A novel infectious disease, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), was detected in Wuhan, China, in December 2019. The disease (COVID-19) spread rapidly, reaching epidemic proportions in China, and has been found in 27 other countries. As of February 27, 2020, over 82,000 cases of COVID-19 were reported, with > 2800 deaths. No specific therapeutics are available, and current management includes travel restrictions, patient isolation, and supportive medical care. There are a number of pharmaceuticals already being tested [1, 2], but a better understanding of the underlying pathobiology is required. In this context, this article will briefly review the rationale for angiotensin-converting enzyme 2 (ACE2) receptor as a specific target.
SARS-CoV-2 and severe acute respiratory syndrome coronavirus (SARS-CoV) use ACE2 receptor to facilitate viral entry into target cells
SARS-CoV-2 has been sequenced [3]. A phylogenetic analysis [3, 4] found a bat origin for the SARS-CoV-2. There is a diversity of possible intermediate hosts for SARS-CoV-2, including pangolins, but not mice and rats [5].
There are many similarities of SARS-CoV-2 with the original SARS-CoV. Using computer modeling, Xu et al. [6] found that the spike proteins of SARS-CoV-2 and SARS-CoV have almost identical 3-D structures in the receptor-binding domain that maintains van der Waals forces. SARS-CoV spike protein has a strong binding affinity to human ACE2, based on biochemical interaction studies and crystal structure analysis [7]. SARS-CoV-2 and SARS-CoV spike proteins share 76.5% identity in amino acid sequences [6] and, importantly, the SARS-CoV-2 and SARS-CoV spike proteins have a high degree of homology [6, 7].
Wan et al. [4] reported that residue 394 (glutamine) in the SARS-CoV-2 receptor-binding domain (RBD), corresponding to residue 479 in SARS-CoV, can be recognized by the critical lysine 31 on the human ACE2 receptor [8]. Further analysis even suggested that SARS-CoV-2 recognizes human ACE2 more efficiently than SARS-CoV increasing the ability of SARS-CoV-2 to transmit from person to person [4]. Thus, the SARS-CoV-2 spike protein was predicted to also have a strong binding affinity to human ACE2.
This similarity with SARS-CoV is critical because ACE2 is a functional SARS-CoV receptor in vitro [9] and in vivo [10]. It is required for host cell entry and subsequent viral replication. Overexpression of human ACE2 enhanced disease severity in a mouse model of SARS-CoV infection, demonstrating that viral entry into cells is a critical step [11]; injecting SARS-CoV spike into mice worsened lung injury. Critically, this injury was attenuated by blocking the renin-angiotensin pathway and depended on ACE2 expression [12]. Thus, for SARS-CoV pathogenesis, ACE2 is not only the entry receptor of the virus but also protects from lung injury. We therefore previously suggested that in contrast to most other coronaviruses, SARS-CoV became highly lethal because the virus deregulates a lung protective pathway [10, 12].
Zhou et al. [13] demonstrated that overexpressing ACE2 from different species in HeLa cells with human ACE2, pig ACE2, civet ACE2 (but not mouse ACE2) allowed SARS-CoV-2 infection and replication, thereby directly showing that SARS-CoV-2 uses ACE2 as a cellular entry receptor. They further demonstrated that SARS-CoV-2 does not use other coronavirus receptors such as aminopeptidase N and dipeptidyl peptidase 4 [13]. In summary, the SARS-CoV-2 spike protein directly binds with the host cell surface ACE2 receptor facilitating virus entry and replication.
Enrichment distribution of ACE2 receptor in human alveolar epithelial cells (AEC)
A key question is why the lung appears to be the most vulnerable target organ. One reason is that the vast surface area of the lung makes the lung highly susceptible to inhaled viruses, but there is also a biological factor. Using normal lung tissue from eight adult donors, Zhao et al. [14] demonstrated that 83% of ACE2-expressing cells were alveolar epithelial type II cells (AECII), suggesting that these cells can serve as a reservoir for viral invasion. In addition, gene ontology enrichment analysis showed that the ACE2-expressing AECII have high levels of multiple viral process-related genes, including regulatory genes for viral processes, viral life cycle, viral assembly, and viral genome replication [14], suggesting that the ACE2-expressing AECII facilitate coronaviral replication in the lung.
