et al., 2012; Ritch et al., 2012; Idris, Thorndyke, & Brown, 2013). Interestingly, analysis of both human (3144 amino acids) and D. discoideum HTT (3095 amino acids) using the Calmodulin Target Database predicts a single CaM‐binding motif (1–8‐14) in each protein located toward the C‐terminal region of the protein (D. discoideum HTT, 1885LDLRKKQLLRLLSL1896; human HTT, 2538LKALDTRFGRKLSII2552). As discussed above (see Section VIII), CaM can be detected in the cytosol but is enriched on membranes of the CV system. D. discoideum cells that lack HTT under conditions of hypoosmotic stress die by rapid cell swelling followed by complete lysis within 3 h of exposure to water (Myre et al., 2011). Importantly, metabolite abnormalities in HD brains might result from defects in osmoregulation (Patassini et al., 2015). In addition, transient hyperosmotic treatment of human neuroblastoma SH‐SY5Y cell lines stably expressing mutant truncated huntingtin (Q88) significantly increases the number of cells with aggregates (Chun et al., 2002). Curiously, microarray analysis of D. discoideum cells where the orthologous huntingtin gene has been knocked out by homologous recombination (htt‐null) identified a fivefold increase (P < 0.003) in the expression of cmbB; a sixfold decrease (P < 0.009) in cmbC; a fourfold decrease (P < 0.004) in symA, a CaMBP transporter protein; and a 2.5‐fold increase (P < 0.05) in CaM during chemotaxis (M.A. Myre, F.J.J. Chain & J.F. Gusella, unpublished data). Moreover, in parental cells, the immunolocalization of CaM can be seen in the cytosol and on the membranes of the CV system. However, in htt‐null cells CaM cannot be detected on the CV system and is found solely in the cytosol (Fig. 3) (M.A. Myre, A.L. Lumsden & J.F. Gusella, unpublished data). Taken together, these data suggest that the interdependence of CaM and HTT needs to be studied in more detail not only in D. discoideum but also in mammalian cells with relevance to HD.
Figure 3.
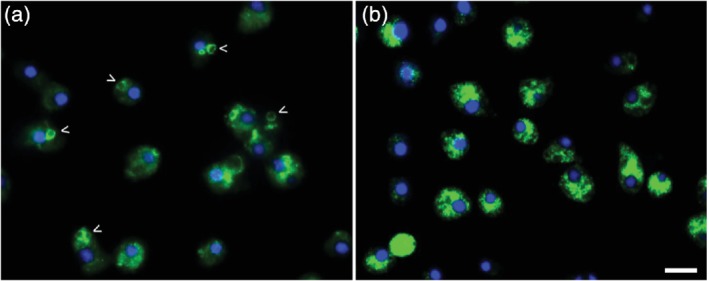
Mislocalization of calmodulin (CaM) in htt‐null cells. (A) Parental and (B) htt‐null cells were probed using mouse monoclonal anti‐CaM antibodies (clone IF11 Sigma) followed by goat anti‐mouse Alexa Fluor 488 secondary antibodies. Arrows denote CaM on the membranes of the CV system. CV system vacuoles are completely absent in mutant cells. DAPI (4′,6‐diamidino‐2‐phenylindole) was used to visualize nuclei (blue). Scale bar, 10 m. M.A. Myre, unpublished data).
Although the fine details of how divalent cations mediate the diversity of events underlying cell movement are still under analysis, Ca2+ signal transduction is essential for cAMP‐mediated chemotaxis in D. discoideum (e.g. Unterweger & Schlatterer, 1995; Schaloske et al., 2007). The role of CaM in chemotaxis was demonstrated by Gauthier & O'Day (2001) who also revealed a diversity of CaMBPs, a group of which were linked to either folic acid‐ or cAMP‐mediated chemotaxis. CaM‐dependent protein phosphorylation coincided with these chemotactic events (Gauthier & O'Day, 2001). At the time, the specific proteins that were phosphorylated and the CaMBPs involved remained to be identified. More critically, chemotaxis is a complex cellular mechanism and the exact components that were CaM‐dependent were unknown.
Other potential CaMBPs have been shown to be involved in chemotaxis but their ability to bind CaM remains to be studied. Probably the best example of this is the IQGAP family of proteins. IQGAP proteins have been linked to chemotaxis and other life cycle events (Catalano & O'Day, 2008). Named for the presence of IQ motifs, four members of the IQGAP family exist in D. discoideum: DdIQGAP1 (RgaA), DdIQGAP2 (GapA) and DdIQGAP3 (IqgC) (Shannon, 2012). From yeast to mammals, IQGAP proteins mediate multiple cellular processes including cell adhesion, cytokinesis, exocytosis and cell motility. Like its mammalian counterparts, D. discoideum IQGAP1 (RgaA) has two putative IQ motifs (114IQELKRNLVSEVRR127; 193LQTEPKYLAGLVYL206) as well as a Ca2+‐dependent 1–16 motif (267MIITYNKRKQGTDYLKAVIG286; Catalano & O'Day, 2008). GapA and IqgC are essential for cytokinesis. All three proteins have critical but overlapping functions in chemotaxis apparently acting through their association with F‐actin and cortexillins I and II. The CaM‐binding ability of the IQ motifs has not been verified. Coupled with the absence of F‐actin binding domains in the proteins, this leaves many questions unanswered about these potential CaMBPs.
X. CALCIUM‐MEDIATED CHEMOTAXIS
Scherer et al. (2010) revealed that steep gradients of Ca2+ could direct the positive chemotaxis of D. discoideum cells. The authors showed the gradients had to be extremely steep, contrasting with the actual levels of Ca2+ present during normal chemotaxis and cellular orientation in increasing spatial gradients of cAMP. In support of this, htt‐null cells failed to undergo both K+‐facilitated chemotaxis in spatial gradients of the major chemoattractant cAMP, and chemotaxis up a spatial gradient of Ca2+, but behaved normally in Ca2+‐facilitated cAMP‐mediated chemotaxis and Ca2+‐dependent flow‐directed motility (Wessels et al., 2014). While the significance of this mode of chemotaxis requires further study, it is possible that rather than being a true form of chemotaxis, non‐biological high levels of Ca2+ may simply activate the downstream Ca2+/CaM‐mediated pathways normally associated with cAMP‐mediated chemotaxis. Any differences between cAMP‐mediated and Ca2+‐mediated chemotaxis could be due to the extremely high levels of Ca2+ activating non‐canonical events or other factors. For example, the requirement for the plasma membrane transporter IplA in Ca2+‐mediated but not cAMP‐mediated chemotaxis could simply be due to the inability of Ca2+ gradients to be effective in iplA mutants, which would not allow the uptake of Ca2+ into cells. Regardless, the role of CaM, a primary Ca2+ target, in Ca2+‐mediated chemotaxis was not evaluated.
XI. BLEBBING MOTILITY
Under certain conditions, D. discoideum uses a blebbing‐based form of cell motility during chemotaxis (Langridge & Kay, 2006; Zatulovskiy et al., 2014). Blebs are small membrane protrusions whose formation has been shown to be dependent on Ca2+, CaM, the CaMBP ADP‐ribosylation factor 6 (ARF6) and phospholipase C in other species (Yanase et al., 2010). These signalling components regulate myosin II‐based contractility. The role of PI3K and myosin II in bleb formation was subsequently validated in D. discoideum (Zatulovskiy et al., 2014). However, they dismissed the role of Ca2+ because iplA − cells did not show altered blebbing. This conclusion may have been premature since previous research has shown that iplA − cells still undergo altered but functional Ca2+ signalling during chemotaxis (Schaloske et al., 2005; Lusche et al., 2012). Thus, it will be critical for future work to resolve this apparently contradictory conclusion.
XII. EXTRACELLULAR MATRIX AND SECRETED PROTEINS
In D. discoideum, CaM is detected extracellularly during all stages of the asexual life cycle where it is thought to function as a modulator of growth, cAMP‐mediated chemotaxis, and proteolysis through its interaction with specific CaMBPs (Suarez et al., 2011; O'Day et al., 2012). The functions of eCaM during growth were discussed above (see Section V). Bakthavatsalam & Gomer (2010) used mass spectrometry to reveal CaM, as well as several putative and established CaMBPs including Cad1, puromycin‐sensitive aminopeptidase B (PsaB), Cbp4a, and CmbC, in conditioned buffer harvested from parental AX2 cells undergoing multicellular development. However, none of these proteins was discussed or studied further (Bakthavatsalam & Gomer, 2010). Huber (2017) used a modified protocol to confirm the secretion of CaM during the early stages of multicellular development of AX3 cells, another parental strain commonly used in D. discoideum research. In addition to Cad1 and PsaB, the latter work also revealed the presence of the CaMBPs PsaA, PgkA and RgaA in conditioned starvation buffer. Together, these results validate the secretion of CaM and CaMBPs during D. discoideum multicellular development.
Once secreted, proteins can either diffuse away, bind to the cell surface to modulate cellular processes, or be incorporated into the ECM (Huber & O'Day, 2017). The cells that comprise the D. discoideum slug are surrounded by a slime sheath that functions as a primitive ECM in the organism. Huber & O'Day (2015) used mass spectrometry to reveal the proteins contained within the sheath ECM of two wild‐type strains of D. discoideum, NC4 and WS380B. Several putative and established CaMBPs were detected in the slime sheath of both strains including Cad1, Ca2+‐dependent cell adhesion protein 2 (Cad2), MhcA (myosin heavy chain A), Cbp4b, and Ca2+‐binding protein G (CbpG) (Huber & O'Day, 2015). In addition, RgaA, PsaB, and CmbC were detected in NC4 ECM, while PsaA, CbpA (CBP1), and Ca2+‐dependent cell‐to‐cell adhesion protein 3 (Cad3) were detected in WS380B ECM. While proteomics‐based analyses and western blotting support the secretion of CaM during multicellular development, and there is some evidence from immunolocalization experiments to support the presence of CaM in the ECM, mass spectroscopy did not detect CaM in the sheath ECM (Bakthavatsalam & Gomer, 2010; Suarez et al., 2011; O'Day et al., 2012; Huber & O'Day, 2015; Huber, 2017). These observations indicate that CaM might not reside long within the ECM, may be retained but undetectable due to its binding to CaMBPs, or may be present at levels that are too low for immunolocalization. They may also reflect the fact that the CaM secretion studies were performed using parental axenic strains of D. discoideum (AX2, AX3), while the sheath ECM proteomics study was performed using wild‐type strains isolated from nature (NC4, WS380B). Nonetheless, these results suggest that CaMBPs function in the ECM to regulate cellular events during the mid‐to‐late stages of D. discoideum development.
