Abstract
Molecular diagnostics of infectious diseases, in particular, nucleic-acid-based methods, are the fastest growing field in clinical laboratory diagnostics. These applications are stepwise replacing or complementing culture-based, biochemical, and immunological assays in microbiology laboratories. The first-generation nucleic acid assays were monoparametric such as conventional tests, determining only a single parameter. Improvements and new approaches in technology now open the possibility for the development of multiparameter assays using microarrays, multiplex nucleic acid amplification techniques, or mass spectrometry, while the introduction of closed-tube systems has resulted in rapid microbial diagnostics with a subsequently reduced contamination risk. Whereas the first assays were focused on the detection and identification of microbial pathogens, these new technologies paved the way for the parallel determination of multiple antibiotic resistance determinants or to perform microbial epidemiology and surveillance on a genetic level.
Keywords: Molecular diagnostics, Real-time PCR, Nucleic acid amplification technique, Microarray, Sequencing, Mass spectrometry, Antibiotic resistance
Introduction
Detection, identification, and drug susceptibility testing of microbial pathogens represent the key duty of microbial diagnostics in medicine. Antibiotic susceptibility testing especially provides important information towards adequate treatment decisions. Also of great importance is the epidemiological genotyping of isolated microorganisms with respect to monitoring and surveillance of routes of infection. This is an essential task in order to develop strategies to prevent or successfully treat infections, both in the community and the health care facilities. Recent progress and extensive research on microbes as well as the development of new nucleic-acid-based methodologies have resulted in the increasing use of molecular assays in clinical laboratory with several commercial tests available. We mainly focus this review on nucleic-acid-based molecular techniques for identification and resistance determination in clinical bacteriology, giving a brief overview of currently used modern bacterial diagnostics and providing an outlook on future technologies, especially dealing with the multiparametric detection of infectious disease-related determinants. When appropriate, we will also mention examples from other fields of clinical microbiology, e.g., clinical virology.
Identification and drug-susceptibility testing of microorganisms
Phenotype-based methods
One of the oldest but still very important methods in clinical bacteriology is the detection of human pathogens by direct microscopic examination of the specimen. Many different staining procedures are available (Gram stain, Giemsa stain, Ziehl–Neelsen stain), providing a first rough classification of the detected organism. However, the final characterization and identification still relies on phenotypic properties of the organism after culturing on appropriate media. The introduction of automated laboratory systems such as the blood culture systems BacTec (Becton Dickinson, Franklin Lakes, NJ, USA) and BacT/Alert (bioMérieux, Marcy l’Etoile, France) have increased the quality and efficiency, meeting the high requirements for standardization. But also for identification and antimicrobial drug-susceptibility testing, automated systems such as the VITEK (bioMérieux), PHOENIX (Becton Dickinson), or Microscan WalkAway (Siemens Healthcare Diagnostics, Munich, Germany) are well established. All these systems are based on miniaturization of conventional methods, using biochemical properties for identification and microdilution for susceptibility testing. However, these sophisticated systems still need, starting from a pure culture, several hours for identification and up to 18 h for susceptibility testing. The turnaround time of a specimen therefore still requires minimally 24 h, but usually 48 h or more are necessary [1].
The development of immunoassays allowed, for the first time, a rapid detection and identification of microorganisms without culturing. The assays either detect the presence of specific antibodies raised in response to a pathogenic antigen or detect the antigen itself. The technique is available in various formats, such as enzyme immunoassays, immunofluorescence assays, latex agglutination assays, line immunoassays or lateral-flow immunoassays, respectively. Direct antigen testing in clinical samples provide rapid and specific identification results, however, antigen detection still suffers from a lack of sensitivity and requires comparable large amounts of the respective antigen. In most cases, screening for specific antibodies as a response of the human immune systems to the respective pathogen is used for serodiagnosis and allows the differentiation of acute and past infection. Although continuously being improved in the past, serology can be ineffective for early diagnosis of infection due to the time lag before seroconversion [2]. With exception of molecular tests such as nucleic acid amplification techniques (NAT), direct microscopy of the specimen, culturing, and direct antigen detection represent the sole possibilities to detect an acute infection as early as possible, facing either a lack of sensitivity, specificity or long time periods until results are available.
Genotype-based methods
Culture-based identification and immunological assays use the phenotypic characteristics of a microorganism. However, these identification criteria can be influenced by in vitro testing conditions and may lead to a misinterpretation and subsequent misidentification [3]. Molecular techniques, in particular nucleic-acid-based identification methods take advantage of the use of stable genotypic characteristics [4]. The bacterial genome provides conserved regions for species identification and phylogenetic relationships [5], whereas genes encoding virulence factors or toxins can be useful for defining pathogenicity of an organism [6–8].
Culture confirmation or direct detection of microorganisms can be performed by direct hybridization assays using labeled oligonucleotide probes. Probe hybridization is useful for identifying slow-growing organisms after isolation in culture using either liquid or solid media. Using stringent reaction conditions, these probe-based assays show high specificity. Direct hybridization assays for bacterial identification require a large number of target cells, resulting in a certain lack of sensitivity. This disadvantage can be partially circumvented by targeting rRNA molecules, which are present in a high copy number per cell [9, 10].
Fluorescent in situ hybridization (FISH) is an attractive method for rapid detection and identification of bacteria or fungi directly from slide smears. This technology has the speed and ease-of-use of conventional staining methods combined with the specificity of molecular methods. Hybridization with fluorescent-labeled probes that target rRNA is performed on smears, using fluorescence microscopy for detection [11–14].
The introduction of NATs that enable the amplification of a few target molecules has provided new tools for rapid, specific and sensitive detection, identification, and resistance testing of microorganisms starting from sample material without culturing. The polymerase chain reaction (PCR) is the technique most used in research or diagnostics of nucleic acids. For almost all clinical relevant human pathogens home-made assays have been described, targeting either suited genes for identification, virulence factors for pathogenicity determination or important resistance determinants [15]. The starting point of commercial assays was mainly focused on the detection of viruses. In contrast, commercial assays only slowly entered the bacteriology laboratory and, at present, are mainly designed for detection of fastidious or uncultivable organisms such as Chlamydia and Mycoplasma species. Alternative DNA amplification techniques such as the ligase chain reaction (LCR) have been used for bacterial diagnostics. Due to certain disadvantages, LCR plays almost no role anymore [16]. RNA amplification techniques such as nucleic-acid-sequence-based amplification (NASBA) and transcription-mediated amplification (TMA) are well established [17–21]. Specific and sensitive detection of rRNA may be advantageous due to the high copy number present in target organisms. PCR is based on the detection of intact DNA rather than intact viable cells and, therefore, a positive reaction may arise from either dead or alive cells. Specific amplification of mRNA can be used for the detection of living pathogenic germs [22]. However, rapid RNAse-mediated degradation of mRNA requires careful and special specimen processing.
The classical way for the analysis of amplified nucleic acid fragments is agarose gel electrophoresis by using the fragment length as an indicator for identification. The specificity of the primers used influences the accuracy of the test result. Insufficient quality of the nucleic acids or the presence of non-target background DNA can influence specific annealing of the primers, resulting in an unspecific amplification, which can lead to misinterpretation of the result. Therefore, most of the amplification-based assays use specific hybridization probes for specific detection and identification of the amplified target molecules.
