Abstract
With the rapid development of molecular diagnostic techniques, there is a growing need for quality controls and standards with favorable properties to monitor the entire detection process. In this study, we describe a novel method to produce armored hepatitis B virus (HBV) and human papillomavirus (HPV) DNA for use in nucleic acid tests, which was confirmed to be stable, homogeneous, noninfectious, nuclease resistant, and safe for shipping. We demonstrated that MS2 bacteriophage could successfully package double-stranded DNA of 1.3-, 3-, 3.5-, and 6.5-kb length into viral capsids with high reassembly efficiency. This is the first application of RNA bacteriophage MS2 as a platform to encapsulate double-stranded DNA, forming virus-like particles (VLPs) which were indistinguishable from native MS2 capsids in size and morphology. Moreover, by analyzing the interaction mechanism of pac site and the MS2 coat protein (CP), we found that in addition to the recognized initiation signal TR-RNA, TR-DNA can also trigger spontaneous reassembly of CP dimers, providing a more convenient and feasible method of assembly. In conclusion, this straightforward and reliable manufacturing approach makes armored DNA an ideal control and standard for use in clinical laboratory tests and diagnostics, possessing prospects for broad application, especially providing a new platform for the production of quality controls for DNA viruses.
Electronic supplementary material
The online version of this article (doi:10.1007/s00253-015-6664-4) contains supplementary material, which is available to authorized users.
Keywords: Armored double-stranded DNA, MS2 bacteriophage, Encapsulation, Virus-like particles, Quality controls
Introduction
In recent years, there has been an increase in the detection methods for quantification of nucleic acids (Datta et al. 2014; Wang et al. 2014b), especially the use of real-time PCR assays (Cai et al. 2014; dos Santos Ade et al. 2014; Wang et al. 2014a) that provide an easy and reliable routine for clinical laboratory tests. Possessing an outstanding quality control or standard is important to ensure precise results and to exclude inaccurate outcomes.
Commonly used types of controls or standards for DNA virus assays include naked plasmid DNA, virus-positive patient specimens, specific cell lines, and armored DNA. Naked plasmid DNA contains the full virus genome, such as HBV (Gotsch et al. 2007; Stocher and Berg 2004) or HPV (Kuebler et al. 2007), and is often used to create amplification standards that allow direct comparisons between laboratories. Plasmid-derived controls are easy to produce, noninfectious to humans, and suitable for transport, but they are likely to degrade owing to the bare, unprotected DNA that is incorporated. Additionally, plasmid DNA cannot serve as a control for the whole real-time PCR process because nucleic acid extraction is not performed. Another alternative is positive patient specimen, a natural clinical sample that does allow for control of the entire detection process. However, these samples need to be screened to exclude hemolysis, lipoidemia, jaundice, or blood-borne pathogens, such as human immune deficiency virus (HIV), hepatitis C virus (HCV), or Treponema pallidum. Further, this type of control is time and labor consuming to prepare, infectious, and it is hard to manufacture and ship. Moreover, the viral load can be significantly reduced after repeated freezing and thawing. As an example, HPV controls from liquid-based cytology (LBC) (Fagan et al. 2010) samples of patients are difficult to obtain in large quantities, and often contain multiple infections with different HPV genotypes at low concentrations. Cervical cancer cell lines, another commonly used HPV controls, are hard to guarantee the uniformity of samples due to the effect of the sedimentation of the cells in the process of sample packing. In addition, obtaining cell lines other than HPV-16 and HPV-18 genomic DNA included is often infeasible. Armored DNA prepared from bacteriophage is yet another choice. At present, there are three kinds of DNA phages that have been applied to the manufacture of DNA controls and standards: filamentous phage (Gotsch et al. 2007), T7 bacteriophage (Masker et al. 1978), and lambda phage (Stocher and Berg 2004). Filamentous phage is not a proper choice due to its single-stranded structure that does not mimic the characteristics (i.e., secondary structure) (Gotsch et al. 2007) of double-stranded DNA which is important in the early stages of real-time PCR assay. T7 bacteriophage is generally not chosen because of its relatively lower packaging efficiency. Armored DNA produced from lambda phage is less stable in plasma than other phages; so, it cannot be widely used (Walkerpeach and Pasloske 2004). Based on these issues, there is a serious need to construct better controls and standards for viral assays to overcome the limitations of these existing ones.
In this study, we took advantage of MS2, a single-stranded RNA bacteriophage, to prepare armored DNA in a novel way. This new construct is superior to the earlier DNA phages in terms of its structural features and application for detection. Historically, MS2 bacteriophage has been used extensively to produce armored RNA as controls or standards for RNA viruses, such as HIV (Pasloske et al. 1998), HCV (Konnick et al. 2005), severe acute respiratory syndrome coronavirus (SARS-CoV) (Bressler and Nolte 2004), avian influenza virus H7N9 (Sun et al. 2013), and long chimeric RNA sequences (Wei et al. 2008b), because they are nuclease-resistant and stable. These studies were based on cloning the gene sequences of coat protein (CP) and the conserved exogenous RNA virus into a vector for expression in E. coli or Saccharomyces cerevisiae. Virus-like particles (VLPs) self-assemble from 180 sequence identical monomers (~14 kD) into a ~28-nm diameter icosahedral protein capsid, through the interaction between CP and the pac site. Armored DNA in our research was prepared in a different way in vitro. We dissociated MS2 into CP dimers first, and then reassembled them into VLPs at the presence of pac site under specific conditions, making use of the same reassembly mechanism as traditional approaches. Furthermore, the size and morphology of MS2 capsids before and after dissociation have no difference. This similarity is important, as the interior surface modification of MS2 (Obermeyer et al. 2014; Wu et al. 2009) based on the technique has been widely used, achieving great achievements such as drug-targeted delivery for cancer treatments (Ashley et al. 2011; Wu et al. 1995), development of MRI contrast agents for imaging (Datta et al. 2008; Garimella et al. 2011), and encapsulation of various molecules such as antisense oligonucleotides (Wu et al. 2005), enzymes (Glasgow et al. 2012), organic fluorophores (Capehart et al. 2013), quantum dots, doxorubicin, and siRNAs (Ashley et al. 2011).
In this article, we constructed armored DNA with improved properties using MS2 capsids packaging 1.3-, 3-, 3.5-, and 6.5-kb HBV or HPV DNA sequences, which are the typical representatives of various types of controls for DNA viruses. This is the first successful encapsulation of double-stranded DNA by RNA bacteriophage. These constructs can be used as excellent controls or standards in routine clinical laboratory tests and diagnosis.
