Abstract
Intrinsic disorder (i.e., lack of a unique 3-D structure) is a common phenomenon, and many biologically active proteins are disordered as a whole, or contain long disordered regions. These intrinsically disordered proteins/regions constitute a significant part of all proteomes, and their functional repertoire is complementary to functions of ordered proteins. In fact, intrinsic disorder represents an important driving force for many specific functions. An illustrative example of such disorder-centric functional class is RNA-binding proteins. In this study, we present the results of comprehensive bioinformatics analyses of the abundance and roles of intrinsic disorder in 3,411 ribosomal proteins from 32 species. We show that many ribosomal proteins are intrinsically disordered or hybrid proteins that contain ordered and disordered domains. Predicted globular domains of many ribosomal proteins contain noticeable regions of intrinsic disorder. We also show that disorder in ribosomal proteins has different characteristics compared to other proteins that interact with RNA and DNA including overall abundance, evolutionary conservation, and involvement in protein–protein interactions. Furthermore, intrinsic disorder is not only abundant in the ribosomal proteins, but we demonstrate that it is absolutely necessary for their various functions.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-013-1446-6) contains supplementary material, which is available to authorized users.
Keywords: Intrinsically disordered protein, Moonlighting protein, Protein–protein interaction, Protein–RNA interaction, Ribosomal proteins
Introduction
It is accepted now that many biologically active proteins do not have a unique 3-D structure as a whole or in part [1–5]. These intrinsically disordered proteins (IDPs) and intrinsically disordered protein regions (IDPRs) possess highly flexible structures and exist as conformational dynamic ensembles characterized by different degree and depth of disorderedness [2, 4, 6–10]. IDPs/IDPRs are highly abundant in virtually any given proteome [1, 3, 5, 11]. Biological functions of IDPs, which are typically involved in regulation, signaling, and control pathways [12–14], represent a crucial complementation to the functional repertoire of ordered proteins [15–18].
Intrinsic disorder was shown to be very common in RNA- and DNA-binding proteins [4, 8, 9, 19]. The results of the analysis of the Saccharomyces genome suggested that proteins containing disorder are over-represented in the cell’s nucleus and are likely to be involved in the regulation of transcription and cell signaling [3]. Systematic bioinformatics studies revealed a significant prevalence of intrinsic disorder in transcription factors [20–22]. For example, analysis of 401 human transcription factors showed that IDPRs occupy ~50 % of the entire sequence of human transcription factors [22].
Multiple functions are associated with the RNA-binding proteins, which are believed to determine RNA fate from synthesis to decay [23]. For example, intrinsically disordered C-terminal domain allows La protein to interact productively with a diversity of noncoding RNA precursors, protect these RNAs from nucleases and affect folding, maturation, and ribonucleoprotein assembly [24]. Other intrinsically disordered RNA-binding proteins often act as specific RNA chaperones, assisting in the structural rearrangements of RNA molecules [25]. An illustrative example of such disordered RNA chaperones are viral core proteins from different Flaviviridae genera [26], bunyavirus nucleocapsid protein [27], hantavirus nucleocapsid protein [28], and potentially core proteins of Pestivirus [29].
In addition to the RNA chaperone activity, many RNA-binding proteins possess a multitude of intrinsic disorder-dependent functions. For example, serine/arginine-rich (SR) splicing factors that play an important role during several steps of RNA metabolism and are involved in constitutive and alternative splicing, were shown to be IDPs [30]. Intrinsic disorder in a small RNA-binding protein, the HIV-1 transcriptional regulator Tat, is essential for viral gene expression and replication, as well as for the ability of Tat to interact with a large number of proteins within both infected and non-infected cells [31, 32]. The intrinsically disordered SARS-CoV nucleocapsid protein binds to the viral RNA genome, forms the ribonucleoprotein core, and is involved in several important functions in the viral life cycle [33]. The intrinsic disorder is used by the stem-loop binding protein (SLBP) for the regulation of histone mRNAs, since the disordered N-terminal domain of SLBP contains signals for mRNA translation and histone mRNA import [34]. Intrinsic disorder in SBP2, which is the SECIS Binding Protein 2 that specifically interacts with a stem-loop structure in the 3′ UTR RNA (the SECIS element), is important for the co-translational incorporation of selenocysteine into selenoproteins at a reprogrammed UGA codon [35].
The ribosome is a large ribonucleoprotein catalyzing protein translation. Although the ribosomes are responsible for the synthesis of proteins across all kingdoms of life, and although their core functions are mRNA decoding and catalysis of the peptide bond formation [36], other translation-related processes (such as initiation, termination, and regulation) are quite different in different domains of life [37, 38]. Since the eukaryotic ribosomes are directly involved in many eukaryote-specific cellular processes, they are at least 40 % larger than their bacterial counterparts due to the presence of additional ribosomal RNA (rRNA) elements called expansion segments and extra ribosomal proteins [39]. In prokaryotes, there are 70S ribosomes, with small and large subunits of 30S and 50S, respectively. The small 30S subunit contains a 16S ribosomal RNA (rRNA) and 21 proteins, whereas in the large 50S subunit there are two rRNAs (5S and 23S) and 31 proteins. The eukaryotic 80S ribosome consists of a small (40S) and a large (60S) subunit. In the 40S small subunit, there is a single 18S rRNA and 33 proteins. The eukaryotic 60S subunit is composed of three rRNAs (5S rRNA, 28S rRNA, and 5.8S rRNA) and 46 proteins [40]. Of the 79 eukaryotic ribosomal proteins, 32 have no homologs in the bacterial or archaeal ribosomes, and those that do have homologs possess long eukaryote-specific extensions [41].
Ribosomal proteins represent an interesting and important category of RNA-binding IDPs due to their unique functional and structural properties. In addition to being a crucial part of the ribosome, many ribosomal proteins are involved in translational regulation via binding to operator sites located on their own messenger RNA [42]. Based on the analysis of the crystal structures of the ribosome subunits, it was discovered that almost half of the ribosomal proteins have globular domains with long extensions that penetrate deeply into the ribosome particle’s core [43–50]. It was indicated that these extensions are disordered in solution but still play a key role in ribosomal assembly [49, 51–53]. In fact, the hypothesis is that the long basic extensions of ribosomal proteins (e.g., L3, L4, L13, L20, L22, and L24) can penetrate deeply into the ribosome subunit cores, undergo disorder-order transition individually or co-fold with their RNA, therefore facilitating the proper rRNA folding [49]. It was also indicated that different extensions do not play a similar role in the assembly of the ribosome subunits in vivo and might have some other functions [49].
Although the fact that in their non-bound forms many ribosomal proteins are either completely disordered or contain long disordered regions has been known for a long time (e.g., ribosomal proteins were included in the early bioinformatics studies dedicated to the sequence peculiarities [4] and functional repertoire of IDPs [19]), the abundance and functional roles of intrinsic disorder in these proteins have never been the subject of focused large-scale bioinformatics analysis. Our study fills this gap by reporting the results of the bioinformatics analysis of 3,411 ribosomal proteins from 32 species. We show here that intrinsic disorder is very common among all the analyzed ribosomal proteins, that it has unique characteristics which differentiate it from the disorder in other RNA- and DNA-binding proteins, and that it plays a role in the various functions of these important RNA-binding proteins.
Materials and methods
Dataset of ribosomal proteins
We collected 3,438 proteins from the Ribosomal Protein Gene Database (RPG) [54] on Nov 7, 2011. This set includes proteins from 24 species in Eukaryota, four in Archaea, and four in Bacteria, respectively. We excluded 27 small peptides with <30 amino acids because they could not be predicted by MFDp [55]. The final dataset, named RPG_3411, is summarized in Table S1.
Datasets of RNA- and DNA-binding proteins
We also collected a representative subset of RNA- and DNA-interacting proteins from a current release of UniProt [56] for the same set of species as in the RPG dataset. Next, for each species we selected at random a subset of RNA- and DNA-interacting proteins to match the number of ribosomal chains. The corresponding sets of DNA-binding, RNA-binding, and the ribosomal proteins are summarized in Table S1. This allowed us to represent a wide spectrum of the nucleic acid interacting chains while keeping the dataset sizes at a level that allows complete computational analysis. The combined set of RNA/DNA-binding chains includes 3,084 proteins; this number is slightly lower than the size of RPG set since some proteins interact with both RNA and DNA and a couple of species (Fusarium graminearum and Rhizopus oryzae) had fewer DNA/RNA–interacting proteins annotated in UniProt than the corresponding number of ribosomal chains in the RPG dataset.
Evaluation of the surface and interface areas
The solvent-accessible surface area (ASA) for all the ribosomal proteins of the eukaryotic ribosome (PDB ID: 3U5C and 3U5E [57]) was calculated using an in-house program based on the double cubic lattice algorithm [58] as implemented in the BALL library [59]. The ASA of a protein is calculated with a probe radius of 1.4 Å. The interface area buried by a complex is defined as the difference between the surface area of the complex and the sum of the surface areas of two partners, where the indicated chain is considered as one partner and the remainder of the subunit (including the rRNA) is taken as the other partner: interface ASA = ASApartner 1 + ASApartner 2 − ASAcomplex. As observed by a reviewer, the ASA of the bound structures of IDPRs are not a measure of the ASA of free IDPRs. Nonetheless, these calculations are useful in distinguishing the unbound order/disorder state of components of a complex structure using the Nussinov’s plot [60].
Nussinov’s plot
According to Gunasekaran et al. [60], the per-residue ASA versus per-residue interface ASA clearly distinguishes between the two classes of proteins, with monomers in the two-state complexes being characterized by extended shapes and larger interface areas, and with monomers in the three-state complexes being more globular and compact. See text below for explanation of two-state and three-complexes. In fact, in the per-residue ASA versus the per-residue interface ASA plot (Nussinov’s plot), the two-state and three-state complexes occupy very different areas, with the disordered proteins (that form complexes in a two-state mechanism) being distributed sparsely over a broad area in the top-right part of the plot, suggesting that disordered proteins opt for extended shapes and larger interface areas, and with ordered proteins (that from complexes in a three-state mechanism) being condensed in the small area at the bottom-left corner of the plot, suggesting that these proteins are more globular and compact in their bound form [60]. Furthermore, it was also pointed out that since the maxima of per-residue surface and interface areas for stable monomers lie around 80 Å2, the line connecting these two extreme values in the per-residue surface area versus the per-residue interface area plot represents a natural boundary separating ordered and disordered proteins forming three-state and two-state complexes, respectively [60]. Here, ordered proteins were systematically located below this boundary, and the disordered proteins were widely spread above the boundary [60].
Identification of likely disorder-to-order transition regions
The Nussinov plot is useful when the proteins of a complex are completely ordered or disordered, but can give ambiguous results when proteins contain both ordered and disordered regions. For these structures, a method to segment each protein of a complex into likely ordered and likely disordered segments would resolve the ambiguity. We base such a method on a similar principle used for the Nussinov plot, the complex structures of IDPRs will have a higher ASA than the structures of ordered regions. The idea behind the method is framed in terms of structural context: a residue with a low ASA is likely in a context in which it is folded and a residue with a high ASA has likely been removed from a context in which it folds. In context (IC) and out of context (OC) residues were modeled using a discrete finite automaton (DFA) with two states. Each state is characterized by the emission probability distributions of the ASA of each residue type—alanine, cysteine, aspartic acid, etc. The ASA distribution of IC residues was calculated directly from a sequence unique set of 4,725 monomer X-ray structures from PDB. The ASA distribution of OC residues was estimated from the same set of structures, but considering only a short sequence window around each residue when calculating the ASA, i.e., the ASA of each residue is calculated out of the context of the monomer structure. ASA distributions were discretized using the method of Fayyad and Irani [61]. A window size of 11 was selected based on convergence of the IC and OC distributions with varying window size (data not shown). Transition probabilities for the DFA were selected to correspond with an average IC region length of 200 residues and an average OC region length of 20 residues. Classification of IC/OC was made by calculating the OC posterior probability using the forward/backward algorithm. For ribosomal proteins, posteriors were calculated from ASAs calculated on the isolated protein structures.
Amino acid composition analysis
Amino acid compositional analysis was carried out using Composition Profiler [62] (http://www.cprofiler.org) using the PDB Select 25 [63] and the DisProt [64] datasets as reference for ordered and disordered proteins, respectively. Enrichment or depletion in each amino acid type was expressed as (C x − C order)/C order, i.e., the normalized excess of a given residue’s content in a query dataset (C x) relative to the corresponding value in the dataset of ordered proteins (C order).
Search for potential globular domains in 3,438 ribosomal proteins
Potential globular domains in ribosomal proteins were identified using the GlobPlot server (http://globplot.embl.de/), which is a popular predictor based on a running sum of the propensity for amino acids to be in an ordered or disordered state [65]. GlobPlot is a computationally efficient Web service that allows the user to plot the tendency within the query protein for order/globularity and disorder [65] and was recently evaluated to provide competitive predictive performance [66].
Computational evaluation of disorder
The disorder was predicted with MFDp method [55], which is a consensus-based predictor that was recently shown to provide strong and competitive predictive quality [67, 68]. MFDp predictions were used to calculate the disorder content (fraction of disordered residues), the number of disordered segments, and the number of long disordered segments that consists of at least 30 consecutive disordered amino acids; such long segments were found to be implicated in protein–protein recognition [69]. We only counted the disordered segments with at least four consecutive disordered residues, which is consistent with other reports [67, 70]. We also assumed that a given domain is considered to be disordered if it includes at least one disordered region with at least four consecutive disordered residues, and to be significantly disordered if at least half of its residues are disordered.
We also used the DisCon method [71] to predict the overall content (fraction of the disordered residues) in the protein chains. DisCon provides more accurate disorder content predictions when compared with MFDp and several other recent disorder predictors [71], but it does not predict the disorder at the residue level, contrary to MFDp. The residue-level predictions allow for a more insightful analysis, including an investigation into the number and size of the predicted disordered segments. In addition to DisCon, two binary disorder classifiers, charge-hydropathy (CH) plot [4, 72] and cumulative distribution function (CDF) plot [72, 73], as well as their combination known as CH–CDF analysis [73–75], were used.
