Abstract
The generation of the B cell response upon vaccination is characterized by the induction of different functional and phenotypic subpopulations and is strongly dependent on the vaccine formulation, including the adjuvant used. Here, we have profiled the different B cell subsets elicited upon vaccination, using machine learning methods for interpreting high‐dimensional flow cytometry data sets. The B cell response elicited by an adjuvanted vaccine formulation, compared to the antigen alone, was characterized using two automated methods based on clustering (FlowSOM) and dimensional reduction (t‐SNE) approaches. The clustering method identified, based on multiple marker expression, different B cell populations, including plasmablasts, plasma cells, germinal center B cells and their subsets, while this profiling was more difficult with t‐SNE analysis. When undefined phenotypes were detected, their characterization could be improved by integrating the t‐SNE spatial visualization of cells with the FlowSOM clusters. The frequency of some cellular subsets, in particular plasma cells, was significantly higher in lymph nodes of mice primed with the adjuvanted formulation compared to antigen alone. Thanks to this automatic data analysis it was possible to identify, in an unbiased way, different B cell populations and also intermediate stages of cell differentiation elicited by immunization, thus providing a signature of B cell recall response that can be hardly obtained with the classical bidimensional gating analysis. © 2019 The Authors. Cytometry Part A published by Wiley Periodicals, Inc. on behalf of International Society for Advancement of Cytometry.
Keywords: machine learning methods, B cells, multiparametric flow cytometry, vaccination, adjuvants, computational data analysis, dimensionality reduction, clustering, bioinformatics
Born more than 50 years ago, as recently celebrated 1, flow cytometry is still one of the leader technologies in immunology and cell biology. Multiple parameters of cells mixed in heterogeneous samples can be quickly and simultaneously detected during their flow in a stream through photonic detectors. The progress of the technology has led to the development of instruments capable of measuring more than 30 parameters on large number of cells, promoting the necessity of developing advanced mathematical approaches for their analysis. Flow cytometric analysis of cell subsets has traditionally been performed with “manual gating” based on the measurement of two parameters visualized on bidimensional plots. This approach is still one of the most used by flow cytometrists and allows the detection of multiple populations among mixed cell samples but is inevitably biased by the operator choices and limited in the discovery of yet undefined populations. Indeed, when many parameters are investigated, is not feasible to visualize all the possible bidimensional combinations of marker expression, and only a subjective gating strategy can be followed. Moreover, the coexpression of more than two markers on the surface of the same cells can be obtained only by the Boolean approach, but the graphical output is not easy and the number of all possible combinations exponentially increases with the increase of parameters. High‐throughput flow cytometry leads to the paradox that we routinely generate more data than the amount that we are able to fully analyze and interpret, thus losing many of the acquired information. This leads to the need of novel bioinformatics tools capable of clustering cells on the base of their simultaneous marker expression in an unbiased way 2.
Flow cytometric data analysis includes data preprocessing, data exploration, visualization of results, and statistical tests. The two most used approaches to explore and visualize such kind of data are dimensionality reduction and unsupervised clustering. The first one allows to display high‐dimensional data in a lower‐dimensional space, using two or three surrogate dimensions where each cell is represented as a dot. Frequently used tools in flow cytometry are based on t‐distributed stochastic neighbor embedding algorithm (t‐SNE) 3, such as vi‐SNE 4, ACCENSE 5, or Rtsne (the version available as R package), which aims to find a lower‐dimensional projection that strongly preserves the similarity in the original, high‐dimensional space 6. t‐SNE method has been shown to work very well with flow cytometric data, enabling to dissect different cell types within heterogeneous samples, and to compare similarities between different samples 4.
Algorithms based on an unsupervised clustering approach stratify cells with similar marker profiles in clusters, which can subsequently be interpreted as cell populations. These clustering packages include tools such as FlowMeans 7, flowClust 8, and FlowSOM 9. FlowSOM is considered one of the best high‐performance algorithms in automated identification of cell subsets showing an extremely fast runtime 10. It has also been used for characterizing both the cell phenotype and the cellular functionality, such as the simultaneous production of intracellular cytokine and degranulation 11, 12, 13, 14. The FlowSOM algorithm is based on a self‐organizing map (SOM), where similar cells are assigned to the same node. In order to reduce the final number of clusters, the nodes of the SOM are usually grouped into metaclusters with a hierarchical clustering algorithm, as the one implemented in ConsensusClusterPlus package 15. The metaclustering process groups together nodes with similar markers expression, thus increasing performance in population identification 10.
Deep analysis of both T and B responses after vaccination can be obtained with multiparametric flow cytometry, measuring the frequency, the phenotype, and the functional features of antigen‐specific cells 16, 17, 18, 19, 20, 21, 22. The dissection of the mechanisms of immune memory generation and reactivation upon antigen/pathogen encounter is fundamental for understanding the processes that drive protective immune responses 23, 24, 25. Nevertheless, the spatial and temporal profiling of B cell response dynamics is complex as different organs are involved in the course of the immune response. Moreover, activation of B cells leads to the generation of subtypes of cells with different functions, such as plasmablasts, short‐or long‐lived plasma cells (PCs), memory cells 26. The presence of the adjuvant, especially in the primary immunization, allows to enhance and modulate the immune responses to vaccination 13, 27, 28 and ensures the induction of long‐lived immunological memory necessary for protection 29.
Here we performed an automated analysis of multiparametric flow cytometric data using two machine learning methods, one based on clustering (FlowSOM) and the other on dimensional reduction (t‐SNE), to automatically profile all the possible B cell subsets elicited by vaccination. The automated methods used allow to characterize the B cell subtypes in un unbiased way, defining different phenotypes on the basis of the simultaneous expression of multiple surface markers. The chimeric antigen H56, a promising vaccine candidate against Mycobacterium tuberculosis 30 was used as model vaccine antigen, and administered by the parenteral route alone or combined with the CAF01 adjuvant, a liposome system capable to elicit both humoral and cellular responses 18, 31. Boosting was performed with a lower dose of antigen alone, according to an immunization schedule already tested 13. The analysis was aimed to identify, in an automated and unbiased way, different B cell subtypes elicited by booster immunization and determine their frequency among mice primed with or without the adjuvant component.
