Abstract
Effective antiviral immunity depends on accurate recognition of viral RNAs by the innate immune system. Double-stranded RNA (dsRNA) often accumulates in virally infected cells and was initially considered a unique viral signature that was sufficient to initiate antiviral response through dsRNA receptors and dsRNA-dependent effectors such as Toll-like receptor 3, retinoic acid inducible gene-1, protein kinase RNA-activated and oligoadenylate synthetase. However, dsRNA is also present in many cellular RNAs, raising a question of how these receptors and effectors discriminate between viral and cellular dsRNAs. Accumulating evidence suggests that innate immune sensors detect not only dsRNA structure but also other and often multiple features of RNA such as length, sequence, cellular location, post-transcriptional processing and modification, which are divergent between viral and cellular RNAs. This review summarizes recent findings on the substrate specificities of a few selected dsRNA-dependent effectors and receptors, which have revealed more complex mechanisms involved in cellular discrimination between self and non-self RNA.
Keywords: Antiviral innate immunity, Double-stranded RNA, PKR, OAS, ADAR, TLR3, RIG-I, MDA5
Introduction
A successful host defense against viral infection depends on both accurate recognition of viral invasion by germ-line encoded pattern recognition receptors (PRRs) and proper functioning of innate immune effectors to suppress viral replication (Fig. 1) [1, 2]. Recognition of invariant virus-associated molecular patterns by PRRs activates signaling pathways to generate antiviral cytokines including, but not limited to, the type I interferons (e.g., IFNα/β). These cytokines in turn stimulate the expression of a series of interferon-stimulated genes (ISGs), such as antiviral effector proteins, to establish antiviral states in infected and neighboring cells (Fig. 1) [1, 3], and activate appropriate adaptive immune response [4]. In vertebrates, several of these receptors and effectors regulate their signaling activity and effector functions, respectively, in a manner dependent on viral RNA binding (Fig. 1) [1, 2]. Such viral RNA-specific PRRs include the Toll-like receptors (TLRs) 3 and 7–8 and the retinoic acid inducible gene-1 (RIG-I)-like receptors (RLRs), RIG-I and melanoma differentiation-associated gene 5 (MDA5) [2]. Viral RNA-dependent antiviral effectors include protein kinase RNA-activated (PKR), oligoadenylate synthetase (OAS) and adenosine deaminase acting on RNA (ADAR) [5]. With the exception of TLR7 and 8, which recognize viral single-stranded RNAs (ssRNAs), these cellular receptors and effectors were shown to recognize double-stranded RNA (dsRNA). As dsRNA structure had been thought to be a unique feature of viral RNAs, it had been widely accepted that dsRNA binding alone is sufficient to activate their respective antiviral functions.
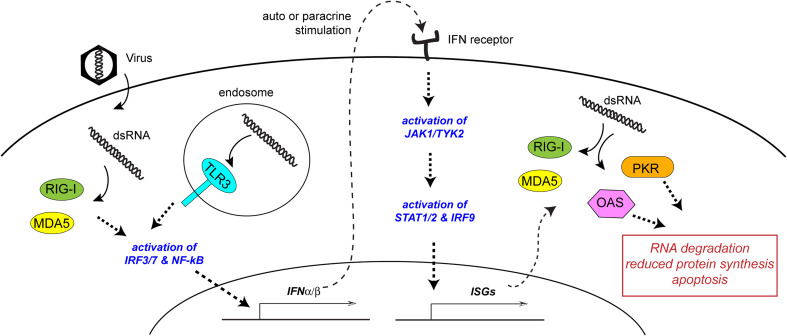
Fig. 1.
Schematic diagram of cellular responses to viral dsRNA. Upon viral infection, viral dsRNA is recognized by pattern recognition receptors (PRRs) such as RIG-I, MDA5, and TLR3, which stimulate expression of type I interferons (e.g., IFNα/β) via IRF3/7 or NF-κB pathways. Expressed IFNα/β cytokines are secreted into the extracellular space and stimulate interferon receptors in an auto or paracrine manner, which in turn activates the JAK/STAT signaling pathway to up-regulate expression of interferon-stimulated genes (ISGs). ISGs include PRRs such as RIG-I and MDA5, as well as antiviral effector proteins such as OAS and PKR, which suppress global protein synthesis and establish the antiviral state
The dsRNA duplex adopts an A-form helix that is distinct from the typical B-form helix of dsDNA. The major groove of dsRNA is narrower and deeper than that of dsDNA (4 vs. 11–12 Å width), whereas the minor groove of dsRNA is wider and shallower than dsDNA (10–11 vs. 6 Å width)[6]. This distinct configuration of the phosphate backbone of dsRNA along with the unique 2′ hydroxyl groups exposed in the minor groove can be specifically recognized by conserved protein motifs such as dsRNA binding domain (dsRBD) motifs in PKR and ADAR [7, 8]. In particular, the narrow major groove, which contains sequence-specific information and is a common site of interaction between protein and dsDNA [9], does not allow insertion of protein to interact with dsRNA bases. Accordingly, protein–dsRNA interaction is largely mediated by the minor groove, which contains degenerate sequence information, and the phosphate backbone, thus is generally RNA sequence-independent [7].
Recent studies on the human transcriptome revealed that dsRNAs, originally thought not to be expressed in the cell, are generated in the form of secondary structures in pre and mature micro or small interfering RNAs (miRNAs or siRNAs) [10, 11], and in the form of long duplexes formed by inverted repeat sequence elements [12, 13] or sense–antisense hybrids [14, 15]. These observations raise a question about the viral selection mechanism of receptors and effectors previously thought to discriminate between viral and cellular origin solely on the basis of presence of dsRNA structure. Recent data suggests that viral RNA receptors and effectors can recognize other features of RNA, in addition to duplex structure such as 5′ or 3′ functional groups, post-transcriptional modification, length, tertiary structure, and in some cases, sequence [18–23]. This sensitivity to multiple other features of RNA is likely important for robust and accurate discrimination between cellular and viral dsRNAs. It also provides an explanation for how some cellular or viral dsRNAs act as antagonists rather than agonists for dsRNA receptors and effectors [16, 17]. It remains to be addressed, however, how these proteins interact with dsRNA in a manner that allows for simultaneous recognition of multiple, seemingly disparate features of RNA to bring about self versus non-self discrimination. In this review, we will discuss multi-layered aspects of RNA specificity and the structural and biochemical mechanisms of dsRNA-dependent effectors (PKR, ADAR, and OAS) and receptors (TLR3, RIG-I, and MDA5). More detailed reviews on the biological functions of each of these proteins can be found elsewhere.
Protein kinase RNA-activated (PKR)
PKR is a cytoplasmic Ser/Thr protein kinase that is up-regulated by type I interferons and plays an important role in the establishment of an antiviral and antiproliferative cell state in response to viral infection [24]. PKR consists of two N-terminal tandem dsRBDs and a C-terminal catalytic kinase domain [11]. In the absence of dsRNA, the kinase domain is in the autorepressed, monomeric state [25]. Binding of dsRNA leads to a conformational change in PKR, which is believed to release the catalytic domain from the autoinhibitory dsRBDs. Binding to dsRNA also brings multiple PKR molecules into close proximity, which enables trans-phosphorylation of PKR at several Ser or Thr residues throughout the protein [26–28]. Phosphorylated PKR then dissociates from dsRNA [29], and functions as a constitutively active kinase that in turn phosphorylates serine 51 of eukaryotic translational initiation factor (eIF2α) and suppresses global protein synthesis by blocking translation initiation (Fig. 2a) [24].
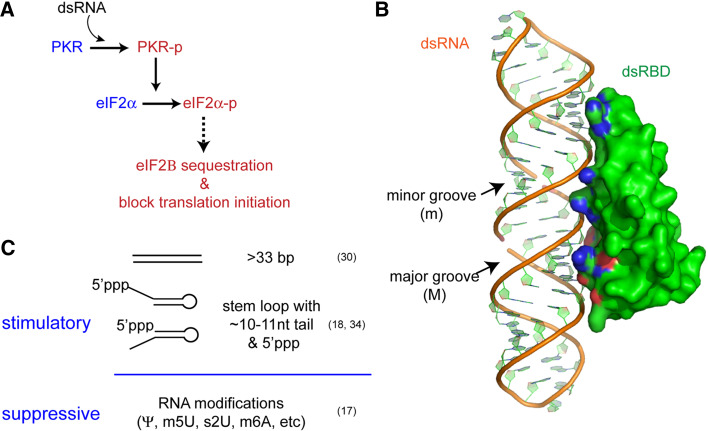
Fig. 2.
a Schematic of dsRNA-dependent effector functions of PKR. b Structure of ribonuclease III dsRBD in complex with dsRNA (PDB: 2EZ6) [34]. No structure is currently available for PKR dsRBD in complex with dsRNA. Protein residues interacting with dsRNA are colored blue for basic residues and red for acidic residues. The minor and major grooves are indicated by m and M, respectively. c Summary of PKR-stimulatory and suppressive features of RNA
How PKR specifically recognizes dsRNA has been extensively studied using a variety of biophysical and biochemical methods. High affinity binding of PKR requires cooperative actions of the two dsRBDs [30] and a minimum dsRNA length of ~16 bp [31]. Stimulation of the kinase activity of PKR, however, requires ~33-bp dsRNA, which is consistent with the minimal length required for dimerization [32]. No atomic level structure is currently available for PKR dsRBDs in complex with dsRNA, but structures of other homologous dsRBDs provide some insight into the dsRNA recognition mechanism of PKR. Crystal structures of dsRBD of RNaseIII and RNA-binding protein A (Xlrbpa) showed that each dsRBD binds to one face of dsRNA spanning ~16 bp (Fig. 2b) [33, 34]. The primary contacts are made with the phosphate backbone and 2′ hydroxyl groups of the minor groove. No protein structure is inserted into the major groove, consistent with sequence-independent recognition of dsRNA. Many of the residues in RNaseIII and Xlrbpa dsRBDs that interact with dsRNA are conserved in PKR dsRBD, suggesting the possibility of a similar dsRNA–dsRBD interaction mode. The relative orientation of the two dsRBDs of PKR is as yet unclear. The long linker (~20 amino acid) between the two dsRBDs are disordered in the absence of dsRNA, but undergoes a conformational change upon binding to 23-bp hairpin RNA as evidenced from the large chemical shift [30]. This could be possibly due to direct binding of RNA to the linker region or an indirect conformational change propagated from dsRBDs upon interaction with dsRNA.
