Abstract
This study aimed at demonstrating the antiviral activity of Lactobacillus gasseri CMUL57 (L. gasseri CMUL57), L. acidophilus CMUL67 and L. plantarum CMUL140 against herpes simplex type 2 (HSV-2) and Coxsackievirus B4E2 (CVB4E2), which are enveloped and naked viruses, respectively. These lactobacilli were non-cytotoxic and were able to reduce the cytopathic effect induced by HSV-2 in Vero cell monolayers. However, lactobacilli were not active against CVB4E2. Tested lactobacilli displayed anti-HSV-2 activity when they were co-incubated with the virus prior to inoculating the mixture to Vero cell monolayers. The detection of HSV-2 DNA by PCR in pellets of bacteria/virus mixtures let us to hypothesize that anti-HSV-2 activity of lactobacilli resulted from the viruses’ entrapment. This study showed the capabilities of vaginal lactobacilli to inhibit enveloped viruses such as HSV-2.
Keywords: Antiviral, Interaction virus and mammalian cells, Lactobacillus, Anti-herpetic activity
Introduction
Herpes simplex virus (HSV)-2 belongs to the α-Ηerpesvirinae subfamily, and its activation is triggered by a primary and localized infection in genital areas. Latent virus particles rest as covalently closed circular DNA (cccDNA) in the neurons of the peripheral nervous system. Upon reactivation, the virus migrates down through the axon to peripheral tissue, leading to localized lesions of the skin and mucus membranes (Field and Vere-Hodge 2013). HSV-2 is the most prevalent infection worldwide (Caldeira et al. 2013), and in recent years its recurrence has remarkably increased (Richman et al. 2010).
Overall, anti-HSV therapy is largely exerted by acyclovir (ACV), which is now replaced by its pro-drug valacyclovir (VACV) and famciclovir (FCV). ACV is a synthetic nucleoside analogue, converted by the virus into an active form. The virus uses the active form of ACV instead of the nucleoside to synthesize DNA, a critical component of viral replication. Incorporation of active ACV into new viral DNA stops production of the viral genomic DNA. Resistance to drugs such as ACV has been increasingly reported. These observations underscore the importance of discovering new therapeutic tools for the treatment of HSV infections (Bacon et al. 2003).
Anti-HSV compounds of which their efficiency was validated in vitro are continuously reported in the literature. Todorov et al. (2010) have shown that a bacteriocin produced by Enterococcus faecium ST5Ha can inhibit the HSV-1 and has no toxicity to the tissue cells. Drannik et al. (2013) described the anti-HSV activity of trappin-2 and elafin, serine protease inhibitor. Chono et al. (2012) demonstrated the anti-HSV activity of ASP2151 by targeting the helicase–primase complex of HSV-1 and HSV-2. Besides, many studies outlined the antiviral activity of probiotic strains. Related to this, Botić et al. (2007) unveiled the potential activity of probiotic lactobacilli against vesicular stomatitis viruses (VSV). This study shows that antiviral activity is due to the direct interaction between probiotic strains and enveloped virus. Attempts to use probiotics as antiviral agents may be a promising alternative (Chono et al. 2012). Probiotic lactic acid bacteria (LAB) and bifidobacteria support immune system’s function and balance and contribute to immuno-modulatory effects in combating microbial pathogens, including viruses. The herpes infection is recurrent and can be reactivated at any time (Field and Vere-Hodge 2013). Acyclovir can be potent only in the reactivation stage, and for these reasons the prevention of this type of infection is of major importance in the sector of public health (Caldeira et al. 2013). Taken together, these data indicate that the need of alternative strategies and probiotics could offer various advantages. The vaginal microflora usually designed as vaginal lactobacilli is the first line of defence against a large variety of microbes (Al Kassaa et al. 2014a, b). The inhibitory potency of vaginal lactobacilli can be exerted through different mechanisms including the aggregation and production of anti-microbial compounds (Al Kassaa et al. 2014b). Related to this, lactic acid, hydrogen peroxide and bacteriocins constitute the main anti-pathogen arsenal produced by vaginal lactobacilli (Al Kassaa et al. 2014a). Conti et al. reported that the anti-HSV-2 activity of L. brevis was originated by a molecule other than lactic acid and H2O2 (Conti et al. 2009). Mastromarino et al. (2011) showed that another strain of L. brevis exerted anti-HSV-2 activity by a heat-resistant and non-protein cell surface component. Subsequent to these findings (Mastromarino et al. 2011), Khani et al. (2012) showed that L. rhamnosus enhanced macrophage viability in elimination of HSV-1, while Zabihoullahi et al. (2012) reported that supernatants from different lactobacilli inhibited HSV replication.