Materials and Methods
Mice
Seven‐week‐old female C57BL/6 mice, purchased from Charles River (Lecco, Italy) were housed under specific pathogen‐free conditions in the animal facility of the Laboratory of Molecular Microbiology and Biotechnology (LA.M.M.B.), Department of Medical Biotechnologies at University of Siena. This study was carried out in accordance with national guidelines (Decreto Legislativo 26/2014). The protocol was approved by the Italian Ministry of Health (authorization n° 1004/2015‐PR, 22 September 2015).
Immunizations and Cell Preparation
Groups of eight mice were immunized by the subcutaneous route at the base of the tail, with the chimeric tuberculosis vaccine antigen H56 (2 μg/mouse; Statens Serum Institut, Denmark), alone or combined with the adjuvant CAF01 (250 μg dimethyldioctadecylammonium [DDA] and 50 μg trehalosedibehenate [TDB]/mouse; Statens Serum Institut, Denmark), and boosted with a lower dose of H56 antigen (0.5 μg/mouse). Draining lymph nodes (sub iliac, medial, and external) were collected five days after booster immunization. Samples were mashed onto 70 μm nylon screens (Sefar Italia, Italy) and washed two times in RPMI medium (Lonza, Belgium) supplemented with 100 U/ml penicillin/streptomycin and 10% fetal bovine serum (GIBCO, USA).
Flow Cytometric Staining
Samples were incubated for 30 min at 4°C in Fc‐blocking solution (complete medium with 5 μg/ml of CD16/CD32 mAb [clone 93; eBioscience, CA]). Cells were washed and surface stained with optimal dilutions of BUV395‐conjugated anti‐CD3 (clone 145‐2C11), BUV737‐conjugated anti‐CD19 (clone1D3), BV510‐conjugated anti‐IgD (11‐26c.2a), AF700‐conjugated anti‐CD45R (B220, clone RA3‐6B2), FITC‐conjugated anti‐GL7 (clone GL‐7), PE‐Cy7‐conjugated anti‐CD95 (clone Jo2), BV421‐conjugated anti‐TACI (clone 8F10), PE‐conjugated anti‐CD138 (clone 281‐2), BB700‐conjugated anti‐CD38 (clone 90/CD38), BV650‐conjugated anti‐IgM (clone II/41), BV605‐conjugated anti‐IgG1 (clone A85‐1), PE‐CF594‐conjugated anti‐CXCR4 (clone 2B11/CXCR4), and AF647‐conjugated anti‐CD73 (cloneTY/239), all from BD Biosciences. Nonviable cells were excluded from the analysis with the LIVE/DEAD Fixable Near‐IR Dead Cell Stain Kit, according to the manufacturer instruction (Invitrogen, USA) and single cells were discriminated using scatter area (SSC‐A) versus scatter width (SSC‐W) parameters. Antibody optimal dilution was assessed with the Staining Index formula [(MFI positive ‐ MFI negative)/2 x rSD]. Fluorescence‐minus‐one controls were used to detect the background fluorescence for each fluorochrome. About 7 × 105 cells were stored for each sample and acquired on LSRFortessa X20 flow cytometer (BD Biosciences).
Data Preprocessing
The B cell population analyzed in our data set was gated on live, singlet, CD3−, IgD−, CD19+ lymphocytes using FlowJo v10 (TreeStar, USA). The analysis was then carried on using R (v3.5.2) platform, an open‐source software environment for statistical analysis, computation, and visualization. Flow cytometry standard (FCS) files were exported as uncompensated data in R environment as a single flowSet object (list of FCS), that was then compensated with FlowCore package v1.48.1 32 and logicle transformed 33 using the estimateLogicle function for automatic parameters selection for each fluorescence marker. Events out of 0.1–99.9 percentile range of each marker were considered as outlier and removed. As each marker has a different fluorescence expression range, that could differently contribute to the analysis 34, a scaling factor, calculated to normalize marker expression, was defined for each marker as μ/(Max‐Min) where μ is the average of all marker expression ranges and Max‐Min is the expression range for each single marker.
FlowSOM
Clustering analysis of flowSet was performed following the FlowSOM function pipeline (package v1.14.1). Grid size of 10 × 10 was selected as optimal for the identification of rare subpopulations. Scaling factors defined in preprocessing were imported in FlowSOM function as “importance” parameter. Similar nodes were merged together (metaclustering step) setting number of metaclusters = 15. The Euclidean distance was used in both the FlowSOM clustering and metaclustering. Thresholds to bisect positive and negative cells for each marker expression were automatically set with flowDensity package. Two thresholds were set for CD38 marker, in order to separate negative from intermediate cells, and intermediate from very bright cells. FlowSOM results were displayed as an heatmap reporting the percentage of positive cells for each marker within the metacluster.
Rtsne
t‐Distributed Stochastic Neighbor Embedding was performed with Rtsne package v0.15. Data from each FCS were concatenated in a single matrix object and multiplied by the scaling factors defined in preprocessing. Rtsne function was run setting perplexity = 30, selected as optimal value in a range between 5 and 50 (Supporting Information Fig. S1) 3.
Statistical Analysis
Mann–Whitney test was used for assessing statistical difference between groups of mice receiving priming with antigen alone or antigen with adjuvant. P‐values were corrected for multiple test with Benjamini‐Hochberg False Discovery Rate 35. Statistical significance was defined as adjusted P‐value (FDR) < 0.05. Validation for the FlowSOM and Mann–Whitney U test was performed with bootstrapping 36. ith bootstrap sample was generated randomly sorting 250 cells (with replacement) from ith sample. Sample variability was taken in account employing the method of leaving one mice out from the bootstrapping analysis. The process was repeated 1,000 times and only metaclusters that were significant (FDR < 0.05) in >50% of times were discussed. Hungarian method 37 was used to match FlowSOM metaclusters with metaclusters identified in each bootstrap iteration. A distance matrix, reporting one clustering by row and the other one by columns, was calculated so that each element in the matrix represents the sum of the cells in both clusters subtracted by twice the cells detected in the intersection of the two clusters. The value = 0 indicated the maximum correspondence between the two analyzed clusters (since means that all elements are in the intersection), while the higher the value the more different were the two clusters. The “solve_LSAP” function from clue package was used to identify the optimal assignment of rows to columns with minimum cost (setting argument maximum = FALSE).