XIII. A MATRICELLULAR CAMBP THAT REGULATES CELL MOTILITY
The best characterized extracellular CaMBP in D. discoideum is CyrA, which was first isolated using the CaM‐binding overlay technique (CaMBOT; O'Day, 2003; O'Day & Huber, 2013). CyrA contains an identified CaMBD and binds to CaM in both the presence and absence of Ca2+ (Suarez et al., 2011; Huber, Suarez, & O'Day, 2012). Prior to being secreted, CyrA localizes to the ER, particularly its perinuclear component (Huber et al., 2012). Secretion of CyrA requires the release of intracellular Ca2+ as well as active CaM, PI3K and phospholipase A2 (PLA2). While CyrA is secreted throughout the D. discoideum life cycle, secretion is highest during the slug stage of multicellular development where it localizes to the slug sheath ECM (Suarez et al., 2011; Huber et al., 2012).
A body of work has established CyrA as the first matricellular protein identified in a eukaryotic microbe. Matricellular proteins localize to the ECM and contain binding sites for both the ECM and cell surface, however they do not directly contribute to the organization or physical properties of the ECM (Murphy‐Ullrich & Sage, 2014). Matricellular proteins are also expressed at high levels during development, associate with extracellular proteases and growth factors, and modulate cellular processes by binding to the cell surface and initiating intracellular signal transduction (Murphy‐Ullrich & Sage, 2014). As will be discussed below, CyrA has all these attributes, which allows it to be classified as a matricellular protein in D. discoideum.
During growth and development, full‐length CyrA (63 kDa) is cleaved into two major C‐terminal products of molecular weights 45 kDa and 40 kDa (Suarez et al., 2011; Huber et al., 2012). Cleavage of CyrA is developmentally regulated and modulated by CaM, which protects CyrA from proteolysis (Suarez et al., 2011). CyrA also contains four tandem EGF‐like repeats in its C‐terminus, which are present in both CyrA cleavage products. In mammalian matricellular proteins, EGF‐like repeats modulate cell motility by binding to the cell surface (Iyer et al., 2008). Interestingly, a synthetic peptide (DdEGFL1), equivalent in sequence to the first 18 amino acids of the first EGF‐like repeat of CyrA, functions extracellularly to increase both random cell motility and cAMP‐mediated chemotaxis in a variety of wild‐type and parental strains of D. discoideum (e.g. NC4, AX2, AX3, KAX3, DH1) (Huber & O'Day, 2009, 2011a, 2011b; Suarez et al., 2011; Nikolaeva, Huber, & O'Day, 2012; Huber & O'Day, 2012a). The role of CyrA in the regulation of cell motility was validated by showing that overexpression of the protein increases cAMP‐mediated chemotaxis (Huber et al., 2012). Unlike cAMP, DdEGFL1 does not function as a chemoattractant during the early stages of multicellular development (Nikolaeva et al., 2012). However, the peptide has been detected on the membranes of cells capped with concanavalin A suggesting that a receptor exists for EGF‐like repeats in D. discoideum (Huber et al., 2012). Furthermore, the dose‐dependent enhancement of motility by DdEGFL1 increases as cells are starved for longer periods of time suggesting that starvation increases the expression or presentation of a receptor that binds EGF‐like repeats (Suarez et al., 2011). Finally, treatment of cells with DdEGFL1 inhibits CyrA proteolysis suggesting that this domain plays a key role in regulating the cleavage of CyrA via a negative feedback mechanism (Suarez et al., 2011).
DdEGFL1 functions independently of the pathway regulating the response of cells to cAMP, since the peptide increases the movement of cells lacking the cAMP receptors CarA and CarC, as well as cells that lack the G‐proteins coupled to these receptors (Huber & O'Day, 2011a). Instead, the peptide activates a novel pathway involving Ca2+, CaM, RasG, PI3K, and PLA2, which leads to increased amounts of actin and myosin II heavy chain in the cytoskeleton (Huber & O'Day, 2009, 2011a, 2013). DdEGFL1 also sustains the threonine phosphorylation of the cytoskeletal protein vinculin B (VinB) during the early stages of D. discoideum development (Huber & O'Day, 2009, 2012a). In addition, the 45 kDa CyrA cleavage product has been detected in VinB immunoprecipitates, thus providing a link between the increased cell motility and a specific cytoskeletal component. Finally, the cytoskeletal proteins talin B and paxillin B, as well as tyrosine kinase activity mediated by protein kinase A (PKA) signalling, are also involved in translating the DdEGFL1 signal into an increase in cell motility (Huber & O'Day, 2012a). In total, these findings show that CyrA, and possibly other secreted CaMBPs, play an important role in regulating cell motility in D. discoideum.
XIV. CELL DIFFERENTIATION AND MORPHOGENESIS
In D. discoideum (see Section I), multicellular morphogenesis and differentiation generates a fruiting body consisting of a mass of viable spores supported by a stalk of dead cells. During the multicellular slug stage, the prestalk cells A, AB and O (pstA, pstB, pstO) mainly reside at the front of the slug and prespore cells at the rear (Fig. 4). A number of ALCs are also localized within this prespore region. The pstA, pstAB and pstO cells are defined by their unique patterns of expression of certain genes (e.g. ecmA, ecmB). The importance of Ca2+, ecmB and the regulatory role of differentiation inducing factor‐1 (DIF‐1) in stalk cell differentiation has been well documented. The endogenous morphogen DIF‐1 and specific pharmacological treatments induce ecmB expression and stalk cell differentiation in vitro through the elevation of intracellular Ca2+ levels (e.g. Williams et al., 1987; Kubohara & Okamoto, 1994; Kay, Flatman, & Thompson, 1999; Kubohara et al., 2007; Saito, Kato, & Kay, 2008). In vivo intracellular Ca2+ levels are significantly higher in prestalk versus prespore cells and the elevation of Ca2+ levels generates slugs with increased prestalk zones that produce stalky fruiting bodies that are deficient in spores (Abe & Maeda, 1989; Baskar et al., 2000). An early pharmacological study showed that CaM antagonism inhibited Ca2+‐induced ecmB expression in vitro (Verkerke‐van Wijk et al., 1998). Poloz & O'Day (2010) followed this up by showing that Ca2+ and CaM regulate ecmB expression as well as pstAB/pstB cell differentiation in normal slugs in vivo. Furthermore, Ca2+‐based pharmacological treatments had differential effects on ecmB expression and cell differentiation in the anterior versus posterior zones of slugs. For example, increased intracellular levels of Ca2+ increased the number of ecmB‐expressing cells in the anterior region of slugs, while decreasing intracellular Ca2+ levels or antagonizing CaM led to an increase in the number of ecmB‐expressing cells in the posterior.
Figure 4.

Calmodulin and calmodulin‐binding proteins linked to stalk cell and spore differentiation in Dictyostelium discoideum. The stalk cells and spores were stained with Calcofluor. ALC, anterior‐like cell; BAPTA, 1,2‐Bis(2‐aminophenoxy)ethane‐N,N,N′,N′‐tetraacetic acid tetrakis(acetoxymethyl ester); CaM, calmodulin; cAMP, cyclic AMP; DIF‐1, differentiation inducing factor‐1; EGTA, ethylene glycol‐bis(β‐aminoethyl ether)‐N,N,N′,N′‐tetraacetic acid; PsaA, puromycin‐sensitive aminopeptidase; W‐7, N‐(6‐aminohexyl)‐5‐chloro‐1‐naphthalenesulfonamide.
The central role of Ca2+/CaM signalling in stalk cell differentiation was further emphasized by results that came from an unexpected direction. For almost half a century, it has been known that colchicine induces developmental defects in Hydra, Drosophila melanogaster, Xenopus laevis and other species, typically altering the pattern and polarity of the whole organism or of specific structures (reviewed in O'Day & Durston, O'Day & Durston, 1978; Poloz & O'Day, 2012). Similarly, in D. discoideum, colchicine inhibits slug formation while inducing disorganized mounds of stalk cells and inhibiting spore formation (O'Day & Durston, O'Day & Durston, 1978). The developmental effects were identified by the altered pattern of neutral red staining, a vital‐dye prestalk marker in D. discoideum. Poloz & O'Day et al. (2012) revealed that, contrary to the expectations of previous researchers, the effects of colchicine were not due to the disruption of microtubules but rather were caused by an increase in intracellular Ca2+ levels that in turn activated CaM. They then showed in greater detail that colchicine affects cell motility, disrupts morphogenesis, inhibits spore cell differentiation and induces stalk cell differentiation through a Ca2+ and CaM‐dependent signal transduction pathway.
In keeping with this, colchicine‐treated cells could be rescued by lowering intracellular Ca2+ levels with BAPTA‐AM (1,2‐Bis(2‐aminophenoxy)ethane‐N,N,N′,N′‐tetraacetic acid tetrakis(acetoxymethyl ester)) or extracellular levels with EGTA (ethylene glycol‐bis(β‐aminoethyl ether)‐N,N,N′,N′‐tetraacetic acid) or by antagonizing CaM using W‐7 (Fig. 4). Rescued cells could complete development to form fruiting bodies with normal stalks and viable spores. Colchicine also induced the expression of stalk‐specific ecmB in both wild‐type cells and a DIF‐1 (dmtA −) mutant, revealing that this induction was DIF‐1 independent. In addition, colchicine inhibited the expression of spore‐specific genes (cotB, spiA), fitting with the absence of spores in colchicine‐treated cultures. Since stalk cell differentiation bears similarities to ACD, colchicine‐induced stalk cell formation could be useful for autophagy research in D. discoideum (Chen & Schapp, Chen & Schaap, 2012; Schaap, 2016).
While CaM primarily functions during stalk differentiation, there is an indirect relationship between certain CaMBPs and spore differentiation. As discussed above (see Section IV), the cell‐cycle‐linked nuclear/nucleolar CaMBP NumA1 binds to various proteins. D. discoideum PsaA is mainly a nuclear protein that associates with NumA1 in the nucleoplasm, but not the nucleolus, during interphase (Catalano et al., 2011). The enzyme is also detectable at low levels in the cytoplasm. Overexpression of full‐length PsaA inhibits spore differentiation leading to increased stalk cell formation. By contrast, deletion of a critical NLS resulted in cytoplasmic localization but, as predicted, no nuclear localization. This altered locational distribution resulted in the differentiation of spores and the absence of stalk cells (Fig. 4). By contrast, the inhibition of aminopeptidase activity with bestatin had a similar effect as treatment of slugs with colchicine, inducing cells to differentiate into stalk cells. Bestatin methyl ester (BME) is an inhibitor of Zn2+‐binding aminopeptidases (see Poloz, Catalano, & O'Day, 2012).