This study aimed at highlighting the anti-HSV-2 potential of vaginal lactobacilli recently isolated from a cohort of Lebanese women. This report focuses, especially on L. gasseri CMUL57, which is resulting hereafter as able to inactivate the enveloped HSV-2 but not the naked enterovirus CVB4E2. Of course, the vaginal sampling and research outlines were authorized by the ethical committee of Lebanese university as well as Lebanese public health authorities.
Materials and methods
It should be pointed out that all the experiments developed in this study were performed in triplicate and in the independent occasions.
Growth conditions of vaginal lactobacilli
The strains used in this study were L. gasseri CMUL57, L. acidophilus CMUL67 and L. plantarum CMUL140 (Al Kassaa et al. 2014a). These strains were grown in MRS broth (De Man et al. 1960) and incubated for 24 h at 37 °C, under anaerobic conditions using AnaeroPack (BioMérieux, France). The cells were harvested (1500 g, 15 min, 4 °C), and the pellets were washed twice with phosphate buffer solution (PBS) (pH 8.0). The number of cells was adjusted to 108 CFU/ml by using the turbidity (600 nm) and counting method on MRS (De Man et al. 1960) agar plate. Staphylococcus aureus (S. aureus) strain was grown in brain heart infusion (Biokar, France) at 37 °C for 18 h. The number of colonies was determined on Baird–Parker agar (Biokar, France).
Cells lineage
The Vero cells (kidney epithelial from African green monkey) (ATCC CCL-81) were grown in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, USA), supplemented with 10 % foetal calf serum (Gibco, USA), and 1 % l-glutamine (Gibco, USA), and incubated at 37 °C in the presence of 5 % CO2 in tissue culture flasks to a confluency of approximately 80 %. Further, 2.5 × 104 cells/ml were seeded in 96-well culture cell microplates and incubated under the same conditions to 80 % confluency.
Cell viability
The cell viability test was carried out using a commercialized kit “UptiBlue” (Uptima, Interchim, France). Briefly, the Vero cells were seeded at 2.5 × 104 cells/well on 96-well microtitre plates and cultured until reaching 80 % of confluency. Afterwards, the Vero cell monolayers were washed twice with PBS and various lactobacilli strains were added on the Vero cell monolayers and incubated at 37 °C with 5 % CO2 for 2 h to insure the bacterial adhesion. After bacterial adhesion, the Vero cell monolayers were washed three times with PBS followed by addition of 100 µl of UptiBlue reagent. This mixture was incubated for 4 h at 37 °C with 5 % CO2. Absorbance was measured on an EIA reader (Lab Systems; MTX Labs, Vienna, VA, USA) at a test wavelength of 570 nm and a reference wavelength of 595 nm. The percentages of cell reductions (cell viability) were determined using the following formula:
where O.D(C + B) is the absorbance measured with a given concentration of Lactobacillus strains adhered on Vero cells, O.D(B) is the absorbance measured for the control well containing bacteria without Vero cells, and O.D(C) is the absorbance measured for control untreated mock-infected cells.
Cytotoxicity and adhesion of lactobacilli
The possible cytotoxic effect of lactobacilli was measured after 4, 24, 32 and 48 h of bacterial adhesion by using the UptiBlue kit. Therefore, Vero cells at 80 % of confluency were incubated with 100 µl of tenfold serial dilutions (108 CFU/ml to 104 CFU/ml) and cultured up to 48 h before measuring cell viability. In addition, the presence of bacteria was measured as previously described (Jacobsen et al. 1999). Briefly, a cell suspension containing 105 cells in 4-ml complete DMEM medium was transferred to each well of a six-well tissue culture plate. When the cells reached 80 % confluency, the medium was removed 24 h prior to the adhesion assay and the cells were supplemented with DMEM, without antibiotics. The monolayer cells were washed twice with 3 ml of PBS. An aliquot of 2 ml of DMEM (without serum and antibiotics) was added to each well and incubated for 30 min at 37 °C. Bacterial cells, at 108 CFU/ml, were resuspended in 1 ml of DMEM medium (without serum and antibiotics) and added to different wells. The plates were incubated for 2 h at 37 °C in the presence of 5 % CO2. The monolayer cells were washed five times with sterile PBS, and the adhesion score was determined by counting the adhered bacteria within 20 random microscopic fields after Giemsa staining (Jacobsen et al. 1999). Bacteria were grouped into three categories: non-adhesive (≤40 bacteria), adhesive (41–100 bacteria) and highly adhesive (>100 bacteria). The percentages of adhesion were determined by plating MRS agar plate followed by colony counting after 48 h of incubation. The adhered bacteria were harvested together with the eukaryotic cells by trypsination; the mixture was serially diluted and streaked onto MRS agar plates (Jacobsen et al. 1999).