Real-time PCR technologies
The development of real-time PCR assays using fluorescent-labeled probes in combination with amplification technologies have paved the way for molecular assays in clinical microbiology. There are many different real-time PCR instruments and detection probe formats available. In this review, not all different probe formats (TaqMan hydrolysis probes, molecular beacons, Scorpion probes, minor groove binding probes, and many more) [23–26] and detection principles will be explained, since detailed information about real-time PCR technology and detection formats are frequently provided in publications and reviews [27]. Some of these technologies rely on the principle of fluorescence resonance energy transfer (FRET) [28, 29]. FRET occurs when two fluorophores are in close proximity to each other and the energy from an excited donor is transferred to an acceptor [23–34] (Fig. 1).
Fig. 1.
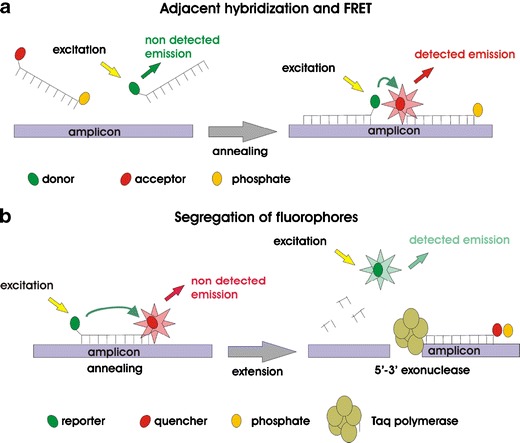
Different detection principles of fluorescent-labeled hybridization probes. a Fluorescence resonance energy transfer (FRET); head-to-tail arrangement of two probes. b TaqMan probes; 5′–3′ exonuclease activity of Taq polymerase leads to a fluorescence signal
The development of closed-tube systems in real-time PCR technologies significantly reduced the risk of contamination, strongly contributing to the strict requirements in clinical diagnostics. One of the main advantages using real-time fluorescence measuring is the ability to quantify and genotype genetic variations such as single-nucleotide polymorphisms (SNPs) using a set of specific probes for each possible SNP nucleotide within the same assay.
Quantification by real-time PCR is well established in virology, e.g., Cytomegalovirus (CMV) monitoring of hemato-oncologic patients during neutropenic stage after stem cell transplantation, Human Immunodeficiency Virus (HIV), and Hepatitis C Virus (HCV) [35–39]. Also, the determination of bacterial load has been established [40, 41], potentially allowing the monitoring of antimicrobial therapy response or discrimination among infection and colonization (Fig. 2). Cystic fibrosis (CF) patients suffer from chronic lung infections, which lead to deterioration of lung function. The most important pathogen in adult CF patients is Pseudomonas aeruginosa. Early diagnosis of P. aeruginosa at the colonizing stage is of great importance for prompt adequate antibiotic treatment aimed at the eradication of the pathogen or delay in the onset of chronic infection. PCR-based screening for P. aeruginosa colonization onset is gaining on average 4.5 months over conventional culture screening [42]. Also, severely burn patients, which frequently get infected with P. aeruginosa, can profit from the early detection and quantification of P. aeruginosa. The development of a quantitative PCR for the detection of P. aeruginosa in wound specimen showed that this method can provide results within 1 h concomitant with a minimal hands-on-time, allowing early antibiotic treatment decisions to be made [43]. The correlation between bacterial load and severity of infection was also shown for meningococcal disease [44].
Fig. 2.
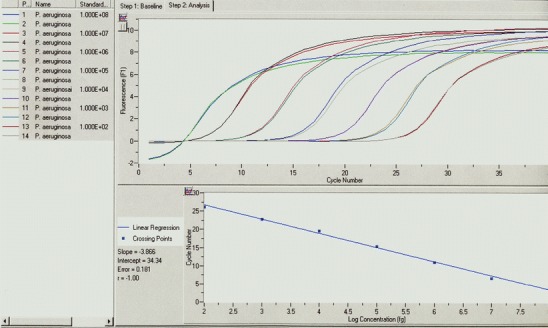
Quantification of Pseudomonas aeruginosa by real-time LightCycler PCR using standard curve analysis. In the upper part of the diagram, fluorescence signals of a 1:10 dilution series of defined P. aeruginosa genome equivalents (1 × 108 to 1 × 102) are displayed (measurement in duplicates). Based on the correlation of crossing-point CP (PCR cycle which is significantly different to the background fluorescence noise) to P. aeruginosa DNA concentration, a standard curve was calculated (lower part of the diagram). The linear correlation of this standard curve (theoretical slope = −3.33; determined slope = −3.86) allows the quantification of an unknown sample based on the CP of the respective sample
As the example shows, the first real-time PCR assays in clinical bacteriology still stick to the principle of monoparametric diagnostics—one analyte, one assay. One step advanced was the introduction of methicillin-resistant Staphylococcus aureus (MRSA) real-time PCR assays (Cepheid Xpert MRSA, BD GeneOhm MRSA), in which for the first time species identification, namely S. aureus, and resistance determination, namely methicillin resistance by detection of the underlying mecA gene cassettes SSCmec, were combined in a single assay [45–49]. A possible upcoming assay using this approach might be the detection of the highly virulent Clostridium difficile ribotype O27 directly from stool specimen [50].
Recently, the first multiparametric PCR assays for the diagnosis of bloodstream infections were released, identifying up to 40 different bacterial and fungal pathogens directly from whole blood (SeptiFast Roche Diagnostics, Mannheim, Germany; VYOO SIRS-Lab, Jena, Germany; SepsiTest Blood Molzyme, Bremen, Germany). Bloodstream infections are a major cause of morbidity and mortality worldwide [51]. So far, the automated blood culture systems are considered as the gold standard, detecting microbial growth based on detectable carbon dioxide production. Despite improvements in growth media and instrumentation, blood culture is too slow and has a poor diagnostic sensitivity and specificity. It is left to hope that amplification-based methods for direct microbial detection in whole blood have the potential to an increased diagnostic sensitivity, specificity, and shorter time to result, which is an absolute critical point in the adequate treatment of sepsis [52]. All above-mentioned assays use the principle of multiplex PCR, meaning the parallel amplification of different targets in one tube using a specific primer pair for each determinant. Whereas the SeptiFast system relies on real-time PCR LightCycler technology using fluorescence and melting curve analysis, the VYOO and SepsiTest systems perform a conventional multiplex PCR with subsequent identification of specific PCR fragments by agarose gel electrophoresis or alternatively by hybridization on microarrays. All systems put an enormous effort on the development of a reliable sample preparation, being crucial for these assay formats. The use of DNA-free reagents and equipment, together with methods for a decrease of human background DNA (Looxster from SIRS-Lab and MolYsis from Molzyme) were implemented [52, 53]. The diagnosis of bloodstream infections by molecular methods was just recently reviewed by Klouche et al. [54].
Sequencing-based technologies
During the last decade, DNA sequencing technologies made an enormous progress regarding throughput capability, cost per reaction and user-friendliness. DNA sequencing has become established in the routine laboratory. Automation was made possible by PCR-based sequencing reactions and capillary electrophoresis. So far, sequencing is used for the identification of organisms that are difficult to identify using conventional methods or to detect and identify uncultivable organisms. From an epidemiological and phylogenetical point of view, 16S and 23S ribosomal genes are the target of choice for taxonomy studies, representing the present gold standard [55–58]. Many ribosomal sequences are available in open access databases and can be used for producing alignments and subsequent species assignment.