Materials and methods
Expression, purification, and disassembly of the MS2 bacteriophage
Primers MCF (5′-CGGGATCCTGGCTATCGCTGTAGGTAGCC-3′) and MCR (5′-CCCAAGCTTATGGCCGGCGTCTATTAGTAG-3′) were used to amplify the sequences of MS2 CP and maturase genes from the pMS27 plasmid (Peabody 1997), which was kindly provided by D. S. Peabody, containing nucleotides (nt) 81–1749 of the MS2 phage (GeneBank accession no. V00642). The underlined sequences are BamHI and HindIII restriction enzyme sites, respectively. PCR products of 1.7 kb were gel purified, digested with BamHI and HindIII, and ligated to the vector pACYCDuet-1 (Fig. S1) to generate the recombinant plasmid. The inserted sequences were identified by sequencing and blast searching in the NCBI databases. It is worth mentioning that the second multiple cloning site of the vector contained neither exogenous sequences nor the pac site. The plasmid was transformed into E. coli strain BL21 (DE3), and then, MS2 phages were expressed by adding 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to the bacterial cultures and incubation for 12 h. The cells were harvested by centrifugation and lysed by sonication. The MS2 VLPs were purified by Sephacryl S-200 gel exclusion chromatography (BioLogic DuoFlow chromatography system, BIO RAD; USA) after the supernatant was incubated with RNase A and DNase I at 37 °C for 30 min to eliminate the E. coli genome. The dissociation method we used was performed according to Wu et al. (1995). Briefly, 1 mL of the MS2 phage (4 mg mL−1) was incubated with 2-mL cold glacial acetic acid on ice for 30 min, then centrifuged at 6600×g for 20 min at 4 °C to remove precipitated maturase and nucleic acid. The supernatant was dialyzed against 1 mM acetic acid using dialysis tubing closures (7 kDa molecular weight cutoffs [MWCO]), with buffer changed three times every 4 h. The protein solution was then applied to Superdex 75 Prep Grade (GE Healthcare; USA) on BioLogic DuoFlow chromatography system using 50 mM Tris with 100 mM NaCl. Fractions which were shown to be the dimers by SDS-PAGE were collected, concentrated, and kept on ice for use at the same day.
Overlapping extension PCR for viral DNA sequences
We amplified the most conserved regions of HBV, HPV-16, and HPV-18. The target sequence of HBV (GeneBank accession no. NC_003977) was 1350-bp long, including the C (nt 1211–2440, 630 bp) and S (nt 121–840, 720 bp) regions, which were amplified from a national HBV DNA serum standard (GBW09150) by overlapping extension PCR (Fig. S2). Using the same method, we overlapped the E6 (nt 26–820, 795 bp), L1 (nt 6151–7069, 919 bp), and E1 regions (nt 1103–2807, 1705 bp) of HPV-16 (GeneBank accession no. K0718), forming a target sequence of 3.5 kb in length. Similarly, we overlapped the E6 (nt 101–1342, 1283 bp), L1 (nt 6265–71246, 860 bp), and E1 regions (nt 2047–2987, 941 bp) of HPV-18 (GeneBank accession no. X05015), forming a target sequence of 3 kb in length. The PCR templates were the plasmids that were purchased from the American Type Culture Collection (Manassas, VA, USA), containing the full genome of virus, e.g., HPV-16 (ATCC No. 45113) and HPV-18 (ATCC No. 45152). Finally, we overlapped the 3.5-kb HPV-16 and 3-kb HPV-18 DNA sequences to form a 6.5-kb HPV sequence. The HPV primer sets, 18-E6F and 168 over-E6F, and 168 over-E1R and HPV 16-E1R, were used in the first round of overlapping extension PCR. In the second round, HPV 18-E6F and HPV 16-E1R were used for a final extension to obtain the 6.5-kb target sequence. The primers are listed in Table S1. For the C and S regions of HBV, E6, L1, and E1 regions of HPV-16 and HPV-18, PCR conditions were the same. The reaction was performed in a 50-μl volume, containing primer star (2*mix, TaKaRa; Japan) 25 μl, forward and reverse primer 1 μl, respectively, template 1 μl, and double distilled water 22 μl, at 94 °C for 3 min; 35 cycles of 10 s at 98 °C, 10 s at 55 °C, and 15 s at 72 °C; and 10 min at 72 °C. For the sequences of 1.3, 3, 3.5, and 6.5 kb in length, PCR conditions were the same by using one step overlapping extension method. The reaction was performed in a 50-μl volume, containing primer star (2*mix) 25 μl, corresponding forward and reverse primer 1 μl, respectively, corresponding templates 2 μl, respectively, and double distilled water 19 μl, at 94 °C for 3 min; 35 cycles of 10 s at 98 °C, 15 s at 55 °C, and 60 s at 72 °C; and 10 min at 72 °C. All amplified sequences were confirmed by sequencing and blast searching in the NCBI databases to ensure the correctness.
Modification of viral DNA sequences with pac site
To prepare DNA-pac site conjugates, the amine modified pac site (TR-RNA and TR-DNA, both included, synthesized by Integrated DNA technologies; USA) dissolved in DNase and RNase free water, adding a 50-fold molar excess of sulfosuccinimidyl-4-(N-maleimidomethyl)cyclohexane-1-carboxylate (Sulfo-SMCC, Thermo Fisher Scientific; USA), which is an amine-to-sulfhydryl crosslinker, for 2 h at 4 °C. A centrifugal filter device (3 kDa MWCO, Thermo Fisher Scientific; USA) was used to remove excess crosslinkers. Sulfhydryl-modified primers were purchased to amplify the DNA sequences which had been purified from gel and diluted, obtaining the PCR products of corresponding lengths. To avoid the sulfhydryl being oxidized, the DNA products were reduced by exposure to 5 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP, Thermo Fisher Scientific; USA) for 2 h at 4 °C and desalted using a polyacrylamide desalting column (50 kDa MWCO, Thermo Fisher Scientific; USA). The sulfhydryl-modified DNA products, involving 1.3 kb HBV DNA, 3.5 kb HPV-16 DNA, 3 kb HPV-18 DNA, and 6.5 kb HPV DNA, were immediately incubated with a 100-fold molar excess of the SMCC-activated pac site for 2 h at 4 °C. Additionally, a small amount of human placenta DNA (Roche; Cat No. 11691112001) was added to the three kinds of HPV DNA in advance to mimic human placenta DNA background. Unreacted pac sites were removed by a centrifugal filter device (50 kDa MWCO), and two forms of DNA-pac site conjugates (eight in total) were resuspended in 50 mM Tris with 100 mM NaCl.