Virus
HSV-2 was isolated at the Laboratory of Virology (CHRU Lille) from a patient with vaginal infection and was cultured on Vero cell line. CVB4E2 is member of the human enterovirus B species of the Enterovirus genus that can be grown on HEp2 cell line and on Vero cell line as well (Sane et al. 2013). The supernatants containing the virus were collected from the flask when the cytopathic effect (CPE) was observed by inverted light microscopy (Olympus, USA). The viral titres of supernatants were determined by the Spaerman–Karber TCID50 (50 % tissue culture infectious dose) titration method (Landry et al. 2002).
Plaque reduction assay (PRA)
The plaque reduction assay was carried out as previously described (Zabihollahi et al. 2012) with some modifications. Briefly, the Vero cells were placed into 24-well culture plates (Nunc, Denmark) at a concentration of 1.7 × 105 cells per well and incubated for 24 h (in antibiotic free medium) until 98 % of confluency. The Vero cells were inoculated with 100 PFU HSV-2 or 100 TCID50/ml CVB4E2 and incubated for 4 h. Afterwards, the cell monolayers were washed and were overlaid with DMEM, supplemented with 1.5 % (w/v) agarose (Invitrogen, UK), 5 % FBS, 100 IU/ml penicillin and 100 μg/ml streptomycin (Gibco, USA). After 72 h of incubation at 37 °C in 5 % CO2 atmosphere, the overlay medium was removed and the Vero cell monolayers washed twice with PBS, followed by fixing with pure methanol for 30 min. Counts of the viral plaques was done after staining with 0.5 % crystal violet (Landry et al. 2002).
qPCR analysis
The DNA extraction was carried out using the QIAamp MinElute Virus Spin Kit (Qiagen, Germany). The primers and probes targeting HSV-2 glycoprotein B were designed using the Primer Express 2.0 software (Applied Biosystems, USA). The primer sequences were FHSV-2 5′-CGCACCTGCGGGAAATC-3, RHSV-2 5′-GCGGGCACACGTAAA-3′ and TaqMan probe sequence (HSV-2 probe): (VIC) 5′-AGGTCGAGAACGCC-3′ (MGB). The qPCR was realized on ABI 7500 (Applied Biosystems, USA) machine using the following PCR program: 2 min/50 °C (1 cycle), 10 min at 95 °C (1 cycle) and 45 cycles of (15 s/95 °C, 1 min/60 °C). TaqMan Universal Master Mix (2×) (Applied Biosystems, USA) and 0.04 μM of primers and probe was used in the qPCRs. DNA extracted from HSV-2 reference strain ATCC VR-734D was used as control of qPCR.
Results
Lactobacilli can inhibit HSV-2 but not CVB4E2
The viability of Vero cells decreased to 81, 88, 91 and 25 %, after 24 h of incubation in the presence of 107 CFU/ml of L. gasseri CMUL57, L. acidophilus CMUL67 or L. plantarum CMUL140 and S. aureus ATCC33862, respectively. When the concentration of lactobacilli increased up to 108 CFU/ml after 24 h of incubation, the viability of Vero cells was 85, 83 and 78 % for CMUL57, CMUL67 and CMUL140, respectively. However, after 48 h of incubation, the cell viability diminished to 65 and 17 %, in the presence of lactobacilli strains and S. aureus ATCC33862, respectively (Fig. 1). Taken together, these data indicate that the viability of Vero cells was particularly decreasing along time of incubation. The efficiency of adhesion was measured after 2 h of incubation and reached about 80 % for lactobacilli, and 34 % for S. aureus ATCC33862 (Table 1).
Fig. 1.
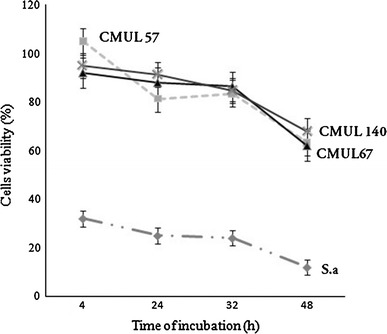
Viability of Vero cells in the presence of vaginal lactobacilli CMUL57, CMUL67, CMUL140 and Staphylococcus aureus (S. aureus) ATCC33862 at 107 CFU/ml. The cell viability was measured using UptiBlue, and the results were expressed as the percentage of viability compared to the negative control (100 % viable Vero cell monolayers). The results were the means (±SD) of at least three independent experiments
Table 1.
