Abstract
Current “fast-acting” insulin analogues contain amino acid modifications meant to inhibit dimer formation and shift the equilibrium of association states toward the monomeric state. However, the insulin monomer is highly unstable and current formulation techniques require insulin to primarily exist as hexamers to prevent aggregation into inactive and immunogenic amyloids. Insulin formulation excipients have thus been traditionally selected to promote insulin association into the hexameric form to enhance formulation stability. This study exploits a novel excipient for the supramolecular PEGylation of insulin analogues, including aspart and lispro, to enhance the stability and maximize the prevalence of insulin monomers in formulation. Using multiple techniques, it is demonstrated that judicious choice of formulation excipients (tonicity agents and parenteral preservatives) enables insulin analogue formulations with 70–80% monomer and supramolecular PEGylation imbued stability under stressed aging for over 100 h without altering the insulin association state. Comparatively, commercial “fast-acting” formulations contain less than 1% monomer and remain stable for only 10 h under the same stressed aging conditions. This simple and effective formulation approach shows promise for next-generation ultrafast insulin formulations with a short duration of action that can reduce the risk of post-prandial hypoglycemia in the treatment of diabetes.
Keywords: diabetes, drug delivery, insulin, PEGylation, polymers, supramolecular
1. Introduction
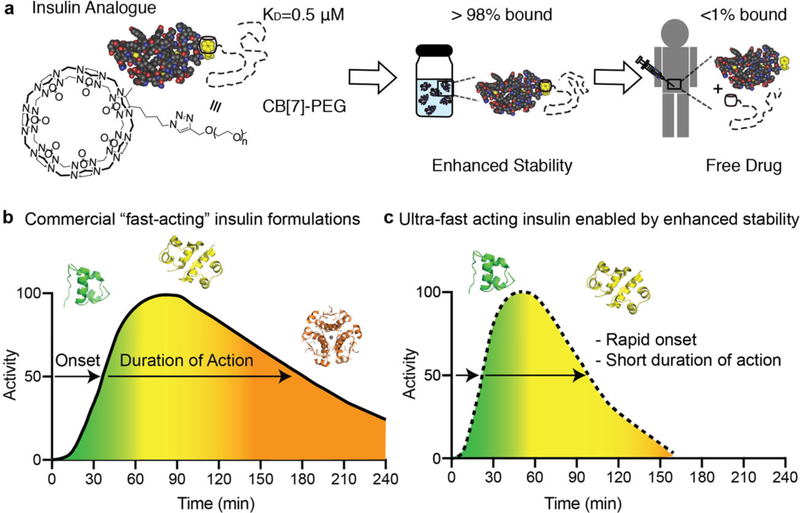
Over 40 million patients live with Type 1 diabetes worldwide.[1] The loss of insulin production in these patients results in an inability to process glucose without exogenous insulin delivery through daily subcutaneous injections, or by infusion with a pump into the subcutaneous tissue.[2,3] Maintaining tight glycemic control is critical to prevent severe long-term side effects, such as kidney disease, heart disease, vision loss, and limb loss.[4] Patients must deliver a carefully calculated bolus of insulin at mealtimes to reduce blood glucose excursions; however, insulin pharmacokinetics following subcutaneous administration are poorly matched to physiologic post-prandial requirements.[3,5] Even current commercial “rapid-acting” insulin analogue formulations, Humalog (insulin lispro), Novolog (insulin aspart) and Apidra (insulin glulisine), exhibit delayed onset of action of ≈20–30 min, peak action at ≈60–90 min and a total duration of action of ≈3–4 h.[6–10] These insulin formulations contain a mixture of hexamers, dimers, and monomers, which, upon subcutaneous injection, dissociate and are absorbed at different rates (Figure 1). This differential absorption results in the rapid onset but long duration of action of these formulations (Figure 1b).[11–13] A completely monomeric insulin formulation would enable faster onset, and reduced duration of action, which is the next step in creating an ultrafast acting insulin formulation (Figure 1c).
Figure 1.
Schematic of contributions of insulin association state on absorption kinetics. a), Non-covalent PEGylation is enabled by the strong specific binding between CB[7]-PEG and the N-terminal phenylalanine on insulin (KD =0.5 μM). Under high concentrations in formulation insulin will be greater than 98% bound, but after dilution upon subcutaneous injection less than 1% of CB[7]-PEG will remain associated with insulin. b,c) Schematic illustration of insulin action over time following subcutaneous administration, whereby insulin association states in formulation dictate insulin activity at various time points. Commercial “fast-acting” insulin formulations comprise a mixture of insulin association states (i.e., hexamers, dimers, and monomers) that result in extended duration of insulin action when delivered subcutaneously. Insulin monomers are absorbed in approximately 5–10 min, dimers are absorbed in 20–30 min, and hexamers can take 1–2 h and cause prolonged insulin action (b). By eliminating insulin hexamers from formulations and shifting the equilibrium to a higher ratio of monomers, future formulations could achieve ultrafast onset and shorter duration of action, which would improve meal-time insulin responses and reduce post-prandial glucose excursions (c).
Current “fast-acting” insulin analogues inhibit dimer formation and shift the equilibrium toward the monomeric state through amino acid modifications. However, the insulin monomer is unstable and rapidly aggregates to form amyloid fibrils.[14–16] Insulin is primarily formulated as hexamers to prevent insulin aggregation.[14] Novolog (aspart) and Humalog (lispro) are formulated in sodium phosphate buffer with a threefold molar excess of zinc ion, relative to the insulin hexamer. Formulation with zinc stabilizes the hexameric state in the T6 formation,[17] and dissociation of the hexamer is known to be the rate-limiting step for subcutaneous absorption and onset of action. In contrast, Apidra (glulisine) is a zinc-free formulation and is formulated with the surfactant polysorbate 20 as a stabilizing agent. Apidra demonstrates slightly faster onset of action, but overall similar control of glucose levels in vivo to Novolog and Humalog, indicating that the removal of zinc alone is not enough to achieve an ultrafast acting monomeric insulin formulation.[18]
Excipients, the inactive ingredients in drug formulations, perform a number of functions and can facilitate improved protein stability, solubility, and absorption.[19,20] Formulations for insulin analogues contain multiple excipients including tonicity agents, preservatives, and stabilizing agents, which are selected to enhance insulin stability. Glycerol or sodium chloride are commonly added to insulin formulations as tonicity agents, whereas phenol and/or meta-cresol are added as parenteral preservatives.[21] The inclusion of glycerol, a tonicity agent used in both Humalog and Novolog formulations, has been shown to increase insulin stability in formulation.[22] Moreover, in addition to anti-microbial properties, phenol and meta-cresol stabilize the R6 insulin hexamer by forming hydrogen bonds between dimers.[17,23] This suggests that even in the absence of zinc, the phenolic preservatives may contribute to higher order insulin structures that may slow absorption from the subcutaneous space.