Search for potential functional sites
We predicted function of the disordered segments based on a local pairwise alignment against functionally annotated disordered segments collected from DisProt 5.9 [64]. We aligned each of the 7,548 disordered segments extracted from the RPG_3411 dataset into a set of 775 disordered segments collected from DisProt database that have functional annotation. We calculated alignment using the Smith–Waterman algorithm [76] using the EMBOSS implementation with default parameters (gap_open = 10, gap_extend = 0.5, and blosum62 matrix). We defined sequence similarity as the number of identical residues in the local alignment divided by the length of the local alignment or the length of the shorter of the two being aligned segments, whichever is larger. We transferred the annotation if the similarity was >0.8; this means that some of the segments may be annotated with multiple functions. The value of the threshold was chosen to assume high similarity even in cases of alignment to a short segment, i.e., for the shortest segments of five residues at least four amino acids have to be matched. Consequently, we successfully annotated 911 disordered segments with 26 functions that are listed in Table S2. These annotations were used to discuss difference of the functional roles between short and long disordered segments in the ribosomal proteins.
We used MoRFpred method [77], which is a leading predictor of molecular recognition features (MoRF), to annotate MoRF regions. MoRFs are short (5–25 amino acids) disordered regions with which undergo disorder-to-order transition upon binding to protein partners and are implicated in signaling and regulatory functions [2, 78–80]. Following Mohan et al. [80], we grouped MoRF regions into α-MoRFs (that fold into α-helices), β-MoRFs (that fold into β-strands), γ-MoRFs (coils) and complex-MoRFs (mixture of different secondary structure), based on the secondary structure predicted with PSI-PRED [81].
Calculation of sequence conservation
We also report sequence conservation for the ordered residues, the disordered residues and the residues in long (with at least 30 consecutive disordered amino acids) disordered segments. The conservation was quantified with relative entropy [82] that was calculated from the Weighted Observed Percentages (WOP) profiles generated by PSI-BLAST [83]. PSI-BLAST was run with default parameters (-j 3, -h 0.001) against the NCBI’s non-redundant (nr) protein database, which was filtered using PFILT [84] to remove low-complexity regions, trans-membrane regions and coiled-coil regions. The use of the relative entropy is motivated by work in [82] that suggests that it leads to more biologically relevant results compared to some other conservation scores and the fact that it was recently applied to investigate disorder in histones [85] and to identify nucleotide-binding residues [86] and catalytic sites [87].
Results
Abundance of intrinsic disorder in ribosomal proteins as evidenced from the crystal structure of the eukaryotic ribosome
Bioinformatics analysis of the full-length ribosomal proteins from Saccharomyces cerevisiae
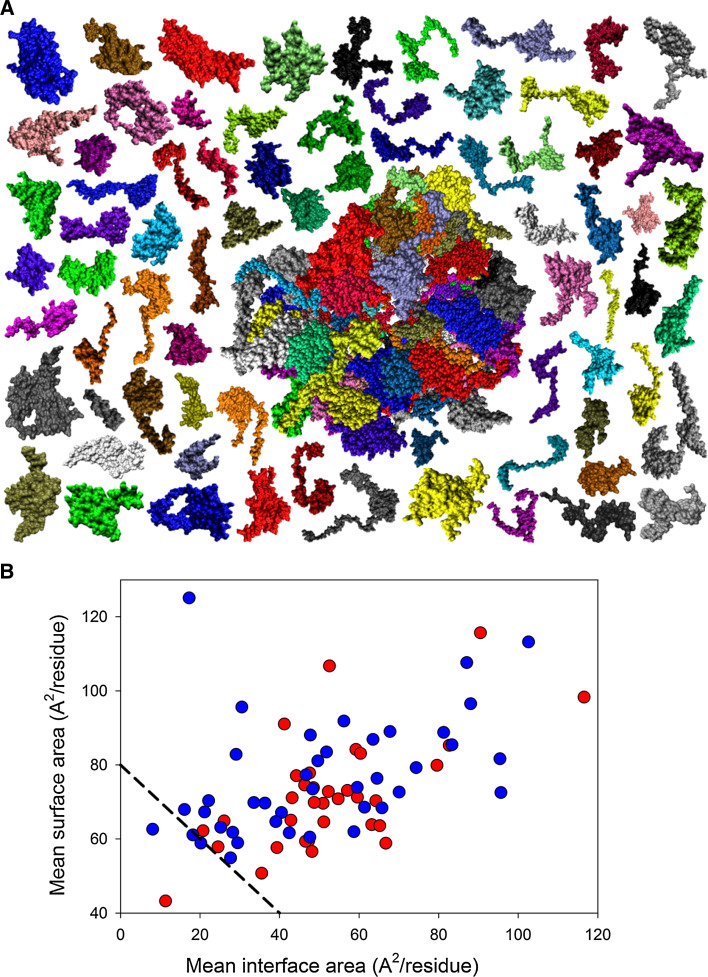
Figure 1a represents the results of the computational disassembly of protein components of the eukaryotic ribosome from the yeast S. cerevisiae and shows that the complex structure of this important nucleoprotein relies on the intrinsic disorder of ribosomal proteins. In fact, even simple visual inspection of the individual ribosomal proteins clearly shows that almost all of them possess very unusual shapes, which are not consistent with simple globular structure. These peculiar shapes suggest that many ribosomal proteins form the so-called two-state (or disordered) complexes, where the monomers unfold upon complex separation. Therefore, individual chains in such complexes are disordered in their unbound forms and fold at complex formation. This behavior is different from that of the so-called three-state (or ordered) complexes, individual chains of which are independently folded even in the unbound state [88, 89].
Fig. 1.
a Computational disassembly of the eukaryotic ribosome from the yeast Saccharomyces cerevisiae (PDB ID: 3U5C and 3U5E; [57]). Structure of the proteinaceous component of the ribosome is shown at the center of the plot as a large complex, and structures of the individual ribosomal proteins are positioned around this central complex. The figure clearly shows that there are almost no ribosomal proteins with simple globular shape, and many of them contain long protrusions or extensions. b Plot of per-residue surface versus per-residue interface areas. Surface and interface area normalized by the number of residues in each chain for the ribosomal proteins were estimated as described in [60]. Proteins of the 40S and 60S subunits are shown by red and blue circles, respectively. A boundary separating ordered and disordered complexes is shown as a black dashed line
As it was mentioned, Nussinov’s plot, where the per-residue surface area is plotted versus per-residue interface area for protein complexes, can distinguish between these two classes of proteins, with monomers in the two-state complexes being characterized by extended shapes and larger interface areas, and with monomers in the three-state complexes being more globular and compact [60]. In fact, the two-state and three-state complexes occupy very different areas in the Nussinov’s plot, with the disordered proteins (that form complexes in a two-state mechanism) being distributed sparsely over a broad area in the top-right part of the plot (above the boundary), suggesting that disordered proteins opt for extended shapes and larger interface areas, and with ordered proteins (that from complexes in a three-state mechanism) being condensed in the small area at the bottom-left corner of the plot (below the boundary, suggesting that these proteins are more globular and compact in their bound form [60].
In agreement with these observations, Fig. 1b shows that almost all ribosomal proteins from the eukaryotic ribosome are located above the order–disorder boundary suggested by Gunasekaran et al. [60]. There are only two clear exceptions to this rule, the protein RACK1 found in the small ribosomal subunit and the ribosomal protein L11 of the large subunit. Five more proteins touch the boundary, with two proteins from the 60S subunit, L3 and L9, being located slightly below the line, and three proteins (L23-A, S1-A, and S12) being found right above the boundary. It is important to note here that although RACK1 is considered to be a component of the small (40S) ribosomal subunit S. cerevisiae, it is not a typical ribosomal protein, being classified as 40S-associated protein. In fact, RACK1 is the guanine nucleotide-binding protein subunit β-like protein, also known as the receptor of activated protein kinase C1 RACK1. This protein is located at the head of the 40S ribosomal subunit in the vicinity of the mRNA exit channel [90]. It acts as a scaffold protein recruiting some other proteins to the ribosome and is involved in the negative regulation of translation of a specific subset of proteins [90]. Since the absolute majority of the yeast ribosomal proteins is located above the boundary of the Nussinov’s plot, these observations suggest that almost all of them belong to the category of proteins participating in the formation of two-state complexes. In other words, the vast majority of ribosomal proteins are mostly unstructured in their unbound state but fold to a different degree upon the ribosome formation. In fact, the hypothesis on the mostly unfolded nature of unbound ribosomal proteins is in agreement with earlier experimental studies which showed that many individual ribosomal proteins do not possess ordered structure in their non-bound forms or at least contain long disordered regions [4, 91–104]. The conclusion on the different degree of folding in bound state follows from the visual inspection of protein structures shown in Fig. 1a suggesting that many ribosomal proteins are folded to different degrees and possess both globular and non-globular domains in their bound forms (see below for more detailed analysis of this phenomenon). Furthermore, analysis of the yeast ribosome crystal structure revealed that many ribosomal proteins contained long stretches of residues with missing electron density. These regions of missing electron density correspond to protein segments that retain high conformational flexibility in their bound forms precluding them from being detected in the crystallography experiments. Some of these regions with missing electron density, which can be found in REMARK 465: MISSING RESIDUES section of corresponding PDB entries, are (in the 40S subunit of the ribosome, PDB ID: 3U5C): residues 208–252 in S0-A, residues 1–19 and 334–355 in S1-A, residues 1–33 and 251–254 in S2, residues 226–240 in S3, residues 1–19 in S5, residues 227–236 in S6-A, residues 124–134 in S8-A, residues 187–197 in S9-A, residues 1–19 in S12, residues 1–10 in S14-A, residues 1–7 and 132–142 in S15, residues 90–94 and 127–136 in S17-A, residues 1–14 in S20, residues 1–35 and 106–107 in S25-A, residues 99–119 in S26-A, residues 1–81 in S31, residues 1–8 and 142–273 in suppressor protein STM1. In the 60S subunit of the yeast ribosome, PDB ID: 3U5E, the proteins with long regions of missing electron density are L6-A (residues 110–128), L7-A (residues 1–22), L8-A (residues 1–23), L10 (residues 103–111), L22-A (residues 1–8 and 109–121), L24-A (residues 99–155), L25 (residues 1–21), L30 (residues 1–8), L34-A (residues 114–121), and L40 (residues 1–76).
Identification of likely disorder-to-order transitioning regions within the ribosomal proteins from S. cerevisiae
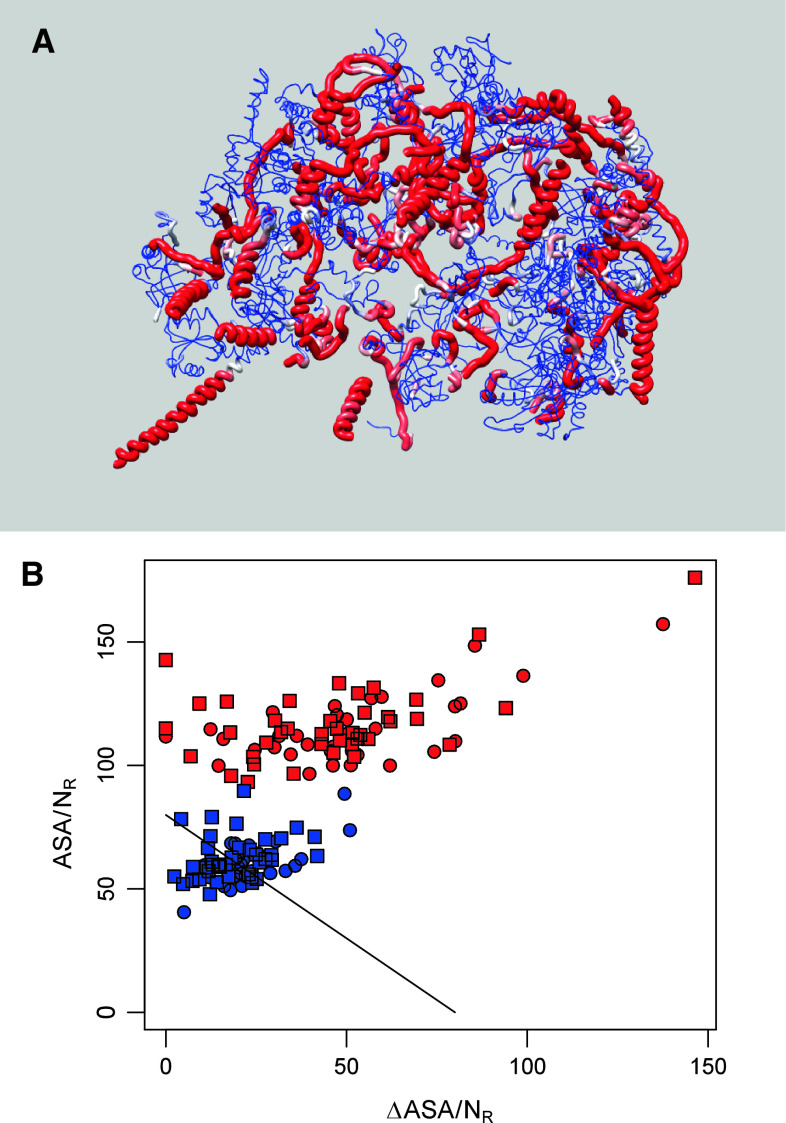
Visual analysis of individual ribosomal proteins in Fig. 1a reveals that many of these proteins have a structured (often globular) domain that might fold independently to binding to the rRNA or other ribosomal proteins and also possess long non-globular domains that are used for interactions with binding partners too. To find how this morphological heterogeneity might affect disorder-to-order transitions, we put together a statistical method for separating extended and collapsed regions based on accessible surface area analysis. The method is based on a discrete finite automaton (DFA) with two states, where one modeled on residues from intact proteins and another modeled on residues from local fragments (see “Materials and methods”). Each residue type is treated separately. The DFA analysis provided a probability that each residue is in/out of context (IC/OC; i.e., the probability that the residues is included or not included in globular structure) and all the ribosome proteins were split into IC and OC residues. Figure 2a represents results of this analysis by showing all 60S proteins with OC are mapped to radius and color. Here, color and width of ribbon corresponds to the OC posterior probability, where regions with a high probability are red and wide and regions with a low probability are blue and thin. This figure agrees well with other data and shows that many ribosomal proteins has long regions with OC residues; i.e., regions not involved in globular structures. Next, we calculated the Nussinov’s plot for each set of residues separately for each protein. The results of this analysis are shown in Fig. 2b, where data for IC and OC regions of 40S (circles) and 60S (squares) ribosomal proteins are shown by blue and red symbols, respectively. Figure 2b illustrates that all OC regions are clearly disordered in their unbound state and undergo binding-induced folding. Also, many globular domains are disordered when unbound. Although many IC regions seem to be ordered prior to binding, the vast majority of points corresponding to these regions/domains are clustered in close proximity to the order–disorder boundary suggested by Gunasekaran et al. [60]. Therefore, the results of these analyses suggest that many ribosomal proteins are entirely disordered in the unbound form and a noticeable portion of their globular domains is formed as a result of binding to rRNA or other ribosomal proteins.