Lactobacilli score and percentage of adhesion on Vero cells after 2 h of incubation
| Bacteria at 107 CFU/ml | Adhesion scorea | % of adhesion |
|---|---|---|
| L. gasseri CMUL57 | 382 ± 22.5 | 81.2 ± 3.8 |
| L. acidophilus CMUL67 | 435 ± 32.1 | 80.2 ± 7.9 |
| L. plantarum CMUL140 | 299 ± 22.3 | 81.0 ± 7.6 |
| S. aureus ATCC33862 | 187 ± 17.5 | 28.3 ± 5.3 |
aThe scores were calculated based on the number of bacteria per 20 microscopic field. The results are the mean ± SD of three independent experiments
The addition of lactobacilli strains to Vero cell monolayers followed by their challenge with 100 PFU/ml HSV-2 reduced the cell death by 28, 16 and 17 %, respectively, compared with the control that contained Vero cells without bacteria and challenged with HSV-2 (Fig. 2). The results of 2-h lactobacilli contact were quietly different to those obtained after 24-h contact (Fig. 2). There was no significant reduction in Vero cells’ death when they were challenged with 100 TCID50/ml CVB4E2 (Fig. 2). No impact on viability of Vero cells was observed when L. gasseri CMUL57 was replaced either by L. acidophilus CMUL67 or by L. plantarum CMUL140 (Fig. 2). However, when Vero cells were infected with HSV-2 or CVB4E2 and then challenged with lactobacilli at 107 CFU/ml, the observed cytotoxicity remained unchanged (data not shown).
Fig. 2.
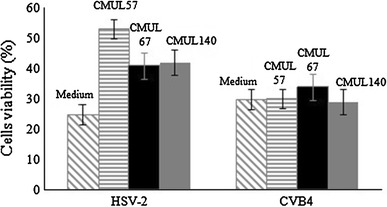
Antiviral activity of live lactobacilli strains on HSV-2- and CVB4E2-infected Vero cells. After removing the unbound bacteria, Vero cells were incubated for a further 24 h before viral infection by HSV-2 or CVB4E2. The results show the cell viability assay obtained after 48 h of infection by the viruses. The results are expressed as the percentage of cell survival, in comparison with the controls represented by the wells containing the untreated Vero cells (medium). The data are the means (±SD) of at least three independent experiments
The co-incubation of L. gasseri CMUL57, L. acidophilus CMUL67 or L. plantarum CMUL140 strain with HSV-2 on Vero cell monolayers has increased the cells viability by 65, 35 and 15 %, respectively, as compared to the control with 22 % viability, after exposure to the virus (Fig. 3a). In case of L. gasseri CMUL57, the increase in post-treatment Vero cell viability was lactobacilli cell concentration dependent (Fig. 3b).
Fig. 3.
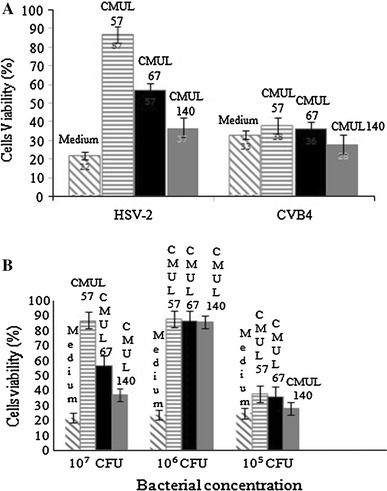
Antiviral activity of live vaginal lactobacilli CMUL57, CMUL67 and CMUL140 on HSV-2- or CVB4E2-infected Vero cells. The antiviral effect was examined when lactobacilli were applied on the Vero cell together with the viruses (co-incubation) (a), taking into account the bacterial concentrations (b). Results were collected after 48 h of HSV-2 challenge and expressed as percentage of cell survival, compared to the control wells (medium) containing cells which were challenged with the virus in the absence of lactobacilli. The data are the means (±SD) of at least three independent experiments
When the Vero cell monolayers were inoculated with CVB4E2 at 100 TCID50/ml and lactobacilli at 107 CFU/ml, the CVB4E2-induced cytopathic effect was not modified. Inhibition of the HSV-2-induced cytopathic effect by L. gasseri CMUL57 was accompanied by a reduction in viral progeny as compared to the control (102 TCID50/ml vs. 105 TCID50/ml P = 0.02). However, there was no reduction in viral progeny with strains L. acidophilus CMUL67 (104 TCID50/ml; P = 0.15) and L. plantarum CMUL140 (1.5 104 TCID50/ml; P = 0.2) (Fig. 4).
Fig. 4.