Formulating monomeric insulin requires new excipients, which do not promote the R6 hexamer, but still imbue insulin with sufficient stability to prevent aggregation and denaturation over time.[24,25] Covalent PEGylation has been successful as a strategy to stabilize insulin in formulation;[26–28] however, the extended pharmacokinetics in vivo associated with PEGylation is not desirable for rapid acting insulins.[29] Recent research has demonstrated non-covalent modification of proteins as a strategy to enhance their stability in formulation.[30,31] In particular, conjugation of a polyethylene glycol (PEG) chain to a cucubit[7]uril (CB[7]) creates a tool for non-covalent PEGylation using host-guest binding with the excipient cucurbit[7]uril-poly(ethylene glycol) (CB[7]-PEG). CB[7]-PEG has strong binding affinities for terminal aromatic amino acids such the N-terminal phenylalanine found on insulin making it an ideal candidate for host-guest binding.[32–35] The dynamic binding of CB[7]-PEG to insulin is promising as a strategy for stabilizing insulin without promoting the insulin hexamer.
Understanding the excipient choices in current commercial formulations is a critical first step in designing the next generation of ultrafast acting insulin formulations that have potential to more closely mimic endogenous pharmacokinetics. In this study, we aim to engineer a stable monomeric insulin formulation through selection of formulation excipients that promote the monomer state. We demonstrate that CB[7]-PEG can be used to stabilize these formulations and that the insulin/CB[7]-PEG complex has a faster diffusion rate than the insulin hexamer.
2. Results and Discussion
2.1. Insulin Association State by Size Exclusion Chromatography with Multiple Angle Light Scattering
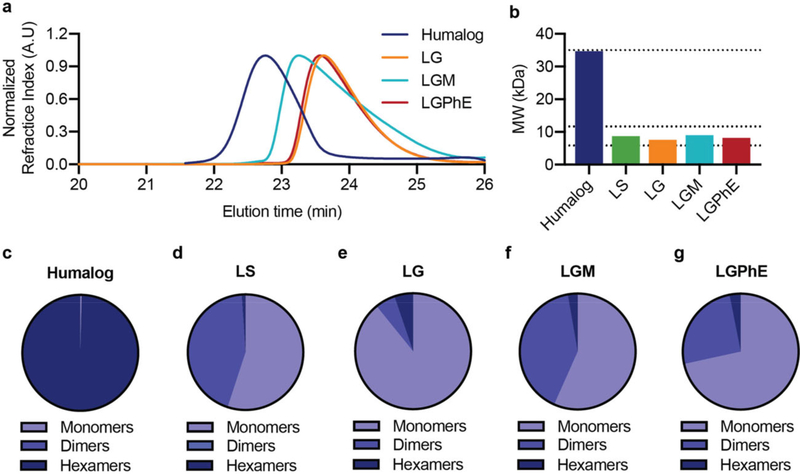
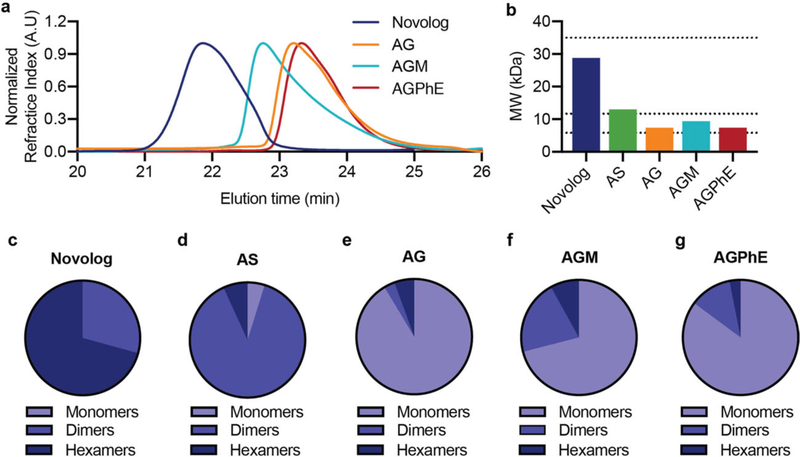
Excipient choice is important, because as in the case of Apidra (glulisine), the absence of zinc is not sufficient to result in an ultrafast acting insulin formulation. To engineer an insulin formulation that promotes the monomer state, the effects of excipients on insulin association state under zinc-free conditions must be understood. Characterization by size exclusion chromatography with multiple angle light scattering (SEC-MALS) allows us to determine insulin molecular weight in formulation and estimate the fraction of hexamers, dimers, and monomers under formulation conditions.[36–40] In these studies, insulin analogues lispro and aspart were formulated with ethylenediaminetetraacetic acid (EDTA) to remove formulation zinc. EDTA forms strong complexes with zinc (KD ≈10×10−18 M)[41,42] and addition of one molar equivalent of EDTA relative to the zinc ion in insulin formulations rapidly sequesters the zinc, preventing it from interacting with the insulin and thus disrupting the insulin hexamer in solution. Zinc-free insulin in pure water is completely monomeric (Mn = 5.7 kDa). However, it becomes a mixture of monomers, dimers, and hexamers with the addition of buffering salts. To select for a mostly monomeric insulin formulation, we evaluated the effect of tonicity agents and preservatives on insulin association state. Formulation details are listed in Tables S1 and S2, Supporting Information. As hypothesized, all zinc-free formulations had decreased hexamer composition compared to either commercial Humalog (Mn = 34.7 kDa; 99.6% hexamer) or commercial Novolog (Mn = 28.8 kDa; 70.7% hexamer). Zinc-free formulations were primarily a mixture of monomers and dimers with small amounts of hexamer (Figures 2 and 3; Figure S1, Supporting Information).
