Abstract
We used a progressive elimination strategy to identify oocyte-specific WEE2 kinase inhibitors for potential non-hormonal contraceptives that target meiosis. Beginning with an in-house library of over 300,000 compounds, virtual high throughput screening identified 57 WEE2 inhibitors with preferential predicted binding over the somatic variant WEE1. Seven compounds were further evaluated in vitro by enzyme-linked immunosorbent assay to measure biochemical inhibition on WEE1 and WEE2 phosphorylation of CDK1. To assess specificity, we evaluated WEE2-mediated inhibition of meiosis using in vitro oocyte fertilization, and WEE1-mediated inhibition of mitosis using a somatic cell proliferation assay. Our results from these assays identified three candidates for further development: 6-(2,6-dichlorophenyl)-2-((4-(2-(diethylamino)ethoxy) phenyl)amino)-8-methylpyrido[2,3-d]pyrimidin-7(8H)-one (2), 6-(2,6-dichlorophenyl)-8-methyl-2-((4-morpholinophenyl) amino)pyrido[2,3-d]pyrimidin-7(8H)-one (12), and 3-((6-(2,6-dichlorophenyl)-8-methyl-7-oxo-7,8-dihydropyrido[2,3-d]pyrimidin-2-yl)amino)benzoic acid (16).
Keywords: Cell cycle, Inhibitor, Meiosis, Oocyte, WEE kinase
Introduction
In somatic cells, WEE1, a serine/threonine protein kinase, is a critical negative regulator of the eukaryotic cell cycle that functions as a checkpoint to ensure adequate cell growth has been achieved before entry into mitosis.[1] By phosphorylating tyrosine 15 on cyclin dependent kinase 1 (CDK1, also known as the cell division cycle 2 or CDC2) located in the cell nucleus, WEE1 inhibits CDK1 from complexing with the regulatory subunit, cyclin B, and prevents formation of M-phase promoting factor (MPF), transiently arresting cell cycle progression at G2/M.[2]
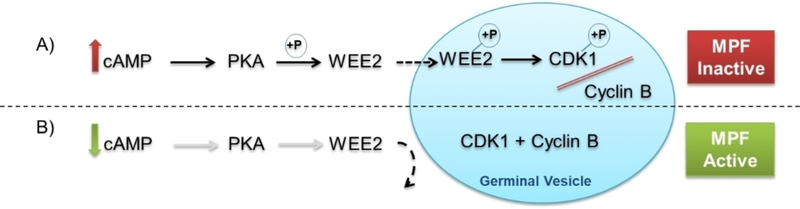
A homolog to WEE1, WEE2 (WEE1 homolog 2, WEE1B), was identified and found to be abundant in the testis and oocytes. Specifically, WEE2 plays a dual regulatory role in oocyte meiosis by preventing premature resumption prior to ovulation and permitting metaphase II exit at fertilization.[3] Mutations in the WEE2 gene are associated with female infertility and have been identified in women who experience recurrent fertilization failure.[4] During prophase I of meiosis, premature oocytes in growing ovarian follicles are arrested in the germinal vesicle (GV) nuclear configuration until recruited for ovulation. This arrest is maintained by elevated levels of cAMP which activate protein kinase A (PKA). PKA in turn phosphorylates cytoplasmic WEE2 allowing it to translocate into the GV (tetraploid nucleus of the egg). Within the GV, WEE2 regulates CDK1 similar in action to WEE1 and prevents complexing with cyclin B, inactivating MPF (sometimes referred to as maturation promoting factor in regards to meiosis). When MPF is inactive, it blocks chromosome reduction to a haploid configuration (Figure 1A).[5]
Figure 1.
A) Maintenance of meiosis arrest in the prophase I oocyte is facilitated by elevated cAMP. Active PKA phosphorylates WEE2 which in turn translocates into the germinal vesicle (GV) to phosphorylate CDK1, a form that prevents complexing with cyclin B and formation of active MPF. B) A decline in cAMP inactivates PKA and the phosphorylating activity on WEE2 so that it is unable to enter the GV. Without an inhibitory phosphate in place, CDK1 complexes with cyclin B to form MPF and drive meiosis resumption.
