Abstract
A recent parameter identification technique, the local lagged adapted generalized method of moments, is used to identify the time-dependent disease transmission rate and time-dependent noise for the stochastic susceptible, exposed, infectious, temporarily immune, susceptible disease model (SEIRS) with vital rates. The stochasticity appears in the model due to fluctuations in the time-dependent transmission rate of the disease. All other parameter values are assumed to be fixed, known constants. The method is demonstrated with US influenza data from the 2004–2005 through 2016–2017 influenza seasons. The transmission rate and noise intensity stochastically work together to generate the yearly peaks in infections. The local lagged adapted generalized method of moments is tested for forecasting ability. Forecasts are made for the 2016–2017 influenza season and for infection data in year 2017. The forecast method qualitatively matches a single influenza season. Confidence intervals are given for possible future infectious levels.
Keywords: Compartment disease model, Stochastic disease model, Local lagged adapted generalized method of moments, Time-dependent transmission rate
Introduction
Knowledge about the transmission of an infectious disease can help control the spread of disease, potentially mitigating disease related deaths and economic losses. The transmission rate is time-dependent and depends on many disparate factors, such as weather, social behavior, and strain-specific factors. While some disease parameters, such as the latency period, can be estimated from laboratory experiments or surveys of infected populations, the transmission rate is difficult to estimate. In this work, we describe a new parameter identification technique that can be used to determine the transmission rate given infection data.
Parameter identification studies the inverse problem ‘under what conditions can observations of a modeled system be used to identify the value of model parameters?’. Focusing on an epidemic model for influenza, ‘how can infection data be used to identify an unknown transmission rate?’. In epidemic models, the observed infected data appears (almost) directly in the model, as a model state. However, the theory of parameter identification applies more broadly to observations that are instead some (possibly unknown) function of the model states. When parameters are unidentifiable, using identifiability techniques may uncover combinations of dependent parameters. Similarly, in the ideal scenario of noise-free data that is modeled with the stated model, even if the parameter values can be theoretically uniquely determined, computational limitation may prevent identifiability and may reveal dependent parameter combinations.
Epidemic SIR-type models are typically non-linear and, as such, model parameters are difficult to identify. (This is in contrast to linear models for which many analytical techniques exist (Godfrey and DiStephano 1987)). The best known identification techniques for non-linear deterministic models and constant parameter values use Taylor series (Gunn et al. 1997; Pohjanpalo 1978), similarity transformations (Chappell et al. 1990; Evans et al. 2002; Vajda et al. 1989), or differential algebra (Audoly et al. 2001; Eisenberg 2013; Eisenberg et al. 2013; Ljung and Glad 1994). For time-dependent parameters, differential algebra approaches can also be used (Hadeler 2011; Mummert 2013; Pollicott et al. 2012), as well as modulating functions methods (method of moment functionals) (Ungarala et al. 2013).
Parameter estimation for stochastic models, also called model calibration, is a type of statistical inference. Well-known estimation procedures for stochastic models are least squares estimation (Banks et al. 2014; Escobar 2012), maximum likelihood estimation (Heijmans and Magnus 1986; Bishwal 2008), Kalman filtering (linear quadratic estimation) (Cazelles and Chau 1995; Julier and Uhlmann 2004; Kalman 1960) and the generalized method of moments (Hansen 1982; Jeisman 2005; Hurn et al. 2007). Some of these techniques can be extended to estimate time-varying parameters, for example using recursive least squares (with a forgetting factor) (Escobar 2012) or an expectation maximization algorithm paired with Kalman filtering (Olama et al. 2009). We direct interested readers to Bishwal (2008) for a compilation of well-known stochastic differential equations and parameter estimation using the maximum likelihood estimate and the Bayes estimate for both continuous and discrete observations.
Parameter estimation for stochastic epidemic models has a rich history. Some early works include Bailey (1953) (and the references within), Becker (1976) using smallpox data, and Longini et al. (1982, 1988) focusing on influenza. Becker (1989) provides a summary of the current techniques for statistical analysis of epidemic models at the time it was published.
More recently, increased computational power has allowed investigation of more complicated and more timely models. In particular, Bayesian inference aided typically by Markov Chain Monte Carlo (MCMC) algorithms allows estimation of model parameter (posterior) distributions. See O’Neill (2002) for a methodological overview. (Here we focus on epidemic models, but Bayesian techniques can be applied more broadly, such as in health applications including designing clinical trials (Berry and Stangl 2018), ecological modeling (Hobbs and Hooten 2015), and economics (Greenberg 2013)). These techniques are applicable to estimate fixed (see O’Neill and Becker 2001) or time varying parameters estimates (see Arnold and Lloyd 2018). The Bayesian approach naturally allows for data imputation via inclusion of missing data as an additional parameter (O’Neill and Roberts 1999; Cauchemez and Ferguson 2008). Data imputation can also be done with other techniques, such as expectation maximization (Becker 1997; Meng and van Dyk 1997).
Modern parameter estimation techniques and increased computational power allow exploration of epidemics as they occur. See for example the dynamics and analysis of bovine spongiform encephalopathy (BSE) epidemic in cattle in Great Britain (Anderson et al. 1996), the dynamics of the Foot and Mouth Disease (FMD) (Keeling et al. 2001) and Ferguson et al. (2001), pH1N1 (Fraser et al. 2009; Yang et al. 2009), estimation analysis of pandemic risk of Middle East Respiratory Syndrome coronavirus (MERS) (Breban et al. 2013), and Ebola (Rivers et al. 2014). (Deterministic models can also be used to study epidemics in real-time. Examples include SARS (Lipsitch et al. 2003), cholera (Tuite et al. 2011), and Ebola (Fisman et al. 2014)). See Cauchemez et al. (2006a) for proof of concept of real-time estimation techniques.
We direct readers interested in statistical epidemic models to the papers of Britton (2010) and Allen (2017), and for modeling and parameter estimation to the books of Andersson and Britton (2000) and Becker (2015).
In addition to identifying model parameters, it is equally important to understand and quantify the uncertainty in these parameter estimates. For simple linear regression with normally distributed errors, the standard error can be calculated directly, leading to confidence intervals for the parameter estimates. For more complicated stochastic models, several techniques exist to estimate the standard error of the resulting parameter estimates, including bootstrapping and asymptotic theory (Efron 1979; Banks et al. 2010). We direct interested readers to Banks (Banks et al. 2014) for a thorough review of uncertainty in modeling, including uncertainty propagation through time due to uncertainty in the model formulation and uncertainty in measurement error during data collection (the observation process); standard error calculations are included.
Here we describe a recent parameter identification technique for time-dependent parameters in stochastic dynamic models (Otunuga 2014; Otunuga et al. 2017, 2019), the local lagged adapted generalized method of moments (LLGMM), an extension of the generalized method of moments (Hansen 1982) (see also Sect. 3). The LLGMM was developed in the context of energy commodity spot prices, which are subject to response time delay and random environmental perturbations. The main advance of the LLGMM is to use some number of the past state values to construct local moment equations which are used estimate model parameters. In real world dynamic modeling problems, current and future states of continuous time dynamic processes can be influenced by the past state history. Therefore, the LLGMM technique is applicable to dynamic processes in the biological, financial, physical, chemical, and social sciences.
The LLGMM method is composed of the following components: (1) development of a stochastic mathematical model of continuous time dynamic process, (2) construction of an equivalent time series model, (3) development of generalized method of moment/observation equations, (4) introduction of a conceptual and computational parameter estimation scheme, (5) introduction of a conceptual and computational state estimation scheme, and (6) derivation of -best sub-optimal state and parameter estimates. In this work, we use the Monte-Carlo method and implicit Euler scheme (Kloeden and Platen 1995) for stochastic differential equation (SDE) to construct local moments equations and describe theoretical parameter estimation procedure for the SDE.