Results
FlowSOM and t‐SNE were used as tools for the identification of B cell subsets in multiparametric data sets derived from cells isolated from draining lymph nodes of mice primed with adjuvanted or non‐adjuvanted vaccine and boosted with antigen alone (Fig. 1). Multidimensional analysis of B cells was based on the expression of B220 (pan‐B cell marker in the mouse) 38, CD38 (highly expressed on mature and memory B cells, while it is reduced to an intermediate expression in germinal center B cells) 39, GL7 and CD95 (coexpressed by B cells involved in the GC reaction) 27, CD73 (memory B cells) 40, IgG1 and IgM (B cell receptors (BCR) in switched or unswitched B cells, respectively) 41, TACI and CD138 (coexpressed by terminally differentiated PCs) 42 and CXCR4 (chemokine receptor involved in long‐lived PCs homing into bone marrow niches) 43. The flow of the process (Fig. 1) was based on an initial manual gating strategy performed with FlowJo for excluding non‐viable cells, doublets, T lymphocytes, and a large part of naïve B cells (IgD+) and selecting CD19+ B cells (Fig. 1). After a data preprocessing step, including compensation and logicle transformation, the analysis of B cell subsets was performed with the automated tools FlowSOM and t‐SNE, capable of simultaneously considering the expression of all markers on single cells to identify distinct phenotypic subsets. The data‐driven analysis allowed the identification of B cell subtypes elicited in response to immunization with adjuvanted versus nonadjuvanted vaccine formulations (Fig. 1).
Figure 1.
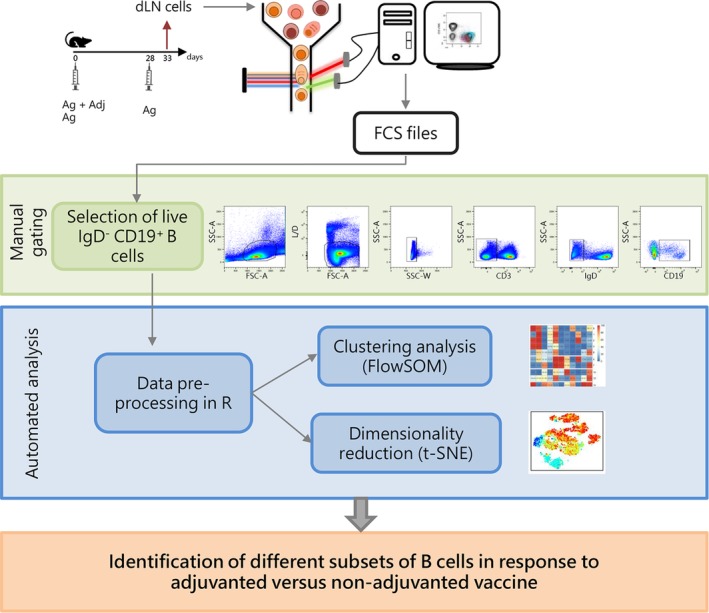
Experimental design and workflow of data analysis. C57BL/6 mice were subcutaneously primed with antigen alone (Ag) or combined with the CAF01 adjuvant (Ag + Adj) and boosted with antigen alone 28 days later. Draining lymph nodes (sub iliac, medial, and external) were collected five days after booster immunization. Cells collected were stained with antibodies in order to identify different B cells subsets, and analyzed with flow cytometry. B cells were sequentially gated as lymphocytes live single CD3− IgD− CD19dim/high cells in FlowJo v10 as shown in dot‐plots. Gated B cells were exported in R environment as uncompensated flowSet, an R object that includes all FCS files. In the preprocessing step, data were compensated, logicle transformed, and scaled. The flowSet was analyzed with both clustering algorithm (FlowSOM) and dimensionality reduction (t‐SNE) in order to identify, on the basis of surface markers expression, different B cell subsets induced by vaccination. [Color figure can be viewed at https://wileyonlinelibrary.com]
Automatic B Cell Subset Identification with FlowSOM
CD19+ B cells identified in each single FCS file were merged into a single flowSet object that was imported in R environment. In this way, it was possible to visualize the data of the two groups of mice in the same plot. Fluorescent signals were compensated and transformed with logicle transformation. The data set was analyzed with a clustering approach, aiming to automatically identify clusters of similar cells. FlowSOM uses an algorithm based on a SOM built assigning similar cells to nodes 9. In a second optional step, similar nodes are metaclustered together with a hierarchical process 15. The metaclustering step facilitates the interpretation of the results, as the high number of nodes in the SOM cannot be easily associated with different cell populations. In order to ensure the identification of all the relevant cell subpopulations, the number of metaclusters was set at 15, a value that exceeds the expected cell subtypes 6. The FlowSOM analysis, reported as SOM grid or minimum spanning tree, in which each node was represented by a star chart with all parameters inside and different B cell subsets were displayed as colored metaclusters, is shown in Supporting Information Figure S2. However, as the visualization of our data set appeared unclear due to the high number of parameters, we adopted an heatmap in which each metacluster was reported as a row and surface markers as columns, and the percentages of cells positive for each marker inside the cluster was visualized as color‐scale from blue (0% of positive cells) to red (100%) (Fig. 2A). Positive and negative cells were defined using the threshold estimated with FlowDensity package. As the expression of CD38 through different B cells subtypes varies from intermediate to very high levels 39 cells were classified as bright (CD38high) and intermediate (CD38dim) subpopulations (Fig. 2A).
Figure 2.