How does PKR avoid recognition of other cellular dsRNAs? It was previously proposed that the ~33-bp requirement prevents cellular RNAs containing short duplex regions, such as miRNAs or siRNAs, from activating PKR. However, recent studies revealed more complex RNA selectivity of PKR beyond recognition of a simple dsRNA structure (Fig. 2c). The 5′ untranslated region (UTR) of IFN-γ mRNA folds into a complex tertiary structure that forms a coaxially stacked 33-bp stem, which then activates PKR [35]. In addition, an RNA library selection experiment revealed that a short (~16-bp) stem-loop of ssRNA can activate PKR as well as a perfect duplex RNA longer than 33 bp [36]. Unlike long dsRNA, these short stem-loops require a ~10-nt single-stranded tail at either the 5′ or 3′ end and a triphosphate group at the 5′ end to activate PKR [20]. In addition, some post-transcriptional modifications, such as pseudouridine and 5-methyluridine, which are known to preserve the RNA secondary structure, abolish or diminish the PKR-stimulatory activity of ssRNA containing short stem-loop [19]. As most cellular RNAs undergo extensive post-transcriptional modifications and 5′ processing in the nucleus, which removes the 5′ triphosphate group present in all nascent transcripts, the sensitivity of PKR to the 5′ triphosphate group and modified nucleotides provides an explanation for how PKR avoids inappropriate activation by cellular ssRNAs with secondary structures. However, it remains to be understood how PKR utilizes dsRBDs to recognize ssRNA tails and the 5′ triphosphate group, how it discriminates among ssRNAs on the basis of nucleotide modification and whether it undergoes similar dimerization upon binding to hairpin-containing ssRNA as with dsRNA.
Adenosine deaminase acting on RNA (ADAR)
ADAR is an RNA-modifying enzyme that converts adenosine to inosine (I) within dsRNA by hydrolytic deamination (Fig. 3a) [37]. This A-to-I conversion is one of many processes commonly referred to as RNA editing. ADAR recognizes dsRNA structure using one to three dsRBDs depending on the organism and the isotype [38]. Mammals express three isotypes of ADAR isotypes 1 and 2 display editing activity and are ubiquitously expressed, whereas isotype 3 lacks a demonstrable editing activity and its expression is limited to the central nervous system [39]. Adenosine editing by ADAR1 and 2 can occur in either a site-specific or non-specific manner depending on the target dsRNA structure (Fig. 3b) [40–42]. In a perfect duplex RNAs, A-to-I editing is non-specific, and can occur for up to ~50 % of adenosines [41, 43]. On the other hand, for an imperfect duplex RNA with bulges or mismatches, A-to-I editing occurs in a more restricted manner that is often sequence-dependent [44, 45]. The best-studied examples of those site-specific targets include the Q/R and R/G sites within mRNAs encoding glutamate receptors (GluR) and serotonin receptor (5-HT2cR) [46, 47]. Since inosine pairs with cytidine, ADAR-mediated editing results in an inosine–uridine mismatch, which decreases the stability of the duplex [48]. Inosine is also read as guanosine by the cellular ribosome, spliceosome, and viral RNA-dependent RNA polymerase, and thus A-to-I editing can alter protein-coding potential, mRNA splicing pattern, and can be propagated into viral genomes (Fig. 3a) [37, 49].
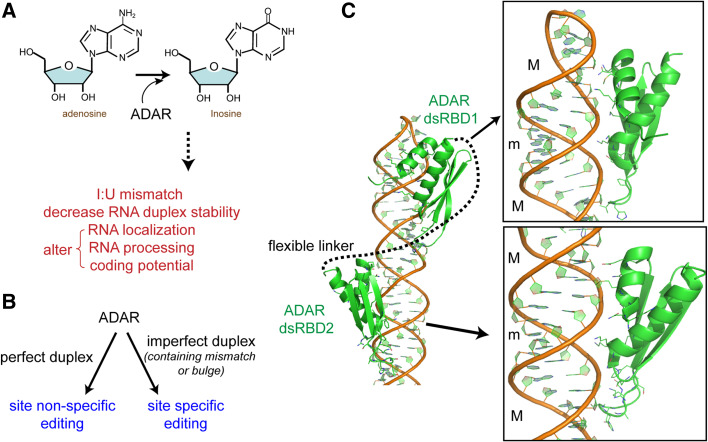
Fig. 3.
a Schematic of dsRNA-dependent effector functions of ADAR. b Summary of RNA specificity of ADAR. Examples of site-specific editing targets include the Q/R and R/G sites of GluR-B pre-mRNA. c Structure of ADAR2 dsRBDs in complex with a RNA stem-loop containing the R/G editing site of the GluR-2 pre-mRNA (PDB: 2L3J [73]). Protein residues interacting with dsRNA are shown in a stick representation and the flexible linker connecting between the two dsRBDs is represented by a dotted line. The minor and major grooves are indicated by m and M, respectively
The effect of RNA editing by ADAR on virus and antiviral immunity appears complex and depends greatly on specific cell types and viruses [50]. A splice variant of ADAR1, p150, is up-regulated upon interferon stimulation and was shown to suppress replication of measles virus and virus-induced cytotoxicity [51]. Although the precise mechanism for the antiviral function of p150 is not clear, extensive mutations of A-to-G and U-to-C were observed in the measles virus genome [52]. Similar antiviral effects of p150 were observed with influenza A, Newcastle disease, and Sendai viruses [51]. However, not all viruses are negatively affected by ADAR1 as positive effects have also been observed [53–55]. For example, site-selective editing of hepatitis D virus mRNA by ADAR is essential for proper synthesis of a viral protein, HDAg-L [56, 57]. In the case of vesicular stomatitis virus, ADAR helps viral replication in a manner independent of its editing activity, but rather by antagonizing PKR [53, 54]. In addition, recent studies showed that inosine containing dsRNAs can inhibit activation of IRF3 and therefore the downstream interferon signaling pathway, possibly by functioning as competitive inhibitors of RIG-I and MDA5 [17]. While it is clear that ADAR is involved in determining the fate of host–virus interaction, how exactly ADAR exerts pro- and anti-viral effects and how these seemingly opposing functions are coordinated remain to be investigated in the future studies.
ADAR-mediated RNA editing is not limited to viral RNAs, but also occurs relatively frequently for cellular RNAs as evidenced by recent transcriptome analyses [37, 58, 59]. The examples include ~70-nt-long pre-miRNAs [58, 60] and inverted Alu elements, which fold into a near-perfect duplex of ~250–300 bp [12, 61]. The precise biological consequences of these RNA-editing events are still incompletely understood, but are likely to be multifaceted through their effects on RNA stability, function, and subcellular localization [37, 49, 62–65]. It is also tempting to speculate that editing could prevent aberrant activation of the dsRNA-dependent innate immune system by disrupting the long duplex structures present among the cellular RNAs. Interestingly, deletion of ADAR1 in hematopoietic stem cells has been shown to increase the level of type I and II interferons [66], which could be possibly due to the inability of the ADAR knock-out cells to disrupt cellular RNA duplex structures. In whole organisms, ADAR deficiency was shown to cause developmental abnormality in vertebrates [67–70] and behavioral defects in invertebrates [71, 72], consistent with an essential and versatile role of ADAR in cellular RNA metabolism.
How does ADAR recognize a specific site on an imperfect duplex RNA while promiscuously modifying a perfect dsRNA? A recent NMR structure of the two dsRBDs of ADAR2 in complex with one of the target sites (R/G site) within GluR-2 mRNA provided an important insight into the sequence-specific dsRNA recognition by dsRBD [73]. In this structure, the overall interaction between dsRNA and ADAR dsRBD was similar to that seen with other homologous dsRBDs [33, 34, 74, 75], i.e., each dsRBD binds to one face of dsRNA, forming an interaction with two successive minor grooves of dsRNA (Fig. 3c). The structure also revealed, however, several unexpected contacts between protein residues and edges of bases in the minor groove at or near the site of mismatch (Fig. 3c). Although these interactions appear to depend on the presence of a mismatch and thus are unlikely to occur in a perfect duplex, the structure provides an intriguing example of the potential of the dsRNA minor groove in sequence-dependent interaction with proteins and demonstrates the versatility of dsRBDs in identifying structural irregularities embedded within a dsRNA.
Oligoadenylate synthetase (OAS)
OAS belongs to a family of template-independent RNA polymerases, which includes the eukaryotic polyadenosine polymerase (PAP) and the class I CCA-adding enzyme (CCA) from Archaea [76]. Mammals express four types of OAS among which three isoforms, OAS1, OAS2, and OAS3, are likely to have evolved from gene duplication. These three members of OAS display an enzymatic activity of linking two ATP molecules (donor and acceptor) via a 2′ and 5′ phosphodiester bond to synthesize 2′,5′-linked oligoadenylates [pxA(2′p5′A)n; x = 1–3; n >2] [77–79]. This 2′,5′-linked oligoadenylate then functions as a cofactor to activate a latent ribonuclease, RNase-L. RNase-L degrades both viral and cellular ssRNAs, such as ribosomal RNAs and mRNAs, with little sequence specificity (typically after UU or UA sites), which results in inhibition of global protein synthesis (Fig. 4a) [80–82]. In a normal, resting state, the level of 2′,5′-oligoadenylate is tightly regulated by the enzymes 5′-phosphatase and 2′-phosphodiesterase, which inactivates and degrades 2′,5′-oligoadenylates, respectively [83, 84]. During viral infection, however, the level of OAS is transiently up-regulated by interferon, which results in transient activation of RNase-L and suppression of viral replication [79, 82, 85, 86].