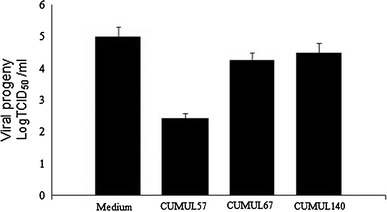
Reduction in viral (HSV-2) progeny by lactobacilli strains. The vaginal lactobacilli were added at the same time as the viruses. HSV-2 and CVB4E2 were used to challenge Vero cells and then incubated for 6 h. The unbound virus particles were removed by three washings. Results were recorded after 48 h of incubation and represented by the virus titration and expressed as logTCID50/ml, compared to the control cells (medium) which were not treated with lactobacilli. The data (±SD) are the means of at least three independent experiments
Adsorption of HSV-2 to L. gasseri CMUL57
In case of L. gasseri CMUL57 (Fig. 5a), the DMEM supernatant (pH 4.8), neutralized supernatant (pH 6.9), and supernatant treated with protease and with catalase to degrade proteins and eliminate possible activity due to the presence of H2O2 caused 0.4, 0.1, 0.14 and 0.21 log reduction in the number of viral particles, respectively. The DMEM lactobacilli supernatants were mixed and incubated with viruses prior to infection step. Further, L. gasseri CMUL57-, L. acidophilus CMUL67- and L. plantarum CMUL140-derived neutralized supernatants enhanced slightly the viability of Vero cell monolayer inoculated with 100 PFU HSV-2 with about 9.5, 5.4 and 2.4 %, respectively, due to reduction in viral infectious particles. These data are normalized to HSV-2 alone used as a control (Table 2). This very weak inhibition prompted us to investigate whether the anti-HSV-2 activity of lactobacilli was exerted through an interaction between bacteria and HSV-2 particles. Thus, L. gasseri CMUL57 at 108 CFU/ml or Dulbecco’s Modified Eagle’s Medium (DMEM, negative control) were incubated for 2 h at 37 °C in CO2 atmosphere in the presence of HSV-2 (104 PFU/ml) or CVB4E2 (103 TCID50/ml). Afterwards, supernatant was collected by centrifugation (20,000 g, 3 min, 4 °C) and assayed on Vero cells. After the treatment, the number of infectious HSV-2 particles was significantly reduced (15,000 vs. 100,000 PFU/ml P = 0.01), whereas that of CVB4E2 remained unchanged as compared to the control (9.8 102 TCID50/ml) (Fig. 5b, c). To be noted that the pellet was washed three times with DMEM in order to eliminate any free residual viral particles in the mixture. After each washing, the supernatants were assayed on Vero cells monolayers to evaluate the level of free infectious particles. After the final washing step, DNA was extracted from the pellet and the final supernatant to test by qPCR the presence of the viral particles trapped with L. gasseri CMUL57. Thereof, the cycle threshold of the control assay was 26.2 ± 0.8 for 1000 PFU, and that of the pellet was 24.2 ± 0.9 (Fig. 5d). In contrast, the detection of HSV-2 DNA in the final washing supernatant was almost negative (Fig. 5d).
Fig. 5.
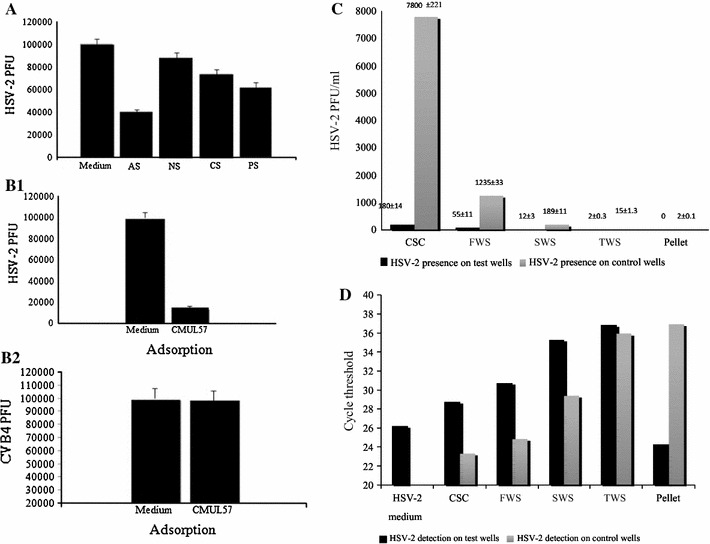
Putative mechanism of anti-HSV-2 activity of L. gasseri CMUL57. a Anti-HSV-2 activity of crude cell-free supernatant (AS) (pH 4.5), neutralized supernatant (NS) (pH 6.8), supernatant treated with catalase (CS) or with protease (PS). The data (±SD) are the means of at least three independent experiments. Adsorption of HSV-2 (b) and CVB4E2 (c) on CMUL57 strain. The data (± SD) are the means of at least three independent experiments. d Detection for infectious particles in the washing supernatants of the CMUL57 strain and HSV-2 mixture was determined using the PRA method. e The real-time PCR method was used to detect the presence of “trapped” viruses in the mixture pellet and residual viral particles in washing supernatants. CSC crude supernatant of co-incubation, FWS first washing supernatant, SWS second washing supernatant, TWS third washing supernatant. The control wells refer to the wells containing HSV-2 only, whereas the test wells refer to the wells containing HSV-2 co-incubated with CMUL57 strain
Table 2.
