Abstract
This study showed that the most of the coronaviruses (CoVs) detected in Brazilian wild birds clustered with the mouse hepatitis virus A59 strain, belonging to the BetaCoV group. Furthermore, CoV detected in two different bird species, Amazona vinacea and Brotogeris tirica, clustered with a CoV isolated from Sparrow (SpaCoV HKU17) belonging to a monophyletic group related with the CoVs isolated from swines (PorCoV HKU15), both belonging to the DeltaCoV genus, previously unreported in South America. Considering the risk of inter-species host switching and further adaptation to new hosts, detection in bird species of CoVs closely related to mammal CoVs should warn for the potential emergence of new threatening viruses.
Electronic supplementary material
The online version of this article (doi:10.1007/s00239-015-9693-9) contains supplementary material, which is available to authorized users.
Keywords: Coronavirus, Betacoronavirus, Deltacoronavirus, Wild birds, Evolution
Coronaviruses (CoVs), belonging to the order Nidovirales, family Coronaviridae, and subfamily Coronavirinae, are grouped into the AlphaCoV, BetaCoV, GammaCoV, and DeltaCoV genera (Woo et al. 2011). Alpha- and BetaCoVs infect or cause disease in mammals, while GammaCoV are almost exclusively found in birds, except for the CoV detected in beluga whale and bottlenose dolphin (Mihindukulasuriya et al. 2008; Woo et al. 2010). The fourth genus (DeltaCoV) was described recently and has already been identified in pigs and birds (Woo et al. 2011).
In this research, 368 cloacal and tracheal swabs from 33 bird species were collected in different regions belonging to the States of São Paulo (Bosque dos Jequitibás, Campinas-SP; Rio Claro-SP; Rehabilitation center [CRAS] Parque Ecológico do Tietê, São Paulo-SP) and Mato Grosso do Sul (CRAS Campo Grande-MS), Brazil. The viral RNA was extracted (QIAmp Viral RNA Mini kit, QIAGEN, Hilden, Germany), and the cDNA synthesis was performed following the manufacturer’s protocol (Applied Biosystems, Foster City, USA).
A PanCoV PCR selective essay was performed targeting a 440 bp of RNA-dependent RNA polymerase (RdRp) gene (Chu et al. 2011). Nineteen samples (5.2 %) distributed in 11 bird species were positive for CoV. The positive CoV species are as follows: Amazona vinacea, Ara ararauna, Asio clamator, Brotogeris tirica, Colaptes campestris, Columba livia, Coragyps atratus, Megascops choliba, Pitangus sulphuratus, Pyroderus scutatus, and Rupornis magnirostris (See Supplementary Table S1).
Different CoV sequences were used to represent the four CoV genera of the family Coronaviridae. Phylogenetic inference was reconstructed and implemented in FastTree 2.1.7 software (Price et al. 2010) using the Maximum-Likelihood (ML) method with the General Time-Reversible (GTR) model of nucleotide evolution with 20 rates of gamma distribution and 1.000 Shimodaira–Hasegawa-like (SH-like) values.
Our results showed that most of the samples clustered with the mouse hepatitis virus A59 strain (MHV A59) belonging to the BetaCoV group. Mice are the natural hosts of MHV A59, which is associated to hepatotropic and neurotropic diseases (Eriksson et al. 2008). Besides, two of our samples that were, respectively, detected in A. vinacea (Parrot-breasted-purple) and B. tirica (Plain Parakeet) clustered with CoV isolated from Sparrow (SpaCoV HKU17) belonging to a monophyletic group related with the CoVs isolated from swines (PorCoV HKU15), both belonging to the DeltaCoV genus (Fig. 1), previously unreported in South America. PorCoV HKU15 was detected in April 2014 in the USA (Wang et al. 2014), in 2015, which was directly associated to play a key role in diarrheic disease in swine; no treatment or vaccines are currently available (Hu et al. 2015; Ma et al. 2015).
Fig. 1.
