Abstract
Background
As of 2015 thousands of refugees are being hosted in temporary refugee camps in Greece. Displaced populations, travelling and living under poor conditions with limited access to healthcare are at a high risk of exposure to vector borne disease (VBD). This study sought to evaluate the risk for VBD transmission within refugee camps in Greece by analyzing the mosquito and sand fly populations present, in light of designing effective and efficient context specific vector and disease control programs.
Methods
A vector/pathogen surveillance network targeting mosquitoes and sand flies was deployed in four temporary refugee camps in Greece. Sample collections were conducted bi-weekly during June–September 2017 with the use of Centers for Disease Control (CDC) light traps and oviposition traps. Using conventional and molecular diagnostic tools we investigated the mosquito/sand fly species composition, population dynamics, pathogen infection rates, and insecticide resistance status in the major vector species.
Results
Important disease vectors including Anopheles sacharovi, Culex pipiens, Aedes albopictus and the Leishmania vectors Phlebotomus neglectus, P. perfiliewi and P. tobbi were recorded in the study refugee camps. No mosquito pathogens (Plasmodium parasites, flaviviruses) were detected in the analysed samples yet high sand fly Leishmania infection rates are reported. Culex pipiens mosquitoes displayed relatively high knock down resistance (kdr) mutation allelic frequencies (ranging from 41.0 to 63.3%) while kdr mutations were also detected in Ae. albopictus populations, but not in Anopheles and sand fly specimens. No diflubenzuron (DFB) mutations were detected in any of the mosquito species analysed.
Conclusions
Important disease vectors and pathogens in vectors (Leishmania spp.) were recorded in the refugee camps indicating a situational risk factor for disease transmission. The Cx. pipiens and Ae. albopictus kdr mutation frequencies recorded pose a potential threat against the effectiveness of pyrethroid insecticides in these settings. In contrast, pyrethroids appear suitable for the control of Anopheles mosquitoes and sand flies and DFB for Cx. pipiens and Ae. albopictus larvicide applications. Targeted actions ensuring adequate living conditions and the establishment of integrated vector-borne disease surveillance programs in refugee settlements are essential for protecting refugee populations against VBDs.
Keywords: Refugee camp, Mosquito, Sand fly, Insecticide resistance
Background
From 2015 onwards a major refugee crisis has been developing on Europe’s doorstep. Political instability, totalitarian state-governance, poverty and armed conflict have led to mass displacement of populations (refugees/asylum seekers and immigrants) mainly from African and Asian countries seeking refuge in Europe. Greece is amongst the European countries most widely affected by the ongoing crisis. In 2015 and 2016 over 1 000 000 refugees/immigrants arrived in Greece. Currently over 135 000 refugees are hosted in the country [1, 2], the majority in temporary accommodation sites/refugee camps with varied levels of infrastructures, housing conditions and amenities. Displaced populations travelling and living under poor conditions with limited access to healthcare are at a high risk of exposure to vector borne disease (VBD) infections [3–5]. The risk for VBD transmission is augmented in a context of host country disease endemicity (or high prevalence of disease vectors), inadequate or absent vector control in host regions, and VBD endemicity in the refugee travel route regions and countries of origin.
Greece displays an intense history of VBDs, posing a situational risk factor for disease transmission among vulnerable refugee populations. West Nile virus (WNV) lineage two outbreaks, attributed to Culex pipiens mosquitoes [6, 7] have taken place from 2010 onwards with the largest occurring in 2018, resulting in 311 cases and 47 deaths [8]. Autochthonous malaria transmission, though eradicated in the 1970s re-appeared in 2009 reaching a peak of 42 locally acquired cases in 2011 [9]. From 2009 onwards imported and locally acquired Plasmodium vivax and Plasmodium falciparum cases are reported every year. An added public health threat and a considerable nuisance problem is the wide spread distribution of the invasive mosquito Aedes albopictus, a known vector of Chinkungunya and dengue virus [10]. Last but not least, visceral leishmaniasis, a lethal disease if left untreated, vectored by Phlebotomine sand flies is endemic in nearly all geographical areas of the country with Leishmania infantum implicated as the predominant etiological agent [11]. The disease is most prevalent in dogs with a few sporadic human cases every year [12].
In the absence of protective human vaccines against most VBDs (including malaria, WNV and leishmaniasis), protection from the diseases largely relies on vector control programs utilizing chemical insecticides to manage vector populations [13]. The larvicides Bacilus thuringiensis israelensis (B.t.i.), diflubenzuron (DFB) and pyrethroid based adulticides constitute the main mosquito control insecticide regime in Greece under the current regulatory policies, with the latter insecticides (pyrethroids) also applied in a number of refugee camps.
A worldwide problem associated with the extensive use of a limited number of available insecticides in vector and agricultural pest control is the development of insecticide resistance, threatening the effectiveness of control efforts. Resistance is primarily conferred by point mutations on the insecticide’s target molecule (target site resistance) and/or increased activity of detoxification enzymes (metabolic resistance) [14]. Insecticide resistance has been reported in important vector species in Greece and neighbouring countries. Voltage gated sodium channel (VGSC) mutations (kdr mutations) associated with pyrethroid resistance have been detected at high frequencies (reaching 88.3%) in Cx. pipiens mosquitoes from Northern and Central Greece [15]. Presence of the kdr mutation L1014F was recently also monitored in sand fly populations from Turkey and Greece (including specimens sampled from Souda refugee camp in Chios island) and was detected in the major Leishmania vector Phlebotomus papatasi in Turkey [16]. Striking DFB resistance, attributed to the chitin synthase (CHS) mutations I1043L and I1043F, was reported in Cx. pipiens populations from Northern Italy in 2016–2017 [17, 18]. While the VGSC mutations V1016G, I1532T, F1534C/L associated with pyrethroid resistance in Ae. albopictus have been recorded in Ae. albopictus populations from Italy and Greece where the mutation 1534C was detected at a frequency of 24.2% in the Athens population [19, 20].
Integrated vector/pathogen surveillance is of utmost importance in epidemiological settings of increased public health significance such as refugee camps. Refugee settlements, whether temporary or permanent, represent unique ecological settings which inevitably affect and are affected by the surrounding ecosystem. Within these unique, vulnerable communities, the status of arthropod vectors remains unknown. Fine scale entomological information on vector species composition and seasonal abundance, vector insecticide resistance status and the presence of arthropod-borne pathogens is essential information in order to evaluate the risk for VBD transmission within these communities and design effective and efficient context specific vector and disease control programs (evidence based control). A vector/pathogen surveillance network targeting mosquitoes (Diptera: Culicidae) and sand flies (Diptera: Psychodidae: Phlebotominae) was deployed in temporary refugee camps in Greece in 2017. Preliminary findings of the study relating to sand fly species composition and Leishmania circulation were communicated in reference [21]. The present study compiles the full data produced by the surveillance network relating to sand fly and mosquito species composition, population dynamics, pathogen infection rates, and insecticide resistance status in four major refugee camps in Greece.