Just prior to ovulation, a luteinizing hormone surge triggers a signal cascade in the dominant antral follicle that reduces cAMP levels within the oocyte leading to deactivation of PKA. No longer actively phosphorylated by PKA, WEE2 cannot translocate into the GV, thus eliminating inhibitory suppression against CDK1 resulting in active MPF (Figure 1B). This initiates a cascade of events including GV breakdown (GVBD), resumption of meiosis (oocyte maturation), and progression to metaphase II where the now mature oocyte arrests again to await fertilization.[5b]
While WEE2 is critical for maintaining meiotic arrest, it is also essential for exit of metaphase II arrest in response to sperm penetration of the egg. At fertilization, calcium-calmodulin-dependent kinase II (CaMKII) responds to a rise in sperm-induced intracellular calcium oscillations and phosphorylates WEE2. This permits reactivation of the CDK1 phosphorylation pathway resulting in decreased MPF activity. With MPF activity decreased, pronucleus formation occurs and mitotic embryonic cleavage initiates. Without deactivation of MPF, the oocyte does not respond to sperm penetration, no pronuclei form, and fertilization fails.[3a,6]
Due to the role it plays prior to fertilization, WEE2 represents a novel contraceptive target that has been demonstrated in mice and nonhuman primates to be oocyte-specific and non-hormonal in action.[7] The dual regulatory pathways of WEE2 – meiosis inhibition at both the start of oocyte maturation and at the start of fertilization – are a desirable attribute for drug development as it presents multiple options for preventing conception. Previous studies have established that overexpression of WEE2 in somatic cells will arrest the mitotic cell cycle,[3b] so a WEE2 analog aimed at preventing oocyte maturation is not practicable for development into a contraceptive. Therefore, we propose the development of WEE2 kinase inhibitors as selective non-hormonal contraceptive agents that block fertilization.
Kinases are known for their “druggability”, that is, their potential to have a small molecule drug bind and have an effect on the protein’s activity. There are 518 known human kinases, and the phylogenetic kinome has been mapped and includes seven distinct branches.[8] While there have been some forays into allosteric binding modulators, most efforts to date utilize the ATP-binding site and exploit protein-specific amino acid sequences to result in compounds that are more selective for certain members of the kinome over others.
One such method for identifying ligands that bind to a protein’s ATP-binding site is high throughput screening (HTS). HTS of small molecule compound libraries has become a well-known method for rapidly completing expansive biological or chemical assay screening campaigns in drug discovery over the last quarter century. Another potential method to rapidly identify chemical matter is in silico screening (known as virtual HTS or vHTS), which uses computational assessments of binding rather than biochemical assays. vHTS utilizes structure-based drug design to determine how well a small molecule can fit a known ligand binding pocket of a target protein (receptor), taking into account not only steric issues but electronics as well in predicting low energy binding conformations.
We therefore hypothesized that through vHTS we would be able to discover compounds that are selective for WEE2 over the closely related WEE1 and through progressive in situ functional and in vitro biological assays we would be able to identify candidates for further development into selective WEE2 inhibitors. These inhibitors would represent a novel resource for identifying non-hormonal contraceptive candidates.
Results and Discussion
At the time the research efforts contained herein were initiated, a crystal structure of WEE2 had not been solved. Additionally, production and purification of WEE2 protein had not yet been established. Therefore, a homology model of WEE2 was generated based on a solved crystal structure of WEE1 with inhibitor PD352396 (PDB:3BI6) using Schrodinger’s molecular modeling suite Maestro, which allows for determination of ligand fit in the binding pocket and assigns a quantitative docking score to each ligand binding pose so that output can be ranked according to best predicted binding. The Institute for Therapeutics Discovery and Development (ITDD) at the University of Minnesota has access to an in-house library which consists of 300,000 compounds. The ligands were prepared for docking, including desalting, generation of ionization states, stereoisomers where possible, and tautomers, generating an initial set of 400,000 compounds.