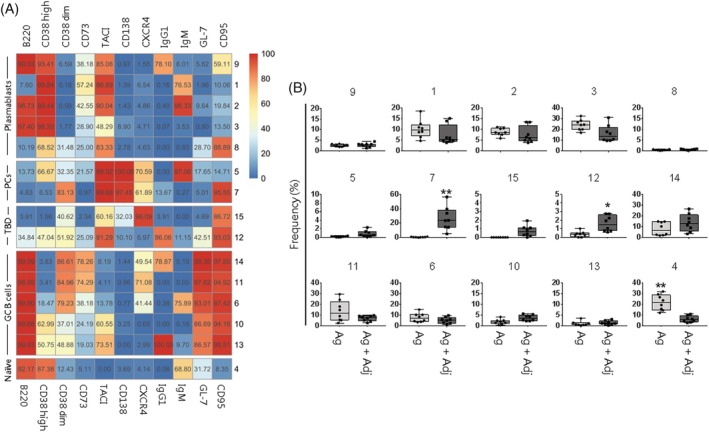
FlowSOM analysis and metacluster frequency in mice immunized with adjuvanted or non‐adjuvanted vaccines. (A) Fifteen metaclusters from FlowSOM analysis were visualized as heatmap. Each row represents a different metacluster, while columns represent analyzed markers. The percentage of cells expressing a marker within a metacluster is reported and visualized with a color scale from blue (0%) to red (100%). B cell populations were indicated on the left (PCs, plasma cells; TBD, to be defined; GC, germinal center). (B) Box and whiskers plots showing the percentage of metaclusters in mice primed with antigen alone (Ag) or antigen and adjuvant (Ag + Adj). Values from individual animals were reported as circles. Mann–Whitney test corrected for multiple test (Benjamini‐Hochberg method) was used for assessing statistical differences between groups (*FDR < 0.05 or **FDR < 0.01). Only metaclusters that were significant (FDR < 0.05) in >50% of bootstrapping analysis were shown as significant. [Color figure can be viewed at https://wileyonlinelibrary.com]
Different stages of plasmablasts were identified in metaclusters 9, 1, 2, 3, and 8 in which cells were still CD38high, frequently B220+ and BCR+, and showed already the upregulation of TACI but not yet of CD138, two markers indicative of terminally differentiated PCs (Fig. 2A). PCs were identified into metaclusters 5 and 7 according to the coexpression of CD138 and TACI, and the loss of B220. Cells of metacluster 7 lost the sIg expression and showed an intermediate expression of CD38, suggesting a more advanced terminal differentiation to PCs compared to metacluster 5. Both metaclusters also expressed high levels of CXCR4 (Fig. 2A). Metaclusters 15 and 12 represented possible stages of differentiating cells, that have already downregulated B220 and CD38, expressed TACI and CD95, with CXCR4 (metacluster 15) or IgG1 (metacluser 12).
GC B cells, defined as CD95+ GL‐7+ B220+ (metaclusters 14, 11, 6, 10, and 13) were subdivided into five different subsets. In some cases cells expressed CD38dim (metaclusters 11, 14, and 6), but differed for the surface expression of immunoglobulins (Ig), as they were IgG1+ (metacluster 14), IgM+ (metacluster 6) or surface Ig (sIg) negative (metacluster 11). Most of cells in metaclusters 11 and 14 expressed also the memory marker CD73. Differently, many cells into metaclusters 10 and 13 were CD38high, TACI+ and IgG1+ (metacluster 13) (Fig. 2A). The different subsets identified, differing for chemokine receptor and BCR expression, can be indicative of the dynamic processes taking place inside the germinal center, that involves cells in the stage of centroblasts or centrocytes. Finally, more undifferentiated cells were included in metacluster 4, in which cells still expressed B220, CD38high, and IgM (in more than 68% of cells), a profile typical of mature naïve B cells (Fig. 2A).
Modulation of the Reactivated B Cell Subtypes According to the Vaccine Formulation
In order to analyze the distribution of B cell populations in mice primed with or without the adjuvant component, the frequencies of metaclusters shown in the heatmap (Fig. 2A) were visualized as boxplots (Fig. 2B). This analysis was obtained extracting the frequencies of identified metaclusters from FCS file of individual animals. Mice primed with adjuvanted formulation showed a significant higher percentage of terminally differentiated PCs grouped in metacluster 7 (23.7% versus 0.4% median frequency in mice immunized with antigen alone; FDR < 0.01), and reactivated B cells of metacluster 12 (1.5% versus 0.3%; FDR < 0.05). The stronger reactivation of B cells in mice immunized with adjuvanted vaccine formulation was also confirmed by the significant lower frequency of mature naïve IgM+ B cells detected in metacluster 4 compared to mice primed with antigen alone (5.5% versus 21.4% in antigen alone group FDR < 0.01). Metaclusters 4 and 7 were confirmed in 100% of bootstrapping resampling, while metacluster 12 was significant in 60% of repetitions. Bootstrapping resampling was performed by leaving one mouse out from the data set in order to take into account the sample variability.
Automated B Cell Subset Identification with t‐SNE
The other automated method commonly used for multiparametric flow cytometry data analysis is the dimensional reduction approach, that allows to display high‐dimensional data in a lower‐dimensional space, using two or three surrogate dimensions. Our data set was analyzed with the t‐SNE algorithm, that grouped cells in distinct areas with continent‐like structure, in which the expression of each parameter was visualized with color code from dark blue (lowest expression) to dark red (highest expression) (Fig. 3). The spatial distribution of different subsets allowed to identify PCs and GC B cells (Fig. 3). PCs were grouped in the upper right region of the plot as B220− CD138+ TACI+. Some of these cells were also positive for CXCR4 (Fig. 3). GC B cells, localized in the upper left side of the graph, were identified as B220+ GL7+ CD95+ CD38dim (Fig. 3). Many of these cells were also positive for the memory marker CD73, and IgG1 (Fig. 3). Naïve B cells, representing the central region of the plot, expressed B220, high level of CD38 and partially IgM (Fig. 3). Overall, the t‐SNE visualization gave a good overview of each marker distribution in dimension‐reduced data space, but it was complex to delineate the coexpression of multiple markers on the identified cell subpopulations. Moreover, the exact quantification of the subpopulations identified with t‐SNE in the different groups, as performed with FlowSOM metaclusters (Fig. 2B), can be obtained only after a subsequent step of automated clustering (for example with the DBSCAN algorithm of ACCENSE 5) or manual gating with FlowJo. Nevertheless, gating on the t‐SNE map followed by phenotypic analysis can lead to over‐fragmentation of immunophenotypes that complicates, rather than simplifies, multidimensional data analysis 44.
Figure 3.
