Graphical abstract
Keywords: MERS-CoV, SARS-Cov, 3CLpro, Coronavirus, Pyrazolone
Abstract
Severe acute respiratory syndrome (SARS) led to a life-threatening form of atypical pneumonia in late 2002. Following that, Middle East Respiratory Syndrome (MERS-CoV) has recently emerged, killing about 36% of patients infected globally, mainly in Saudi Arabia and South Korea. Based on a scaffold we reported for inhibiting neuraminidase (NA), we synthesized the analogues and identified compounds with low micromolar inhibitory activity against 3CLpro of SARS-CoV and MERS-CoV. Docking studies show that a carboxylate present at either R1 or R4 destabilizes the oxyanion hole in the 3CLpro. Interestingly, 3f, 3g and 3m could inhibit both NA and 3CLpro and serve as a starting point to develop broad-spectrum antiviral agents.
1. Introduction
Severe acute respiratory syndrome (SARS) led to a life-threatening form of atypical pneumonia in late 2002. In March 2003, the causative agent was identified and named as SARS coronavirus (SARS-CoV). SARS-CoV belongs to the genus Coronaviridae, and is an enveloped, positive-stranded virus with ∼30,000 nucleotides.1 Viral genome encodes two polyproteins, pp1a (∼490 kDa) and pp1ab (∼790 kDa). 3C-like protease (3CLpro), the main protease, and papain-like protease (PLpro) cleave these polyproteins to generate non-structural proteins essential for the viral replication.2, 3 Due to its vital role in replication, 3CLpro is an attractive drug target. Many inhibitors were discovered from high throughput screening and structure-based rational design.4, 5, 6, 7, 8, 9, 10
Although the SARS-CoV infection died down soon, another human CoV associated with the Middle East Respiratory Syndrome (MERS-CoV) has recently emerged, killing about 36% (584 of 1621) of patients infected globally, mainly in Saudi Arabia and South Korea.11 Due to the similar maturation pathway, MERS-CoV 3CLpro is also regarded as a target for developing anti-viral drug. The previously reported SARS-CoV 3CLpro inhibitors cannot potently inhibit MERS-CoV 3CLpro without modifications, due to the subtle structural differences in their active sites.12 Though tremendous efforts have been made to develop inhibitors, therapeutic interventions for the continuous outbreaks of these deadly CoVs, are yet to reach the market.13, 14
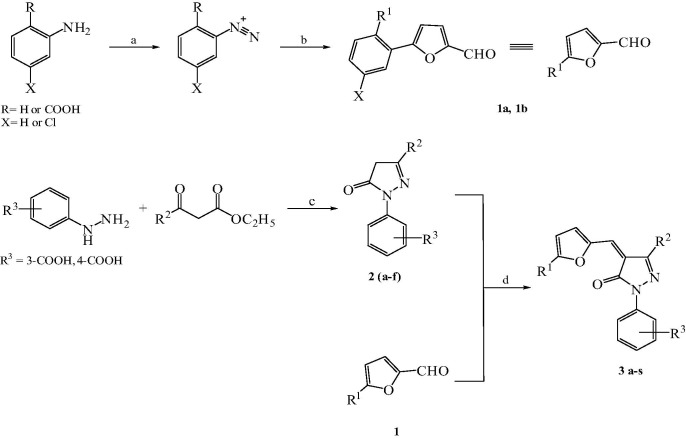
While the origin of SARS dates back to early 2003, influenza has a century-old history of affecting humans. Influenza virus is an enveloped RNA virus that belongs to the orthomyxoviridae family.15 It has caused four major pandemics in the last century, namely, 1918 (‘Spanish’ flu, H1N1), 1957 (‘Asian’ flu, H2N2), 1968 (‘Hong Kong’ flu, H3N2), and 1977 (‘Russian’ flu, H1N1).16 Spanish flu pandemic claimed about 50 million lives worldwide.17 Recent outbreak of H7N9 in China along with A/Shanghai/1/2013 H7N9 virus with R292K mutation is a serious concern.18, 19 One of the most accessible targets is neuraminidase (NA), which is essential for the release of viral particle from the cell surface and has been the target for the marketed drugs oseltamivir and zanamivir. We recently reported new inhibitors of both N1 and N2 type NAs and also showed their anti-viral activities in the cell-based assay.20 Due to their structural similarity to our previously discovered SARS-CoV 3CLpro inhibitors,21 we screened these NA inhibitors on the SARS-CoV and MERS-CoV 3CLpro and synthesized the analogues of hits to establish the structure–activity-relationship (SAR) on these 3CLpro as reported herein (Table 1 ).
Table 1.
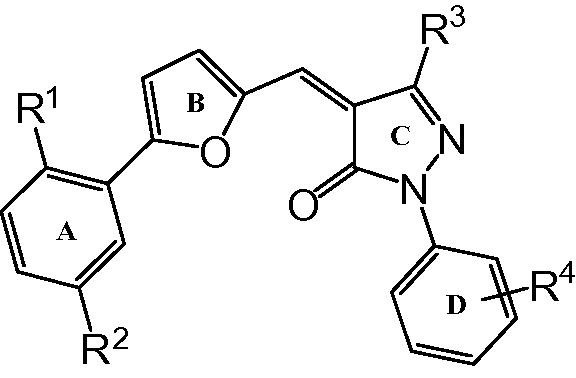
| Sl. No. | R1 | R2 | R3 | R4 | SARS IC50 (μM) | MERS IC50 (μM) |
|---|---|---|---|---|---|---|
| 3a | COOH | Cl | CH3 | 3-COOH | >50 | >50 |
| 3b | COOH | Cl | CF3 | 3-COOH | >50 | >50 |
| 3c | COOH | H | CF3 | 3-COOH | >50 | >50 |
| 3d | COOH | Cl | Ph | 3-COOH | 44.7 ± 5.1 | >50 |
| 3e | COOH | Cl | CH3 | H | >50 | >50 |
| 3f | COOH | Cl | Ph | H | 16.4 ± 0.7 | 12.2 ± 2.2 |
| 3g | COOH | Cl | Ph | 4-F | 20.2 ± 0.3 | 10.1 ± 1.8 |
| 3h | COOH | Cl | Ph | 4-CH(CH3)2 | 6.0 ± 1.2 | 7.3 ± 2.1 |
| 3i | COOH | Cl | Ph | 4-CH(CH3)3 | 5.8 ± 1.5 | 7.4 ± 2.2 |
| 3j | H | H | Ph | H | >50 | >50 |
| 3k | H | H | Ph | COOH | 6.4 ± 1.2 | 5.8 ± 1.6 |
| 3l | H | H | CH3 | COOH | >50 | >50 |
| 3m | COOH | H | Ph | H | 41.2 ± 9.5 | 30.3 ± 4.5 |
| 3n | COOH | H | Ph | 4-F | 37.5 ± 0.7 | >50 |
| 3o | COOH | H | Ph | 4-CH(CH3)2 | 11.7 ± 2.5 | 8.9 ± 1.8 |
| 3p | COOH | H | Ph | 4-CH(CH3)3 | 8.6 ± 2.1 | 7.7 ± 2.2 |
| 3q | COOH | H | Ph | 4-CN | 18.7 ± 4.5 | 9.6 ± 1.6 |
| 3r | COOH | H | Ph | 4-OCH3 | 30.7 ± 5.8 | 8.9 ± 1.8 |
| 3s | No ring A | Ph | COOH | >50 | >50 | |
All the compounds were synthesized as described in Scheme 1 . The pyrazolone core was synthesized by the condensation of substituted hydrazines with various β-ketoesters. The substituted aldehydes were synthesized by Meerwein arylation of substituted diazonium salts with furfural.22 Knoevenagel condensation of substituted aldehydes with various pyrazolones in the presence of a catalytic amount of sodium acetate in acetic acid yielded the final compounds (50–88%). Chromatographic purification was not required for most of the compounds as pure compounds could be obtained by methanolic wash.
Scheme 1.
Reagents and conditions: (a) NaNO2/HCl, 0 °C; (b) furfural, CuCl2·H2O, rt, 48 h; (c) AcOH, reflux, 24 h; (d) AcONa–AcOH, reflux, 3 h.