Fig. 2.
Foldability of globular and extended domains of ribosomal proteins from the yeast Saccharomyces cerevisiae. a Worm representation of 60S proteins. Color and width of ribbon corresponds to the OC posterior probability, where regions with a high probability are red and wide and regions with a low probability are blue and thin. b Nussinov’s plot of ΔASA against the ASA for the IC (blue) and OC (red) residues of 40S (circles) and 50S (squares) proteins
Contact order analysis of the ribosomal proteins from S. cerevisiae
Figure 3 represents the results of the contact order analysis of the conformations adopted by ribosomal proteins in their bound states. The contact order values were computed for proteins from the eukaryotic ribosome (PDB IDs: 3U5C and 3U5E) based on a recent definition of the residue–residue contacts [105], where two residues are assumed in contact if their C β atoms (except for G where we use Cα atoms) are separated by <8Å. The plot shown in Fig. 3 is the asymmetric bimodal distribution with the bigger peaks corresponding to the structures with lower contact order (in the ranges of 0.05–0.10 and 0.10–0.15 for the small and large ribosomal subunits, respectively) and much smaller peaks corresponding to the structures with the relatively high contact order (in the range of 0.20–0.25). One should remember that the low contact order values could be indicative of an elongated structure or low density packing of residues in a globular structure. However, the analysis of structures of the eukaryotic ribosomal proteins with low contact order clearly shows that they possess highly extended structures (e.g., chains R and b of the 60S subunit and chains e and h of the 40S subunit) or have highly asymmetric hybrid structures containing relatively small globular domains and disproportionally long extended regions (chain f of the 40S ribosomal subunit). On the other hand, proteins with high contact order are characterized by the presence of large globular domains and short extended protrusions.
Fig. 3.

Contact order values for proteins from the eukaryotic ribosome (PDB IDs: 3U5C and 3U5E). The figure includes three distributions of the contact order values: for all chains combined (black line), for 3U5C (green line), and for 3U5E (red line). The chains’ identifiers from these proteins that have contact order values in a given interval are listed above the x-axis. Illustrative examples of structures of the ribosomal proteins with low contact order [chains e, f, and h in the crystal structure of the 40S subunit (PDB ID: 5U3C), and chains R and b in the crystal structure of the 60S subunit (PDB ID: 3E5E)] and the ribosomal proteins with relatively high contact order [chains U and c in the crystal structure of the 40S subunit (PDB ID: 5U3C), and chains c, d, f, and o in the crystal structure of the 60S subunit (PDB ID: 3E5E)] are shown on the sides of the plot
Some peculiarities of the amino acid compositions of ribosomal proteins
Amino acid compositions of the full-length ribosomal proteins
Analysis of the amino acid composition biases can provide interesting information on the nature of a protein. For example, the amino acid compositions of extended IDPs are characterized by some global biases, where low mean hydropathy is combined with high mean net charge. These global biases determine the highly unstructured and extended state of these proteins, since high net charge leads to strong electrostatic repulsion, and low hydropathy prevents efficient compaction [4]. In agreement with these global observations, IDPs were shown to be significantly depleted in so-called order-promoting amino acids, C, W, I, Y, F, L, H, V, and N, and substantially enriched in disorder-promoting residues, A, G, R, T, S, K, Q, E, and P [8, 15, 62, 106, 107]. We use a computational tool, Composition Profiler [62], to investigate the compositional biases in ribosomal proteins. This approach is based on the calculation of a normalized composition of a given protein or protein dataset in the (C s − C order)/C order form, where C s is a content of a given residue in a query (ribosomal) protein or dataset, and C order is the corresponding value for the set of ordered proteins from PDB Select 25 [63]. Figure 4a shows that, in comparison with typical ordered proteins, ribosomal proteins from all three domains of life are depleted in the major order-promoting amino acids, C, W, F, Y, L, V, H, and N, and are enriched in some disorder-promoting residues, particularly R, K, G (except for eukaryotic ribosomal proteins), A (except for archaeal ribosomal proteins), and E (except for eukaryotic ribosomal proteins). Obviously, the enrichment in positively charged R and K residues is determined by the functional need for the ribosomal proteins to interact with negatively charged rRNA. This high lysine-arginine content also defines the unusually high pI values reported for the majority of the ribosomal proteins (average pI ~ 10.1). Overall, the pronounced depletion in bulky hydrophobic and aromatic amino acids and enrichment in polar and charge residues may define the low propensity of ribosomal proteins for autonomous (or partner-independent) folding. On the other hand, there are several interesting compositional biases for the ribosomal proteins that differentiate them from the typical IDPs. These biases include some enrichment in the order-promoting amino acids I and V, and the noticeable depletion in the content of disorder-promoting residues T, D, Q, and S.
Fig. 4.

Fractional difference in the amino acid composition between the different members of the family of ribosomal proteins from Bacteria (green bars), Archaea (red bars), and Eukaryota (yellow bars) and a set of completely ordered proteins calculated for each amino acid residue (compositional profiles). The fractional differences were evaluated for the full-length ribosomal proteins (a) and for extended (b) and globular domains (c). The fractional difference was calculated as (C x − C order)/C order, where C x is the content of a given amino acid in a query set, and C order is the corresponding content in the dataset of fully ordered proteins. Composition profile of typical intrinsically disordered proteins from the DisProt database is shown for comparison (black bars). Positive bars correspond to residues found more abundantly in ribosomal proteins, whereas negative bars show residues, in which ribosomal proteins are depleted. Amino acid types were ranked according to their increasing disorder-promoting potential [15]. Panel d shows enrichment of amino acid M in the functions assumed by disordered regions that are considered in this work. We considered 26 functions from Table S2 that were annotated using DisProt database (as explained in the “Materials and methods” section); to assure statistically sound results 13 functions that have at least 20 annotated segments are shown. The fractional difference was calculated for M for the 13 functions that are sorted alphabetically on the x-axis. Positive bars correspond to function (disordered segments annotated with a given function) found with high counts of M while negative bars show functions where M is depleted. Panels e and f compare the amino acid compositions of the ribosomal, RNA- and DNA-binding proteins. In e, the fractional difference was calculated as (C x − C order)/C order, where C x is the content of a given amino acid in a query set, and C order is the corresponding content in the dataset of fully ordered proteins. In f, the compositions of the RNA- and DNA-binding proteins are compared with the general amino acid composition of the ribosomal proteins. Here, the normalized compositions of the RNA- and DNA-binding proteins are evaluated in the (C s − C ribosomal)/C ribosomal form, with C s being a content of a given residue in a dataset of the RNA- or DNA-binding proteins), and C ribosomal being the corresponding value for ribosomal proteins. In both plots, composition profiles of typical intrinsically disordered proteins from the DisProt database are shown for comparison (black bars)
Compositions of globular domains and extended regions
We analyzed peculiarities of the amino acid compositions of globular and disordered domains predicted using the GlobPlot server. Figure 4b shows that all non-globular regions of the ribosomal proteins clearly possess compositions typical for the IDPs/IDPRs, being enriched in major disorder-promoting residues and depleted in order-promoting residues. On the other hand, Fig. 4c illustrates that predicted globular domains possess amino acid biases consistent with the idea that they might contain a significant amount of disorder. In fact, in many respects, the composition profiles of globular domains resemble profiles calculated for the full-length ribosomal proteins. These domains are depleted in all order-promoting residues except for isoleucine and are enriched in some disorder-promoting residues (e.g., G, A, K, and E). Figure 4d provides further analysis of amino acid methionine that we found to be substantially enriched in extended regions (Fig. 4b) while being moderately depleted in globular domains (Fig. 4c). We studied the enrichment/depletion of this residue type over all segments with functional annotations (as explained in the “Materials and methods” section); we consider 13 functions that are possessed by at least 20 annotated sequences. We found that enrichment in methionine is associated with several functions carried out by disordered regions, such as polymerization, transactivation, autoregulation, regulation of apoptosis, and interactions with RNA and metals.
Overall characterization of the intrinsic disorder in ribosomal, RNA-, and DNA-binding proteins
Ribosomal proteins are important components of the ribonucleoprotein machine, the ribosome, where they specifically interact with rRNA and other ribosomal proteins. Therefore, it was interesting to compare the various behaviors of the ribosomal protein group (RPG) with those of general RNA- and DNA-binding proteins. To this end, representative sample sets of RNA- and DNA-binding proteins were assembled as described in the “Materials and methods” section and these three datasets were used in the subsequent studies.
Figure 4e and f represent the comparison of amino acid compositions of the ribosomal proteins, RNA- and DNA-binding proteins. In Fig. 4e, the normalized amino acid compositions of these three classes of nucleic acid-binding proteins are shown. Here, the normalized compositions were calculated as described above; i.e., in the (C s − C order)/C order form, where C s is a content of a given residue in a query dataset (IDPs, ribosomal, RNA- and DNA-binding proteins), and C order is the corresponding value for the set of ordered proteins from PDB Select 25 [63]. This figure shows that all nucleic acid-binding proteins are characterized by comparable depletion in order-promoting residues. As far as disorder-promoting residues are concerned, while the RNA- and DNA-binding proteins generally follow the trend typical for the IDPs, being moderately enriched in major disorder-promoting residues, the ribosomal proteins are quite different. Two major features strike the eye—substantial enrichment of the ribosomal proteins in R and K compensated by noticeable depletion in D, Q, S, and E residues. To get a better understanding of the amino acid composition biases of the RNA- and DNA-binding proteins relative to the ribosomal proteins, we evaluated their normalized compositions in the (C s − C ribosomal)/C ribosomal form, with C s being a content of a given residue in a dataset of the RNA- or DNA-binding proteins), and C ribosomal being the corresponding value for ribosomal proteins. Results of this analysis are shown in Fig. 4f, which re-emphasizes the relative depletion of the RNA- and DNA-binding proteins in N, D, Q, S, E, and P and their depletion in V, R, A, and K. Generally, data shown in Fig. 4e and f suggest that the RNA- and DNA-binding proteins are closer to each other than to the ribosomal proteins.
The average disorder content (i.e., the fraction of disordered residues) in the ribosomal protein group (RPG) ranges between 36 and 37.4 % across the three domains of life, see Fig. 5. This is substantially higher than the overall disorder content in various proteomes, which was estimated to be 18.9, 5.7, and 3.8 % for Eukaryota, Bacteria, and Archaea, respectively [3]. Our results indicate similar levels of disorder in the three domains of life and across the 32 considered species, with the lowest content at over 28 %. Figure 5 also shows that between 2.5 and 23.2 % of ribosomal proteins across the 32 species are fully disordered, with the largest average fraction (11.7 %) of fully disordered chains being found in the bacterial species.
Fig. 5.
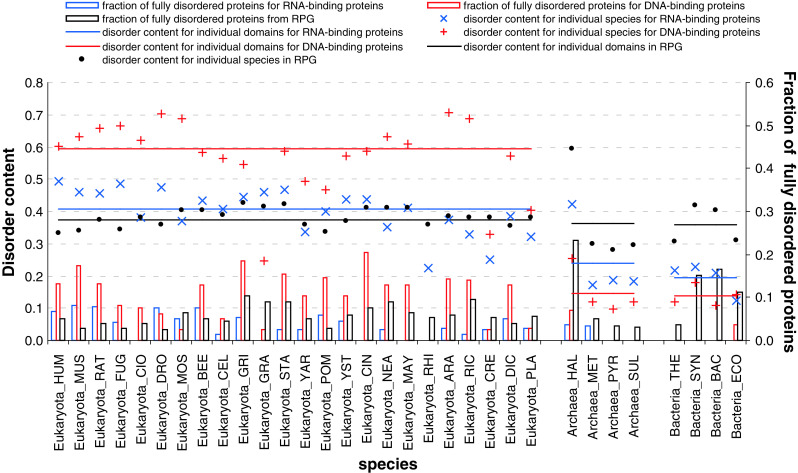
Disorder content (crosses and lines) and fraction of fully disordered proteins (black bars) in different species and domains of life for the ribosomal, DNA-, and RNA-binding proteins. The species, which are shown on the x-axis, are grouped into the Eukaryota, Archaea, and Bacteria domains
This behavior of ribosomal proteins is rather different from that of DNA- and RNA-binding proteins. In fact, disorder in DNA- and RNA-binding proteins is unevenly distributed among the three domains of life, with proteins from Eukaryotes being substantially more disordered than corresponding proteins from Archaea and Bacteria. Interestingly, the overall disorder contents of eukaryotic ribosomal and RNA-binding proteins are rather similar (~37 and 41 %, respectively) whereas eukaryotic DNA-binding proteins possess more disorder (~60 %). However, in Archaea and Bacteria, the situation is reversed, and ribosomal proteins are more disordered than RNA- and DNA-binding proteins (see Fig. 5). Fully disordered eukaryotic ribosomal proteins are somewhat more abundant than fully disordered RNA-binding proteins and noticeably less abundant than fully disordered DNA-binding proteins. In Archaea and Bacteria, fully disordered chains are essentially more abundant among the ribosomal proteins than among the corresponding RNA- and DNA-binding proteins.