Encapsulation of various DNA sequences within MS2 viral capsids
DNA-pac site conjugates (about 10 μM) were incubated with a 10-fold molar excess of disassembled MS2 CP for 3 h at room temperature, then at 4 °C for 40–48 h. The solution was then centrifuged at 8000×g for 5 min to retrieve supernatant, and then, PEG 6000 (10 % w/v) and NaCl (1 M) were added. The solution was set on a magnetic stirrer at 4 °C for 2–4 h to fully dissolve, and then centrifuged at 10,000×g for 30 min. The pellet was resuspended in 50 mM Tris with 100 mM NaCl before incubating with RNase A and DNase I at 37 °C for 2 h to eliminate the nucleic acids that were not encapsulated in the capsids.
Determination of the capsid reassembly
First, the reassembly efficiency of MS2 capsids was determined by size exclusion chromatography (SEC) on a Bio-Gel A-1.5-m gel column using a rate of 0.5 mL min−1. Reassembled capsids were then concentrated and analyzed by transmission electron microscopy (TEM) and dynamic light scattering (DLS). Samples for TEM images were absorbed on formvar and carbon-coated grids for 1 min and then stained with 1 % (w/v) phosphotungstic acid (pH 6.8) for 1 min. Grids were air-dried and observed under a Tecnai 12 transmission electron microscope (FEI, Eindhoven, Netherlands). DLS measurements were carried out using an ALV/DLS/SLS5022F light-scattering apparatus that was equipped with a 22-mW He-Ne laser (632.8-nm wavelength). The temperature was controlled at 25 °C ± 0.5 °C. To prepare dust-free solutions for measurements, the solutions were filtered through a 0.22-μm membrane of a hydrophilic PVDF filter into cells. The cells were then placed within a refractive index matching bath of filtered toluene during the measurements. The scattering angle was 90°. Finally, after incubating with RNase A and DNase I at 37 °C for 12 h, the concentrated intact capsids were assayed via native agarose gel electrophoresis and then stained with Coomassie Brilliant Blue R-250 (routine method) to enable visualization of the protein content.
PCR determination of the length of the encapsulated DNA
DNA was extracted from two kinds of purified VLPs (TR-RNA and TR-DNA as the initiation signals), using QIAamp Viral Mini Kit (Qiagen, Germany). The amplified PCR products were analyzed by agarose gel electrophoresis. In every round of PCR, a positive control was involved, whose template was the plasmid containing the corresponding length of DNA sequences (1.3, 3, 3.5, and 6.5 kb).
Identification of armored HBV DNA by real-time PCR
The armored HBV DNA was diluted with newborn calf serum by 10-fold serial dilutions to obtain 10–105-fold diluted samples. The WHO international standard for HBV DNA (National Institute Biological Standards and Control [NIBSC], 97/746) was used for quantification. Six different and available commercial kits (Shanghai Kehua, Shanghai Haoyuan, Shanghai Zhijiang, Hunan Shengxiang, Shenzhen Qiagen, and Shenzhen Daan; China) were used in the real-time PCR assay. The results were recorded and compared. Then, the five dilutions (5 × 107, 5 × 106, 5 × 105, 5 × 104, and 5 × 103 IU mL−1) were quantified to perform a linear analysis to show the relationship between the concentrations and Ct values. Triplicate samples and negative controls (newborn calf serum) for each dilution were evaluated during the run, and the values were recorded and averaged.
Identification of armored HPV DNA by real-time PCR and other assays
The armored HPV DNA (3, 3.5, and 6.5 kb) were 10-fold serially diluted to obtain 10–105-fold diluted samples. At first, the samples were identified by Hybrid Capture 2 (HC2) High-Risk HPV DNA Kit (Qiagen, Germany), which has been regarded as a leader in HPV DNA detection in the past 10 years and is still most commonly used. According to specifications, the sample is considered positive when the ratio of relative light units/cutoff (RLU/CO) is above 1. All the diluted samples were detected by PCR-reverse dot hybridization (PCR-RDB)-based HPV genotyping assay, using the commercial kits HybriBio (Chaozhou, China) and Yaneng (Shenzhen, China), which occupied a leading position in proficiency test for HPV genotyping of China in 2013 (Zhang et al. 2013). At last, four different and available commercial kits (Shanghai Kehua, Shanghai Zhijiang, Shenzhen Qiagen, and Zhongshan Daan; China) were applied to real-time PCR. Triplicate samples and negative controls for each dilution were evaluated during the run.
Stability of the armored DNA
After the armored HBV DNA was quantified, it was diluted with newborn calf serum to yield 1000 IU mL−1 and 200,000 IU mL−1 in 0.5 mL for each aliquot of an individual time-point sample. The samples were then incubated at 4 °C, 37 °C, and room temperature, and then removed and stored at each time point at −80 °C until all of the samples were collected. They were quantified, in duplicate, by real-time PCR, with newborn calf serum used as a negative control in each run.
The armored HPV DNA was examined for its stability in phosphate buffer. Aliquots of 0.5 mL were prepared in 102- and 105-fold dilutions to obtain two concentration types for each sample (9.6 × 106 and 1.1 × 104 IU mL−1 for armored HPV-18 DNA, 6.6 × 106 and 5.8 × 103 IU mL−1for armored HPV-16 DNA, and 3.2 × 106 and 3.0 × 103 IU mL−1 for armored HPV DNA of 6.5 kb, quantified by the international standards for HPV-16 (NIBSC code: 06/202, UK) and HPV-18 (NIBSC code: 06/206, UK). The following steps were the same as that of armored HBV DNA, with double distilled water was used as a negative control in each run. In addition, all armored DNA samples were frozen at −20 °C and thawed to room temperature five times, and then quantified, in duplicate, using the same method.
Results
TR-RNA or TR-DNA-driven reassembly of MS2 CP enables encapsulation of DNA sequences of various lengths within capsids
Our goal was to encapsulate virus double-stranded DNA into MS2 capsids by covalent coupling to the MS2 assembly-initiation signal, namely pac site—TR-RNA or TR-DNA. An appropriate crosslinker was chosen to conjugate the DNA products to pac site, which was modified with a 3′ amine moiety. We used sulfhydryl-modified primers to amplify gel purified and diluted DNA sequences, obtaining the corresponding length products (1.3 kb HBV DNA, 3.5 kb HPV-16 DNA, 3 kb HPV-18 DNA, and 6.5 kb HPV DNA) by means of PCR for multiple rounds. The molar ratio of DNA products to pac site was determined to be 1:100, and then, the conjugates were added to the 10-fold molar excess of disassembled MS2 CP to give rise to the self-assembly of capsids, as outlined in Fig. 1a. At last, the mixed solution was purified and concentrated to obtain the MS2 capsids that encapsulated double-stranded DNA of various lengths.
Fig. 1.