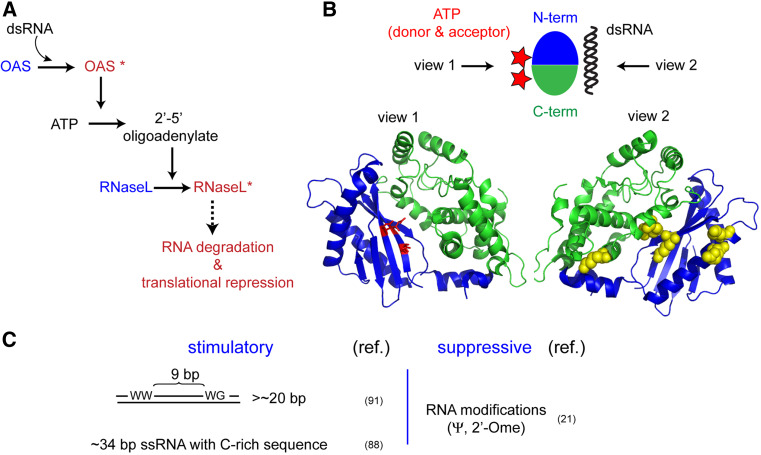
Fig. 4.
a Schematic of dsRNA-dependent effector functions of OAS. Active states of OAS and RNase-L are indicated by an asterisk. The precise molecular nature of their active states is as yet unclear. b Structure of OAS free of dsRNA or ATP (PDB: 1PX5 [87]) in two opposite views. No structure is available in complex with ATP or dsRNA. Protein residues proposed to interact with the donor ATP molecule and dsRNA are shown as red sticks and yellow spheres, respectively. c Summary of RNA specificity of OAS. ‘W’ stands for A or U
Unlike PKR or ADAR, OAS does not harbor dsRBDs. The crystal structure of OAS revealed a single globular domain, composed of the N-terminal and C-terminal lobes [87]. Although the structure was obtained without either a donor or an acceptor ATP molecule bound, comparison of the active site of OAS with that of PAP or CCA led to the proposal that the donor ATP binds at the interface between the two lobes (Fig. 4b) [87, 88]. It is as yet unclear how the acceptor ATP and dsRNA bind, and how dsRNA binding stimulates the catalytic activity of OAS. Based on the location of the positively charged groove on the OAS surface, it was proposed that dsRNA binds across the N and C terminal domains on the opposite side of the ATP binding surface (Fig. 4b) [87]. Interestingly, OAS can bind to multiple types of nucleic acids, including non-activating ssRNA with little or no secondary structure [89]. Based on these data, a two-step activation model has been proposed where OAS non-specifically binds to nucleic acids through electrostatic interactions, but only upon dsRNA binding can it undergo a conformational change, possibly involving a rotation of the N- and C-terminal halves, to form a functional active site [79].
Is the dsRNA structure sufficient to activate OAS? Binding of OAS to dsRNA requires a minimum length of ~18–20 bp dsRNA, but a higher enzymatic activity was observed when stimulated with longer dsRNA in certain reaction conditions [90], suggesting the possibility of oligomer formation [91]. Unlike sequence-independent recognition of perfect dsRNA by dsRBD, activation of OAS requires two consensus sequence motifs on dsRNA that are separated by one full turn of the A-helix (Fig. 4c) [92]. In addition, the OAS activity is significantly affected by base modifications, such as pseudouridine and 2′-O methylation [23, 92], which are the two most common modifications in cellular RNAs [93]. It has been proposed that OAS binds to one face of dsRNA forming a direct contact with two consecutive minor grooves [92], much like dsRBDs of PKR and ADAR. This model explains the separation of the two sequence motifs necessary for OAS activation, and the sensitivity of OAS to 2′-O methylation. However, this model does not explain how OAS detects pseudouridine modification, which affects the major groove. Adding to this complexity are the findings that ssRNA aptamers with little secondary structure and cellular as well as viral mRNAs can efficiently activate OAS [89, 94, 95]. Comprehensive understanding of the molecular mechanism by which OAS recognizes diverse, dissimilar RNAs (Fig. 4c) to regulate its catalytic function awaits structures of OAS in complex with agonist dsRNA and ssRNA.
Toll-like receptor 3 (TLR3)
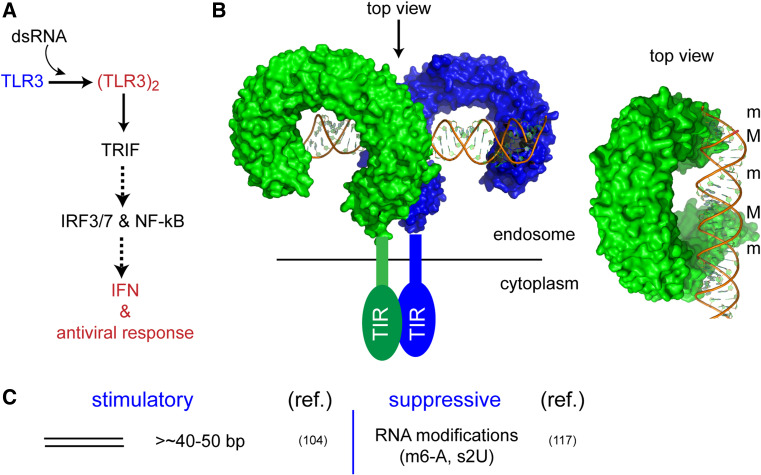
Members of the Toll-like receptor (TLR) family are type I integral membrane receptors that recognize various pathogen-associated molecular patterns (PAMP) originated from viruses, fungi, bacteria, and protozoa, and activate appropriate innate immune responses [96, 97]. So far, 15 subfamilies of TLRs have been identified in vertebrates [98]. They share a similar domain structure, which consists of a ligand-binding ectodomain containing 19–25 tandem copies of leucine-rich repeats (LRRs). The ectodomain is linked by a single transmembrane helix to an intracellular Toll-like/interleukin-1 (IL-1) receptor (TIR) domain that is involved in activation of the cellular signaling pathways [98]. Each TLR is specialized in recognition of distinct PAMPs among which TLR3, 7–9 recognize foreign nucleic acids [97]. TLR7 and TLR8 recognize virus-derived ssRNA [99–101], while TLR9 recognizes microbial non-methylated CpG-containing DNA [102]. TLR3 is the only TLR that recognizes virus-derived dsRNA and its synthetic analogue, polyriboinosinic:polyribocytidylic acid (polyI:C)[103]. Interestingly, these nucleic acid-sensitive TLRs are primarily localized in endosomal compartments, whereas other TLRs are on the cell surface.
Binding of dsRNA by TLR3 occurs via cooperative dimerization of the ectodomain, which triggers dimerization of TIR across the endosomal membrane [104, 105]. Dimerized TIR then recruits TIR-containing adapter-inducing interferon-β (TRIF), which in turn activates antiviral signaling pathways (Fig. 5a) [106]. Forced dimerization of TLR3 ectodomain via α-TLR3 polyclonal antibodies is sufficient to activate signaling, whereas blocking dimerization via mutations of the dimer interface abrogates signaling, suggesting that dimerization is the key mechanism for dsRNA-dependent signal activation [104, 105].
Fig. 5.
a Schematic of dsRNA recognition and antiviral signal activation by TLR3. b Structure of TLR3 bound to dsRNA (PDB: 3CIY [109]) with a schematic depiction of the cytoplasmic TIR domain across the endosomal membrane. The minor and major grooves are indicated by m and M, respectively. c Summary of TLR3-stimulatory and suppressive features of RNA
The crystal structure of the TLR3 ectodomain resembles a long solenoid bent into the shape of a horseshoe, with each turn of the solenoid corresponding to a single LRR sequence from a total of 23 leucine-rich repeats, an architecture that is shared in all the structures of TLRs reported to date (Fig. 5b) [107, 108]. The structure of TLR3 in complex with dsRNA revealed little conformational change in either protein or RNA upon complex formation (Fig. 5b) [109]. TLR3 interacts with dsRNA largely through the minor groove and the nearby phosphate backbone. Consistent with prior biochemical data, the structure showed that dsRNA binding induces dimerization of the ectodomain, where the dimer symmetry coincides with the twofold symmetry of dsRNA (Fig. 5b) [109]. Dimerization involves direct protein–protein contacts between the C-terminal regions, which likely play an important role in dimerization of the cytoplasmic TIR domain (Fig. 5b). Each TLR3 molecule interacts with dsRNA via two distinct surface patches, one at the N-terminus and the other close to the C-terminus of the ectodomain. Interestingly, both patches of dsRNA-binding regions contain few basic residues such as arginine or lysine, but instead are composed of several indispensable histidine residues. Protonation of the histidine imidazole groups under acidic conditions, as expected in the endosome, would allow ionic interactions between TLR3 and the negatively charged phosphate backbone of dsRNA, which accounts for the requirement of low pH for high affinity interaction with dsRNA [105].
Three mechanisms have been proposed to be responsible for specific recognition of viral dsRNAs against cellular dsRNAs by TLR3. First, the endosomal location of the TLR3 ectodomain restricts access of cellular RNAs to TLR3 or other endosomal TLRs [110]. The exact mechanism by which viral dsRNA gains access to the endosomal space is not clear, but possibly through phagocytosis of released dsRNAs from virally infected or dying cells [97, 111]. According to this mechanism, recognition of viral RNA through TLR3 does not require viral infection of the cells expressing TLR3, and thus referred to as “extrinsic sensing” mechanism as opposed to “intrinsic sensing” by cytoplasmic receptors, such as RIG-I and MDA5 [4]. This extrinsic sensing, however, implies that cellular dsRNAs from dying cells must avoid recognition by TLR3 through alternative mechanisms [112]. Degradation of cellular nucleic acids by cellular or extracellular nucleases during apoptosis or post cell death appears to play an important role in preventing aberrant activation of TLR7 and 9, and protects the host from developing autoimmunity [97, 110]. Similar mechanisms may also apply to TLR3, but whether and how cellular dsRNA is selectively degraded in comparison to viral dsRNA remains unclear [113].