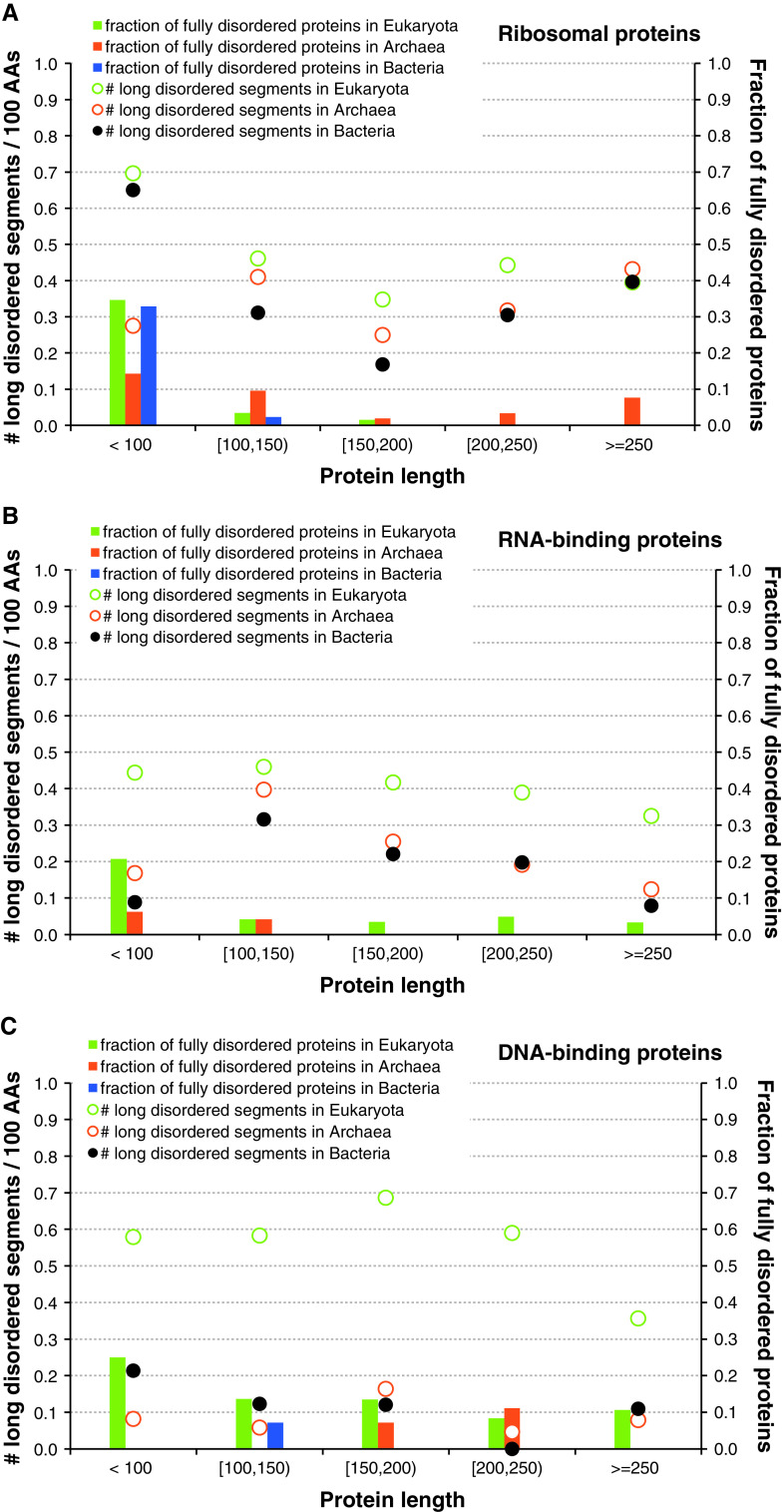
Figure S1 reveals that, on average, ribosomal proteins have between 1.4 (in Eukaryota) and ~1.5 (in Bacteria and Archaea) disordered segments per 100 residues (we normalize by unit of length to allow direct comparison to longer DNA- and RNA-binding chains), including 0.3–0.4 long disordered segments (>30 amino acids) per 100 residues. Therefore, according to all these parameters, ribosomal proteins are substantially more disordered than RNA- or DNA-binding proteins. This is an interesting observation since ribosomal proteins are typically significantly shorter than RNA- and DNA-binding proteins (see Figure S1).
We further analyze the distribution of the disordered regions across chains with different length, see Fig. 6. While in Archaea the number of long disordered segments in ribosomal proteins increases linearly with the length of the protein chain, we observe increased number of disordered segments for short chains in Eukaryota and Bacteria (see Fig. 6a). Furthermore, short (<100 amino acids) fully disordered ribosomal proteins are relatively common in Eukaryota and Bacteria, where about 1/3 of short chains are fully disordered. In contrast, Archaea has some longer fully disordered chains. This is due to the inclusion of Halobacterium salinarum (HAL) that has the highest disorder content (59.3 %), which stems from the fact that it has the largest fraction (23.2 %) of fully disordered proteins among all considered species, see Fig. 5. Overall, our analysis implies that small ribosomal proteins in Eukaryota and Bacteria are enriched in disorder, when compared with the ribosomal proteins in Archaea. These behaviors are different from trends observed for the DNA- and RNA-binding proteins, which typically possess less disorder-related features than ribosomal proteins, except for the eukaryotic DNA-binding proteins, and whose disorder attributes decrease with the protein length (see Fig. 6b, c).
Fig. 6.
The number of long disordered segments (30 or more residues) per protein (y-axis on the left; hollow points) and the fraction of fully disordered protein (y-axis on the right; solid bars) against protein length (x-axis) across the three domains of life in ribosomal (a), RNA- (b), and DNA-binding proteins (c)
Characterization of the domains in ribosomal, RNA-, and DNA-binding proteins
Application of the GlobPlot and MFDp tools to the set of 3,438 ribosomal proteins revealed that 412 proteins (12.0 %) were predicted without globular domains, 502 proteins (14.6 %) were predicted not to have disordered regions, whereas the remaining proteins were predicted to be hybrid proteins that contained both globular and disordered domains. Figure 7a shows that in all three kingdoms of life, most ribosomal proteins with globular domains are single domain proteins (in ~60 % proteins, >95 % residues are included in a GlobPlot predicted domain). However, more detailed analysis of globular domains using the MFDp tool showed that many of them contained disordered regions and some are predicted to be entirely disordered (see Fig. 7b). Figure 7c shows that almost all globular domains contain at least one disordered region with at least four consecutive disordered residues, and ~20 % of domains were significantly disordered, containing at least half disordered residues.
Fig. 7.

Characterization of the globular domains in ribosomal proteins. Globular domains were predicted using the GlobPlot server (http://globplot.embl.de/). a The distribution of fraction of amino acids in domain per protein. b The distribution of disorder content per domain. c The fraction of disordered domains (hollow and solid circles, respectively; y-axis on the left) and the average length of disordered (red and orange bars) and ordered domains (dark and bright green bars; y-axis on the right). Domains were assumed to be disordered when they contain at least one disordered region with at least four consecutive disordered residues (def_1) or when at least half of their residues are disordered (def_2)
Figure 8 represents the results of CH–CDF analysis of ribosomal proteins and provides further support to their highly disordered nature. In this plot, the coordinates of each spot are calculated as a distance of the corresponding protein in the CH-plot (charge–hydropathy plot) from the boundary (Y-coordinate) and an average distance of the respective cumulative distribution function (CDF) curve from the CDF boundary (X-coordinate) [73–75]. The quadrants of CDF-CH phase space correspond to the following expectations: Q1, proteins predicted to be disordered by CH-plots, but ordered by CDFs; Q2, ordered proteins; Q3, proteins predicted to be disordered by CDFs, but compact by CH-plots (i.e., putative molten globules or proteins with alternating ordered and disordered regions); Q4, proteins predicted to be disordered by both methods (i.e., proteins with extended disorder). Although these classifications could be questionable for large, multidomain proteins, they provide relatively unbiased description of ribosomal proteins, which are typically small proteins.
Fig. 8.
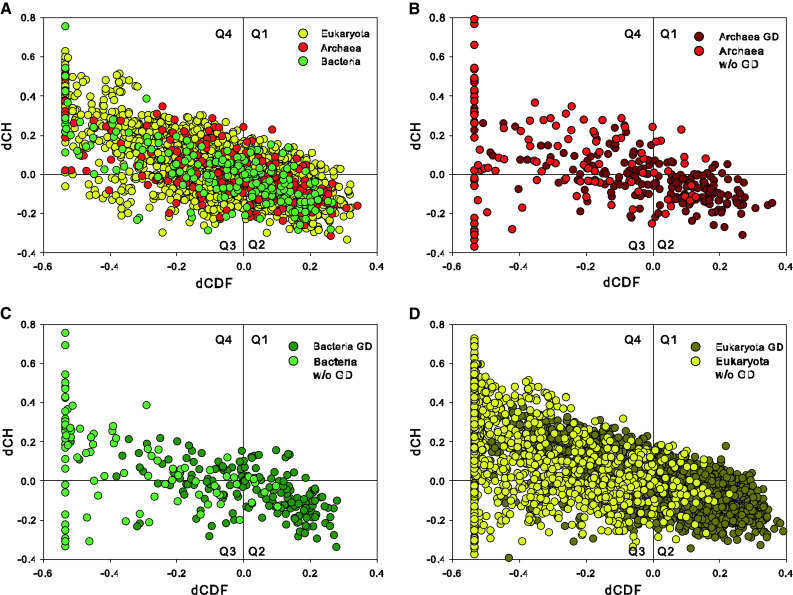
a Evaluation of the abundance of intrinsic disorder in ribosomal proteins from the three domains of life, Bacteria (green circles), Archaea (red circles), and Eukaryota (yellow circles), in the form of a CH–CDF plot [74, 75]. b CH–CDF plot for archaeal ribosomal proteins that are split on globular (dark red) and non-globular domains (red). c CH–CDF plot for bacterial ribosomal proteins that are split on globular (dark green) and non-globular domains (green). d CH–CDF plot for eukaryotic ribosomal proteins that are split on globular (dark yellow) and non-globular domains (yellow)
Figure 8a shows that many full-length ribosomal proteins are predicted to be disordered as a whole, with >60 % of all ribosomal proteins being found in Q1, Q3, and Q4, and being therefore expected to behave as native molten globules, native coils, or native pre-molten globules in their unbound states. The distribution of archaeal, bacterial, and eukaryotic proteins between the four quadrants of the CH–CDF plot is as follows: Archaea, 9.2 % (Q1), 37.2 % (Q2), 17.6 % (Q3), and 36.0 % (Q4); Bacteria, 11.7 % (Q1), 35.5 % (Q2), 15.4 % (Q3), and 37.4 % (Q4); and Eukaryota, 17.1 % (Q1), 30.6 % (Q2), 14.1 % (Q3), and 38.2 % (Q4). Therefore, ribosomal proteins from different life domains are different in their disorder propensities, and can be sorted as Archaea > Bacteria > Eukaryota by the number of ordered proteins in their Q2 quadrants. There is also an unusual bias in the number of ribosomal proteins populating Q1, which is typically considered as a quadrant containing rare proteins [75]. In fact, our analysis shows that between 9 and 17 % of ribosomal proteins are found in Q1, whereas only 2.5 % proteins from entire mouse proteome are in this quadrant. Earlier, it was pointed out that Q1 proteins might have functions related to interaction with RNA, with four of the five distinctive GO terms found for these proteins dealing with RNA binding and modification [75]. By the CH analysis, these Q1 proteins are highly charged, and this feature may be related to their ability to interact with RNA [75].
Figure 9b, c, and d represent CH–CDF plots for globular and non-globular domains of ribosomal proteins from the three kingdoms of life. Results of this analysis are further summarized in Table S3, which shows that non-globular domains are systematically predicted to be mostly disordered and that many GlobPlot identified globular domains are expected to be disordered. In fact, quadrants Q3 and Q4 of the CH–CDF plots that typically correspond to the disordered proteins/domains/regions contain 15.5 % (Q3) and 27.8 % (Q4) of predicted archaeal globular domains, 16.8 % (Q3) and 21.8 % (Q4) of predicted bacterial globular domains, and 10.8 % (Q3) and 32.3 % (Q4) of GlobPlot predicted eukaryotic globular domains. Table S3 also shows that 21.7, 19.2, and 9.9 % of archaeal, bacterial, and eukaryotic ribosomal proteins were predicted to be devoid of globular domains.
Fig. 9.
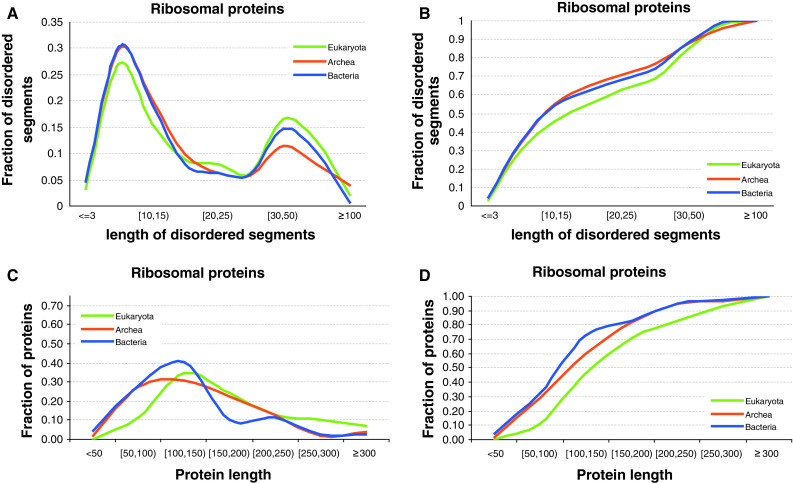
Distribution of the length of the disordered segments across the three domains of life of ribosomal proteins (a) and the corresponding cumulative distribution (b). Length distributions of corresponding ribosomal proteins (c) with its cumulative distribution (d)
All these data clearly show that intrinsic disorder is very common in ribosomal proteins form all three kingdoms of life.
Functional analysis of disordered segments in ribosomal proteins
Distributions of the sizes of the disordered segments in ribosomal proteins across the three domains of life are shown in Fig. 9a. Interestingly, we observe that the sizes follow bimodal distribution with a relatively large number of short segments (between 4 and 15 amino acids) and with a second peak for longer fragments (between 25 and 100 amino acids). Figure 9b represents the overall ribosomal protein length distributions and shows that these proteins are relatively short and possess the average length of about 100–150 residues.
Since intrinsically disordered regions have a bimodal length distribution, we analyze the function for two classes of the disordered segments: short segments with <30 amino acids, and long with at least 30 amino acids. For ribosomal proteins, we consider 26 functions, which are annotated based on sequence alignment into the functionally characterized disordered segments from the DisProt database (as explained in the “Materials and methods” section), which are summarized in Table S2. We exclude functions with <20 annotations for both short and long disordered segments.
Figure 10 compares the annotations of the 13 remaining predicted (using alignment) functions between the short and long disordered segments of ribosomal proteins. The results reveal that disorder in ribosomal proteins plays several important roles, from facilitating the protein–protein, protein–DNA, protein–RNA, and protein–other–ligand interactions, to involvement in metal binding, post-translational modifications, and implementation of linkers and intra-protein interactions. Overall, both long and short disordered segments are equally implicated in several functions, including interactions with proteins, DNA, and ligands. The short segments are predominant in a larger number of functions, including RNA and metal binding, auto-regulatory functions, transactivation, polymerization, apoptosis, and are more prevalent in the post-translational modification sites. At the same time, the long disordered segments more often serve as linkers and play a strong role in intra-protein interactions. Our analysis provides useful clues that can be used to narrow down potential functions of IDPs and IDPRs, especially knowing the size of the corresponding segments, in ribosomal chains that currently lack functional annotations.
Fig. 10.