Figure 2.
Association state of lispro with different formulation excipients. Zinc-free insulin lispro association states when formulated in i) phosphate buffer, sodium chloride (0.9%) (LS), ii) phosphate buffer with glycerol (2.6%) (LG), iii) phosphate buffer with glycerol (2.6%) and meta-cresol (0.315%) (LGM), and iv) phosphate buffer with glycerol (2.6%) and phenoxyethanol (0.85%) (LGPhE). Formulations were compared against a formulation of commercial Humalog. a) SEC-MALS elution profiles. b) Number-averaged molecular weight of the distribution of insulin lispro association states. c–g) Ratio of monomers, dimers, and hexamers in each formulation.
Figure 3.
Association state of aspart with different formulation excipients. Zinc-free insulin aspart association states when formulated in i) phosphate buffer, sodium chloride (0.9%) (AS), ii) phosphate buffer with glycerol (2.6%) (AG), iii) phosphate buffer with glycerol (2.6%) and metacresol (0.315%) (AGM), and iv) phosphate buffer with glycerol (2.6%) and phenoxyethanol (0.85%) (AGPhE). Formulations were compared against a formulation of commercial Novolog. a) SEC-MALS elution profiles. b) Number-averaged molecular weight of the distribution of insulin aspart association states. c–g) Ratio of monomers, dimers, and hexamers in each formulation.
Glycerol is the most commonly used tonicity agent in insulin formulations followed closely by sodium chloride. Zinc-free lispro or zinc-free aspart were formulated in phosphate buffer (pH = 7.4) with either sodium chloride (0.9%) or glycerol (2.6%). Glycerol demonstrated a lower number average molecular weight and higher percentage of monomers for both lispro (Mn = 7.6 kDa; 89.2% monomer) and aspart (Mn = 7.4 kDa; 91.4% monomer) compared to sodium chloride formulations for lispro (Mn =8.7 kDa; 55.0% monomer) and aspart (Mn =13 kDa; 4.8% monomer) (Figures 2 and 3; Figure S1, Supporting Information). Therefore, glycerol was maintained as the tonicity agent when evaluating the effect of preservatives on insulin association state.
While glycerol alone showed the highest proportion of monomers in formulation, insulin requires an antimicrobial preservative, because it is a multidose formulation. There are only several commercially used parenteral preservatives, the most common of which are benzyl alcohol, chloro-butanol, meta-cresol, phenol, methyl paraben, propyl paraben, phenoxyethanol, and thimerosal.[21] Of these anti-microbial agents, only phenol and meta-cresol are currently used in commercial insulin formulations.[21] However, phenolic preservatives are recognized for their promotion of R6 insulin hexamer formation through hydrogen bonding between the hydroxyl group on phenol and the insulin dimeric pocket.[43,44] To create a monomeric insulin formulation, it is imperative to reduce phenol promoted R6 hexamer formation; thus, it is necessary to identify a different parenteral preservative for insulin formulations. Other research has indicated that ethanol may play a role in disrupting the insulin dimer, thus promoting the insulin monomer.[45] This led to the selection of phenoxyethanol as an alternative anti-microbial agent. We hypothesized that the ethanol chain off of the phenolic ring will create a steric hindrance that disrupts the intermolecular forces leading to R6 hexamer formation and that the ethanol side chain could further promote monomer formation through the disruption of the insulin dimer. Indeed, after glycerol alone, phenoxyethanol demonstrated the lowest number average molecular weight (Mn) and highest ratio of insulin monomers in both lispro (Mn =8.2 kDa; 71.5% monomer) and aspart formulations (Mn = 7.4 kDa; 85.2% monomer) (Figures 2 and 3). In contrast, formulation of meta-cresol had increased number average molecular weight compared to phenoxyethanol for lispro (Mn = 9.0 kDa; 56.7% monomer) and aspart (Mn =9.4 kDa; 71.0% monomer).
Work by Gast et al. has shown that phenol shows a stronger affinity for hexamer formation than meta-cresol.[13] Therefore, it is expected that phenoxyethanol would also have a higher monomer content than a phenol based formulation. While the proportion of hexamers between formulations with phenoxyethanol and meta-cresol were similar, meta-cresol formulations had a higher percentage of hexamers and dimers combined. Since the R6 hexamer is a dynamic structure held together by Intermolecular forces, insulin that originates as a hexamer may dissociate and appear in the dimer form in the SEC-MALS.
2.2. Stability of Monomeric Insulin with CB[7]-PEG
As insulin monomer content increases, formulation stability becomes more challenging and alternative stabilizing excipients are needed to increase insulin stability in the absence of zinc. The current commercial zinc-free insulin analogue, Apidra, contains both meta-cresol and the surfactant polysorbate 20, which aid formulation stability, but meta-cresol promotes the R6 hexamer. Non-covalent PEGylation has potential as a stabilizing excipient that will not promote hexamer formation and will leave insulin monomers unmodified to act in vivo. Previous studies have shown that non-covalent PEGylation with (CB[7]-PEG) imbues long-term insulin stability without reducing insulin activity in vivo. Lispro formulated with glycerol/phenoxyethanol (LGPhE) and three molar equivalents CB[7]-PEG with respect to insulin demonstrated equal in vitro bioactivity compared to commercial Humalog in a dose response assay of protein kinase B (AKT) phosphorylation (Humalog EC50 = 1 × 10−4 mg mL−1; LGPhE EC50 = 6 × 10−5 mg mL−1) (Figure S2, Supporting Information). CB[7] has a binding affinity of 0.54 μM to insulin aspart (Figure S3, Supporting Information), such that under typical formulation concentrations, the CB[7]-PEG/insulin complex will be greater than 98% bound, but immediately upon dilution following subcutaneous administration, less than 1% of the CB[7]-PEG will remain associated with insulin.