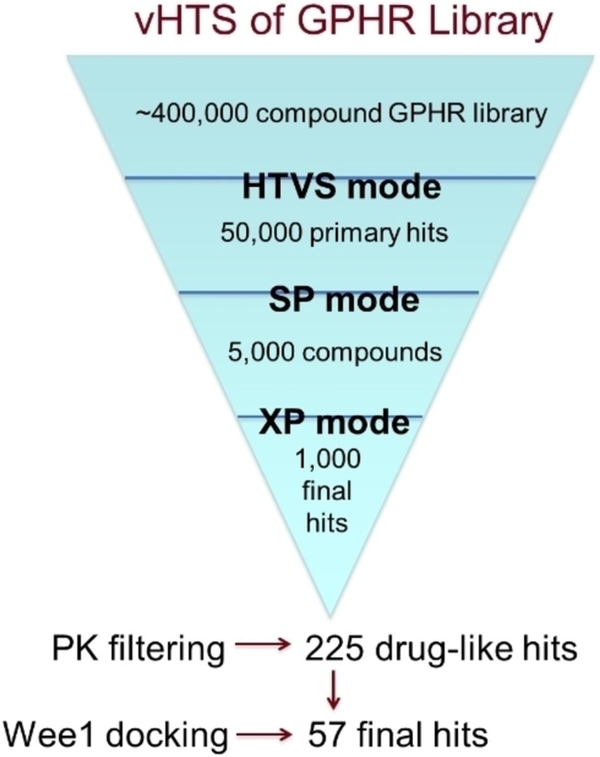
To prepare the protein, hydrogens and disulfide bonds were added. Coordination to metals, and hydrogen bonding to water were allowed during energy minimization. Initially, HTVS (high throughput virtual screening) mode was used to cull the group to 50,000 primary hits (hit rate of 12.5%). The more negative a docking score is, the better the predicted binding; e.g. a ligand with docking score −11.0 is predicted to be a more tightly bound ligand than one with a score of −9.5. These primary hits with the most negative scores were subjected to standard precision (SP) docking simulation, resulting in 5,000 secondary hits (hit rate of 10%; Figure 2). The docking scores at this stage were in the range of −10.70 to −8.30. Those were then submitted to docking using XP (extra precision) mode, and 1,000 final hits were selected (docking scores ranged from −13.61 to −8.77). At this point, it could be said that these compounds were predicted to fit well into the ATP-binding site of WEE2, but that did not qualify the suitability for these compounds to be moved forward in the drug discovery process. A QikProp in silico pharmacokinetic (PK) assessment was performed on these compounds, and a variety of PK filters were then applied: molecular weight <500, * (i.e. non-drug-like alerts)=0, logP octanol/water <5, H-donors <5, H-acceptors <10, rotatable bonds <10, CNS score <1, % predicted oral absorption >80%. The final filtered data set consisted of 225 compounds that had docked well into the WEE2 binding site and were expected to have good potential for being bioavailable.
Figure 2.
Virtual screen funnel to final hits. Using predictive docking simulations, pharmacokinetic filters, and comparative docking scores to WEE1, an initial 400,000 compounds were reduced to a selected 57 with expected inhibitory activity against WEE2
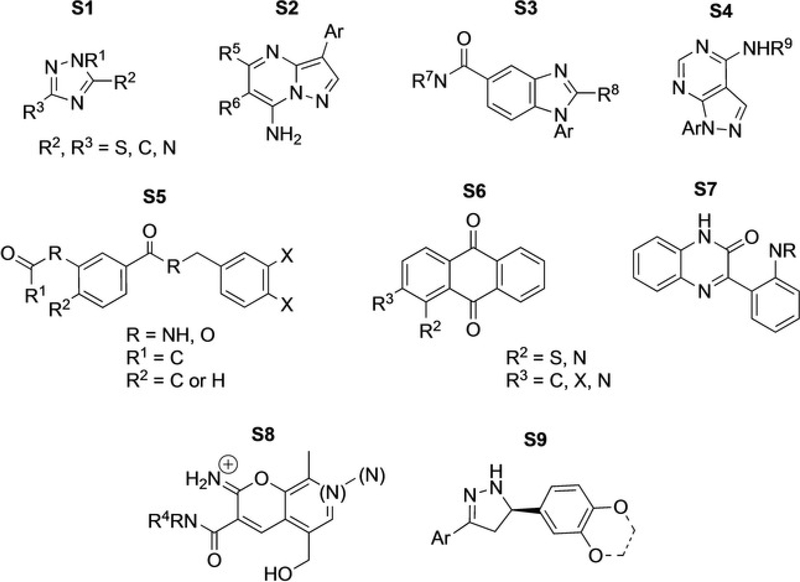
As ultimately the goal of this research is to find potent and selective compounds for WEE2, these 225 compounds were then docked into the WEE1 crystal structure. The WEE2-WEE1 docking scores were paired, and the 57 compounds whose ΔWEE2-WEE1 scores were greater than 20% were selected as compounds that could potentially be selective for WEE2 over WEE1. While, in general, docking scores can fluctuate slightly between various runs, the overall order of predicted binding potential is usually static. As such, we utilized docking scores as an additional method of molecular triage. These 57 compounds were then assessed for structural similarity and 9 common scaffolds were identified (Figure 3). Identification of common scaffolds rather than singletons allows purchase of compounds that are more likely to be true hits in the experimental assays. The top hits were later re-evaluated and the docking scores and modes were confirmed using the recently solved crystal structure of WEE2 (PDB ID: 5VDK).[9]
Figure 3.
Common scaffolds identified as WEE2 hits among the 57 selected compounds following vHTS.