Anti-CPE activity of lactobacilli supernatants after neutralization (pH = 6.8)
| Cell-free supernatant of | pH | Antiviral activity | |
|---|---|---|---|
| HSV-2 inhibition | P value | ||
| Cells viability (%) ± SD | <0.05 | ||
| L. gasseri CMUL57 | 4.8 | 92.1 ± 4.5 | 0.04 |
| L. acidophilus CMUL67 | 5.1 | 88.0 ± 6.2 | 0.07 |
| L. plantarum CMUL140 | 4.0 | 85.0 ± 2.8 | 0.1 |
| HSV-2 control | 82.6 ± 2.0 | ||
Percentages of survival of Vero cells compared to the untreated cells (control). The data are the means of at least three independent experiments
The bacterial pellet suspected to be containing HSV-2 infectious particles, as well as the washing supernatants were serially diluted (10−1 to 10−5) and added to Vero cell monolayers. Afterwards, a PRA method was carried out to examine these samples’ infectivity, by incubating them for 5 days at 37 °C in 5 % CO2. After the incubation, no viral infection was observed for the supernatant of the final washing and for the bacterial pellet (Fig. 5e).
Discussion
Vaginal lactobacilli play crucial role in prevention of urogenital infections such as bacterial vaginosis, vaginal candidiasis and viral infections such as those of HSV-2 (Conti et al. 2009). HSV-2 is one of the most common sexually transmitted infections, acting further as a potential risk factor for acquiring other sexually transmitted infections (Chopra et al. 2013). Here, we demonstrate the anti-HSV-2 potential of L. gasseri CMUL57, L. acidophilus CMUL67 and L. plantarum CMUL140 that were recently isolated from human vagina and characterized for their large antagonism and probiotic characteristics (Al Kassaa et al. 2014a). The highest anti-HSV-2 activity was observed for L. gasseri CMUL57. Recently, Zabihoullahi et al. (2012) showed the anti-HSV-2 activity of another vaginal L. gasseri strain. These two independent studies utilized live and non-live L. gasseri, respectively, and revealed the capabilities of these lactobacilli to inhibit HSV-2. Further, the absence of cytotoxicity of LAB probiotics is a prerequisite before undertaking any antiviral study. L. gasseri CMUL57, L. acidophilus CMUL67 and L. plantarum CMUL140 used in this study are shown to be not cytotoxic for Vero cells. Safety of Bifidobacterium adolescentis SPM 0214 was established on Vero cells (An et al. 2012), and that of E. faecium on porcine macrophage cell line (Wang et al. 2013). Remarkably, virus inactivation by LAB probiotics has unveiled at least two main pathways (Al Kassaa et al. 2014b). The first one is linked to the production of inhibitory metabolites such as lactic acid, bacteriocins and hydrogen peroxide that are endowed with antiviral activity (Conti et al. 2009). Here, this possibility is discarded because the assays conducted with the supernatants gathered from L. gasseri CMUL57, L. acidophilus CMUL67 and L. plantarum CMUL140 cultures revealed either slight or absence of anti-HSV-2 activity. However, the anti-HSV-2 activity attributed to molecules of a other than H2O2 and lactic acid bacteria was reported for L. brevis CD2, L. salivarius FV2 and L. plantarum FV9 isolated from human vagina (Conti et al. 2009). The impact of lactic acid produced by vaginal lactobacilli was shown to interfere with the negative charges of the viral envelopes, enhancing their sensitivity and provoking ease virus inactivation (Tao et al. 1997).
The second possibility of virus inactivation by LAB probiotics is thought to be exerted through a physical contact between bacterial cells and virus envelope (Al Kassaa et al. 2014b). The data obtained here agree with this concept, as we show that L. gasseri CMUL57 strain is active against enveloped virus (HSV-2), but not against naked one (CBV4E2). Interestingly, co-incubation of HSV-2 and L. gasseri CMUL57 experiments as well as the qPCR one let us to think that HSV-2 particles were indeed present in the bacterial pellet, and the absence of their infectivity could result from trapping/binding mechanism. In direct line, Tao et al. (1997) showed the capabilities of lactobacilli to trap human immunodeficiency virus (HIV) by binding the mannose sugar-rich “dome” of their attachment glycoprotein gp120. Ivec et al. (2007) reported the trapping and adsorption of vesicular stomatitis virus (VSV) by Lactobacillus and Bifidobacterium species. Lai et al. (2009) indicated that the trapping of HIV by vaginal lactobacilli rendered the diffusion of HIV very slower than in the normal conditions. Recently, Chai et al. (2013) showed the attachment of the enteropathogenic coronavirus to the surface of E. faecium, which might be a means to trap viruses and prevent infection. Wang et al. showed that E. faecium inhibited influenza viruses by mechanisms including a direct physical interaction and strengthening of innate defence at the cellular level (Wang et al. 2013). The use of LAB probiotics as antiviral agents was reported as promising approach for aquaculture (Lakshmi et al. 2013) and poultry industry (Seo et al. 2012) because viral diseases cause an enormous loss in the production in shrimp farms (Lai et al. 2009), and avian influenza virus occurrence is responsible of severe losses to the poultry industry (Seo et al. 2012).