Fraction of short (4–30 amino acids) and long (over 30 amino acids) disordered segments for a given function; x-axis represents the 13 considered functions sorted by the decreasing number of short segments
The results of the predictions of potential binding sites were validated against the functions of known components. To this end, the predicted binding sites of proteins in the yeast ribosome were compared to the ribosome structure to determine whether regions predicted to be involved in binding of proteins and RNA actually perform these functions. Potential protein–protein interaction sites were predicted in 13 proteins that are found in the crystal structure of the yeast ribosome: S8-A (residues 119–150), S17-A (residues 1–5), S19-A (residues 1–5), S20 (residues 1–23), S26-A (residues 83–119), S27-A (residues 78–82), L4-A (residues 1–19), L10 (residues 217–220), L18-A (residues 140–186), L22-A (residues 101–121), L28 (residues 89–93), L31-A (residues 108–113), and L40-A (residues 35–38). RNA-binding site was predicted in L31-A (residues 108–113). Analysis of the crystal structures of the yeast ribosomal subunits revealed that there is a reasonably good correlation between the predicted and actual binding sites, since many predicted protein–protein interaction sites of the yeast ribosomal proteins either coincided, or overlapped, or were located in the close proximity to the actual binding sites. For example, in the crystal structure of the small ribosomal subunit, residues 117 and 149–153 of S8-A are involved in interaction with S11-A; N-terminal residues 8, 12, 15–16 and 18–19 of the S17-A interact with protein S3; residues 6–12 of S19-A are at the interface with S16-A; residues 25–29 of S20 bind to S3; S26-A interacts with S14-A and S2 via residues 42–71 and 59–70, respectively; S27-A is engaged in binding to S13 and S7-A via residue 82. In the crystal structure of the 60S ribosome, residues 28–33 of L4-A protein interact with residues 123–133 of L18-A; region containing residues 206–221 of L10 is at the interface with L5; besides being involved in interaction with L4-A, residues 164–172 of L18-A bind to L13-A; L28 binds to L13-A via region containing residues 96–111; the interaction between L40-A and L9-A is secured by residues 77–91. The fact that the predicted binding sites of L22-A and L31-A were not involved in interaction with other ribosomal proteins does not necessarily mean wrong prediction, since these regions (as well as predicted binding regions of other yeast ribosomal proteins) can be engaged in binding to non-ribosomal proteins.
MoRF regions in ribosomal, RNA-, and DNA-binding proteins
The most prevalent function of disorder in ribosomal proteins is facilitation of protein–protein interactions. Figure 11 shows that well over 30 % of the functionally annotated disordered segments in ribosomal proteins are implicated in these binding events. This motivates our analysis of MoRFs regions [2, 78–80], which are defined as short disordered regions that undergo disorder-to-order transition upon binding to protein partners and fold into mostly helical (α-MoRFs), strand (β-MoRFs), coil (ι-MoRFs) and complex (complex-MoRFs, which combine multiple secondary structure) secondary structures. Figure 11a demonstrates that there are on average about 0.85 MoRFs per 100 residues (we normalize by unit of length to allow direct comparison to longer DNA- and RNA-binding chains) in eukaryotic ribosomal proteins, including a large fraction of α-MoRF and ι-MoRF and relatively lower numbers of complex- and β-MoRFs. The complex-MoRFs, ι-MoRFs, and α-MoRFs are similarly abundant in ribosomal chains from the three domains of life, while bacterial and archaeal ribosomal proteins are enriched in β-MoRFs. Both RNA- (Fig. 11b) and DNA-binding proteins (Fig. 11c) have fewer MoRF regions per 100 residues, and are characterized by rather different distributions of the overall abundance of MoRFs (which vary more widely between species) and their split into α-, β-, ι-, and complex-MoRFs between eukaryotic, archaeal and bacterial proteins, particularly for DNA-binding chains that are depleted in β-MoRFs. This suggests that MoRF regions in the ribosomal chains may be involved in different types of protein–protein interactions across different domains.
Fig. 11.

Number of MoRFs per protein, shown using stacked bars, across different species and domains. The bars are subdivided using colors that correspond to different MoRF types. The solid lines show a cumulative (over MoRF types located below the line) average number of a given MoRF type for each of the three domains. The species, which are shown on the x-axis, are grouped into Eukaryota, Archaea, and Bacteria domains. Plots a, b, and c correspond to ribosomal, and RNA- and DNA-binding proteins, respectively
Evolutionary conservation of disorder in ribosomal proteins
Next, we investigate evolutionary conservation of intrinsic disorder in ribosomal proteins. The conservation is quantified using the relative entropy computed from the Weighted Observed Percentages (WOP) profiles generated by PSI-BLAST (as explained in the “Materials and methods” section). Higher values of the relative entropy indicate a higher degree of conservation. Figure 12 shows that ribosomal, RNA-, and DNA-binding proteins in Bacteria are characterized by higher levels of conservation when compared with the Archaea and Eukaryota. This can also be observed in Fig. 13 where we compare conservation between disordered and ordered residues. Besides the overall trend that shows higher conservation in Bacteria, our results show that disordered residues are more conserved when compared with the structured parts of the ribosomal proteins (see Fig. 13a). This is true for all species in Eukaryota and Archaea, while in Bacteria the disordered and ordered residues have similarly high conservation. Moreover, we show that residues located in long disordered segments of ribosomal proteins are more conserved than the overall population of both disordered and ordered amino acids across all three domains of life. In eukaryotic RNA-binding proteins, the situation is reversed and ordered regions are more conserved (Fig. 13b), whereas eukaryotic DNA-binding proteins are characterized by the higher conservation of long disordered and ordered regions (see Fig. 13c). This suggests that disorder plays important role in all the kingdom of life from the evolutionary perspective, particularly in ribosomal proteins where it is characterized by higher conservation levels.
Fig. 12.

Distribution of the average relative entropy, which quantifies evolutionary conservation, for the proteins from Eukaryota, Archaea, and Bacteria. Plots a, b, and c correspond to ribosomal, and RNA- and DNA-binding proteins, respectively
Fig. 13.
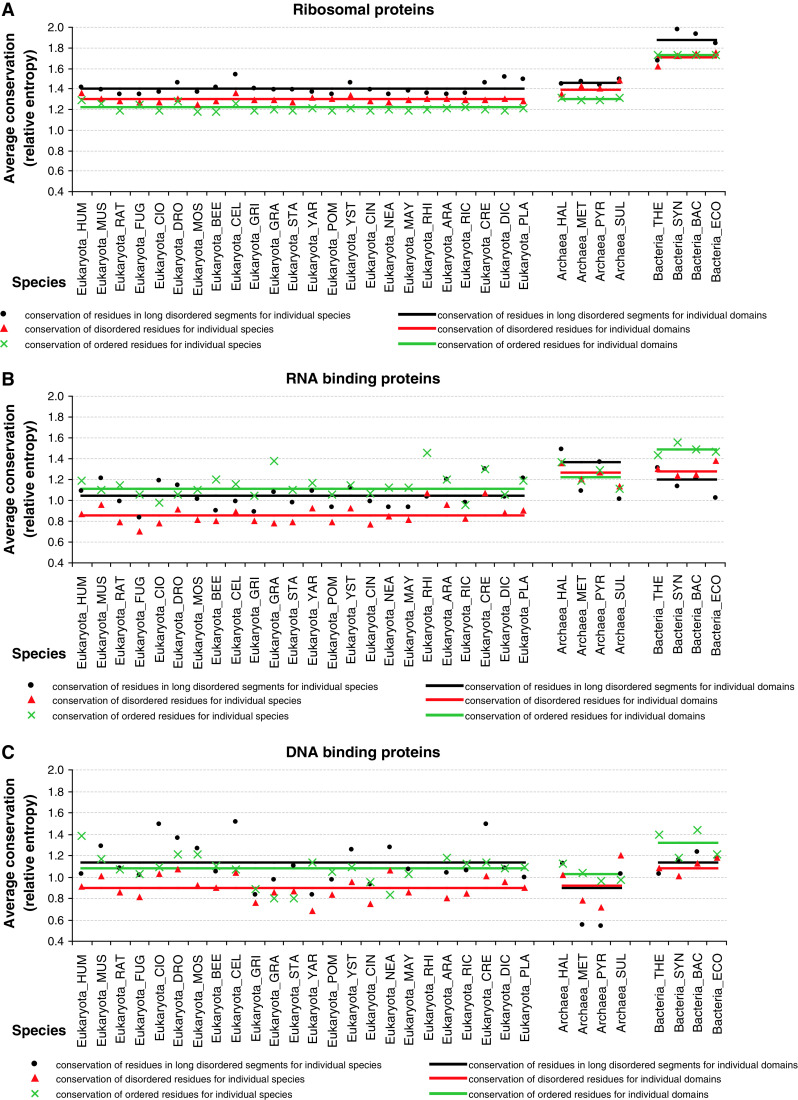
The average relative entropy that quantifies evolutionary conservation across different species and domains. Blue points/lines, green triangles/lines, and orange crosses/lines denote the average relative entropy of disordered residues in long disordered segments, all disordered residues, and ordered residues, respectively. The species, which are shown on the x-axis, are grouped into Eukaryota, Archaea, and Bacteria domains. Plots a, b, and c correspond to ribosomal, RNA- and DNA-binding proteins, respectively
Orthology and disorder in ribosomal proteins
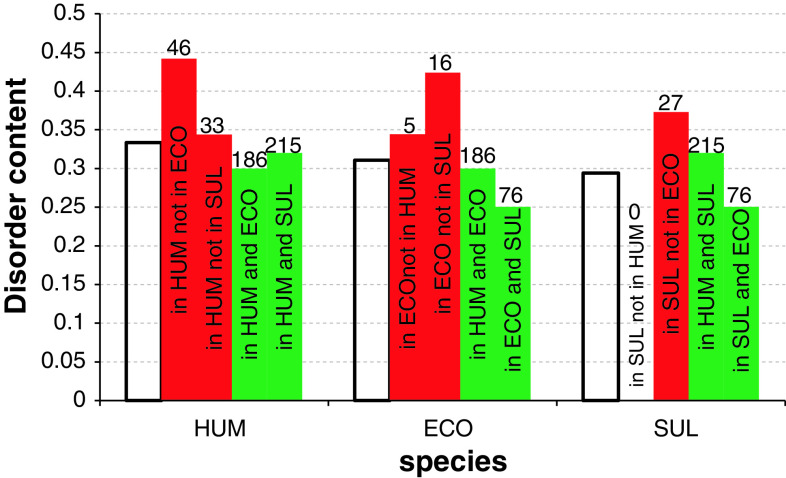
Using a representative organism from each kingdom of life (H. sapiens, E. coli, and S. tokodaii) we annotated proteins for all pairs of the selected species as either orthologous or non-orthologous using the data available in RPG database [54]. The selected bacterial and archaeal species have the largest proteomes in their respective sets of species. The overall disorder content in the three species and the content for their orthologous or non-orthologous proteins is summarized in Fig. 14. We observe that the orthologous chains are characterized by lower amounts of disorder compared to the amount of disorder for the corresponding non-orthologous proteins. This trend is true across all three proteomes, which suggests that disorder may play a role in specializing and adjusting the ribosome for a particular kingdom of life.
Fig. 14.
Comparison of disorder content between orthologous (green bars) and non-orthologous (red bars) proteins across all pairs of the selected species from the three kingdoms of life, including H. sapiens (HOMO), E. coli (ECO), and S. tokodaii (SUL). The hollow bars denote the overall, for a given species, disorder content. The numbers above the bars indicate the corresponding count of orthologous and non-orthologous chains
Discussion
Commonness and peculiarities of intrinsic disorder in the ribosomal proteins
We are showing in this study that intrinsic disorder is widely spread among the ribosomal proteins from all three kingdoms of life. This conclusion is in line with the results of the analysis of crystal structure of the eukaryotic ribosome from the yeast S. cerevisiae that revealed that many ribosomal proteins contain regions of intrinsic disorder, which are seen as regions with missing electron density [57]. Many ribosomal proteins contain IDPRs that are at least 8 residues long, with some IDPRs can be as long as 94 residues. The illustrative examples of such proteins are listed in Supplementary Materials. We also point out that many of the eukaryotic core proteins contain eukaryote-specific extensions that interact with the rRNA expansion segments in 60S subunit. For example, the conserved proteins that are associated with the polypeptide exit tunnel, L22, L4, L23, and L29 all contain very long extensions, up to 140 Å in the case of L4, that reach the periphery of 60S [57]. Another protein with a very unusual configuration is L24e whose N-terminal domain resides in the 60S subunit whereas the C-terminal domain reaches to the back of the 40S subunit due to the presence of a long flexible linker that protrudes deep into the side of the 40S body [57].
Visual analysis of the crystal structures of individual ribosomal proteins revealed that many of them possess very unusual morphologies inconsistent with simple globular structures suggesting that these structures are likely to be formed as a result of binding-induced folding (see Fig. 1a). This hypothesis is supported by the computational analysis of these structures in the form of Nussinov’s plot, where the vast majority of eukaryotic ribosomal proteins is found above the order–disorder boundary suggested by Gunasekaran et al. [60]. In order to understand whether the globular domains seen for many ribosomal proteins are independent folding units or are formed due to the binding-induced disorder-order transitions, we developed a tool (discrete finite automaton, DFA) to computationally separate proteins with known 3D-structure into globular domains and non-globular parts. The subsequent Nussinov’s plot analysis showed that many globular domains were formed due to binding to other components of the ribosome (Fig. 2). These findings provide very important support to the hypothesis that many eukaryotic ribosomal proteins are mostly disordered in their unbound states.
To understand how general this statement is, we next analyzed a large dataset of ribosomal proteins from all kingdoms of life. Application of various computational tools unequivocally showed that disorder is very common in all the ribosomal proteins and that many potential globular domains still possess noticeable levels of disorder (see Figs. 4, 5, 6, 7, 8). Since disorder is reliably predicted using computational tools developed based on the disorder-related data from large databases (e.g., PDB), one can conclude that disordered regions of ribosomal proteins are generally similar in their properties to disordered regions of many other proteins observed in several large databanks.
The ribosome is a ribonucleoprotein machine whose proteins are involved in interactions with both proteins and RNA. To understand how ribosomal proteins differ from other nucleic acid binding proteins, we compared some of their disorder-related features with disorder characteristics of large randomly selected sets of RNA- and DNA-binding proteins. Data shown in Figs. 4, 5, 9, 11, 12, and 13 suggest that disorder in ribosomal proteins, especially its functional roles, and evolutionary features are different from those aspects of disorder in DNA- and RNA-binding proteins. It is likely that some of these differences are related to the functional uniqueness of ribosomal proteins, many of which are involved in multiple simultaneous binding events, being involved in interactions, not only with RNA, but also with neighboring proteins. Some of the reasons for the abundance of disorder in ribosomal proteins are considered in the next several paragraphs.