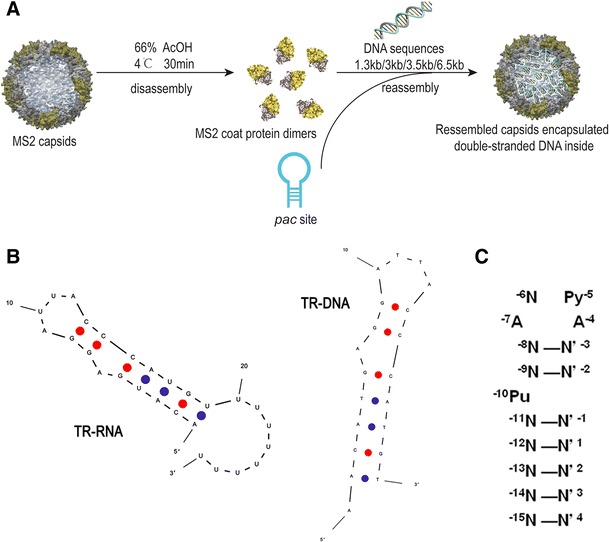
Overall strategy for construction of armored DNA by MS2 bacteriophage. a Wild-type MS2 was disassembled into CP dimers when adding into acetic acid. By mixing with DNA-pac site conjugates for incubation after centrifugation to remove maturase and genome RNA, reassembled capsids were formed. b Two forms of pac site used in the study, TR-RNA and TR-DNA. They were hairpin structures and can both trigger reassembly of MS2 CP. In particular, TR-RNA used also had a 3′ extension sequence of eight uridines to ensure that the encapsulated DNA sequences did not interfere with pac site-CP recognition. c The stem-loop structure of pac site that bind to MS2 CP. Operator consensus sequence where Py = pyramiding, Pu = purine and N–N′ represents any Watson-Crick base pair (Rowsell et al. 1998)
Determination of the capsid reassembly
Incubation of MS2 CP dimers with the 28 nt TR-RNA or the 20 nt TR-DNA (Fig. 1b) induced reassembly of the capsids. As shown in Fig. 2a, the intact MS2 VLPs started to elute at about the 55–60th min and the unreacted dimers at about the 110–120th min when using a rate of 0.5 mL min−1. The reassembly efficiency of the capsids triggered by pac site of different concentrations was recorded (Fig. 2b). Compared to TR-DNA, TR-RNA induced the maximum reassembly efficiency at an extremely low concentration, but both decreased in output at higher levels. Wild-type MS2 capsids had a diameter of 28.2 ± 0.4 nm by DLS (Fig. 2d), while the reassembled capsids were found to have diameters of 27.1 ± 0.3 nm, 27.6 ± 0.5 nm, 28.1 ± 0.9 nm, and 29.4 ± 0.6 nm for 1.3 kb HBV DNA, 3.5 kb HPV-16 DNA, 3 kb HPV-18 DNA, and 6.5 kb HPV DNA inside, respectively. Along with the TEM images (Fig. 2c) for VLPs packaging double-stranded DNA of different lengths, there was little difference in size and morphology among them. SDS-PAGE of the eight reassembled VLPs showed a single band of about 14 kD, compared with the wild-type MS2 that also contained miscellaneous proteins of about 55 kD–70 kD (Fig. 3c). Therefore, all the experimental results illustrated successful reassembly.
Fig. 2.
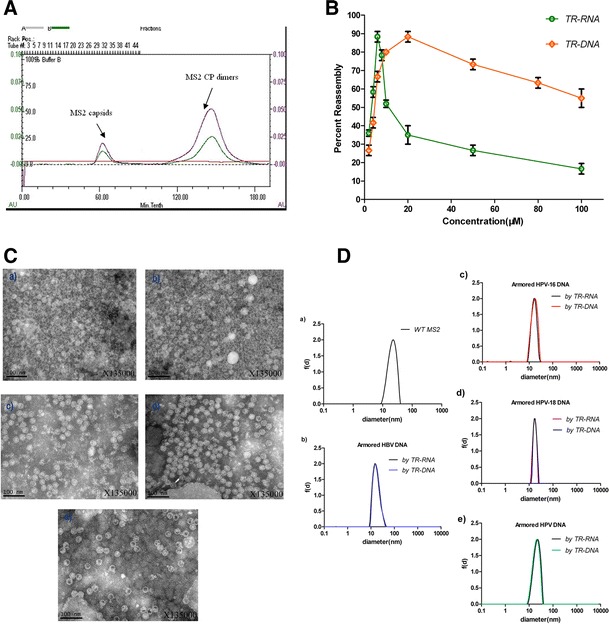
Identification of the capsid reassembly. a SEC traces of reassembled MS2 capsids and CP dimers. b Reassembly efficiency of MS2 capsids with increasing concentrations of TR-RNA or TR-DNA. c TEM images of a wild-type MS2, and reassembled MS2 capsids encapsulating b HBV DNA sequences of 1.3kb, c HPV-16 DNA sequences of 3.5 kb, d HPV-18 DNA sequences of 3 kb, and e HPV DNA sequences of 6.5 kb. d DLS results of the wild-type MS2 and reassembled capsids whose reassembly was induced by TR-RNA or TR-DNA, in comparison
Fig. 3.