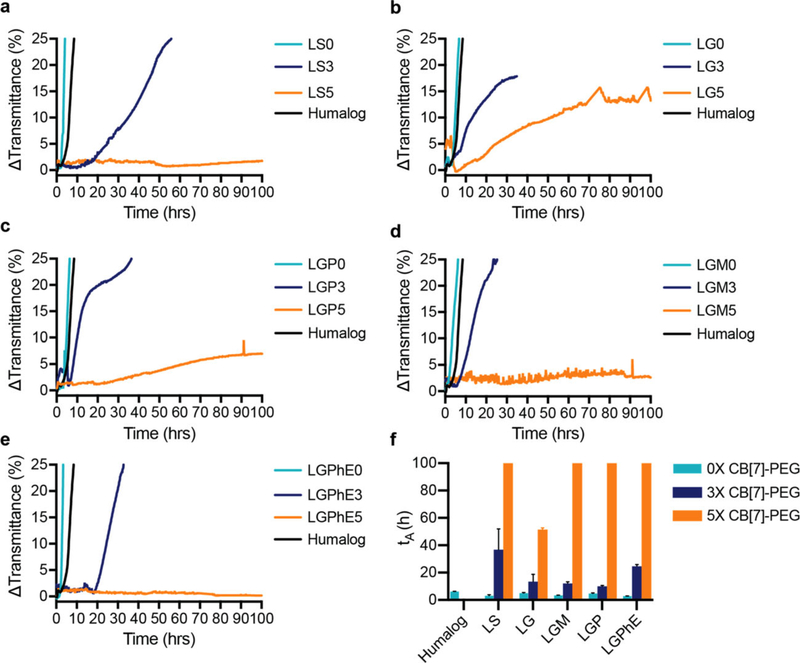
Here, we demonstrate the utility of non-covalent PEGylation with CB[7]-PEG as a strategy to stabilize monomeric insulin long-term under a variety of formulation conditions. Zinc-free insulin lispro and aspart were formulated in phosphate buffer with either i) saline (LS), ii) glycerol (LG), iii) glycerol/phenol (LGP), iv) glycerol/meta-cresol (LGM), or v) glycerol/phenoxyethanol (LGPhE) to evaluate the combination of excipients providing greatest stability (Figure 4). CB[7]-PEG was added to formulations in excess to insulin at either three molar equivalents or five molar equivalents. Glycerol was chosen as a tonicity agent for combination with preservatives (including phenol, meta-cresol, phenoxyethanol) due to the increased affinity for the insulin monomer in the presence of glycerol compared to sodium chloride. Zinc-free lispro under all formulation conditions was less stable than the commercial Humalog formulation, thus demonstrating the need for additional stabilizing agents. A threefold excess of CB[7]-PEG to insulin resulted in increased stability, which ranged from 1.5-fold increase in the LGP3 formulation to a sixfold increase in the LS3 formulation. Addition of fivefold excess of CB[7]-PEG to lispro extended lispro stability over 100 h in both of the LS5 and LGPhE5 formulations.
Figure 4.
Formulation with CB[7]-PEG stabilizes zinc-free lispro. In vitro stability of insulin lispro under different formulation conditions with a molar ratio of CB[7]-PEG:Lispro of 0:1 (teal), 3:1 (blue), and 5:1 (orange) against a commercial Humalog control (black). a) Lispro in phosphate buffer with saline (0.9%). b) Lispro in phosphate buffer with glycerol (2.6%). c) Lispro in phosphate buffer with glycerol (2.6%) and phenol (0.25%). d) Lispro in phosphate buffer with glycerol (2.6%) and meta-cresol (0.315%). e) Lispro in phosphate buffer with glycerol (2.6%) and phenoxyethanol (0.85%). f) Comparison of stability by aggregation times (tA), defined as the time to a change in transmittance (λ=540 nm) of 10% or greater following stressed aging (i.e., continuous agitation at 37° C). Data shown are average transmittance traces for n =3 samples per group and error bars ((f) only) are standard deviation.
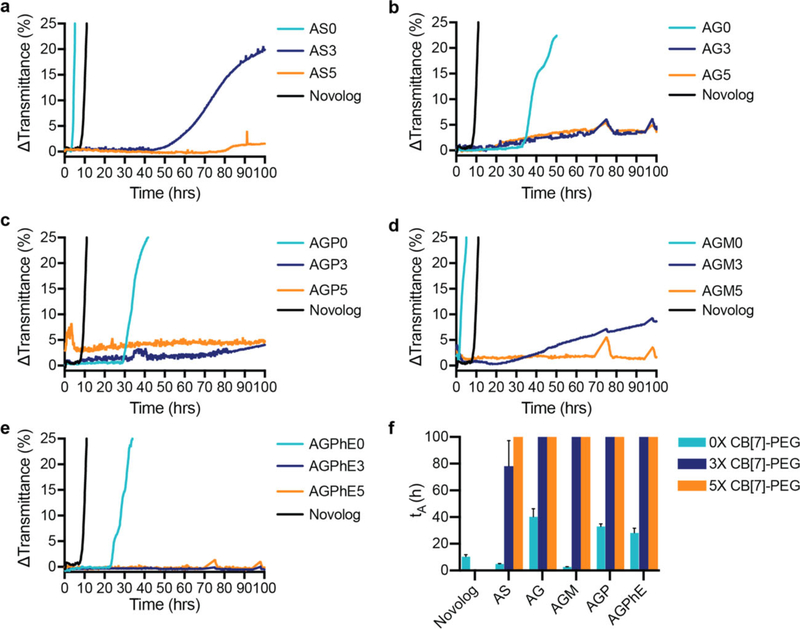
Zinc-free aspart was formulated under the same conditions as the zinc-free lispro: i) saline (AS), ii) glycerol (AG), iii) glycerol/phenol (AGP), iv) glycerol/meta-cresol (AGM), or v) glycerol/phenoxyethanol (AGPhE) to evaluate stability (Figure 5). Zinc-free aspart formulations were overall more stable than lispro formulations and the AG0, AGP0, and AGPhE0 formulations were more stable than the commercial Novolog formulation without the addition of CB[7]-PEG. A threefold excess of CB[7]-PEG stabilized aspart over 100 h in all formulation conditions except AS3. All formulations were stable over 100 h with the fivefold excess of CB[7]-PEG. Moreover, no difference in blood glucose depletion was observed in diabetic rats when animals were dosed with i) Novolog or ii) Zinc-free aspart formulated with CB[7]-PEG5k (5 equiv. with respect to insulin), indicating that formulation of monomeric insulin with CB[7]-PEG does not alter bioactivity or pharmacodynamics in vivo (Figure S4, Supporting Information).