2. Results and discussion
Absence of NOE signal between the olefinic and R3 phenyl protons in NMR confirmed pyrazolones as a single E-isomer.20 However, methyl substitution at R3 gave geometrical isomers (E/Z) that was inseparable due to rapid interconversion in solution.20
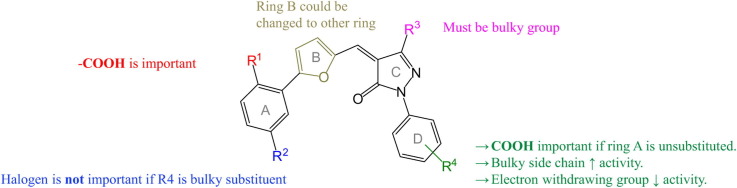
Assay results confirmed that bulkier phenyl group at R3 was essential for the inhibitory activity as compounds 3a–c, 3e and 3l with CH3 or CF3 at this position were unable to inhibit SARS 3CLpro. Carboxylate moiety seemed to be an essential component as compound 3j without it at R1 and R4 was inactive. Removal of R4 carboxylate from 3d (IC50 = 44.7 ± 5.1 μM) but retaining it at R1 resulted in 3f (IC50 = 16.4 ± 0.7 μM) with improved activity. Isosteric replacement of hydrogen by fluorine, 3g (IC50 = 20.2 ± 0.3 μM), caused a slight drop in activity. However, introducing more lipophilic substitution at R4 improved activity as seen in case of 3h (IC50 = 6.0 ± 1.2 μM) and 3i (IC50 = 5.8 ± 1.5 μM) which had about 3-fold improvement in activity in comparison with 3f. In contrast, compound 3k (IC50 = 6.4 ± 1.2 μM) with carboxylate only at R4 was 3-fold and 7-fold more potent compared with 3f and 3m respectively. From these results we conclude that carboxylate moiety is an important pharmacophore and its presence either at R1 or R4 is critical for the activity as 3k was as equipotent as 3h and 3i. Chlorine at R2 slightly changed the activity as we observed 2-fold decrease in the activity of 3m, 3n, 3o and 3p as compared with 3f, 3g, 3h and 3i respectively. Substituent on ring D was also important. While compounds with electron-withdrawing F or CN (3g, 3q) did not show significant difference in activity, electron-donating methoxy group (3r) on ring D caused 2-fold drop in activity as compared with 3f without any substituent.
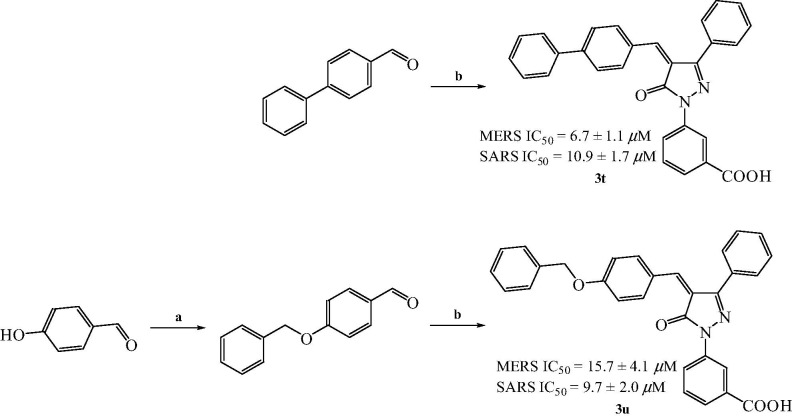
To study the effect of ring B (furan ring), we replaced it with aromatic system to obtain 3t (Scheme 2 ). There was no significant change in the activity of 3t (IC50 = 10.9 ± 1.7 μM) as compared with 3k (IC50 = 6.4 ± 1.2 μM). Ring A of 3k was critical as absence of ring A abolished the activity. We then replaced the rigid link between ring A and B with an ether linkage (3u) to study the effect. Interestingly, the flexible ether linkage on 3u (IC50 = 9.7 ± 2.0 μM) (Scheme 2) did not alter activity as compared with 3k (IC50 = 6.4 ± 1.2 μM).
Scheme 2.
Reagents and conditions: (a) Benzyl bromide, dry DMF, 0 °C, 3 h; (b) 2f, AcONa–AcOH, reflux, 3 h.
From this SAR study, we reach a conclusion that pharmacophores phenyl at R3 and carboxylate either at R1 or R4 are essential for the activity. As modification of ring A and B is tolerated well, this can be further altered to enhance the activity of the compounds.
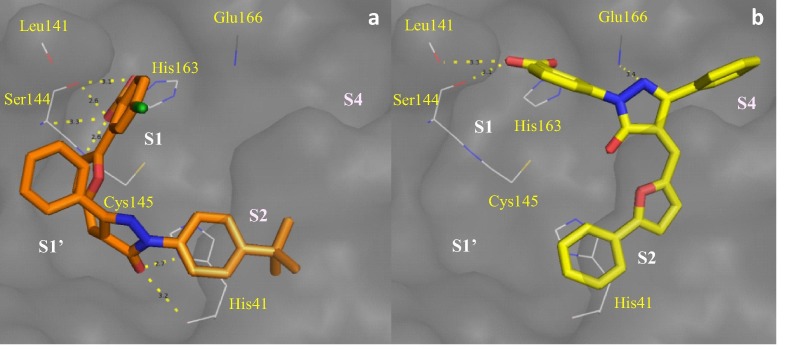
Observed inhibitory activity against SARS 3CLpro was rationalized by docking simulation using ligand bound crystal structure (PDB ID: 2ALV). The docking simulation option (Accurate docking) of iGemdock v2.1 was used to generate 20 solutions. To rationalize the inhibitory effect of these molecules, we must first understand amino acids that constitute the active site of 3CLpro. Active site of 3CLpro can be divided into subsites S1–S6. Subsite S1 is made of vital catalytic residue Cys145 which forms catalytic dyad with His41 to process the polyproteins at eleven sites comprising of conserved Gln followed by small amino acids like Ser, Ala or Gly.23 Other vital component of S1 subsite is the oxyanion hole, formed by the interaction of C-terminal carboxylate anion of the conserved Gln with Gly143, Ser144 and Cys145, which stabilizes the transition state during proteolysis.24, 25 Glu166 at the entrance of the pocket interacts via H-bond with Nɛ2 of the conserved Gln.24 The S2 and S4 subsites contain hydrophobic and bulky side chains like Val, Leu or Phe. Subsites S5 and S6 are near the surface of the active site and has little role in the substrate binding.
Docking (Fig. 1 a) shows that compound 3i with 5.8 μM IC50 binds to SARS 3CLpro at S1, S1′ and S2 subsites to obstruct substrate binding. At S1 subsite, R1 carboxylate of 3i, as a representative of 3f–3i and 3o–3p, forms H-bonds with Gly143, Ser144 and Cys145 which are the vital residues forming the oxyanion hole. This prevents efficient cleavage of substrate by obstructing stabilization of the tetrahedral intermediate formed during the transition state. The carboxylate moiety of these compounds makes an additional H-bond with His163 at the S1 subsite which is responsible for the specificity of protease towards conserved Gln residue. The furan ring interacts with the hydrophobic side chain of Leu27 in S1′ subsite (not shown).
Figure 1.
Docking of 3i (a) with carboxylate at R1 and 3k (b) with carboxylate at R4 on SARS 3CLpro (PDB ID: 2ALV) in order for carboxylate to form H-bonds with S1 subsite.
Due to the hydrophobic nature of S2 subsite, additional alkyl substitution on ring D of 3h, 3i, 3o and 3p enhances interaction with Met49 and Gln189 through hydrophobic contact. This additional contact seems to be responsible for 3-fold enhanced inhibition of 3h and 3i compared with 3f and 3g. In addition to hydrophobic interaction, carbonyl moiety of pyrazolone core in 3f–3i and 3o–3p interacts with His41 through H-bond to destabilize catalytic dyad. Removal of R2 chlorine, comparing 3f and 3g with 3m and 3n respectively, unfavorably reorients the molecule to lose H-bonding with His41, Gly143 and Cys145 (Fig. S1). This explains the observed 2-fold drop in activity on removal of chlorine.
Unlike other compounds in the series, 3k does not seem to occupy S1′ subsite (Fig. 1b). While the carboxylate moiety at R4 position interacts with Leu141, Ser144 and His163, R3 phenyl group prefers the hydrophobic pocket S4. Ring A and B being hydrophobic sit well in S2 subsite. Ring A seems to form a weak π–π stacking with His41 and thus destabilizes the catalytic dyad. Nitrogen atom in the pyrazolone core seems to interact with Glu166 located at the entrance of S1 pocket to prevent binding with Gln of the substrate. Modification of 3k to 3t and 3u did not have significant effect as they prefer binding to similar conformation (not shown).
Since 3CLpro of both MERS-CoV and SARS-CoV are structurally similar, we expressed MERS-CoV 3CLpro and tested our compounds on it. All SARS-CoV 3CLpro inhibitors were found to inhibit MERS-CoV 3CLpro effectively at low micromolar concentrations. Compounds with bulkier groups, electron-withdrawing/donating or hydrophobic, substituents on ring D had IC50 around 10 μM. Similar to SARS 3CLpro, presence of carboxylate moiety seemed essential for inhibiting MERS 3CLpro. Compound 3k with the carboxylate moiety on ring D was the best inhibitor with IC50 of 5.8 ± 1.6 μM.