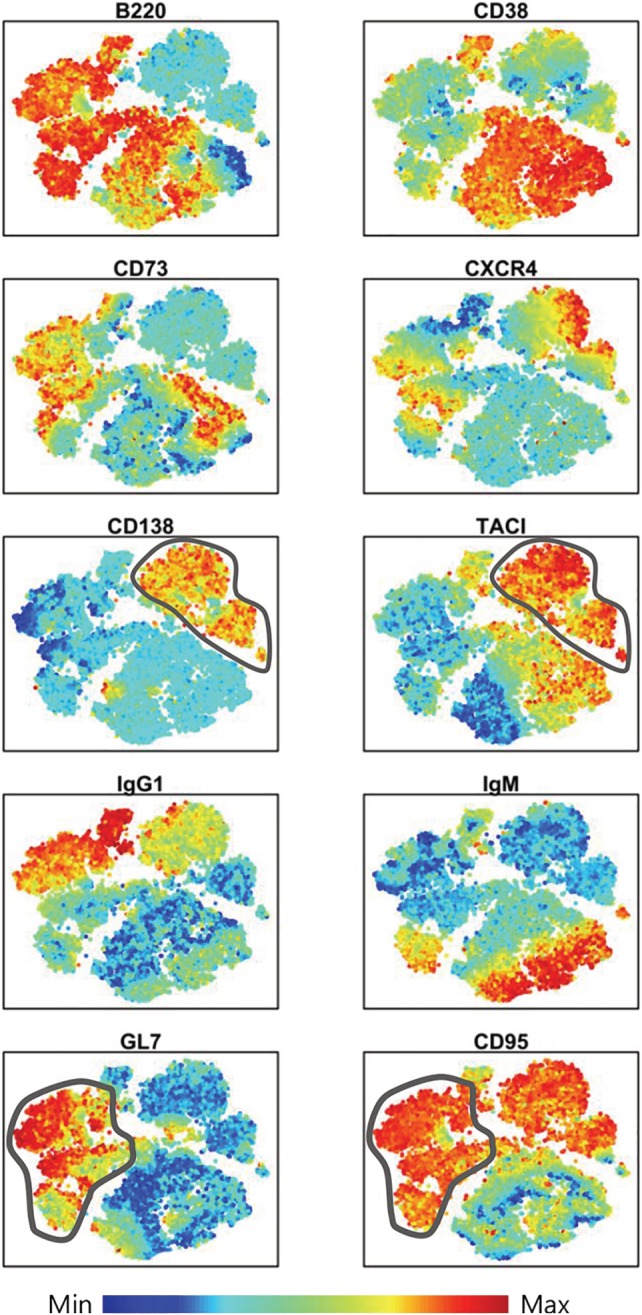
Surface markers distribution in t‐SNE dimensional reduced space. Cells were visualized in a dimensionally reduced space grouped by their immunophenotype similarities. Relative antigen expression was visualized by the color tone (from blue to red). Gray lines surrounded PCs (CD138+ TACI+) in upper right region and GC B cells (GL7+ CD95+) in left side of t‐SNE map. [Color figure can be viewed at https://wileyonlinelibrary.com]
In order to evaluate if the two methods defined the same type of cellular subsets, cells were shown in the t‐SNE dimensionally reduced space colored according to FlowSOM metaclusters (Fig. 4). The metaclustered cells were uniformly arranged into the distinct t‐SNE areas with a strong agreement of markers expression, as shown for example by the localization of PCs identified into metacluster 7, that corresponded to the t‐SNE region positive for CD138 and TACI markers (Fig. 4).
Figure 4.
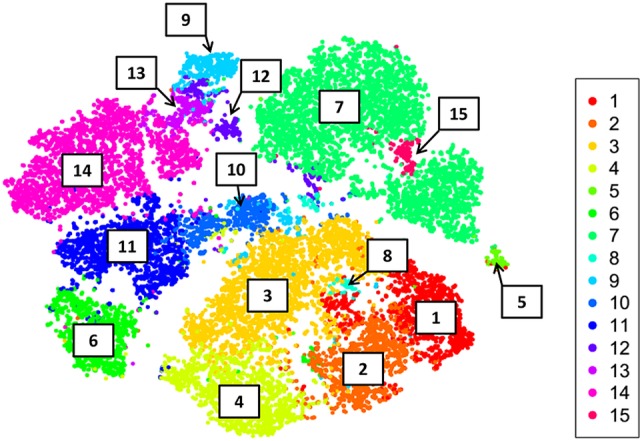
FlowSOM clusters in t‐SNE dimensional reduced space. The analysis outputs obtained with the two computational tools were overlaid, and cells displayed as single point in t‐SNE map were colored according to FlowSOM metaclusters labeled from 1 to 15. [Color figure can be viewed at https://wileyonlinelibrary.com]
As t‐SNE preserves the local structure so that points that are close to one another in the high‐dimensional data set will tend to be close to one another also in the dimensionally reduced space, the localization of metaclusters 12 and 15 provided additional information on these cells that showed a less defined phenotype. Metacluster 15 was localized next to terminally differentiated PCs of metacluster 7 suggesting a possible transient stage of cell differentiation toward this subtype of cells, while cells of metacluster 12 were close to metaclusters 9 and 13. The integration of the two methods on a single graph, roughly provides also information on the relative abundance of the identified metaclusters within the analyzed sample (Fig. 4).
Discussion
In this study we employed automated computational tools for data‐driven analysis of cells elicited by vaccination with an adjuvanted vaccine formulation compared to antigen alone. Computational analysis of multiparametric flow cytometric data performed employing clustering approach (FlowSOM) allowed to group B cells into different subsets on the basis of their marker expression, thus identifying phenotypes otherwise difficult to detect with the classical bidimensional gating. The analysis provided also a signature of B cell recall response specific for the vaccine formulation used. The dimensionality reduction (t‐SNE) tool was less efficient in defining cell subsets that differed for only one or two markers, therefore the identification of intermediate stage of differentiating cells and the evaluation of multiple markers coexpression on the identified cell subpopulations were complex. Nevertheless, the integration of the t‐SNE spatial visualization of cells with the FlowSOM clusters could help in characterizing less defined phenotypes.
The FlowSOM approach clusters cellular subsets on the basis of multiple markers coexpression, but due to the high number of markers included in the staining, it is not straightforward the visualization of data inside the nodes. To overcome this limitation, we changed the FlowSOM cluster visualization from a grid to an heatmap, in which the percentage of positive cells, calculated respect to an automatically set threshold value, was reported giving the possibility to better identify the phenotype of clustered cells. Nevertheless, with this type of data analysis, the information on the fluorescence intensity, that is, the amount of marker expression on the cell surface, is lost. This can be a limit for markers, such as the CD38 molecule, that can be negatively, intermediately, or very brightly expressed according to the cell subtype. For this reason, we considered two different thresholds for the CD38 expression, one that distinguished negative from intermediate and another separating intermediate from high expressing cells. The analysis confirmed that the intermediate and bright expression of CD38 correlated with different cell subsets.
The number of clusters to be included into the FlowSOM analysis is a very important choice, as a high number could lead to a better purity but also to the fragmentation of results in too many groups 9. We used the FlowSOM default setting to determine the number of nodes, while we increased the metacluster number to 15, a relatively high value that exceeds the expected cell subtypes. This value allowed the identification of particular subsets of cell (such as IgM+ plasmacells, centrocytes within the germinal center B cells and subsets of plasmablasts) that would be missed with a lower number of metaclusters.