In addition to endosomal access, TLR3 also requires dsRNA to be longer than ~40 bp for robust stimulation [105]. This length requirement would help avoid inappropriate recognition of cellular ssRNAs with short hairpin structures or mature siRNAs or miRNAs. The crystal structure of the TLR3:dsRNA complex provides a mechanistic explanation for the length restriction, as a dimeric TLR3 spans ~40 bp dsRNA, with each monomer occupying ~20 bp [109]. Longer dsRNAs of ~100 bp were shown to stimulate TLR3 more robustly, possibly suggesting a weak lateral association between TLR3 dimers along dsRNA [105]. In support of lateral clustering of TLR3, neutralizing Fab fragments, which bind to the TLR3 ectodomain in a manner that could disrupt its lateral clustering, were shown to inhibit the signaling activity of TLR3 without disrupting its dsRNA binding or dimerization activity [114]. In an apparent contradiction to the importance of dimerization or oligomerization, recent studies showed that exogenously introduced 21-bp siRNA can also stimulate TLR3 [115, 116], suggesting that low-affinity interaction with short dsRNA can be compensated for by high dose of RNA. It is possible that TLR3 can still dimerize on 21-bp dsRNA, albeit inefficiently, in the same manner as on 40-bp dsRNA or through an alternative binding mode [117]. These observations suggest that dsRNA length is not an absolute criterion used by TLR3 for self and non-self discrimination, but rather a relative condition that is dependent on and can be scaled by the abundance of RNA and receptors in the cell.
Finally, dsRNA recognition by TLR3 is suppressed by the presence of modified nucleotides in RNA [118]. Modified nucleotides such as N 6-methyladenosine and 2-thiouridine ablate the interferon signaling activity of TLR3, whereas pseudouridine and 5-methyluridine have more minor effects on TLR3. Interestingly, in vitro transcribed or mitochondrial RNAs, but not cytoplasmic RNAs from mammalian cell extracts, can activate the innate immune response in dendritic cells [112, 118]. Considering that mitochondrial RNAs contain a low level of modified nucleotides in comparison to cytoplasmic cellular RNAs [93], these observations suggest that nucleotide modification provides an additional physicochemical specificity for TLR3 to efficiently discriminate between self and non-self dsRNAs (Fig. 5c).
Retinoic acid-inducible gene-I (RIG-I)
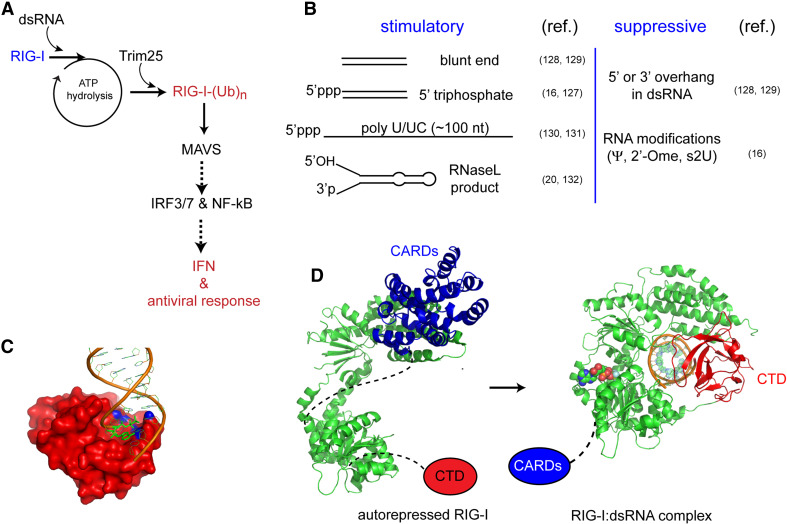
RIG-I-like receptors, which include RIG-I, MDA5, and LGP2, represent another antiviral PRR pathway parallel to that of TLRs 3 and 7–9. While nucleic acid-specific TLRs are functional in the endosome, RIG-I-like receptors are located in the cytoplasm and directly sense viral RNAs in the infected cell (“intrinsic sensing”) [119, 120]. RIG-I and MDA5 share a common domain architecture consisting of two tandem caspase activation recruitment domains (CARDs), which interact with the downstream signaling adaptor, mitochondrial antiviral-signaling protein (MAVS); a central DExD/H motif helicase domain responsible for RNA-dependent ATP hydrolysis; and a C-terminal domain (CTD) that binds to dsRNA [120–123]. LGP2 also has a similar domain architecture to RIG-I and MDA5, but lacks the CARD domain [120]. Accordingly, LGP2 does not possess an immune signaling activity by itself, but is thought to up- and down-regulate the signaling activities of MDA5 and RIG-I, respectively [124, 125]. Exactly how RIG-I and MDA5 relay antiviral signals to MAVS is currently poorly understood, but several recent studies collectively propose the following series of events during RIG-I signaling. Upon viral RNA binding, RIG-I hydrolyzes ATP and the second CARD domain becomes covalently conjugated with K63-linked polyubiquitin by Trim25 [126]. The ubiquitinated RIG-I CARD domain then self-oligomerizes, interacts with CARD of MAVS, and triggers formation of filamentous oligomers of MAVS CARD on the mitochondrial surface [127, 128]. This oligomeric form of MAVS CARD then recruits downstream signaling molecules such as TRAF2 and 3, which in turn activate IRF3/7 or NF-κB signaling pathways in the interferon antiviral response (Fig. 6a) [128].
Fig. 6.
a Schematic of dsRNA recognition and antiviral signal activation by RIG-I. b Summary of RIG-I-stimulatory and suppressive features of RNA. c Structure of RIG-I CTD in complex with dsRNA containing the 5′ triphosphate group (5′ppp) and blunt end (PDB: 3LRR [137]). The nucleotide at the 5′ end (green) is bound by positively charged residues (blue) in the 5′ppp binding pocket. d Structure of RIG-I before and after dsRNA binding (PDB: 4A2W [141] and 3TMI [139], respectively). Dotted lines and ovals indicate flexible linkers and disordered domains, respectively, which are not represented in the crystal structure
The RNA selectivity of RIG-I appears to be complex and has been much debated over the last several years (Fig. 6b). It was first identified as a receptor stimulated by a dsRNA mimic, polyI:C, and thus thought to recognize simple dsRNA structure [119]. Later studies revealed, however, that the 5′ triphosphate group and blunt end of RNA are important for viral recognition of short (~20–25 bp) dsRNA by RIG-I [18, 129–131]. RIG-I was also reported to recognize long (>100 nt) ssRNA with a 5′ triphosphate group, such as the polyU/UC region of the HCV genomic RNA, in a sequence- and length-dependent manner [132, 133]. In addition, RNA cleavage products produced by RNase L, which contain the 5′ hydroxyl and 3′ monophosphate group, can also activate RIG-I [22, 134]. As with PKR, OAS, and TLR3, modified nucleotides (pseudouridine, 2-thio-uridine and 2′-O-methyl-uridine) suppress RIG-I stimulation by RNA [133].
More detailed biochemical and biophysical studies revealed that the CTD, which displays little similarity to any previously characterized RNA binding proteins, is responsible for recognition of the 5′ triphosphate group and blunt end of dsRNA [135, 136]. Structures of the CTD bound to dsRNA with the 5′ triphosphate group and blunt end revealed that a conserved, essential phenylalanine in the CTD forms a face-to-face contact with the blunt-end bases [137, 138]. The 5′ triphosphate group forms electrostatic interactions with a cluster of lysine residues (Fig. 6c). The combination of pi-stacking and electrostatic interactions provides an explanation for the observed preference of RIG-I for dsRNA ends. Recently, three groups have independently determined crystal structures of isolated helicase domain or helicase-CTD of RIG-I in complex with blunt-ended dsRNA [139–141]. The interaction between the CTD and dsRNA is similarly preserved in the helicase-CTD–dsRNA structure, but helicase wraps around dsRNA, forming additional contacts with the RNA phosphate backbone (Fig. 6d). The most striking feature of these structures was a long, previously unrecognized “pincer” domain, which connects between the helicase subdomains (helicase 1, helicase 2, helicase 2i), possibly coordinating RNA binding, ATP hydrolysis, and a conformational change for signaling.
Despite the advances in structural and biochemical understanding of RIG-I, several issues remain unresolved in RNA detection and signaling mechanism. First, it is as yet unclear how RIG-I CARD transmits signals to MAVS. Between CARD and the helicase domain is a ~50-amino-acid linker with no predicted secondary or tertiary structure. The structure of full-length RIG-I showed that CARD interacts tightly with the helicase domain in the autorepressed state [141]. Small-angle X-ray scattering (SAXS) analysis suggests that upon binding to dsRNA, CARD is dissociated from the helicase domain and is placed near the pincer domain [139], but whether this conformation represents the “active”, signaling-competent conformation and how it is affected by K63-linked polyubiquitination requires future investigation. Second, RIG-I was proposed to form a higher-order oligomer upon viral infection, as judged by native gel analyses and atomic force microscopy [125, 142], but the crystal structures and other biochemical analyses provide little evidence for such oligomerization, and instead indicate that RIG-I functions as a monomer [139–141]. Earlier studies suggest dimerization of RIG-I upon dsRNA binding [136], but this is likely due to RIG-I binding to two ends of dsRNA, rather than via direct protein–protein interactions. Thirdly, it is still unclear how ATP hydrolysis regulates RIG-I conformation or its signaling activity. Mutations of the active site abrogated the signaling activity of RIG-I without altering its RNA binding activity [143], which led to the proposal that ATP hydrolysis is a conformational “switch” to convert the autorepressed conformation to the signaling competent form. In support of this proposal, the ATP hydrolysis activity in vitro has been shown to correlate with the interferon stimulatory activity in the cell [130]. If ATP hydrolysis indeed serves as a conformational switch, then one could, in principle, be able to lock the conformation of RIG-I into the “active” state using a certain ATP analog. On the other hand, it is possible that dynamic, repetitive ATP hydrolysis is important for signaling, possibly through a mechanism involving the observed activity of RIG-I to translocate along dsRNA [144]. Fourth, while the structures provide a good explanation for the recognition of the 5′ triphosphate group and blunt end of dsRNA, it is unclear how RIG-I can also recognize other features of RNA, such as poly-U/UC sequence in ssRNA and RNase-L-degradation products. As with other dsRNA receptors discussed above, understanding the molecular mechanisms for the diverse RNA selectivity of RIG-I would require additional structural and biochemical analyses in the future.