Expression of the ACE2 receptor is also found in many extrapulmonary tissues including heart, kidney, endothelium, and intestine [15–19]. Importantly, ACE2 is highly expressed on the luminal surface of intestinal epithelial cells, functioning as a co-receptor for nutrient uptake, in particular for amino acid resorption from food [20]. We therefore predict that the intestine might also be a major entry site for SARS-CoV-2 and that the infection might have been initiated by eating food from the Wuhan market, the putative site of the outbreak. Whether SARS-CoV-2 can indeed infect the human gut epithelium has important implications for fecal–oral transmission and containment of viral spread. ACE2 tissue distribution in other organs could explain the multi-organ dysfunction observed in patients [21–23]. Of note, however, according to the Centers for Disease Control and Prevention [24], whether a person can get COVID-19 by touching surfaces or objects that have virus on them and then touching mucus membranes is yet to be confirmed.
Potential approaches to address ACE2-mediated COVID-19
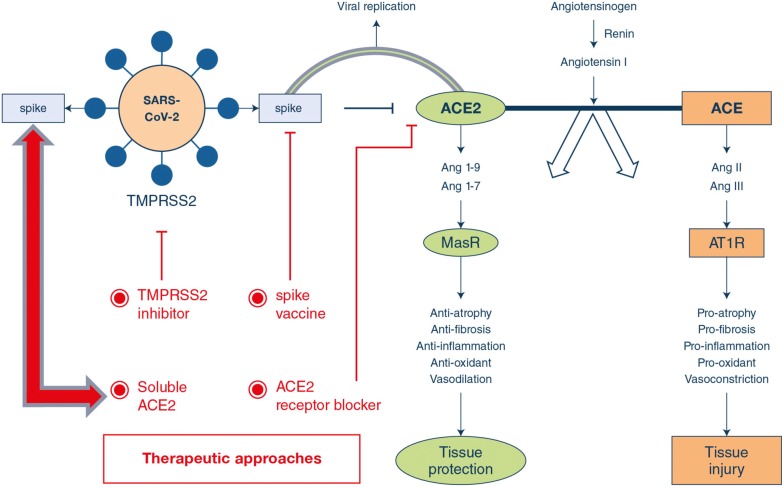
There are several potential therapeutic approaches (Fig. 1).
-
Spike protein-based vaccine.
Development of a spike1 subunit protein-based vaccine may rely on the fact that ACE2 is the SARS-CoV-2 receptor. Cell lines that facilitate viral replication in the presence of ACE2 may be most efficient in large-scale vaccine production.
-
Inhibition of transmembrane protease serine 2 (TMPRSS2) activity.
Hoffman et al. [25] recently demonstrated that initial spike protein priming by transmembrane protease serine 2 (TMPRSS2) is essential for entry and viral spread of SARS-CoV-2 through interaction with the ACE2 receptor [26, 27]. The serine protease inhibitor camostat mesylate, approved in Japan to treat unrelated diseases, has been shown to block TMPRSS2 activity [28, 29] and is thus an interesting candidate.
-
Blocking ACE2 receptor.
The interaction sites between ACE2 and SARS-CoV have been identified at the atomic level and from studies to date should also hold true for interactions between ACE2 and SARS-CoV-2. Thus, one could target this interaction site with antibodies or small molecules.
-
Delivering excessive soluble form of ACE2.
Kuba et al. [10] demonstrated in mice that SARS-CoV downregulates ACE2 protein (but not ACE) by binding its spike protein, contributing to severe lung injury. This suggests that excessive ACE2 may competitively bind with SARS-CoV-2 not only to neutralize the virus but also rescue cellular ACE2 activity which negatively regulates the renin-angiotensin system (RAS) to protect the lung from injury [12, 30]. Indeed, enhanced ACE activity and decreased ACE2 availability contribute to lung injury during acid- and ventilator-induced lung injury [12, 31, 32]. Thus, treatment with a soluble form of ACE2 itself may exert dual functions: (1) slow viral entry into cells and hence viral spread [7, 9] and (2) protect the lung from injury [10, 12, 31, 32].