Overall, this study shows that HSV-2 envelope is a key element in the inactivation by vaginal L. gasseri CMUL57. After the in vivo studies have been established, L. gasseri CMUL57 could be recommended as vaginal probiotic to preventing HSV-2 infection in healthy women, or to decreasing the period of treatment in the case of infected women.
Acknowledgments
I.A.K. was a recipient of Ph.D. fellowship from AZM Center of Biotechnology (Lebanon) and Lille 1 University (France).
Conflict of interest
The authors declare that there is no conflict of interest.
References
- Al Kassaa I, Hamze M, Hober D, Monzer H, Chihib NE, Drider D. Identification of vaginal lactobacilli with potential probiotic properties isolated from women in North Lebanon. Microb Ecol. 2014;67:722–734. doi: 10.1007/s00248-014-0384-7. [DOI] [PubMed] [Google Scholar]
- Al Kassaa I, Hober D, Hamze M, Chihib NE, Drider D. Antiviral potential of lactic acid bacteria and their bacteriocins. Probiotics Antimicrob Prot. 2014;6:177–185. doi: 10.1007/s12602-014-9162-6. [DOI] [PubMed] [Google Scholar]
- An HM, Lee do K, Kim JR, Lee SW, Cha MK, Lee KO, Ha NJ. Antiviral activity of Bifidobacteriumadolescentis SPM 0214 against herpes simplex virus type 1. Arch Pharm Res. 2012;35:1665–1671. doi: 10.1007/s12272-012-0918-9. [DOI] [PubMed] [Google Scholar]
- Bacon TH, Levin MJ, Leary JJ, Sarisky RT, Sutton D. Herpes simplex virus resistance to acyclovir and penciclovir after two decades of antiviral therapy. Clin Microbiol Rev. 2003;16:114–128. doi: 10.1128/CMR.16.1.114-128.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botić T, Klingberg TD, Weingartl H, Cencic A. A novel eukaryotic cell culture model to study antiviral activity of potential probiotic bacteria. Int J Food Microbiol. 2007;115:227–234. doi: 10.1016/j.ijfoodmicro.2006.10.044. [DOI] [PubMed] [Google Scholar]
- Caldeira TD, Gonçalves CV, Oliveira GR, Fonseca TV, Gonçalves R, Amaral CT, Hora VP, Martinez AM. Prevalence of herpes simplex virus type 2 and risk factors associated with this infection in women in southern Brazil. Rev Inst Med Trop Sao Paulo. 2013;55:315–321. doi: 10.1590/S0036-46652013000500004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai W, Burwinkel M, Wang Z, Palissa C, Esch B, Twardziok S, Rieger J, Wrede P, Schmidt MF. Antiviral effects of a probiotic Enterococcusfaecium strain against transmissible gastroenteritis coronavirus. Arch Virol. 2013;158:799–807. doi: 10.1007/s00705-012-1543-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chono K, Katsumata K, Kontani T, Shiraki K, Suzuki H. Characterization of virus strains resistant to the herpes virus helicase–primase inhibitor ASP2151 (Amenamevir) Biochem Pharmacol. 2012;84:459–467. doi: 10.1016/j.bcp.2012.05.020. [DOI] [PubMed] [Google Scholar]
- Chopra S, Devi P, Devi B. Herpes simplex virus 2: a boon to develop other sexually transmitted infections. J Indian Med Assoc. 2013;111:236–238. [PubMed] [Google Scholar]
- Conti C, Malacrino C, Mastromarino P. Inhibition of herpes simplex virus type 2 by vaginal lactobacilli. J Physiol Pharmacol. 2009;6:19–26. [PubMed] [Google Scholar]
- De Man JC, Rogosa M, Sharpe ME. A medium for the cultivation of lactobacilli. J Appl Microbiol. 1960;63:551–558. [Google Scholar]
- Drannik AG, Nag K, Sallenave JM, Rosenthal KL. Antiviral activity of trappin-2 and elafin in vitro and in vivo against genital herpes. J Virol. 2013;87:7526–7538. doi: 10.1128/JVI.02243-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field HJ, Vere-Hodge R. A recent developments in anti-herpesvirus drugs. Br Med Bull. 2013;106:213–249. doi: 10.1093/bmb/ldt011. [DOI] [PubMed] [Google Scholar]
- Ivec M, Botić T, Koren S, Jakobsen M, Weingartl H, Cencic A. Interactions of macrophages with probiotic bacteria lead to increased antiviral response against vesicular stomatitis virus. Antivir Res. 2007;75:266–274. doi: 10.1016/j.antiviral.2007.03.013. [DOI] [PubMed] [Google Scholar]