Melanoma differentiation-associated gene 5 (MDA5)
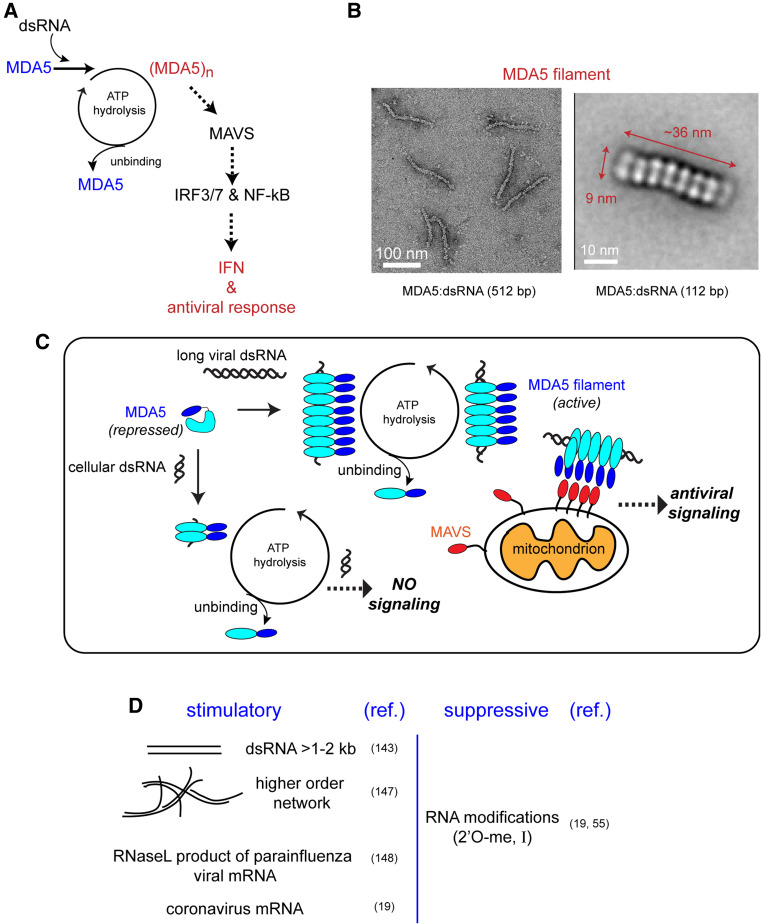
Our understanding of MDA5 lags significantly behind that of RIG-I, despite the conserved domain architecture and the shared signaling adaptor, MAVS (Fig. 7a). The molecular features of RNA recognized by MDA5 have long remained enigmatic, but a pioneering work by Kato et al. [145] revealed that dsRNA length is the major determinant that allows MDA5 to distinguish between cellular and viral dsRNAs. While RIG-I prefers short dsRNAs, MDA5-mediated signaling positively correlates with the length of dsRNA in the range of ~1–7 kb [145]. The length discrimination at this scale distinguishes MDA5 from that of other dsRNA sensors such as PKR, OAS, and TLR3.
Fig. 7.
a Schematic of RNA recognition and antiviral signal activation by MDA5. b Electron micrograph and 2D-averaged image of the MDA5 filament formed on 512 and 112 bp dsRNA, respectively [146]. c Proposed model of dsRNA length-dependent signaling by MDA5. Filaments formed on short dsRNA disassemble rapidly during ATP hydrolysis, while filaments on longer dsRNA can undergo continuous cycles of filament assembly and disassembly, during which it activates the downstream antiviral signaling pathway through MAVS. d Summary of MDA5-stimulatory and suppressive features of RNA
We have recently shown that human MDA5 forms filamentous oligomers, with the appearance of stacked rings along the length of the dsRNA duplex (Fig. 7b). Each ring in the MDA5 filament is reminiscent of that of a single RIG-I monomer bound to dsRNA [146]. A similar filamentous structure was observed with mouse MDA5, suggesting that this oligomerization property is conserved in MDA5 [147]. The MDA5 filament is distinct from a “beads-on-a-string” type of oligomers as evidenced by the high cooperativity in dsRNA binding and its high affinity for long dsRNA far beyond the linear combination of monomer interactions [146]. MDA5 filament formation was shown to correlate with ATP hydrolysis and signaling potential, also suggesting its functional importance [146]. For example, MDA5 binds to various types of nucleic acids with comparable affinities, but only dsRNA binding triggers filament formation and ATP hydrolysis in vitro [146] and signaling in vivo [145]. No ATP hydrolysis was observed with the genomic ssRNA from EMCV, which contains a complex secondary structure in the internal ribosome entry site (IRES) located within the 5′ UTR [146]. This result is consistent with the observation that viral replication is required for MDA5 activation by murine norovirus [148], and suggests that MDA5 can discriminate between a perfect and an imperfect duplex containing bulges and mismatches.
In seeming contradiction to the positive role of ATP in antiviral signaling, ATP hydrolysis by MDA5 triggers its dissociation from dsRNA and consequent filament disassembly [146, 147]. Although a single MDA5 monomer bound to dsRNA is sufficient to hydrolyze ATP and there is no coordination of ATP hydrolysis between neighboring molecules within a filament, ATP-driven filament disassembly does occur in a manner dependent upon neighboring MDA5 molecules [146]. Incorporation of catalytically inactive mutants within a filament stabilizes dsRNA bound wild-type MDA5 without diminishing its ATP hydrolysis activity [146]. In addition, MDA5 dissociation is inversely proportional to the length of dsRNA [146], suggesting some ordered disassembly mechanism such as sequential dissociation of MDA5 from filament ends. Examination of filament disassembly intermediates by electron microscopy showed apparent internal breaks upon ATP hydrolysis [147]. This could be interpreted as disassembly of MDA5 in small fragments from internal regions of a filament. Another possibility is that MDA5 does not form a single continuous filament on dsRNA, and these apparent internal breaks represent boundaries between independent filaments (propagated from independent nuclei), each of which undergoes a separate end-disassembly process. More detailed biophysical and biochemical analyses of the filament assembly and disassembly processes are required to understand the complexities of filament dynamics.
The dynamic instability of the MDA5 filament and thus the transient nature of the interaction between MDA5 and dsRNA is intriguing in comparison to the conventional view of a stable receptor–ligand interaction, and raises several questions as to the RNA recognition and signaling mechanisms. First, how does MDA5 utilize its dynamic assembly and disassembly processes to measure the length of dsRNA? Although an answer to this question still awaits a comprehensive understanding of the filament dynamics, an insight can be obtained from the finding that MDA5 dissociates from dsRNA at a rate inversely proportional to the length of dsRNA [146]. It is plausible to speculate that ATP hydrolysis mediated instability of the filament serves as a mechanism to discriminate against short dsRNA, whereas longer dsRNA gains a competitive advantage from the delayed dissociation (Fig. 7c). The dynamic nature of the MDA5 filament also raises a question as to how MDA5 interacts with MAVS during the repetitive cycle of filament assembly and disassembly. Since not every ATP hydrolysis event triggers dissociation of MDA5 from dsRNA, it is tempting to speculate that signal activation is largely mediated by MDA5 molecules that are able to hydrolyze ATP, while remaining bound to dsRNA. Testing this hypothesis would require in vitro reconstitution of the MDA5:MAVS signaling complex using purified proteins.
Are dsRNA structure and length the only features recognized by MDA5? Examination of the immunoreactivity of gel-fractionated total RNAs extracted from virally infected cells showed that viral RNA species resistant to electrophoresis are potent stimulators of MDA5-mediated interferon signaling [149]. It was proposed that these RNA species contain a network of ssRNA and dsRNA regions, as would be expected for viral replicative intermediates, which led to a proposal that higher order RNA structure, instead of perfect duplex, stimulates MDA5 [149]. However, the precise identity and structure of the stimulatory RNA from this study remain to be further investigated. Is MDA5 recognition dependent on RNA sequence and modification? Biochemical analyses of RNA binding [146] and dsRNA-dependent ATP hydrolysis activity (unpublished result) revealed sequence independent recognition of dsRNA by MDA5. However, a recent study showed that MDA5 recognizes a specific, ~430-nucleotide region of a parainfluenza viral mRNA in an RNase-L-dependent manner [150]. MDA5 was also reported to be sensitive to certain types of RNA modifications. In a recent study of Coronavirus, 2′-O methylation of the 5′ penultimate nucleotide of the viral mRNAs, a modification present in most cellular mRNAs, prevented activation of MDA5-mediated interferon signaling [21]. However, unlike PKR and RIG-I, MDA5 is insensitive to the 5′ functional group of dsRNA [151]. It would be interesting to test how other RNA modifications, such as pseudouridine and N 6-methyladenosine, affect the RNA binding and signaling activity of MDA5. All together, these studies suggest a more complex picture of MDA5 RNA specificity than a simple recognition of dsRNA length and structure (Fig. 7c). Future studies are necessary to determine the precise identities of the stimulatory RNAs, and dissect the importance of sequence, secondary structure, and position and type of nucleotide modification for MDA5 activation.
DHX9 and DDX1
Since the discovery of RLRs, several other helicases have been identified that are involved in viral dsRNA sensing in the cytoplasm. These helicases include DHX9 (a.k.a. RNA helicase A) and DDX1, which were proposed to recognize dsRNA (and also dsDNA for DHX9) in a manner independent of RIG-I and MDA5 [152–154]. DHX9 was shown to utilize two distinct domains to bind to dsRNA and DNA, and bifurcate the downstream signal via MAVS and Myd88, respectively [152, 153]. DDX1 was shown to bind to dsRNA mimic, polyI:C, and form a complex with two other helicases, DDX21 and DHX36, which interact with TRIF and activate the antiviral response [154]. Interestingly, both DHX9 and DDX1 were previously implicated in diverse cellular functions other than viral nucleic acid detection. For example, DHX9 was proposed to be involved in gene regulation of cellular RNAs through remodeling of ribonucleoprotein particles during translation [155] and DDX1 was shown to up-regulate NF-κB-mediated transcriptional activity [156]. Exactly how they coordinate these multiple divergent biological functions and whether they recognize any additional features of RNA beyond a simple duplex structure remain to be investigated.