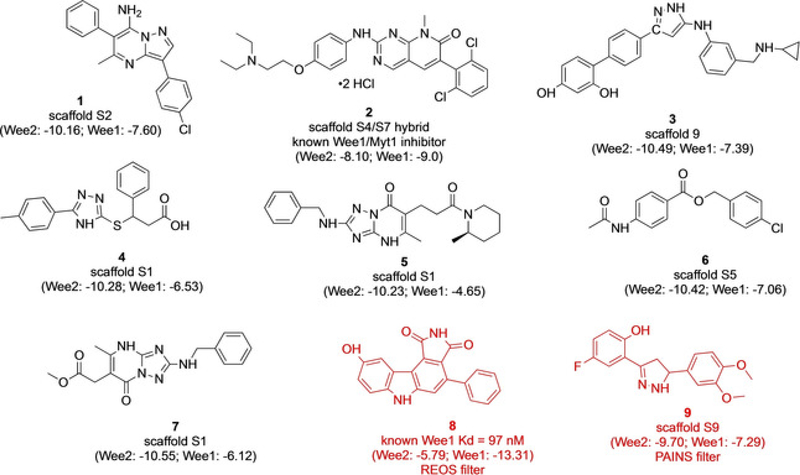
Various in silico overlays of the fragment building blocks in the binding pocket allowed for novel larger scaffolds to be proposed (Figure 4). A similarity search was then conducted based on the proposed combinations. Nine commercial compounds were identified as containing our exact fragment hits or very similar moieties (Figure 5). The parent scaffold(s) from which the compounds arose are noted, as are the WEE2 and WEE1 docking scores. REOS and PAINS filters[8] later identified 8 and 9 as potentially problematic compounds (indicated in red) due to known flagged moieties predicting assay interference.
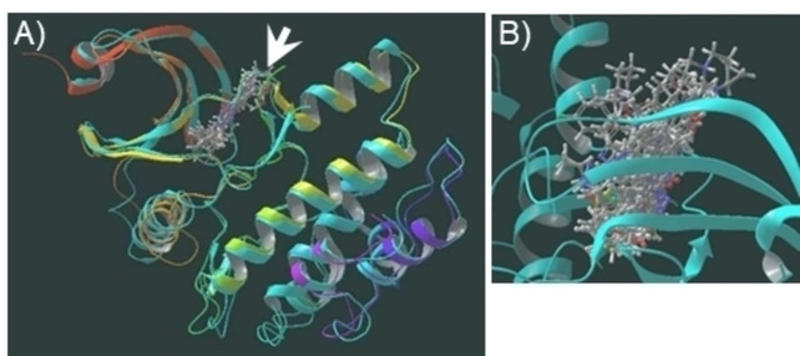
Figure 4.
A) Overlay of WEE2 homology model (turquoise ribbons) and experimentally-obtained crystal structure of WEE2 (rainbow ribbons) with ligands docked into binding site (arrow). B) Close up of binding illustrates how overlay of compounds can identify areas of binding pocket that some ligands reach while others do not.
Figure 5.
Commercial compounds from similarity search of common scaffold hits for WEE2. Compounds 1–7 were purchased for biological assays. Compounds 8 and 9 (shown in red) were considered potentially problematic due to predicted assay interference from known flagged moieties.
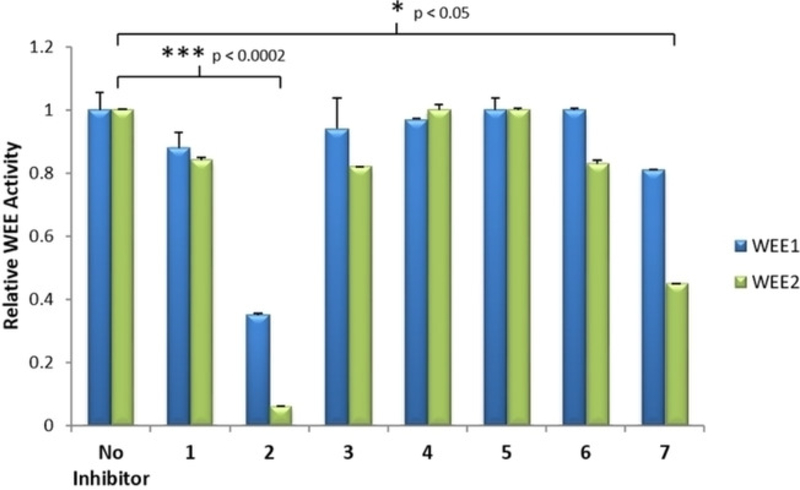
Initial enzyme-linked immunosorbent assay (ELISA) on the identified seven commercial compounds indicated that 2 (PD-166285) and 7 (ChemBridge-7932176) showed promise for significant functional inhibitory activity against WEE2 when compared to the controls (Figure 6).
Figure 6.
ELISA assay to screen inhibitors for activity against WEE1 and WEE2 in vitro. Antibodies bind phosphorylated CDC2 and undergo a chromogenic reaction to provide a semi-quantitative measurement of WEE phosphorylating activity in the presence of 1 μM of inhibitor. Error bars represent the standard error mean. Significant changes in WEE2 activity by an inhibitor compared to the no inhibitor control determined by Student’s t-test.