We demonstrate the local lagged adapted generalized method of moments parameter identification procedure (LLGMM) for the time-dependent transmission rate in a stochastic susceptible-exposed-infectious-recovered-susceptible (SEIRS) epidemic model, using US influenza data from 2004–2017. We assume the data is noise-free. The deterministic and corresponding stochastic SEIRS models are described in Sect. 2. The LLGMM is described in Section 3 and its application to the SEIRS model in Sect. 3.1. The identification procedure is applied to US influenza data in Sect. 4. The technique is evaluated with goodness-of-fit measures described in Sect. 4.1. The LLGMM is evaluated for forecasting ability in Sect. 4.2.
SEIRS model
The deterministic susceptible-exposed-infectious-recovered-susceptible (SEIRS) model with vital rates (Fig. 1) is given by the equations
| 1 |
where S(t), E(t), I(t), R(t) are the fractions of the total population that are susceptible, infected but not infectious (exposed), infectious, and temporarily immune (temporarily recovered), respectively, at time . We assume every individual is in one of these compartments at with . Adding the equations shows for all t. The transmission rate is , assumed to be a positive, bounded and continuous function of time t. We also assume that all susceptible individuals are exposed at the same transmission rate, . The parameters , , , and are each positive constants. Individuals are exposed for time , infectious for , and temporarily immune for . The demographic birth and death rate is ; there are no disease related deaths. We direct readers interested in a review of epidemiological compartment modeling with systems of differential equations to Edelstein-Keshet (2005), Kong et al. (2015) and Vynnycky and White (2010).
Fig. 1.
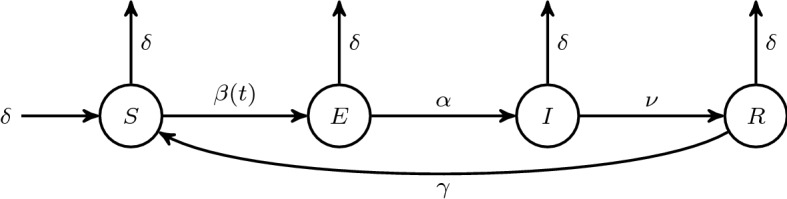
SEIRS model schematic with time-dependent transmission rate
We assume that the transmission rate is a function of time and is influenced by a Gaussian white noise process causing the rate to fluctuate around the function . The function corresponds to known influences such as periodic weather patterns or school terms; could be constant. According to Méndez et al. (2012), external fluctuations may be caused by variability in the number of contacts between infected and susceptible individuals and such random variations can be modeled by a white noise. Thus, we assume that external noise appears multiplicatively in the SEIRS model as follows
| 2 |
and is able to modify the dynamical behavior of the model. The transmission rate fluctuates rapidly so that we can model as a Gaussian white noise term with mean 0 and , where W(t) is the standard Wiener process defined on a complete filtered probability space , is measurable and is right continuous. The noise intensity, , of the white noise due to variations in infectivity is a function of time, measuring the amplitude of fluctuations in the transmission rate. We also assume that is -measurable and independent of . By substituting (2) into (1), we get a stochastic differential equation, and in particular a Langevin equation. By construction, the resulting stochastic model will be an adapted (non-anticipating) process.
The Itô approach on stochastic differential equation depends on Markovian and Martingale properties. These properties do not obey the traditional chain rule. Whereas, the Stratonovich approach obeys the traditional chain rule and allows white noise to be treated as a regular derivative of a Brownian or Wiener process. West et al. (1979) recommend that physical scientists avoid Itô calculus and claims that Stratonovich calculus is appropriate for Langevin equations with both internal and external noise. In their work, Moon and Wettlaufer (2014) suggests that in most areas of physical science where white noise is defined in terms of a -function autocorrelation, Stratonovich calculus is preferred due to the fluctuation-dissipation theorem. Also, Méndez et al. (2012) claim that any dynamics describing systems with real noise should be interpreted using Stratonovich equations. For these reasons, by substituting (2) into (1), we extend the resulting SEIRS model with vital rates and stochasticity to the Stratonovich equations and obtain
| 3 |
where is the Stratonovich integral. The formation of as in equation (2) and interpretation of the stochastic dynamic system of equations as a Stratonovich system of equations follow the work of Méndez et al. (2012), who investigated a stochastic susceptible-infectious-susceptible (SIS) epidemic model. We direct readers interested in a review of stochastic modeling with systems of differential equations to Ladde and Ladde (2013).
The SEIRS stochastic differential equation (3) has a unique (up to equivalence) global solution and the solution will remain within whenever it starts from there. This follows from Witbooi (2017), since the model presented here is a special case of the one presented there. (See also Parra et al. 2017 and Zhang and Teng 2007).
We assume that all model parameter values are known constants, except and . Also that the I(t) data is known and noise-free at discrete time points and can be modeled with the SEIRS model. The goal is to identify the time-dependent parameters and , specifically and at each time when I is known.
The LLGMM parameter identification procedure
The local lagged adapted generalized method of moments (LLGMM) builds on the generalized method of moments (GMM) (Hansen 1982) and we begin with a brief review of GMM. (See DeGroot and Schervish (DeGroot and Schervish 2011) for a review of the method of moments and other parameter estimation techniques.) Using the classical method of moments, one parameter is estimated by equating the model estimated expectation of y and the data estimated expectation of y. Assuming that y is generated by the model, the model estimated expectation is the population mean, while the data estimated expectation is the sample mean. The generalized method of moments constructs the moment conditions, also called orthogonality conditions, more generally as some function of the model parameters and data. The parameters are estimated by minimizing a certain norm of the sample averages of the moment conditions. If the model contains more than one parameter to be identified, the second moment, and higher if needed, would be used to construct the required moment conditions. Note that the minimization process in the GMM method uses the entire data set to estimate the constant parameters.
The LLGMM identifies time-dependent parameters using a limited number of past data points to form the moment conditions, not the entire data set. Thus the method is lagged. Also, the number of points used varies for each time estimate. Thus the method is local and adaptive.
Let
be an Itô stochastic model for y, with time-dependent parameter . The LLGMM parameter identification procedure estimates the expectation of using an average of more than one past data point. The number of past data points used at a particular time point in the LLGMM is allowed to vary between 2 and some fixed maximum M. By defining as the local admissible sampled data observation size at time , the parameters and states at time are estimated using past data on the interval . The expected value of is estimated from the data as
and from the stochastic model as
where for each time . Here, is the filtration process up to time and for simplicity we assume is constant. We call M the maximum delay constant and the delay constant; may also be thought of as the observation size. We derive appropriate moment equations, for example by equating the two expectations above, then solve for the parameter estimate of at time using the well known integral mean value theorem (Khalili and Vasiliu 2010). The computed time-dependent parameter depends on the number of past data points used in the average, so the parameter is more accurately expressed as .
In describing the LLGMM technique so far, we have at each time point, , an average of past data points. The value of the delay constant is determined using the following procedure. Fix m between 2 and M. At each time point, estimate in the stochastic model and use the value to generate a time series for y, say, . Compare the model output at time to the known value of , for example using sum of square distances. Since the model is stochastic, repeat the comparison some large number of times and compute the mean sum of square distances. Repeat for all m. Select the m value with mean model output closest to the known value. Record the value as . Evaluate the state and parameter, and , at as and , respectively.
SEIRS parameter estimation derivation
In this section, we detail how the LLGMM technique, explained in general terms above, is used to estimate the state, parameters, and observation size for the SEIRS model. The method description is organized following the six components of the LLGMM scheme outlined in the Introduction (Sect. 1).
Development of an Itô stochastic differential equations model
We use the Stratonovich-Itô conversion theorem given in Bernardi et al. (2001) and Kloeden and Platen (1995) to convert the Stratonovich dynamical model (3) to its Itô equivalent. We give the theorem below without proof.