Figure 5.
Formulation with CB[7]-PEG stabilizes zinc-free aspart. In vitro stability of insulin aspart under different formulation conditions with a molar ratio of CB[7]-PEG:Aspart of 0:1 (teal), 3:1 (blue), and 5:1 (orange) against a commercial Novolog control (black). a) Aspart in phosphate buffer with saline (0.9%). b) Aspart in phosphate buffer with glycerol (2.6%). c) Aspart in phosphate buffer with glycerol (2.6%) and phenol (0.25%). d) Aspart in phosphate buffer with glycerol (2.6%) and meta-cresol (0.315%). e) Aspart in phosphate buffer with glycerol (2.6%) and phenoxyethanol (0.85%). f) Comparison of stability by aggregation times (tA), defined as the time to a change in transmittance (λ = 540 nm) of 10% or greater following stressed aging (i.e., continuous agitation at 37 °C). Data shown are average transmittance traces for n =3 samples per group and error bars ((f) only) are standard deviation.
2.3. Insulin Monomer Diffusion by Diffusion-Ordered NMR Spectroscopy
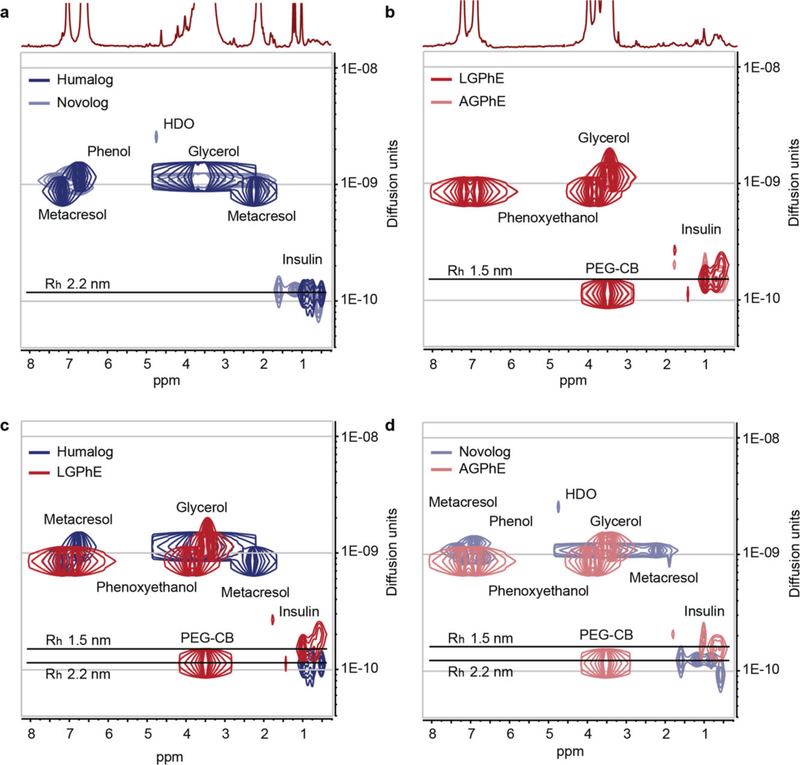
Diffusion-ordered NMR spectroscopy (DOSY) was used to provide insight into the size and diffusion characteristics of insulin lispro and aspart under LGPhE and AGPhE formulation conditions in the presence of CB[7]-PEG (Figure 6). Formulations comprising CB[7]-PEG could not be assessed with SEC-MALS on account of confounding alterations to the retention time of the insulin species on the chromatography column and in the light scattering. Formulating monomeric insulin analogues with CB[7]-PEG is expected to negligibly increase the diffusivity and corresponding hydrodynamic radius of the insulin in formulation compared to the insulin monomer alone on account of the highly dynamic nature of the CB[7]-insulin binding interaction. The diffusivities of the insulin molecules in zinc-free formulations of either LGPhE or AGPhE were compared against commercial Humalog and Novolog formulations. Humalog and Novolog both exhibited insulin diffusivities of 1.13 × 10−10 m2 s−1, corresponding to hydrodynamic radii of 2.2 nm, which is consistent with reported literature values.[46,47] By using EDTA to remove zinc from commercial Novolog, the hydrodynamic radius remains unchanged at 2.2 nm, demonstrating that the absence of zinc alone is not sufficient to alter the insulin association state (Figure S5, Supporting Information). In contrast, the insulin molecules in zinc-free LGPhE and AGPhE formulations comprising CB[7]-PEG demonstrated increased diffusivities of 1.60 × 10−10 m2 s−1, corresponding to a hydrodynamic radius of 1.5 nm, which is significantly smaller than commercial insulin analogue formulations and approximately the same size as the reported literature value for the insulin monomer (Figure 6b).[46] These observations suggest that while CB[7]-PEG stabilizes the monomeric form of insulin in formulation, the highly dynamic binding of CB[7]-PEG to insulin negligibly alters the diffusivity of the insulin molecule.
Figure 6.