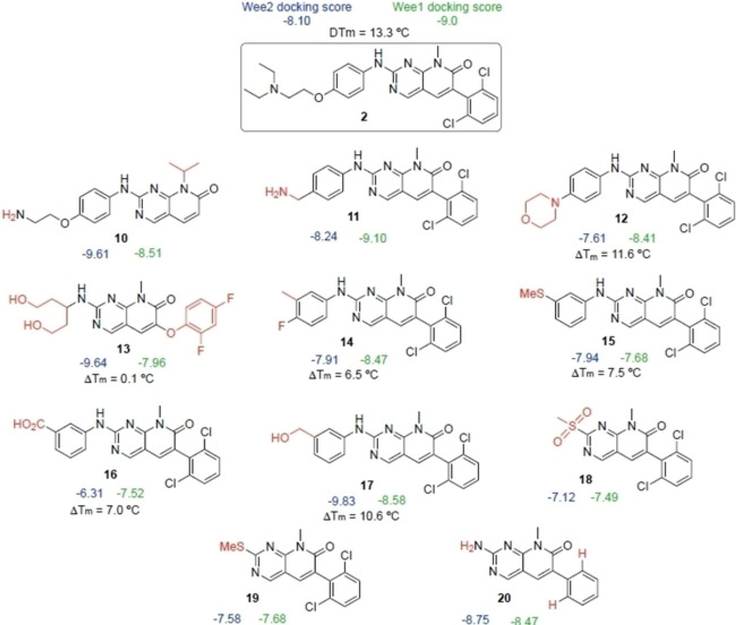
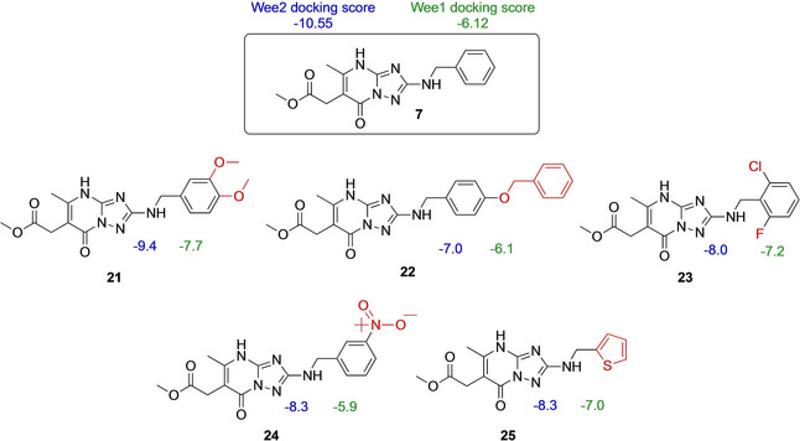
Compound 2 reduced WEE1-catalyzed phosphorylation of CDK1 in situ by 65% compared to controls and had an even more profound effect on WEE2 almost completely eliminating activity (94% reduction; p <0.0002). In comparison, 7 was not as strong of an inhibitor against WEE1 as there was only a 19% loss in function, but there was noticeable inhibition against WEE2 (55% reduced phosphorylating activity; p <0.05) suggesting both compounds possessed the desired inhibitory characteristics for the oocyte specific variant of the kinase. The mechanism of action for compound 2 is a broad spectrum protein tyrosine kinase inhibitor and is effective against Src and FGRF but was developed as a WEE1 inhibitor that functions as a cellular radiosensitizer.[10] In certain tumor and cancer cell lines, 2 is used to nullify the G2 checkpoint and prevent transient mitotic arrest despite radiation induced injury which leads to cell death.[11] In comparison, compound 7 is available in select screening libraries, though there is a dearth of literature reports on its activity or selectivity. Similar scaffolds, however, were reported to inhibit HMG-CoA reductase, an enzyme involved in mammalian cholesterol pathways.[12] Due to their initial activity in our assays, compounds 2 and 7 were selected as parent compounds for SAR by commerce. A similarity search was performed for 2 and 7, and commercially available compounds were selected. These two families of compounds were then subjected to docking studies with both of the WEE kinases to determine if any structural moieties might favor WEE2 over WEE1 (Figures 7 and 8).
Figure 7.
Docking scores of compound 2 analogs as determined by vHTS
Figure 8.
Docking scores of compound 7 analogs as determined by vHTS.