Materials and methods
Study sites
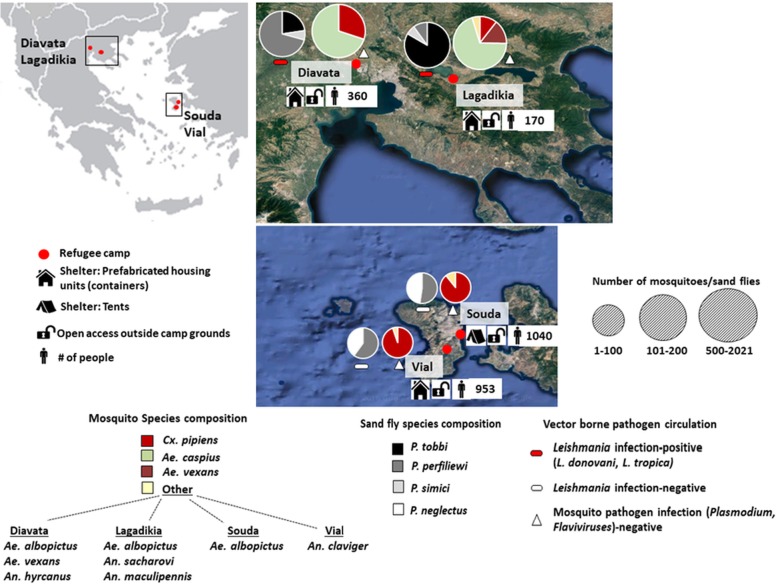
Four refugee camps were surveyed in 2017: Lagadikia and Diavata camps (open reception facilities), situated in Thessaloniki regional Unit, Central Macedonia, Northern Greece; Vial camp (reception and identification centre) and Souda camp (open reception facility), located on the island of Chios, Northeastern Aegean island complex. Lagadikia camp is situated 46 km east of Thessaloniki city in an agricultural zone and at 5 km from Lake Koroneia. Diavata camp is situated in a peri-urban setting 7 km away from the agricultural land (20 000 ha of rice-fields) of West Thessaloniki and 2 km away from Gallikos river bed. Vial camp is located in an inland agricultural area of the island with no major water bodies, and Souda camp (currently dismantled) was established in the moat of Chios Castle in Chios town, next to the island’s main harbour. The average size of the four study camps was 1.75 ha. In 2017 the four camps hosted populations mainly from Syria, Afghanistan and Iraq [22]. In the summer of 2017 stray dogs were consistently present in the Thessaloniki camps (Diavata, Lagadikia). Shelter and occupancy information for 2017 [22] is included in Fig. 1.
Fig. 1.
Vector composition and vector borne pathogen detection in Refugee camps, Greece In each refugee camp site - 2 chart pies are depicted. From left to right (in each site); Sand fly species composition chart pie, followed by Mosquito species composition chart pie. Sand fly species data displayed corresponds to male and female(*) adult collections, mosquito species data corresponds to adult female collections (under representation of Aedes albopictus specimens/egg collections not included). Mosquito and sand fly species recorded are depicted with different colours. The size of each chart pie correlates to the number of sand flies (♂/♀) and mosquitoes (♀) collected in each site. Absence/presence of mosquito and sand fly pathogens(**) is depicted with different symbols. Coloured symbols denote infection presence/detection, white colour denotes absence, in the analysed samples. Camp descriptors including shelter type, access status and number of occupants are displayed for the year 2017 [22]. (*)The female sand fly specimens identification and (**)Leishmania detection was conducted in [21]
Vector surveillance and sample handling
Entomological surveillance was conducted bi-weekly from June to September 2017 with the use of Centers for Disease Control (CDC) light traps baited with dry ice (targeting adult mosquitoes and sand flies) and oviposition traps (for surveillance of the container breeder Ae. albopictus). Overall seven sampling events were conducted in each refugee camp with the deployment of three CDC light traps (from 18.00 pm–8.00 am) and four oviposition traps (renewed every 14 days) per site per sampling event. In Lagadikia and Diavata camps (Thessaloniki), all sampling events were conducted within the camp grounds whereas in Souda and Vial camps (Chios) the majority of sampling events (five out of seven in each camp) were conducted in a buffer zone of < 200 m around the camp perimeter.
Mosquitoes collected with CDC light traps were either killed with a twenty minute exposure period at − 20 °C, morphologically identified to species level [13, 23] and stored in 70% ethanol (for use in molecular diagnostic assays, e.g. species identification, resistance mutations and Plasmodium parasite detection), or anesthetized with a cold shock and following morphological identification stored in RNA later (for Flavivirus detection). Mosquito eggs collected with the oviposition traps were stored at − 20 °C. Sand fly handling, described in reference [21] included dissection of the head from the body in the female wild caught specimens and storage of all body parts in ethanol.
DNA and RNA extraction
Genomic DNA was extracted from single female adult mosquitoes and from pools of mosquito eggs (five eggs per pool) using the DNAzol protocol according to the manufacturer’s instructions (Invitrogen, Carlsbad, USA). Total RNA was isolated from Cx. pipiens mosquito pools (5–10 mosquitoes per pool) using the TRI reagent protocol (Invitrogen, Carlsbad, USA). The purity and concentration of the total extracted RNA from each pool was determined using a NanoDrop 2000c spectrophotometer (Thermo Scientific, Waltham, Massachusetts) and its integrity was confirmed with 1% agarose gel electrophoresis. DNA extraction from individual female sand fly specimens and sand fly species molecular identification was conducted and reported in reference [21]. DNA was extracted using the NucleoSpin Tissue kit (Macherey-Nagel, Düren, Germany) according to the manufacturer’s instructions. Female sand flies were identified to species using a multiplex diagnostic assay based on species-specific single-nucleotide polymorphisms of the nuclear 18S rRNA gene [24]. The male sand flies were identified morphologically to species level.
Mosquito species molecular identification
A subset of mosquitoes belonging to the Cx. pipiens complex were identified to species with a PCR diagnostic assay which relies on polymorphisms in the second intron of the acetylcholinesterase-2 (AChE2) locus as described in reference [25]. Primers specific for Cx. pipiens, Cx. quinquefasciatus, Cx. pallens, Cx. australicus, and Cx. torrentium were included in the assay (Table 1). DNA fragments were analyzed by electrophoresis on a 1.5% agarose gel.
Table 1.
Molecular assays used in the study: primers and PCR conditions
| Species | Assay | Primers (5′ → 3′) | PCR condition | Reference | |
|---|---|---|---|---|---|
| Insecticide resistance assays | Culex pipiens | VGSC (1014F)/kdr - allele specific PCR |
Cgd1: GTGGAACTTCACCGACTTC Cgd2: GCAAGGCTAAGAAAAGGTTAAG Cgd3: CCACCGTAGTGATAGGAAATTTA Cgd4: CCACCGTAGTGATAGGAAATTTT |
95 °C 5 min, 40 cycles × (94 °C 30 s, 48 °C 30 s, 72 °C 1 min), 72 °C 10 min | [26] |
| Cx. pipiens | CHS (1043 L) - allele specific PCR |
External_F: GCAGTCCTTCGGCGATCTT External_R: GAACAGTCCGGCGATGGATA ATC_ R: AACAGCAAGTACATAGACGGGAT CTC_ F: GGCTTGATCTACCTGCTGTCTC |