Currently, Sanger-based sequencing technology is the most common sequencing method. Nevertheless, other technologies such as pyrosequencing, single-cell sequencing and other currently described massively parallel DNA sequencing approaches are very promising and powerful alternatives for future applications in clinical microbiology [59–66]. We are not able to explain all of these fascinating technologies in this review and would like to pick emulsion PCR coupled with pyrosequencing as just one example out of many different sequencing techniques available (Fig. 3). Single-nucleotide polymorphism analysis represents one of the fields pyrosequencing exhibits excellent performance. It is based on an enzyme-cascade system with online monitoring of light produced as a result of incorporation of nucleotides [67]. The method is accurate, user-friendly, and large data can be generated in a short time. State-of-the-art systems can sequence up to 20 million nucleotides in 4 h [68–72]. These rapid results are an advantage for diagnostic applications where time-to-result is essential, such as sepsis or meningitis [73]. However, due to the large amount of generated data, powerful interpretation software is required, stressing the need for an equally advancing progress in bioinformatics.
Fig. 3.
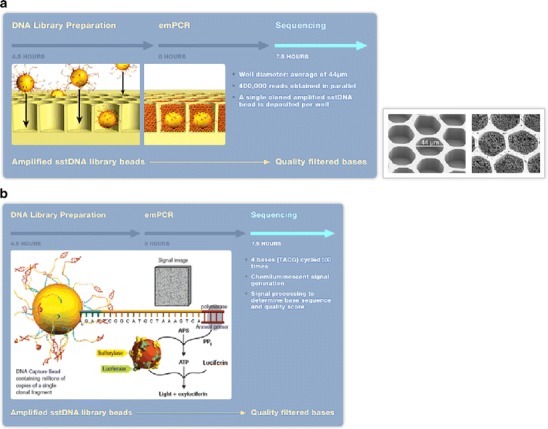
Emulsion PCR DNA sequencing technology on Genome Sequencer 20 and FLX Systems (Roche Diagnostics). a Emulsion PCR: single-stranded DNA (ssDNA) library beads are added to the “DNA bead incubation mix” (containing DNA polymerase) and are layered with “enzyme beads” (containing sulfurylase and luciferase) onto the PicoTiterPlate device. The device is centrifuged to deposit the beads into the wells. The layer of Enzyme Beads ensures that the DNA beads remain positioned in the wells during the sequencing reaction. The bead-deposition process maximizes the number of wells that contain a single amplified library bead (avoiding more than one sstDNA library bead per well). b Pyrosequencing: the loaded PicoTiterPlate device is placed into the Genome Sequencer FLX Instrument. The fluidics sub-system flows sequencing reagents (containing buffers and nucleotides) across the wells of the plate. Nucleotides are flowed sequentially in a fixed order across the PicoTiterPlate device during a sequencing run. During the nucleotide flow, each of the hundreds of thousands of beads with millions of copies of DNA is sequenced in parallel. If a nucleotide complementary to the template strand is flowed into a well, the polymerase extends the existing DNA strand by adding nucleotide(s). Addition of one (or more) nucleotide(s) results in a reaction that generates a light signal that is recorded by the CCD camera in the Instrument. The signal strength is proportional to the number of nucleotides, for example, homopolymer stretches, incorporated in a single-nucleotide flow. Provided and reprinted by permission from Roche Diagnostics, Mannheim, Germany
Mass spectrometry
In the beginning, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) was applied in the analysis of whole bacteria or proteins. Sequencing of bacterial nucleic acids using MALDI-TOF MS has been introduced and further improved. Base-specific cleaved nucleic acids fragments are generated by different methods and subsequently analyzed, and the resulting mass patterns are compared with reference spectra for sequence and species determination, respectively. At this time, mass spectrometry for microbial genotyping is still in its infancy. However, large diagnostic centers already use MALDI-TOF MS for bacterial identification (Fig. 4). This technology has the potential to become the method of choice for high-throughput testing in clinical microbiology, providing accurate, fast, automatable, and cost-efficient results, respectively [74–77].
Fig. 4.
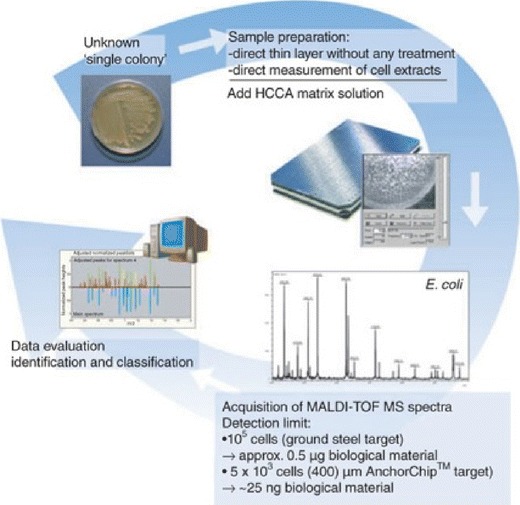
General workflow for ClinProt BioTyper (Bruker Daltonics, Billerica, MA, USA) microorganism identification and classification using MALDI-TOF MS. Reprinted by permission from [76]
Array-based technologies
Standard solid-phase hybridization use specific hybridization probes attached to a solid support to detect labeled target molecules in solution. Initially, in assays for detection and identification of single parameters, specific probes were immobilized in the wells of a microtiter plate [77]. Later, macroarrays of fixed multiple probes at specific locations on nitrocellulose or nylon membranes were developed, with the different probes being applied as dots (dot-blots) or as lines (line probe assay LIPA) [78–80]. A variety of different LIPA assays is available; e.g. for the identification of Mycobacterium tuberculosis complex and atypical Mycobacteria [81], in combination with the determination of the most common genetic resistance determinants against rifampin and isoniazid [82]. Also, LIPAs for the determination of vancomycin-resistant Enterococci (VRE), MRSA, C. difficile Toxin A/B, Neisseria meningitidis, entero-hemorrhagic E. coli (EHEC), extended-spectrum beta-lactamases (ESBL) and metallo-beta-lactamases (MBL), as well as a set of Gram-positive and -negative pathogens in positive blood cultures are on the market (DNA-Strips, Hain Lifescience, Nehren, Germany and hyplex Tests, BAG Health Care, Lich, Germany).
During the last decade, the development of microarrays has emerged in research, focused mainly on gene expression studies. Compared to macroarrays, microarrays are miniaturized versions with spot sizes usually less than 200 to 300 µm in diameter. Macroarrays usually are limited to fewer than 100 probes, DNA microarrays can vary from low-density arrays carrying a few hundred to a thousand probes, to high-density arrays containing tens of thousands to millions of spots. Different labeling and detection methods can be used in conjunction with array-based technologies, ranging from fluorescence over silver precipitation to measurable enzyme-mediated substrate conversion. The use of microarrays in diagnostic applications for microbiology is still in its beginning; nevertheless, the number of publications is increasing [83–85]. Thereby, various etiologic agents were targeted using different approaches regarding the target molecules and surface chemistry applied. Peptide/protein arrays were described for the detection of HIV/HCV and M. tuberculosis [86, 87], antigen microarrays for the diagnosis of blood culture negative endocarditis [88], respectively. DNA microarrays were applied e.g. in the diagnosis of bacterial meningitis, showing more accurate and rapid results as cerebrospinal fluid (CSF) and blood culture results [89], as well as for acute respiratory tract infections in children caused by viruses [90], and other fields, even without PCR amplification [91, 92]. The first commercially available microarray-based assays suitable for routine applications were released to the market like the PapilloCheck HPV Screening Test (Greiner Bio-One GmbH, Frickenhausen, Germany). The microarray-based PapilloCheck HPV Screening Test showed similar results compared to the Linear Array HPV Genotyping test and Amplicor HPV test (both Roche Diagnostics), however, studies showed that there is, e.g., an influence of the nucleic acid isolation method used and therefore a need for standardization of these genotyping methods [93–95].