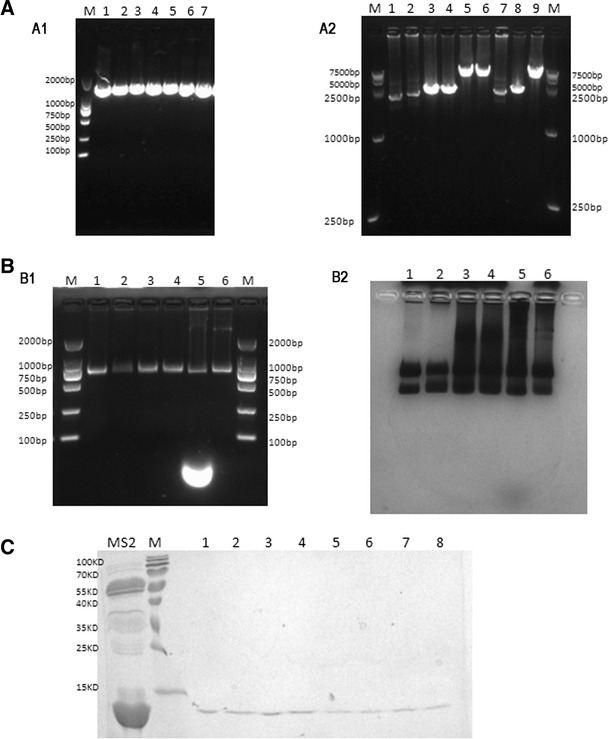
Identification of the armored DNA. a PCR products of armored HBV DNA were analyzed on agarose gel electrophoresis. Lane M DNA marker; lanes1–3 PCR templates were the nucleic acids extracted from armored HBV DNA of 104, 105, and 106-fold dilutions, respectively, whose reassembly was triggered by TR-RNA; lanes 4–6 PCR templates were the nucleic acids extracted from armored HBV DNA of 104, 105, and 106-fold dilutions, respectively, whose reassembly was triggered by TR-DNA; lane 7 positive control—the template used was the plasmid containing HBV DNA sequences of 1.3 kb. a2 PCR products of armored HPV DNA were analyzed on agarose gel electrophoresis. Lane M DNA marker; lanes 1–2 PCR templates were the nucleic acids extracted from armored HPV-18 DNA of 106-fold dilution, whose reassembly was triggered by TR-RNA and TR-DNA, respectively; lanes 3–4 PCR templates were the nucleic acids extracted from armored HPV-16 DNA of 106-fold dilution, whose reassembly was triggered by TR-RNA and TR-DNA, respectively; lanes 5–6 PCR templates were the nucleic acids extracted from armored HPV DNA of 106-fold dilution, whose reassembly was triggered by TR-RNA and TR-DNA, respectively; lanes 7–9 positive controls—the templates used were plasmids containing 3.5-kb HPV-16 DNA sequences, 3-kb HPV-18 DNA sequences, and 6.5-kb HPV DNA sequences, respectively. b1 Results of native agarose gel electrophoresis for purified MS2 capsids. Lane M DNA marker; lanes 1–4 armored HBV DNA, armored HPV-18 DNA, armored HPV-16 DNA, and armored HPV DNA, respectively, incubated with RNase A and DNase I at 37 °C for 12 h; lane 5 wild-type MS2 without digestion; lane 6 wild-type MS2 incubated with RNase A and DNase I at 37 °C for 12 h. b2 The gel was then stained with Coomassie Brilliant Blue R250 for visualization of the protein contents at the same position. c SDS-PAGE results for reassembled MS2 capsid and wild-type MS2. Lanes 1, 3, 5, and 7 MS2 VLPs containing 1.3-kb HBV DNA sequences, 3.5-kb HPV-16 DNA sequences, 3-kb HPV-18 DNA sequences, and 6.5-kb HPV DNA sequences, respectively, whose reassembly was triggered by TR-RNA; lanes 2, 4, 6, and 8 MS2 capsid containing 1.3-kb HBV DNA sequences, 3.5-kb HPV-16 DNA sequences, 3-kb HPV-18 DNA sequences, and 6.5-kb HPV DNA sequences, respectively, whose reassembly was triggered by TR-DNA; Lane MS2 wild-type MS2 before disassembly, whose band was not specific
Identification of DNA sequences inside the MS2 capsids by various assays
The PCR amplification products of the nucleic acid extracted from purified VLPs were all of the correct length, i.e., 1.3, 3, 3.5, and 6.5 kb (Fig. 3a). All the VLPs encapsulating DNA of different lengths showed the same electrophoretic mobility on the agarose gel, including the wild-type MS2, and the protein content could be observed at the same position when the gel was stained with Coomassie Brilliant Blue R250 (Fig. 3b). These indicated that DNA sequences of different lengths were all successfully packaged into the MS2 capsids.
The performance of armored HBV DNA as a calibrator for real-time PCR assays was evaluated. The quantitative results of six available commercial kits for the five samples by 10-fold serial dilutions were compared, and the concentrations of 103-fold diluted sample were 5.02 × 106, 5.14 × 106, 5.08 × 106, 5.07 × 106, 5.15 × 106, and 5.10 × 106 IU mL−1 for the six kits. As shown in Fig. 4a, the relationship between the concentrations and Ct values of the five dilutions was defined, and the r 2 value (0.9996) was calculated by linear regression analysis (Fig. 4e). This suggests that armored HBV DNA can function as a calibration standard or control for the clinical quantification of HBV loads.
Fig. 4.
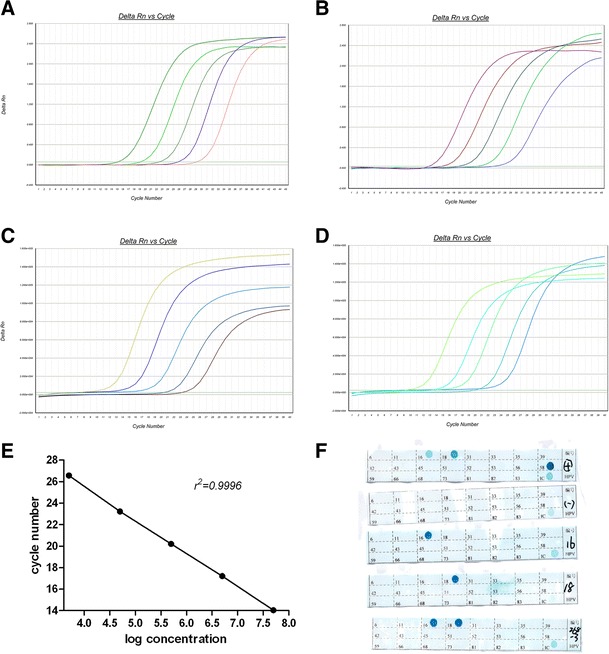
a–d Amplification curves of five serial 10-fold diluted samples for armored HBV DNA, HPV-16 DNA, HPV-18 DNA, and HPV DNA, respectively, by real-time PCR, using the commercial kit of Hunan Shengxiang. e Linear analysis for armored HBV DNA of serial 10-fold dilutions (5 × 107, 5 × 106, 5 × 105, 5 × 104, 5 × 103 IU mL−1) in newborn calf serum by real-time PCR, calibrated by the WHO international standard for HBV DNA. f The results for PCR-RDB assay by Yaneng commercial kit, with 16, 18, and 268 representing the 106-fold-diluted samples of armored HPV-16 DNA, HPV-18 DNA, and HPV DNA, respectively. The appearance of blue dots indicated that the samples contained DNA of the corresponding HPV genotype. Positive and negative controls of the kit monitored the whole experimental process, as well as the IC (internal control)
The results for HC2 of armored HPV DNA by 10-fold serial dilutions are shown in Table 1. The results for PCR-RDB assay with 103-fold diluted samples by the Yaneng kit are shown in Fig. 4f. Ct values of two adjacent dilution degree samples of armored HPV DNA were about 3.2, verified by real-time PCR (Fig. 4b, d). The outcomes above suggested that all samples were determined positive by commonly used commercial assays and kits. Thus, armored HPV DNA shows promise as a control in clinical tests.
Table 1.