For the compounds with carboxylate on ring A and unsubstituted or substituted ring D with less bulkier group like fluorine, removal of chlorine atom caused significant loss of activity by comparing 3f (12.2 ± 2.2 μM) with 3m (30.3 ± 4.5 μM) or fluoro-substituted 3g (10.1 ± 1.8 μM) with 3n (>50 μM). However, in the presence of bulkier substituents, 3o (8.9 ± 1.8 μM) and 3p (7.7 ± 2.2 μM), lost activity of 3m and 3n was regained. Removal of chlorine from compounds with bulkier ring D had insignificant effect on inhibition as 3h (7.3 ± 2.1 μM) and 3i (7.4 ± 2.2 μM) showed activity similar to 3o (8.9 ± 1.8 μM) and 3p (7.7 ± 2.2 μM), respectively.
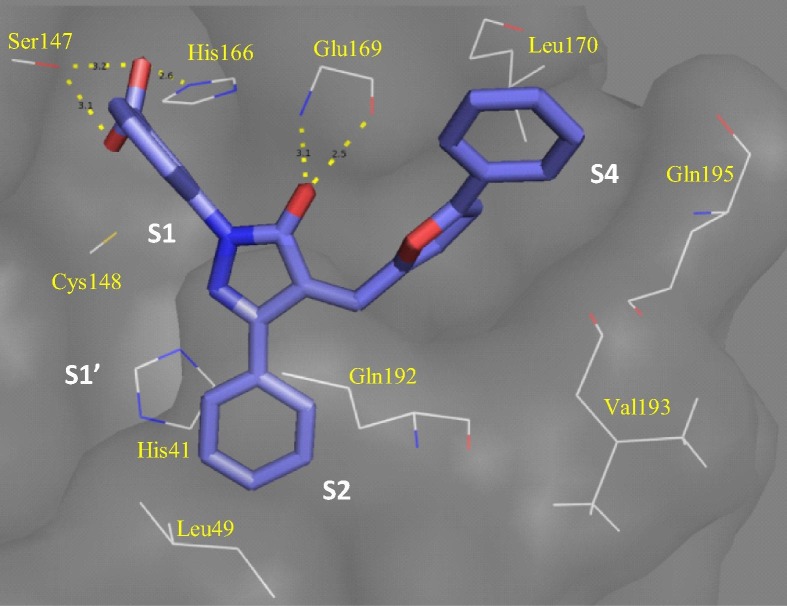
The recently solved crystal structure (PDB ID: 4YLU) of MERS-CoV 3CLpro gave insight into the structural difference between the two 3CLpro.12 Replacement of residue Thr25 in SARS 3CLpro with Met25 in case of MERS 3CLpro has shrunk S2 subsite to prevent latter from accommodating bulkier groups. Unlike SARS-CoV 3CLpro which accommodates ring A and B of 3k, smaller S2 subsite of MERS-CoV 3CLpro accommodates phenyl moiety at R3 (Fig. 2 ). This smaller size of S2 makes those molecules with the R3 phenyl group fit snugly into the active site of MERS-CoV 3CLpro and should be the reason behind most of the active compounds being equipotent. In S2 subsite, R3 interacts with His41 via π–π stacking interaction to perturb the catalytic dyad. At S1 subsite our best inhibitor 3k (Fig. 2), with its carboxylate moiety at ring D, interacts with Ser147 to destabilize oxyanion hole. Carbonyl moiety in pyrazolone core also interacts with Glu169 at the entrance of S2 subsite. At S2 subsite, its R3 phenyl formed π–π stacking with His41 to perturb catalytic dyad. Ring A and B which are primarily hydrophobic interact with the side chains of Leu170, Val193 and Gln195 at S4 subsite. Compound 3f with unsubstituted phenyl group at R3 prefers hydrophobic S2. The ring C forms T-shaped π–π stacking with His41 and destabilizes the oxyanion hole (Fig. S2a). Removal of chlorine atom caused 3-fold loss in activity of 3m as compared with 3f due to loss of T-shaped π–π stacking with His41 (Fig. S2b). In addition, ring D interacts with hydrophobic side chain of Met25. However, when ring D is substituted with bulkier groups, removal of chlorine did not make a significant change in activity as both the molecules maintain similar interactions at S1, S2 and S4 subsites (not shown).
Figure 2.
Docking of 3k on MERS-CoV 3CLpro that has smaller S2 site (PDB ID: 4YLU).
In comparison with inhibiting SARS 3CLpro, most of the active compounds are more potent on MERS 3CLpro. In case of 3g, 3q and 3r, the differences in IC50 reach 2 to 3-fold. The smaller size of S2 in MERS 3CLpro should accommodate the R3 phenyl group and therefore, the substituted ring D is forced to occupy and make better interaction with the S3 and/or S4 subsites. This point is further illustrated with Figure S3 where the electron density to reflect the shape and size of the active sites and the transparent carbon skeleton of the inhibitor are presented.
It is interesting that compounds 3f, 3g and 3m could inhibit H5N1 neuraminidase (NA) with IC50 of 2.8, 2.9, and 13.7 μM, respectively20 and two 3CLpro at low-micromolar concentrations. Although NA and 3CLpro are not homologous proteins, we observed that they share similar arrangement of the electron-rich amino acid residues in the active sites. While NA contains arginine triad, 3CLpro contains cysteine–histidine dyad in the active site and are essential for substrate processing. These active sites are occupied by mainly two pharmacophores, namely carboxylate and phenyl ring. While carboxylate present at R1 interacts with the vital arginine residues in case of H5N1 NA, 20 it destabilizes the oxyanion hole of MERS 3CLpro (Fig. S4). Another pharmacophore, R3 phenyl interacts with Trp403 and His41 by π–π stacking inside the active site of NA and 3CLpro respectively.
3. Conclusion
Based on a common scaffold, we have optimized the NA inhibitors as inhibitors of 3CLpro of SARS-CoV and MERS-CoV. Thus, we have discovered broad-spectrum inhibitors effective against the drug targets of both coronaviruses and avian influenza virus. To the best of our knowledge there is no report regarding common inhibitors of these targets from coronavirus and influenza virus in the literature. While there is still no drug for recently emerged MERS-CoV infection, our report raises a possibility of modifying the known inhibitors of NA to inhibit 3CLpro and vice versa. This approach was tried on MERS by using clinically approved drugs.26, 27, 28 With the escalating cost of drug discovery, concept of drug repurposing is gaining importance. Developing an anti-viral agent with broad-spectrum activity might be advantageous to overcome financial hurdles in the drug discovery. We shall look into modification of the molecules keeping the pharmacophores intact as it seems to be vital for broad-spectrum activity. Compounds with a pyrazole ring surrounded by three hydrophobic groups were also reported to inhibit 3C proteases of human picornaviruses, such as rhinovirus, enterovirus, and coxsackievirus, in addition to 3CLpro of coronaviruses.29 Efforts are being made to co-crystallize these compounds with the enzymes. Cell-based assay would be needed to facilitate further development into more potent inhibitors which could have clinically usefulness.
4. Experimental section
All commercial reagents (Sigma–Aldrich, Acros, Alfa-Aesar) were used as provided and all solvents were of the highest purity. Proton nuclear magnetic resonance (1H NMR) spectra were recorded on a 400 MHz or 500 MHz spectrometer. Carbon nuclear magnetic resonance (13C NMR) spectra were recorded on a 100 MHz or 125 MHz spectrometer. Chemical shifts (δ) are reported in parts per million (ppm) relative to solvent, and the signals are described as br (broad), s (singlet), d (doublet), dd (doublet of doublets), dddd = doublet of doublets of doublets of doublets, t (triplet), and m (multiplet). Coupling constants (J values) are given in Hz. High Resolution Mass Spectrometry (HRMS) was obtained by Bruker BioTOF II mass spectrometer and ESI-TOF-MS spectra were recorded. Reaction was monitored on Silica gel 60 F254 thin layer plates (TLC) from Merck.