Conclusions
The distinct backbone arrangement of dsRNA, in comparison to dsDNA or ssRNA, has supported a conventional model that dsRNA structure, much like lipopolysaccharides and other bacterial-specific chemical structures, is recognized by innate immune receptors as a unique molecular signature of viruses. However, recent studies of the human transcriptome have revealed a prevalence and diversity of non-coding RNAs, many of which contain a range of secondary structures, varying from an imperfect duplex of ~21 bp in miRNAs to a near-perfect duplex of ~250–300 bp in inverted repeat elements. In parallel, the innate immune sensors initially thought to recognize the dsRNA structure are now known to detect and regulate its antiviral activity in a manner dependent on other, and often multiple, features of RNA, such as nucleotide modification, 5′ functional groups, bulges, mismatches, and sequences. While the list of these additional immune-stimulatory and suppressive RNA features is expanding, our understanding of the versatile selectivity of the receptors and effectors at the level of molecular structure, thermodynamics and kinetics remains rudimentary. Future structural and biochemical studies would help us understand the underlying molecular principle, and perhaps identify combinatorial rules that could be used in therapeutic RNAs for either immune suppression or activation [157, 158].
Abbreviations
- ADAR
Adenosine deaminase acting on RNA
- CARD
Caspase activation and recruitment domain
- DsRNA
Double-stranded RNA
- SsRNA
Single-stranded RNA
- DsRBD
DsRNA binding domain
- eIF2
Eukaryotic initiation factor 2
- IFN
Interferon
- ISG
Interferon-stimulated gene
- LGP2
Laboratory of genetics and physiology-2
- MAVS
Mitochondrial antiviral signaling protein
- MDA5
Melanoma differentiation-associated gene 5
- MiRN
Micro RNA
- OAS
Oligoadenylate synthetase
- polyI:C
Polyriboinosinic:polyribocytidylic acid
- PKR
Protein kinase RNA-activated
- RIG-I
Retinoic acid inducible gene-1
- RNase-L
Ribonuclease L
- RLR
RIG-I-like receptor
- SAXS
Small-angle X-ray scattering
- SiRNA
Small interfering RNA
- TIR
Toll/interleukin-1 receptor
- TLR
Toll-like receptor
- TRIF
TIR-containing adaptor inducing interferon-β
- UTR
Untranslated region
References
- 1.Sadler AJ, Williams BRG. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pichlmair A. Innate recognition of viruses. Immunity. 2007;27:370–383. doi: 10.1016/j.immuni.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 3.Borden EC, Williams BR. Interferon-stimulated genes and their protein products: what and how? J Interferon Cytokine Res. 2011;31:1–4. doi: 10.1089/jir.2010.0129. [DOI] [PubMed] [Google Scholar]
- 4.Iwasaki A, Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science. 2010;327:291–295. doi: 10.1126/science.1183021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saenger W (1984) Principles of nucleic acid structure. Springer, New York, p 113
- 7.Doyle M, Jantsch MF. New and old roles of the double-stranded RNA-binding domain. J Struct Biol. 2003;140:147–153. doi: 10.1016/s1047-8477(02)00544-0. [DOI] [PubMed] [Google Scholar]
- 8.Saunders LR, Barber GN (2003) The dsRNA binding protein family: critical roles, diverse cellular functions. FASEB 961–983 [DOI] [PubMed]
- 9.Pabo CO, Sauer RT. Protein-DNA recognition. Ann Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- 10.Watanabe T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 11.Chiang HR, et al. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010;24:992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2(12):e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Leung FCC. A study on genomic distribution and sequence features of human long inverted repeats reveals species-specific intronic inverted repeats. FEBS J. 2009;276:1986–1998. doi: 10.1111/j.1742-4658.2009.06930.x. [DOI] [PubMed] [Google Scholar]
- 14.Yelin R, et al. Widespread occurrence of antisense transcription in the human genome. Nat Biotechol. 2003;21:379–386. doi: 10.1038/nbt808. [DOI] [PubMed] [Google Scholar]
- 15.Rosok O, Sioud M. Systematic identification of sense-antisense transcripts in mammalian cells. Nat Biotechnol. 2004;22:104–108. doi: 10.1038/nbt925. [DOI] [PubMed] [Google Scholar]
- 16.McKenna SA, et al. Viral dsRNA inhibitors prevent self-association and autophosphorylation of PKR. J Mol Biol. 2007;372:103–113. doi: 10.1016/j.jmb.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vitali P, Scadden ADJ. Double-stranded RNAs containing multiple IU pairs are sufficient to suppress interferon induction and apoptosis. Nat Struct Mol Biol. 2010;17(9):1043–1050. doi: 10.1038/nsmb.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hornung V, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 19.Nallagatla SR, Bevilacqua PC. Nucleoside modifications modulate activation of the protein kinase PKR in an RNA structure-specific manner. RNA. 2008;14:1201–1203. doi: 10.1261/rna.1007408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nallagatla SR, et al. 5′-Triphosphate-dependent activation of PKR by RNAs with short stem-loops. Science. 2007;318:1455–1458. doi: 10.1126/science.1147347. [DOI] [PubMed] [Google Scholar]
- 21.Zust R, et al. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol. 2011;12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malathi K, et al. RNase L releases a small RNA from HCV RNA that refolds into a potent PAMP. RNA. 2010;16:2108–2119. doi: 10.1261/rna.2244210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson BR, et al. Nucleoside modifications in RNA limit activation of 2′-5′-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res. 2011;39:9329–9338. doi: 10.1093/nar/gkr586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia MA, et al. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol Mol Biol Rev. 2006;70:1032–1060. doi: 10.1128/MMBR.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lemaire PA, Cole JL. Mechanism of PKR activation by dsRNA. J Mol Biol. 2008;381:351–360. doi: 10.1016/j.jmb.2008.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfeller CK, et al. Protein kinase PKR and RNA adenosine deaminase ADAR1: new roles for old players as modulators of the interferon response. Curr Opin Immunol. 2011;23:573–582. doi: 10.1016/j.coi.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKenna SA, et al. Biophysical and biochemical investigations of dsRNA-activated kinase PKR. Methods Enzym. 2007;430:373–396. doi: 10.1016/S0076-6879(07)30014-1. [DOI] [PubMed] [Google Scholar]
- 28.Cole JL. Analysis of PKR activation using analytical ultracentrifugation. Macromol Biosci. 2010;10:703–713. doi: 10.1002/mabi.201000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lemaire PA, Lary J, Cole JL. Mechanism of PKR activation: dimerization and kinase activation in the absence of double-stranded RNA. J Mol Biol. 2005;345:81–90. doi: 10.1016/j.jmb.2004.10.031. [DOI] [PubMed] [Google Scholar]
- 30.Kim I, Liu CW, Puglisi JD. Specific recognition of HIV TAR RNA by the dsRNA binding domains (dsRBD1–dsRBD2) of PKR. J Mol Biol. 2006;358:430–442. doi: 10.1016/j.jmb.2006.01.099. [DOI] [PubMed] [Google Scholar]
- 31.Bevilacqua PC, Cech TR. Minor-groove recognition of double-stranded RNA by the double-stranded RNA-binding domain from the RNA-activated protein kinase PKR. Biochemistry. 1996;35:9983–9994. doi: 10.1021/bi9607259. [DOI] [PubMed] [Google Scholar]
- 32.Manche L, et al. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol Cell Biol. 1992;12:5238–5248. doi: 10.1128/mcb.12.11.5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryter JM, Schultz SC. Molecular basis of double-stranded RNA-protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 1998;17:7505–7513. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gan J, et al. Structural insight into the mechanism of double-stranded RNA processing by ribonuclease III. Cell. 2006;124:355–366. doi: 10.1016/j.cell.2005.11.034. [DOI] [PubMed] [Google Scholar]
- 35.Cohen-Chalamish S, et al. Dynamic refolding of IFN-gamma mRNA enables it to function as PKR activator and translation template. Nat Chem Biol. 2009;5:896–903. doi: 10.1038/nchembio.234. [DOI] [PubMed] [Google Scholar]
- 36.Zheng X, Bevilacqua PC. Activation of the protein kinase PKR by short double-stranded RNAs with single-stranded tails. RNA. 2004;10:1934–1945. doi: 10.1261/rna.7150804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hundley HA, Bass BL. ADAR editing in double-stranded UTRs and other noncoding RNA sequences. Trends Biochem Sci. 2010;35:377–383. doi: 10.1016/j.tibs.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keegan LP, et al. Adenosine deaminases acting on RNA (ADARs): RNA-editing enzymes. Genome Biol. 2004;5:209. doi: 10.1186/gb-2004-5-2-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen CX, et al. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA. 2000;6:755–767. doi: 10.1017/s1355838200000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wong SK, Sato S, Lazinski DW. Substrate recognition by ADAR1 and ADAR2. RNA. 2001;7:846–858. doi: 10.1017/s135583820101007x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lehmann KA, Bass BL. Double-stranded RNA adenosine deaminases ADAR1 and ADAR2 have overlapping specificities. Biochemistry. 2000;39:12875–12884. doi: 10.1021/bi001383g. [DOI] [PubMed] [Google Scholar]
- 42.Wahlstedt H, Ohman M. Site-selective versus promiscuous A-to-I editing. Wiley Interdiscip Rev RNA. 2011;2:761–771. doi: 10.1002/wrna.89. [DOI] [PubMed] [Google Scholar]
- 43.Polson AG, Bass BL. Preferential selection of adenosines for modification by double-stranded RNA adenosine deaminase. EMBO J. 1994;13:5701–5711. doi: 10.1002/j.1460-2075.1994.tb06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lehmann KA, Bass BL. The importance of internal loops within RNA substrates of ADAR1. J Mol Biol. 1999;291:1–13. doi: 10.1006/jmbi.1999.2914. [DOI] [PubMed] [Google Scholar]
- 45.Dawson TR, Sansam CL, Emeson RB. Structure and sequence determinants required for the RNA editing of ADAR2 substrates. J Biol Chem. 2004;279:4941–4951. doi: 10.1074/jbc.M310068200. [DOI] [PubMed] [Google Scholar]
- 46.Higuchi M, et al. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell. 1993;75:1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- 47.Burns CM, et al. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 48.Bass BL, Weintraub H. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell. 1988;55:1089–1098. doi: 10.1016/0092-8674(88)90253-x. [DOI] [PubMed] [Google Scholar]