95 °C 5 min, 28 cycles × (95 °C 30 s, 68 °C 30 s, 72 °C 1 min), 72 °C 5 min | [17] | |
| Cx. pipiens | CHS (1043 M) - PCR and RFLP |
Diagnostic I1043M_R: CCCAGGAGACGACGTTCAG Diagnostic I1043M_F: GCCTGTCTCCATCCGCAAG |
95 °C 5 min, 30 cycles × (95 °C 30 s, 60 °C 30 s, 72 °C 30 s),72 °C 5 min | [17] | |
| Aedes albopictus | VGSC (1016G)/kdr - PCR and sequencing |
F1016: TTCACCGACTTCATGCACTC R1016: CGCAATCTGGCTTGTTAACTT |
95° 3 min, 40 cycles × (94 °C 30 s, 55 °C 30 s, 72 °C 1 min), 72 °C 5 min | – | |
| Ae. albopictus | VGSC (1532 T,1534C)/kdr - PCR and sequencing |
For: GAGAACTCGCCGATGAACTT Rev.: GACGACGAAATCGAACAGGT Rev. for sequencing: AGCTTTCAGCGGCTTCTTC |
95 °C 3 min, 40 cycles × (94 °C 30 s, 57 °C 30 s, 72 °C 1 min), 72 °C 5 min | – | |
| Ae. albopictus | CHS (1043 L/M) - PCR and sequencing |
kkv F3: TCGGAAGTCCTTCGGCTTAT TC kkv R3: TGGATACTTCAATGGAACCTTCC |
95 °C 5 min, 40 cycles × (94 °C 30 s, 55 °C 30 s, 72 °C 1 min), 72 °C 10 min | – | |
| Anopheles (An. sacharovi, An. hyrcanus, An. maculipennis, An. claviger) | VGSC (1014F/C/S) / kdr - PCR and sequencing |
KDRF: GGMGAATGGATYGAATCMATGTGGGA KDR R2: GATGAACCRAAATTKGACAAAAGCAA |
94 °C 5 min, 35 cycles × (94 °C 1 min, 50 °C 2 min, 72 °C 2 min), 72 °C 2 min | – | |
| Sand flies (Phlebotomus perfiliewi, P. tobbi, P. simici, P. neglectus) | VGSC (1011, 1014, 1016, and 1020) / PCR and sequencing |
Vssc8F: AATGTGGGATTGCATGCTGG Vssc1bR: CGTATCATTGTCTGCAGTTGGT |
95 °C 5 min, 36 cycles × (94 °C 45 s, 51 °C 50 s, 72 °C 50 s),72 °C 7 min | [27] | |
| Species id assays | Cx. pipiens complex | AChE2- PCR diagnostic assay |
ACEquin: CCTTCTTGAATGGCTGTGGCA ACEpall: ATGGTGGAGACGCATGACG ACEpip: GGAAACAACGACGTATGTACT ACEtorr: TGCCTGTGCTACCAGTGATGTT B1246s: TGGAGCCTCCTCTTCACGG |
94 °C 5 min, 35 cycles × (94 °C 30 s, 55 °C 30 s, 72 °C 1 min), 72 °C 5 min | [25] |
| Cx. pipiens biotype | CQ11 - allele specific PCR |
pipRev: CATGTTGAGCTTCGGTGAA molRev: CCCTCCAGTAAGGTATCAAC biotype-forward: GATCCTAGCAAGCGAGAAC |
95 °C 5 min, 40 cycles × (94 °C 30 s, 54 °C 30 s, 72 °C 40 s), 72 °C 5 min | [28] | |
| Anopheles | ITS2 - PCR and sequencing |
5.8 s primer: TGTGAACTGCAGGACACATG 28 s primer: ATGCTTAAATTTAGGGGGTA |
94 °C 2 min, 40 cycles × (94 °C 30 s, 53 °C 30 s, 72 °C 50 s), 72 °C 10 min | [29] | |
| Anopheles | COI - PCR and sequencing |
Cl -N-2191: CCCGGTAAAATTAAAATATAAACTTC Cl-J-1718: GCAGGATTTGGAAATTGATTAGTTCC |
94 °C 2 min, 40 cycles × (94 °C 30 s, 50 °C 30 s, 72 °C 50 s), 72 °C 10 min | [29] | |
| Ae. albopictus | ITS2 - PCR and sequencing |
5.8 s primer: TGTGAACTGCAGGACACATG 28 s primer: ATGCTTAAATTTAGGGGGTA |
94 °C 10 min, 40 cycles × (94 °C 1 min, 50 °C 1 min, 72 °C 1 min), 72 °C 10 min | [30] | |
| Sand flies | Direct multiplex PCR |
Forward: TGCGGTTAAAACGTTCGTAG perf: TAAACCCACAAATCAGAAT sim: CTTTCATTAAATTTAGCCTGCC papat: TAACATAAGGGGCGTATTAATATGCTTT tob: GCGTRTTAACAATAGAGTCCATTAAA |
95 °C 5 min, 32 cycles × (94 °C 40 s, 54 °C 30 s, 72 °C 1 min), 72 °C 10 min | [24] | |
| Pathogen detection assays | Plasmodium (P. vivax, P. falciparum) | PCR 1 (Nested PCR) |
rPLU5: CCTGTTGTTGCCTTAAACTTC rPLU6: TTAAAATTGTTGCAGTTAAAACG |
95 °C 5 min, 24 cycles × (94 °C 1 min, 57 °C 2 min,72 °C 2 min), 72 °C 5 min | [31] |
| PCR 2 (Nested PCR) |
VIV-1: CGCTTCTAGCTTAATCCACATAACTGATAC VIV-2: ACTTCCAAGCCGAAGCAAAGAAAGTCCTTA FAL-1: TTAAACTGGTTTGGAAAACCAAATATATT FAL-2: ACACAATGAACTCAATCATGACTACCCGTC |
95 °C 5 min, 30 cycles × (94 °C 1 min, 57 °C 2 min, 72 °C 2 min), 72 °C 5 min | [31] | ||
| Flavivirus detection | Conventional one-step reverse transcriptase-PCR (RT-PCR) |
Forward: TACAACATGATGGGAAAGAGAGAGAA Reverse: GTGTCCCAGCCGGCGGTGTCATCAGC |
50 °C 30 min, 95 °C 3 min, 40 cycles × (95 °C 15 s, 55 °C 20 s, 72 °C 60 s), 72 °C 10 min | [32] | |
| West Nile virus detection | Multiplex Real Time one step RT-PCR TaqMan assay |
Forward: GTGATCCATGTAAGCCCTCAGAA Reverse: GTCTGACATTGGGCTTTGAAGTTA FAM probe (MGB): AGGACCCCACATGTT HEX probe (MGB): AGGACCCCACGTGCT |
50 °C 15 min, 95 °C 3 min, 45 cycles × (95 °C 3 s and 60 °C 30 s) | [33, 34] |
VGSC Voltage gated sodium channel; CHS Chitin synthase; kdr Knock down resistance; AChE2 Acetylcholinesterase-2; CQ11 Microsatellite locus; ITS2 Internal transcribed spacer two; COI Cytochrome oxidase subunit 1; [REF] References
Cx. pipiens mosquitoes molecularly identified to species were also identified to biotype (pipiens, molestus, hybrid) with the use of a PCR diagnostic assay [28] which relies on polymorphisms of the 5′ flanking region of the microsatellite locus CQ11 specific for pipiens and molestus alleles. Primers are described in Table 1. DNA fragments were analyzed by electrophoresis on a 2% agarose gel.
In order to verify the species identification in samples morphologically identified as Ae. albopictus and in eggs collected from the oviposition traps, the nuclear ribosomal spacer gene, internal transcribed spacer two (ITS2) was amplified [30]. Adult specimens were analysed individually while eggs were pooled prior to DNA extraction. DNA fragments were analyzed by electrophoresis on a 1.5% agarose gel, (primers described in Table 1).
All Anopheles mosquitoes collected were identified to species with ITS2 and cytochrome oxidase subunit 1 (COI) product sequencing and BLAST analysis.
DNA fragments were analyzed by electrophoresis on a 1.5% agarose gel (primers in Table 1).
Prior to sequencing the PCR products were purified using a commercially available kit (Macherey Nagel, Dueren, Germany). Species identification of mosquitoes belonging to the Anopheles maculipennis complex was further confirmed with restriction fragment length polymorphism (RFLP) assays [29] based on ITS2 and COI product restriction sites for the restriction enzymes AluI and HpaII, respectively.
Pathogen detection
Monitoring of P. vivax and P. falciparum infections in Anopheles mosquitoes was conducted with a genus- and species-specific nested polymerase chain reaction [31]. Genomic DNA extracted from Anopheles specimens infected with P. vivax and P. falciparum were used as positive controls (primers in Table 1).