Theorem 1
(Bernardi et al. 2001 and Kloeden and Platen 1995) The Itô stochastic differential equation (SDE)
| 4 |
defined componentwise as
having the same solution as the N-dimensional Stratonovich SDE with dimensional Wiener process
| 5 |
has drift coefficient f(t, X) that is defined in terms of , componentwise, by
| 6 |
To apply the LLGMM to the SEIRS model, we derive the corresponding Itô equations from system (3) using Theorem 1 as
| 7 |
where the initial conditions , , , and are each between 0 and 1. Together they satisfy . As it is well known, the Itô model (7) rests upon the Markovian and Martingale properties.
Construction of the time series model and generalized method of moment equations
As part of the components of the LLGMM scheme, we construct the time series model from the Itô equations (7) and develop generalized method of moment equations (orthogonality conditions) used to estimate parameters and .
From the Itô Lemma,
where is the quadratic variation symbol. Multiplying S and the dE equation from (7) gives,
| 8 |
To estimate the parameters and at time , we first integrate (8) on the interval to give
and generate moment equations from the integral equation. From the continuity of and positivity of S(t) and I(t), it follows from the integral mean value theorem (Khalili and Vasiliu 2010) that there exist , with such that
Discretizing the above equation and applying expectation gives
| 9 |
where , , , and is defined as the estimate of at time using delay constant .
Conceptual and computational parameter estimation scheme
We use the moment equation in (9) to derive parameter estimate for . Let be a parameter estimate of that is estimated using past data points at time . It follows from (9) that
| 10 |
Using the variance of , the second parameter can be estimated similarly as
| 11 |
The parameter estimates for the transmission rate and noise intensity are well-defined. If , then there are no infectious individuals during the time period and . From the discretized equations governing the disease spread (7), one can verify that with no infectious individuals there are also no exposed individuals. In particular, the disease spread has ended and at that time we no longer estimate the transmission rate. Also note, we do not have the case where . If this happens, then it follows from the discretized equation governing S in (7) that for , that is, for . In (11), if , then there is no disease transmission at time and therefore the noise intensity in the transmission rate is also zero.
We assume that the infection data is known at each point and that the data can be modeled with the SEIRS model. Using a simple discretization of the deterministic model equations (1) and the known initial conditions, it is possible to identify , , and . In particular, R can be determined from (1) as
E can be determined as
and finally . These values are used as the known data values in the LLGMM estimation.
Conceptual and computational state estimation scheme
When using a traditional explicit forward Euler-type discretization to construct solutions to the SEIRS model, we encountered negative solutions. Such unrealistic negative solutions is a common numerical artifact in solutions of systems with stochasticity. Schurz (1996) developed an implicit Euler method to ensure nonnegativity of solutions to SDE. Solutions to the SEIRS model are nonnegative (Sect. 2). Therefore, to estimate the state value at time , we discretize the Itô equations using the implicit Euler scheme described in Schurz (1996). (See also Cyganowski and Grune 2001.) Namely, for a stochastic model with time-dependent parameters , using an Itô interpretation,
the implicit Euler scheme gives that satisfies
for , where for sample size N, for L simulations in the Monte-Carlo method. For the SEIRS model, represents , the transmission rate and noise intensity described in (10) and (11), respectively. The simulated state for the l-th simulation at time using past data set is . The solution can be determined iteratively. A value of is used when solving I; a value of is used for S, E, and R.
Derivation of -best sub-optimal state and parameter estimates
Finally, there is a need to find, among the estimated values at time , the value closest to the known real value. Specifically, the correct delay constant must be determined. Let , , and be the simulated susceptible, exposed, infectious and recovered estimate for the l-th simulation at time using past data set. We take the average
| 12 |
as the simulated value of S(t), E(t), I(t) and R(t) at time using past data set. Define
| 13 |
as the quadratic mean square error between the known data and the averaged realizations . For any arbitrary small positive number and for each time , we define the -sub-optimal admissible set of at as
| 14 |
If gives the minimum value for , then we record as . If condition (14) is not met at time , then the value of where the minimum is attained is recorded as . The -best sub-optimal parameter and state estimates at time are now recorded as and , respectively.
Case study influenza
We apply the LLGMM to the SEIRS epidemic model and influenza data. The influenza data ‘Influenza Positive Tests Reported to CDC by Public Health Laboratories’ was collected from the Center for Disease Control and Prevention (CDC) Flu View for the thirteen influenza seasons 2004–2005 through 2016–2017.1 The number of positive tests is given weekly starting from week 40 of 2004 through week 39 of 2017. Our goal is to use the CDC data in the SEIRS model as a proxy for the infectious class I, however, there is an obvious mismatch between the number of infectious and the number of positive tests. We make several adjustments to the data to account for the main differences—underreporting by the laboratories and underreporting due to people who do not seek health care. The details are described in the next paragraph.
Following the general adjustment procedure in Bresee et al. (2013), we multiply the number of positive tests by 2.74 to account for underreporting by the laboratories. Additionally, we divide by 44.1556% to account for individuals not seeking medical care. This value is the weighted average of the percentage of individuals who seek medical care by age group (weighted by percent of each age group in the U.S. population) as listed in Bresee et al. (2013). Finally, the data is scaled so that the total attack rate for each season is within known estimates. We use the weighted average of the estimates computed by Molinari et al. (2007), by age group. The resulting attack rate is 8.44%. (This matches well with WHO estimates of 5–10% of adults and 20–30% of children are infected with influenza each year worldwide,2 when these estimates are weighted by US population age group, the attack rate is estimated at 8.867–15.156%.) After the underreporting and non-medical seeking adjustments, we determine the seasonal attack rates. A scale factor is determined to guarantee that the smallest attack rate is at the minimum 8.44% rate. The scale factor is determined by dividing the estimated attack rate (8.44% of the total population) by the minimum number infected over one season. This scale factor is applied to all influenza data; in particular, the adjusted data is multiplied by 231.53. We assume the total U.S. population is fixed at 311,749,110 people. The adjusted number of positive tests is divided by the total population to get the fraction of the population infectious each week ( in the model).
The CDC influenza data is given weekly, so that week. According to the CDC,3 influenza has a latency period between 1 and 4 days, with an average of 2 days, and an infectious period of 5 to 7 days after symptoms appear and 1 day before symptoms. Therefore, with t measured in weeks, and should be selected in the ranges [1.75, 7] and [0.875, 1.4] per week, respectively. The annual US birth rates for the years 2004–2016 range from 13.42 to 14.18 per 1000, while the death rates range from 8.14 to 8.39 per 1000.4 Therefore, should be selected in the range [0.000157, 0.000277]. No good information is known about how long an individual remains temporarily immune from influenza after having the disease. Xu et al. (2010), suggested that prior infection with the 1918 Spanish flu provided protection during the 2009 influenza pandemic, a span of almost 100 years. However, here we consider all influenza types as one ‘disease’, and given the conglomeration of virus strains, we assume it is possible for individuals to become infected each year. (In fact, it is possible to become infected more than once a year, though this is rare.) Kucharski et al. (2015), estimate that adults 30 years or older become infected with influenza approximately twice each decade. Therefore, could range from around 0.000192, corresponding to 100 years temporarily immune, through to 0.04, corresponding to 1/2 year, though likely the value should be closer to 0.00385, corresponding to 5 years, than to the 100 year value. A complete list of values used in the simulation is given in Table 1.
Table 1.