Why is intrinsic disorder so common in the ribosomal proteins?
Functional viewpoint: protein–rRNA and protein–protein interactions on the ribosome
Being components of a large ribonucleoprotein complex, ribosomal proteins are obviously involved in interaction with both RNA and other proteins. Their ability to bind to RNA is determined by high positive charge. In general, ribosomal proteins are very basic (average pI ~ 10.1), suggesting that a general function of these proteins may be to counteract the negative charges of the phosphate residues in the rRNA backbone. In agreement with this hypothesis, many ribosomal proteins were shown to serve as RNA chaperones and therefore play crucial roles during the ribosome assembly [108, 109]. The only exceptions to this rule are S1 and S6 in the small subunit and the L7/L12 proteins in the large subunit, none of which have significant contacts with RNA, being predominantly engaged in the protein–protein interactions. Here, L7/L12 interact directly with L10 to form the pentameric L10 × (L7/L12)4 or heptameric L10 × (L7/L12)6 complex, S6 makes extensive contact with S18, and S1 interacts with S21, S11, and S18 [109].
Many ribosomal proteins possess complex structures and are often characterized by a tadpole-like shape (see Fig. 1) containing a globular domain, which is generally located on the surface of the ribosome, and a long extended region that penetrates into the ribosome’s interior. In fact, all S-proteins (except S4 and S15) and about 50 % of the L-proteins possess such extensions, which have distinctive amino acid compositions, containing multiple Gly residues to allow flexibility and tight packing, and which are rich in basic amino acids to interact with rRNA [109]. In fact, the content of the basic amino acids Arg/Lys in the extensions of the large subunit ribosomal proteins (27 %) noticeably exceeds that of the globular parts (19 %). As a result, these extensions that constitute only ~20 % of the protein mass of the large subunit are responsible for burying of ~50 % of total RNA surface area [109]. It was pointed out that some ribosomal proteins, when studied in isolation, contain globular regions, whereas their extended tails are typically not observed in the isolated structures [109], suggesting that these regions undergo disorder-to-order transitions induced by interaction with rRNA. Among the most extreme examples are the long extensions of L2 and L3 that reach towards the peptidyl-transferase center. S12 has an extremely long extension that starts from the globular domain located adjacent to the decoding center on the intersubunit side of the small subunit and reaches all the way to the back or solvent side of the 30S subunit where it interacts with S8 and S17. Thus, S12 provides an illustrative example of the “penetrator” binding mode, where a significant part of an IDP penetrates deep inside the structure of its binding partner [110]. Also, the 61 amino acid ribosomal protein S14 is completely devoid of any globular domain [109]. Therefore, IDPRs of many ribosomal proteins are important foldable regions that serve to ensure the formation of a correctly folded rRNA state during the ribosome assembly process and also support the correct conformation of the rRNA in the final assembled complex [109].
Besides the aforementioned intensive contacts with rRNA, several ribosomal proteins are involved in a well-developed network of protein–protein interactions. For example, a tight heterodimeric complex is formed by S6 and S18 proteins on the outer edge of the platform of the small subunit, whereas at the back of the 30S head, S3, S10, and S14 form a tight complex, and in the large subunit there are previously mentioned pentameric L10 × (L7/L12)4 or heptameric L10 × (L7/L12)6 protein complexes [109]. Formation of these tight protein–protein complexes may also involve disorder-to-order transition, at least in some parts of the interacting proteins.
Functional viewpoint: specific on-ribosome functions
It was recognized long ago that some ribosomal proteins are mostly essential for the assembly of the ribonucleoprotein particle and are dispensable for function after the ribosomal subunits are fully assembled [111], suggesting that the major function of these “dispensable” proteins (e.g., S16, L15, L16, L20, and L24) in the assembled ribosome could be to improve the ribosome stability. Furthermore, there are several ribosomal proteins that are not essential for the translational function of the ribosome, the hypothesis based on the observations E. coli strains lacking S6, S9, S13, S17, S20, L1, L9, L11, L15, L19, L24, L27 to L30, and L33 are viable [109, 112, 113]. Since the subject of the on-ribosome functions of the ribosomal proteins was covered in a recent in-depth review [109], we are simply listing some of these functions in the Supplementary Materials. Interested readers are encouraged to look for the original review, where the functional roles of many ribosomal proteins were considered in great detail [109].
All of these functions are relying on multiple interactions with various partners, suggesting that ribosomal proteins can be considered to be ribosomal hubs. Earlier, it was shown that binding promiscuity of hubs benefit from the use of intrinsic disorder in one of the two ways, where one disordered region can bind to many different partners and many disordered region can bind to one partner [13, 114–119].
Functional viewpoint: moonlighting or off-ribosome functions
The core ribosome functions, i.e., the precise interaction of mRNA codon with tRNA anticodon and the catalysis of peptide bond formation, are carried out by rRNA molecules of the small and the large ribosomal subunits, respectively. Therefore, the major or core on-ribosome functions of ribosomal proteins are to assist in rRNA folding (i.e., to serve as RNA chaperones) and function, to assist in the ribosome assembly, and to be involved in related protein–protein, protein–rRNA, protein–mRNA, and protein–tRNA interactions. On the other hand, many ribosomal proteins have been shown to be involved in some extra-ribosomal or auxiliary functions, thereby being involved in moonlighting activities. In agreement with this hypothesis, numerous extra-ribosomal functions have been assigned to ribosomal proteins [120–124]. It was even stated recently that “moonlighting is particularly widespread among ribosomal proteins, many of which have extra-ribosomal employment” [122]. Even the first systematic analysis of this subject (which was performed in 1996) revealed that ribosomal proteins might have up to 30 extra-ribosomal functions [120]. Recently, it was emphasized that the numerous extra-ribosomal functions of ribosomal proteins reported in the literature so far can be grouped into two major categories: 1. Control the balance among the ribosomal components; and 2. Control nucleolar stress or aberrant ribosomal synthesis, leading to cell cycle arrest or apoptosis [124]. Some of the extra-ribosomal functions of ribosomal proteins within the ribosome system were already described above (e.g., see notes for S1, L1, and L4) and are covered in great detail in a recent review [124]. In E. coli, these extra-ribosomal activities include the L4-mediated inhibition of translation of the S10 operon that encodes 11 different ribosomal proteins including L4 itself [42] and binding of L4 to RNase E thereby leading to the modulation of its acitivity, resulting in the stress-related changes in the mRNA composition [125]. It was emphasized that, among other regulatory ribosomal proteins, L4 occupies a unique position due to its ability to regulate both the transcription and the translation of the L4-mRNA-containing S10 operon [126–128]. Furthermore, via a comprehensive analysis of deletion and point mutants, these two functions of L4 were assigned to different regions of this protein [129]. In fact, although the C-terminal region of L4 (residues 171–201) was shown to be crucial for the L4-mediated autogenous control, it was not involved in the incorporation of this protein to the ribosome. On the other hand, the central region of L4 (residues 67–103) was involved in the ribosome assembly but did not play a significant role in the regulatory L4 functions [129]. Curiously, the last third of the regulatory C-terminal fragment of L4 is predicted to be highly disordered, whereas the central region required for the ribosome assembly is expected to be mostly disordered throughout its entire length.
In Eukaryotes, L30 inhibits splicing by binding to its own transcript [130], S14 controls the splicing of the transcript of one of its genes [131], L2 controls the level of its mRNA through accelerated turnover [132], S13 binds to the first intron of its transcript to inhibit splicing [133, 134], and L12 controls its own synthesis by inhibiting the splicing of its own mRNA [135]. In addition to these roles in the control of the balance among ribosomal components during the ribosome synthesis, the established off-ribosome functions of ribosomal proteins are related to the surveillance of the ribosome assembly, as well as numerous roles in development, apoptosis, and cancer [124]. It is very likely that the ability of ribosomal proteins to act off the ribosome can be attributed to their intrinsically disordered nature. This hypothesis is in agreement with a recent analysis that showed that the structural malleability characteristic for the IDPs/IDPRs is strongly associated with the ability of proteins to moonlight [121].
Evolutionary viewpoint
Ribosomes are intricate subjects for evolutionary analysis, since they are found in all living cells where they are absolutely necessary for protein biosynthesis. It was pointed out that although ribosomal proteins are generally highly conserved within the different domains of life, there is a noticeable difference between the ribosomal proteins of Bacteria, Archaea, and Eukaryota [41, 109]. In fact, only ~30% of bacterial, eukaryotic and archaeal ribosomal proteins are considered to be orthologous. An additional 30 % of the ribosomal proteins are in common between the archaeal and eukaryotic ribosomes. However, no proteins are exclusively in common between the bacterial and archaeal ribosomes or between the bacterial and eukaryotic ribosomes, thus supporting the theory that the separation of the common ancestor of Archaea and Eukarya form the Bacteria happened before the Archaea and Eukarya become separated [41, 109]. The high sequence conservation detected in several ribosomal proteins (especially those critical for ribosomal function and assembly) indicates their functional importance.
On the other hand, the ribosomes with their unique ribozymatic activities support the validity of the “RNA world” theory, according to which the biosphere once was dominated by organisms in which RNA was used for both information storage and catalysis [136]. Based on this hypothesis and on the assumption that during the evolution of enzymatic activity, catalysis was transferred from RNA to ribonucleoprotein to protein, it was proposed that the first proteins to come into being were RNA chaperones [137, 138]. In fact, it is rather obvious that the first proteins should be short and unfolded polypeptides [139], since the chance for the spontaneous appearance of a polypeptide chain capable of folding into a unique 3-D structure is extremely low. Furthermore, the first biological functions of these disordered primordial polypeptides are also obvious—they have to be involved in interactions with ribozymes to stabilize their unstable and prone-to-misfold structure. In fact, it is well known that the single-stranded RNAs are flexible macromolecules and can fold into a wide variety of alternative conformations. However, for a given ribozyme, only one given conformation is functionally relevant. Therefore, in order for a given RNA to reach the biologically relevant conformation and not be trapped in one of the many structurally available but functionally incorrect structures, a special mechanism for assisted RNA folding should be implemented [140]. Currently, this special mechanism mostly relies on RNA chaperone proteins [140]. Therefore, it is reasonable to hypothesize that ancient polypeptides would serve as first RNA chaperones, which via their interactions with primordial RNAs would assist in productive folding of the ancient ribozymes and also would stabilize the biologically active structures of those ribozymes. Since many ribosomal proteins are intrinsically disordered RNA chaperones, the ribosome can clearly be considered as a living fossil that represents a snapshot of one of the early stages of prehistoric development.
In conclusion, this paper presents the results of comprehensive computational analyses of ribosomal proteins and shows that the vast majority of these important RNA-binding proteins are IDPs with ribosome-specific sequence features. We also show that intrinsic disorder is very important for various biological functions of ribosomal proteins, being commonly used in numerous interactions of any given ribosomal protein with its various binding partners of different nature, such as other ribosomal proteins, RNA, and proteins from the translational machinery. The intrinsically disordered nature of ribosomal proteins is highly conserved in different domains of life, indicating that the lack of rigid structure, the resulting ability of ribosomal proteins to interact with various binding partners and be involved in the wide spectrum of moonlighting activities represent a strong evolutionary advantage. Therefore, careful consideration and appreciation of intrinsic disorder are crucial for better understanding of structure and conformational behavior of ribosomal proteins, their promiscuity, molecular mechanisms of their numerous extra-ribosomal functions, and mechanisms underlying regulation and control of these very important proteins.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was supported in part by the Programs of the Russian Academy of Sciences for the “Molecular and Cellular Biology” (to V.N.U), the Alberta Innovates Graduate Scholarship in Omics (to Z.P.), the Killam Memorial Scholarship (to M.J.M.), and the Natural Sciences and Engineering Research Council (NSERC) Discovery grant (to L.K.).