Conventional one-step reverse transcriptase-PCR (RT-PCR) was performed to assess the presence of flaviviruses based on the method described in reference [32] and detection of WNV was performed by a multiplex real time one step RT-PCR TaqMan assay that simultaneously detects and differentiates WNV-lineage 1 and WNV-lineage 2, as described in references [33, 34] (primers in Table 1). All the Cx. pipiens specimens which were transported live to the laboratory (n = 40, from Diavata and Lagadikia camp) were pooled per camp site and included in the Flavivirus/WNV detection analysis. Leishmania detection and Leishmania species identification were conducted in reference [21]. Leishmania presence was evaluated by amplification of a 300–350-bp fragment of the Leishmania DNA ribosomal internal transcribed spacer one (ITS1) [35]. Leishmania species was identified with a series of assays including; an ITS1 RFLP assay [36], ITS1 product sequencing and BLAST analysis, a species-specific PCR assay for the cysteine protease b gene [37] and amplification/sequencing of a segment of the heat-shock protein 70 [38].
Insecticide resistance genotyping
Presence and frequency of kdr mutation L1014F (TTA to TTT) on the VGSC was monitored in Cx. pipiens mosquitoes with an allele specific PCR assay as described in reference [26] (primers in Table 1). DNA fragments of the VGSC gene flanking the codon 1014 in Anopheles specimens (An. sacharovi, An. maculipennis, An. hyrcanus, An. claviger) and the codons 1016, 1532, 1534 in Ae. albopictus samples were amplified, purified and sequenced (primers in Table 1). In sand flies, a DNA fragment of the VGSC gene (flanking the codons 1011, 1014, 1016, and 1020) was amplified, purified (Macherey Nagel, Dueren, Germany commercial kit) and sequenced (primers in Table 1) as described in reference [27].
The presence of the chitin synthase C-terminus target site resistance mutations I1043L and I1043M were monitored in Cx. pipiens with an allele specific PCR assay and a PCR-RFLP diagnostic assay, respectively, as described in reference [17] (primers in Table 1). A sequence of Ae. albopictus CHS gene flanking the codon 1043 was amplified and sequenced for the detection of DFB resistance mutations.
Results
Vector species population structure and dynamics
Mosquito and sand fly surveillance conducted in the refugee camps resulted in the collection of important proven and suspected disease vectors (Fig. 1, Table 2).
Table 2.
Mosquito and sand fly population structure in the study refugee camps
| Mosquitoes | Sand flies | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refugee camp | Culex | Aedes | Anopheles | Phlebotomus | |||||||||||
| Cx. pipiens | Ae. albopictus | Ae. caspius | Ae. vexans | An. sacharovi | An. maculipennis | An. hyrcanus | An. claviger | Total | P. tobbi | P. simici | P. perfiliewi | P. neglectus | Total | ||
| (A) | (E) | ||||||||||||||
| Diavata | 606 (31) | - | 64 (15) | 1409 | 2 | - | - | 4 (4) | - | 2085 | 36 (15) | 11 (7) | 114 (45) | - | 161 |
| Lagadikia | 44 (25) | 8 (8) | 75 (15) | 402 | 62 | 4 (4) | 6 (6) | - | - | 601 | 128 (45) | 10 (6) | 15 (9) | - | 153 |
| Souda | 84 (18) | 9 (9) | 23 (15) | - | - | - | - | - | - | 116 | - | - | 13 (11) | 11 (10) | 24 |
| Vial | 18 (12) | - | - | - | - | - | - | - | 1 (1) | 19 | - | - | 33 (24) | 24 (16) | 57 |
The primary WNV vector Cx. pipiens was amongst the most dominant mosquito species in all four camps. Biotype analysis in a sub-sample of the Cx. pipiens samples detected the Cx. pipiens pipiens biotype as the most dominant in Diavata camp while Cx. pipiens molestus was the most dominant in Lagadikia, Souda and Vial camp. The hybrid Cx. pipiens pipiens/molestus was recorded in all four camps (Table 3).
Table 3.
Culex pipiens biotype composition in the study refugee camps
| Culex pipiens biotype n(%) | |||||
|---|---|---|---|---|---|
| Region | Camp | N | pipiens | molestus | hybrid |
| Thessaloniki | Diavata | 29 | 19 (65.5) | 2 (6.9) | 8 (27.6) |
| Lagadikia | 24 | 2 (8.3) | 19 (79.2) | 3 (12.5) | |
| Chios | Souda | 15 | 0 (0.0) | 11 (73.3) | 4 (26.7) |
| Vial | 9 | 1 (11.1) | 5 (55.6) | 3 (33.3) | |
N is the number of samples tested per camp, n = the number of samples identified for each biotype, [%] = the corresponding frequency of each biotype per camp
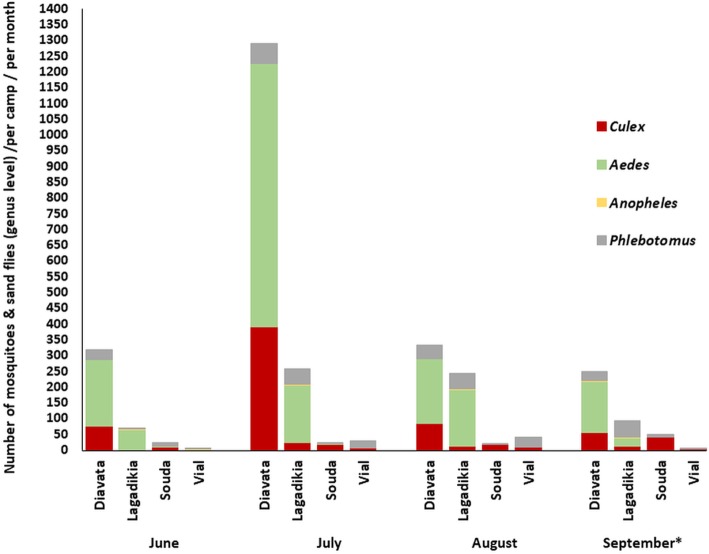
In Diavata camp where Cx. pipiens was highly abundant the populations peaked in July (Fig. 2).
Fig. 2.
Mosquito and sand fly (genus level) population dynamics in Refugee camps, Greece. Sand fly species data displayed corresponds to male and female adult collections, mosquito species data corresponds to adult female collections. The mosquito and sand fly genera recorded are depicted with different colours. (*) one sample collection was conducted in September (Week 1–2) (vs) two sample collections (Week 1–2, Week 3–4) in June, July, August
Ae. albopictus was detected in Diavata, Lagadikia (Thessaloniki) and Souda camp (Chios). In Lagadikia and Diavata camp Ae. albopictus eggs were first collected in the last week of July. Oviposition collections in Vial camp (Chios) were halted because traps were knocked down by animal activity.
The malaria vector An. sacharovi and the potential vector An. maculipennis s.s. were recorded in Lagadikia camp, albeit all at very low numbers while An. hyrcanus was found in Diavata camp. A single An. claviger specimen was collected from Vial camp (Chios).
The nuisance species Ae. caspius was by far the most dominant species in Diavata and Lagadikia camp (Thessaloniki) (Fig. 1, Table 1) composing 70 and 74% of the mosquito population, respectively, while Ae. vexans was also reported in both camps.
The sand fly species recorded were P. perfiliewi, P. tobbi and P. simici in Diavata and Lagadikia camps (Thessaloniki) and P. neglectus, P. perfiliewi in Souda and Vial camps (Chios). P. perfiliewi was the dominant species in Lagadikia, Souda and Vial camp and P. tobbi was the most abundant species in Diavata camp (Fig. 1, Table 1). In Lagadikia and Diavata (Thessaloniki) the sand fly populations showed very low activity in June but from then onwards population numbers increased, maintaining a relatively steady activity, with the highest population numbers in Lagadikia camp noted in early September (Fig. 2).
Pathogen detection
All Cx. pipiens pools analysed were negative for Flavivirus/WNV presence. All Anopheles samples collected were negative for P. vivax and P. falciparum infections. Leishmania donovani and Leishmania tropica DNA was detected in P. perfiliewi, P. tobbi and P. simici specimens from Lagadikia and Diavata camps (Thessaloniki) [21].