Cad1 displays differential localization, being predominantly a cell surface protein in prestalk cells but localizing internally in prespore cells (Sriskanthadevan et al., 2011). Cad1 is transported and inserted into the cell surface via the CV system. Experiments with chimeras of parental and knockout (cadA −) cells revealed that Cad1 fulfils a primary function in cell sorting. Although Cad1 is a proven CaMBP as discussed above (see Section VIII), the role of CaM in the Cad1‐mediated events discussed here was not investigated.
XV. PSEUDOPLASMODIUM REGENERATION
The regeneration of any multicellular eukaryotic structure involves some basic developmental events: wound healing, cell de‐differentiation (loss of cell‐type specific characteristics), re‐establishment of pattern and polarity, and cell re‐differentiation (specialization of cells and tissues) (Alibardi, 2017; Wells & Watt, 2018). The existence of stem cells can serve as a resupply unit of multipotent, pluripotent or totipotent cells that can supplement the population of de‐differentiated cells. While D. discoideum appears to lack 'stem cells', it does carry out the other regenerative events and processes: wound healing, de‐differentiation, and re‐differentiation with the re‐establishment of a normal pattern and polarity. That said, currently the regeneration of pseudoplasmodia is one of the least explored areas of research in D. discoideum.
Extensive early research into D. discoideum regeneration was designed to determine if the pattern and polarity of the multicellular slug was due to cell differentiation or sorting of prespore (rear) and prestalk (front) cells (Bonner, 1951). Along with accumulated evidence and based on cutting and grafting experiments by Francis & O'Day (Francis & O'Day, 1971), it was argued that cell differentiation/re‐differentiation as well as cell sorting were at play, thus fitting with models of animal development and regeneration, which extends to early metazoans like sponges. The results of these regeneration experiments fit with the argument that re‐differentiation must occur in D. discoideum, while the presence of various subpopulations of developing cell types indicates that at least some cell sorting occurs.
As shown in Fig. 4, the front of the slug is inhabited by future stalk cells while the rear is primarily comprised of prespore cells. If the tip is cut off and the slug is forced to culminate immediately, then a disproportional fruiting body is formed from each segment. The front will be stalky with a small spore mass, while the rear will have a thin stalk with an enlarged spore mass. However, letting the cut pieces migrate before fruiting allows the normal proportioning to be regenerated. These events coincide with cell‐specific gene expression patterns within each segment that are dependent on CaM function.
Ca2+/CaM regulate ecmB expression as well as pstAB/pstB cell differentiation in regenerating slugs (Poloz & O'Day, 2010). During regeneration, treatments that decrease intracellular Ca2+ or antagonize CaM also decrease ecmB expression and the differentiation of pstAB/pstB cells. The accumulated evidence suggests that CaM has no effect on slug migration but does function in the resultant morphology of regenerating slugs. CaM inhibition generates long thin slugs regenerated from the prespore end and short, wider front‐end slugs (Poloz & O'Day, 2010). However, the reasons for these morphological differences remain to be studied. CaM was also shown to function in other aspects of regeneration such as the rate of recovery of and cell motility in regenerating pseudoplasmodia. Understanding the biomolecular events and the role of CaM in D. discoideum regeneration could provide valuable evolutionary insights into this process.
XVI. SPORE GERMINATION
In D. discoideum, spore germination begins a new life cycle. In nature, the dormant spores become activated by the presence of an appropriate microbial food source. In the laboratory, they can be induced to germinate through the addition of an endogenous autoactivator or by the controlled application of heat (Cotter & Raper, 1966; Cotter et al., 1992). Spore germination consists of four stages: activation, lag, swelling and amoeba emergence. Three of these are active transition phases (activation, swelling, emergence) characterized by specific transcriptional events (Xu et al., 2004). CaM antagonists inhibit autoactivation but not heat‐induced activation, however both types of induced swelling and emergence are CaM dependent (Lydan & Cotter, 1994; Lydan, Cotter, & O'Day, 1994a, 1994b). While a diverse population of Ca2+‐dependent and ‐independent CaMBPs are present during both types of induced germination, a single unidentified Ca2+‐dependent CaMBP of 64 kDa (possibly CyrA) was linked specifically to autoactivated germination (Lydan et al., 1994a). In addition, emergence was characterized by the loss of a 52 kDa Ca2+‐independent CaMBP. Despite numerous changes in phosphoproteins specifically associated with either type of activation, they could not be linked to the activity of a CaM‐dependent kinase (Lydan et al., 1994b).
As discussed above, IplA is a CaMBP involved in blebbing motility (see Sections VII and XI). Interestingly, inositol 1,4,5‐trisphosphate, the activating substrate for IplA, also causes the autoactivation of spores (Lydan & Cotter, 1994), whereas inhibitors of Ca2+ efflux from intracellular stores inhibit autoactivation. Another phenotype observed in autophagy mutants was the lack of spores, suggesting defective physiological processes that control spore formation and dormancy (Otto et al., 2003). This may be due to a lack of autophagic vacuoles found in dormant spores and during the germination process (Cotter et al., 1992). Increased levels of intracellular Ca2+ promote the actions of the CaM‐dependent kinases (CaMKs) that are involved in the initiation of autophagy (Høyer‐Hansen et al., 2007; Pfisterer et al., 2011; Evankovich et al., 2012). These include CaMKs I, II and IV which act to promote autophagy and autophagosome formation in human cells (Pfisterer et al., 2011; Evankovich et al., 2012; Li et al., 2017). In addition, CaM‐dependent phosphorylation does occur in response to starvation in D. discoideum, which has orthologs of CaMKI and CaMKII (Gauthier & O'Day, 2001; Goldberg et al., 2006). Although CaM‐dependent phosphorylation overlaps with starvation in D. discoideum, more work in this area is required to discern its role in autophagy and spore germination.
A histidine kinase (DhkB) was shown to regulate spore germination but its ability to phosphorylate other proteins was not studied nor is there any evidence this kinase is activated by CaM in other species (Zinda & Singleton, 1998). Previous work performed a transcriptomics analysis of heat shock and dimethyl sulfoxide‐induced spore germination to monitor changes in global transcription patterns (Xu et al., 2004). A cursory analysis of their raw data reveals the differential expression of CaM and a number of verified and putative CaMBPs including CaMBP38, CmbB, CmbC and Myo1C (Fig. 5). In addition, CaMKI and a non‐specified Ca2+/CaM‐dependent protein kinase were also expressed. While CanA plays a role in spore differentiation as detailed above (see Sections III & VIII), its role in spore germination has not been studied nor was its presence detected in the transcriptional analysis (Horn & Gross, 1996; Xu et al., 2004). Clearly a detailed analysis of the data of Xu et al. (2004) in relation to the function of CaM during the different stages of germination could be enlightening. Despite an enthusiastic start into the role of CaM in spore germination, there has been no apparent subsequent research into this topic.
Figure 5.
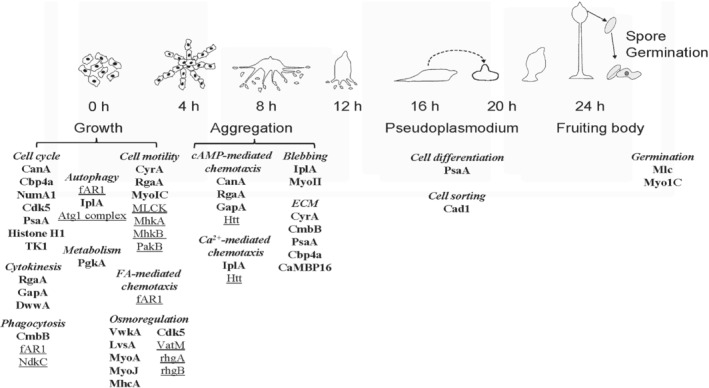
Calmodulin‐binding proteins (CaMBPs) linked to specific events during asexual development in Dictyostelium discoideum. Names in bold indicate experimentally proven CaMBPs; underlined names are putative CaMBPs containing calmodulin binding domains (CaMBDs). Atg1 complex, autophagy‐related 1 complex; Cad1, Ca2+‐dependent, cell‐to‐cell adhesion molecule 1; CaMBP16, calmodulin binding protein 16; cAMP, cyclic 3′,5′ adenosine monophosphate; CanA, calcineurin A; Cbp‐4a, calcium‐binding protein 4a; Cdk5, cyclin‐dependent kinase 5; CmbB, calmodulin‐binding protein 46; CyrA, cysteine‐rich protein A; DwwA, Dictyostelium WW domain‐containing protein A; ECM, extracellular matrix; FA, folic acid; fAR1, colic acid receptor 1; GapA, IQGAP family member DdIQGAP2; H1, Histone H1; Htt, huntingtin protein; IplA, inositol 1,4,5‐trisphosphate receptor, LvsA, BEACH domain‐containing protein; MhkA, myosin heavy chain kinase A; MhkB, myosin heavy chain kinase B; Mlc, myosin light chain; MLCK, myosin light chain kinase; MlhcA, myosin‐2 heavy chain; MyoIC, myosin‐1C; MyoII, myosin II; MyoA, myosin A; MyoJ, myosin J; NdkC, nucleotide diphosphate kinase C; NumA1, nucleomorphin A1; PakB, p21‐activated protein kinase; PAT1, Ca2+‐ATPase; PgkA, phosphoglycerate kinase; PsaA, puromycin‐sensitive aminopeptidase; RgaA, ras GTPase‐activating‐like protein A; rhgA, Rhesus‐like glycoprotein A; rhgB, Rhesus‐like glycoprotein B; TK1, thymidine kinase; VatM, V‐ATPase subunit M; VwkA, Von Willebrand factor kinase A.
XVII. CONCLUSIONS
CaM carries out critical functions for diverse events during the asexual development of D. discoideum (Fig. 1). Coupled with this, specific CaMBPs have been identified that are either linked with or critical to many of those events (Fig. 5). This makes D. discoideum one of the most comprehensively studied eukaryotes in terms of Ca2+ signalling and CaM function.
While not detailed here, putative and experimentally defined CaMBDs of multiple other CaMBPs have been identified (Catalano & O'Day, 2008). To date, the CaMBDs that have been identified primarily fall into canonical motif subclasses. Only a few have been shown to be non‐canonical motifs. Does this suggest some evolutionary distinction in CaM‐binding across species? Did canonical CaMBDs appear first during evolution?