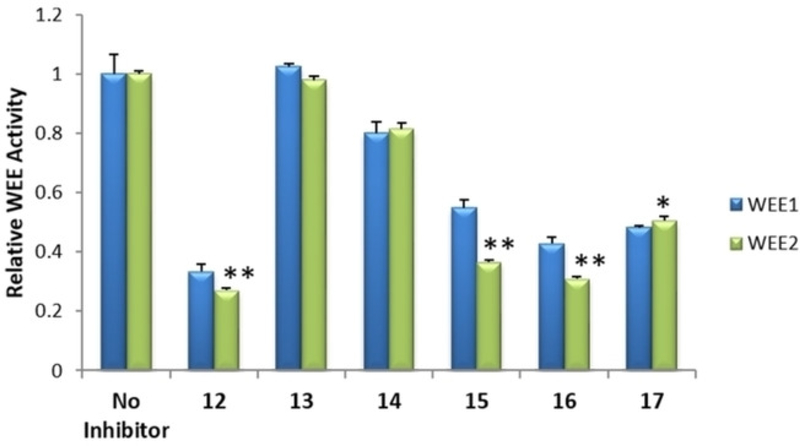
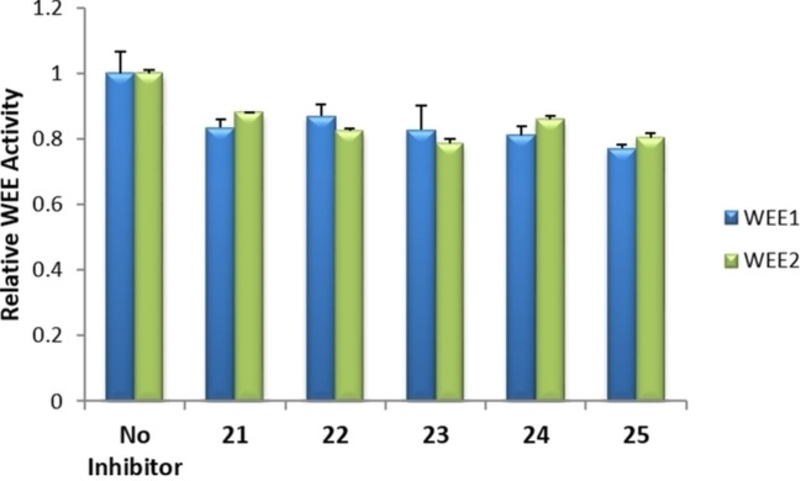
Additional ELISA were performed with selected 2 and 7 analogs, 11 in total, to identify those with functional inhibitory activity against WEE2 kinase. Selection was based on commercial availability and acquisition investment. Among the analogs of 2, four were determined to induce a significant decrease in WEE2 activity when compared to the uninhibited controls while none of the analogs of 7 appeared to inhibit either WEE kinase (Figures 9 and 10). Overall, 17 significantly decreased WEE2 activity by 50% (p <0.01) while 12, 15, and 16 induced a 73, 64, and 70% reduction (respectively; p < 0.005) in WEE2 phosphorylation activity against CDK1.
Figure 9.
ELISA with 1 μM analogs to compound 2. Assay detects phosphorylated CDC2 as a measurement of functional inhibitory activity in vitro against WEE1 and WEE2. Error bars represent the standard error mean. Significant changes in WEE2 activity by an inhibitor compared to no inhibitor control determined by Student’s t-test with * p<0.01 and **p<0.005.
Figure 10.
ELISA with 1 μM analogs to compound 7 measuring inhibitory activity in vitro against WEE1 and WEE2. Error bars represent the standard error mean. Significant changes in WEE2 activity by an inhibitor compared to the control was not detected by Student’s t-test.
Having identified candidate compounds with functional activity against WEE2, biological activity was evaluated by assessing in vitro inhibition of exit from metaphase II of meiosis. To perform this analysis, bovine oocytes were used. Human and bovine WEE2 protein share a 73% homology and WEE kinase functions are conserved across species.[13] Additionally, bovine oocytes are a good experimental model for meiosis studies as they share similar ovarian cycle length and endocrine activity with women and undergo similar timings of meiosis resumption, fertilization, and embryonic cleavage.[14] Immature GV stage bovine oocytes were cultured for 20 hours with inhibitors during in vitro oocyte maturation. At this stage little effect from the inhibitors is expected on meiosis as physiological inhibition of WEE2 already occurs during this time through inactivation of PKA. However, this pre-culture permitted adequate exposure time of the oocytes to the compounds prior to in vitro fertilization which was performed in the presence of the same compound and concentration for an additional 24 hours. This allowed for targeting WEE2 at a critical time when it must be active for the transition from meiosis to mitosis to occur. The ideal WEE2 inhibitor would prevent oocyte exit from metaphase II arrest leading to failed fertilization and a lack of embryonic cleavage (mitosis) that can be used as a direct functional assessment of inhibitor potency (Figure 11).
Figure 11.