Footnotes
Z. Peng and C. J. Oldfield contributed equally to this study.
References
- 1.Dunker AK, Obradovic Z, Romero P, Garner EC, Brown CJ. Intrinsic protein disorder in complete genomes. Genome Inf Ser Workshop Genome Inf. 2000;11:161–171. [PubMed] [Google Scholar]
- 2.Uversky VN, Dunker AK. Understanding protein non-folding. Biochim Biophys Acta. 2010;1804(6):1231–1264. doi: 10.1016/j.bbapap.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337(3):635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Uversky VN, Gillespie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 2000;41(3):415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 5.Xue B, Dunker AK, Uversky VN. Orderly order in protein intrinsic disorder distribution: disorder in 3500 proteomes from viruses and the three domains of life. J Biomol Struct Dyn. 2012;30(2):137–149. doi: 10.1080/07391102.2012.675145. [DOI] [PubMed] [Google Scholar]
- 6.Dunker AK, Garner E, Guilliot S, Romero P, Albrecht K, Hart J, Obradovic Z, Kissinger C, Villafranca JE. Protein disorder and the evolution of molecular recognition: theory, predictions and observations. Pac Symp Biocomput. 1998;3:473–484. [PubMed] [Google Scholar]
- 7.Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293(2):321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 8.Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, Ausio J, Nissen MS, Reeves R, Kang C, Kissinger CR, Bailey RW, Griswold MD, Chiu W, Garner EC, Obradovic Z. Intrinsically disordered protein. J Mol Graph Model. 2001;19(1):26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 9.Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci. 2002;27(10):527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 10.Daughdrill GW, Pielak GJ, Uversky VN, Cortese MS, Dunker AK. Natively disordered proteins. In: Buchner J, Kiefhaber T, editors. Handbook of protein folding. Weinheim: Wiley-VCH Verlag, GmbH & Co. KGaA; 2005. pp. 271–353. [Google Scholar]
- 11.Uversky VN. The mysterious unfoldome: structureless, underappreciated, yet vital part of any given proteome. J Biomed Biotechnol. 2010;2010:568068. doi: 10.1155/2010/568068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol. 2002;323(3):573–584. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- 13.Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets: the roles of intrinsic disorder in protein interaction networks. FEBS J. 2005;272(20):5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 14.Uversky VN, Oldfield CJ, Dunker AK. Showing your ID: intrinsic disorder as an ID for recognition, regulation and cell signaling. J Mol Recognit. 2005;18(5):343–384. doi: 10.1002/jmr.747. [DOI] [PubMed] [Google Scholar]
- 15.Radivojac P, Iakoucheva LM, Oldfield CJ, Obradovic Z, Uversky VN, Dunker AK. Intrinsic disorder and functional proteomics. Biophys J. 2007;92(5):1439–1456. doi: 10.1529/biophysj.106.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vucetic S, Xie H, Iakoucheva LM, Oldfield CJ, Dunker AK, Obradovic Z, Uversky VN. Functional anthology of intrinsic disorder. 2. Cellular components, domains, technical terms, developmental processes, and coding sequence diversities correlated with long disordered regions. J Proteome Res. 2007;6(5):1899–1916. doi: 10.1021/pr060393m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie H, Vucetic S, Iakoucheva LM, Oldfield CJ, Dunker AK, Uversky VN, Obradovic Z. Functional anthology of intrinsic disorder. 1. Biological processes and functions of proteins with long disordered regions. J Proteome Res. 2007;6(5):1882–1898. doi: 10.1021/pr060392u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie H, Vucetic S, Iakoucheva LM, Oldfield CJ, Dunker AK, Obradovic Z, Uversky VN. Functional anthology of intrinsic disorder. 3. Ligands, post-translational modifications, and diseases associated with intrinsically disordered proteins. J Proteome Res. 2007;6(5):1917–1932. doi: 10.1021/pr060394e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41(21):6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- 20.Liu J, Perumal NB, Oldfield CJ, Su EW, Uversky VN, Dunker AK. Intrinsic disorder in transcription factors. Biochemistry. 2006;45(22):6873–6888. doi: 10.1021/bi0602718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhalla J, Storchan GB, MacCarthy CM, Uversky VN, Tcherkasskaya O. Local flexibility in molecular function paradigm. Mol Cell Proteomics. 2006;5(7):1212–1223. doi: 10.1074/mcp.M500315-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Minezaki Y, Homma K, Kinjo AR, Nishikawa K. Human transcription factors contain a high fraction of intrinsically disordered regions essential for transcriptional regulation. J Mol Biol. 2006;359(4):1137–1149. doi: 10.1016/j.jmb.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 23.Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, Krijgsveld J, Hentze MW. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149(6):1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 24.Kucera NJ, Hodsdon ME, Wolin SL. An intrinsically disordered C terminus allows the La protein to assist the biogenesis of diverse noncoding RNA precursors. Proc Natl Acad Sci USA. 2011;108(4):1308–1313. doi: 10.1073/pnas.1017085108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tompa P, Csermely P. The role of structural disorder in the function of RNA and protein chaperones. FASEB J. 2004;18(11):1169–1175. doi: 10.1096/fj.04-1584rev. [DOI] [PubMed] [Google Scholar]
- 26.Ivanyi-Nagy R, Lavergne JP, Gabus C, Ficheux D, Darlix JL. RNA chaperoning and intrinsic disorder in the core proteins of Flaviviridae. Nucleic Acids Res. 2008;36(3):712–725. doi: 10.1093/nar/gkm1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mir MA, Panganiban AT. The bunyavirus nucleocapsid protein is an RNA chaperone: possible roles in viral RNA panhandle formation and genome replication. RNA. 2006;12(2):272–282. doi: 10.1261/rna.2101906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mir MA, Panganiban AT. Characterization of the RNA chaperone activity of hantavirus nucleocapsid protein. J Virol. 2006;80(13):6276–6285. doi: 10.1128/JVI.00147-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murray CL, Marcotrigiano J, Rice CM. Bovine viral diarrhea virus core is an intrinsically disordered protein that binds RNA. J Virol. 2008;82(3):1294–1304. doi: 10.1128/JVI.01815-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haynes C, Iakoucheva LM. Serine/arginine-rich splicing factors belong to a class of intrinsically disordered proteins. Nucleic Acids Res. 2006;34(1):305–312. doi: 10.1093/nar/gkj424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shojania S, O’Neil JD. Intrinsic disorder and function of the HIV-1 Tat protein. Protein Pept Lett. 2011;17(8):999–1011. doi: 10.2174/092986610791498993. [DOI] [PubMed] [Google Scholar]
- 32.Xue B, Mizianty MJ, Kurgan L, Uversky VN. Protein intrinsic disorder as a flexible armor and a weapon of HIV-1. Cell Mol Life Sci. 2012;69(8):1211–1259. doi: 10.1007/s00018-011-0859-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang CK, Sue SC, Yu TH, Hsieh CM, Tsai CK, Chiang YC, Lee SJ, Hsiao HH, Wu WJ, Chang WL, Lin CH, Huang TH. Modular organization of SARS coronavirus nucleocapsid protein. J Biomed Sci. 2006;13(1):59–72. doi: 10.1007/s11373-005-9035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thapar R, Mueller GA, Marzluff WF. The N-terminal domain of the Drosophila histone mRNA binding protein, SLBP, is intrinsically disordered with nascent helical structure. Biochemistry. 2004;43(29):9390–9400. doi: 10.1021/bi036314r. [DOI] [PubMed] [Google Scholar]
- 35.Olieric V, Wolff P, Takeuchi A, Bec G, Birck C, Vitorino M, Kieffer B, Beniaminov A, Cavigiolio G, Theil E, Allmang C, Krol A, Dumas P. SECIS-binding protein 2, a key player in selenoprotein synthesis, is an intrinsically disordered protein. Biochimie. 2009;91(8):1003–1009. doi: 10.1016/j.biochi.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 36.Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461(7268):1234–1242. doi: 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- 37.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11(2):113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136(4):731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spahn CM, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J. Structure of the 80S ribosome from Saccharomyces cerevisiae—tRNA-ribosome and subunit-subunit interactions. Cell. 2001;107(3):373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- 40.Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science. 2011;334(6058):941–948. doi: 10.1126/science.1211204. [DOI] [PubMed] [Google Scholar]
- 41.Lecompte O, Ripp R, Thierry JC, Moras D, Poch O. Comparative analysis of ribosomal proteins in complete genomes: an example of reductive evolution at the domain scale. Nucleic Acids Res. 2002;30(24):5382–5390. doi: 10.1093/nar/gkf693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zengel JM, Lindahl L. Diverse mechanisms for regulating ribosomal protein synthesis in Escherichia coli . Prog Nucleic Acid Res Mol Biol. 1994;47:331–370. doi: 10.1016/s0079-6603(08)60256-1. [DOI] [PubMed] [Google Scholar]
- 43.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289(5481):905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 44.Wimberly BT, Brodersen DE, Clemons WM, Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407(6802):327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 45.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292(5518):883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 46.Harms J, Schluenzen F, Zarivach R, Bashan A, Gat S, Agmon I, Bartels H, Franceschi F, Yonath A. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 2001;107(5):679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- 47.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310(5749):827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 48.Selmer M, Dunham CM, Murphy FVt, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313(5795):1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 49.Timsit Y, Acosta Z, Allemand F, Chiaruttini C, Springer M. The role of disordered ribosomal protein extensions in the early steps of eubacterial 50 S ribosomal subunit assembly. Int J Mol Sci. 2009;10(3):817–834. doi: 10.3390/ijms10030817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2011;330(6008):1203–1209. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- 51.Garrett RA. Structure and role of eubacterial ribosomal proteins. Horiz Biochem Biophys. 1983;7:101–138. [PubMed] [Google Scholar]
- 52.Brodersen DE, Clemons WM, Jr, Carter AP, Wimberly BT, Ramakrishnan V. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 S RNA. J Mol Biol. 2002;316(3):725–768. doi: 10.1006/jmbi.2001.5359. [DOI] [PubMed] [Google Scholar]
- 53.Klein DJ, Moore PB, Steitz TA. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J Mol Biol. 2004;340(1):141–177. doi: 10.1016/j.jmb.2004.03.076. [DOI] [PubMed] [Google Scholar]
- 54.Nakao A, Yoshihama M, Kenmochi N. RPG: the Ribosomal Protein Gene database. Nucleic Acids Res. 2004;32(Database issue):D168–D170. doi: 10.1093/nar/gkh004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mizianty MJ, Stach W, Chen K, Kedarisetti KD, Disfani FM, Kurgan L. Improved sequence-based prediction of disordered regions with multilayer fusion of multiple information sources. Bioinformatics. 2010;26(18):i489–i496. doi: 10.1093/bioinformatics/btq373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.UniProt Consortium Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2012;40(Database issue):D71–D75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011;334(6062):1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 58.Eisenhaber F, Lijnzaad P, Argos P, Sander C, Scharf M. The double cubic lattice method—efficient approaches to numerical-integration of surface-area and volume and to dot surface contouring of molecular assemblies. J Comput Chem. 1995;16(3):273–284. [Google Scholar]
- 59.Kohlbacher O, Lenhof HP. BALL–rapid software prototyping in computational molecular biology. Biochemicals algorithms library. Bioinformatics. 2000;16(9):815–824. doi: 10.1093/bioinformatics/16.9.815. [DOI] [PubMed] [Google Scholar]
- 60.Gunasekaran K, Tsai CJ, Nussinov R. Analysis of ordered and disordered protein complexes reveals structural features discriminating between stable and unstable monomers. J Mol Biol. 2004;341(5):1327–1341. doi: 10.1016/j.jmb.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 61.Fayyad UM, Irani KB. Multi-interval discretization of continuous-valued attributes for classification learning. In: Bajcsy R, editor. The 13th International Joint Conference on Uncertainty in Artificial Intelligence, Chambery, France, 1993. Menlo Park: Morgan-Kaufmann; 1993. pp. 1022–1027. [Google Scholar]
- 62.Vacic V, Uversky VN, Dunker AK, Lonardi S. Composition profiler: a tool for discovery and visualization of amino acid composition differences. BMC Bioinforma. 2007;8:211. doi: 10.1186/1471-2105-8-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sickmeier M, Hamilton JA, LeGall T, Vacic V, Cortese MS, Tantos A, Szabo B, Tompa P, Chen J, Uversky VN, Obradovic Z, Dunker AK. DisProt: the database of disordered proteins. Nucleic Acids Res. 2007;35(Database issue):D786–D793. doi: 10.1093/nar/gkl893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Linding R, Russell RB, Neduva V, Gibson TJ. GlobPlot: exploring protein sequences for globularity and disorder. Nucleic Acids Res. 2003;31(13):3701–3708. doi: 10.1093/nar/gkg519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li BQ, Hu LL, Chen L, Feng KY, Cai YD, Chou KC. Prediction of protein domain with mRMR feature selection and analysis. PLoS One. 2012;7(6):e39308. doi: 10.1371/journal.pone.0039308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Monastyrskyy B, Fidelis K, Moult J, Tramontano A, Kryshtafovych A. Evaluation of disorder predictions in CASP9. Proteins. 2011;79(Suppl 10):107–118. doi: 10.1002/prot.23161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Peng ZL, Kurgan L. Comprehensive comparative assessment of in-silico predictors of disordered regions. Curr Protein Pept Sci. 2012;13:6–18. doi: 10.2174/138920312799277938. [DOI] [PubMed] [Google Scholar]
- 69.Tompa P, Fuxreiter M, Oldfield CJ, Simon I, Dunker AK, Uversky VN. Close encounters of the third kind: disordered domains and the interactions of proteins. Bioessays. 2009;31(3):328–335. doi: 10.1002/bies.200800151. [DOI] [PubMed] [Google Scholar]
- 70.Noivirt-Brik O, Prilusky J, Sussman JL. Assessment of disorder predictions in CASP8. Proteins. 2009;77(Suppl 9):210–216. doi: 10.1002/prot.22586. [DOI] [PubMed] [Google Scholar]
- 71.Mizianty MJ, Zhang T, Xue B, Zhou Y, Dunker AK, Uversky VN, Kurgan L. In-silico prediction of disorder content using hybrid sequence representation. BMC Bioinforma. 2011;12:245. doi: 10.1186/1471-2105-12-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Oldfield CJ, Cheng Y, Cortese MS, Brown CJ, Uversky VN, Dunker AK. Comparing and combining predictors of mostly disordered proteins. Biochemistry. 2005;44(6):1989–2000. doi: 10.1021/bi047993o. [DOI] [PubMed] [Google Scholar]