The ratio RLU/CO of armored HPV DNA for HC2 assay
| Sample ID | 1a | 2a | 3a | 4a | 5a |
|---|---|---|---|---|---|
| armored HPV-16 DNA | 1191.95 | 317.71 | 151.27 | 39.42 | 4.35 |
| armored HPV-18 DNA | 842.04 | 134.13 | 59.48 | 16.15 | 3.58 |
| armored HPV DNA | 1041.37 | 286.23 | 116.91 | 33.75 | 5.38 |
aFive samples by 10-fold serial dilution
Stability of the armored DNA
The armored HBV DNA of high and low concentrations in newborn calf serum was stable at 4 °C for at least 6 months, and at 37 °C and room temperature for 2 months. The average value for the high concentration samples at 4 °C was 194,984 IU mL−1 (5.29 log10; range, 178,006–201,977 IU mL−1), and the coefficient of variation (CV) was 8.1 %. The average value for the low concentration samples at 4 °C was 871 IU mL−1 (2.94 log10; range, 720–1016 IU mL−1), and the CV was 13.2 % (Fig. 5a).
Fig. 5.
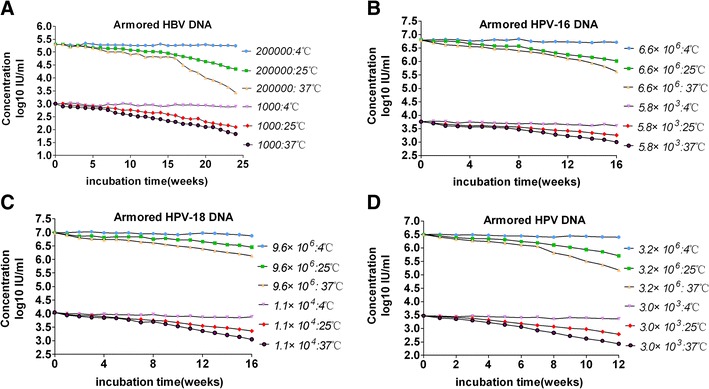
Stability of the armored DNA. a Armored HBV DNA in newborn calf serum was stable at 4 °C for at least 6 months. b–c Armored HPV-16 DNA and HPV-18 DNA in phosphate buffer were stable at 4 °C for at least 4 months. d Armored HPV DNA in phosphate buffer was stable at 4 °C for at least 3 months
The armored HPV-16 DNA of two concentrations in phosphate buffer was stable at 4 °C for at least 4 months, and at 37 °C and room temperature for 2 months. The average value for the high concentration samples at 4 °C was 6.02 × 106 IU mL−1 (6.78 log10; range, 5,867,620–6,604,356 IU mL−1), and the CV was 9.3 %. The average value for the low concentration samples at 4 °C was 5.13 × 103 IU mL−1 (3.71 log10; range, 4900–5920 IU mL−1), and the CV was 11.6 % (Fig. 5b).
The armored HPV-18 DNA of two concentrations in phosphate buffer was stable at 4 °C for at least 4 months, and at 37 °C and room temperature for 2 months. The average value for the high concentration samples at 4 °C was 9.12 × 106 IU mL−1 (6.96 log10; range, 8,830,210–9,540,620 IU mL−1), and the CV was 16.3 %. The average value for the low concentration samples at 4 °C was 9.12 × 103 IU mL−1 (3.96 log10; range, 8600–10,920 IU mL-1), and the CV was 19.6 % (Fig. 5c).
The armored HPV DNA of two concentrations in phosphate buffer was stable at 4 °C for at least 3 months, and at 37 °C and room temperature for 1 month. The average value for the high concentration samples at 4 °C was 2.82 × 106 IU mL−1 (6.45log10; range, 2,616,341–3,205,769 IU mL−1), and the CV was 13.8 %. The average value for the low concentration samples at 4 °C was 2.57 × 103 IU mL−1 (3.41 log10; range, 2,386–2,988 IU mL−1), and the CV was 21.5 % (Fig. 5d). Additionally, all the armored DNA samples were stable after being frozen and thawed five times (data not shown).
Discussion
In this work, we encapsulated double-stranded DNA of different lengths into MS2 capsids by covalent coupling them to initiation signals, TR-RNA, or TR-DNA. Both of these signals can induce reassembly of MS2 CP dimers and attain a maximum efficiency of 90 %, but the concentration of TR-RNA used in the experiment was much lower. For the DNA fragments of the same length packaged into the capsids, the assembly efficiency reached the maximum in the presence of 5 μM TR-RNA or 20 μM TR-DNA, and both were followed by a decrease in yield at high concentrations (Fig. 2b). The difference between the concentrations of TR-RNA and TR-DNA that led to the maximum assembly efficiency was determined by the specific interactions between the CP and 2’-OH groups on TR-RNA (Valegard et al. 1994). For the DNA sequences of different lengths encapsulated into the capsids, the efficiency was similar when using a certain concentration of pac site.
Many researchers have produced armored RNA that is nuclease-resistant by the MS2 bacteriophage (Huang et al. 2006; Konnick et al. 2005; Pasloske et al. 1998; Sun et al. 2013; Wei et al. 2008a, b; Yu et al. 2008; Zhan et al. 2009). Our team has made continuous progress in this aspect. We have demonstrated that by applying a one-plasmid expression system with two C-variant pac sites (Wei et al. 2008a), a two-plasmid expression system with one C-variant pac site (Wei et al. 2008b), and a one-plasmid double-expression system with three C-variant pac sites (Zhan et al. 2009), 1981-base, 2248-base, and 3034-base chimeric RNA can be successfully packaged into MS2 capsids. Due to the limitations of the existing armored DNA manufactured with commonly used DNA phages, we used MS2, a single-strand RNA bacteriophage, to prepare armored DNA in a distinct approach (Fig. 1a). Previous studies have clarified the ability of MS2 CP to assemble spontaneously in the presence of the so-called pac site (Pickett and Peabody 1993; Wu et al. 1995), i.e., translational repressor, a stem-loop structure in the RNA. In this way, attachment of the pac site to the molecule of interest can induce it be packaged within the capsids when mixed with CP dimers. Taking advantage of this reassembly characteristic in vitro, we tested the encapsulation of much longer DNA fragments than that of armored RNA, which had a maximum length of about 3 kb in the protein shell by the plasmid expression system (Zhan et al. 2009). It is necessary to know that the nucleic acid packaging capacity is mostly restricted by the viral vector; thus, the length of the packaged DNA is generally similar to that of the viral genome. However, without the limit of a particular gene delivery vector, MS2 capsid was able to encapsulate DNA sequences up to 6.5 kb in our study, which was nearly twice as long as the 3.5 kb of genomic MS2 DNA. The reason we attempted to package such long sequence is that it would be beneficial for some armored DNA in various detection assays and kits. On the other hand, we were pleased to see the maximum packaging capability of MS2 capsids by this routine. Based on these results, we suppose that much longer DNA sequences can be encapsulated in the MS2 capsids, when the current method is used.