SEIRS model parameter values used in application of the LLGMM parameter identification technique to the SEIRS model and influenza data. 100 simulations are used to determine delay constant (observation size) ; for all times j, . Simulation values selected from within the ranges determined from the data sources; see text for complete description of parameter ranges. The other simulation values are assumptions
| Parameter | Description | Simulation Value | Source |
|---|---|---|---|
| Latency rate () | 3.5 | ||
| Recovery rate () | 1 | ||
| Loss of immunity rate () | 0.0078 | Xu et al. (2010); Kucharski et al. (2015) | |
| Birth / death rate () | 0.0002 | ||
| Initial time, week 40 of 2004 | 0 | ||
| I(0) | Initial condition | 10 | , then scaled |
| E(0), R(0) | Initial conditions | 0 | |
| S(0) | Initial condition | ||
| , | Initial conditions | 0 | |
| M | Maximum allowed past data points | 4, 13, 26, 52 weeks | |
| Combined data scaling value | Bresee et al. (2013); Molinari et al. (2007) |
The scaled influenza data and simulated infectious values are shown in the top panel of Fig. 2, assuming a maximum time delay (observation size) of weeks (1 year).
Fig. 2.
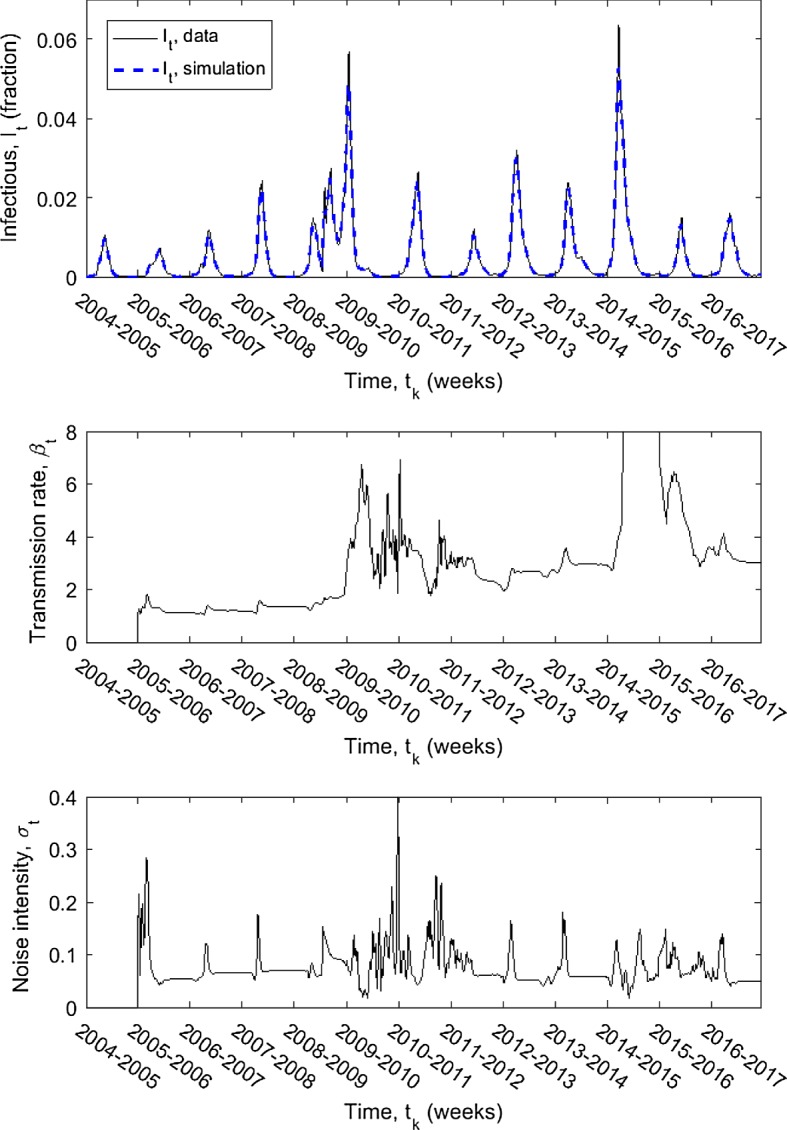
Scaled influenza data (solid) I(t) collected from the CDC Flu View for the thirteen influenza seasons 2004–2005 through 2016–2017 and simulated infectious values (dashed) I(t) with maximum time delay weeks (1 year). Time-dependent transmission rate and noise intensity for the simulated values; both are zero for the initial delay period of weeks. reaches a maximum value of 455.60 during the 2014–2015 influenza season
The ‘scaled influenza data’ is the number of positive tests reported to the CDC for thirteen influenza season scaled to account for underreporting, individuals not seeking medical care, and to guarantee the attack rate is in known estimates, as detailed at the start of Sect. 4. The ‘simulated infectious values’ are computed using the data by applying the LLGMM allowing up to one year of past observations () and assuming the data is well-described by an SEIRS model, as derived in Sect. 3.1.
The algorithm does a very good job of capturing the data. The only discrepancies lie at the peaks of the largest infections. The middle and bottom panels show the time-dependent transmission rate and noise intensity, respectively. Both the transmission rate and the noise intensity show modest fluctuations each season (note the difference in scales of the two functions). Together these influence the peak and timing of the infections. The large peak in the 2009–2010 influenza season can be seen as a significant influence by the transmission rate; the noise intensity shows fluctuations only after the peak. The large peak in the 2014–2015 season can be seen as a very significant peak in the transmission rate, but no corresponding peak in the noise intensity. Similar behavior is seen with all of the delay constants M tested (Table 1).
Figure 3 shows the number of past data values used in the LLGMM procedure, as determined by the computation itself. Notice that the algorithm seems to prefer using many past data points, except directly after the two large outbreaks during the 2009–2010 and 2014–2015 seasons. This is an artifact of the high delay constant of . As the maximum allowable number of past data points is decreased, the number of large spikes in decreases. In particular, with a maximum delay constant the algorithm uses the minimum value of 2 around 75% of the time, with the value of 2 is used around 50% of the time, and with around 25%. Each of the spikes in the determined delay constant comes around the largest peaks in infections. The larger the allowed maximum delay, the more peaks appear in the delay function.
Fig. 3.
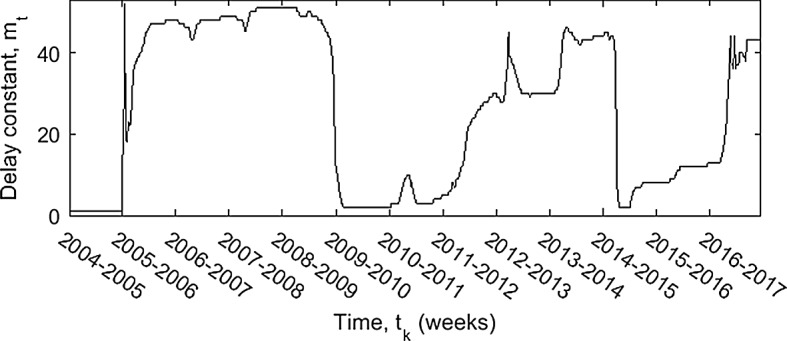
Time-dependent delay constant determined using the LLGMM procedure, with maximum delay weeks (1 year). The delay constant is zero for the initial delay period of weeks. Its minimum value after that is 2
Goodness-of-fit measures
We find the goodness-of-fit measures for the simulated infectious values following Czellar et al. (2007). We compute the root mean square error of the simulated path , the variability using the average median absolute deviation , and the average median bias :
| 15 |
where is the data and is a simulated data set for each at the times . A small indicates that the estimated parameter values can generate simulated paths similar to the actual data set. A small indicates that there is small variability among all simulated paths.
The goodness-of-fit measures are computed for the LLGMM procedure with maximum delay constant , and 52, using pseudo-data sets (Table 2). The fit measures show that using a higher delay constant (more past data points) allows for a better fit, though visually the infectious curves show almost no differences. In order to statistically compare the estimation results using different delay constant, we estimate the statistics , , and defined in (15).