The t‐SNE analysis returned a good visualization of each marker distribution in the dimension‐reduced data space at single cell level, but the analysis of the coexpression of two and more markers on the identified grouped cells was difficult. t‐SNE analysis results therefore highly and rapidly informative in heterogeneous samples in which different cell types are well separated in the dimensionally reduced data space, but it is less efficacious in separating cell subtypes, in which many parameters are coexpressed on the same cells.
Our data set included not only terminally differentiated cells, but also transient stages of differentiation, that can be identified with automatic data analysis while are hardly identified with the classical bidimensional gating analysis. Indeed, computational tools were able to further subdivide these B cell populations based on the expression of additional cell markers, thus providing important information on activation and developmental state of B cells in different immunization conditions.
Upon the second contact with antigen, reactivated memory B cells differentiate into proliferating plasmablasts from which can originate both terminally differentiated antibody‐secreting PCs, mainly found in the extrafollicular areas of secondary lymphoid tissues, and memory‐GC reactivating cells, which can then give rise to further memory B cells and long‐lived PC with improved antigen‐affinity 24. These two arms of the B cell response—the extrafollicular short‐lived PC, with the rapid antibody secretion, and the GC reaction with the production of memory cells and long‐lived PCs as terminal fates—are the main effector cells. The differentiation of B cells into plasmablasts and PCs (collectively termed antibody‐secreting cells [ASCs]) is essential for the production of the protective antibodies 45, therefore their measurements can provide data to inform the induction of humoral response upon vaccination.
Plasmablasts, indicating proliferating cells were identified in Metaclusters 9, 1, and 2. This subset of cells still expressed the BCR and B220, together with TACI, while CD138 was not yet expressed. Also cells of Metaclusters 1 and 8 were included into the plasmablast population, as they still expressed high levels of CD38 46. Terminally differentiated PCs, generally identified as B220− CD138+ TACI+ cells, were included into metaclusters 5 and 7 according to the coexpression of CD138 and TACI. Cells of metacluster 7 lost the sIg expression and showed an intermediate expression of CD38, suggesting a more advanced terminal differentiation to PCs compared to metacluster 5. They also expressed high levels of CXCR4, a receptor required intrinsically within PCs for homing into the bone marrow 42, thus suggesting a possible phenotype of long‐lived PC. These cells were significantly higher in mice immunized with the CAF01‐adjuvanted vaccine compared to the group primed with the antigen alone. Metaclusters 12 and 15 could represent transient stages, not clearly identifiable. The integration of the FlowSOM metaclusters into the t‐SNE map suggests that cells from metacluster 12 could be a subset of reactivated plasmablasts while cells in metacluster 15 could include cells committed to a PC fate yet, as in the t‐SNE map they were localized next to terminally differentiated PCs.
Within B220+ GL7+ CD95+ GC B cells were indeed identified five different subgroups, that differently expressed the chemokine receptor CXCR4, the surface BCR, the memory CD73 marker and the CD38 (metaclusters 6, 10, 11, 13, and 14). The GC follicle is polarized into a so‐called dark zone of rapidly dividing centroblasts processing antibody maturation and a “light zone” of small non‐dividing centrocytes that undergo selection based on the affinity of their surface antibody 47. In mice, proliferating centroblasts in the dark zone express the CXCR4 receptor 48. The expression of CXCR4 was observed in more than 71% of metacluster 11 cells, and correlated with the absence of sIg (IgG1− IgM−), the presence of the CD73 memory marker and the intermediate expression of CD38, suggesting the identification of centroblasts B cells. More than 40% of cells grouped in metaclusters 6 and 14 were also CXCR4 positive, and they mutually expressed IgM (metacluster 6) or the switched IgG (metacluster 14). Possibly, these cells were trafficking between the dark and the light zone of the GC, where the B cells re‐express the BCR on their surface.
Recent insights into B‐cell memory development pointed to two pathways of memory cell formation, in which CD38+ GL7+ precursors were identified as source of both GC‐independent and GC‐dependent memory B cells. These precursors were capable of differentiating directly into memory B cells, mainly of the IgM+ variety, without passing through a GC cell intermediate stage, or alternatively into GC B cells, some of which then became memory B cells 49. A good candidate to mark GC‐derived but not GC‐independent memory B cells was the surface marker CD73, a nucleotidase that plays a key role in Ig class switch recombination 50. Based on these suggestions, we can speculate that metaclusters 11 and 14 are compatible with GC‐dependent memory B cells (B220+ CD38dim CD73+ IgM− GL7+ CD95+) while cells in metacluster 6 could be part of an extra‐follicular differentiation pathway (B220+ CD38dim CD73− IgM+ GL7+ CD95+). Finally, undifferentiated cells (B220+ CD38high and mostly IgM+) were grouped in metacluster 4, and were significantly higher in mice immunized with antigen alone.
The automated analysis of our data set allowed to identify in an unbiased way cellular phenotypes, that included not only terminally differentiated cells but also transient stages of differentiation, providing important information on activation and developmental state of B cells in different immunization conditions. The assessment of the frequency, phenotype, and function of lymphocytes represents a powerful tool in the characterization of the vaccine immune response in addition to the classical measures of humoral immunity.
Supporting information
MIFlowCyt MIFlowCyt‐Compliant Items
Supplementary Figure 1 Setting of t‐SNE perplexity t‐SNE analysis changing perplexity from 5 to 50 values. The value selected for the analysis was 30.
Supplementary Figure 2 FlowSOM visualization of clustering analysis.
Acknowledgments
The authors would like to thank Prof. Simone Furini for the helpful discussion concerning data analysis and the Staten Serum Institute for providing H56 and CAF01 reagents. This study has been carried out with financial support from the Commission of the European Communities, Horizon 2020 Framework Programme, grant number 730964 (TRANSVAC2).