4.1. Preparation of substituted furfural
2-Amino-4-chloro benzoic acid (1.0 equiv) was diazotized with NaNO2/HCl at 0–5 °C and to this mixture was gradually added furfural (1.2 equiv) in acetone while maintaining temperature around 0–5 °C followed by addition of copper(II) chloride (0.3 equiv) in water at once. The reaction mixture was maintained below 5 °C for 1 h and then allowed to gradually attain room temperature. The reaction was continued at room temperature for 24 h and the precipitate obtained was filtered and washed with methanol–water mixture to obtain pale yellow compound.20
4.1.1. 4-Chloro-2-(5-formylfuran-2-yl)benzoic acid (1a)
1H NMR (400 MHz, CDCl3): δ 9.643 (s, 1H), 7.894 (d, J = 8.8 Hz, 1H), 7.680 (d, J = 2.0 Hz, 1H), dd, J = 8.4 Hz, 2.0 Hz, 1H), 7.314 (d, J = 4.0 Hz, 1H), 6.821 (d, J = 3.6 Hz, 1H). 13C NMR (100 MHz, CDCl3): 178.0 (CH), 168.9 (C), 156.5 (C), 152.8 (C), 136.9 (C), 131.1 (CH), 130.2 (C), 130.1 (C), 129.2 (CH), 128.7 (CH), 123.2 (C), 111.6 (CH). HRMS (m/z): [MH−] calcd for C12H16ClO4: 248.9966. Found 248.9967 (4.14 g, 57%).
4.1.2. 2-(5-Formylfuran-2-yl)benzoic acid (1b)
1H NMR (400 MHz, CDCl3): δ 9.627 (s, CHO), 7.939 (dd, J = 7.5 Hz, 1.2 Hz, 1H), 7.692 (dd, J = 7.6 Hz, 1.2 Hz, 1H), 7.617 (td, J = 7.6 Hz, 1.6 Hz, 1H), 7.525 (td, J = 7.6 Hz, 1.6 Hz, 1H), 7.318 (d, J = 3.6 Hz, 1H). 13C NMR (100 MHz, CDCl3): 177.8 (CH), 171.8 (C), 157.9 (C), 152.6 (C), 132.2 (CH), 130.1 (CH), 129.9 (CH), 129.7 (CH), 129.6 (C), 129.4 (C), 122.7 (C), 111.4 (CH). HRMS (m/z): [MH+] calcd for C12H9O4: 217.0501. Found 217.0566 (1.6 g, 20%).
4.2. General procedure for synthesis of pyrazolones
To 1 equiv of β-ketoester in 50 ml of acetic acid was added 1 equiv of substituted phenylhydrazine (for HCl salt 1 equiv of triethylamine was added). The content was refluxed for 24–36 h and the contents cooled and solvent was removed in vacuo. To the precipitate in flask was added ethylacetate to suspend the product and was then filtered to obtain pure compound. Thus obtained product was dried to yield substituted pyrazolone. (Note: In some case product isolated was tautomer due to keto-enol tautomerism.)20
4.2.1. 2,5-Diphenyl-2,4-dihydro-3H-pyrazol-3-one (2a)
1H NMR (500 MHz, CDCl3): δ 8.020 (d, J = 8.45 Hz, 2H), 7.769 (m, 2H), 7.470 (m, 5H), 7.254 (t, J = 7.5 Hz, 1H), 3.760 (s, 2H). 13C NMR (125 MHz, CDCl3): 170.1 (C), 154.5 (C), 138.1 (C), 130.8 (C), 130.6 (CH), 128.8 (CH), 128.7 (CH), 125.9 (CH), 125.1 (CH), 118.9 (CH), 39.43 (CH2). HRMS (m/z): [MH−] calcd for C15H11N2O: 235.0882. Found 235.0880 (6.12 g, 75%).
4.2.2. 3-(3-Methyl-5-oxo-4,5-dihydro-1H-pyrazol-1-yl)benzoic acid (2b)
1H NMR (400 MHz, MeOD): δ 8.256 (s, 1H), 7.878 (t, J = 8.0 Hz, 2H), 7.537 (t, J = 8.0 Hz, 2H), 2.200 (s, 3H). 13C NMR (100 MHz, MeOD): 169.0 (C), 151.7 (C), 138.8 (C), 133.1 (C), 130.3 (CH), 129.9 (C), 128.1 (CH), 127.0 (C), 126.6 (CH), 124.0 (C), 123.3 (CH), 120.9 (C). HRMS (m/z): [MH+] calcd for C11H10N2O3: 218.0697. Found 218.0764 (0.99 g, 74%).
4.2.3. 2-(4-Methoxyphenyl)-5-phenyl-2,4-dihydro-3H-pyrazol-3-one (2c)
1H NMR (400 MHz, CDCl3): δ 7.842–7.811 (m, 2H), 7.744–7.720 (m, 2H), 7.435–7.419 (m, 3H), 6.955–6.915 (m, 2H). 13C NMR (100 MHz, CDCl3): 169.9 (C), 157.1 (C), 154.5 (C), 131.4 (C), 130.9 (C), 130.5 (CH), 128.8 (CH), 125.8 (CH), 120.9 (CH), 114.0 (CH), 55.43 (-OCH3), 39.38 (CH2). HRMS (m/z): [MH+] calcd for C16H15N2O2: 267.1134. Found 267.1128.
4.2.4. 4-(5-Oxo-3-phenyl-4,5-dihydro-1H-pyrazol-1-yl)benzonitrile (2d)
1H NMR (400 MHz, CDCl3): δ 8.193–8.168 (m, 2H), 7.780–7.756 (m, 2H), 7.712–7.697 (m, 2H), 7.493–7.449 (m, 3H). 13C NMR (100 MHz, CDCl3): 170.5 (C), 155.6 (C), 141.5 (C), 133.1 (CH), 131.3 (CH), 130.3 (C), 129.0 (CH), 126.1 (CH), 118.8 (CH), 118.4 (C), 108.0 (C), 39.72 (CH2). HRMS (m/z): [MH+] calcd for C16H11N3O: 262.0980. Found 262.0896.
4.2.5. 3-(5-Oxo-3-phenyl-4,5-dihydro-1H-pyrazol-1-yl)benzoic acid (2f)
1H NMR (400 MHz, DMSO-d 6): δ 13.133 (br, s, COOH), 12.023 (br, s, OH), 8.430 (t, J = 2.2 Hz, 1H), 8.108–8.082 (m, 1H), 7.844 (dd, J = 9.05 Hz, 1.45 Hz, 1H), 7.621 (t, J = 9.9 Hz, 1H), 7.433 (t, J = 9.4 Hz, 2H), 7.348 (t, J = 9.1 Hz, 1H), 6.040 (s, 1H). 13C NMR (100 MHz, DMSO-d 6): 167.3 (C), 154.5 (C), 150.5 (C), 139.5 (C), 133.6 (C), 132.1 (C), 129.8 (CH), 129.1 (CH), 128.5 (CH), 126.7 (CH), 125.7 (CH), 125.3 (CH), 121.7 (CH), 85.8 (CH). HRMS (m/z): [MH+] calcd for C16H13N2O3: 281.0926. Found 281.0917 (1.34 g, 49%).
4.3. Preparation of substituted pyrazolone (3a–3s)
Equimolar amount of 1 and 2 were taken in acetic acid and to this was added catalytic amount of sodium acetate (0.1 equiv). As the contents were refluxed the product started to precipitate. The reaction was continued for 3 h and the solvent was removed in vacuo followed by the addition of methanol–water mixture and filtering crude compound which was again thoroughly washed with methanol–water mixture to remove the impurities and any traces of starting material to get final compounds. Some compounds were purified using flash column using ethylacetate: hexane: acetic acid as solvent system.20
4.3.1. 2-(5-((1-(3-Carboxyphenyl)-3-methyl-5-oxo-1,5-dihydro-4H-pyrazol-4-ylidene)methyl)furan-2-yl)-4-chlorobenzoic acid (3a)
1H NMR (500 MHz, DMSO-d 6): δ 13.18 (s, COOH), 8.707 (d, J = 4.0 Hz, 1H), 8.574 (t, J = 2.0 Hz, 1H), 8.199 (ddd, J = 8.1 Hz, 2.5 Hz, 1.0 Hz, 1H), 7.982 (d, J = 2.0 Hz, 1H), 7.769–7.752 (m, 2H), 7.688 (s, 1H), 7.646 (d, J = 2.5 Hz, 8.0 Hz, 1H), 7.591–7.559 (m, 1H), 7.301 (d, J = 4.0 Hz, 1H). 2.531 (s, 3H). 13C NMR (125 MHz, DMSO-d 6): 168.9 (C), 167.5 (C), 162.2 (C), 157.1 (C), 151.6 (C), 151.0 (C), 138.9 (C), 136.2 (C), 132.3 (C), 132.0 (C), 131.6 (CH), 131.1 (C), 130.4 (C), 130.2 (CH), 130.0 (CH), 129.8 (C), 129.4 (C), 128.7 (CH), 127.1 (CH), 125.5 (CH), 122.3 (CH), 122.1 (C), 119.0 (CH), 115.0 (CH). HRMS (m/z): [MH−] calcd for C23H14ClN2O6: 449.0546. Found 449.0542 (0.216 g, 66%).