Vector insecticide resistance status
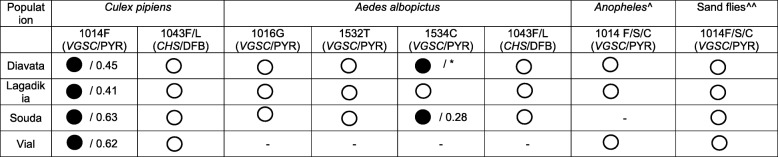
The pyrethroid resistance mutation L1014F, was analysed in a total of 80 Cx. pipiens mosquitoes and was recorded at an allele frequency ranging from 41.0 to 63.3% (Table 4). Homozygotes for the resistance mutation were detected in all camps (Additional file 1: Table S1). In a total of 46 female Cx. pipiens mosquitoes, no CHS mutations were detected (Table 4).
Table 4.
Insecticide resistance status of the main vectors sampled in the refugee camps
 = Detection of Target site mutations/resistant allele frequency
= Detection of Target site mutations/resistant allele frequency
 = no resistance mechanisms detected
= no resistance mechanisms detected
- = no samples analysed; * = Both 1534F (susceptible allele) and 1534C were found in eggs (pooled samples) from Diavata / no precise information regarding mutation frequency; VGSC/PYR Pyrethroid resistant mutation(s) in the Voltage gated sodium channel gene; CHS/DFB Diflubenzuron resistant mutations in the Chitin synthase 1 gene; ^ Anopheles species analysed: An. sacharovi, An. maculipennis, An. hyrcanus; ^^ Sand fly species analysed: P. perfiliewi, P. tobbi, P. simici, P. neglectus; kdr analysis in sand flies from Souda camp was conducted in [16]
Fourty five Ae. albopictus eggs and 17 Ae. albopictus adults were screened for the presence of pyrethroid and DFB resistance mutations at the corresponding sites 1016, 1532, 1534 (VGSC) and 1043 (CHS). All samples were homozygous for the susceptible – wild type alleles 1043I, 1016 V and 1532 T. The mutation 1534C was detected in Souda camp at an allele frequency of 28.0%, corresponding to three heterozygote specimens’ 1534F/C and a single homozygote 1534C/C. Both 1534F (susceptible allele) and 1534C were found in a pool of eggs from Diavata (Table 4).
A fragment of the VGSC gene was analyzed in a subset (between 14 and 24 specimens per camp) of the collected sand flies including all sand fly species detected. No pyrethroid resistance mutations were detected. Likewise no pyrethroid resistance mutations at codon 1014 were reported in the Anopheles specimens analyzed (Table 4).
Discussion
Over the last five years, hundreds of thousands of refugees have sought refuge/asylum in Greece and other European countries [39]. Travelling and often living under poor conditions with limited access to healthcare, these displaced populations are vulnerable to VBDs, especially when travelling or hosted in VBD endemic regions. Entomological monitoring in four major refugee camps in Greece revealed the presence of competent disease vectors at varied levels of abundance, including vectors infected with pathogens and resistant to insecticides.
Presence of important disease vectors in the refugee camps
Mosquito and sand fly species analyses revealed the presence of important proven and suspected disease vectors including the mosquito species; Cx. pipiens, Ae. albopictus, An. sacharovi and the sand flies [21] P. tobbi, P. perfiliewi, P. simici and P. neglectus. Even though there were differences in vector abundance among the four camp sites, Cx. pipiens s.l. mosquitoes (WNV vectors) were ubiquitous, followed by Ae. albopictus (dengue & Chikungunya vector) and An. maculipennis s.l. (malaria vector). The Leishmania vectors P. perfiliewi and P. tobbi were also dominant in the camp sites [21]. The observed seasonality of the recorded mosquito and sand fly vectors is in accordance with available literature from the region [11, 15] with July–September being the period with maximum activity.
Vector population structure in relation to camp locations
High Cx. pipiens numbers were recorded in Diavata camp, Thessaloniki (n = 606), compared to the other camps, which may be attributed to the camp’s location. The camp is situated in a peri-urban area of intense industrial activity and is in close proximity to highly proliferative agricultural breeding sites (rice fields). The former possibly providing a plethora of Cx. pipiens breeding sites adjacent to the camp grounds and the latter resulting in the production of high Cx. pipiens numbers, potentially drawn towards the camp site. The highest densities of sand flies were detected in Diavata and Lagkadikia camps (Thessaloniki), with the latter being located adjacent to animal facilities (< 150 m), which are known prolific sand fly breeding sites [11]. When selecting sites for hosting refugee populations, analyses on the vector breeding potential of the candidate sites as well as their proximity in areas with a history of vector borne diseases is of high importance. Risk assessment models incorporating data on the presence/absence of confirmed and potential breeding sites, as well as historic entomological and disease surveillance data may guide the selection of appropriate sites, minimizing the potential risk for VBD transmission [40, 41]. Furthermore, the development of geo-spatial information platforms featuring real-time entomological and epidemiological data on vectors and the pathogens they transmit from the broader refugee camp regions will potentially provide well-informed risk assessments and evidence based guidance for targeted vector control.
Presence of vector-borne pathogens
Even though all Culex samples tested were negative for WNV only a small number of mosquitoes were screened and, thus, may not be representative of the actual WNV pathogen circulation levels of WNV in mosquitoes within and around the survey camp sites. Consecutive follow up analyses on a larger number of specimens is essential for assessing the potential/ongoing risk for WNV transmission. To our knowledge no WNV cases have been reported to date in the study refugee camps. However, the high Cx. pipiens numbers and hybrid biotype representation (Cx. pipiens molestus/pipiens hybrids were detected at a rate of 12.5 to 33.3%) recorded in the study camp sites in conjunction with the WNV disease history in Greece [42] pose a risk for the resident camp populations and the surrounding communities. Low numbers of Anopheles mosquitoes were recorded in the refugee camps and no Plasmodium parasites were detected in the Anopheles samples. Nevertheless, the presence of competent malaria vectors such as An. sacharovi within the camp grounds indicates risk for malaria transmission upon the presence of infected hosts. The recent history of autochtonous malaria transmission in Northern Greece and the fact that many refugees/immigrants start their journeys or travel through malaria endemic regions [43] highlights the need for systematic pathogen monitoring in all camps.
High L. donovani and L. tropica infection rates were reported in P. perfiliewi, P. tobbi and P. simici sand flies from Lagadikia and Diavata camps (Thessaloniki), indicating high levels of parasite circulation [21]. Parasite circulation (of the two rare Leishmania species in Greece) appeared to be focal in the two camps (absence of parasite detection outside the camp sites) [21], indicating a minimized threat for disease transmission to the surrounding communities. The absence of Leishmania parasites in the analyzed sand fly samples collected nearby the Vial and Souda camps (Chios) does not prove absence of parasite circulation in these study sites. The low numbers of sand flies tested and the fact that the sampling events in Chios took place outside the camp grounds (in contrast to the camps in Thessaloniki where all samplings were conducted within) may have decreased the sampling efficiency for pathogen detection, pin pointing the need for follow up monitoring studies within the same as well as additional camp sites.
Parasite transmission from sand fly to host depends on a number of factors including the average rate of biting [44] which is primarily associated with the host exposure to sand flies seeking blood meals and is augmented in a context of overcrowding and inadequate sheltering (risk factors). These risk factors have been associated with leishmaniasis outbreaks in refugee camps/communities in Lebanon [45, 46] and are also present to an extent in the study and other refugee camps (e.g. Moria camp on Lesvos island, Greece currently hosting over 15 000 refugees while originally designed to host 3000) raising further concerns regarding disease transmission. The number of people hosted/accommodated in Lagadikia, Diavata and Vial camp doubled in 2018 compared to 2017 (Souda camp was dismantled in October 2017), with 30 and 1176 refugees/asylum seekers sleeping in tents and rub halls (inadequate vector proofing) in Lagadikia and Vial camp respectively [47].