Table 2.
Goodness of fit measures for the full simulation, a forecasted data set assuming data through the end of year 2016, and a forecasted data set assuming data through the end of the 2015–2016 influenza season (week 39 of 2016). Maximum delay constants M were considered for 4 weeks (1 month), 13 weeks (3 months), 26 weeks (6 months), and 52 weeks (1 year). When forecasting, the first delay constant is that used for the simulated values; the second is used in the forecasted values. As the delay constant increases, the root mean square error ( value) of the full simulation result decreases significantly. Likewise, the average median bias () decreases significantly as the delay constant increases. The table shows that the smallest bias of the simulation is derived using delay constant weeks. The accuracy of the simulation is the best with weeks (it gives the smallest value). The same conclusion applies to the forecast estimates derived assuming data through the end of year 2016. The fit measures show that using a higher delay constant (more past data points) allows for a better fit while forecasting for a year period. Using delay constant weeks reduces the variability () in the simulation best, followed by delay constants and weeks. The measure is 0 (see rows 5–8) because the simulated infectious data is estimated using . The results in the last four row (forecasted 2016–2017 data results assuming data through the end of 2015–2016) suggest that using small delay constant gives a better result while forecasting for a long period of time
| Delay constant(s) | ||||
|---|---|---|---|---|
| Full simulation | 4 | 0.0055 | ||
| Full simulation | 13 | 0.0046 | ||
| Full simulation | 26 | 0.0037 | ||
| Full simulation | 52 | 0.0016 | ||
| Forecast 2017 data | 52; 4 | 0 | 0.0045 | |
| Forecast 2017 data | 52; 13 | 0 | 0.0045 | |
| Forecast 2017 data | 52; 26 | 0 | 0.0045 | |
| Forecast 2017 data | 52; 52 | 0 | 0.0044 | |
| Forecast 2016–2017 data | 52; 4 | 0 | 0.0041 | |
| Forecast 2016–2017 data | 52; 13 | 0 | 0.0042 | |
| Forecast 2016–2017 data | 52; 26 | 0 | 0.0041 | |
| Forecast 2016–2017 data | 52; 52 | 0 | 0.0043 |
Forecasting
We test the efficacy of the LLGMM estimations in forecasting US influenza infection levels. We assume infection data is known up to some time, and thus S, E, and R are also known. Future data values are determined using the LLGMM technique in the following way.
Each of S, E, I, R, , and are estimated from the past data values iteratively using the implicit Euler method. These values depend on the random term and so we simulate each 100 times and compute the averages. These averages are summed and compared with the known value 1. At each time , the value of is determined as the value with sum closest to 1.
We perform two forecasts, one assuming data through the end of the 2015–2016 influenza season and forecasting the values for the entire 2016–2017 season (Fig. 4) and the second assuming data through the end of 2016 and forecasting the values for weeks 1 through 29 of 2017 (Fig. 5). The 95% confidence intervals are given around the forecasted values; the confidence intervals grow as the number of data points in the estimation increases. In both cases, the forecast shows a good qualitative fit.
Fig. 4.
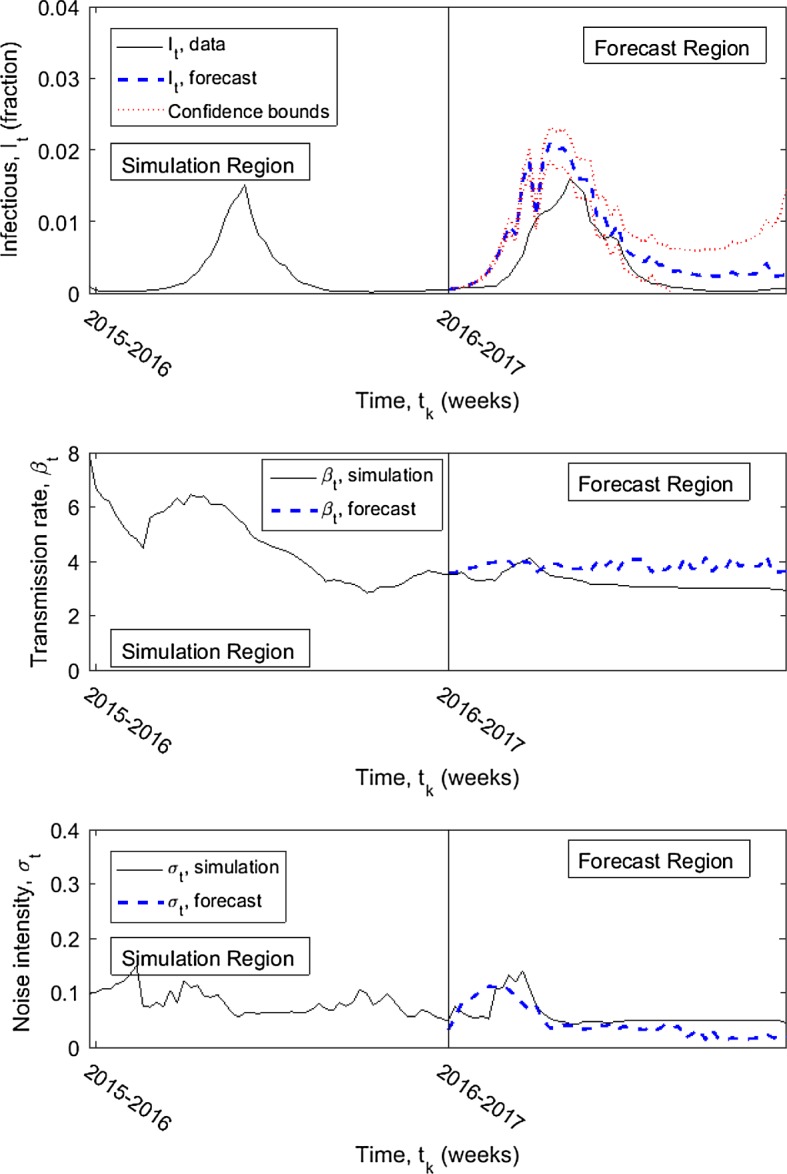
Forecasted infectious influenza data (thick blue dashed in forecast region) for year 2017 compared with the known data (solid) and simulation carried out assuming data through the end of the 2015–2016 influenza season and forecasting the values for the entire 2016–2017 season; upper and lower 95% confidence intervals (red dotted). Known infection data is shown for one year prior to forecasting. Simulated transmission rate and noise intensity also shown with forecasted values. Simulated data is generated with maximum delay constant (1 year); forecasted data is generated with (3 months) (color figure online)
Fig. 5.