Great potential in the application of DNA microarrays lies in resistance genotyping, since phenotypic antimicrobial resistance is very often the result of a complex network of different underlying genetic alterations, requiring complex and parallel detection methods [96]. Antimicrobial resistance, especially the threat arising from multidrug-resistant pathogens, is one of the most important and urgent problems in clinical bacteriology, for which the usage of microarrays has already demonstrated its suitability in general [97–101].
P. aeruginosa is one of the most important pathogens causing nosocomial infections, in particular, nosocomial pneumonia, showing a high capability for the development of multidrug resistance, thereby expressing an enormous array of different underlying genetic resistance determinants. We described a DNA microarray for genotyping antibiotic resistance in P. aeruginosa, targeting various resistance mechanisms such as multidrug efflux transporters, porin loss, target alterations due to mutations (SNPs and small insertions/deletions), and resistance-mediating genes present on plasmids or other mobile genetic elements, respectively (Fig. 5). In this assay, 36 different resistance-related DNA fragments are amplified in parallel, which encode a total of 388 different genetic resistance determinants. The genotype/phenotype correlation was almost 90%. In addition, the genotype-based prediction rate was about 10–15% higher as initially observed by phenotyping, which correlated well with the known induction phenomenon of phenotypically expressed resistance under therapy [102]. Furthermore, for other complex resistance problems such as detection of hundreds of different ESBL variants in tem, oxa, shv, and ctx-m genes, or MRSA and other clinical relevant resistances in Staphylococci, DNA microarray assays were published [103, 104] All these assays, although needing further optimization regarding implementation in routine clinical laboratories, are characterized by comparable short time requirements (4–5 h) compared to conventional methods. Besides the fact that resistance-related information is crucial for the adequate treatment of life-threatening infections, the more determinants assigned to the same organism are genotyped at the same time, the obtained information can be used concomitant for epidemiological surveillance. Although the described P. aeruginosa microarray represents the status of a research approach, it is currently in the process of partial integration into a complete diagnostic device including sample preparation, multiplex PCR amplification, array hybridization and result readout.
Fig. 5.
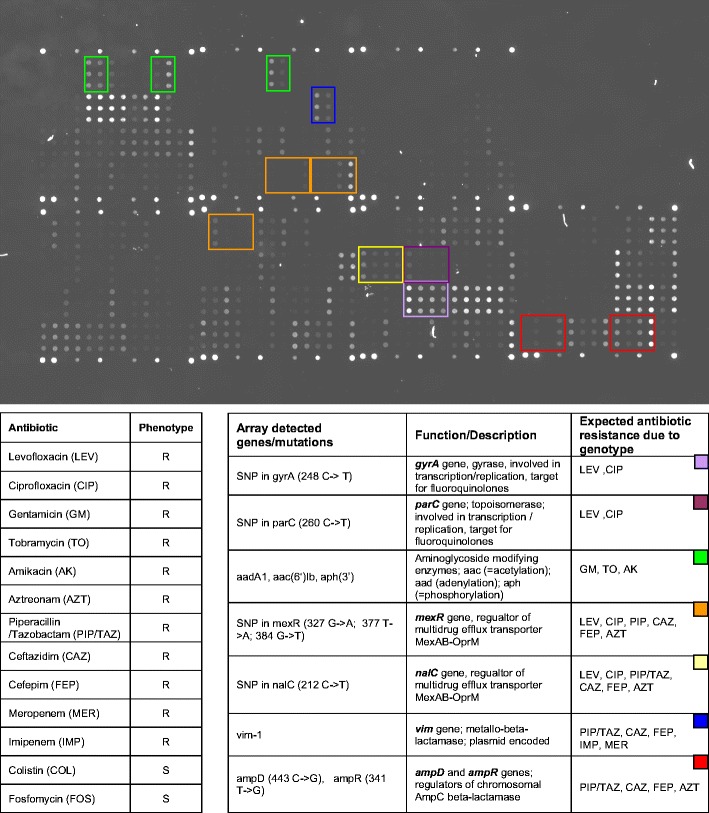
DNA microarray hybridization pattern of a multidrug-resistant P. aeruginosa clinical isolate. Compared to reference strain P. aeruginosa PAO1 (www.pseudomonas.com), which exhibits no resistance against therapy relevant anti-pseudomonal antibiotics, this P. aeruginosa strain harbors various genes or mutations assigned to antibiotic resistance. Single-nucleotide polymorphisms were detected in gyrA, parC, mexR, nalC, ampD, and ampR gene, respectively. Additionally, specific fluorescence signals above the cut-off indicated the presence of a vim-1, aac(6′)-Ib, aph(3′) and aadA1 gene (all positions are highlighted by colored frames). The observed phenotypical resistance was in complete accordance with the resistance determinants detected by DNA microarray genotyping. As indicated in the table, different resistance mechanisms rendering the same antibiotics as resistant (e.g. target alteration in gyrase and topoisomerase as well as efflux for fluoroquinolones, or chromosomal and plasmid encoded beta-lactamases as well as efflux for penicillins, cephalosporines, and carbapenems) were present at the same time, demonstrating the complex underlying genotype of phenotypically expressed resistance
High specificity, high sensitivity, short time requirement, and user-friendliness are the most important requirements for microbial diagnostic assays. The use of a three-dimensional membrane structure, providing a 500-fold increase in reaction surface compared with a flat two-dimensional surface, allows more targets to bind and enhances probe-target interaction. The ability to measure temperature variation and continuous monitoring of the reaction using fluorescence technology allows melting-curve analysis instead of end-point detection [105–107].
Suspension arrays are based on the coupling of oligonucleotide probes to microbeads that are color-coded using different ratios of two fluorescent dyes. A third dye is used for generating labeled target DNA, which is subsequently hybridized in suspension with a set of different beads, each carrying a different probe. The bead mixture is sorted by flow cytometry based on their internal colors and hybridized samples produce a fluorescent signal (Fig. 6) [108]. A panel for the detection of multiple viral strains and subtypes using this technology was approved for respiratory disease diagnostics by the FDA (xTAG Respiratory Viral Panel (RVP), Luminex, Austin, Texas, USA) [109–111] and others diseases like meningococcal infection [112, 113]. Currently, the development of sepsis diagnostics using this technology is in progress.
Fig. 6.