Notably, a recombinant human ACE2 (rhACE2; APN01, GSK2586881) has been found to be safe, with no negative hemodynamic effects in healthy volunteers and in a small cohort of patients with ARDS [33–35]. The administration of APN01 rapidly decreased levels of its proteolytic target peptide angiotensin II, with a trend to lower plasma IL-6 concentrations. Our previous work on SARS-CoV pathogenesis makes ACE2 a rational and scientifically validated therapeutic target for the current COVID-19 pandemic. The availability of recombinant ACE2 was the impetus to assemble a multinational team of intensivists, scientists, and biotech to rapidly initiate a pilot trial of rhACE2 in patients with severe COVID-19 (Clinicaltrials.gov #NCT04287686).
Fig. 1.
Potential approaches to address ACE2-mediated COVID-19 following SARS-CoV-2 infection. The finding that SARS-CoV-2 and SARS-CoV use the ACE2 receptor for cell entry has important implications for understanding SARS-CoV-2 transmissibility and pathogenesis. SARS-CoV and likely SARS-CoV-2 lead to downregulation of the ACE2 receptor, but not ACE, through binding of the spike protein with ACE2. This leads to viral entry and replication, as well as severe lung injury. Potential therapeutic approaches include a SARS-CoV-2 spike protein-based vaccine; a transmembrane protease serine 2 (TMPRSS2) inhibitor to block the priming of the spike protein; blocking the surface ACE2 receptor by using anti-ACE2 antibody or peptides; and a soluble form of ACE2 which should slow viral entry into cells through competitively binding with SARS-CoV-2 and hence decrease viral spread as well as protecting the lung from injury through its unique enzymatic function. MasR—mitochondrial assembly receptor, AT1R—Ang II type 1 receptor
Acknowledgements
This study was funded in part by the following agencies; the Canadian Institutes of Health Research FDN143285; National Nature Science Foundation of China (81270125, 81490530, 2018GZR0201002); and Clinical Innovation Research Program of Guangzhou Regenerative Medicine and Health Guangdong Laboratory (2018GZR0201002).
Compliance with ethical standards
Conflicts of interest
Josef Penninger is the founder and a shareholder of Apeiron, the company that makes rhACE2. Arthur Slutsky has been a paid consultant for Apeiron. No other conflicts of interested have been reported.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.https://www.clinicaltrialsarena.com/news/company-news/gilead-coronavirus-remdesivir-trial/
- 2.Li G, De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat Rev Drug Discov. 2020 doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 3.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020 doi: 10.1016/s0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS. J Virol. 2020 doi: 10.1128/jvi.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.https://www.nature.com/articles/d41586-020-00364-2
- 6.Xu X, Chen P, Wang J, Feng J, Zhou H, Li X, Zhong W, Hao P. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its Spike protein for risk of human transmission. Sci China Life Sci. 2020 doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li F, Li W, Farzan M, Harrison SC. Structure of SARS coronavirus Spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 8.Wu KL, Peng GQ, Wilken M, Geraghty RJ, Li F. Mechanisms of host receptor adaptation by severe acute respiratory syndrome coronavirus. J Biol Chem. 2012;287:8904–8911. doi: 10.1074/jbc.M111.325803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H, Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuba K, Imai Y, Rao S, Gao H, Guo F, Guan B, Huan Y, Yang P, Zhang Y, Deng W, Bao L, Zhang B, Liu G, Wang Z, Chappell M, Liu Y, Zheng D, Leibbrandt A, Wada T, Slutsky AS, Liu D, Qin C, Jiang C, Penninger JM. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus–induced lung injury. Nat Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang XH, Deng W, Tong Z, Liu YX, Zhang LF, Zhu H, Gao H, Huang L, Liu YL, Ma CM, Xu YF, Ding MX, Deng HK, Qin C. Mice transgenic from human angiotensin-converting enzyme 2 provide a model for SARS coronavirus infection. Comput Med. 2007;57(5):450–459. [PubMed] [Google Scholar]
- 12.Imai Y, Kuba K, Rao S, Huan Y, Guo F, Guan B, Yang P, Sarao R, Wada T, Leong-Poi H, Crackower MA, Fukamizu A, Hui CC, Hein L, Uhlig S, Slutsky AS, Jiang C, Penniger JM. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436:112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020 doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Y, Zhao Z, Wang Y, Zhou Y, Ma Y, Zuo W (2020) Single-cell RNA expression profiling of ACE2, the putative receptor of Wuhan COVID-19. 10.1101/2020.01.26.919985
- 15.Crackower MA, Sarao R, Oudit GY, Yagil C, Kozieradzki I, Scanga SE, Oliveira-dos-Santos AJ, da Costa J, Zhang L, Pei Y, Scholey J, Ferrario CM, Manoukian AS, Chappell MC, Backx PH, Yagil Y, Penninger JM. Angiotensin-converting enzyme 2 is an essential regulator of heart function. Nature. 2002;417(6891):822–828. doi: 10.1038/nature00786. [DOI] [PubMed] [Google Scholar]
- 16.Danilczyk U, Sarao R, Remy C, Benabbas C, Stange G, Richter A, Arya S, Pospisilik JA, Singer D, Camargo SM, Makrides V, Ramadan T, Verrey F, Wagner CA, Penninger JM. Essential role for collectrin in renal amino acid transport. Nature. 2006;444(7122):1088–1091. doi: 10.1038/nature05475. [DOI] [PubMed] [Google Scholar]
- 17.Gu J, Gong E, Zhang B, Zheng J, Gao Z, Zhong Y, Zou W, Zhan J, Wang S, Xie Z, Zhuang H, Wu B, Zhong H, Shao H, Fang W, Gao D, Pei F, Li X, He Z, Xu D, Shi X, Anderson VM, Leong AS. Multiple organ infection and the pathogenesis of SARS. J Exp Med. 2005;202(3):415–424. doi: 10.1084/jem.20050828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding Y, He L, Zhang Q, Huang Z, Che X, Hou J, Wang H, Shen H, Qiu L, Li Z, Geng J, Cai J, Han J, Li X, Kang W, Weng D, Liang P, Jiang S. Organ distribution of severe acute respiratory syndrome (SARS) associated coronavirus (SARS-CoV) in SARS patients: implications for pathogenesis and virus transmission pathways. J Pathol. 2004;203:622–630. doi: 10.1002/path.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamming I, Timens W, Bulthuis MLC, Lely AT, Navis GJ, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus: a first step in understanding SARS pathogenesis. J Pathol. 2004;203:631–663. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hashimoto T, Perlot T, Rehman A, Trichereau J, Ishiguro H, Paolino M, Sigl V, Hanada T, Hanada R, Lipinski S, Wild B, Camargo SM, Singer D, Richter A, Kuba K, Fukamizu A, Schreiber S, Clevers H, Verrey F, Rosenstiel P, Penninger JM. ACE2 links amino acid malnutrition to microbial ecology andf intestinal inflammation. Nature. 2012;487(7408):477–481. doi: 10.1038/nature11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, Wang B, Xiang H, Cheng Z, Xiong Y, Zhao Y, Li Y, Wang X, Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020 doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020 doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guan W, Ni Z, Hu Y, Liang W, Ou C, He H, Liu L, Shan H, Lei C, Hui DSC, Du B, Li L, Zeng G, Yuen KY, Chen R, Tang C, Wang T, Chen P, Xiang J, Li S, Wang J, Liang Z, Peng Y, Wei L, Liu Y, Hu Y, Peng P, Wang J, Liu J, Chen Z, Li G, Zheng Z, Qiu S, Luo J, Ye C, Zhu S, Zhong N. Clinical characteristics of 2019 novel coronavirus infection in China. medRxiv. 2020 doi: 10.1101/2020.02.06.20020974. [DOI] [Google Scholar]