Essential steps for the prevention and control of leishmaniasis transmission in the refugee camps include systematic active (upon consent) and passive detection of leishmaniasis in the refugee populations, effective treatment of infected patients, detection-treatment and removal of infected dogs from the camp grounds, and the replacement of tents and rubber halls with insect proof shelters. Despite the fact that the leishmaniasis burden is high in refugee populations in the Eastern Mediterranean region [43, 45], refugee population screening and clinical assessment for Leishmania infections is limited in European host countries [48, 49]. On the contrary, a positive step improving living conditions is an accommodation scheme providing rented housing to a small number of vulnerable asylum-seekers and refugees in Greece [50].
Insecticide resistance status of the major disease vectors
The pyrethroid resistance mutation 1534C was detected in Ae. albopictus populations from Souda and Diavata camp at low frequencies, while no kdr mutations were detected in Anopheles mosquitoes and sand fly populations. On the other hand, the pyrethroid resistance mutation L1014F was detected in Cx. pipiens mosquitoes at an allele frequency ranging from 41.0 to 45.2% in the Thessaloniki camps (Lagadikia, Diavata) and 62.4 to 63.3% in Chios camps (Vial, Souda - out of the 27 samples analysed from Chios none were homozygous for the susceptible allele L1014) (Additional file 1: Table S1), indicating the presence of strong selection pressures. In Chios island no large scale mosquito control applications have taken place in recent years while pyrethroids are used extensively for agricultural pest control, possibly imposing strong selection pressure on the local mosquito populations, resulting in the detected mutation frequencies. This is in agreement with recent studies [15, 18, 51] where high kdr, CHS mutation frequencies and neonicotinoid resistant mosquitoes were associated with intensive agricultural insecticide applications. Further analyses investigating the presence/frequency of other polymorphisms (point mutations, locus duplications [52, 53]) at the Cx. pipiens VGSC codon 1014 are required in order to fully assess the 1014 locus contribution to pyrethroid resistance.
Pyrethroid based control strategies (indoor residual spraying, long lasting insecticidal nets, outdoors residual spraying) constitute the primary (or most commonly used) approach for vector management within refugee settlements around the globe [54–56]. Although the operational significance and the possible effect of the kdr mutation frequencies recorded in the Cx. pipiens and Ae. albopictus samples, in terms of pyrethroid suitability/efficiency for their control has to be studied, the high mutation frequencies recorded in the studied camps indicate the need for expanding the currently available methods to include novel strategies as well as novel insecticidal groups [57]. Bioassay data against the major insecticides used and genotyping of other resistance markers (e.g. levels of major detoxification genes) will further enhance our knowledge on the resistance status of the major disease vectors and facilitate the design of effective control and insecticide resistance management programs. No DFB resistance mutations were detected in Cx. pipiens or Ae. albopictus mosquitoes, indicating the suitability of the larvicide for mosquito control in the camp areas. However, as Ae. albopictus is a known container breeder its control via conventional larviciding applications may be extremely challenging. A plethora of container breeding sites were identified in all study camps indicating source reduction through community engagement as an important control method in the camp grounds. Educational campaigns aiming to increase the awareness of refugee populations on vector borne diseases and provide training for appropriate personal protection measures against biting arthropods needs to be integrated in the currently available healthcare support systems.
This study has potential limitations. These include: a) sample bias regarding the Flavivirus/WNV presence as a small number of samples was analyzed, potentially not representative of the actual WNV circulation levels in the mosquitoes, b) sand fly collection bias in the refugee camps in Chios (Vial, Souda) where the majority of collections were conducted outside the camp grounds - potentially reducing the sampling efficiency for Leishmania detection compared to the Thessaloniki camps where all samples were collected within the camp grounds and c) the lack of bioassay data for Cx. pipiens and Ae. albopictus against pyrethroids in order to better evaluate their resistance status and the operational impact of the detected mutation frequencies.
Conclusions
Important disease vectors and pathogens in vectors (Leishmania spp.) were recorded in the refugee camps indicating a situational risk factor for disease transmission. The Cx. pipiens and Ae. albopictus kdr mutation frequencies recorded pose a potential threat against the effectiveness of pyrethroid insecticides in these settings. In contrast, pyrethroids appear suitable for the control of Anopheles mosquitoes and sand flies and DFB for Cx. pipiens and Ae. albopictus larvicide applications. Targeted actions ensuring adequate living conditions and the establishment of integrated vector-borne disease surveillance programs in refugee settlements are essential for protecting refugee populations against VBDs.
Supplementary information
Additional file 1: Table S1. VGSC-1014 genotype and kdr mutation 1014F allele frequency in Culex pipiens populations in the study refugee camps.
Acknowledgements
We thank the Greek Ministry of the Interior and Administrative Reconstruction for facilitating the establishment of our surveillance activities. This work was supported by the General Secretariat for Research and Technology and the Hellenic Foundation for Research and Innovation in the context of the action 1st Proclamation of Scholarships from ELIDEK for PhD Candidates (scholarship code 532).
Abbreviations
- AChE2
Acetylcholinesterase-2
- B.t.i.
Bacilus thuringiensis israelensis
- CDC
Centers for Disease Control
- CHS
Chitin Synthase
- COI
Cytochrome oxidase subunit 1
- DFB
Diflubenzuron
- IRS
Indoor Residual Spraying
- ITS2
Internal transcribed spacer two
- kdr
Knock Down Resistance
- LLINs
Long Lasting Insecticidal Nets
- PCR
Polymerase chain reaction
- VBD
Vector Borne Disease
- VGSC
Voltage Gated Sodium Channel
- WNV
West Nile Virus
Authors’ contributions
EF conducted sample collections, the majority of molecular analyses and made substantial contributions to the conception and design of the work, analysis and interpretation of the data and manuscript preparation. IG conducted molecular analyses in sand fly samples. JS made substantial contributions in the sample collections, sorting and morphological identification of the specimens collected. FT, SB and KM conducted molecular analyses in mosquito samples SK provided access to the refugee camps in Northern Greece and facilitated the surveillance activities. JV substantively revised the work/manuscript. AC made substantial contributions to the design of the work, interpretation of data and substantively revised the work/manuscript. All authors have approved the submitted version
Funding
This work was partly supported by the General Secretariat for Research and Technology (GSRT) and the Hellenic Foundation for Research and Innovation (HFRI) in the context of the action “1st Proclamation of Scholarships from ELIDEK for PhD Candidates”.
Availability of data and materials
All data generated or analysed during this study are included in this published article (and its supplementary information files).
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s40249-020-0635-4.
References
- 1.United Nations High Commissioner for Refugees (UNCHR). UNCHR - Greece. https://www.unhcr.org/greece.html. Accessed 30 Dec 2019.
- 2.United Nations High Commissioner for Refugees (UNCHR). UNCHR – Population statistics. http://popstats.unhcr.org/en/overview#_ga=2.244019333.1697519650.1562076503-639856367.1547638167. Accessed 30 Dec 2019.