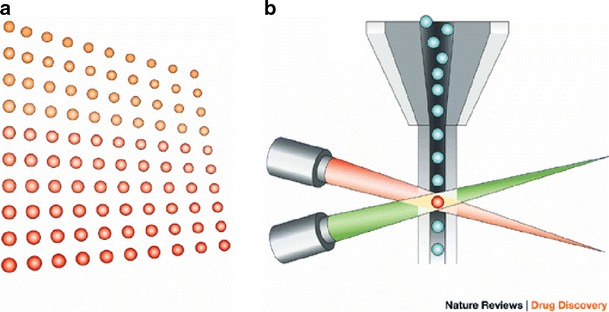
Bead-based Luminex xMAP system. a A set of 100 microspheres, each microsphere having a unique ratio of two fluorescent dyes. b The microspheres are identified individually in a rapidly flowing fluid stream that passes by two laser beams: one reveals the color code of the bead, and one quantifies the biomolecular reaction by measuring the fluorescence intensity of the reporter. Reprinted from [114]
Microfluidics and point-of-care diagnostics
In recent years, the innovation in microfluidics and nanotechnology has made tremendous progress, making these technologies to a cornerstone of future clinical diagnostics [115–119]. DNA and protein functional nanoparticles (FNP), for example, are sphere-like biocompatible materials made of inert silica, metal, or crystals of a nanometer in size. They may be used as hybridization probes in single-nucleotide polymorphism (SNP) screening and to detect biological markers for cancer, infection, and cardiovascular diseases [120, 121]. The precise handling of very small amounts, ranging in the nanoliter to picoliter scale, in microchannels [122–125], combined with sensitive detection methods like electrochemical microarrays [126], predispose these technologies for point-of-care (POC) and lab-on-a-chip (LOC) diagnostics. They allow the fabrication of disposable single-use cartridges, resulting in rapid and affordable diagnostics without the need for sophisticated and expensive laboratory equipment [127–131]. LOC, microfluidics, and nanotechnologies’ advantages such as increased sensitivity, mobility, and efficiency are likely to favor its uptake in private and public health sectors [132]. Further innovations, such as transition to digital microfluidics rather than the present-day concept of continuous flow microfluidics, is expected to attract more attention from diagnostics manufacturers and determine the future of this technology [133].
The LOCs provide short time-to-result information [134], as opposed to the current day-long wait, and can make a vast difference in diagnosis and treatment, not to mention patient outcome. Presently, LOCs are being developed to test sepsis [135, 136], endocarditis [88] HIV, tuberculosis, severe acute respiratory syndrome (SARS), pneumonia [137], malaria, and numerous other infectious diseases.
Conclusions
Already now, the introduction of molecular diagnostic technologies in human genetics and virology has changed the way clinical diagnostics are performed in these fields. The number of diagnostic assays in clinical bacteriology has increased steadily during the last decade. However, conventional microbiology will not be replaced in the immediate future, but multiparameter identification of the most important pathogens using array-based detection technologies or rapid real-time PCR-based assays are becoming common in today’s laboratories. Advancing automation and decreasing costs in molecular microbiology will bring these tools closer to the routine laboratory. The improvement of multiplexing strategies and analysis technologies like MALDI-TOF MS might harbor the potential to cope with the evident problems of modern medicine, especially the steadily increasing threat by multidrug-resistant pathogens. Combined with rapidly advancing innovations in microfluidics, the future trend clearly points towards complex multiparametric assays on miniaturized and automated lab-on-a-chip devices, also suitable for point-of-care diagnostics.
Acknowledgments
The authors would like to thank the German Department for Research and Education (BMBF) and the Robert Bosch Foundation for continuous support.
References
- 1.Mohr O’Hara C, Weinstein MP, Miller JM. In: Manual of clinical microbiology. 8. Murray PR, editor. Washington, DC: ASM; 2003. pp. 185–207. [Google Scholar]
- 2.Andreotti PE, Ludwig GV, Peruski AH, Tuite JJ, Morse SS, Peruski LF. Biotechniques. 2003;35:850–859. doi: 10.2144/03354ss02. [DOI] [PubMed] [Google Scholar]
- 3.Fahr AM, Hammann R. J Clin Microbiol. 1998;36:1464. [Google Scholar]
- 4.Ludwig W. Int J Food Microbiol. 2007;120:225–236. doi: 10.1016/j.ijfoodmicro.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 5.Konstantinidis KT, Ramette A, Tiedje JM. Philos Trans R Soc Lond B Biol Sci. 2006;361:1929–1940. doi: 10.1098/rstb.2006.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Vos D, Lim A, Pirnay JP, et al. J Clin Microbiol. 1997;35:1295–1299. doi: 10.1128/jcm.35.6.1295-1299.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deurenberg RH, Vink C, Driessen C, et al. FEMS Microbiol Lett. 2004;240:225–228. doi: 10.1016/j.femsle.2004.09.031. [DOI] [PubMed] [Google Scholar]
- 8.Burrack LS, Higgins DE. Curr Opin Microbiol. 2007;10:4–9. doi: 10.1016/j.mib.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 9.Bourbeau PP, Heiter BJ, Figdore M. J Clin Microbiol. 1997;35:144–147. doi: 10.1128/jcm.35.1.144-147.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stewart DS, Reineke KF, Tortorello ML. J AOAC Int. 2002;85:395–403. [PubMed] [Google Scholar]
- 11.Stender H. Expert Rev Mol Diagn. 2003;3:649–655. doi: 10.1586/14737159.3.5.649. [DOI] [PubMed] [Google Scholar]
- 12.Yilmaz O, Demiray E. World J Gastroenterol. 2007;13:671–675. doi: 10.3748/wjg.v13.i5.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tenover FC. Clin Infect Dis. 2007;44:418–423. doi: 10.1086/510684. [DOI] [PubMed] [Google Scholar]
- 14.Patel R, Osmon DR, Hanssen AD. Clin Orthop Relat Res. 2005;437:55–58. doi: 10.1097/01.blo.0000175121.73675.fd. [DOI] [PubMed] [Google Scholar]
- 15.Nikkari S, Lopez FA, Lepp PW, et al. Emerg Infect Dis. 2002;8:188–194. doi: 10.3201/eid0802.010150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shafer MA, Moncada J, Boyer CB, Betsinger K, Flinn SD, Schachter J. J Clin Microbiol. 2003;41:4395–4399. doi: 10.1128/JCM.41.9.4395-4399.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D’Souza DH, Jaykus LA. J Appl Microbiol. 2003;95:1343–1350. doi: 10.1046/j.1365-2672.2003.02106.x. [DOI] [PubMed] [Google Scholar]