- 24.https://www.cdc.gov/coronavirus/2019-ncov/about/transmission.html
- 25.Hoffmann M, Kleine-Weber H, Krüger N, Müller M, Drosten C, Pöhlmann S. The novel coronavirus 2019 (COVID-19) uses the SARS-1 coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. bioRxiv. 2020 doi: 10.1101/2020.01.31.929042. [DOI] [Google Scholar]
- 26.Glowacka I, Bertram S, Muller MA, Allen P, Soilleux E, Pfefferle S, Steffen I, Tsegaye TS, He Y, Gnirss K, Niemeyer D, Schneider H, Drosten C, Pöhlmann S. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus Spike protein for membrane fusion and reduces viral control by the humoral immune response. J Virol. 2011;85:4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iwata-Yoshikawa N, Okamura T, Shimizu Y, Hasegawa H, Takeda M, Nagata N. TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. J Virol. 2019 doi: 10.1128/jvi.01815-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawase M, Shirato K, van der Hoek L, Taguchi F, Matsuyama S. Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. J Virol. 2012;86:6537–6654. doi: 10.1128/JVI.00094-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou Y, Vedantham P, Lu K, Agudelo J, Carrion R, Jr, Nunneley JW, Barnard D, Pöhlmann S, McKerrow JH, Renslo AR, Simmons G. Protease inhibitors targeting coronavirus and filovirus entry. Antiviral Res. 2015;116:76–84. doi: 10.1016/j.antiviral.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu L, Yuan K, Phuong HT, Park BM, Kim SH. Angiotensin-(1-5), an active mediator of renin-angiotensin system, stimulates ANP secretion via Mas receptor. Peptides. 2016;86:33–41. doi: 10.1016/j.peptides.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 31.Zhang R, Pan Y, Fanelli V, Wu S, Luo AA, Islam D, Han B, Mao P, Ghazarian M, Zeng W, Spieth PM, Wnag D, Khang J, Mo H, Liu X, Uhlig S, Liu M, Laffey J, Slutsky AS, Li Y, Zhang H. Mechanical stress and the induction of lung fibrosis via the midkine signaling pathway. Am J Respir Crit Care Med. 2015;192:315–323. doi: 10.1164/rccm.201412-2326OC. [DOI] [PubMed] [Google Scholar]
- 32.Wosten-van Asperen RM, Lutter R, Specht PA, Moll GN, van Woensel JB, van der Loos CM, van Goor H, Kamilic J, Florquin S, Bos AP. Acute respiratory distress syndrome leads to reduced ratio of ACE/ACE2 activities and is prevented by angiotensin-(1-7) or an angiotensin II receptor antagonist. J Pathol. 2011;225:618–627. doi: 10.1002/path.2987. [DOI] [PubMed] [Google Scholar]
- 33.Haschke M, Schuster M, Poglitsch M, Loibner H, Salzberg M, Bruggisser M, Penninger J, Krahenbuhl S. Pharmacokinetics and pharmacodynamics of recombinant human angiotensin-converting enzyme 2 in healthy human subjects. Clin Pharmacokinet. 2013;52:783–792. doi: 10.1007/s40262-013-0072-7. [DOI] [PubMed] [Google Scholar]
- 34.Khan A, Benthin C, Zeno B, Albertson TE, Boyd J, Christie JD, Hall R, Poirier G, Ronco JJ, Tidswell M, et al. A pilot clinical trial of recombinant human angiotensin-converting enzyme 2 in acute respiratory distress syndrome. Crit Care. 2017;21:234. doi: 10.1186/s13054-017-1823-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang H, Baker A. Recombinant human ACE2: acing out angiotensin II in ARDS therapy. Crit Care. 2017;21:305. doi: 10.1186/s13054-017-1882-z. [DOI] [PMC free article] [PubMed] [Google Scholar]