- 3.Carballo M, Nerukar A. Migration, refugees, and health risk. Emerg Infect Dis. 2001;7(3 Suppl):556–560. doi: 10.3201/eid0707.017733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eiset AH, Wejse C. Review of infectious diseases in refugees and asylum seekers-current status and going forward. Public Health Rev. 2017;38:22. doi: 10.1186/s40985-017-0065-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castelli F, Sulis G. Migration and infectious diseases. Clin Microbiol Infect. 2017;23(5):283–289. doi: 10.1016/j.cmi.2017.03.012. [DOI] [PubMed] [Google Scholar]
- 6.Mavridis K, Fotakis EA, Kioulos I, Mpellou S, Konstantas S, Varela E, et al. Detection of West Nile virus - lineage 2 in Culex pipiens mosquitoes, associated with disease outbreak in Greece, 2017. Acta Trop. 2018;182:64–68. doi: 10.1016/j.actatropica.2018.02.024. [DOI] [PubMed] [Google Scholar]
- 7.Chaskopoulou A, Dovas CI, Chaintoutis SC, Kashefi J, Koehler P, Papanastassopoulou M. Detection and early warning of West Nile virus circulation in Central Macedonia, Greece, using sentinel chickens and mosquitoes. Vector Borne Zoonotic Dis. 2013;13(10):723–732. doi: 10.1089/vbz.2012.1176. [DOI] [PubMed] [Google Scholar]
- 8.European Centre for Disease Prevention and Control (ECDC) Epidemiological update: West Nile virus transmission season in Europe. 2018. [Google Scholar]
- 9.National Public Health Organization (NPHO) Epidemiological Surveillance Report - Malaria in Greece. 2015. [Google Scholar]
- 10.European Centre for Disease Prevention and Control (ECDC). Aedes albopictus - Factsheet for experts. https://ecdc.europa.eu/en/disease-vectors/facts/mosquito-factsheets/aedes-albopictus. Accessed 30 Dec 2019.
- 11.Chaskopoulou A, Giantsis IA, Demir S, Bon MC. Species composition, activity patterns and blood meal analysis of sand fly populations (Diptera: Psychodidae) in the metropolitan region of Thessaloniki, an endemic focus of canine leishmaniasis. Acta Trop. 2016;158:170–176. doi: 10.1016/j.actatropica.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 12.Ntais P, Sifaki-Pistola D, Christodoulou V, Messaritakis I, Pratlong F, Poupalos G, et al. Leishmaniases in Greece. Am J Trop Med Hyg. 2013;89(5):906–915. doi: 10.4269/ajtmh.13-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Becker N, Petric D, Zgomba M, Boase C, Madon M, Dahl C, et al. Mosquitoes and their control. Berlin: Springer; 2010. [Google Scholar]
- 14.Liu N. Insecticide resistance in mosquitoes: impact, mechanisms, and research directions. Annu Rev Entomol. 2015;60:537–559. doi: 10.1146/annurev-ento-010814-020828. [DOI] [PubMed] [Google Scholar]
- 15.Fotakis EA, Chaskopoulou A, Grigoraki L, Tsiamantas A, Kounadi S, Georgiou L, et al. Analysis of population structure and insecticide resistance in mosquitoes of the genus Culex, Anopheles and Aedes from different environments of Greece with a history of mosquito borne disease transmission. Acta Trop. 2017;174:29–37. doi: 10.1016/j.actatropica.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 16.Fotakis EA, Giantsis IA, Demir S, Vontas J, Chaskopoulou A. Detection of pyrethroid resistance mutations in the major leishmaniasis vector Phlebotomus papatasi. J Med Entomol. 2018;55(5):1225–1230. doi: 10.1093/jme/tjy066. [DOI] [PubMed] [Google Scholar]
- 17.Grigoraki L, Puggioli A, Mavridis K, Douris V, Montanari M, Bellini R, et al. Striking diflubenzuron resistance in Culex pipiens, the prime vector of West Nile virus. Sci Rep. 2017;7(1):11699. doi: 10.1038/s41598-017-12103-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Porretta D, Fotakis EA, Mastrantonio V, Chaskopoulou A, Michaelakis A, Kioulos I, et al. Focal distribution of diflubenzuron resistance mutations in Culex pipiens mosquitoes from northern Italy. Acta Trop. 2019;193:106–112. doi: 10.1016/j.actatropica.2019.02.024. [DOI] [PubMed] [Google Scholar]
- 19.Xu J, Bonizzoni M, Zhong D, Zhou G, Cai S, Li Y, et al. Multi-country survey revealed prevalent and novel F1534S mutation in voltage-gated sodium channel (VGSC) gene in Aedes albopictus. PLoS Negl Trop Dis. 2016;10(5):e0004696. doi: 10.1371/journal.pntd.0004696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pichler V, Malandruccolo C, Serini P, Bellini R, Severini F, Toma L, et al. Phenotypic and genotypic pyrethroid resistance of Aedes albopictus, with focus on the 2017 chikungunya outbreak in Italy. Pest Manag Sci. 2019;75(10):2642–2651. doi: 10.1002/ps.5369. [DOI] [PubMed] [Google Scholar]
- 21.Fotakis EA, Giantsis IA, Avgerinou A, Kourtidis S, Agathaggelidou E, Kapoula C, et al. Identification of Leishmania species in naturally infected sand flies from refugee camps, Greece. Emerg Infect Dis. 2019;25(2):361–364. doi: 10.3201/eid2502.181359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Site Management Support ((SMS). Greece-SMS WG-Site Profiles, 2017. https://data2.unhcr.org/en/documents/details/58471. Accessed 30 Dec 2019.
- 23.Schaffner F, Angel G, Geoffroy B, Hervy J, Rhaiem A, Brunhes J. The Mosquitoes of Europe. https://www.nhbs.com/the-mosquitoes-of-europe-les-moustiques-deurope. Accessed 30 Dec 2019.
- 24.Giantsis IA, Chaskopoulou A, Claude BM. Direct multiplex PCR (dmPCR) for the identification of six phlebotomine sand fly species (Diptera: Psychodidae), including major Leishmania vectors of the Mediterranean. J Econ Entomol. 2017;110(1):245–249. doi: 10.1093/jee/tow269. [DOI] [PubMed] [Google Scholar]
- 25.Smith JL, Fonseca DM. Rapid assays for identification of members of the Culex (Culex) pipiens complex, their hybrids, and other sibling species (Diptera: culicidae) Am J Trop Med Hyg. 2004;70(4):339–345. doi: 10.4269/ajtmh.2004.70.339. [DOI] [PubMed] [Google Scholar]
- 26.Martinez-Torres D, Chevillon C, Brun-Barale A, Bergé J, Pasteur N, Pauron D. Voltage-dependent NA+ channels in pyrethroid-resistant Culex pipiens L. mosquitoes. Pest Sci. 1999;55(10):1012–1020. doi: 10.1002/(SICI)1096-9063(199910)55:10<1012::AID-PS39>3.0.CO;2-5. [DOI] [Google Scholar]
- 27.Gomes B, Purkait B, Deb RM, Rama A, Singh RP, Foster GM, et al. Knockdown resistance mutations predict DDT resistance and pyrethroid tolerance in the visceral leishmaniasis vector Phlebotomus argentipes. PLoS Negl Trop Dis. 2017;11:e0005504. doi: 10.1371/journal.pntd.0005504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bahnck CM, Fonseca DM. Rapid assay to identify the two genetic forms of Culex (Culex) pipiens L. (Diptera: Culicidae) and hybrid populations. Am J Trop Med Hyg. 2006;75:251–255. doi: 10.4269/ajtmh.2006.75.2.0750251. [DOI] [PubMed] [Google Scholar]
- 29.Patsoula E, Samanidou-Voyadjoglou A, Spanakos G, Kremastinou J, Nasioulas G, Vakalis NC. Molecular characterization of the Anopheles maculipennis complex during surveillance for the 2004 Olympic games in Athens. Med Vet Entomol. 2007;21(1):36–43. doi: 10.1111/j.1365-2915.2007.00669.x. [DOI] [PubMed] [Google Scholar]
- 30.Patsoula E, Samanidou-Voyadjoglou A, Spanakos G, Kremastinou J, Nasioulas G, Vakalis NC. Molecular and morphological characterization of Aedes albopictus in northwestern Greece and differentiation from Aedes cretinus and Aedes aegypti. J Med Entomol. 2006;43(1):40–54. doi: 10.1093/jmedent/43.1.40. [DOI] [PubMed] [Google Scholar]