A perplexing aspect of CaM function in D. discoideum is the lack of a large number of Ca2+/CaM‐dependent kinases. In mammals, it has long been known that there is a critical interplay between CanA dephosphorylation and Ca2+/CaM‐dependent kinase phosphorylation of specific proteins (e.g. Levitan, 1994; Penny & Gold, 2018). D. discoideum does have a well characterized CaM‐dependent CanA (Dammann et al., 1996). However, a CaM‐dependent phosphorylation–dephosphorylation interplay has not been identified in D. discoideum in spite of the presence of multiple CaMKs. The 21 CAMK group members consist of both potential Ca2+/CaM‐activated kinase or CAMK1 family members (eight genes) and CaM‐independent kinases (e.g. CAMK‐like RAD53) (Goldberg et al., 2006). An enzyme identified originally as D. discoideum myosin light chain kinase (MLCK) is in fact a CaMK1 family member. To date, only the alpha kinase VwkA has been proved to be activated by Ca2+/CaM which is confusing considering the evolutionary consistency of multiple Ca2+/CaM‐dependent kinases coupled with the central role of CaM in the life cycle of D. discoideum.
There is no doubt that CaM is one of the most perplexing of all regulatory proteins. Despite its central role in a multitude of basic biological processes, the way it binds to its Ca2+‐dependent target proteins is still a comparatively poorly understood process. While multiple pharmacological CaM antagonists exist, none target specific CaMBPs. What will be key in the future is to develop anti‐CaM agents that affect its binding to specific CaMBPs. Already new antagonists have been designed (e.g. Audran et al., 2013). CaM‐ and CaMBP‐specific peptides also are under development. For example, a peptide sequence generated from CaM disrupts its binding to huntingtin reducing its cytotoxicity in differentiated neuroblastoma cells (SH‐SY5Y) and providing neuroprotection in a mouse (R6/2) model (Dudek, Dai, & Muma, 2008, 2010).
The ease of working with D. discoideum and its genetic tractability suggests it could be on the forefront of biomedical research on CaM. For example, D. discoideum has become a powerful model for gathering new insights into proteins linked to Batten disease [the neuronal ceroid lipofuscinoses (NCLs); McLaren, Mathavarajah, & Huber, 2019]. The NCLs are a group of lysosomal storage disorders that are the most common form of neurodegeneration in children. Recent work showed that 11 of the 13 human NCL proteins contain putative CaMBDs (Mathavarajah, McLaren, & Huber, 2018a). The two Batten disease proteins that do not contain putative CaMBDs are potentially linked to CaM through their association with cathepsin L, which contains a putative CaMBD (Mathavarajah et al., 2018a). Since D. discoideum is an established NCL model, this presents an opportunity in the future to study the relationship between CaM and the NCL proteins. Together, these observations highlight the need to study CaM and its role in human disease.
CaM is central to the Ca2+ signalling events that mediate a diverse number of cellular and developmental events in this model eukaryote and, hopefully, future studies will take that one step further to find out how it performs its various jobs. While questions remain, the universal importance of CaM and its binding proteins in all aspects of the life cycle of D. discoideum is firmly established and worthy of more intense analysis.
A recent report has detailed the events involved in CaM binding providing insight into the promiscuous binding mechanism that defines this critical regulatory protein (Westerlund & Delemotte, 2018). In addition, Li et al. (2018) recently defined a short‐linear motif (SLM) feature for predicting CaMBDs. An extensive and detailed review of phospho‐calmodulin function in pathophysiological processes (Villalobo, 2018) has illuminated how post‐translational modifications can alter the function of this essential protein and how it binds to its CaMBPs.
XVIII. ACKNOWLEDGMENTS
This research was supported by various grants: (NSERC Research Grant, D.O.'D.); University of Massachusetts Lowell Grant ‐ L115389 (M.A.M.); National Institutes of Health ‐ Grant: 1R15GM134498‐01 (M.A.M); Natural Sciences and Engineering Research Council of Canada Discovery Grant (R.J.H.); Banting Research Foundation Discovery Award (R.J.H.); Canada Foundation for Innovation (R.J.H.). There are no conflicts of interest.
XIX. REFERENCES
- Abe, T. & Maeda, Y. (1989). The prestalk/prespore differentiation and polarized cell movement in Dictyostelium discoideum slugs. A possible involvement of the intracellular Ca2+‐concentration. Protoplasma 151, 175–178. [Google Scholar]
- Alibardi, L. (2017). Review limb regeneration in humans: dream or reality? Annals of Anatomy 217, 1–6. [DOI] [PubMed] [Google Scholar]
- Annesley, S. J. & Fisher, P. R. (2001). Dictyostelium discoideum – a model for many reasons. Molecular and Cellular Biochemistry 329, 73–91. [DOI] [PubMed] [Google Scholar]
- Annesley, S. J. , Bago, R. , Bosnar, M. H. , Filic, V. , Marinovic, M. , Weber, I. , Mehta, A. & Fisher, P. R. (2011). Dictyostelium discoideum nucleotide diphosphate kinase C plays a negative regulator role in phagocytosis, macropinocytosis and exocytosis. PLoS One 6, e26024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audran, E. , Dagher, R. , Gioria, S. , Tsvetkov, P. O. , Kulikova, A. A. , Didier, B. , Villa, P. , Makarov, A. A. , Kilhoffer, M.‐C. & Haiech, J. (2013). A general framework to characterize inhibitors of calmodulin: use of calmodulin inhibitors to study the interaction between calmodulin and its calmodulin binding domains. Biochimica et Biophysica Acta 1833, 1720–1731. [DOI] [PubMed] [Google Scholar]
- Bakthavatsalam, D. & Gomer, R. H. (2010). The secreted proteome profile of developing Dictyostelium discoideum cells. Proteomics 10, 2556–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskar, R. , Chhabra, P. , Mascarenhas, P. & Nanjundiah, V. (2000). A cell type‐specific effect of calcium on pattern formation and differentiation in Dictyostelium discoideum . International Journal of Developmental Biology 44, 491–498. [PubMed] [Google Scholar]
- Bazari, W. L. & Clarke, M. (1981). Characterization of a novel calmodulin from Dictyostelium discoideum . Journal of Biological Chemistry 256, 3598–3603. [PubMed] [Google Scholar]
- Berchtold, M. W. & Villalobo, A. (2014). The many faces of calmodulin in cell proliferation, programmed cell death, autophagy and cancer. Biochimica et Biophysica Acta 1843, 398–435. [DOI] [PubMed] [Google Scholar]
- Betapudi, V. , Mason, C. , Licate, L. & Egelhoff, T. T. (2005). Identification and characterization of a novel α‐kinase with a von Willebrand factor A‐like motif localized to the contractile vacuole and Golgi complex in Dictyostelium discoideum . Molecular Biology of the Cell 16, 2248–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettcher, B. & Barral, Y. (2013). The cell biology of open and closed mitosis. Nucleus 4, 160–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner, J. T. (1951). The pattern of differentiation in amoeboid slime moulds. American Naturalist 86, 79–89. [Google Scholar]
- Calvo‐Garrido, J. , Carilla‐Latorre, S. , Kubohara, Y. , Santos‐Rodrigo, N. , Mesquita, A. , Soldati, T. , Golstein, P. & Escalante, R. (2010). Autophagy in Dictyostelium: genes and pathways, cell death and infection. Autophagy 6, 686–701. [DOI] [PubMed] [Google Scholar]
- Catalano, A. & O'Day, D. H. (2008). Calmodulin‐binding proteins in the model organism Dictyostelium: a complete and critical review. Cellular Signalling 20, 277–291. [DOI] [PubMed] [Google Scholar]
- Catalano, A. & O'Day, D. H. (2013). Rad53 homologue forkhead‐associated kinase A (FhkA) and Ca2+‐binding protein 4a (CBP4a) are nucleolar proteins that differentially redistribute during mitosis in Dictyostelium . Cell Division 8, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalano, A. , Poloz, Y. & O'Day, D. H. (2011). Dictyostelium puromycin‐sensitive aminopeptidase A is a nucleoplasmic nucleomorphin‐binding protein that relocates to the cytoplasm during mitosis. Histochemistry and Cell Biology 136, 677–688. [DOI] [PubMed] [Google Scholar]
- Chen, Z. H. & Schaap, P. (2012). The prokaryote messenger c‐di‐GMP triggers stalk cell differentiation in Dictyostelium . Nature 488, 680–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, G. , Zhuchenko, O. & Kuspa, A. (2007). Immune‐like phagocyte activity in the social amoeba. Science 317, 678–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin, D. & Means, A. R. (2000). Calmodulin: a prototypical calcium sensor. Trends in Cell Biology 10, 3222–3328. [DOI] [PubMed] [Google Scholar]
- Chun, W. , Lesort, M. , Lee, M. & Johnson, G. V. (2002). Transient osmotic stress facilitates mutant huntingtin aggregation. Neuroreport 13(18), 2543–2546. [DOI] [PubMed] [Google Scholar]
- Clarke, M. , Köhler, J. , Arana, Q. , Liu, T. , Heuser, J. & Gerisch, G. (2002). Dynamics of the vacuolar H+‐ATPase in the contractile vacuole complex and the endosomal pathway of Dictyostelium cells. Journal of Cell Science 115, 2893–2905. [DOI] [PubMed] [Google Scholar]
- Collins, S. R. & Meyer, T. (2011). Evolutionary origins of STIM1 and STIM2 within ancient Ca2+ signaling systems. Trends in Cell Biology 21, 202–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter, D. A. & Raper, K. B. (1966). Spore germination in Dictyostelium discoideum . Proceedings of the National Academy of Sciences, USA 56, 880–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter, D. A. , Sands, T. W. , Virdy, K. J. , North, M. J. , Klein, G. & Satre, M. (1992). Patterning of development in Dictyostelium discoideum: factors regulating growth, differentiation, spore dormancy and germination. Biochemistry and Cell Biology 70, 892–919. [DOI] [PubMed] [Google Scholar]