Encapsulation is based on the specific interaction between the RNA hairpin operator, i.e., pac site, and the MS2 CP, leading to spontaneous assembly of the viral shell and packaging the molecules around them. It has been demonstrated that (Rohrmann and Krueger 1970; Stockley et al. 1994; Witherell et al. 1991) the conserved bases of the MS2 operator are the A-4 and A-7 positions of the single-stranded loop, the −5 position must be a pyrimidine for tight binding, and the mismatched base of −10 position can be a purine (Fig. 1c). Bases of the remaining positions are not so significant that they can be any Watson-Crick base pair (Witherell et al. 1991). The loop and the unpaired A-10 are needed to provide sufficient affinity for the interaction between the MS2 operator and CP (Rowsell et al. 1998). However, it was proved that the sequence-specific complex can still form without an unpaired base at the −10 position (Rowsell et al. 1998), though the interaction was weak. All these above illustrate that TR-DNA can indeed initiate reassembly of the MS2 capsids when mixed with CP dimers, playing a similar role to TR-RNA and making the package more convenient and feasible. Moreover, four kinds of VLPs, which encapsulated different lengths of DNA and had diameters of 27–30 nm (Fig. 2d), were morphologically indistinguishable from wild-type MS2 phages.
As shown in Fig. 5, the stability experiments for temperature and time demonstrated that a series of ambient temperature and storage conditions did not affect the performance of armored DNA in laboratory assays. In comparison, armored HBV DNA produced by lambda bacteriophage was so susceptible in plasma that it was stable for no more than 4 days (Walkerpeach and Pasloske 2004). The steady MS2 phage used in our study solved this problem perfectly, in that this kind of HBV control is stable for at least 6 months at 4 °C and the armored HPV DNA was stable at 4 °C for at least 3 months. Therefore, controls for other HPV genotypes can be manufactured by this routine to construct a sample panel for proficiency testing.
Herein, we describe a novel way to produce armored double-stranded DNA using the MS2 bacteriophage to overcome the weaknesses of current controls or standards for DNA viruses. Armored DNA, which is resistant to nucleases, enables the whole detection process be monitored. This technology provides a practical and potentially scalable way of studying armored DNA to meet the requirements for clinical testing, such as stability, easy extraction by conventional methods, favorable linearity and reproducibility, and comparability of results from different laboratories.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(PDF 298 kb)
Acknowledgments
We thank all the manufacturers of commercial kits for providing the reagents needed in the experiments. This work was supported by AIDS and Hepatitis, and Other Major Infectious Disease Control and Prevention Program of China (No. 2013ZX10004805) and National Natural Science Foundation of China (No. 81171981).
Conflict of interest
We declare no conflicts of interest to this work.
References
- Ashley CE, Carnes EC, Phillips GK, Durfee PN, Buley MD, Lino CA, Padilla DP, Phillips B, Carter MB, Willman CL, Brinker CJ, Caldeira Jdo C, Chackerian B, Wharton W, Peabody DS. Cell-specific delivery of diverse cargos by bacteriophage MS2 virus-like particles. ACS Nano. 2011;5(7):5729–5745. doi: 10.1021/nn201397z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressler AM, Nolte FS. Preclinical evaluation of two real-time, reverse transcription-PCR assays for detection of the severe acute respiratory syndrome coronavirus. J Clin Microbiol. 2004;42(3):987–991. doi: 10.1128/JCM.42.3.987-991.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai SH, Lv FF, Zhang YH, Jiang YG, Peng J. Dynamic comparison between Daan real-time PCR and Cobas TaqMan for quantification of HBV DNA levels in patients with CHB. BMC Infect Dis. 2014;14:85. doi: 10.1186/1471-2334-14-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capehart SL, Coyle MP, Glasgow JE, Francis MB. Controlled integration of gold nanoparticles and organic fluorophores using synthetically modified MS2 viral capsids. J Am Chem Soc. 2013;135(8):3011–3016. doi: 10.1021/ja3078472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A, Hooker JM, Botta M, Francis MB, Aime S, Raymond KN. High relaxivity gadolinium hydroxypyridonate-viral capsid conjugates: nanosized MRI contrast agents. J Am Chem Soc. 2008;130(8):2546–2552. doi: 10.1021/ja0765363. [DOI] [PubMed] [Google Scholar]
- Datta S, Chatterjee S, Veer V. Recent advances in molecular diagnostics of hepatitis B virus. World J Gastroenterol. 2014;20(40):14615–14625. doi: 10.3748/wjg.v20.i40.14615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Santos AO, Souza LF, Borzacov LM, Villalobos-Salcedo JM, Vieira DS. Development of cost-effective real-time PCR test: to detect a wide range of HBV DNA concentrations in the western Amazon region of Brazil. Viro J. 2014;11:16. doi: 10.1186/1743-422X-11-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagan EJ, Moore C, Jenkins C, Rossouw A, Cubie HA, James VL. External quality assessment for molecular detection of human papillomaviruses. J Clin Virol. 2010;48(4):251–254. doi: 10.1016/j.jcv.2010.05.006. [DOI] [PubMed] [Google Scholar]
- Garimella PD, Datta A, Romanini DW, Raymond KN, Francis MB. Multivalent, high-relaxivity MRI contrast agents using rigid cysteine-reactive gadolinium complexes. J Am Chem Soc. 2011;133(37):14704–14709. doi: 10.1021/ja204516p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasgow JE, Capehart SL, Francis MB, Tullman-Ercek D. Osmolyte-mediated encapsulation of proteins inside MS2 viral capsids. ACS Nano. 2012;6(10):8658–8664. doi: 10.1021/nn302183h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotsch A, Schubert A, Bombis A, Wiedmann M, Zauke M, Schorling S. Nuclease-resistant single-stranded DNA controls for nucleic acid amplification assays. J Clin Microbiol. 2007;45(8):2570–2574. doi: 10.1128/JCM.00647-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Cheng Y, Guo Q, Li Q. Preparation of a chimeric armored RNA as a versatile calibrator for multiple virus assays. Clin Chem. 2006;52(7):1446–1448. doi: 10.1373/clinchem.2006.069971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konnick EQ, Williams SM, Ashwood ER, Hillyard DR. Evaluation of the COBAS Hepatitis C Virus (HCV) TaqMan analyte-specific reagent assay and comparison to the COBAS Amplicor HCV Monitor V2.0 and Versant HCV bDNA 3.0 assays. J Clin Microbiol. 2005;43(5):2133–2140. doi: 10.1128/JCM.43.5.2133-2140.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuebler DL, Illingworth A, Blenc AM, Wilbur DC. A peer comparison program for the quality assurance of human papillomavirus DNA detection using the Digene Hybrid Capture II/SurePath method shows excellent analytic interlaboratory correlation. Cancer. 2007;111(5):339–343. doi: 10.1002/cncr.22951. [DOI] [PubMed] [Google Scholar]