4.3.2. 2-(5-((1-(3-Carboxyphenyl)-5-oxo-3-(trifluoromethyl)-1,5-dihydro-4H-pyrazol-4-ylidene)methyl)furan-2-yl)-4-chlorobenzoic acid (3b)
1H NMR (400 MHz, DMSO-d 6): δ 13.38 (s, COOH), 8.871 (d, J = 3.2 Hz, 1H), 8.439 (t, J = 1.6 Hz, 1H), 8.127 (d, J = 8.0 Hz, 1H), 8.079 (d, J = 1.2 Hz, 1H), 7.891 (d, J = 7.6 Hz, 1H), 7.803 (d, J = 8.4 Hz, 1H), 7.718 (m, 3H), 7.437 (d, J = 3.2 Hz, 1H). 13C NMR (100 MHz, DMSO-d 6): 168.8 (C), 167.2 (C), 161.4 (C), 159.7 (C), 150.8 (C), 138.0 (C), 136.3 (C), 132.2 (C), 131.7 (CH), 131.4 (C), 130.6 (CH), 130.0 (CH), 129.1 (CH), 128.9, 127.2 (CH), 123.9 (CH), 120.4 (CH), 116.2 (CH), 115.3 (CH). HRMS(m/z): [MH+] calcd for C23H13ClF3N2O6: 505.0420. Found 505.0409 (0.119 g, 59%).
4.3.3. 2-(5-((1-(3-Carboxyphenyl)-5-oxo-3-(trifluoromethyl)-1,5-dihydro-4H-pyrazol-4-ylidene)methyl)furan-2-yl)benzoic acid (3c): (e/z mixture)
1H NMR (500 MHz, DMSO-d 6): δ 13.29 (s, 2-COOH), 8.128 (d, J = 7.5 Hz, 1H), 7.949 (d, J = 7.5 Hz, 1H), 7.880 (d, J = 8.0 Hz, 1H), 7.775 (d, J = 7.5 Hz, 1H), 7.703–7.606 (m, 4H), 7.359 (d, J = 4.0 Hz, 1H). 13C NMR (125 MHz, DMSO-d 6): 169.6 (C), 167.2 (C), 162.0 (C), 161.5 (C), 150.6 (C), 138.1 (C), 132.8 (C), 132.2 (C), 131.6 (CH), 131.1 (CH), 130.5 (CH), 130.0 (CH), 129.7 (CH), 127.1 (CH), 127.0 (C), 123.9 (CH), 127.1 (C), 127.0 (C), 123.9 (C), 120.4 (CH), 115.4 (CH), 114.5 (C). HRMS (m/z): [MH−] calcd for C23H13F3N2O6: 469.0647. Found 469.0633 (0.020 g, 4%).
4.3.4. 2-(5-((1-(3-Carboxyphenyl)-5-oxo-3-phenyl-1,5-dihydro-4H-pyrazol-4-ylidene)methyl)furan-2-yl)-4-chlorobenzoic acid (3d)
1H NMR (500 MHz, DMSO-d 6): δ 13.29 (s, COOH), 8.800 (d, J = 3.7 Hz, 1H), 8.637 (t, J = 1.9 Hz, 1H), 8.264 (ddd, J = 8.3 Hz, 2.0 Hz, 0.9 Hz, 1H), 7.998 (d, J = 2.1 Hz, 1H), 7.816–7.753 (m, 4H), 7.733 (d, J = 8.3 Hz, 1H), 7.627–7.595 (m, 5H), 7.560 (s, 1H), 7.345 (d, J = 4.0 Hz, 1H). 13C NMR (125 MHz, DMSO-d 6): 168.9 (C), 167.4 (C), 162.3 (C), 157.6 (C), 152.3 (C), 151.0 (C), 138.8 (C), 136.1 (C), 132.0 (C), 131.5 (CH), 131.2 (C), 130.7 (C), 130.5 (CH), 130.1 (CH), 129.8 (CH), 129.6(CH), 129.1 (C), 129.0 (CH), 128.7 (CH), 128.0 (CH), 126.1 (CH), 122.9 (CH), 120.5 (C), 119.5 (CH), 115.3 (CH). HRMS (m/z): [MH+] calcd for C28H18ClN2O6: 513.09. Found 513.0997 (0.145 g, 71%).
4.3.5. 4-Chloro-2-(5-((3-methyl-5-oxo-1-phenyl-1,5-dihydro-4H-pyrazol-4-ylidene)methyl)furan-2-yl)benzoic acid (3e)
1H NMR (400 MHz, DMSO-d 6): 8.690 (d, J = 4.0 Hz, 1H), 7.958–7.912 (m, 3H), 7.779 (d, J = 8.0 Hz, 1H), 7.645 (m, 2H), 7.460 (m, 2H), 7.278 (d, J = 4.0 Hz, 1H), 7.213 (m, 2H), 2.475 (s, 3H –CH3), 2.321 (s, 3H, –CH3). 13C NMR (100 MHz, DMSO-d 6): 168.9, 162.0, 156.9, 151.0, 138.7, 136.2, 132.3, 133.6 (CH), 131.1 (CH), 130.0 (CH), 129.8 (CH), 129.5 (CH), 129.4 (CH), 128.7 (CH), 126.9 (CH), 125.0 (CH), 122.4, 118.6 (CH), 118.3 (CH), 114.9 (CH), 114.6 (CH), 17.73, 13.30 (-CH3), 13.29 (–CH3). HRMS (m/z): [MH−] calcd for C22H14ClN2O4: 405.0642. Found 405.0641.
4.3.6. 4-Chloro-2-(5-((5-oxo-1,3-diphenyl-1,5-dihydro-4H-pyrazol-4-ylidene)methyl)furan-2-yl)benzoic acid (3f)
1H NMR (500 MHz, DMSO-d 6): δ 8.983 (d, J = 4.0 Hz, 1H), 8.193 (d, J = 7.5 Hz, 2H), 7.995(d, J = 1.5 Hz, 1H), 7.922 (d, J = 8.5 Hz, 1H), 7.875–7.857 (m, 2H), 7.707 (s, 1H), 7.681–7.643 (m, 4H), 7.550 (t, J = 8.5 Hz, 3H), 7.338 (d, J = 4.0 Hz, 1H), 7.307 (t, J = 7.0 Hz, 1H). 13C NMR (125 MHz, DMSO-d 6): 169.0 (C), 162.2 (C), 157.5 (C), 152.0 (C), 151.0 (C), 138.6 (C), 136.1 (C), 131.6 (C), 131.5 (CH), 131.2 (CH), 130.8 (C), 130.4 (CH), 130.1 (CH), 129.6 (CH), 129.4 (CH), 129.2 (C), 129.1 (C), 129.0 (CH), 128.7 (CH), 127.7 (C), 125.5 (CH), 120.7 (C), 119.3 (CH), 115.2(CH). HRMS (m/z): [MH+] calcd for C27H18ClN2O4: 469.0961. Found 469.0950 (0.187 g, 88%).
4.3.7. 4-Chloro-2-(5-((1-(4-fluorophenyl)-5-oxo-3-phenyl-1,5-dihydro-4H-pyrazol-4-ylidene)methyl)furan-2-yl)benzoic acid (3g)
1H NMR (400 MHz, DMSO-d 6): δ 13.44 (s, COOH), 8.768 (d, J = 4.0, 1H), 8.010–7.961 (m, 3H), 7.751–7.708 (m, 3H), 7.620–7.581 (m, 4H), 7.531 (s, 1H), 7.335–7.291 (m, 3H). 13C NMR (100 MHz, DMSO-d 6): 169.0 (C), 162.0 (C), 160.9 (C), 158.5 (C), 157.5 (C), 152.0 (C), 151.0 (C), 136.1 (C), 135.1 (C), 131.5 (CH), 131.3 (CH), 131.1 (C), 130.7 (C), 130.4 (CH), 130.1 (CH), 129.6 (CH), 129.1 (CH), 129.0 (C), 128.7 (CH), 127.8 (C), 121.3 (CH), 121.2 (CH), 120.5 (C), 116.2 (CH), 116.0 (CH), 115.2 (CH). HRMS (m/z): [MH−] calcd for C27H15ClFN2O4: 485.0704. Found 485.0691 (0.297 g, 76%).