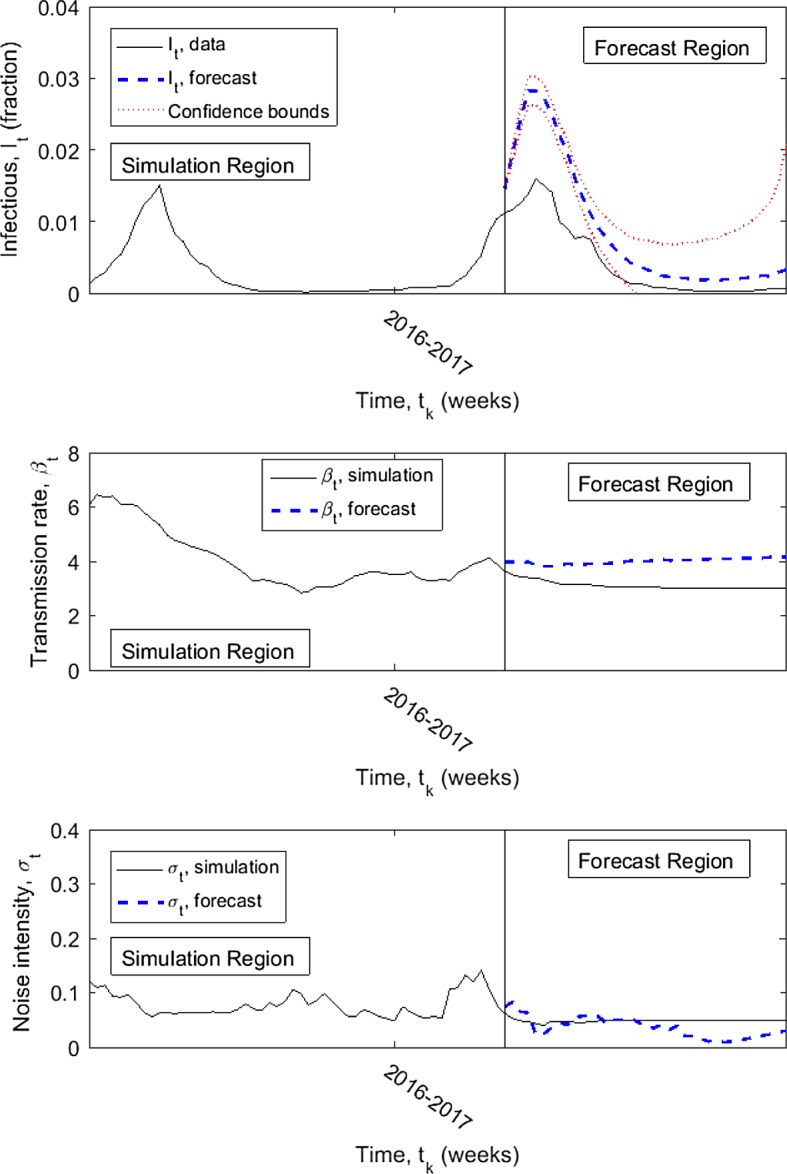
Forecasted infectious influenza data (thick dashed in forecast region) for year 2017 compared with the known data (solid) assuming data through the end of 2016 and forecasting the values for weeks 1 through 29 of 2017; upper and lower 95% confidence intervals (dotted). Known infection data is shown for one year prior to forecasting. Simulated transmission rate and noise intensity also shown with forecasted values. Simulated data is generated with maximum delay constant (1 year); forecasted data is generated with (3 months)
The goodness-of-fit measures (Sect. 4.1) are computed for the forecasted values—forecasting year 2017 only and forecasting the 2016–2017 season (Table 2). The forecasted values begin with assuming the simulated values up to the end of 2016 or the end of the 2015–2016 influenza season, respectively, using a maximum delay constant of weeks. The forecasted values are computed with , and 52, using pseudo-data sets. The measure is 0 because the simulated infectious data is estimated using . Based on the goodness-of-fit values, the forecast for the 2016–2017 influenza season is noticeably better than those for just the year 2017. However, visually no benefit is seen to a higher or lower M value in either forecast.
Discussion
The local lagged adapted generalized method of moments (LLGMM), a parameter identification procedure for stochastic dynamic models, was described and demonstrated with case study of the SEIRS epidemic model. In the case study, influenza data from the US 2004–2005 to 2016–2017 influenza seasons was used to identify both the time-dependent transmission function and the time-dependent noise intensity. The LLGMM provides a new technique for gaining valuable information about the spread of diseases, namely identification of the complicated transmission rate.
As noted in Sect. 2, we follow the basic formation of our stochastic SEIRS model as in Méndez et al. (2012). In particular, the model we consider here captures only the stochastic nature of the transmission function with the Weiner process explicitly in the formation of the function . This is done for two reasons. First, the transmission rate is the most difficult epidemiological parameter to estimate (Anderson and May 1991) and it is highly variable, making it of paramount importance to determine. The latency and recovery period can be measured from data and are well described by a constant value, the population mean. Second, we offer here a proof of concept that the LLGMM is a powerful tool for the inverse problem in epidemiological modeling by focusing on a simple SEIRS model applied to influenza data. Future applications of the LLGMM would include more realistic modeling of influenza outbreaks in the United States. As a case in point, vaccination uptake is highly variable and important to the modern control of influenza outbreaks, therefore influenza models should include vaccination. See for example Kong et al. (2015) who investigate the effect of vaccination on a SEIRA epidemic model with separate juvenile and adult groups. They introduce an inverse method for extracting time-dependent transmission rate from pre-and-post vaccination measles incident data.
The main technique for LLGMM involves computing the expected value of model variables by averaging the values of some number of past data points. We have considered here all reported influenza infections as infections arising from one disease, which is a gross generalization. Each year multiple influenza types, e.g. A and B, and strains, e.g. influenza A H1N1 or H3N2, circulate and are constantly mutating. By averaging the transmission patterns of several past weeks to determine a current transmission rate, we allow explicit computation of assuming previous transmission patterns are influential on the transmission of current week. From a biological view, averaging assumes mutated strains are likely to have similar transmission rates, among other properties.
The last recorded influenza pandemic (pH1N1) occurred during the 2009–2010 season and the data (Fig. 2) clearly show the summer peak followed by the higher peak in November. In the United States the 2014–2015 influenza season was moderately severe, perhaps accounted for by a mutated (drifted) H3N2 strain that did not match the vaccine Appiah et al. (2015). Again, this peak is noticeable in the data. These two high peaks in the fraction of infectious individuals correspond to the largest spikes in the transmission rate , with rapid oscillations only seen for the 2009–2010 season. Extreme oscillations in the noise intensity occur around these two influenza seasons, though only the 2009–2010 season shows a larger than normal noise intensity. The delay constant m shows a sharp decrease around both seasons. Thus, the LLGMM procedure is able to identify that these two seasons are different than the other ‘normal’ influenza seasons. It is also able to capture some fundamental difference between these two extreme seasons.
Our main goal here was to describe the LLGMM technique. We use the SEIRS model and influenza data as a demonstration of the technique. Therefore, we considered only a basic epidemiological model and did only modest adjustments to the CDC influenza data. The technique shows promise for future study given the differences it is able to capture in and between the two ‘non-normal’ influenza seasons, even with the simple model and data adjustments. Researchers with a goal of analyzing trends in the identified transmission rate should consider a more realistic model for influenza, accounting for influential aspects such as vaccination, asymptomatic infection, effects based on age group, and changes in the total population size. More accurate initial conditions and modifications to the CDC data should also be considered.
Footnotes
http://gis.cdc.gov/grasp/fluview/fluportaldashborad.html, accessed 7.5.2017.
https://www.who.int/biologicals/vaccines/influenza/en, accessed 6.28.2016
https://www.cdc.gov/flu/professional/acip/clinical.htm, accessed 6.28.2016
CIA World Factbook, https://www.cia.gov/library/publications/the-world-factbook/, accessed 5.23.2016.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Anna Mummert, Email: mummerta@marshall.edu.