- Dammann, H. , Hellstern, S. , Husain, Q. & Mutzel, R. (1996). Primary structure, expression and developmental regulation of a Dictyostelium calcineurin A homologue. European Journal of Biochemistry 238, 391–399. [DOI] [PubMed] [Google Scholar]
- De La Roche, M. A. , Lee, S. F. & Coté, G. P. (2003). The Dictyostelium class I myosin, myoD, contains a novel light chain that lacks high‐affinity calcium‐binding sites. Biochemical Journal 374, 697–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, F. , Edwards, K. , Shen, Z. , Sun, B. , De Lozanne, A. , Briggs, S. & Firtel, R. A. (2008). Regulation of contractile vacuole formation and activity in Dictyostelium . EMBO Journal 27, 2064–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudek, N. L. , Dai, Y. & Muma, N. A. (2008). Protective effects of interrupting the binding of calmodulin to mutant huntingtin. Journal of Neuropathology and Experimental Neurology 67, 355–365. [DOI] [PubMed] [Google Scholar]
- Dudek, N. L. , Dai, Y. & Muma, N. A. (2010). Neuroprotective effects of calmodulin peptide 76‐121aa: disruption of calmodulin binding to mutant huntingtin. Brain Pathology 20, 176–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn, J. D. , Bosmani, C. , Barisch, C. , Rykov, L. , Lefrancois, L. H. , Carenal‐Munoz, E. , Lopez‐Jimenez, A. T. & Soldati, T. (2018). Eat, prey, live: Dictyostelium discoideum as a model for cell‐autonomous diseases. Frontiers in Immunology 8, 1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichinger, L. , Pachebat, J. A. , Glöckner, G. , Rajandream, M. A. , Sucgang, R. , Berriman, M. , Song, J. , Olsen, R. , Szafranski, K. , Xu, Q. , Tunggal, B. , Kummerfeld, S. , Madera, M. , Konfortov, B. A. , Rivero, F. , Bankier, A. T. , Lehmann, R. , Hamlin, N. , Davies, R. , Gaudet, P. , Fey, P. , Pilcher, K. , Chen, G. , Saunders, D. , Sodergren, E. , Davis, P. , Kerhornou, A. , Nie, X. , Hall, N. , Anjard, C. , Hemphill, L. , Bason, N. , Farbrother, P. , Desany, B. , Just, E. , Morio, T. , Rost, R. , Churcher, C. , Cooper, J. , Haydock, S. , van Driessche, N. , Cronin, A. , Goodhead, I. , Muzny, D. , Mourier, T. , Pain, A. , Lu, M. , Harper, D. , Lindsay, R. , Hauser, H. , James, K. , Quiles, M. , Madan Babu, M. , Saito, T. , Buchrieser, C. , Wardroper, A. , Felder, M. , Thangavelu, M. , Johnson, D. , Knights, A. , Loulseged, H. , Mungall, K. , Oliver, K. , Price, C. , Quail, M. A. , Urushihara, H. , Hernandez, J. , Rabbinowitsch, E. , Steffen, D. , Sanders, M. , Ma, J. , Kohara, Y. , Sharp, S. , Simmonds, M. , Spiegler, S. , Tivey, A. , Sugano, S. , White, B. , Walker, D. , Woodward, J. , Winckler, T. , Tanaka, Y. , Shaulsky, G. , Schleicher, M. , Weinstock, G. , Rosenthal, A. , Cox, E. C. , Chisholm, R. L. , Gibbs, R. , Loomis, W. F. , Platzer, M. , Kay, R. R. , Williams, J. , Dear, P. H. , Noegel, A. A. , Barrell, B. & Kuspa, A. (2005). The genome of the social amoeba Dictyostelium discoideum . Nature 435, 43–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elferlink, J. G. R. , Deierkauf, M. & Riemersma, J. C. (1982). Involvement of calmodulin in granulocyte chemotaxis: the effect of calmodulin inhibitors. Research Communications in Chemical Pathology and Pharmacology 38, 77–84. [PubMed] [Google Scholar]
- Evankovich, J. , Zhang, R. , Cardinal, J. S. , Zhang, L. , Chen, J. , Huang, H. , Beer‐Stolz, D. , Billiar, T. R. , Rosengart, M. R. & Tsung, A. (2012). Calcium/calmodulin‐dependent protein kinase IV limits organ damage in hepatic ischemia‐reperfusion injury through induction of autophagy. American Journal of Physiology‐Gastrointestinal and Liver Physiology 303, 189–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Földvári‐Nagy, L. , Ari, E. , Csermely, P. , Korcsmáros, T. & Vellai, T. (2014). Starvation‐response may not involve Atg1‐dependent autophagy induction in non‐unikont parasites. Scientific Reports 4, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis, D. & O'Day, D. H. (1971). Sorting out in pseudoplasmodia of Dictyostelium discoideum . Journal of Experimental Zoology 176, 265–272. [DOI] [PubMed] [Google Scholar]
- Gauthier, M. L. & O'Day, D. H. (2001). Detection of calmodulin‐binding proteins and calmodulin‐dependent phosphorylation linked to calmodulin‐dependent chemotaxis to folic and cAMP in Dictyostelium . Cellular Signalling 13, 575–584. [DOI] [PubMed] [Google Scholar]
- Gerald, N. J. , Siano, M. & De Lozanne, A. (2002). The Dictyostelium LvsA protein is localized on the contractile vacuole and is required for osmoregulation. Traffic 3, 50–60. [DOI] [PubMed] [Google Scholar]
- Gerisch, G. , Hulser, D. , Malchow, D. & Wick, U. (1975). Cell communication by periodic cyclic‐AMP pulses. Philosophical Transactions of the Royal Society of London B. Biological Sciences 272, 181–192. [DOI] [PubMed] [Google Scholar]
- Goldberg, J. , Manning, G. , Liu, A. , Fey, P. , Pilcher, K. E. , Xu, Y. & Smith, J. L. (2006). The Dictyostelium kinome—analysis of the protein kinases from a simple model organism. PLoS Genetics 2, e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grønlien, H. K. , Stock, C. , Aihara, M. S. , Allen, R. D. & Naitoh, Y. (2002). Relationship between the membrane potential of the contractile vacuole complex and its osmoregulatory activity in Paramecium multimicronucleatum . Journal of Experimental Biology 205, 3261–3270. [DOI] [PubMed] [Google Scholar]
- Hoeflich, K. P. & Ikura, M. (2002). Calmodulin in action: diversity in target recognition and activation mechanisms. Cell 108, 739–742. [DOI] [PubMed] [Google Scholar]
- Horn, F. & Gross, J. (1996). A role for calcineurin in Dictyostelium discoideum development. Differentiation 60, 269–275. [DOI] [PubMed] [Google Scholar]
- Høyer‐Hansen, M. , Bastholm, L. , Szyniarowski, P. , Campanella, M. , Szabadkai, G. , Farkas, T. , Bianchi, K. , Fehrenbacher, N. , Elling, F. , Rizzuto, R. & Mathiasen, I. S. (2007). Control of macroautophagy by calcium, calmodulin‐dependent kinase kinase‐β, and Bcl‐2. Molecular Cell 25, 193–205. [DOI] [PubMed] [Google Scholar]
- Huber, R. J. (2016). Using the social amoeba Dictyostelium discoideum to study the functions of proteins linked to ceroid lipfuscinosis. Journal of Biomedical Science 23, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber, R. J. (2017). Loss of Cln3 impacts protein secretion in the social amoeba Dictyostelium . Cellular Signalling 35, 61–72. [DOI] [PubMed] [Google Scholar]
- Huber, R. J. & O'Day, D. H. (2009). An EGF‐like peptide sequence from Dictyostelium enhances cell motility and chemotaxis. Biochemical Biophysical Research Communications 379, 470–475. [DOI] [PubMed] [Google Scholar]
- Huber, R. J. & O'Day, D. H. (2011a). EGF‐like peptide‐enhanced cell motility in Dictyostelium functions independently of the cAMP‐mediated pathway and requires active Ca2+/calmodulin signalling. Cellular Signalling 23, 731–738. [DOI] [PubMed] [Google Scholar]
- Huber, R. J. & O'Day, D. H. (2011b). Nucleocytoplasmic transfer of cyclin dependent kinase 5 and its binding to puromycin‐sensitive aminopeptidase in Dictyostelium discoideum . Histochemistry and Cell Biology 136, 177–189. [DOI] [PubMed] [Google Scholar]
- Huber, R. J. & O'Day, D. H. (2012a). EGF‐like peptide‐enhanced cell movement in Dictyostelium is mediated by protein kinases and the activity of several cytoskeletal proteins. Cellular Signalling 24, 1770–1580. [DOI] [PubMed] [Google Scholar]
- Huber, R. J. & O'Day, D. H. (2012b). The cyclin‐dependent kinase inhibitor roscovitine inhibits cell proliferation, multicellular development, and Cdk5 nuclear translocation in Dictyostelium discoideum . Journal of Cellular Biochemistry 113, 868–876. [DOI] [PubMed] [Google Scholar]
- Huber, R. J. & O'Day, D. H. (2013). A matricellular protein and EGF‐like repeat signalling in the social amoebozoan Dictyostelium discoideum . Cellular and Molecular Life Sciences 69, 3989–3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber, R. J. & O'Day, D. H. (2015). Proteomic profiling of the extracellular matrix (slime sheath) of Dictyostelium discoideum . Proteomics 15, 3315–3319. [DOI] [PubMed] [Google Scholar]
- Huber, R. J. & O'Day, D. H. (2017). Extracellular matrix dynamics and functions in the social amoeba Dictyostelium: a critical review. Biochimica et Biophysica Acta 1861, 2971–2980. [DOI] [PubMed] [Google Scholar]
- Huber, R. J. , Suarez, A. & O'Day, D. H. (2012). CyrA, a matricellular protein that modulates cell motility in Dictyostelium discoideum . Matrix Biology 3, 271–280. [DOI] [PubMed] [Google Scholar]
- Huber, R. J. , Catalano, A. & O'Day, D. H. (2013). Cyclin‐dependent kinase 5 is a calmodulin‐binding protein that associates with puromycin‐sensitive aminopeptidase in the nucleus of Dictyostelium . Biochimica et Biophysica Acta 1833, 11–20. [DOI] [PubMed] [Google Scholar]
- Idris, M. M. , Thorndyke, M. C. & Brown, E. R. (2013). Evidence for dynamic and multiple roles for huntingtin in Ciona intestinalis . Invertebrate Neuroscience 13, 151–165. [DOI] [PubMed] [Google Scholar]