References
- 1. Robinson JP, Roederer M, HISTORY OF SCIENCE . Flow cytometry strikes gold. Science 2015;350:739–740. 10.1126/science.aad6770. [DOI] [PubMed] [Google Scholar]
- 2. Kvistborg P, Gouttefangeas C, Aghaeepour N, Cazaly A, Chattopadhyay PK, Chan C, Eckl J, Finak G, Hadrup SR, Maecker HT, et al. Thinking outside the gate: Single‐cell assessments in multiple dimensions. Immunity 2015;42:591–592. 10.1016/j.immuni.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. van der Maaten L, Hinton G. Visualizing DATA using t‐SNE. J Mach Learn Res 2008;9:2579–2605. [Google Scholar]
- 4. Amir ED, Davis KL, Tadmor MD, Simonds EF, Levine JH, Bendall SC, Shenfeld DK, Krishnaswamy S, Nolan GP, Pe'er D. viSNE enables visualization of high dimensional single‐cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol 2013;31:545–552. 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shekhar K, Brodin P, Davis MM, Chakraborty AK. Automatic classification of cellular expression by nonlinear stochastic embedding (ACCENSE). Proc Natl Acad Sci U S A 2014;111:202–207. 10.1073/pnas.1321405111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Saeys Y, Gassen SV, Lambrecht BN. Computational flow cytometry: Helping to make sense of high‐dimensional immunology data. Nat Rev Immunol 2016;16:449–462. 10.1038/nri.2016.56. [DOI] [PubMed] [Google Scholar]
- 7. Aghaeepour N, Nikolic R, Hoos HH, Brinkman RR. Rapid cell population identification in flow cytometry data. Cytometry A 2011;79:6–13. 10.1002/cyto.a.21007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lo K, Hahne F, Brinkman RR, Gottardo R. flowClust: A bioconductor package for automated gating of flow cytometry data. BMC Bioinformatics 2009;10:145 10.1186/1471-2105-10-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Van Gassen S, Callebaut B, Van Helden MJ, Lambrecht BN, Demeester P, Dhaene T, Saeys Y. FlowSOM: Using self‐organizing maps for visualization and interpretation of cytometry data. Cytometry A 2015;87:636–645. 10.1002/cyto.a.22625. [DOI] [PubMed] [Google Scholar]
- 10. Weber LM, Robinson MD. Comparison of clustering methods for high‐dimensional single‐cell flow and mass cytometry data. Cytometry A 2016;89:1084–1096. 10.1002/cyto.a.23030. [DOI] [PubMed] [Google Scholar]
- 11. Verhagen FH, Hiddingh S, Rijken R, Pandit A, Leijten E, Olde Nordkamp M, ten Dam‐van Loon NH, Nierkens S, Imhof SM, de Boer JH, et al. High‐dimensional profiling reveals heterogeneity of the Th17 subset and its association with systemic immunomodulatory treatment in non‐infectious uveitis. Front Immunol 2018;9:2519 10.3389/fimmu.2018.02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Collier AJ, Panula SP, Schell JP, Chovanec P, Plaza Reyes A, Petropoulos S, Corcoran AE, Walker R, Douagi I, Lanner F, et al. Comprehensive cell surface protein profiling identifies specific markers of human naive and primed pluripotent states. Cell Stem Cell 2017;20:874–890.e7. 10.1016/j.stem.2017.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ciabattini A, Pettini E, Fiorino F, Lucchesi S, Pastore G, Brunetti J, Santoro F, Andersen P, Bracci L, Pozzi G, et al. Heterologous prime‐boost combinations highlight the crucial role of adjuvant in priming the immune system. Front Immunol 2018;9:380 10.3389/fimmu.2018.00380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Spear TT, Nishimura MI, Simms PE. Comparative exploration of multidimensional flow cytometry software: A model approach evaluating T cell polyfunctional behavior. J Leukoc Biol 2017;102:551–561. 10.1189/jlb.6A0417-140R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wilkerson MD, Hayes DN. ConsensusClusterPlus: A class discovery tool with confidence assessments and item tracking. Bioinformatics 2010;26:1572–1573. 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ciabattini A, Pettini E, Medaglini D. CD4(+) T cell priming as biomarker to study immune response to preventive vaccines. Front Immunol 2013;4:421 10.3389/fimmu.2013.00421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ciabattini A, Prota G, Christensen D, Andersen P, Pozzi G, Medaglini D. Characterization of the antigen‐specific CD4(+) T cell response induced by prime‐boost strategies with CAF01 and CpG adjuvants administered by the intranasal and subcutaneous routes. Front Immunol 2015;6:430 10.3389/fimmu.2015.00430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Prota G, Christensen D, Andersen P, Medaglini D, Ciabattini A. Peptide‐specific T helper cells identified by MHC class II tetramers differentiate into several subtypes upon immunization with CAF01 adjuvanted H56 tuberculosis vaccine formulation. Vaccine 2015;33:6823–6830. 10.1016/j.vaccine.2015.09.024. [DOI] [PubMed] [Google Scholar]
- 19. Ellebedy AH, Jackson KJL, Kissick HT, Nakaya HI, Davis CW, Roskin KM, McElroy AK, Oshansky CM, Elbein R, Thomas S, et al. Defining antigen‐specific plasmablast and memory B cell subsets in human blood after viral infection or vaccination. Nat Immunol 2016;17:1226–1234. 10.1038/ni.3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bolton DL, Roederer M. Flow cytometry and the future of vaccine development. Expert Rev Vaccines 2009;8:779–789. 10.1586/erv.09.41. [DOI] [PubMed] [Google Scholar]
- 21. Furman D, Davis MM. New approaches to understanding the immune response to vaccination and infection. Vaccine 2015;33:5271–5281. 10.1016/j.vaccine.2015.06.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pastore G, Carraro M, Pettini E, Nolfi E, Medaglini D, Ciabattini A. Optimized protocol for the detection of multifunctional epitope‐specific CD4+ T cells combining MHC‐II tetramer and intracellular cytokine staining technologies. Front Immunol 2019;10:2304 10.3389/fimmu.2019.02304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lanzavecchia A. Dissecting human antibody responses: Useful, basic and surprising findings. EMBO Molecular Medicine 2018;10:e8879 10.15252/emmm.201808879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McHeyzer‐Williams LJ, Dufaud C, McHeyzer‐Williams MG, Do Memory B. Cells form secondary germinal centers? Impact of antibody class and quality of memory T‐cell help at recall. Cold Spring Harb Perspect Biol 2018;10 10.1101/cshperspect.a028878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sallusto F, Lanzavecchia A, Araki K, Ahmed R. From vaccines to memory and back. Immunity 2010;33:451–463. 