- 49.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Ann Rev Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Samuel CE. Adenosine deaminases acting on RNA (ADARs) are both antiviral and proviral. Virology. 2011;411:180–193. doi: 10.1016/j.virol.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ward SV, et al. RNA editing enzyme adenosine deaminase is a restriction factor for controlling measles virus replication that also is required for embryogenesis. Proc Natl Acad Sci USA. 2011;108:331–336. doi: 10.1073/pnas.1017241108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cattaneo R, et al. Biased hypermutation and other genetic changes in defective measles viruses in human brain infections. Cell. 1988;55:255–265. doi: 10.1016/0092-8674(88)90048-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nie Y, Hammond GL, Yang JH. Double-stranded RNA deaminase ADAR1 increases host susceptibility to virus infection. J Virol. 2007;81:917–923. doi: 10.1128/JVI.01527-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Z, Wolff KC, Samuel CE. RNA adenosine deaminase ADAR1 deficiency leads to increased activation of protein kinase PKR and reduced vesicular stomatitis virus growth following interferon treatment. Virology. 2010;396:316–322. doi: 10.1016/j.virol.2009.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Casey JL. RNA editing in hepatitis delta virus. Curr Top Microbiol Immunol. 2006;307:67–89. doi: 10.1007/3-540-29802-9_4. [DOI] [PubMed] [Google Scholar]
- 56.Wong SK, Lazinski DW. Replicating hepatitis delta virus RNA is edited in the nucleus by the small form of ADAR1. Proc Natl Acad Sci USA. 2002;99:15118–15123. doi: 10.1073/pnas.232416799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sato S, Cornillez-Ty C, Lazinski DW. By inhibiting replication, the large hepatitis delta antigen can indirectly regulate amber/W editing and its own expression. J Virol. 2004;78:8120–8134. doi: 10.1128/JVI.78.15.8120-8134.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peng Z, et al. Comprehensive analysis of RNA-seq data reveals extensive RNA editing in a human transcriptome. Nat Biotechol. 2012;30:253–260. doi: 10.1038/nbt.2122. [DOI] [PubMed] [Google Scholar]
- 59.Paz-Yaacov N, et al. Adenosine-to-inosine RNA editing shapes transcriptome diversity in primates. Proc Natl Acad Sci USA. 2010;107:12174–12179. doi: 10.1073/pnas.1006183107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alon S et al (2012) Systematic identification of edited microRNAs in the human brain. Genome Res [Epub ahead] [DOI] [PMC free article] [PubMed]
- 61.Carmi S, Borukhov I, Levanon EY. Identification of widespread ultra-edited human RNAs. PLoS Genet. 2011;7:e1002317. doi: 10.1371/journal.pgen.1002317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heale BS, Keegan LP, O’Connell MA. The effect of RNA editing and ADARs on miRNA biogenesis and function. Adv Exp Med Biol. 2010;700:76–84. [PubMed] [Google Scholar]
- 63.Jepson JEC, Reenan RA. RNA editing in regulating gene expression in the brain. Biochim Biophys Acta. 2008;1779:459–470. doi: 10.1016/j.bbagrm.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 64.Chen LL, Carmichael GG. Nuclear editing of mRNA 3′-UTR. Curr Top Microbiol Immunol. 2012;353:111–121. doi: 10.1007/82_2011_149. [DOI] [PubMed] [Google Scholar]
- 65.Wang Q, Carmichael GG. Effects of length and location on the cellular response to double-stranded RNA. Microbiol Mol Biol Rev. 2004;68(3):432–452. doi: 10.1128/MMBR.68.3.432-452.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hartner JC, et al. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat Immunol. 2008;10:109–115. doi: 10.1038/ni.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Q, et al. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science. 2000;290:1765–1768. doi: 10.1126/science.290.5497.1765. [DOI] [PubMed] [Google Scholar]
- 68.Hartner JC, et al. J Biol Chem. 2004;279:4894–4902. doi: 10.1074/jbc.M311347200. [DOI] [PubMed] [Google Scholar]
- 69.Wang Q, et al. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase. J Biol Chem. 2004;279:4952–4961. doi: 10.1074/jbc.M310162200. [DOI] [PubMed] [Google Scholar]
- 70.Higuchi M, et al. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 71.Palladino MJ, et al. A-to-I pre-mRNA editing in Drosophila is primarily involved in adult nervous system function and integrity. Cell. 2000;102:437. doi: 10.1016/s0092-8674(00)00049-0. [DOI] [PubMed] [Google Scholar]
- 72.Tonkin LA, et al. RNA editing by ADARs is important for normal behavior in Caenorhabditis elegans . EMBO J. 2002;21:6025–6035. doi: 10.1093/emboj/cdf607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stefl R, et al. The solution structure of the ADAR2 dsRBM-RNA complex reveals a sequence-specific readout of the minor groove. Cell. 2010;143:225–237. doi: 10.1016/j.cell.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ramos A, et al. RNA recognition by a Staufen double-stranded RNA-binding domain. EMBO J. 2000;19:997–1009. doi: 10.1093/emboj/19.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu H, et al. Structural basis for recognition of the AGNN tetraloop RNA fold by the double-stranded RNA binding domain of Rnt1p RNase III. Proc Natl Acad Sci USA. 2004;101:8307–8312. doi: 10.1073/pnas.0402627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Torralba S, Sojat J, Hartmann R. 2′-5′ oligoadenylate synthetase shares active site architecture with the archaeal CCA-adding enzyme. Cell Mol Life Sci. 2008;65:2613–2620. doi: 10.1007/s00018-008-8164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hovanessian AG, et al. Identification of 69-kd and 100-kd forms of 2–5A synthetase in interferon-treated human cells by specific monoclonal antibodies. EMBO J. 1987;6:1273–1280. doi: 10.1002/j.1460-2075.1987.tb02364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rebouillat D, Marie I, Hovanessian AG. Molecular cloning and characterization of two related and interferon-induced 56-kDa and 30-kDa proteins highly similar to 2–5 oligoadenylate synthetase. Eur J Biochem. 1998;257:319–330. doi: 10.1046/j.1432-1327.1998.2570319.x. [DOI] [PubMed] [Google Scholar]
- 79.Kristiansen H, et al. The oligoadenylate synthetase family: an ancient protein family with multiple antiviral activities. J Interferon Cytokine Res. 2011;31:41–47. doi: 10.1089/jir.2010.0107. [DOI] [PubMed] [Google Scholar]
- 80.Wreschner DH, et al. Interferon action—sequence specificity of the ppp(A2′p)nA-dependent ribonuclease. Nature. 1981;278:414–417. doi: 10.1038/289414a0. [DOI] [PubMed] [Google Scholar]
- 81.Floyd-Smith G, Slattery E, Lengyel P. Interferon action: RNA cleavage pattern of a (2′-5′)oligoadenylate-dependent endonuclease. Science. 1981;212:1030–1032. doi: 10.1126/science.6165080. [DOI] [PubMed] [Google Scholar]
- 82.Chakrabarti A, Jha BK, Silverman RH. New insights into the role of RNase-L in innate immunity. J Interferon Cytokine Res. 2011;31:49–57. doi: 10.1089/jir.2010.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kubota K, et al. Identification of 2′-phosphodiesterase, which plays a role in the 2–5A system regulated by interferon. J Biol Chem. 2004;279:37832–37841. doi: 10.1074/jbc.M400089200. [DOI] [PubMed] [Google Scholar]
- 84.Knight M, et al. Radioimmune, radiobinding and HPLC analysis of 2–5A and related oligonucleotides from intact cells. Nature. 1980;288:189–192. doi: 10.1038/288189a0. [DOI] [PubMed] [Google Scholar]
- 85.Rebouillat D, Hovanessian AG. The human 2,5-oligoadenylate synthetase family: interferon-induced proteins with unique enzymatic properties. J Interferon Cytokine Res. 1999;19:295–308. doi: 10.1089/107999099313992. [DOI] [PubMed] [Google Scholar]
- 86.Schoggins JW, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hartmann R, et al. Crystal structure of the 2′-specific and double-stranded RNA-activated interferon-induced antiviral protein 2′-5′-oligoadenylate synthetase. Mol Cell. 2003;12:1173–1185. doi: 10.1016/s1097-2765(03)00433-7. [DOI] [PubMed] [Google Scholar]
- 88.Sarkar SN, et al. The nature of the catalytic domain of 2′-5′-oligoadenylate synthetases. J Biol Chem. 1999;274:25535–25542. doi: 10.1074/jbc.274.36.25535. [DOI] [PubMed] [Google Scholar]
- 89.Hartmann R, et al. Activation of 2′-5′ oligoadenylate synthetase by single-stranded and double-stranded RNA aptamers. J Biol Chem. 1998;273:3236–3246. doi: 10.1074/jbc.273.6.3236. [DOI] [PubMed] [Google Scholar]
- 90.Desai SY, Sen GC. Effects of varying lengths of double-stranded RNA on binding and activation of 2′-5′-oligoadenylate synthetase. J Interferon Cytokine Res. 1997;17:531–536. doi: 10.1089/jir.1997.17.531. [DOI] [PubMed] [Google Scholar]
- 91.Ghosh A, et al. Enzymatic activity of 2′-5′-oligoadenylate synthetase is impaired by specific mutations that affect oligomerization of the protein. J Biol Chem. 1997;272:33220–33226. doi: 10.1074/jbc.272.52.33220. [DOI] [PubMed] [Google Scholar]
- 92.Kodym R, Kodym E, Story MD. 2′-5′-Oligoadenylate synthetase is activated by a specific RNA sequence motif. Biochem Biophy Res Comm. 2009;388:317–322. doi: 10.1016/j.bbrc.2009.07.167. [DOI] [PubMed] [Google Scholar]
- 93.Grosjean H, Benne R (1998) Modification and editing of RNA. American Society for Microbiology, Washington
- 94.Nilsen TW, et al. Heterogeneous nuclear RNA promotes synthesis of (2′,5′)oligoadenylate and is cleaved by the (2′,5′) oligoadenylate-activated endoribonuclease. Mol Cell Biol. 1982;2:154–160. doi: 10.1128/mcb.2.2.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Molinaro RJ, et al. Selection and cloning of poly(rC)-binding protein 2 and Raf kinase inhibitor protein RNA activators of 2′,5′-oligoadenylate synthetase from prostate cancer cells. Nucleic Acids Res. 2006;34:6684–6695. doi: 10.1093/nar/gkl968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kumagai Y, Takeuchi O, Akira S. Pathogen recognition by innate receptors. J Infect Chemother. 2008;14:86–92. doi: 10.1007/s10156-008-0596-1. [DOI] [PubMed] [Google Scholar]