4.3.8. 4-Chloro-2-(5-((1-(4-isopropylphenyl)-3-methyl-5-oxo-1,5-dihydro-4H-pyrazol-4-ylidene)methyl)furan-2-yl)benzoic acid (3h)
1H NMR (400 MHz, DMSO-d 6): δ 8.788 (d, J = 3.9 Hz, 1H), 7.989 (d, J = 2.4 Hz, 1H), 7.892 (d, J = 8.8 Hz, 2H), 7.757–7.713 (m, 3H), 7.628–7.587 (m, 4H), 7.535 (s, 1H), 7.361–7.326 (m, 3H), 2.957–2.889 (sep, J = 17.1 Hz, 10.1 Hz, 3.3 Hz, 1H), 1.235 (d, J = 6.9 Hz, 6H). 13C NMR (100 MHz, DMSO-d 6): 169.0 (C), 162.0 (C), 157.4 (C), 151.7 (C), 151.0 (C), 145.8 (C), 136.4 (C), 136.1 (C), 131.5 (C), 131.1 (C), 130.8 (C), 130.3 (C), 130.1 (C), 129.6 (C), 129.2 (C), 129.0 (C), 128.7 (C), 127.6 (C), 127.2 (C), 120.1 (C), 119.5 (C), 115.1 (C), 33.45 (CH–CH3), 24.35 (CH–CH 3). HRMS (m/z): [MH−] calcd for C30H22ClN2O4: 509.1268. Found 509.1242 (0.095 g, 47%).
4.3.9. (Z)-2-(5-((1-(4-(tert-Butyl)phenyl)-5-oxo-3-phenyl-1,5-dihydro-4H-pyrazol-4-ylidene)methyl)furan-2-yl)-4-chlorobenzoic acid (3i)
1H NMR (400 MHz, DMSO-d 6): δ 8.794 (d, J = 3.8 Hz, 1H), 7.981 (d, J = 1.6 Hz, 1H), 7.902 (d, J = 8.8 Hz, 1H), 7.757–7.719 (m, 3H), 7.629–7.591 (m, 4H), 7.532 (s, 1H), 7.505 (d, J = 8.8 Hz, 1H), 7.334 (d, J = 3.6 Hz, 1H), 1.311 (s, 1H). 13C NMR (100 MHz, DMSO-d 6): 169.0 (C), 162.0 (C), 157.4 (C), 151.7 (C), 151.0 (C), 148.0 (C), 136.1 (C), 131.5 (CH), 131.1 (C), 131.0 (CH), 130.8 (C), 130.3 (CH), 130.1 (CH), 129.6 (CH), 129.2 (C), 129.0 (CH), 128.7 (CH), 127.6 (CH), 126.1 (C), 120.7 (C), 119.2 (CH), 115.15 (CH). HRMS (m/z): [MH−] calcd for C31H24ClN2O4: 523.1430. Found 523.1421 (0.065 g, 31%).
4.3.10. 2,5-Diphenyl-4-((5-phenylfuran-2-yl)methylene)-2,4-dihydro-3H-pyrazol-3-one (3j)
1H NMR (400 MHz, DMSO-d 6): 8.811 (d, J = 3.2 Hz, 1H), 8.034 (dd, J = 1.0 Hz, 8.4 Hz, 2H), 7.960 (d, J = 5.2 Hz, 2H), 7.766–7.747 (m, 2H), 7.631 (s, 1H), 7.617–7.585 (m, 3H), 7.521–7.441 (m, 6H), 7.243 (t, J = 7.2 Hz, 1H). 13C NMR (100 MHz, DMSO-d 6): 162.5 (C), 160.4 (C), 151.9 (C), 150.7 (C), 138.6 (C), 131.1 (C), 130.8 (CH), 129.8 (CH), 129.6 (CH), 129.0 (CH), 129.0, 128.8 (CH), 128.7 (CH), 128.4 (CH), 125.2 (CH), 125.0 (CH), 120.2 (C), 119.3 (CH), 110.8(CH). HRMS (m/z): [MH−] calcd for C26H19N2O2: 391.1447. Found 391.1442 (0.157 g, 69%).
4.3.11. 3-(5-Oxo-3-phenyl-4-((5-phenylfuran-2-yl)methylene)-4,5-dihydro-1H-pyrazol-1-yl)benzoic acid (3k)
1H NMR (400 MHz, DMSO-d 6): δ 13.14 (s, COOH), 8.783 (d, J = 1.4 Hz, 1H), 8.623 (t, J = 1.8 Hz, 1H), 8.272 (m, 1H), 7.965 (m, 2H), 7.802 (m, 3H), 7.620 (m, 5H), 7.505 (m, 4H). 13C NMR (100 MHz, DMSO-d 6): 167.5 (C), 162.4 (C), 160.8 (C), 152.5 (C), 150.5 (C), 139.0 (C), 132.1 (C), 131.5 (CH), 130.9 (C), 130.7 (CH), 130.4 (CH), 129.7 (CH), 129.6 (CH), 129.3 (CH), 129.1 (CH), 128.8 (CH), 125.9 (CH), 125.8 (CH), 122.9 (CH), 119.4 (CH), 119.2 (C), 112.2 (CH). HRMS (m/z): [MH−] calcd for C27H17N2O4: 433.1188. Found 433.1183 (0.069 g, 55%).
4.3.12. 3-(3-Methyl-5-oxo-4-((5-phenylfuran-2-yl)methylene)-4,5-dihydro-1H-pyrazol-1-yl)benzoic acid (3l): (e/z mixture)
1H NMR (500 MHz, DMSO-d 6): δ 8.699 (d, J = 3.4 Hz, 1H), 8.559 (s, 1H), 8.209 (d, J = 7.0 Hz, 1H), 7.978 (d, J = 7.5 Hz, 2H), 7.925 (d, J = 7.5 Hz, 1H), 7.813 (d, J = 4.0 Hz, 1H), 7.760–7.743 (m, 3H), 7.582–7.531 (m, 5H), 7.503–7.450 (m, 3H), 2.749 (s, [3/2] H), 2.358 (s, 3H). 13C NMR (125 MHz, DMSO-d 6): 162.3 (C), 161.5 (C), 160.1 (C), 151.7 (C), 150.5 (C), 150.0 (C), 148.7 (C), 139.0 (C), 132.0 (C), 131.6 (CH), 130.6 (CH), 130.5 (CH), 130.2 (CH), 129.9 (CH), 129.7 (C), 129.8 (C), 129.0 (C), 128.3 (CH), 127.1 (CH), 125.7 (CH), 125.6 (CH), 125.5 (CH), 122.3 (CH), 122.1 (CH), 121.0 (C), 119.4 (C), 119.0 (CH), 118.7 (CH), 111.9 (CH), 111.7 (CH), 18.32 (CH3), 13.33 (CH3). HRMS (m/z): [MH−] calcd for C22H15N2O4: 371.1032. Found 371.1028 (0.057 g, 53%).
4.3.13. 2-(5-((5-Oxo-1,3-diphenyl-1,5-dihydro-4H-pyrazol-4-ylidene)methyl)furan-2-yl)benzoic acid (3m)
1H NMR (400 MHz, DMSO-d 6): δ 13.29 (s, COOH), 8.833 (d, J = 4.9 Hz, 1H), 8.024 (dd, J = 8.7 Hz, 1.1 Hz, 2H), 7.890 (dd, J = 7.7 Hz, 0.7 Hz, 1H), 7.775 (m, 2H), 7.725 (dd, J = 7.6 Hz, 1.2 Hz, 1H), 7.653 (m, 5H), 7.527 (m, 3H), 7.266 (m, 2H). 13C NMR (100 MHz, DMSO-d 6): 169.8 (C), 162.3 (C), 159.6 (C), 152.0 (C), 150.8 (C), 138.7 (C), 132.6 (C), 131.5 (CH), 131.2 (CH), 130.9 (C), 130.5 (CH), 130.4 (CH), 129.6 (CH), 129.4 (CH), 129.3 (CH), 129.0 (CH), 128.6 (C), 128.1 (CH), 127.2 (C), 125.5 (C), 120.0 (C), 119.2 (C), 114.3 (C). HRMS (m/z): [MH−] calcd for C27H17N2O4: 433.1188. Found 433.174 (0.223 g, 50%).
4.3.14. 2-(5-((1-(4-Fluorophenyl)-5-oxo-3-phenyl-1,5-dihydro-4H-pyrazol-4-ylidene)methyl)furan-2-yl)benzoic acid (3n)
1H NMR (500 MHz, DMSO-d 6): 8.807 (d, J = 3.5 Hz, 1H), 8.021–7.993 (m, 2H), 7.866 (d, J = 7.5 Hz, 1H), 7.750–7.704 (m, 3H), 7.651–7.547 (m, 5H), 7.509 (s, 1H), 7.332 (t, J = 9.0 Hz, 2H), 7.245 (d, J = 3.5 Hz, 1H). 13C NMR (125 MHz, DMSO-d6): 169.9 (C), 162.1 (C), 160.6 (C), 159.6 (C), 152.0 (C), 150.8 (C), 135.1 (C), 132.6 (C), 131.4 (CH), 131.3 (CH), 130.8 (C), 130.4 (CH), 129.6 (CH), 129.5 (CH), 129.3 (C), 129.0 (CH), 128.6 (C), 128.2 (CH), 127.2 (C), 121.3 (CH), 121.2 (CH), 119.8, 116.2 (CH), 116.0 (CH), 114.3 (CH). HRMS (m/z): [MH−] calcd for C27H16FN2O4: 451.1094. Found 451.1095.