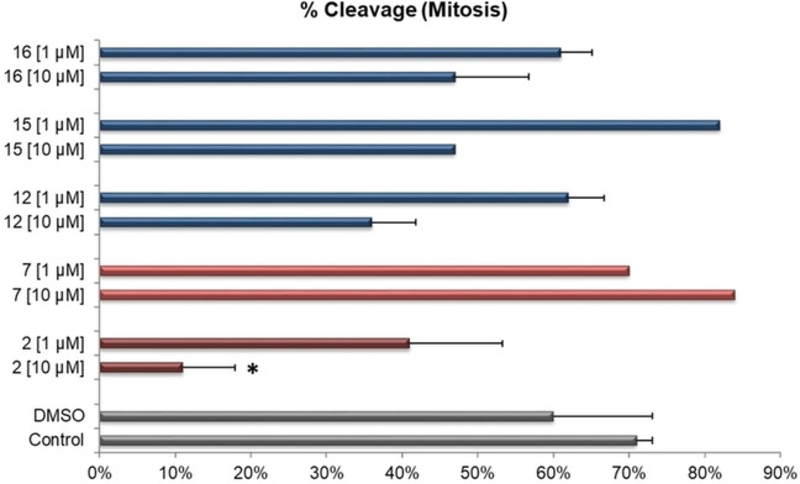
Embryonic cleavage rates following in vitro maturation and fertilization with selected WEE inhibitors (parent compounds represented as red bars, analogs as blue) solubilized in DMSO at either 10 or 1 μM were compared to DMSO only or no further treatment Control (grey bars). Compoud IDs are listed on the y-axis (16, 15, 12, 7, and 2). Error bars indicate +SEM and (*) indicates a significant difference to either control.
Compounds were evaluated at 1 and 10 μM, and the proportion of oocytes which were able to undergo fertilization and initiate embryonic mitotic cleavage were compared to non-treated control or DMSO only controls. At 10 μM, parent compound 2 was the only one to induce a significant inhibition on embryonic development with only 11% (p<0.05) initiating mitotic cleavage while there was a trend for 12 and 16 at 10 μM to decrease overall fertilization rates to 36 and 47%, respectively.
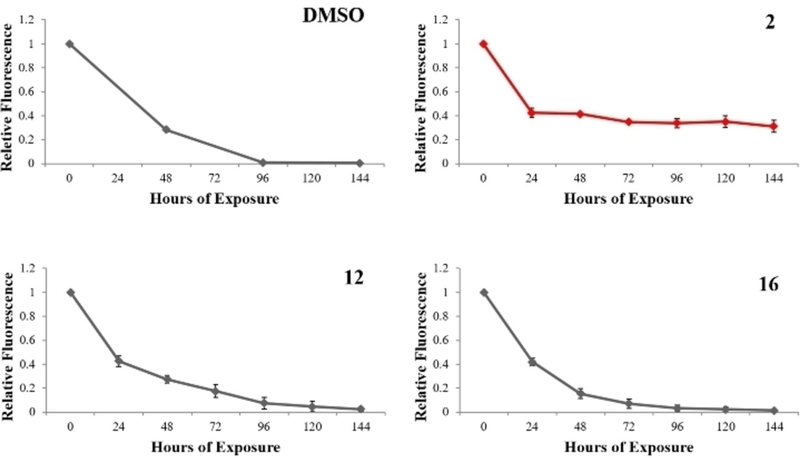
Having identified those compounds with a favorable biological function against WEE2, additional biological activity was determined by culturing selected compounds with HEK 293 cells to evaluate the effects on somatic WEE1. Cells were labeled with a fluorescence tracking dye that decreases in signal with each successive cell division and serves as a measurement of cell proliferation. Cells were cultured over 144 hours with samples taken every 24 hours to measure fluorescent intensity by flow cytometry. In normal and control populations of cells, the relative fluorescence should approach 0 by 96 hours (Figure 12) as is observed in those treated with DMSO alone. Compounds 2, 12, and 16 were evaluated and only 2 appeared to inhibit WEE1 as the cell population arrested after just 24 hours of culture.
Figure 12.
Flow cytometry analysis of cellular proliferation in HEK 293 cells cultured with selected compounds or DMSO only (Control). Cells were exposed to a fluorescent trace dye that becomes diluted with each cell division. Cells cultured with compound 2 were induced into mitotic arrest (red). Error bars indicate one standard deviation.
Given these data it becomes clear that 2 is a potentially strong inhibitor of meiosis that could be used to block the exit from metaphase II and prevent successful fertilization. Not surprisingly, this compound also had an overt biological effect on WEE1 as evident by the ELISA (Figure 6) and cell culture proliferation data (Figure 11). While inhibitors 12 and 16, both analogs of 2, relatively reduced fertilization success, they both had a negligible effect on mitosis suggesting that both of these compounds would also make strong candidates for further development or modification.