Diffusion of insulin analogues in various formulations with CB[7]-PEG. Diffusion-ordered NMR spectroscopy (DOSY) provides insight into the formation of protein/CB[7]-PEG complexes and their rates of diffusion in formulation. Diffusion characteristics demonstrate that lispro and aspart diffuse at a similar rate under both a) commercial Humalog (navy blue) and Novolog (light blue) in the presence of zinc ion and b) LGPhE (red) and AGPhE (pale red) in the presence of CB[7]-PEG (0.6 equiv.). Increased diffusion was observed for insulin c) LGPhE and d) AGPhE formulated with 0.6 equiv. CB[7]-PEG (red) compared to commercial formulation conditions (blue).
3. Conclusion
Excipient choice in insulin formulations is critical in determining insulin association state, stability, and the rate of absorption in vivo. At present, there is a need for insulin formulations that more closely mimic endogenous secretion, which ultimately requires insulin formulations that are more monodisperse and primarily contain insulin monomers. In order to engineer the next generation of monomeric insulin formulations, an understanding of the effect of excipient choices on insulin association state is necessary. In this study, commonly used parenteral preservatives were evaluated for their propensity to increase the ratio of insulin monomers in formulation, and with the assistance of a stabilizing excipient, CB[7]-PEG, which enhances insulin stability. We have identified that a formulation containing glycerol as a tonicity agent and phenoxyethanol as a preservative is the optimal combination to promote the insulin monomer, whereby upward of 85% of the insulin is in a monomer state. This formulation exhibits over tenfold extended stability compared to commercial formulations when formulated with CB[7]-PEG. Moreover, DOSY NMR highlights that CB[7]-PEG binding to insulin does not significantly impact the diffusivity of insulin or its association state. The increased insulin monomer composition in these formulations can potentially enable ultrafast onset of insulin action combined with short duration of action to allow for meal-time responsiveness with reduced post-prandial hypoglycemic events.
4. Experimental Section
Materials
CB[7]-PEG was prepared according to published protocols,[30] with method modification to enable copper “click” chemistry following reported protocols.[48] Novolog (Novo Nordisk) and Humalog (Eli Lilly) were purchased and used as received. Zinc-free lispro and zinc-free aspart were isolated using PD MidiTrap G-10 gravity columns (GE Healthcare) and then concentrated using Amino Ultra 3K centrifugal units (Millipore). All other reagents were purchased from Sigma-Aldrich unless otherwise specified.
SEC-MALS
Number-averaged molecular weight (Mn) and dispersity (Ð = Mw/Mn) of formulations were obtained using SEC carried out using a Dionex Ultimate 3000 instrument (including pump, autosampler, and column compartment) outfitted with a Dawn Heleos II multi angle light scattering detector and a Optilab rEX refractive index detector. The column was a Superose 6 Increase 10/300 GL from GE healthcare. Data were analyzed using Astra 6.0 software.
Zinc-free insulin lispro and aspart were evaluated under the following buffer conditions: i) sodium chloride (0.9%), ii) glycerol (2.6%), iii) glycerol (2.6%) and meta-cresol (0.315%), and iv) glycerol (2.6%) and phenoxyethanol (0.85%); controls consisted of either v) commercial Humalog formulation comprising glycerol (1.6%), meta-cresol (0.315%), dibasic sodium phosphate (0.188%), and zinc (0.00197%), or vi) commercial Novolog formulation comprising glycerol (1.6%), meta-cresol (0.172%), phenol (0.15%), sodium chloride (0.058%), dibasic sodium phosphate (0.125%), and zinc (0.00196%). Insulin lispro and aspart were injected at a concentration of a minimum 36 mg mL−1 protein and a volume of 100 μL. A dn/dc of 0.186 mL g−1 was used for all samples. The resulted in max peak concentrations ranging from 3.0 to 4.3 mg mL−1, depending of protein oligomerization equilibria.
The molar fraction of monomeric, dimeric, and hexameric insulin was determined by fitting experimentally derived number-averaged (Mn) and weight-averaged (Mw) molecular weights (Figure S6, Supporting Information) determined by SEC-MALS to Equations (1) and (2), where m, d, and h, respectively, represent the molar fractions of monomeric, dimeric, and hexameric insulin while I represents the molecular weight of monomeric insulin (5831 g mol−1). The solver was constrained so that m + d + h =1 while m, d, and h remain between 0 and 1.
| (1) |
| (2) |
In Vitro Stability
Methods for aggregation assays for recombinant human insulin were adapted from Webber et al.[30] Briefly, formulation samples were plated at 150 μL per well (n =3 per group) in a clear 96-well plate and sealed with optically clear and thermally stable seal (VWR). The plate was immediately placed into a plate reader and incubated with continuous shaking 37 °C. Absorbance readings were taken every 10 min at 540 nm for 100 h (BioTek SynergyH1 microplate reader). The aggregation of insulin leads to light scattering, which results in reduction of sample transmittance. The time for aggregation was defined as a > 10% increase in transmittance from the transmittance at time zero. Zinc(II) was removed from the insulin lispro and insulin aspart through competitive binding by addition of EDTA, which exhibits a dissociation binding constant approaching attomolar concentrations (KD ≈10−18 m).[41,42] EDTA was added to formulations (4 equiv. with respect to zinc) to sequester zinc from the formulation and then removed using a PD-10 desalting column (GE Healthcare) and then concentrated using Amino Ultra 3K centrifugal units (Millipore). Insulin lispro or aspart concentration was then measured by ELISA (Mercodia, Iso-Insulin ELISA) and then formulation excipients were added. All lispro or aspart formulations were formulated in phosphate buffer (pH =7.4) with the following combinations of excipients i) sodium chloride (0.9%), ii) glycerol (2.6%), iii) glycerol (2.6%) and phenol (0.25%), iv) glycerol (2.6%) and meta-cresol (0.315%), and v) glycerol (2.6%) and phenoxyethanol (0.85%). CB[7]-PEG was added to formulations at either three or five molar equivalents with respect to insulin. Controls included commercial formulations of Novolog (insulin aspart) and Humalog (insulin lispro).