- 73.Xue B, Oldfield CJ, Dunker AK, Uversky VN. CDF it all: consensus prediction of intrinsically disordered proteins based on various cumulative distribution functions. FEBS Lett. 2009;583(9):1469–1474. doi: 10.1016/j.febslet.2009.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mohan A, Sullivan WJ, Jr, Radivojac P, Dunker AK, Uversky VN. Intrinsic disorder in pathogenic and non-pathogenic microbes: discovering and analyzing the unfoldomes of early-branching eukaryotes. Mol Biosyst. 2008;4(4):328–340. doi: 10.1039/b719168e. [DOI] [PubMed] [Google Scholar]
- 75.Huang F, Oldfield C, Meng J, Hsu WL, Xue B, Uversky VN, Romero P, Dunker AK. Subclassifying disordered proteins by the CH-CDF plot method. Pac Symp Biocomput. 2012;17:128–139. [PubMed] [Google Scholar]
- 76.Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol. 1981;147(1):195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 77.Disfani FM, Hsu W-L, Mizianty MJ, Oldfield CJ, Xue B, Dunker AK, Uversky VN, Kurgan L. MoRFpred, a computational tool for sequence-based prediction and characterization of short disorder-to-order transitioning binding regions in proteins. Bioinformatics. 2012;28(12):i75–83. doi: 10.1093/bioinformatics/bts209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Oldfield CJ, Cheng Y, Cortese MS, Romero P, Uversky VN, Dunker AK. Coupled folding and binding with alpha-helix-forming molecular recognition elements. Biochemistry. 2005;44(37):12454–12470. doi: 10.1021/bi050736e. [DOI] [PubMed] [Google Scholar]
- 79.Vacic V, Oldfield CJ, Mohan A, Radivojac P, Cortese MS, Uversky VN, Dunker AK. Characterization of molecular recognition features, MoRFs, and their binding partners. J Proteome Res. 2007;6(6):2351–2366. doi: 10.1021/pr0701411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mohan A, Oldfield CJ, Radivojac P, Vacic V, Cortese MS, Dunker AK, Uversky VN. Analysis of molecular recognition features (MoRFs) J Mol Biol. 2006;362(5):1043–1059. doi: 10.1016/j.jmb.2006.07.087. [DOI] [PubMed] [Google Scholar]
- 81.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292(2):195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 82.Wang K, Samudrala R. Incorporating background frequency improves entropy-based residue conservation measures. BMC Bioinforma. 2006;7:385. doi: 10.1186/1471-2105-7-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jones DT, Swindells MB. Getting the most from PSI-BLAST. Trends Biochem Sci. 2002;27(3):161–164. doi: 10.1016/s0968-0004(01)02039-4. [DOI] [PubMed] [Google Scholar]
- 85.Peng Z, Mizianty MJ, Xue B, Kurgan L, Uversky VN. More than just tails: intrinsic disorder in histone proteins. Mol Biosyst. 2012;8(7):1886–1901. doi: 10.1039/c2mb25102g. [DOI] [PubMed] [Google Scholar]
- 86.Chen K, Mizianty MJ, Kurgan L. Prediction and analysis of nucleotide-binding residues using sequence and sequence-derived structural descriptors. Bioinformatics. 2012;28(3):331–341. doi: 10.1093/bioinformatics/btr657. [DOI] [PubMed] [Google Scholar]
- 87.Johansson F, Toh H. A comparative study of conservation and variation scores. BMC Bioinforma. 2010;11(1):388. doi: 10.1186/1471-2105-11-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Teschke CM, King J. Folding and assembly of oligomeric proteins in Escherichia coli . Curr Opin Biotechnol. 1992;3(5):468–473. doi: 10.1016/0958-1669(92)90073-r. [DOI] [PubMed] [Google Scholar]
- 89.Xu D, Tsai CJ, Nussinov R. Mechanism and evolution of protein dimerization. Protein Sci. 1998;7(3):533–544. doi: 10.1002/pro.5560070301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gerbasi VR, Weaver CM, Hill S, Friedman DB, Link AJ. Yeast Asc1p and mammalian RACK1 are functionally orthologous core 40S ribosomal proteins that repress gene expression. Mol Cell Biol. 2004;24(18):8276–8287. doi: 10.1128/MCB.24.18.8276-8287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Morrison CA, Garrett RA, Bradbury EM. Physical studies on the conformation of ribosomal protein S4 from Escherichia coli . Eur J Biochem. 1977;78(1):153–159. doi: 10.1111/j.1432-1033.1977.tb11724.x. [DOI] [PubMed] [Google Scholar]
- 92.Venyaminov SY, Gudkov AT, Gogia ZV, Tumanova LG (1981) Absorption and circular dichroism spectra of individual proteins from Escherichia coli ribosomes. Scientific Center of Biological Research of the Academy of Sciences of the USSR in Pushchino, Moscow Region, Russia
- 93.van de Ven FJ, de Bruin SH, Hilbers CW. 500-MHz 1H-NMR studies of ribosomal proteins isolated from 70-S ribosomes of Escherichia coli . Eur J Biochem. 1983;134(3):429–438. doi: 10.1111/j.1432-1033.1983.tb07585.x. [DOI] [PubMed] [Google Scholar]
- 94.Sayers EW, Gerstner RB, Draper DE, Torchia DA. Structural preordering in the N-terminal region of ribosomal protein S4 revealed by heteronuclear NMR spectroscopy. Biochemistry. 2000;39(44):13602–13613. doi: 10.1021/bi0013391. [DOI] [PubMed] [Google Scholar]
- 95.Woestenenk EA, Gongadze GM, Shcherbakov DV, Rak AV, Garber MB, Hard T, Berglund H. The solution structure of ribosomal protein L18 from Thermus thermophilus reveals a conserved RNA-binding fold. Biochem J. 2002;363(Pt 3):553–561. doi: 10.1042/0264-6021:3630553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Raibaud S, Lebars I, Guillier M, Chiaruttini C, Bontems F, Rak A, Garber M, Allemand F, Springer M, Dardel F. NMR structure of bacterial ribosomal protein l20: implications for ribosome assembly and translational control. J Mol Biol. 2002;323(1):143–151. doi: 10.1016/s0022-2836(02)00921-x. [DOI] [PubMed] [Google Scholar]
- 97.Ohman A, Rak A, Dontsova M, Garber MB, Hard T. NMR structure of the ribosomal protein L23 from Thermus thermophilus . J Biomol NMR. 2003;26(2):131–137. doi: 10.1023/a:1023502307069. [DOI] [PubMed] [Google Scholar]
- 98.Aramini JM, Huang YJ, Cort JR, Goldsmith-Fischman S, Xiao R, Shih LY, Ho CK, Liu J, Rost B, Honig B, Kennedy MA, Acton TB, Montelione GT. Solution NMR structure of the 30S ribosomal protein S28E from Pyrococcus horikoshii . Protein Sci. 2003;12(12):2823–2830. doi: 10.1110/ps.03359003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wu B, Yee A, Pineda-Lucena A, Semesi A, Ramelot TA, Cort JR, Jung JW, Edwards A, Lee W, Kennedy M, Arrowsmith CH. Solution structure of ribosomal protein S28E from Methanobacterium thermoautotrophicum . Protein Sci. 2003;12(12):2831–2837. doi: 10.1110/ps.03358203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Turner CF, Moore PB. The solution structure of ribosomal protein L18 from Bacillus stearothermophilus . J Mol Biol. 2004;335(3):679–684. doi: 10.1016/j.jmb.2003.11.018. [DOI] [PubMed] [Google Scholar]
- 101.Nishimura M, Yoshida T, Shirouzu M, Terada T, Kuramitsu S, Yokoyama S, Ohkubo T, Kobayashi Y. Solution structure of ribosomal protein L16 from Thermus thermophilus HB8. J Mol Biol. 2004;344(5):1369–1383. doi: 10.1016/j.jmb.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 102.Jeon BY, Jung J, Kim DW, Yee A, Arrowsmith CH, Lee W. Solution structure of TA1092, a ribosomal protein S24e from Thermoplasma acidophilum . Proteins. 2006;64(4):1095–1097. doi: 10.1002/prot.21045. [DOI] [PubMed] [Google Scholar]
- 103.Edmondson SP, Turri J, Smith K, Clark A, Shriver JW. Structure, stability, and flexibility of ribosomal protein L14e from Sulfolobus solfataricus . Biochemistry. 2009;48(24):5553–5562. doi: 10.1021/bi9003205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wu B, Lukin J, Yee A, Lemak A, Semesi A, Ramelot TA, Kennedy MA, Arrowsmith CH. Solution structure of ribosomal protein L40E, a unique C4 zinc finger protein encoded by archaeon Sulfolobus solfataricus . Protein Sci. 2008;17(3):589–596. doi: 10.1110/ps.073273008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Monastyrskyy B, Fidelis K, Tramontano A, Kryshtafovych A. Evaluation of residue-residue contact predictions in CASP9. Proteins. 2011;79(Suppl 10):119–125. doi: 10.1002/prot.23160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Williams RM, Obradovi Z, Mathura V, Braun W, Garner EC, Young J, Takayama S, Brown CJ, Dunker AK. The protein non-folding problem: amino acid determinants of intrinsic order and disorder. Pac Symp Biocomput. 2001;6:89–100. doi: 10.1142/9789814447362_0010. [DOI] [PubMed] [Google Scholar]
- 107.Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins. 2001;42(1):38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 108.Semrad K, Green R, Schroeder R. RNA chaperone activity of large ribosomal subunit proteins from Escherichia coli . RNA. 2004;10(12):1855–1860. doi: 10.1261/rna.7121704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wilson DN, Nierhaus KH. Ribosomal proteins in the spotlight. Crit Rev Biochem Mol Biol. 2005;40(5):243–267. doi: 10.1080/10409230500256523. [DOI] [PubMed] [Google Scholar]
- 110.Uversky VN. Multitude of binding modes attainable by intrinsically disordered proteins: a portrait gallery of disorder-based complexes. Chem Soc Rev. 2011;40(3):1623–1634. doi: 10.1039/c0cs00057d. [DOI] [PubMed] [Google Scholar]
- 111.Nierhaus KH. The assembly of prokaryotic ribosomes. Biochimie. 1991;73(6):739–755. doi: 10.1016/0300-9084(91)90054-5. [DOI] [PubMed] [Google Scholar]
- 112.Dabbs ER. Mutational alterations in 50 proteins of the Escherichia coli ribosome. Mol Gen Genet. 1978;165(1):73–78. doi: 10.1007/BF00270378. [DOI] [PubMed] [Google Scholar]
- 113.Dabbs ER. Mutant studies on the prokaryotic ribosome. In: Hardesty B, Kramer G, editors. Structure, function and genetics of ribosomes. New York: Springer; 1986. pp. 733–748. [Google Scholar]
- 114.Patil A, Nakamura H. Disordered domains and high surface charge confer hubs with the ability to interact with multiple proteins in interaction networks. FEBS Lett. 2006;580(8):2041–2045. doi: 10.1016/j.febslet.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 115.Ekman D, Light S, Bjorklund AK, Elofsson A. What properties characterize the hub proteins of the protein-protein interaction network of Saccharomyces cerevisiae? Genome Biol. 2006;7(6):R45. doi: 10.1186/gb-2006-7-6-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Haynes C, Oldfield CJ, Ji F, Klitgord N, Cusick ME, Radivojac P, Uversky VN, Vidal M, Iakoucheva LM. Intrinsic disorder is a common feature of hub proteins from four eukaryotic interactomes. PLoS Comput Biol. 2006;2(8):e100. doi: 10.1371/journal.pcbi.0020100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dosztanyi Z, Chen J, Dunker AK, Simon I, Tompa P. Disorder and sequence repeats in hub proteins and their implications for network evolution. J Proteome Res. 2006;5(11):2985–2995. doi: 10.1021/pr060171o. [DOI] [PubMed] [Google Scholar]
- 118.Singh GP, Ganapathi M, Dash D. Role of intrinsic disorder in transient interactions of hub proteins. Proteins. 2007;66(4):761–765. doi: 10.1002/prot.21281. [DOI] [PubMed] [Google Scholar]
- 119.Singh GP, Dash D. Intrinsic disorder in yeast transcriptional regulatory network. Proteins. 2007;68(3):602–605. doi: 10.1002/prot.21497. [DOI] [PubMed] [Google Scholar]
- 120.Wool IG. Extraribosomal functions of ribosomal proteins. Trends Biochem Sci. 1996;21(5):164–165. [PubMed] [Google Scholar]
- 121.Tompa P, Szasz C, Buday L. Structural disorder throws new light on moonlighting. Trends Biochem Sci. 2005;30(9):484–489. doi: 10.1016/j.tibs.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 122.Weisberg RA. Transcription by moonlight: structural basis of an extraribosomal activity of ribosomal protein S10. Mol Cell. 2008;32(6):747–748. doi: 10.1016/j.molcel.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lindstrom MS. Emerging functions of ribosomal proteins in gene-specific transcription and translation. Biochem Biophys Res Commun. 2009;379(2):167–170. doi: 10.1016/j.bbrc.2008.12.083. [DOI] [PubMed] [Google Scholar]
- 124.Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins? Mol Cell. 2009;34(1):3–11. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Singh D, Chang SJ, Lin PH, Averina OV, Kaberdin VR, Lin-Chao S. Regulation of ribonuclease E activity by the L4 ribosomal protein of Escherichia coli . Proc Natl Acad Sci USA. 2009;106(3):864–869. doi: 10.1073/pnas.0810205106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Freedman LP, Zengel JM, Archer RH, Lindahl L. Autogenous control of the S10 ribosomal protein operon of Escherichia coli: genetic dissection of transcriptional and posttranscriptional regulation. Proc Natl Acad Sci USA. 1987;84(18):6516–6520. doi: 10.1073/pnas.84.18.6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zengel JM, Lindahl L. Escherichia coli ribosomal protein L4 stimulates transcription termination at a specific site in the leader of the S10 operon independent of L4-mediated inhibition of translation. J Mol Biol. 1990;213(1):67–78. doi: 10.1016/S0022-2836(05)80122-6. [DOI] [PubMed] [Google Scholar]
- 128.Zengel JM, Lindahl L. Ribosomal protein L4 stimulates in vitro termination of transcription at a NusA-dependent terminator in the S10 operon leader. Proc Natl Acad Sci USA. 1990;87(7):2675–2679. doi: 10.1073/pnas.87.7.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Li X, Lindahl L, Zengel JM. Ribosomal protein L4 from Escherichia coli utilizes nonidentical determinants for its structural and regulatory functions. RNA. 1996;2(1):24–37. [PMC free article] [PubMed] [Google Scholar]
- 130.Eng FJ, Warner JR. Structural basis for the regulation of splicing of a yeast messenger RNA. Cell. 1991;65(5):797–804. doi: 10.1016/0092-8674(91)90387-e. [DOI] [PubMed] [Google Scholar]
- 131.Fewell SW, Woolford JL., Jr Ribosomal protein S14 of Saccharomyces cerevisiae regulates its expression by binding to RPS14B pre-mRNA and to 18S rRNA. Mol Cell Biol. 1999;19(1):826–834. doi: 10.1128/mcb.19.1.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Presutti C, Ciafre SA, Bozzoni I. The ribosomal protein L2 in S. cerevisiae controls the level of accumulation of its own mRNA. EMBO J. 1991;10(8):2215–2221. doi: 10.1002/j.1460-2075.1991.tb07757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Malygin AA, Parakhnevitch NM, Ivanov AV, Eperon IC, Karpova GG. Human ribosomal protein S13 regulates expression of its own gene at the splicing step by a feedback mechanism. Nucleic Acids Res. 2007;35(19):6414–6423. doi: 10.1093/nar/gkm701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Parakhnevich NM, Ivanov AV, Malygin AA, Karpova GG. Human ribosomal protein S13 inhibits splicing of the own pre-mRNA. Mol Biol (Mosk) 2007;41(1):51–58. [PubMed] [Google Scholar]
- 135.Mitrovich QM, Anderson P. Unproductively spliced ribosomal protein mRNAs are natural targets of mRNA surveillance in C. elegans . Genes Dev. 2000;14(17):2173–2184. doi: 10.1101/gad.819900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gilbert W. Origin of life: the RNA world. Nature. 1986;319(6055):618. [Google Scholar]
- 137.Jeffares DC, Poole AM, Penny D. Relics from the RNA world. J Mol Evol. 1998;46(1):18–36. doi: 10.1007/pl00006280. [DOI] [PubMed] [Google Scholar]
- 138.Poole AM, Jeffares DC, Penny D. The path from the RNA world. J Mol Evol. 1998;46(1):1–17. doi: 10.1007/pl00006275. [DOI] [PubMed] [Google Scholar]
- 139.Doi N, Yanagawa H. Origins of globular structure in proteins. FEBS Lett. 1998;430(3):150–153. doi: 10.1016/s0014-5793(98)00674-7. [DOI] [PubMed] [Google Scholar]
- 140.Cristofari G, Darlix JL. The ubiquitous nature of RNA chaperone proteins. Prog Nucleic Acid Res Mol Biol. 2002;72:223–268. doi: 10.1016/s0079-6603(02)72071-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.