- Iyer, A. K. , Tran, K. T. , Griffith, L. & Wells, A. (2008). Cell surface restriction of EGFR by a tenascin cytotactin‐encoded EGF‐like repeat is preferential for motility‐related signaling. Journal of Cellular Physiology 214, 504–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, G. , Titus, M. A. & Hammer, J. A. (2009). The Dictyostelium type V myosin MyoJ is responsible for the cortical association and motility of contractile vacuole membranes. Journal of Cell Biology 186, 555–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakuta, S. , Yamaguchi, J. , Suzuki, C. , Sasaki, M. , Kazuno, S. & Uchiyama, Y. (2017). Small GTPase Rab1B is associated with ATG9A vesicles and regulates autophagosome formation. FASEB Journal 31, 3757–3773. [DOI] [PubMed] [Google Scholar]
- Kang, B. N. , Ha, S. G. , Bahaie, N. S. , Hosseinkhani, M. R. , Ge, X. N. , Blumenthal, M. N. , Rao, S. P. & Sriramarao, P. (2013). Regulation of serotonin‐induced trafficking and migration of eosinophils. PLoS One 8, e54840 10.1371/journal.pone.0054840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasri, N. N. , Bultynck, G. , Sienaert, I. , Callewaert, G. , Erneux, C. , Missiaen, L. , Parys, J. B. & De Smedt, H. (2002). The role of calmodulin for inositol 1, 4, 5‐trisphosphate receptor function. Biochimica et Biophysica Acta 1600, 19–31. [DOI] [PubMed] [Google Scholar]
- Kay, R. R. , Flatman, P. & Thompson, C. R. L. (1999). DIF signalling and cell fate. Seminars in Cell and Developmental Biology 10, 577–585. [DOI] [PubMed] [Google Scholar]
- Kim, S. H. & Ryan, T. A. (2013). Balance of calcineurin Aα and CDK5 activities sets release probability at nerve terminals. Journal of Neuroscience 33, 8937–8950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobel‐Höller, K. , Gley, K. , Jochinke, J. , Heider, K. , Fritsch, V. N. , Nguyen, H. V. D. , Lischke, T. , Radek, R. , Baugrass, R. , Mutzel, R. & Thewes, S. (2018). Calcineurin silencing in Dictyostelium discoideum leads to cellular alterations affecting mitochondria, gene expression, and oxidative stress response. Protist 169, 584–602. [DOI] [PubMed] [Google Scholar]
- Konijn, T. M. , van de Meene, J. G. C. , Bonner, J. T. & Barkly, S. (1967). The acrasin activity of adenosine‐3′,5′‐cyclic phosphate. Proceedings of the National Academy of Sciences of the USA 58, 1152–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosta, A. , Roisin‐Bouffay, C. , Luciani, M. F. , Otto, G. P. , Kessin, R. H. & Golstein, P. (2004). Autophagy gene disruption reveals a non‐vacuolar cell death pathway in Dictyostelium . Journal of Biological Chemistry 279, 48404–48409. [DOI] [PubMed] [Google Scholar]
- Kubohara, Y. & Okamoto, K. (1994). Cytoplasmic Ca2+ and H+ concentrations determine cell fate in Dictyostelium discoideum . FASEB Journal 8, 869–874. [DOI] [PubMed] [Google Scholar]
- Kubohara, Y. , Arai, A. , Gokan, N. & Hosaka, K. (2007). Pharmacological evidence that stalk cell differentiation involves increases in the intracellular Ca2+ and H+ concentrations in Dictyostelium discoideum . Development Growth and Differentiation 49, 253–264. [DOI] [PubMed] [Google Scholar]
- Kuwayama, H. , Ecke, M. , Gerisch, G. & Van Haastert, P. J. (1996). Protection against osmotic stress by cGMP‐mediated myosin phosphorylation. Science 271, 207–209. [DOI] [PubMed] [Google Scholar]
- Kwan, W. , Träger, U. , Davalos, D. , Chou, A. , Bouchard, J. , Andre, R. , Miller, A. , Weiss, A. , Giorgini, F. , Cheah, C. , Möller, T. , Stella, N. , Akassoglou, K. , Tabrizi, S. J. & Muchowski, P. J. (2012). Mutant huntingtin impairs immune cell migration in Huntington disease. Journal of Clinical Investigation 122, 4737–4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, D. , Kosta, A. , Luciani, M. F. & Golstein, P. (2008). The inositol 1, 4, 5‐trisphosphate receptor is required to signal autophagic cell death. Molecular Biology of the Cell 19, 691–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelaan, D. N. , Liburd, J. , Yang, Y. , Miller, E. , Chiayat, S. , Crawley, S. W. , Coté, G. P. & Smith, S. P. (2016). Structure of the single‐lobe myosin light chain C in complex with the light chain‐binding domains of myosin‐1C provides insights into the diverent IQ motif recognition. Journal of Biological Chemistry 291, 19606–19617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langridge, P. D. & Kay, R. R. (2006). Blebbing of Dictyostelium cells in response to chemoattractant. Experimental Cell Research 312, 2009–2017. [DOI] [PubMed] [Google Scholar]
- Levine, B. & Klionsky, D. J. (2017). Autophagy wins the 2016 Nobel Prize in physiology or medicine: breakthroughs in baker's yeast fuel advances in biomedical research. Proceedings of the National Academy of Sciences 114, 201–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitan, I. B. (1994). Modulation of ion channels by protein phosphorylation and dephosphorylation. Annual Review of Physiology 56, 193–212. [DOI] [PubMed] [Google Scholar]
- Li, X. , Wu, X. Q. , Deng, R. , Li, D. D. , Tang, J. , Chen, W. D. , Chen, J. H. , Ji, J. , Jiao, L. , Jiang, S. & Yang, F. (2017). CaMKII‐mediated Beclin 1 phosphorylation regulates autophagy that promotes degradation of Id and neuroblastoma cell differentiation. Nature Communications 8, 1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Maleki, M. , Carruthers, N. J. , Stemmer, P. M. , Gnom, A. & Rueda, L. (2018). The predictive performance of short‐linear motif features in the prediction of calmodulin‐binding proteins. BMC Bioinformatics 19(Suppl 14), 410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, T. & Clarke, M. (1996). The vacuolar proton pump of Dictyostelium discoideum: molecular cloning and analysis of the 100 kDa subunit. Journal of Cell Science 109, 1041–1051. [DOI] [PubMed] [Google Scholar]
- Luciani, M.‐F. , Giusti, C. , Marms, B. , Oshima, Y. , Kikuchi, H. , Kubohara, Y. & Golstein, P. (2011). Atg1 allows second‐signaled autophagic cell death in Dictyostelium . Autophagy 7, 501–508. [DOI] [PubMed] [Google Scholar]
- Lusche, D. F. , Wessels, D. , Scherer, A. , Daniels, K. , Kuhl, S. & Soll, D. R. (2012). The IplA Ca2+ channel of Dictyostelium discoideum is necessary for chemotaxis mediated through Ca2+, but not through cAMP, and has a fundamental role in natural aggregation. Journal of Cell Science 125, 1770–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydan, M. A. & Cotter, D. A. (1994). Spore swelling in Dictyostelium is a dynamic process mediated by calmodulin. FEMS Letters 115, 137–142. [Google Scholar]
- Lydan, M. A. , Cotter, D. A. & O'Day, D. H. (1994a). Calmodulin function and calmodulin‐binding proteins during autoactivation and spore germination in Dictyostelium discoideum . Cellular Signalling 6, 751–762. [DOI] [PubMed] [Google Scholar]
- Lydan, M. A. , Cotter, D. A. & O'Day, D. H. (1994b). Stage‐specific changes in protein phosphorylation during spore germination in Dictyostelium: role of calmodulin. Biochemical Biophysical Research Communications 201, 430–435. [DOI] [PubMed] [Google Scholar]
- Malchow, D. , Lusche, D. F. & Schlatterer, C. (2004). A link of Ca 2+ to cAMP oscillations in Dictyostelium: the calmodulin antagonist W‐7 potentiates cAMP relay and transiently inhibits the acidic Ca2+‐store. BMC Developmental Biology 4, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malchow, D. , Lusche, D. F. , Schlatterer, C. , De Lozanne, A. & Muller‐Taubenberger, A. (2006). The contractile vacuole in Ca2+ regulation in Dictyostelium: its essential function for cAMP induced Ca2+ influx. BMC Developmental Biology 6, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcassa, E. , Raimondi, M. , Anwar, T. , Eskelinen, E. L. , Myers, M. P. , Triolo, G. , Schneider, C. & Demarchi, F. (2017). Calpain mobilizes Atg9/Bif‐1 vesicles from Golgi stacks upon autophagy induction by thapsigargin. Biology Open 6, 551–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathavarajah, S. , McLaren, M. D. & Huber, R. J. (2018a). Cln3 function is linked to osmoregulation in a Dictyostelium model of Batten disease. Biochimica et Biophysica Acta 1864, 3559–3573. [DOI] [PubMed] [Google Scholar]
- Mathavarajah, S. , O'Day, D. H. & Huber, R. J. (2018b). Neuronal ceroid lipofuscinoses: connecting calcium signalling through calmodulin. Cell 7, E188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren, M. D. , Mathavarajah, S. & Huber, R. J. (2019). Recent insights into NCL protein function using the model organism Dictyostelium discoideum . Cell 8, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercanti, V. , Blanc, C. , Lefkir, Y. , Cosson, P. & Letourneur, F. (2006). Acidic clusters target transmembrane proteins to the contractile vacuole in Dictyostelium cells. Journal of Cell Science 119, 837–845. [DOI] [PubMed] [Google Scholar]
- Mesquita, A. , Tábara, L. C. , Martinez‐Costa, O. , Santos‐Rodrigo, N. , Vincent, O. & Escalante, R. (2015). Dissecting the function of Atg1 complex in Dictyostelium autophagy reveals a connection with the pentose phosphate pathway enzyme transketolase. Open Biology 5, 150088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesquita, A. , Cardenal‐Munoz, E. , Dominguez, E. , Muñoz‐Braceras, S. , Nunez‐Corcuera, B. , Phillips, B. A. , Tábara, L. C. , Xiong, Q. , Coria, R. , Eichinger, L. & Golstein, P. (2017). Autophagy in Dictyostelium: mechanisms, regulation and disease in a simple biomedical model. Autophagy 13, 24–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messling, S. , Matthias, J. , Xiong, Q. , Fischer, S. & Eichinger, L. (2017). The two Dictyostelium discoideum autophagy 8 proteins have distinct autophagic functions. European Journal of Cell Biology 96, 312–324. [DOI] [PubMed] [Google Scholar]