NMR DOSY
1H 2D DOSY spectra were recorded at a protein concentration (lispro or aspart) of 3.4 mg mL−1 with 50–60% D2O under the following conditions: i) commercial Novolog (glycerol (1.6%), meta-cresol (0.175%), phenol (0.15%), sodium chloride (0.058%), dibasic sodium phosphate (0.125%), zinc (0.00196%)), ii) commercial Humalog (glycerol (1.6%), meta-cresol (0.315%), dibasic sodium phosphate (0.188%), and zinc (0.00196%)), iii) lispro/glycerol/phenoxyethanol (phosphate buffer, glycerol (2.6%), and phenoxyethanol (0.85%)), and iv) aspart/glycerol/phenoxyethanol (phosphate buffer, glycerol (2.6%), and phenoxyethanol (0.85%)). 1D 1H NMR of the insulin/CB[7]-PEG complex showed a broadening of both insulin and CB[7]-PEG signals (Figure S7, Supporting Information). This was exacerbated with an increasing ratio of CB[7]-PEG to insulin (Figure S8, Supporting Information). As such, an optimum ratio of CB[7]-PEG to insulin for DOSY was established to 60 mol% with respect to insulin. A Varian Inova 600 MHz NMR instrument was used to acquire the data. Magnetic field strengths ranged from 2 to 57 G cm−1. The DOSY time and gradient pulse were set at 132 ms (Δ) and 3 ms (δ) respectively. All NMR data were processed by using MestReNova 11.0.4 software.
Statistical Analysis
Insulin stability experiments were conducted with n =3 and the data are shown as the mean transmittance over time. The extracted time to aggregation (tA) was plotted as mean ± standard deviation. Rat studies were performed with n =5–6 rats for each treatment group and blood glucose results were reported as mean blood glucose ± standard deviation. In vitro protein kinase B (AKT) activity assays were performed with n =3 and data are shown as mean ± standard deviation of the relative pAKT normalized by the total AKT ([pAKT]/[AKT]). An EC50 regression (log(agonist) versus response (three parameters)) was plotted and an extra sum of squares F-test (α = 0.05) was performed using GraphPad Prism 8.
Supplementary Material
Acknowledgements
C.L.M. and A.A.A.S. contributed equally to this work. This work was funded in part by NIDDK R01 (NIH grant #R01DK119254) and a Pilot and Feasibility funding from the Stanford Diabetes Research Center (NIH grant #P30DK116074) and the Stanford Child Health Research Institute, as well as the American Diabetes Association Grant (1-18-JDF-011) and a Research Starter Grant from the PhRMA Foundation. C.L.M. was supported by the NSERC Postgraduate Scholarship. A.A.A.S. was funded by grant NNF18OC0030896 from the Novo Nordisk Foundation and the Stanford Bio-X Program, and also funded by the Danish Council of Independent Research (Grant No. DFF5054-00215). J.L.M. is supported by the Department of Defense NDSEG Fellowship and by a Stanford Graduate Fellowship. C.M.M. is supported by a Stanford Graduate Fellowship. The authors also acknowledge the High Throughput Bioscience Center (HTBC) at Stanford Medicine.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
The ORCID identification number(s) for the author(s) of this article can be found under https://doi.org/10.1002/adtp.201900094
Contributor Information
Caitlin L. Maikawa, Department of Bioengineering, Stanford University, Stanford, CA 94305, USA
Anton A. A. Smith, Department of Materials Science and Engineering, Stanford University, Stanford, CA 94305, USA Department of Science and Technology, Aarhus University, 8000 Aarhus, Denmark.
Lei Zou, Department of Chemical and Biomolecular Engineering, University of Notre Dame, Notre Dame, IN 46556, USA.
Catherine M. Meis, Department of Materials Science and Engineering, Stanford University, Stanford, CA 94305, USA
Joseph L. Mann, Department of Materials Science and Engineering, Stanford University, Stanford, CA 94305, USA
Matthew J. Webber, Department of Chemical and Biomolecular Engineering, University of Notre Dame, Notre Dame, IN 46556, USA
Eric A. Appel, Department of Bioengineering, Stanford University, Stanford, CA 94305, USA Department of Materials Science and Engineering, Stanford University, Stanford, CA 94305, USA; Department of Pediatrics (Endocrinology), Stanford University, Stanford, CA 94305, USA.