4.3.15. (Z)-2-(5-((1-(4-(tert-Butyl)phenyl)-5-oxo-3-phenyl-1,5-dihydro-4H-pyrazol-4-ylidene)methyl)furan-2-yl)benzoic acid (3p)
1H NMR (400 MHz, DMSO-d 6): δ 8.834 (d, J = 3.9 Hz, 1H), 7.913 (t, J = 8.8 Hz, 3H), 7.764–7.704 (m, 3H), 7.672–7.495 (m, 9H), 7.263 (d, J = 4.0 Hz, 1H), 1.312 (s, 9H). 13C NMR (100 MHz, DMSO-d 6): 169.8 (C), 162.1 (C), 159.5 (C), 151.7 (C), 150.8 (C), 148.0 (C), 136.2 (C), 132.6 (C), 131.5 (CH), 131.1 (CH), 130.9 (C), 130.4 (CH), 130.3 (CH), 129.6 (CH), 129.5 (CH), 129.3 (CH), 129.0 (CH), 128.0 (CH), 127.3 (C), 126.1 (CH), 120.1 (C), 119.2 (CH), 114.3 (CH). HRMS (m/z): [MH+] calcd for C31H26N2O4: 491.1965. Found 491.1969 (0.080 g, 44%).
4.3.16. 2-(5-((1-(4-Cyanophenyl)-5-oxo-3-phenyl-1,5-dihydro-4H-pyrazol-4-ylidene)methyl)furan-2-yl)benzoic acid (3q)
1H NMR (400 MHz, DMSO-d 6): δ 13.27 (s, COOH), 8.784 (d, J = 3.9 Hz, 1H), 8.249 (d, J = 8.8 Hz, 2H), 7.947 (d, J = 8.8 Hz, 2H), 7.882 (d, J = 7.4 Hz, 1H), 7.771 (m, 2H), 7.722 (d, J = 7.2 Hz, 1H), 7.604 (m, 5H), 7.538 (s, 1H), 7.271 (d, J = 3.9 Hz, 1H). 13C NMR (100 MHz, DMSO-d 6): 169.8 (C), 162.8 (C), 160.1 (C), 153.3 (C), 150.7 (C), 142.1 (C), 134.0 (C), 132.6 (C), 131.8 (C), 131.5 (C), 130.7 (C), 130.5 (C), 129.6 (C), 129.4 (C), 129.1 (C), 128.8 (C), 127.2 (C), 119.3 (C), 118.7 (C), 114.6 (C), 107.0 (C). HRMS(m/z): [MH−] calcd for C28H16N3O4: 458.1146. Found 458.1137 (0.121 g, 66%).
4.3.17. 2-(5-((1-(4-Methoxyphenyl)-5-oxo-3-phenyl-1,5-dihydro-4H-pyrazol-4-ylidene)methyl)furan-2-yl)benzoic acid (3r)
1H NMR (400 MHz, DMSO-d 6): δ 13.26 (s, COOH), 8.832 (d, J = 3.9 Hz, 1H), 7.884 (m, 3H), 7.758 (m, 3H), 7.650 (m, 5H), 7.509 (s, 1H), 7.252 (d, J = 4.0 Hz, 1H), 7.065 (m, 2H), 3.795 (s, 3H). 13C NMR (100 MHz, DMSO-d 6): 169.8 (C), 161.8 (C), 159.4 (C), 157.1 (C), 150.8 (C), 132.6 (C), 131.9 (C), 131.4 (C), 131.0 (C), 130.4 (C), 130.3 (C), 129.6 (C), 129.5 (C), 129.3 (C), 129.0 (C), 127.9 (C), 127.3 (C), 121.3 (C), 120.1 (C), 114.5 (C), 114.2 (C), 55.78 (OCH 3). HRMS(m/z): [MH−] calcd for C28H20N2O5: 463.1299. Found 463.1284 (0.157 g, 85%).
4.3.18. 3-(4-(Furan-2-ylmethylene)-5-oxo-3-phenyl-4,5-dihydro-1H-pyrazol-1-yl)benzoic acid (3s)
1H NMR (500 MHz, DMSO-d 6): δ 13.09 (s, COOH), 8.735 (d, J = 4.0 Hz, 1H), 8.625 (t, J = 2.0 Hz, 1H), 8.289 (s, 1H), 8.260 (dd, J = 8.0 Hz, 1.0 Hz, 1H), 7.817 (d, J = 7.5 Hz, 1H), 7.779–7.761 (m, 2H), 7.625–7.592 (m, 5H), 6.981 (d, J = 3.5 Hz, 1H). 13C NMR (125 MHz, DMSO-d 6): 172.5 (C), 167.5 (C), 162.4 (C), 152.5 (C), 151.7 (CH), 150.7 (C), 138.3 (C), 132.6 (CH), 132.1 (C), 130.6 (C), 130.5 (CH), 129.9 (CH), 129.6 (CH), 129.1 (CH), 126.4 (CH), 126.1 (CH), 123.0 (CH), 120.0 (C), 119.5 (CH), 115.9 (CH). HRMS (m/z): [MH+] calcd for C21H15N2O4: 359.1037. Found 359.1026 (0.094 g, 79%).
4.3.19. (Z)-3-(4-([1,1′-Biphenyl]-4-ylmethylene)-5-oxo-3-phenyl-4,5-dihydro-1H-pyrazol-1-yl)benzoic acid (3t)
1H NMR (400 MHz, DMSO-d 6): δ 13.08 (s, COOH), 8.606 (dd, J = 2.0 Hz, 8.4 Hz, 3H), 8.205 (dd, J = 2.4, 8.4 Hz, 1H), 7.855–7.727 (m, 8H), 7.575–7.543 (m, 4H), 7.492 (dt, J = 1.6 Hz, 7.2 Hz, 2H), 7.4147–7.3942 (m, 1H). 13C NMR (100 MHz, DMSO-d 6): 167.0 (C), 162.3 (C), 162.2 (C), 153.6 (C), 151.4 (C), 149.9 (C), 145.2 (C), 139.2 (C), 138.8 (C), 136.2 (C), 134.5 (C), 132.6 (C), 132.0 (C), 131.3 (C), 130.7 (C), 130.4 (C), 129.7 (C), 128.9 (C), 128.3 (C), 127.9 (C), 126.7 (C), 126.3 (C), 125.4 (C), 122.3 (C), 120.6 (C), 118.8 (C). HRMS (m/z): [MH−] calcd for C29H19N2O3: 443.1401. Found 443.1389 (0.163 g, 52%).
4.3.20. (Z)-3-(4-(4-(Benzyloxy)benzylidene)-5-oxo-3-phenyl-4,5-dihydro-1H-pyrazol-1-yl)benzoic acid (3u)
1H NMR (400 MHz, DMSO-d 6): δ 13.14 (s, COOH), 8.630–8.617 (m, 3H), 8.245–8.216 (m, 1H), 7.801–7.740 (m, 1H), 7.731–7.716 (m, 3H), 7.602–7.563 (m, 4H), 7.482–7.461 (m, 2H), 7.423–7.343 (m, 3H), 7.200 (d, J = 9.2 Hz, 2H), 5.246 (s, 2H). 13C NMR (100 MHz, DMSO-d 6): 167.5 (C), 163.6 (C), 162.6 (C), 153.8 (C), 151.1 (CH), 138.9 (C), 137.7 (CH), 136.7 (C), 132.0 (C), 130.9 (C), 130.4 (CH), 129.8 (CH), 129.5 (CH), 129.4 (CH), 129.0 (CH), 128.6 (CH), 128.4 (CH), 126.5 (C), 126.0 (CH), 123.1 (CH), 122.7 (C), 119.6 (CH), 115.6 (CH), 70.22 (CH2). HRMS (m/z): [MH−] calcd for C30H21N2O4: 473.1507. Found 473.1501 (0.069 g, 53%).