Olusegun M. Otunuga, Email: otunuga@marshall.edu
References
- Allen LJS. A primer on stochastic epidemic models: formulation, numerical simulation, and analysis. Infect Dis Model. 2017;2(2):128–142. doi: 10.1016/j.idm.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RM, May RM. Infectious diseases of humans: dynamics control. New York: Oxford University Press; 1991. [Google Scholar]
- Anderson RM, Donnelly CA, Ferguson NM, Woolhouse MEJ, Watt CJ, Udy HJ, MaWhinney S, Dunstan SP, Southwood TRE, Wilesmith JW, Ryan JBM, Hoinville LJ, Hillerton JE, Austin AR, Wells GAH. Transmission dynamics and epidemiology of BSE in British cattle. Nature. 1996;382:779–788. doi: 10.1038/382779a0. [DOI] [PubMed] [Google Scholar]
- Andersson H, Britton T (2000) Stochastic epidemic models and their statistical analysis, vol 151. Lecture notes in statistics. Springer, New York
- Appiah GD, Blanton L, D’Mello T, Kniss K, Smith S, Mustaquim D, Steffens C, Dhara R, Cohen J, Chaves SS, Bresee J, Wallis T, Xu X, Ela AIA, Gubareva L, Wentworth DE, Katz J, Jernigan D, Brammer L. Influenza activity–United States, 2014–15 season and composition of the 2015–16 influenza vaccine. MMWR Morb Mortal Wkly Rep. 2015;64(21):583–590. [PMC free article] [PubMed] [Google Scholar]
- Arnold AM, Lloyd AL. An approach to periodic, time-varying parameter estimation using nonlinear filtering. Inverse Probl. 2018;34(10):105005. [Google Scholar]
- Audoly S, Bellu G, D’Angio L, Saccomani M, Cobelli C. Global identifiability of nonlinear models of biological systems. IEEE Trans Biomed Eng. 2001;48(1):55–65. doi: 10.1109/10.900248. [DOI] [PubMed] [Google Scholar]
- Banks HT, Holm K, Robbins D (2010) Standard error computations for uncertainty quantification in inverse problems: asymptotic theory vs. bootstrapping. Math Comput Model 52(9–10):1610–1625 [DOI] [PMC free article] [PubMed]
- Banks HT, Hu S, Thompson WC. Modeling and inverse problems in the presence of uncertainty. 1. Boca Raton: CPC Press; 2014. [Google Scholar]
- Bailey NTJ. The total size of a general stochastic epidemic. Biometrika. 1953;40(1–2):177–185. [Google Scholar]
- Becker NG. Estimation for an epidemic model. Biometrics. 1976;32(4):769–777. [PubMed] [Google Scholar]
- Becker NG (1989) Analysis of infectious disease data. monographs on statistics and applied probability. Chapman and Hall/CRC, London
- Becker NG. Uses of the EM algorithm in the analysis of data on HIV/AIDS and other infectious diseases. Stat Methods Med Res. 1997;6(1):24–37. doi: 10.1177/096228029700600103. [DOI] [PubMed] [Google Scholar]
- Becker NG. Modeling to onform infectious disease control. New York: CRC Press; 2015. [Google Scholar]
- Berry DA, Stangl D. Bayesian biostatistics. New York: CRC Press; 2018. [Google Scholar]
- Bernardi C, Madday Y, Blowey JF, Coleman JP, Craig AW. Theory and numerics of differential equations. Berlin: Springer; 2001. [Google Scholar]
- Breban R, Riou J, Fontanet A. Interhuman transmissibility of Middle East respiratory syndrome coronavirus: estimation of pandemic risk. Lancet. 2013;382(9893):694–699. doi: 10.1016/S0140-6736(13)61492-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresee J, Reed C, Kim IK, Finelli L, Fry A, Chaves SS, Burns E, Gargiullo P, Jernigan D, Cox N, Singleton J, Zhai Y, O’Halloran A, Kahn K, Lu P-J, Santibanez TA. Estimated influenza illnesses and hospitalizations averted by influenza vaccination–United States, 2012–13 influenza season. MMWR Morb Mortal Wkly Rep. 2013;62(49):1151–1154. [PMC free article] [PubMed] [Google Scholar]
- Bishwal JPN. Parameter estimation in stochastic differential equations. Berlin: Springer; 2008. [Google Scholar]
- Britton T. Stochastic epidemic models: a survey. Math Biosci. 2010;225(1):24–35. doi: 10.1016/j.mbs.2010.01.006. [DOI] [PubMed] [Google Scholar]
- Cauchemez S, Boëlle PY, Donnelly CA, Ferguson NM, Thomas G, Leung GM, Hedley AJ, Anderson RM, Valleron AJ. Real-time estimates in early detection of SARS. Emerg Infect Dis. 2006;12(1):110–113. doi: 10.3201/eid1201.050593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauchemez S, Ferguson NM. Likelihood-based estimation of continuous-time epidemic models from time-series data: application to measles transmission in London. J R Soc Interface. 2008;5:885–897. doi: 10.1098/rsif.2007.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazelles B, Chau NP. Adaptive dynamic modeling of HIV/AIDS epidemic using extended Kalman filter. J Biol Syst. 1995;3(3):759–768. [Google Scholar]
- Chappell MJ, Godfrey KR, Vajda S. Global identificaiton of the parameters of a nonlinear systems with specified input: a comparison of methods. Math Biosci. 1990;102(1):41–73. doi: 10.1016/0025-5564(90)90055-4. [DOI] [PubMed] [Google Scholar]
- Cyganowski S, Grune L, Kloeden PE. Maple for stochastic differential equations. In: Blowey James F, Coleman John P, Craig Alan W., editors. Theory and numerics of differential equations. Berlin: Springer; 2001. [Google Scholar]
- Czellar V, Karolyi GA, Ronchetti E. Indirect robust estimation of the short-term interest rate process. J Emper Finance. 2007;14(4):546–563. [Google Scholar]
- DeGroot MH, Schervish MJ. Probability and statistics. 4. London: Pearson; 2011. [Google Scholar]
- Edelstein-Keshet L. Mathematical models in biology, classics in applied mathematics. Philadelphia: Society for Industrial and Applied Mathematics; 2005. [Google Scholar]
- Efron B. Bootstrap methods: another look at the jackknife. Ann Stat. 1979;7:1–26. [Google Scholar]
- Eisenberg MC (2013) Generalizing the differential algebra approach to input-output equations in structural identifiability. ArXiv e-prints
- Eisenberg MC, Robertson SL, Tien JH. Identifiability and estimation of multiple transmission pathways in cholera and waterborne disease. J Theor Biol. 2013;324:84–102. doi: 10.1016/j.jtbi.2012.12.021. [DOI] [PubMed] [Google Scholar]
- Escobar J. Time-varying parameter estimation under stochastic perturbations using LSM. J Math Control Inform. 2012;29(2):235–258. doi: 10.1093/imamci/dnr028. [DOI] [Google Scholar]
- Evans ND, Chapman MJ, Chappell MJ, Godfrey KR. Identifiability of uncontrolled nonlinear rational systems. Automatica. 2002;38(10):1799–1805. [Google Scholar]
- Fraser C. Estimating individual and household reproduction numbers in an emerging epidemic. PLoS ONE. 2007;2(8):e758. doi: 10.1371/journal.pone.0000758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser C, Donnelly CA, Cauchemez S, Hanage WP, Van Kerkhove MD, Hollingsworth D, Griffin J, Baggaley RF, Jenkins HE, Lyons EJ, Jombart T, Hinsley WR, Grassly NC, Balloux F, Ghani AC, Ferguson NM, Rambaut A, Pybus OG, Lopez-Gatell H, Alpuche-Aranda CM, Chapela IB, Zavala EP, Guevara DME, Checchi F, Garcia E, Hugonnet S, Roth C. Pandemic potential of a strain of influenza A (H1N1): early findings. Science. 2009;324(5934):1557–1561. doi: 10.1126/science.1176062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson NM, Donnelly CA, Anderson RM. Transmission intensity and impact of control policies on the foot and mouth epidemic in Great Britain. Nature. 2001;413:542–548. doi: 10.1038/35097116. [DOI] [PubMed] [Google Scholar]