- 31.Singh B, Bobogare A, Cox-Singh J, Snounou G, Abdullah M, Rahman HA. A genus- and species- specific nested polymerase chain reaction. Malaria detection assay for epidemiologic studies. Am J Trop Med Hyg. 1999;60(4):687–692. doi: 10.4269/ajtmh.1999.60.687. [DOI] [PubMed] [Google Scholar]
- 32.Kuno G, Chang GJ, Tsuchiya KR, Karabatsos N, Cropp CB. Phylogeny of the genus Flavivirus. J Virol. 1998;72(1):73–83. doi: 10.1128/JVI.72.1.73-83.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Del Amo J, Sotelo E, Fernández-Pinero J, Gallardo C, Llorente F, Agüero M, et al. A novel quantitative multiplex real-time RT-PCR for the simultaneous detection and differentiation of West Nile virus lineages 1 and 2, and of Usutu virus. J Virol Methods. 2013;189:321–327. doi: 10.1016/j.jviromet.2013.02.019. [DOI] [PubMed] [Google Scholar]
- 34.Jimenez-Clavero MA, Aguero M, Rojo G, Gomez-Tejedor C. A new fluorogenic real-time RT-PCR assay for detection of lineage 1 and lineage 2 West Nile viruses. J Vet Diagn Investig. 2006;18:459–462. doi: 10.1177/104063870601800505. [DOI] [PubMed] [Google Scholar]
- 35.Rêgo FD, Rugani JMN, Shimabukuro PHF, Tonelli GB, Quaresma PF, Gontijo CMF. Molecular detection of Leishmania in phlebotomine sand flies (Diptera: Psychodidae) from a cutaneous leishmaniasis focus at Xakriabá indigenous reserve, Brazil. PLoS One. 2015;10:e0122038. doi: 10.1371/journal.pone.0122038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schönian G, Nasereddin A, Dinse N, Schweynoch C, Schallig HDF, Presber W, et al. PCR diagnosis and characterization of Leishmania in local and imported clinical samples. Diagn Microbiol Infect Dis. 2003;47:349–358. doi: 10.1016/S0732-8893(03)00093-2. [DOI] [PubMed] [Google Scholar]
- 37.Hide M, Bañuls AL. Species-specific PCR assay for L. infantum/L. donovani discrimination. Acta Trop. 2006;100:241–245. doi: 10.1016/j.actatropica.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 38.Van der Auwera G, Maes I, De Doncker S, Ravel C, Cnops L, van Esbroeck M, et al. Heat-shock protein 70 gene sequencing for Leishmania species typing in European tropical infectious disease clinics. Euro Surveill. 2013;18:20543. doi: 10.2807/1560-7917.ES2013.18.30.20543. [DOI] [PubMed] [Google Scholar]
- 39.Eurostat. Asylum applications (non-EU) in the EU-28 Member States, 2008–2018. https://ec.europa.eu/eurostat/statistics-explained/index.php/Asylum_statistics. Accessed 30 Dec 2019.
- 40.Longbottom J, Browne AJ, Pigott DM, Sinka ME, Golding N, Hay SI, et al. Mapping the spatial distribution of the Japanese encephalitis vector, Culex tritaeniorhynchus Giles, 1901 (Diptera: Culicidae) within areas of Japanese encephalitis risk. Parasit Vectors. 2017;10(1):148. doi: 10.1186/s13071-017-2086-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coleman M, Hemingway J, Gleave KA, Wiebe A, Gething PW, Moyes CL. Developing global maps of insecticide resistance risk to improve vector control. Malar J. 2017;16(1):86. doi: 10.1186/s12936-017-1733-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.National Public Health Organization (NPHO) Annual epidemiological report for West Nile virus human infection, Greece. 2018. [Google Scholar]
- 43.Isenring E, Fehr J, Gültekin N, Schlagenhauf P. Infectious disease profiles of Syrian and Eritrean migrants presenting in Europe: a systematic review. Travel Med Infect Dis. 2018;25:65–76. doi: 10.1016/j.tmaid.2018.04.014. [DOI] [PubMed] [Google Scholar]
- 44.Torres-Guerrero E, Quintanilla-Cedillo MR, Ruiz-Esmenjaud J, Arenas R. Leishmaniasis: a review. F1000Res. 2017;6:750. doi: 10.12688/f1000research.11120.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alawieh A, Musharrafieh U, Jaber A, Berry A, Ghosn N, Bizri AR. Revisiting leishmaniasis in the time of war: the Syrian conflict and the Lebanese outbreak. Int J Infect Dis. 2014;29:115–119. doi: 10.1016/j.ijid.2014.04.023. [DOI] [PubMed] [Google Scholar]
- 46.Saroufim M, Charafeddine K, Issa G, Khalifeh H, Habib RH, Berry A, et al. Ongoing epidemic of cutaneous leishmaniasis among Syrian refugees, Lebanon. Emerg Infect Dis. 2014;20(10):1712–1715. doi: 10.3201/eid2010.140288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Site Management Support (SMS) Greece-SMS WG-Site Profiles. 2018. [Google Scholar]
- 48.Seedat F, Hargreaves S, Nellums LB, Ouyang J, Brown M, Friedland JS. How effective are approaches to migrant screening for infectious diseases in Europe? A systematic review. Lancet Infect Dis. 2018;18(9):e259–e271. doi: 10.1016/S1473-3099(18)30117-8. [DOI] [PubMed] [Google Scholar]
- 49.Rojek AM, Gkolfinopoulou K, Veizis A, Lambrou A, Castle L, Georgakopoulou T, et al. Clinical assessment is a neglected component of outbreak preparedness: evidence from refugee camps in Greece. BMC Med. 2018;16(1):43. doi: 10.1186/s12916-018-1015-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.United Nations High Commissioner for Refugees (UNCHR). UNCHR – Greece. Accommodation update. https://data2.unhcr.org/en/documents/download/69859. Accessed 30 Dec 2019.
- 51.Mouhamadou CS, de Souza SS, Fodjo BK, Zoh MG, Bli NK, Koudou BG. Evidence of insecticide resistance selection in wild Anopheles coluzzii mosquitoes due to agricultural pesticide use. Infect Dis Poverty. 2019;8(1):64. doi: 10.1186/s40249-019-0572-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Labbé P, Berticat C, Berthomieu A, Unal S, Bernard C, Weill M, et al. Forty years of erratic insecticide resistance evolution in the mosquito Culex pipiens. PLoS Genet. 2007;3(11):e205. doi: 10.1371/journal.pgen.0030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martins WFS, Subramaniam K, Steen K, et al. Detection and quantitation of copy number variation in the voltage-gated sodium channel gene of the mosquito Culex quinquefasciatus. Sci Rep. 2017;7:5821. doi: 10.1038/s41598-017-06080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roll Back Malaria Partnership (RBM Partnership) Vector Control in Humanitarian Emergencies. Basel: Roll Back Malaria Vector Control Working Group; 2017. [Google Scholar]
- 55.Jamali S. Role of pyrethroids in control of malaria amongst refugee population. J Pak Med Assoc. 2011;61(5):486–490. [PubMed] [Google Scholar]
- 56.Graham K, Rehman H, Ahmad M, Kamal M, Khan I, Rowland M. Tents pre-treated with insecticide for malaria control in refugee camps: an entomological evaluation. Malar J. 2004;3:25. doi: 10.1186/1475-2875-3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Corbel V, Durot C, Achee NL, Chandre F, Coulibaly MB, David JP, et al. Second WIN international conference on “integrated approaches and innovative tools for combating insecticide resistance in vectors of arboviruses”, October 2018, Singapore. Parasit Vectors. 2019;12(1):331. doi: 10.1186/s13071-019-3591-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. VGSC-1014 genotype and kdr mutation 1014F allele frequency in Culex pipiens populations in the study refugee camps.
Data Availability Statement
All data generated or analysed during this study are included in this published article (and its supplementary information files).