- Moniakis, J. , Coukell, M. B. & Janiec, A. (1999). Involvement of the Ca2+‐ATPase PAT1 and the contractile vacuole in calcium regulation in Dictyostelium discoideum. Journal of Cell Science 112, 405–414. [DOI] [PubMed] [Google Scholar]
- Muñoz‐Braceras, S. , Calvo, R. & Escalante, R. (2015). TipC and the chorea‐acanthocytosis protein VPS13A regulate autophagy in Dictyostelium and human HeLa cells. Autophagy 11, 918–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy‐Ullrich, J. E. & Sage, E. H. (2014). Revisiting the matricellular concept. Matrix Biology 37, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myre, M. A. & O'Day, D. H. (2002). Nucleomorphin: a novel, acidic, nuclear calmodulin‐binding protein from Dictyostelium that regulates nuclear number. Journal of Biological Chemistry 277, 19735–19744. [DOI] [PubMed] [Google Scholar]
- Myre, M. A. & O'Day, D. H. (2004a). Dictyostelium calcium‐binding protein 4a interacts with Nucleomorphin, a BRCT‐domain protein that regulates nuclear number. Biochemical Biophysical Research Communications 322, 665–671. [DOI] [PubMed] [Google Scholar]
- Myre, M. A. & O'Day, D. H. (2004b). Dictyostelium nucleomorphin is a member of the BRCT‐domain family of cell cycle checkpoint proteins. Biochimica et Biophysica Acta 1675, 192–197. [DOI] [PubMed] [Google Scholar]
- Myre, M. A. & O'Day, D. H. (2005). An N‐terminal nuclear localization sequence but not the calmodulin‐binding domain mediates nuclear localization of nucleomorphin, a protein that regulates nuclear number in Dictyostelium . Biochemical Biophysical Research Communications 332, 157–166. [DOI] [PubMed] [Google Scholar]
- Myre, M. A. , Lumsden, A. L. , Thompson, M. N. , Wasco, W. , MacDonald, M. E. & Gusella, J. F. (2011). Deficiency of huntingtin has pleiotropic effects in the social amoeba Dictyostelium discoideum . PLoS Genetics 7, e1002052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myre, M. A. , Huber, R. J. & O'Day, D. H. (2018). Functional analysis of proteins involved in neurodegeneration using the model organism Dictyostelium: Alzheimer's, Huntington's and Batten disease In Molecular‐Genetic and Statistical Techniques for Behavioral and Neural Research (eds Crusio W. E. and Gerlai R. T.), pp. 491–518. Elsevier, San Diego, CA. [Google Scholar]
- Na, J. , Tunggal, B. & Eichinger, L. (2007). STATc is a key regulator of the transcriptional response to hyperosmotic shock. BMC Genomics 8, 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaeva, I. , Huber, R. J. & O'Day, D. H. (2012). EGF‐like peptide of Dictyostelium discoideum is not a chemoattractant but it does restore folate‐mediated chemotaxis in the presence of signal transduction inhibitors. Peptides 34, 145–149. [DOI] [PubMed] [Google Scholar]
- Nolta, K. V. & Steck, T. L. (1994). Isolation and initial characterization of the bipartite contractile complex from Dictyostelium discoideum . Journal of Biological Chemistry 269, 2225–2233. [PubMed] [Google Scholar]
- O'Day, D. H. (2003). CaMBOT: profiling and characterizing calmodulin binding proteins. Cellular Signalling 15, 347–355. [DOI] [PubMed] [Google Scholar]
- O'Day, D. H. (2019). Proteins of the nucleolus of Dictyostelium discoideum, nucleolar compartmentalization, targeting sequences, protein translocations and binding partners. Cell 8, 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Day, D. H. & Budniak, A. (2015). Nucleocytoplasmic protein translocation during mitosis in the social amoebozoan Dictyostelium discoideum . Biological Reviews 90, 126–141. [DOI] [PubMed] [Google Scholar]
- O'Day, D. H. , Suhre, K. , Myre, M. A. , Chatterjee‐Chakraborty, M. & Chavez, S. E. (2006). Isolation, characterization and bioinformatic analysis of Dictyostelium protein cmbB reveal calmodulin binding properties and a novel tandem IP22 repeat with structural homology to Leucine Rich Repeat (LRR) proteins. Biochemical Biophysical Research Communications 346, 879–888. [DOI] [PubMed] [Google Scholar]
- O'Day, D. H. , Huber, R. J. & Suarez, A. (2012). Extracellular calmodulin regulates growth and cAMP‐mediated chemotaxis in Dictyostelium discoideum . Biochemical Biophysical Research Communications 425, 750–754. [DOI] [PubMed] [Google Scholar]
- O'Day, D. H. & Durston, A. J. (1978). Colchicine induces multiple axis formation and stalk cell differentiation in Dictyostelium discoideum . Journal of Embryology and Experimental Morphology 47, 195–206. [PubMed] [Google Scholar]
- Onikubo, T. , Nicklay, J. J. , Xing, L. , Warren, C. , Anson, B. , Wang, W.‐L. , Burgos, E. S. , Ruff, S. E. , Shabanowitz, J. , Cheng, R. H. , Hunt, D. F. & Schechter, D. (2015). Developmentally regulated post‐translational modification of nucleoplasmin controls histone sequestration and deposition. Cell Reports 10, 1735–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi, A. , Razi, M. , Dooley, H. C. , Robinson, D. , Weston, A. E. , Collinson, L. M. & Tooze, S. A. (2012). Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Molecular Biology of the Cell 23, 1860–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto, G. P. , Wu, M. Y. , Kazgan, N. , Anderson, O. R. & Kessin, R. H. (2003). Macroautophagy is required for multicellular development of the social amoeba Dictyostelium discoideum . Journal of Biological Chemistry 278, 17636–17645. [DOI] [PubMed] [Google Scholar]
- Otto, G. P. , Wu, M. Y. , Kazgan, N. , Anderson, O. R. & Kessin, R. H. (2004). Dictyostelium macroautophagy mutants vary in the severity of their developmental defects. Journal of Biological Chemistry 279, 15621–15629. [DOI] [PubMed] [Google Scholar]
- Oyama, M. (1996). cGMP accumulation induced by hypertonic stress in Dictyostelium discoideum . Journal of Biological Chemistry 271, 5574–5579. [PubMed] [Google Scholar]
- Oyama, M. & Kubota, K. (1997). H+ secretion induced by hypertonic stress in the cellular slime mold Dictystelium discoideum. The . Journal of Biochemistry 122(1), 64–70. [DOI] [PubMed] [Google Scholar]
- Pan, M. , Xu, X. , Chen, Y. & Jin, T. (2016). Identification of a chemoattractant G‐protein coupled receptor for folic acid that controls both chemotaxis and phagocytosis. Developmental Cell 26, 428–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patassini, S. , Begley, P. , Reid, S. J. , Xu, J. , Church, S. J. , Curtis, M. , Dragunow, M. , Waldvogel, H. J. , Unwin, R. D. , Snell, R. G. , Faull, R. L. & Cooper, G. J. (2015). Identification of elevated urea as a severe, ubiquitous metabolic defect in the brain of patients with Huntington's disease. Biochemical Biophysical Research Communications 468, 161–166. [DOI] [PubMed] [Google Scholar]
- Penny, C. J. & Gold, M. G. (2018). Mechanisms for localising calcineurin and CaMKII in dendritic spines. Cellular Signalling 49, 46–58. [DOI] [PubMed] [Google Scholar]
- Peterson, O. H. , Michalak, M. & Verkhratsky, A. (2015). Calcium signaling: past present and future. Cell Calcium 38, 161–169. [DOI] [PubMed] [Google Scholar]
- Pfisterer, S. G. , Mauthe, M. , Codogno, P. & Proikas‐Cezanne, T. (2011). Ca2+/Calmodulin‐dependent Kinase Signaling via CaMKI and AMPK contributes to the regulation of WIPI‐1 at the onset of autophagy. Molecular Pharmacology 80, 1066–1075. [DOI] [PubMed] [Google Scholar]
- Pittman, J. K. (2011). Vacuolar Ca2+ uptake. Cell Calcium 50, 139–146. [DOI] [PubMed] [Google Scholar]
- Plattner, H. (2013). Contractile vacuole complex—its expanding protein inventory. International Review of Cell and Molecular Biology 306, 371–416. [DOI] [PubMed] [Google Scholar]
- Plattner, H. & Verkhratsky, A. (2015). Evolution of calcium signaling. Cell Calcium 57, 121–122. [DOI] [PubMed] [Google Scholar]
- Poloz, Y. & O'Day, D. H. (2010). Ca+2 signaling regulates ecmB expression, cell differentiation and slug regeneration in Dictyostelium . Differentiation 84, 163–175. [DOI] [PubMed] [Google Scholar]
- Poloz, Y. & O'Day, D. H. (2012). Colchicine affects cell motility, pattern formation and stalk cell differentiation in Dictyostelium by altering calcium signalling. Differentiation 83, 185–190. [DOI] [PubMed] [Google Scholar]
- Poloz, Y. , Catalano, A. & O'Day, D. H. (2012). Bestatin inhibits cell growth, division and differentiation in Dictyostelium through inhibition of puromycin sensitive aminopeptidase. Eukaryotic Cell 11, 545–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritch, J. J. , Valencia, A. , Alexander, J. , Sapp, E. , Gatune, L. , Sangrey, G. R. , Sinha, S. , Scherber, C. M. , Zeitlin, S. , Sadri‐Vakili, G. , Irimia, D. , Difiglia, M. & Kegel, K. B. (2012). Multiple phenotypes in Huntington disease mouse neural stem cells. Molecular and Cell Neuroscience 50, 70–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosel, D. , Puta, F. , Blahuskova, A. , Smykal, P. & Folk, P. (2000). Molecular characterization of a calmodulin‐like Dictyostelium protein CalB. FEBS Letters 473, 323–327. [DOI] [PubMed] [Google Scholar]
- Saito, T. , Kato, A. & Kay, R. R. (2008). DIF‐1 induces the basal disc of the Dictyostelium fruiting body. Developmental Biology 317, 444–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaap, P. (2016). Secreted c‐di‐GMP induces stalk cell differentiation in the eukaryote Dictyostelium discoideum . Journal of Bacteriology 198, 27–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaloske, R. H. , Lusche, D. F. , Bezares‐Roder, K. , Happle, K. , Malchow, D. & Schlatterer, C. (2005). Ca2+ regulation in the absence of the iplA gene product in Dictyostelium discoideum . BMC Cell Biology 6, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaloske, R. H. , Blaesius, D. , Schlatterer, C. & Lusche, D. F. (2007). Arachidonic acid is a chemoattractant for Dictyostelium discoideum cells. Journal of Biosciences 32, 1281–1289. [DOI] [PubMed] [Google Scholar]