- 18.Piersimoni C, Scarparo C, Piccoli P, et al. J Clin Microbiol. 2002;40:4138–4142. doi: 10.1128/JCM.40.11.4138-4142.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marangi M, Di Tullio R, Mens PF, Martinelli D, Fazio V, Angarano G, Schallig HD, Giangaspero A, Scotto G. Malar J. 2009;8:12. doi: 10.1186/1475-2875-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore C, Corden S, Sinha J, Jones R. J Virol Methods. 2008;153:84–89. doi: 10.1016/j.jviromet.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 21.Loens K, Beck T, Ursi D, Overdijk M, Sillekens P, Goossens H, Ieven M. J Microbiol Methods. 2008;73:257–262. doi: 10.1016/j.mimet.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 22.Rijpens NP, Herman LM. J AOAC Int. 2002;85:984–995. [PubMed] [Google Scholar]
- 23.De Clerck E, Van Mol K, Jannes G, Rossau R, De Vos P. Lett Appl Microbiol. 2004;39:109–115. doi: 10.1111/j.1472-765X.2004.01550.x. [DOI] [PubMed] [Google Scholar]
- 24.Cheung PY, Chan CW, Wong W, Cheung TL, Kam KM. Lett Appl Microbiol. 2004;39:509–515. doi: 10.1111/j.1472-765X.2004.01609.x. [DOI] [PubMed] [Google Scholar]
- 25.Templeton KE, Scheltinga SA, van der Zee A, et al. J Clin Microbiol. 2003;41:4121–4126. doi: 10.1128/JCM.41.9.4121-4126.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodriguez-Lazaro D, Lloyd J, Herrewegh A, et al. FEMS Microbiol Lett. 2004;237:119–126. doi: 10.1111/j.1574-6968.2004.tb09686.x. [DOI] [PubMed] [Google Scholar]
- 27.Espy MJ, Uhl JR, Sloan LM, Buckwalter SP, et al. Clin Microbiol Rev. 2006;19:165–256. doi: 10.1128/CMR.19.1.165-256.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsai MT, Cheng YH, Liu YN, Liao NC, Lu WW, Kung SH. Antimicrob Agents Chemother. 2009;53:748–755. doi: 10.1128/AAC.00841-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Intapan PM, Thanchomnang T, Lulitanond V, Maleewong W. J Med Entomol. 2009;46:158–164. doi: 10.1603/033.046.0119. [DOI] [PubMed] [Google Scholar]
- 30.Burggraf S, Reischl U, Malik N, Bollwein M, Naumann L, Olgemoller B. J Clin Microbiol. 2005;43:1564–1569. doi: 10.1128/JCM.43.4.1564-1569.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raggam RB, Leitner E, Berg J, Muhlbauer G, Marth E, Kessler HH. J Mol Diagn. 2005;7:133–138. doi: 10.1016/S1525-1578(10)60019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sloan LM, Uhl JR, Vetter EA, et al. J Clin Microbiol. 2004;42:2636–2643. doi: 10.1128/JCM.42.6.2636-2643.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ng CT, Gilchrist CA, Lane A, Roy S, Hague R, Houpt ER. J Clin Microbiol. 2005;43:1256–1260. doi: 10.1128/JCM.43.3.1256-1260.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Massart S, De Clercq D, Salmon M, Dickburt C, Jijakli MH. J Microbiol Methods. 2005;60:73–82. doi: 10.1016/j.mimet.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 35.Tang W, Elmore SH, Fan H, Thorne LB, Gulley ML. Diagn Mol Pathol. 2008;17:166–73. doi: 10.1097/PDM.0b013e3181599242. [DOI] [PubMed] [Google Scholar]
- 36.Schaade L, Kockelkorn P, Ritter K, Kleines M. J Clin Microbiol. 2000;38:4006–4009. doi: 10.1128/jcm.38.11.4006-4009.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Detmer J, Lagier R, Flynn J, Zayati C, Kolberg J, Collins M, Urdea M, Sanchez-Pescador R. J Clin Microbiol. 1996;34:901–907. doi: 10.1128/jcm.34.4.901-907.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sizmann D, Boeck C, Boelter J, Fischer D, Miethke M, Nicolaus S, Zadak M, Babiel R. J Clin Virol. 2007;38:326–333. doi: 10.1016/j.jcv.2006.12.021. [DOI] [PubMed] [Google Scholar]
- 39.Hölzl G, Stöcher M, Leb V, Stekel H, Berg J. J Clin Microbiol. 2003;41:1248–1251. doi: 10.1128/JCM.41.3.1248-1251.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li K, Chen B, Zhou Y, Huang R, Liang Y, Wang Q, Xiao Z, Xiao J. J Microbiol Methods. 2009;76:289–94. doi: 10.1016/j.mimet.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 41.Matsuda K, Tsuji H, Asahara T, Kado Y, Nomoto K. Appl Environ Microbiol. 2007;73:32–39. doi: 10.1128/AEM.01224-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu J, Moore JE, Murphy PG, Millar BC, Elborn JS. Ann Clin Microbiol Antimicrob. 2004;3:21–25. doi: 10.1186/1476-0711-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pirnay JP, De Vos D, Duinslaeger L, et al. Crit Care. 2000;4:255–261. doi: 10.1186/cc702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Darton T, Guiver M, Naylor S, Jack DL, Kaczmarski EB, Borrow R, Read RC. Clin Infect Dis. 2009;48:587–594. doi: 10.1086/596707. [DOI] [PubMed] [Google Scholar]
- 45.Rossney AS, Herra CM, Brennan GI, Morgan PM, O’Connell B. J Clin Microbiol. 2008;46:3285–3290. doi: 10.1128/JCM.02487-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rossney AS, Herra CM, Fitzgibbon MM, Morgan PM, Lawrence MJ, O’Connell B. Eur J Clin Microbiol Infect Dis. 2007;26:459–466. doi: 10.1007/s10096-007-0303-7. [DOI] [PubMed] [Google Scholar]
- 47.Palavecino E. Methods Mol Biol. 2007;391:1–19. doi: 10.1007/978-1-59745-468-1_1. [DOI] [PubMed] [Google Scholar]
- 48.Carroll KC. Mol Diagn Ther. 2008;12:15–24. doi: 10.1007/BF03256265. [DOI] [PubMed] [Google Scholar]
- 49.Stamper PD, Cai M, Howard T, Speser S, Carroll KC. J Clin Microbiol. 2007;45:2191–2196. doi: 10.1128/JCM.00552-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.MacCannell DR, Louie TJ, Gregson DB, Laverdiere M, Labbe A, Laing F, Henwick S. J Clin Microbiol. 2006;44:2147–2152. doi: 10.1128/JCM.02563-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beekmann SE, Dickema DJ, Chapin KC, Doern GV. J Clin Microbiol. 2003;41:3119–3125. doi: 10.1128/JCM.41.7.3119-3125.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peters RPH, van Agtmael MA, Danner SA, Savelkoul PHM, Vandenbroucke-Grauls CMJE. Lancet Infect Dis. 2004;4:751–760. doi: 10.1016/S1473-3099(04)01205-8. [DOI] [PubMed] [Google Scholar]
- 53.Millar BC, Xu J, Moore JE. J Clin Microbiol. 2002;40:1575–1580. doi: 10.1128/JCM.40.5.1575-1580.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klouche M, Schröder U. Clin Chem Lab Med. 2008;46:888–908. doi: 10.1515/CCLM.2008.157. [DOI] [PubMed] [Google Scholar]
- 55.Horz HP, Scheer S, Huenger F, Vianna ME, Conrads G. J Microbiol Methods. 2008;72:98–102. doi: 10.1016/j.mimet.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 56.Cai H, Archambault M, Prescott JF. J Vet Diagn Invest. 2003;15:465–469. doi: 10.1177/104063870301500511. [DOI] [PubMed] [Google Scholar]