10.1016/j.immuni.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McHeyzer‐Williams M, Okitsu S, Wang N, McHeyzer‐Williams L. Molecular programming of B cell memory. Nat Rev Immunol 2011;12:24–34. 10.1038/nri3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ciabattini A, Pettini E, Fiorino F, Pastore G, Andersen P, Pozzi G, Medaglini D. Modulation of primary immune response by different vaccine adjuvants. Front Immunol 2016;7:427 10.3389/fimmu.2016.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Santoro F, Pettini E, Kazmin D, Ciabattini A, Fiorino F, Gilfillan GD, Evenroed IM, Andersen P, Pozzi G, Medaglini D. Transcriptomics of the vaccine immune response: Priming with adjuvant modulates recall innate responses after boosting. Front Immunol 2018;9:1248 10.3389/fimmu.2018.01248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Reed SG, Orr MT, Fox CB. Key roles of adjuvants in modern vaccines. Nat Med 2013;19:1597–1608. 10.1038/nm.3409. [DOI] [PubMed] [Google Scholar]
- 30. Aagaard C, Hoang T, Dietrich J, Cardona P‐J, Izzo A, Dolganov G, Schoolnik GK, Cassidy JP, Billeskov R, Andersen P. A multistage tuberculosis vaccine that confers efficient protection before and after exposure. Nat Med 2011;17:189–194. 10.1038/nm.2285. [DOI] [PubMed] [Google Scholar]
- 31. Agger EM, Rosenkrands I, Hansen J, Brahimi K, Vandahl BS, Aagaard C, Werninghaus K, Kirschning C, Lang R, Christensen D, et al. Cationic liposomes formulated with synthetic mycobacterial Cordfactor (CAF01): A versatile adjuvant for vaccines with different immunological requirements. PLOS ONE 2008;3:e3116 10.1371/journal.pone.0003116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hahne F, LeMeur N, Brinkman RR, Ellis B, Haaland P, Sarkar D, Spidlen J, Strain E, Gentleman R. flowCore: A bioconductor package for high throughput flow cytometry. BMC Bioinformatics 2009;10:106 10.1186/1471-2105-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Parks DR, Roederer M, Moore WA. A new “Logicle” display method avoids deceptive effects of logarithmic scaling for low signals and compensated data. Cytometry A 2006;69A:541–551. 10.1002/cyto.a.20258. [DOI] [PubMed] [Google Scholar]
- 34. Qian Y, Wei C, Eun‐Hyung Lee F, Campbell J, Halliley J, Lee JA, Cai J, Kong YM, Sadat E, Thomson E, et al. Elucidation of seventeen human peripheral blood B‐cell subsets and quantification of the tetanus response using a density‐based method for the automated identification of cell populations in multidimensional flow cytometry data. Cytometry B Clin Cytom 2010;78(Suppl 1):S69–S82. 10.1002/cyto.b.20554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Benjamini Y, Hochberg Y. Controlling the False discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B Methodol 1995;57:289–300. [Google Scholar]
- 36. Efron B. Bootstrap methods: Another look at the Jackknife. Ann Statist 1979;7:1–26. 10.1214/aos/1176344552. [DOI] [Google Scholar]
- 37. Kuhn HW. The Hungarian method for the assignment problem. Nav Res Logist Q 1955;2:83–97. 10.1002/nav.3800020109. [DOI] [Google Scholar]
- 38. Coffman RL, Weissman IL. B220: A B cell‐specific member of the T200 glycoprotein family. Nature 1981;289:681 10.1038/289681a0. [DOI] [PubMed] [Google Scholar]
- 39. Vences‐Catalán F, Santos‐Argumedo L. CD38 through the life of a murine B lymphocyte. IUBMB Life 2011;63:840–846. 10.1002/iub.549. [DOI] [PubMed] [Google Scholar]
- 40. Pape KA, Jenkins MK, Do Memory B. Cells form secondary germinal Centers? It depends. Cold Spring Harb Perspect Biol 2018;10 10.1101/cshperspect.a029116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stavnezer J, Guikema JEJ, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol 2008;26:261–292. 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Brynjolfsson SF, Persson Berg L, Olsen Ekerhult T, Rimkute I, Wick M‐J, Mårtensson I‐L, Grimsholm O. Long‐lived plasma cells in mice and men. Front Immunol 2018;9:2673 10.3389/fimmu.2018.02673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cheng Q, Khodadadi L, Taddeo A, Klotsche J, Hoyer BF, Radbruch A, Hiepe F. CXCR4‐CXCL12 interaction is important for plasma cell homing and survival in NZB/W mice. Eur J Immunol 2018;48:1020–1029. 10.1002/eji.201747023. [DOI] [PubMed] [Google Scholar]
- 44. Mazza EMC, Brummelman J, Alvisi G, Roberto A, Paoli FD, Zanon V, Colombo F, Roederer M, Lugli E. Background fluorescence and spreading error are major contributors of variability in high‐dimensional flow cytometry data visualization by t‐distributed stochastic neighboring embedding. Cytometry A 2018;93:785–792. 10.1002/cyto.a.23566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tellier J, Nutt SL. Standing out from the crowd: How to identify plasma cells. Eur J Immunol 2017;47:1276–1279. 10.1002/eji.201747168. [DOI] [PubMed] [Google Scholar]
- 46. Oliver AM, Martin F, Kearney JF. Mouse CD38 is down‐regulated on germinal center B cells and mature plasma cells. J Immunol 1997;158:1108–1115. [PubMed] [Google Scholar]
- 47. MacLennan IC. Germinal centers. Annu Rev Immunol 1994;12:117–139. 10.1146/annurev.iy.12.040194.001001. [DOI] [PubMed] [Google Scholar]
- 48. Allen CDC, Ansel KM, Low C, Lesley R, Tamamura H, Fujii N, Cyster JG. Germinal center dark and light zone organization is mediated by CXCR4 and CXCR5. Nat Immunol 2004;5:943–952. 10.1038/ni1100. [DOI] [PubMed] [Google Scholar]
- 49. Taylor JJ, Pape KA, Jenkins MK. A germinal center‐independent pathway generates unswitched memory B cells early in the primary response. J Exp Med 2012;209:597–606. 10.1084/jem.20111696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Anderson SM, Tomayko MM, Ahuja A, Haberman AM, Shlomchik MJ. New markers for murine memory B cells that define mutated and unmutated subsets. J Exp Med 2007;204:2103–2114. 10.1084/jem.20062571. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MIFlowCyt MIFlowCyt‐Compliant Items
Supplementary Figure 1 Setting of t‐SNE perplexity t‐SNE analysis changing perplexity from 5 to 50 values. The value selected for the analysis was 30.
Supplementary Figure 2 FlowSOM visualization of clustering analysis.