References
- [1].WHO, Diabetes: Key Facts, World Health Organization, Geneva: 2017. [Google Scholar]
- [2].Mo R, Jiang T, Di J, Tai W, Gu Z, Chem. Soc. Rev 2014, 43, 3595. [DOI] [PubMed] [Google Scholar]
- [3].Heise T, Zijlstra E, Nosek L, Rikte T, Haahr H, Diabetes, Obes. Metab 2017, 19, 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Maahs DM, West NA, Lawrence JM, Mayer-Davis EJ, Endocrinol. Metab. Clin. North Am 2010, 39, 481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Senior P, Hramiak I, Can. J. Diabetes 2019, 43, 515. [DOI] [PubMed] [Google Scholar]
- [6].Heptulla RA, Rodriguez LM, Bomgaars L, Mason K, Haymond MW, Diabetes 2005, 54, A110. [DOI] [PubMed] [Google Scholar]
- [7].Ryan GJ, Jobe LJ, Martin R, Clin. Ther 2005, 27, 1500. [DOI] [PubMed] [Google Scholar]
- [8].Want LL, Ratner R, Int. J. Clin. Pract 2006, 60, 1522. [DOI] [PubMed] [Google Scholar]
- [9].Jones MC, Am. Fam. Physician 2007, 75, 1831. [PubMed] [Google Scholar]
- [10].Taraghdari ZB, Imani R, Mohabatpour F, Macromol. Biosci 2019, 19, e1800458. [DOI] [PubMed] [Google Scholar]
- [11].Mathieu C, Gillard P, Benhalima K, Nat. Rev. Endocrinol 2017, 13, 385. [DOI] [PubMed] [Google Scholar]
- [12].Holleman F, Hoekstra JBL, Engl N J. Med 1997, 337, 176. [DOI] [PubMed] [Google Scholar]
- [13].Gast K, Schüler A, Wolff M, Thalhammer A, Berchtold H, Nagel N, Lenherr G, Hauck G, Seckler R, Pharm. Res 2017, 34, 2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hua QX, Weiss MA, J. Biol. Chem 2004, 279, 21449. [DOI] [PubMed] [Google Scholar]
- [15].Woods RJ, Alarcón J, McVey E, Pettis JR, J. Diabetes Sci. Technol 2012, 6, 265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Xu Y, Yan Y, Seeman D, Sun L, Dubin PL, Langmuir 2012, 28, 579. [DOI] [PubMed] [Google Scholar]
- [17].Weiss MA, Vitam. Horm 2009, 80, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Home PD, Diabetes, Obes. Metab 2012, 14, 780. [DOI] [PubMed] [Google Scholar]
- [19].Kamerzell TJ, Esfandiary R, Joshi SB, Middaugh CR, Volkin DB, Adv. Drug Delivery Rev 2011, 63, 1118. [DOI] [PubMed] [Google Scholar]
- [20].Rayaprolu BM, Strawser JJ, Anyarambhatla G, Drug Dev. Ind. Pharm 2018, 44, 1565. [DOI] [PubMed] [Google Scholar]
- [21].Meyer BK, Ni A, Hu B, Shi L, J. Pharm. Sci 2007, 96, 3155. [DOI] [PubMed] [Google Scholar]
- [22].Saha S, Deep S, Phys. Chem. Chem. Phys 2016, 18, 18934. [DOI] [PubMed] [Google Scholar]
- [23].Derewenda U, Derewenda Z, Dodson EJ, Dodson GG, Reynolds CD, Smith GD, Sparks C, Swenson D, Nature 1989, 338, 594. [DOI] [PubMed] [Google Scholar]
- [24].Manning MC, Chou DK, Murphy BM, Payne RW, Katayama DS, Pharm. Res 2010, 27, 544. [DOI] [PubMed] [Google Scholar]
- [25].Mitragotri S, Burke PA, Langer R, Nat. Rev. Drug Discovery 2014, 13, 655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Yang C, Lu D, Liu Z, Biochemistry 2011, 50, 2585. [DOI] [PubMed] [Google Scholar]
- [27].Guerreiro LH, Guterres MFAN, Melo-Ferreira B, Erthal LCS, Rosa M. d. S., Lourenço D, Tinoco P, Lima LMTR, AAPS Pharm SciTech 2013, 14, 1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Sisnande T, Guerreiro LH, Braga RR, Jotha-Mattos L, Erthal LCS, Tinoco P, Ferreira BM, Lima LMTR, PLoS One 2015, 10, e0138803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Veronese FM, Mero A, BioDrugs 2008, 22, 315. [DOI] [PubMed] [Google Scholar]
- [30].Webber MJ, Appel EA, Vinciguerra B, Cortinas AB, Thapa LS, Jhunjhunwala S, Isaacs L, Langer R, Anderson DG, Proc. Natl. Acad. Sci. U. S. A 2016, 113, 14189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hirotsu T, Higashi T, Abu Hashim II, Misumi S, Wada K, Motoyama K, Arima H, Mol. Pharmaceutics 2017, 14, 368. [DOI] [PubMed] [Google Scholar]
- [32].Bush M, Bouley N, Urbach AR, J. Am. Chem. Soc 2005, 127, 14511. [DOI] [PubMed] [Google Scholar]
- [33].Heitmann LM, Taylor AB, Hart PJ, Urbach AR, J. Am. Chem. Soc 2006, 128, 12574. [DOI] [PubMed] [Google Scholar]
- [34].Rajgariah P, Urbach AR, J. Inclusion Phenom. Macrocyclic Chem 2008, 62, 251. [Google Scholar]
- [35].Reczek JJ, Kennedy AA, Halbert BT, Urbach AR, J. Am. Chem. Soc 2009, 131, 2408. [DOI] [PubMed] [Google Scholar]
- [36].Jonassen I, Havelund S, Hoeg-Jensen T, Steensgaard DB, Wahlund PO, Ribel U, Pharm. Res 2012, 29, 2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Attri AK, Fernández C, Minton AP, Biophys. Chem 2010, 148, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Attri AK, Fernández C, Minton AP, Biophys. Chem 2010, 148, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Jensen MH, Wahlund P-O, Jacobsen JK, Vestergaard B, van de Weert M, Havelund S, J. Chromatogr. B 2011, 879, 2945. [DOI] [PubMed] [Google Scholar]
- [40].Ludwig DB, Webb JN, Fernández C, Carpenter JF, Randolph TW, Biotechnol. Bioeng 2011, 108, 2359. [DOI] [PubMed] [Google Scholar]
- [41].Berthon G, Handbook of Metal-Ligand Interactions in Biological Fluids: Bioinorganic Chemistry, Marcel Dekker, New York: 1995. [Google Scholar]
- [42].Waters RS, Bryden NA, Patterson KY, Veillon C, Anderson RA, Biol. Trace Elem. Res 2001, 83, 207. [DOI] [PubMed] [Google Scholar]
- [43].Whittingham JL, Chaudhuri S, Dodson EJ, Moody PCE, Dodson GG, Biochemistry 1995, 34, 15553. [DOI] [PubMed] [Google Scholar]
- [44].Smith GD, Ciszak E, Pangborn W, Protein Sci. 1996, 5, 1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Banerjee P, Mondal S, Bagchi B, J. Chem. Phys 2019, 150, 084902. [DOI] [PubMed] [Google Scholar]
- [46].Patil SM, Keire DA, Chen K, AAPS J. 2017, 19, 1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Adams GG, Meal A, Morgan PS, Alzahrani QE, Zobel H, Lithgo R, Kok MS, Besong DTM, Jiwani SI, Ballance S, Harding SE, Chayen N, Gillis RB, PLoS One 2018, 13, e0195010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Zou L, Braegelman AS, Webber MJ, ACS Appl. Mater. Interfaces 2019, 11, 5695. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.