- Scherer, A. , Kuhl, S. , Wessels, D. , Lusche, D. F. , Raisley, B. & Soll, D. R. (2010). Ca2+ chemotaxis in Dictyostelium discoideum . Journal of Cell Science 123, 2756–3767. [DOI] [PubMed] [Google Scholar]
- Schlatterer, C. & Malchow, D. (1993). Intracellular Gunaosine‐5′‐O‐(3‐thiotriphosphate) blocks chemotactic motility of Dictyostelium discoideum amoebae. Cell Motility and Cytoskeleton 25, 298–307. [DOI] [PubMed] [Google Scholar]
- Schönemann, B. , Bledowski, A. , Sehring, I. M. & Plattner, H. (2013). A set of SNARE proteins in the contractile vacuole complex of Paramecium regulates cellular calcium tolerance and also contributes to organelle biogenesis. Cell Calcium 53, 204–216. [DOI] [PubMed] [Google Scholar]
- Shannon, K. B. (2012). IQGAP family members in yeast, Dictyostelium and mammalian cells. International Journal of Cell Biology 2012, 894817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, S. K. , Brock, D. A. , Ammann, R. R. , DeShazo, T. , Khosla, M. , Gomer, R. H. & Weeks, G. (2002). The Cdk5 homolog, Crp, regulates endocytosis and secretion in Dictyostelium and is necessary for both growth and differentiation. Developmental Biology 247, 1–10. [DOI] [PubMed] [Google Scholar]
- Sillo, A. , Bloomfield, G. , Balest, A. , Balbo, A. , Pergolizzi, B. , Peracino, B. , Skelton, J. , Ivens, A. & Bozzaro, S. (2008). Genome‐wide transcriptional changes induced by phagocytosis of growth on bacteria in Dictyostelium . BMC Genomics 9, 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriskanthadevan, S. , Brar, S. K. , Manoharan, K. & Siu, C. H. (2009). Ca2+‐calmodulin interacts with DdCAD‐1 and promotes DdCAD‐1 transport by contractile vacuoles in Dictyostelium cells. FEBS Journal 280, 1795–1806. [DOI] [PubMed] [Google Scholar]
- Sriskanthadevan, S. , Zhu, Y. , Manoharan, K. , Yang, C. & Siu, C. H. (2011). The cell adhesion molecule DdCAD‐1 regulates morphogenesis through differential spatiotemporal expression in Dictyostelium discoideum . Development 138, 2487–2497. [DOI] [PubMed] [Google Scholar]
- Suarez, A. , Huber, R. , Myre, M. & O'Day, D. H. (2011). An extracellular matrix, calmodulin‐binding protein with EGF‐like repeats that enhance cell motility. Cellular Signalling 23, 1197–1206. [DOI] [PubMed] [Google Scholar]
- Swer, P. B. , Lohia, R. & Saran, S. (2014). Analysis of rapamycin induced autophagy in Dictyostelium discoideum . Indian Journal of Experimental Biology 52, 295–304. [PubMed] [Google Scholar]
- Tanaka, Y. , Itakura, R. , Amagai, A. & Maeda, Y. (1998). The signals for starvation response are transduced through elevated [Ca2+] in Dictyostelium cells. Experimental Cell Research 240, 340–348. [DOI] [PubMed] [Google Scholar]
- Tatischeff, I. (2019). Dictyostelium: a model for studying the extracellular vesicle messengers involved in human health and disease. Cell 8, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekinay, T. , Wu, M. Y. , Otto, G. P. , Anderson, O. R. & Kessin, R. H. (2006). Function of the Dictyostelium discoideum Atg1 kinase during autophagy and development. Eukaryotic Cell 5, 1797–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thewes, S. , Schubert, S. K. , Park, K. & Mutzel, R. (2016). Stress and development in Dictyostelium discoideum: the involvement of the catalytic calcineurin subunit. Journal of Basic Microbiology 54, 606–612. [DOI] [PubMed] [Google Scholar]
- Tirtom, N. E. , Okuno, D. , Nakano, M. , Yokoyama, K. & Noji, H. (2013). Mechanical modulation of ATP‐binding affinity of V1‐ATPase. Journal of Biological Chemistry 288, 619–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung, S. M. , Ünal, C. , Ley, A. , Peña, C. , Tunggal, B. , Noegel, A. A. , Krut, O. , Steinert, M. & Eichinger, L. (2010). Loss of Dictyostelium ATG9 results in a pleiotropic phenotype affecting growth, development, phagocytosis and clearance and replication of Legionella pneumophila . Cellular Microbiology 12, 765–780. [DOI] [PubMed] [Google Scholar]
- Unterweger, N. & Schlatterer, C. (1995). Introduction of calcium buffers into the cytosol of Dictyostelium discoideum amoebae alters cell morphology and inhibits chemotaxis. Cell Calcium 17, 97–110. [DOI] [PubMed] [Google Scholar]
- Urrutia, J. , Aguado, A. , Muguruza‐Montero, A. , Nunez, E. , Malo, C. , Casis, O. & Villarroel, A. (2019). The crossroad of ion channels and calmodulin in disease. International Journal of Molecular Science 20, 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Driel, R. (1981). Binding of the chemoattractant folic acid by Dictyostelium cells. European Journal of Biochemistry 115, 391–395. [DOI] [PubMed] [Google Scholar]
- Van Driessche, N. , Shaw, C. , Katoh, M. , Morio, T. , Scugang, R. , Ibarra, M. , Kuyayama, H. , Saito, T. , Urushihara, H. , Maeda, M. , Takeuchi, I. , Ochiai, H. , Eaton, W. , Tollet, J. , Halter, J. , Kuspa, A. , Tanaka, Y. & Shaulsky, G. (2002). A transcriptional profile of multicellular development in Dictyostelium discoideum . Development 129, 1543–1552. [DOI] [PubMed] [Google Scholar]
- Verkerke‐van Wijk, I. , Brandt, R. , Bosman, L. & Schaap, P. (1998). Two distinct signaling pathways mediate DIF induction of prestalk gene expression in Dictyostelium . Experimental Cell Research 245, 179–185. [DOI] [PubMed] [Google Scholar]
- Villalobo, A. (2018). The multifunctional role of phospho‐calmodulin in pathophysiological processes. Biochemical Journal 475, 4011–4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, K. K. , Villalobo, A. & Roufogalis, B. D. (1989). Calmodulin‐binding proteins as calpain substrates. Biochemical Journal 262, 693–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D. , Epstein, D. , Khalaf, O. , Srinivasan, S. , Williamson, W. R. , Fayyazuddin, A. , Quiocho, F. & Hiesinger, P. R. (2014). Ca2+–Calmodulin regulates SNARE assembly and spontaneous neurotransmitter release via v‐ATPase subunit V0a1. Journal of Cell Biology 205, 21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb, S. E. & Miller, A. (2003). Calcium: calcium signaling during embryonic development. Nature Reviews Molecular Cell Biology 4, 539–551. [DOI] [PubMed] [Google Scholar]
- Wells, J. M. & Watt, F. M. (2018). Diverse mechanisms for endogenous regeneration and repair in mammalian organs. Nature 557, 322–328. [DOI] [PubMed] [Google Scholar]
- Wessels, D. , Lusche, D. F. , Scherer, A. , Kuhl, S. , Myre, M. A. & Soll, D. R. (2014). Huntingtin regulates Ca(2+) chemotaxis and K(+)‐facilitated cAMP chemotaxis, in conjunction with the monovalent cation/H(+) exchanger Nhe1, in a model developmental system: insights into its possible role in Huntington's disease. Developmental Biology 394, 24–38. [DOI] [PubMed] [Google Scholar]
- Westerlund, A. M. & Delemotte, L. (2018). Effect of Ca2+ on the promiscuous target protein binding of calmodulin. PLoS Computational Biology 14, e10067072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, J. G. , Ceccarelli, A. , McRobbie, S. , Mahbubani, H. , Kay, R. R. , Early, A. , Berks, M. & Jermyn, K. A. (1987). Direct induction of Dictyostelium prestalk gene expression by DIF provides evidence that DIF is a morphogen. Cell 49, 185–192. [DOI] [PubMed] [Google Scholar]
- Xu, O. , Ibarra, M. , Mahdeo, C. , Sha, D. , Huang, E. , Kuspa, A. , Cotter, D. & Shaulsky, G. (2004). Transcriptional transitions during Dictyostelium spore germination. Eukaryotic Cell 3, 1101–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, H. , Kakuta, S. , Watanabe, T. M. , Kitamura, A. , Sekito, T. , Kondo‐Kakuta, C. , Ichikawa, R. , Kinjo, M. & Ohsumi, Y. (2012). Atg9 vesicles are an important membrane source during early steps of autophagosome formation. Journal of Cell Biology 198, 219–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanase, Y. , Carvou, N. , Frohman, M. A. & Cockcroft, S. (2010). Reversible bleb formation in mast cells stimulated with antigen is Ca2+/calmodulin‐dependent and bleb size is regulated by ARF6. Biochemical Journal 425, 179–193. [DOI] [PubMed] [Google Scholar]
- Yap, K. L. , Kim, J. , Truong, K. , Sherman, M. , Yuan, T. & Ikura, M. (2000). Calmodulin target database. Journal of Structural and Functional Genomics 1, 8–14. [DOI] [PubMed] [Google Scholar]
- Zatulovskiy, E. , Tyson, R. , Breschneider, T. & Kay, R. R. (2014). Bleb‐driven chemotaxis of Dictyostelium cells. Journal of Cell Biology 204, 1027–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y. , Lewis, T. L. , Robinson, L. J. , Brundage, K. M. , Schafer, R. , Martin, K. H. , Blair, H. C. , Soboloff, J. & Barnett, J. B. (2011). The role of calcium release activated calcium channels in osteoclast differentiation. Journal of Cellular Physiology 226, 1082–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Q. & Clarke, M. (1992). Association of calmodulin and an unconventional myosin with the contractile vacuole complex of Dictyostelium discoideum . Journal of Cell Biology 118, 347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, S. , Chim, S. M. , Cheng, T. , Ang, E. , Ng, B. , Lim, B. , Chen, K. , Qiu, H. , Tickner, J. , Xu, H. , Pavlos, N. & Xu, J. (2016). Calmodulin interacts with Rab3D and modulates osteoclastic bone resorption. Nature Scientific Reports 6, 37963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinda, M. J. & Singleton, C. K. (1998). The hybrid histidine kinase dhkB regulates spore germination in Dictyostelium discoideum . Developmental Biology 196, 171–183. [DOI] [PubMed] [Google Scholar]