- 57.Woo PC, Ng KH, Lau SK, et al. J Clin Microbiol. 2003;41:1996–2001. doi: 10.1128/JCM.41.5.1996-2001.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsai JC, Hsueh PR, Lin HM, Chang HJ, Ho SW, Teng LJ. J Clin Microbiol. 2005;43:235–241. doi: 10.1128/JCM.43.1.235-241.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blow N. Nature Methods. 2008;5:267–272. doi: 10.1038/nmeth0308-267. [DOI] [PubMed] [Google Scholar]
- 60.Bonetta L. Nature Methods. 2006;3:141–146. doi: 10.1038/nmeth0206-141. [DOI] [Google Scholar]
- 61.Chi KR. Nature Methods. 2008;5:11–14. doi: 10.1038/nmeth1154. [DOI] [PubMed] [Google Scholar]
- 62.Graveley BR. Nature. 2008;453:1197–1198. doi: 10.1038/4531197b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schuster SC. Nature Methods. 2008;5:16–18. doi: 10.1038/nmeth1156. [DOI] [PubMed] [Google Scholar]
- 64.Shendure J, Porreca GJ, Reppas NB, Lin X, McCutcheon JP, Rosenbaum AM, et al. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 65.Von Bubnoff A. Cell. 2008;132:721–723. doi: 10.1016/j.cell.2008.02.028. [DOI] [PubMed] [Google Scholar]
- 66.Wold B, Myers RM. Nature Methods. 2008;5:19–21. doi: 10.1038/nmeth1157. [DOI] [PubMed] [Google Scholar]
- 67.Drancourt M, Roux V, Fournier PE, Raoult D. J Clin Microbiol. 2004;42:497–504. doi: 10.1128/JCM.42.2.497-504.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Diggle MA, Clarke SC. Mol Biotechnol. 2004;28:129–137. doi: 10.1385/MB:28:2:129. [DOI] [PubMed] [Google Scholar]
- 69.Fan J, Chee MS, Gunderson KL. Nat Rev Gen. 2006;7:632–644. doi: 10.1038/nrg1901. [DOI] [PubMed] [Google Scholar]
- 70.Poland H. Nat Genet. 2006;38:513–514. doi: 10.1038/ng0506-513. [DOI] [PubMed] [Google Scholar]
- 71.Margulies M, Eghold M, et al. Nature. 2005;437:326–327. doi: 10.1038/nature03959. [DOI] [PubMed] [Google Scholar]
- 72.Meyer M, Stenzel U, Myles S, Prüfer K, Hofreiter M. Nuc Ac Res. 2007;35:e97. doi: 10.1093/nar/gkm566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jordan JA, Butchko AR, Durso MB. J Mol Diagn. 2005;7:105–110. doi: 10.1016/S1525-1578(10)60015-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.von Wintzingerode F, Bocker S, Schlotelburg C, et al. Proc Natl Acad Sci USA. 2002;99:7039–7044. doi: 10.1073/pnas.102165899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lowe CA, Diggle MA, Clarke SC. Br J Biomed Sci. 2004;61:8–10. doi: 10.1080/09674845.2004.11732638. [DOI] [PubMed] [Google Scholar]
- 76.Maier T, Klepel S, Renner U, Kostrzewa M (2006) Nature Methods 3
- 77.Van der Pol B, Quinn TC, Gaydos CA, et al. J Clin Microbiol. 2000;38:1105–1112. doi: 10.1128/jcm.38.3.1105-1112.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tortoli E, Mariottini A, Mazzarelli G. J Clin Microbiol. 2003;41:4418–4420. doi: 10.1128/JCM.41.9.4418-4420.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jardi R, Rodriguez-Frias F, Tabernero D, Homs M, Schaper M, Esteban R, Buti M. J Clin Microbiol. 2009;47:485–488. doi: 10.1128/JCM.01678-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Spinillo A, Dal Bello B, Gardella B, Roccio M, Dacco’ MD, Silini EM. Gynecol Oncol. 2009;113:115–9. doi: 10.1016/j.ygyno.2008.12.037. [DOI] [PubMed] [Google Scholar]
- 81.Yoshida S, Suzuki K, Tsuyuguchi K, Iwamoto T, Tomita M, Okada M, Sakatani M. Kekkaku. 2009;84:15–21. [PubMed] [Google Scholar]
- 82.Shah NS, Lan NT, Huyen MN, Laserson K, Iademarco MF, Binkin N, Wells C, Varma JK. Int J Tuberc Lung Dis. 2009;13:247–252. [PubMed] [Google Scholar]
- 83.Anthony RM, Brown TJ, French GL. J Clin Microbiol. 2000;38:781–788. doi: 10.1128/jcm.38.2.781-788.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu YT. Infect Disord Drug Targets. 2008;8:183–188. doi: 10.2174/1871526510808030183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mikhailovich V, Gryadunov D, Kolchinsky A, Makarov AA, Zasedatelev A. Bioessays. 2008;30:673–682. doi: 10.1002/bies.20781. [DOI] [PubMed] [Google Scholar]
- 86.Burgess ST, Kenyon F, O’Looney N, Ross AJ, Kwan MC, Beattie JS, Petrik J, Ghazal P, Campbell CJ. Anal Biochem. 2008;382:9–15. doi: 10.1016/j.ab.2008.07.017. [DOI] [PubMed] [Google Scholar]
- 87.Gaseitsiwe S, Valentini D, Mahdavifar S, Magalhaes I, Hoft DF, Zerweck J, Schutkowski M, Andersson J, Reilly M, Maeurer MJ. PLoS ONE. 2008;3:e3840. doi: 10.1371/journal.pone.0003840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gouriet F, Samson L, Delaage M, Mainardi JL, Meconi S, Drancourt M, Raoult D. Clin Microbiol Infect. 2008;14:1112–1118. doi: 10.1111/j.1469-0691.2008.02094.x. [DOI] [PubMed] [Google Scholar]
- 89.Ben RJ, Kung S, Chang FY, Lu JJ, Feng NH, Hsieh YD. J Formos Med Assoc. 2008;107:448–453. doi: 10.1016/S0929-6646(08)60152-7. [DOI] [PubMed] [Google Scholar]
- 90.Chiu CY, Urisman A, Greenhow TL, Rouskin S, Yagi S, Schnurr D, Wright C, Drew WL, Wang D, Weintrub PS, Derisi JL, Ganem D. J Pediatr. 2008;153:76–83. doi: 10.1016/j.jpeds.2007.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jung YK, Kim TW, Jung C, Cho DY, Park HG. Small. 2008;4:1778–1784. doi: 10.1002/smll.200800947. [DOI] [PubMed] [Google Scholar]
- 92.Guenther S, Groth I, Schierack P, Grabley S, Munder T. J Basic Microbiol. 2008;48:315–318. doi: 10.1002/jobm.200800038. [DOI] [PubMed] [Google Scholar]
- 93.Iqbal J, Hanel F, Ruryk A, Limmon GV, Tretiakov AM, Durst M, et al. Int J Med Microbiol. 2008;26:13–20. doi: 10.4103/0255-0857.38851. [DOI] [PubMed] [Google Scholar]
- 94.Dalstein V, Merlin S, Bali C, Saunier M, Dachez R, Ronsin C. J Virol Methods. 2009;156:77–83. doi: 10.1016/j.jviromet.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 95.Koidl C, Bozic M, Hadzisejdic I, Grahovac M, Grahovac B, Kranewitter W, Marth E, Kessler HH. Am J Obstet Gynecol. 2008;199:144e1–144e6. doi: 10.1016/j.ajog.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 96.Fluit AC, Visser MR, Schmitz FJ. Clin Microbiol Rev. 2001;14:836–871. doi: 10.1128/CMR.14.4.836-871.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Levy SB, Marshall B. Nat Med Suppl. 2004;10:122–129. doi: 10.1038/nm1145. [DOI] [PubMed] [Google Scholar]
- 98.Martinez JL, Baquero F. Clin Microbiol Rev. 2002;15:647–679. doi: 10.1128/CMR.15.4.647-679.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Baquero F, Coque TM, Canton R. ASM News. 2003;69:547–552. [Google Scholar]