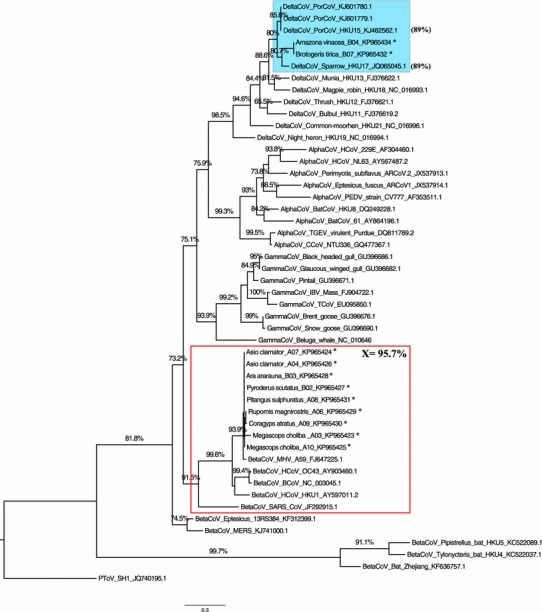
Molecular phylogenetic inference with Alpha-, Beta-, Gamma-, and DeltaCoV groups by ML method under GTR evolutionary model for the partial RdRp gene (267 bp). The colors represent CoVs detected in wild birds grouped with BetaCoV (red) and DeltaCoV (highlighted in blue) groups. The asterisk represents the bird samples from which the DNA sequences were obtained. The number within parenthesis shows the nucleotide identities of PorCoV HKU15 and SpaCoV HKU17 (both DeltaCoVs) with the CoV detected in Amazona vinacea. The letter X represents the arithmetic mean of nucleotide identities between the CoVs detected in the study and the MHV A59. The GenBank accession numbers of sequences deposited (from study) and retrieved from the database are shown in the tree. PorCoV porcine coronavirus; HCoV human coronavirus; PEDV porcine epidemic diarrhea virus; BatCoV bat coronavirus; TGEV transmissible gastroenteritis coronavirus; CCoV canine coronavirus; IBV infectious bronchitis virus; TCoV turkey coronavirus; MHV mouse hepatitis virus; BCoV bovine coronavirus; SARS severe acute respiratory syndrome; MERS middle east respiratory syndrome coronavirus; PToV (outgroup) porcine torovirus. The numbers above the branches represent the SH-like support values of 1000 replicates. Only values greater than or equal to 70 % are presented. Online version in color
Finally, we highlighted in this study the evolutionary proximity of CoVs detected in wild birds with Beta- and DeltaCoVs, both isolated from mammal and involved in the pathogenicity of their hosts. We believe that these findings may be useful for the understanding of CoVs’ evolutionary dynamics. Considering the risk of inter-species host switching and further adaptation to new hosts, detection in bird species of CoVs closely related to mammal CoVs should warn for the potential emergence of new threatening viruses.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank the teams from Bosque dos Jequitibás; Rio Claro-SP; CRAS Parque Ecológico do Tietê-SP; and Campo Grande-MS. This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), Grants 2013/03922-6 and 2011/50919-5.
References
- Chu DKW, Leung CYH, Gilbert M, Joyner PH, Ng EM, Tse TM, Guan Y, Peiris JS, Poon LL. Avian coronavirus in wild aquatic birds. J Virol. 2011;85:12815–12820. doi: 10.1128/JVI.05838-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson KK, Cervantes-Barraga L, Ludewig B, Thiel V. Mouse hepatitis virus liver pathology is dependent on ADP-ribose-1-phosphatase, a viral function conserved in the alpha-like supergroup. J Virol. 2008;82:12325–12334. doi: 10.1128/JVI.02082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Jung K, Vlasova AN, Chepngeno J, Lu Z, Wang Q, Saif LJ. Isolation and characterization of porcine deltacoronavirus from pigs with diarrhea in the United States. J Clin Microbiol. 2015;53:1537–1548. doi: 10.1128/JCM.00031-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Zhang Y, Liang X, Lou F, Oglesbee M, Krakowka S, Li J. Origin, evolution, and virulence of porcine deltacoronaviruses in the United States. mBio. 2015;6:e00064-15. doi: 10.1128/mBio.00064-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihindukulasuriya KA, Wu G, St Leger J, Nordhausen RW, Wang D. Identification of a novel coronavirus from a beluga whale by using a panviral microarray. J Virol. 2008;82:5084–5088. doi: 10.1128/JVI.02722-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Byrum B, Zhang Y. Porcine coronavirus HKU15 detected in 9 US States, 2014. Emerg Infect Dis. 2014;20:1594–1595. doi: 10.3201/eid2009.140756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo PCY, Huang Y, Lau SKP, Yuen KY. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2:1804–1820. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo PCY, Lau SKP, Lam CSF, Lau CCY, Tsang AKL, Lau JHN, Bai R, Teng JL, Tsang CC, Wang M, Zheng BJ, Chan KH, Yuen KY. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. 2011;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