4.4. Expression and purification of SARS-CoV and MERS-CoV 3CLpro
The expression and purification of SARS-CoV followed our reported procedure.30 For expression of MERS-CoV 3CLpro, the Factor Xa cleavage site (IEGR) and the 3CLpro (accession KJ361502.1, Ser3248–Gln3553) DNA sequence was synthesized and cloned into the pET32 expression vector by Mission Biotech. Company (Taiwan) and was transformed into Escherichia coli BL21 (DE3). A 10 ml overnight culture of a single transformant was used to inoculate 1L of fresh LB medium containing 100 μg/ml ampicillin, The cells were grown at 37 °C to A600 = 0.8 and induced with 0.4 mM isopropyl-β-thiogalactopyranoside (IPTG) for 22 h at 16 °C. The cells were harvested by centrifugation at 7000×g for 15 min and the pellet was suspended in lysis buffer (12 mM Tris–HCl, 120 mM NaCl, 0.1 mM EDTA, and 5 mM DTT, pH 7.5). A French-press instrument (Constant Cell Disruption System) was used to disrupt the cells at 20000 psi and centrifuged at 20,000×g for 1 h to discard the debris. The cell-free extract was loaded onto Ni–NTA column which was equilibrated with lysis buffer containing 5 mM imidazole. After exhaustive washing with lysis buffer, the imidazole concentration of the washing buffer was increased to 30 mM. The protein eluted by lysis buffer with 300 mM imidazole were dialyzed against lysis buffer to removed imidazole and then Factor Xa was added to the fusion proteins to a final concentration of 1% (w/w) and incubated at 16 °C for 24 h to remove the His-tag. Subsequently, the processed MERS-CoV 3CLpro was passed through a Ni–NTA column for purification. The protein concentration was determined by the protein assay kit (BioRad, USA) and BSA was used as standard.
4.5. Measurement of IC50
A fluorometric assay30 was utilized to determine the inhibition constants of the prepared samples. Fluorogenic peptide, Dabcyl-KTSAVLQSGFRKME-Edans, was used as the substrate, and the enhanced fluorescence due to the cleavage of this substrate catalyzed by the 3CLpro was monitored at 538 nm with excitation at 355 nm. The IC50 value of the individual sample was measured in a reaction mixture containing 50 nM SARS 3CLpro or 300 nM MERS-Cov 3CLpro and 10 μM of the fluorogenic substrate in 20 mM Bis–Tris (pH 7.0).
Acknowledgement
This work was supported by Academia Sinica, Taiwan.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.bmc.2016.05.013.
Supplementary data
Supplementary Figures S1–S3.
References and notes
- 1.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Science. 2003;300:1394. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 2.Thiel V., Herold J., Schelle B., Siddell S.G. J. Virol. 2001;75:6676. doi: 10.1128/JVI.75.14.6676-6681.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thiel V., Ivanov K.A., Putics A., Hertzig T., Schelle B., Bayer S., Weissbrich B., Snijder E.J., Rabenau H., Doerr H.W., Gorbalenya A.E., Ziebuhr J. J. Gen. Virol. 2003;84:2305. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 4.Kuo C.J., Liang P.H. ChemBioEng. Rev. 2015;2:118. [Google Scholar]
- 5.Kumar V., Jung Y.S., Liang P.H. Expert Opin. Ther. Pat. 2008;23:1337. doi: 10.1517/13543776.2013.823159. [DOI] [PubMed] [Google Scholar]
- 6.Baez-Santos Y.M., St John S.E., Mesecar A.D. Antivir. Res. 2015;115:21. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramajayam R., Tan K.P., Liang P.H. Biochem. Soc. Trans. 2011;39:1371. doi: 10.1042/BST0391371. [DOI] [PubMed] [Google Scholar]
- 8.Zhao Q., Weber E., Yang H. Recent Pat. Antiinfect. Drug Discov. 2013;8:150. doi: 10.2174/1574891x113089990017. [DOI] [PubMed] [Google Scholar]
- 9.Tong T.R. Expert Opin. Ther. Pat. 2009;19:415. doi: 10.1517/13543770802600698. [DOI] [PubMed] [Google Scholar]
- 10.Hilgenfeld R., Peiris M. Antivir. Res. 2013;100:286. doi: 10.1016/j.antiviral.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.WHO, Middle East respiratory syndrome coronavirus (MERS-CoV), http://www.who.int/mediacentre/factsheets/mers-cov/en/ (10.05.16).
- 12.Tomar S., Johnston M.L., St John S.E., Osswald H.L., Nyalapatla P.R., Paul L.N., Ghosh A.K., Denison M.R., Mesecar A.D. J. Biol. Chem. 2015;290:19403. doi: 10.1074/jbc.M115.651463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kilianski A., Baker S.C. Antivir. Res. 2014;101:105. doi: 10.1016/j.antiviral.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barnard D.L., Kumaki Y. Future Virol. 2011;6:615. doi: 10.2217/fvl.11.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krug R.M., Lamb R.A. 4th ed. Lippincott Williams and Wilkins; Philadelphia: 2001. Orthomyxoviridae: The Viruses and their Replication. [Google Scholar]
- 16.Watanabe Y., Ibrahim M.S., Suzuki Y., Ikuta K. Trends Microbiol. 2012;20:11. doi: 10.1016/j.tim.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 17.Taubenberger J.K., Morens D.M. Emer. Infect. Dis. 1918;12:15. doi: 10.3201/eid1201.050979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao R.B., Cao B., Hu Y.W., Feng Z.J., Wang D.Y., Hu W.F., Chen J., Jie Z.J., Qiu H.B., Xu K., Xu X.W., Lu H.Z., Zhu W.F., Gao Z.C., Xiang N.J., Shen Y.Z., He Z.B., Gu Y., Zhang Z.Y., Yang Y., Zhao X., Zhou L., Li X.D., Zou S.M., Zhang Y., Li X.Y., Yang L., Guo J.F., Dong J., Li Q., Dong L.B., Zhu Y., Bai T., Wang S.W., Hao P., Yang W.Z., Zhang Y.P., Han J., Yu H.J., Li D.X., Gao G.F., Wu G.Z., Wang Y., Yuan Z.H., Shu Y.L. N. Engl. J. Med. 2013;368:1888. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 19.CDC. Avian Influenza A (H7N9) Virus. http://www.cdc.gov/flu/avianflu/h7n9-virus.htm (accessed 10.05.16).
- 20.Kumar V., Chang C.K., Tan K.P., Jung Y.S., Chen S.H., Cheng Y.S., Liang P.H. Org. Lett. 2014;16:5060. doi: 10.1021/ol502410x. [DOI] [PubMed] [Google Scholar]
- 21.Ramajayam R., Tan K.P., Liu H.G., Liang P.H. Bioorg. Med. Chem. 2010;18:7849. doi: 10.1016/j.bmc.2010.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Racane L., Tralic-Kulenovic V., Boykin D.W., Karminski-Zamola G. Molecules. 2003;8:342. doi: 10.3390/11050325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan K., Wei P., Feng Q., Chen S., Huang C., Ma L., Lai B., Pei J., Liu Y., Chen J., Lai L. J. Biol. Chem. 2004;279:1637. doi: 10.1074/jbc.M310875200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsu M.F., Kuo C.J., Chang K.T., Chang H.C., Chou C.C., Ko T.P., Shr H.L., Chang G.G., Wang A.H.J., Liang P.H. J. Biol. Chem. 2005;280:31257. doi: 10.1074/jbc.M502577200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu T.C., Zhang Y., Li L.W., Wang K.F., Chen S., Chen J., Ding J.P., Jiang H.L., Shen X. Virology. 2009;388:324. doi: 10.1016/j.virol.2009.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sisk J.M., Frieman M.B. ACS Infect. Dis. 2015;1:401. doi: 10.1021/acsinfecdis.5b00089. [DOI] [PubMed] [Google Scholar]
- 27.de Wilde A.H., Jochmans D., Posthuma C.C., Zevenhoven-Dobbe J.C., van Nieuwkoop S., Bestebroer T.M., van den Hoogen B.G., Neyts J., Snijder E.J. Antimicrob. Agents Chemother. 2014;58:4875. doi: 10.1128/AAC.03011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dyall J., Coleman C.M., Hart B.J., Venkataraman T., Holbrook M.R., Kindrachuk J., Johnson R.F., Olinger G.G., Jahrling P.B., Laidlaw M., Johansen L.M., Lear-Rooney C.M., Glass P.J., Hensley L.E., Frieman M.B. Antimicrob. Agents Chemother. 2014;58:4885. doi: 10.1128/AAC.03036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuo C.J., Liu H.G., Lo Y.K., Seong C.M., Lee K.I., Jung Y.S., Liang P.H. FEBS Lett. 2009;583:549. doi: 10.1016/j.febslet.2008.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuo C.J., Chi Y.H., Hsu J.T., Liang P.H. Biochem. Biophys. Res. Commun. 2004;318:862. doi: 10.1016/j.bbrc.2004.04.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–S3.