- Jacobsen CN, Rosenfeldt-Nielsen V, Hayford AE, Møller PL, Michaelsen KF, Paerregaard A, Sandström B, Tvede M, Jakobsen M. Screening of probiotic activities of forty-seven strains of Lactobacillus spp. by in vitro techniques and evaluation of the colonization ability of five selected strains in humans. Appl Environ Microbiol. 1999;65:4949–4956. doi: 10.1128/aem.65.11.4949-4956.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khani S, Motamedifar M, Golmoghaddam H, Hosseini HM, Hashemizadeh Z. In vitro study of the effect of a probiotic bacterium Lactobacillusrhamnosus against herpes simplex virus type 1. Braz J Infect Dis. 2012;16:129–135. doi: 10.1016/S1413-8670(12)70293-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai SK, Hida K, Shukair S, Wang YY, Figueiredo A, Cone R, Hope TJ, Hanes J. Human immunodeficiency virus type 1 is trapped by acidic but not by neutralized human cervico vaginal mucus. J Virol. 2009;83:11196–11200. doi: 10.1128/JVI.01899-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmi B, Viswanath B, Sai Gopal DV (2013) Probiotics as antiviral agents in shrimp aquaculture. J Pathog 2013:424123. doi:10.1155/2013/424123 [DOI] [PMC free article] [PubMed]
- Landry ML, Stanat S, Biron K, Brambilla D, Britt W, Jokela J, Chou S, Drew WL, Erice A, Gilliam B, Lurain N, Manischewitz J, Miner R, Nokta M, Reichelderfer P, Spector S, Weinberg A, Yen-Lieberman B, Crumpacker C. A standardized plaque reduction assay for determination of drug susceptibilities of cytomegalovirus clinical isolates. Antimicrob Agents Chemother. 2002;44:688–692. doi: 10.1128/AAC.44.3.688-692.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastromarino P, Cacciotti F, Masci A, Mosca L. Antiviral activity of Lactobacillusbrevis towards herpes simplex virus type 2: role of cell wall associated components. Anaerobe. 2011;17:334–336. doi: 10.1016/j.anaerobe.2011.04.022. [DOI] [PubMed] [Google Scholar]
- Richman DD, Whitley RJ, Hayden FG. Clinical virology. 3. Washington, DC: ASM Press; 2010. [Google Scholar]
- Sane F, Caloone D, Gmyr V, Engelmann I, Belaich S, Kerr-Conte J, Pattou F, Desailloud R, Hober D. Coxsackievirus B4 can infect human pancreas ductal cells and persist in ductal-like cell cultures which results in inhibition of Pdx1 expression and disturbed formation of islet-like cell aggregates. Cell Mol Life Sci. 2013;70:4169–4180. doi: 10.1007/s00018-013-1383-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo BJ, Rather IA, Kumar VJ, Choi UH, Moon MR, Lim JH, Park YH. Evaluation of Leuconostocmesenteroides YML003 as a probiotic against low-pathogenic avian influenza (H9N2) virus in chickens. J Appl Microbiol. 2012;113:163–171. doi: 10.1111/j.1365-2672.2012.05326.x. [DOI] [PubMed] [Google Scholar]
- Tao L, Pavlova SI, Mou SM, Ma WG, Kiliç AO. Analysis of lactobacillus products for phages and bacteriocins that inhibit vaginal lactobacilli. Infect Dis Obstet Gynecol. 1997;5:244–251. doi: 10.1155/S1064744997000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todorov SD, Wachsman M, Tomé E, Dousset X, Destro MT, Dicks LM, Franco BD, Vaz-Velho M, Drider D. Characterisation of an antiviral pediocin-like bacteriocin produced by Enterococcus faecium. Food Microbiol. 2010;27:869–879. doi: 10.1016/j.fm.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Wang Z, Chai W, Burwinkel M, Twardziok S, Wrede P, Palissa C, Esch B, Schmidt MF. Inhibitory influence of Enterococcusfaecium on the propagation of swine influenza A virus in vitro. PLoS ONE. 2013;8:e53043. doi: 10.1371/journal.pone.0053043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabihollahi R, Motevaseli E, Sadat SM, Azizi-Saraji AR, Asaadi-Dalaie S, Modarressi MH. Inhibition of HIV and HSV infection by vaginal lactobacilli in vitro and in vivo. Daru J Pharm Sci. 2012;20:53. doi: 10.1186/2008-2231-20-53. [DOI] [PMC free article] [PubMed] [Google Scholar]