Conclusions
WEE2 is an oocyte specific kinase and represents a novel non-hormonal target for contraceptive drug development. Using virtual high throughput in silico screening, compounds were identified with predicted specificity to inhibit WEE2 over the somatic counterpart, WEE1. Selected compounds were then evaluated in vitro to confirm inhibitory activity of the drug to target WEE2 and biological activity was confirmed on both meiotic and mitotic pathways. Through these assays, candidates were filtered from 300,000 down to three based on structure and function. Ultimately, 2 and its analogs 12 and 16 were identified as candidates for further drug development and evaluation as non-hormonal contraceptives. Additionally, since these studies were initiated, the structure of WEE2 has been solved, confirming the principal conclusions using the homology model described in this study.[9]
Supplementary Material
Acknowledgements
Research reported in the publication was fully supported the by the Bill & Melinda Gates Foundation under award number OPP1178119, the National Institutes of Health under award number P51OD011092 and by the Eunice Kennedy Shriver National Institute of Child Health and Human Development under award numbers U54HD055744 and U01HD076542 (GIG). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This work would not have been possible without the ONPRC Endocrine Technologies Core, Flow Cytometry Support Core, Surgical Services Unit and the Division of Comparative Medicine.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Supporting Information Summary
The supporting information file contains physical data for each compound and the corresponding assigned number as well as detailed protocols for the ELISA, in vitro oocyte fertilization, and flow cytometry experiments.
References
- [1] a).Nurse P, Thuriaux P, Genetics 1980, 96, 627–637; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Parker LL, Piwnica-Worms H, Science 1992, 257, 1955–1957. [DOI] [PubMed] [Google Scholar]
- [2] a).Moser BA, Russell P, Curr. Opin. Microbiol 2000, 3, 631–636; [DOI] [PubMed] [Google Scholar]; b) Han SJ, Conti M, Cell Cycle 2006, 5, 227–231. [DOI] [PubMed] [Google Scholar]
- [3] a).Oh JS, Susor A, Conti M, Science 2011, 332, 462–465; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Nakanishi M, Ando H, Watanabe N, Kitamura K, Ito K, Okayama H, Miyamoto T, Agui T, Sasaki M, Genes Cells 2000, 5, 839–847. [DOI] [PubMed] [Google Scholar]
- [4] a).Dai J, Zheng W, Dai C, Guo J, Lu C, Gong F, Li Y, Zhou Q, Lu G, Lin G, Fertil. Steril 2019, 111, 510–518; [DOI] [PubMed] [Google Scholar]; b) Zhao S, Chen T, Yu M, Bian Y, Cao Y, Ning Y, Su S, Zhang J, Zhao S, Fertil. Steril 2019, 111, 519–526. [DOI] [PubMed] [Google Scholar]
- [5] a).Solc P, Schultz RM, Motlik J, Mol. Hum. Reprod 2010, 16, 654–664; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Oh JS, Han SJ, Conti M, The J Cell Biol. 2010, 188, 199–207; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Jaffe LA, Egbert JR, Annu. Rev. Physiol 2017, 79, 10.11–10.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Marangos P, FitzHarris G, Carroll J, Development 2003, 130, 1461–1472. [DOI] [PubMed] [Google Scholar]
- [7] a).Han SJ, Chen R, Paronetto MP, Conti M, Curr. Biol 2005, 15, 1670–1676; [DOI] [PubMed] [Google Scholar]; b) Hanna CB, Yao S, Patta MC, Jensen JT, Wu X, Biol. Reprod 2010, 82, 1190–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S, Science 2002, 298, 1912–1934. [DOI] [PubMed] [Google Scholar]
- [9].Zhu J-Y, Cuellar RA, Berndt N, Lee HE, Olesen SH, Martin MP, Jensen JT, Georg GI, Schönbrunn E, J. Med. Chem 2017, 60, 7863–7875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wang Y, Li J, Booher RN, Kraker A, Lawrence T, Leopold WR, Sun Y, Cancer Res. 2001, 61, 8211–8217. [PubMed] [Google Scholar]
- [11].Hashimoto O, Shinkawa M, Torimura T, Nakamura T, Selvendiran K, Sakamoto M, Koga H, Ueno T, Sata M, BMC Cancer 2006, 6, 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Son M, Baek A, Sakkiah S, Park C, John S, Lee KW, PloS ONE 2013, 8, e83496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Harashima H, Dissmeyer N, Schnittger A, Trends Cell Biol. 2013, 23, 345–356. [DOI] [PubMed] [Google Scholar]
- [14].Woclawek-Potocka I, Rawinska P, Kowalczyk-Zieba I, Boruszewska D, Sinderewicz E, Wasniewski T, Skarzynski DJ, Mediators Inflamm. 2014, 2014, 649702. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.