- Masker WE, Kuemmerle NB, Allison DP. In vitro packaging of bacteriophate T7 DNA synthesized in vitro. J Virol. 1978;27(1):149–163. doi: 10.1128/jvi.27.1.149-163.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermeyer AC, Capehart SL, Jarman JB, Francis MB. Multivalent viral capsids with internal cargo for fibrin imaging. PLoS One. 2014;9(6):e100678. doi: 10.1371/journal.pone.0100678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasloske BL, Walkerpeach CR, Obermoeller RD, Winkler M, DuBois DB. Armored RNA technology for production of ribonuclease-resistant viral RNA controls and standards. J Clin Microbiol. 1998;36(12):3590–3594. doi: 10.1128/jcm.36.12.3590-3594.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peabody DS. Role of the coat protein-RNA interaction in the life cycle of bacteriophage MS2. Mol Gen Genet. 1997;254(4):358–364. doi: 10.1007/s004380050427. [DOI] [PubMed] [Google Scholar]
- Pickett GG, Peabody DS. Encapsidation of heterologous RNAs by bacteriophage MS2 coat protein. Nucleic Acids Res. 1993;21(19):4621–4626. doi: 10.1093/nar/21.19.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrmann GF, Krueger RG. The self-assembly of RNA free protein subunits from bacteriophage MS-2. Biochem Biophys Res Commun. 1970;38(3):406–413. doi: 10.1016/0006-291X(70)90728-X. [DOI] [PubMed] [Google Scholar]
- Rowsell S, Stonehouse NJ, Convery MA, Adams CJ, Ellington AD, Hirao I, Peabody DS, Stockley PG, Phillips SE. Crystal structures of a series of RNA aptamers complexed to the same protein target. Nat Struct Biol. 1998;5(11):970–975. doi: 10.1038/2946. [DOI] [PubMed] [Google Scholar]
- Stocher M, Berg J. Internal control DNA for PCR assays introduced into lambda phage particles exhibits nuclease resistance. Clin Chem. 2004;50(11):2163–2166. doi: 10.1373/clinchem.2004.035519. [DOI] [PubMed] [Google Scholar]
- Stockley PG, Stonehouse NJ, Valegard K. Molecular mechanism of RNA phage morphogenesis. Int J Biochem. 1994;26(10-11):1249–1260. doi: 10.1016/0020-711X(94)90094-9. [DOI] [PubMed] [Google Scholar]
- Sun Y, Jia T, Sun Y, Han Y, Wang L, Zhang R, Zhang K, Lin G, Xie J, Li J. External quality assessment for Avian Influenza A (H7N9) Virus detection using armored RNA. J Clin Microbiol. 2013;51(12):4055–4059. doi: 10.1128/JCM.02018-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valegard K, Murray JB, Stockley PG, Stonehouse NJ, Liljas L. Crystal structure of an RNA bacteriophage coat protein-operator complex. Nature. 1994;371(6498):623–626. doi: 10.1038/371623a0. [DOI] [PubMed] [Google Scholar]
- Walkerpeach CR, Pasloske BL. DNA bacteriophage as controls for clinical viral testing. Clin Chem. 2004;50(11):1970–1971. doi: 10.1373/clinchem.2004.039776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Liang H, Zeng Y, Lin J, Liu C, Jiang L, Yang B, Ou Q. Establishment of a novel two-probe real-time PCR for simultaneously quantification of hepatitis B virus DNA and distinguishing genotype B from non-B genotypes. Clin Chim Acta. 2014;437:168–174. doi: 10.1016/j.cca.2014.07.021. [DOI] [PubMed] [Google Scholar]
- Wang Y, Liu Y, Ding Y, Sun N, Gong Y, Gao S. A PCR-based microwell-plate hybrid capture assay for high-risk human papillomavirus. Arch Viro. 2014;159(12):3365–3370. doi: 10.1007/s00705-014-2186-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei B, Wei Y, Zhang K, Yang C, Wang J, Xu R, Zhan S, Lin G, Wang W, Liu M, Wang L, Zhang R, Li J. Construction of armored RNA containing long-size chimeric RNA by increasing the number and affinity of the pac site in exogenous rna and sequence coding coat protein of the MS2 bacteriophage. Intervirology. 2008;51(2):144–150. doi: 10.1159/000141707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Yang C, Wei B, Huang J, Wang L, Meng S, Zhang R, Li J. RNase-resistant virus-like particles containing long chimeric RNA sequences produced by two-plasmid coexpression system. J Clin Microbiol. 2008;46(5):1734–1740. doi: 10.1128/JCM.02248-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witherell GW, Gott JM, Uhlenbeck OC. Specific interaction between RNA phage coat proteins and RNA. Prog Nucleic Acid Res Mol Biol. 1991;40:185–220. doi: 10.1016/S0079-6603(08)60842-9. [DOI] [PubMed] [Google Scholar]
- Wu M, Brown WL, Stockley PG. Cell-specific delivery of bacteriophage-encapsidated ricin A chain. Bioconjug Chem. 1995;6(5):587–595. doi: 10.1021/bc00035a013. [DOI] [PubMed] [Google Scholar]
- Wu M, Sherwin T, Brown WL, Stockley PG. Delivery of antisense oligonucleotides to leukemia cells by RNA bacteriophage capsids. Nanomedicine. 2005;1(1):67–76. doi: 10.1016/j.nano.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Wu W, Hsiao SC, Carrico ZM, Francis MB. Genome-free viral capsids as multivalent carriers for taxol delivery. Angew Chem Int Ed Engl. 2009;48(50):9493–9497. doi: 10.1002/anie.200902426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XF, Pan JC, Ye R, Xiang HQ, Kou Y, Huang ZC. Preparation of armored RNA as a control for multiplex real-time reverse transcription-PCR detection of influenza virus and severe acute respiratory syndrome coronavirus. J Clin Microbiol. 2008;46(3):837–841. doi: 10.1128/JCM.01904-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan S, Li J, Xu R, Wang L, Zhang K, Zhang R. Armored long RNA controls or standards for branched DNA assay for detection of human immunodeficiency virus type 1. J Clin Microbiol. 2009;47(8):2571–2576. doi: 10.1128/JCM.00232-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Huang J, Wang P, Sun Y, Zhang K, Xie J, Gao S, Wang L, Li J. Proficiency test for human papillomavirus genotyping in china. Intervirology. 2013;56(5):295–301. doi: 10.1159/000351620. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 298 kb)