- 100.Westin L, Miller C, Vollmer D, Canter D, Radtkey R, Nerenberg M, O’Connell JP. J Clin Microbiol. 2001;39:1097–1104. doi: 10.1128/JCM.39.3.1097-1104.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Monecke S, Jatzwauk L, Weber S, Slickers P, Ehricht R. Clin Microbiol Infect. 2008;14:534–545. doi: 10.1111/j.1469-0691.2008.01986.x. [DOI] [PubMed] [Google Scholar]
- 102.Weile J, Schmid RD, Bachmann TT, Susa M, Knabbe C. Diagn Microbiol Infect Dis. 2007;59:325–338. doi: 10.1016/j.diagmicrobio.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 103.Grimm V, Ezaki S, Susa M, Knabbe C, Schmid RD, Bachmann TT. J Clin Microbiol. 2004;42:3766–3774. doi: 10.1128/JCM.42.8.3766-3774.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Strommenger B, Schmidt C, Werner G, Roessle-Lorch B, Bachmann TT, Witte W. Mol Cell Probes. 2007;21:161–170. doi: 10.1016/j.mcp.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 105.van Beuningen R, van Damme H, Boender P, Bastiaensen N, Chan A, Kievits T. Clin Chem. 2001;47:1931–1933. [Google Scholar]
- 106.Anthony RM, Schuitema AR, Oskam L, Klatser PR. J Microbiol Methods. 2005;60:47–54. doi: 10.1016/j.mimet.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 107.Liu WT, Wu JH, Li ES, Selamat ES. Appl Environ Microbiol. 2005;71:6453–6457. doi: 10.1128/AEM.71.10.6453-6457.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dunbar SA, Vander Zee CA, Oliver KG, Karem KL, Jacobson JW. J Microbiol Methods. 2003;53:245–252. doi: 10.1016/S0167-7012(03)00028-9. [DOI] [PubMed] [Google Scholar]
- 109.Mahony J, Chong S, Merante F, Yaghoubian S, Sinha T, Lisle C, Janeczko R. J Clin Microbiol. 2007;45:2965–2970. doi: 10.1128/JCM.02436-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pabbaraju K, Tokaryk KL, Wong S, Fox JD. J Clin Microbiol. 2008;46:3056–3062. doi: 10.1128/JCM.00878-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Krunic N, Yager TD, Himsworth D, Merante F, Yaghoubian S, Janeczko R. J Clin Virol. 2007;40(Suppl 1):S39–S46. doi: 10.1016/S1386-6532(07)70009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Beran O, Lawrence DA, Andersen N, Dzupova O, Kalmusova J, Musilek M, Holub M (2009) Sequential analysis of biomarkers in cerebrospinal fluid and serum during invasive meningococcal disease. Eur J Clin Microbiol Infect Dis. doi:10.1007/s10096-009-0708-6 [DOI] [PMC free article] [PubMed]
- 113.Bøving MK, Pedersen LN, Møller JK. J Clin Microbiol. 2000;47:908–913. doi: 10.1128/JCM.01966-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Braeckmans K, De Smedt SC, Leblans M, Pauwels R, Demeester J. Nat Rev Drug Dis. 2002;1:447–456. doi: 10.1038/nrd817. [DOI] [PubMed] [Google Scholar]
- 115.Malamud D, Bau H, Niedbala S, Corstjens P. Adv Dent Res. 2005;18:12–16. doi: 10.1177/154407370501800104. [DOI] [PubMed] [Google Scholar]
- 116.Smoot LM, Smoot JC, Smidt H, Noble PA, Könneke M, McMurry ZA, Stahl DA. Adv Dent Res. 2005;18:6–11. doi: 10.1177/154407370501800103. [DOI] [PubMed] [Google Scholar]
- 117.Lin FY, Sabri M, Erickson D, Alirezaie J, Li D, Sherman PM. Analyst. 2004;129:823–828. doi: 10.1039/b409222h. [DOI] [PubMed] [Google Scholar]
- 118.Marsh P, Cardy DL. Methods Mol Biol. 2004;266:167–189. doi: 10.1385/1-59259-763-7:167. [DOI] [PubMed] [Google Scholar]
- 119.Anthony RM, Brown TJ, French GL. Expert Rev Mol Diagn. 2001;1:30–38. doi: 10.1586/14737159.1.1.30. [DOI] [PubMed] [Google Scholar]
- 120.Pedroso S, Guillen IA. Comb Chem High Throughput Screen. 2006;9:389–397. doi: 10.2174/138620706777452438. [DOI] [PubMed] [Google Scholar]
- 121.Gu H, Xu K, Xu C, Xu B. Chem Commun (Camb) 2006;9:941–949. doi: 10.1039/b514130c. [DOI] [PubMed] [Google Scholar]
- 122.Basu M, Seggerson S, Henshaw J, Jiang J, del A Cordona R, Lefave C, Boyle PJ, Miller A, Pugia M, Basu S. Glycoconj J. 2004;21:487–496. doi: 10.1007/s10719-004-5539-1. [DOI] [PubMed] [Google Scholar]
- 123.Prow TW, Kotov NA, Lvov YM, Rijnbrand R, Leary JF. J Mol Histol. 2004;35:555–564. doi: 10.1007/s10735-004-2196-4. [DOI] [PubMed] [Google Scholar]
- 124.Service RF. Science. 2002;298:2322–2323. doi: 10.1126/science.298.5602.2322. [DOI] [PubMed] [Google Scholar]
- 125.Tang S, Zhao J, Storhoff JJ, Norris PJ, Little RF, Yarchoan R, Stramer SL, Patno T, Domanus M, Dhar A, Mirkin CA, Hewlett IK. J Acquir Immune Defic Syndr. 2007;46:231–237. doi: 10.1097/QAI.0b013e31814a554b. [DOI] [PubMed] [Google Scholar]
- 126.Liao JC, Mastali M, Gau V, Suchard MA, Møller AK, Bruckner DA, Babbitt JT, Li Y, Gornbein J, Landaw EM, McCabe ER, Churchill BM, Haake DA. J Clin Microbiol. 2006;44:561–570. doi: 10.1128/JCM.44.2.561-570.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Schulte TH, Bardell RL, Weigl BH. Clinica Chimica Acta. 2002;321:1–10. doi: 10.1016/S0009-8981(02)00093-1. [DOI] [PubMed] [Google Scholar]
- 128.Klapperich C. J Vis Exp. 2008;12:665. doi: 10.3791/665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Myers FB, Lee LP. Lab Chip. 2008;8:2015–2031. doi: 10.1039/b812343h. [DOI] [PubMed] [Google Scholar]
- 130.Weigl B, Domingo G, Labarre P, Gerlach J. Lab Chip. 2008;8:1999–2014. doi: 10.1039/b811314a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sato K, Mawatari K, Kitamori T. Lab Chip. 2008;8:1992–1998. doi: 10.1039/b814098g. [DOI] [PubMed] [Google Scholar]
- 132.Robertson BH, Nicholson JK. Annu Rev Public Health. 2005;26:281–302. doi: 10.1146/annurev.publhealth.26.021304.144522. [DOI] [PubMed] [Google Scholar]
- 133.Srinivasan V, Pamula VK, Fair RB. Lab Chip. 2004;4:310–315. doi: 10.1039/b403341h. [DOI] [PubMed] [Google Scholar]
- 134.Herr AE, Hatch AV, Throckmorton DJ, Tran HM, Brennan JS, Giannobile WV, Singh AK. Proc Natl Acad Sci USA. 2007;104:5268–5273. doi: 10.1073/pnas.0607254104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Figueiredo Costa S. Shock. 2008;30(Suppl 1):23–29. doi: 10.1097/SHK.0b013e3181818990. [DOI] [PubMed] [Google Scholar]
- 136.Andrade SS, Bispo PJ, Gales AC. Shock. 2008;30(Suppl 1):41–46. doi: 10.1097/SHK.0b013e3181819f6c. [DOI] [PubMed] [Google Scholar]
- 137.Charles PG. Curr Opin Pulm Med. 2008;14:176–182. doi: 10.1097/MCP.0b013e3282f7642f. [DOI] [PubMed] [Google Scholar]