- 97.Blasius AL, Beutler B. Intracellular Toll-like receptors. Immunity. 2010;32:305–315. doi: 10.1016/j.immuni.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 98.Temperley ND, et al. Evolution of the chicken Toll-like receptor gene family: a story of gene gain and gene loss. BMC Genomics. 2008;9:62. doi: 10.1186/1471-2164-9-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Diebold SS, et al. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 100.Heil F, et al. Species-specific recognition of single-stranded RNA via Toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 101.Lund JM, et al. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc Natl Acad Sci USA. 2004;101:5598–5603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hemmi H, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 103.Alexopoulou L, et al. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 104.Wang Y, et al. Dimerization of Toll-like receptor 3 (TLR3) is required for ligand binding. J Biol Chem. 2010;285:36836–36841. doi: 10.1074/jbc.M110.167973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Leonard JN, et al. The TLR3 signaling complex forms by cooperative receptor dimerization. Proc Natl Acad Sci USA. 2008;105(1):258–263. doi: 10.1073/pnas.0710779105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Oshiumi H, et al. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-bold beta induction. Nat Immunol. 2003;4:161–167. doi: 10.1038/ni886. [DOI] [PubMed] [Google Scholar]
- 107.Bell JK, et al. The molecular structure of the Toll-like receptor 3 ligand-binding domain. Proc Natl Acad Sci USA. 2005;102:10976–10980. doi: 10.1073/pnas.0505077102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jim MS, Lee J-O. Structures of the Toll-like receptor family and its ligand complexes. Immunity. 2008;29:182–191. doi: 10.1016/j.immuni.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 109.Liu L, et al. Structural basis of Toll-like receptor 3 signaling with double-stranded RNA. Science. 2008;320:379–381. doi: 10.1126/science.1155406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sioud M. Innate sensing of self and non-self RNAs by Toll-like receptors. Trends Mol Med. 2006;12:167–176. doi: 10.1016/j.molmed.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 111.Schroder M, Bowie AG. TLR3 in antiviral immunity: key player or bystander? Trends Immunol. 2005;26:462–468. doi: 10.1016/j.it.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 112.Kariko K, et al. mRNA is an endogenous ligand for Toll-like receptor 3. J Biol Chem. 2004;279:12542–12550. doi: 10.1074/jbc.M310175200. [DOI] [PubMed] [Google Scholar]
- 113.Cavassani KA, et al. TLR3 is an endogenous sensor of tissue necrosis during acute inflammatory events. J Exp Med. 2008;205:2609–2621. doi: 10.1084/jem.20081370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Duffy K.E et al (2012) Lateral clustering of TLR3:dsRNA signaling units revealed by TLR3ecd:3Fabs quaternary structure. J Mol Biol [Epub ahead] [DOI] [PMC free article] [PubMed]
- 115.Kleinman ME, et al. Sequence- and target-independent angiogenesis suppression by siRNA via TLR3. Nature. 2008;452:591–597. doi: 10.1038/nature06765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Karikó K, et al. Small interfering RNAs mediate sequence independent gene suppression and induce immune activation by signaling through Toll-like receptor 3. J Immunol. 2004;172:6545–6549. doi: 10.4049/jimmunol.172.11.6545. [DOI] [PubMed] [Google Scholar]
- 117.Pirher N, et al. A second binding site for double-stranded RNA in TLR3 and consequences for interferon activation. Nat Struc Mol Biol. 2008;15:761–763. doi: 10.1038/nsmb.1453. [DOI] [PubMed] [Google Scholar]
- 118.Kariko K, et al. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 119.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5(7):730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 120.Yoneyama M, et al. Shared and unique functions of the DExD/H-Box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 121.Seth RB, et al. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF3. Cell. 2005;122:699–782. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 122.Xu LG, et al. VISA is an adapter protein required for virus-triggered IFN-b signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 123.Kawai T, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6(10):981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 124.Satoh T, et al. LGP2 is a positive regulator of RIG-I and MDA5-mediated antiviral responses. Proc Natl Acad Sci USA. 2010;107:1512–1517. doi: 10.1073/pnas.0912986107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Saito T, Hirai R, Loo YM, Owen D, Johnson CL, Sinha SC, Akira S, Fujita T, Gale M., Jr Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci USA. 2007;104(2):582–587. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gack MU, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 127.Jiang X, et al. Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity. 2012;36:959–973. doi: 10.1016/j.immuni.2012.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hou F, et al. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146:1–14. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pichlmair A, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 130.Schlee M, et al. Recognition of 5′triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31(1):25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Marques JT, et al. A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nat Biotechnol. 2006;24:559–565. doi: 10.1038/nbt1205. [DOI] [PubMed] [Google Scholar]
- 132.Saito T, et al. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Uzri D, Gehrke L. Nucleotide sequences and modifications that determine RIG-I/RNA binding and signaling activities. J Virol. 2009;83:4174–4184. doi: 10.1128/JVI.02449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Malathi K, et al. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 2007;448:816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Takahasi K, et al. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol Cell. 2008;29(4):428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 136.Cui S, et al. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol Cell. 2008;29(2):169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 137.Lu C, et al. The structural basis of 5′ triphosphate double-stranded RNA recognition by RIG-I C-terminal domain. Structure. 2010;18:1032–1043. doi: 10.1016/j.str.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wang Y, et al. Structural and functional insights into 5′-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat Struct Mol Biol. 2010;17(7):781–787. doi: 10.1038/nsmb.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jiang F, et al. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011;479(7373):423–427. doi: 10.1038/nature10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Luo D, et al. Structural insights into RNA recognition by RIG-I. Cell. 2011;147:409–422. doi: 10.1016/j.cell.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kowalinski E, et al. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- 142.Binder M, et al. Molecular mechanism of signal perception and integration by the innate immune sensor retinoic acid inducible gene-I. J Biol Chem. 2011;286:27278–27287. doi: 10.1074/jbc.M111.256974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bamming D, Horvath CM. Regulation of signal transduction by enzymatically inactive antiviral RNA helicase proteins MDA5, RIG-I, and LGP2. J Biol Chem. 2009;284(15):9700–9712. doi: 10.1074/jbc.M807365200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Myong S, et al. Cytosolic viral sensor RIG-I Is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323:1070–1074. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kato H, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205(7):1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Peisley A, et al. Cooperative assembly and dynamic disassembly of MDA5 filaments for viral dsRNA recognition. Proc Natl Acad Sci USA. 2011;108(52):21010–21015. doi: 10.1073/pnas.1113651108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Berke IC, Modis Y. MDA5 cooperatively forms dimers and ATP-sensitive filaments upon binding double-stranded RNA. EMBO J. 2012;7:1714–1726. doi: 10.1038/emboj.2012.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.McCartney SA, et al. MDA-5 recognition of a murine norovirus. PLoS Pathog. 2008;4(7):e1000108. doi: 10.1371/journal.ppat.1000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Pichlmair A, et al. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J Virol. 2009;83(20):10761–10769. doi: 10.1128/JVI.00770-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Luthra P, et al. Activation of IFN-β expression by a viral mRNA through RNase L and MDA5. Proc Natl Acad Sci USA. 2011;108:2118–2123. doi: 10.1073/pnas.1012409108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 152.Kim T, et al. Aspartate-glutamate-alanine-histidine box motif (DEAH)/RNA helicase A helicases sense microbial DNA in human plasmacytoid dendritic cells. Proc Natl Acad Sci USA. 2010;107:15181–15186. doi: 10.1073/pnas.1006539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhang Z, et al. DHX9 pairs with IPS-1 to sense double-stranded RNA in myeloid dendritic cells. J Immunol. 2011;187:4501–4508. doi: 10.4049/jimmunol.1101307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhang Z, et al. DDX1, DDX21, and DHX36 helicases form a complex with the adaptor molecule TRIF to sense dsRNA in dendritic cell. Immunity. 2011;34:866–878. doi: 10.1016/j.immuni.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Jin J, et al. Evidence that Lin28 stimulates translation by recruiting RNA helicase A to polysomes. Nucleic Acids Res. 2011;39:3724–3734. doi: 10.1093/nar/gkq1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ishaq M, et al. The DEAD-box RNA helicase DDX1 interacts with RelA and enhances nuclear factor kappaB-mediated transcription. J Cell Biochem. 2009;106:297–305. doi: 10.1002/jcb.22004. [DOI] [PubMed] [Google Scholar]
- 157.Hennessy EJ, Parker AE, O’Neill LAJ. Targeting Toll-like receptors: emerging therapeutics? Nat Rev Drug Discov. 2010;9:293–307. doi: 10.1038/nrd3203. [DOI] [PubMed] [Google Scholar]
- 158.Ireton RC, Gale M., Jr RIG-I like receptors in antiviral immunity and therapeutic applications. Viruses. 2011;3:906–919. doi: 10.3390/v3060906. [DOI] [PMC free article] [PubMed] [Google Scholar]