- Fisman D, Khoo E, Tuite A. Early epidemic dynamics of the West African 2014 Ebola outbreak: estimates derived with a simple two-parameter model. PLoS Curr. 2014 doi: 10.1371/currents.outbreaks.89c0d3783f36958d96ebbae97348d571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg E. Introduction to bayesian econometrics. 2. Cambridge: Cambridge University Press; 2013. [Google Scholar]
- Godfrey KR, DiStephano JJ., III . Identifiability of model parameters. In: Walter E, editor. Identifiability of parmetric models. Amsterdam: Elsevier; 1987. [Google Scholar]
- Gunn RN, Cahppell MJ, Cunningham VJ (1997) Reparameterization of unidentifiable systems using the Taylor series approach. In: Linkens DA, Carson E (eds), Proceedings of the third IFAC symposium on modelling and control in biomedical systems, Pergamon, Oxford, vol 7, pp 252
- Hadeler K. Parameter identification in epidemic models. Math Biosci. 2011;229:185–189. doi: 10.1016/j.mbs.2010.12.004. [DOI] [PubMed] [Google Scholar]
- Hansen LP. Large sample properties of generalized method of moments estimators. Econometrica. 1982;50(4):1029–1054. [Google Scholar]
- Heijmans RDH, Magnus JR. Consistent maximum-likelihood estimation with dependent observations. The general (non-normal) case and the normal case. J Econ. 1986;32:253–285. [Google Scholar]
- Hobbs NT, Hooten MB. Bayesian models: a statistical primer for ecologists. Princeton: Princeton University Press; 2015. [Google Scholar]
- Hurn S, Jeisman J, Lindsay K. Seeing the wood for the trees: a critical evaluation method to estimate the parameters of stochastic differential equations. J Financ Econ. 2007;5(3):390–455. [Google Scholar]
- Jeisman J (2005) Estimation of the parameters of stochastic differential equations, Ph.D. Dissertation, Queensland University of Technology
- Julier SJ, Uhlmann JK. Unscented filtering and nonlinear estimation. Proc IEEE. 2004;92(3):401–422. [Google Scholar]
- Kalman RE. A new approach to linear filtering and prediction problems. J Basic Eng. 1960;82(1):35–45. [Google Scholar]
- Keeling MJ, Wollhouse MEJ, Shaw DJ, Matthews L, Chase-Topping M, Haydon DT, Cornell SJ, Kappey J, Wilesmith J, Grenfell BT. Dynamics of the 2001 UK foot and mouth epidemic: stochastic dispersal in a heterogeneous landscape. Science. 2001;294(5543):813–817. doi: 10.1126/science.1065973. [DOI] [PubMed] [Google Scholar]
- Khalili P, Vasiliu D. An extension of the mean value theorem for integrals. Int J Math Educ Sci Technol. 2010;41(5):707–710. [Google Scholar]
- Kloeden PE, Platen E. Numerical solution of stochastic differential equations. New York: Springer; 1995. [Google Scholar]
- Kong JD, Jin C, Wang H. The inverse method for a childhood infectious disease model with its application to pre-vaccination and post-vaccination measles data. Bull Math Biol. 2015;77:2231–2263. doi: 10.1007/s11538-015-0121-5. [DOI] [PubMed] [Google Scholar]
- Kucharski AJ, Lessler J, Read JM, Zhu H, Jiang CQ, Guan Y, Cummings DAT, Riley S. Estimating the life course of influenza A(H3N2) antibody responses from cross-sectional data. PLoS Biol. 2015;13(3):1–16. doi: 10.1371/journal.pbio.1002082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladde AG, Ladde GS. An introduction to differential equations: stochastic modeling, methods and analysis. Singapore: World Scientific Publishing; 2013. [Google Scholar]
- Lipsitch M, Cohen T, Cooper B, Robins JM, Ma S, James L, Gopalakrishna G, Chew SK, Tan CC, Samore MH, Fisman D, Murray M. Transmission dynamics and control of severe acute respiratory syndrome. Science. 2003;300(5627):1966–1970. doi: 10.1126/science.1086616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljung L, Glad T. On global identifiability for arbitrary model parameterizations. Automatica. 1994;30(2):265–276. [Google Scholar]
- Longini IM, Koopman JS, Monto AS, Fox JP. Estimating household and community transmission parameters for influenza. Am J Epidemiol. 1982;115(5):736–751. doi: 10.1093/oxfordjournals.aje.a113356. [DOI] [PubMed] [Google Scholar]
- Longini IM, Koopman JS, Haber M, Cotsonis GA. Statistical inference for infectious diseases. Am J Epidemiol. 1988;128(4):845–859. doi: 10.1093/oxfordjournals.aje.a115038. [DOI] [PubMed] [Google Scholar]
- Méndez V, Campos D, Horsthemke W. Stochastic fluctuations of the transmission rate in the susceptible-infected-susceptible epidemic model. Phys Rev E. 2012;86:011919. doi: 10.1103/PhysRevE.86.011919. [DOI] [PubMed] [Google Scholar]
- Meng XL, van Dyk D. The EM Algorithm: an old folk-song sung to a fast new tune. J R Statist Soc B. 1997;59(3):511–567. [Google Scholar]
- Molinari NAM, Ortega-Sanchez IR, Messonniere ML, Thompson WW, Wortley PM, Weintraub E, Bridges CB. The annual impact of seasonal influenza in the US: measure disease burden and costs. Vaccine. 2007;25:5086–5096. doi: 10.1016/j.vaccine.2007.03.046. [DOI] [PubMed] [Google Scholar]
- Moon W, Wettlaufer JS. On the interpretation of Stratonovich calculus. New J Phys. 2014;16(055017):1–13. [Google Scholar]
- Mummert A. Studying the recovery procedure for the time-dependent transmission rate(s) in epidemic models. J Math Biol. 2013;67:483–507. doi: 10.1007/s00285-012-0558-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olama MM, Djouadi SM, Charalambous CD. Stochastic differential equations for modeling, estimation and identification of mobile-to-mobile communication channels. IEEE Trans Wireless Commun. 2009;8(4):1754–1763. [Google Scholar]
- O’Neill PD, Roberts GO. Bayesian inference for partially observed stochastic epidemics. J R Statist Soc A. 1999;162(1):121–129. [Google Scholar]
- O’Neill PD, Becker NG. Inference for an epidemic when susceptibility varies. Biostatistics. 2001;1(1):99–108. doi: 10.1093/biostatistics/2.1.99. [DOI] [PubMed] [Google Scholar]
- O’Neill PD. A tutorial introduction to Bayesian inference for stochastic epidemic models using Markov chain Monte Carlo methods. Math Biosci. 2002;180(1–2):103–114. doi: 10.1016/s0025-5564(02)00109-8. [DOI] [PubMed] [Google Scholar]
- Otunuga OM (2014) Stochastic modeling and analysis of energy commodity spot price processes, Ph.D. thesis, University of South Florida
- Otunuga OM, Ladde GS, Ladde NS. Local lagged adapted generalized method of moments and applications. Stoch Anal Appl. 2017;35(1):100–143. [Google Scholar]
- Otunuga OM, Ladde GS, Ladde NS. Local lagged adapted generalized method of moments and applications: an innovative estimation and forecasting approach and its applications. Time Ser Econ. 2019 doi: 10.1515/jtse-2016-0024. [DOI] [Google Scholar]
- Parra GG, Arenas AJ, Cogollo MR. Positivity and boundedness of solutions for a stochastic seasonal epidemiological model for respiratory syncytial virus (RSV) Ingeniería y Ciencia. 2017;13(25):95–121. [Google Scholar]
- Pohjanpalo H. System identifiability based on the power series expansion of the solution. Math Biosci. 1978;41(1–2):21–33. [Google Scholar]
- Pollicott M, Wang H, Weiss H. Extracting the time-dependent transmission rate from infection data via solution of an inverse ODE problem. J Biol Dyn. 2012;6:509–523. doi: 10.1080/17513758.2011.645510. [DOI] [PubMed] [Google Scholar]
- Rivers CM, Lofgren ET, Marathe M, Eubank S, Lewis BL (2014) Modeling the impact of interventions on an epidemic of Ebola in Sierra Leone and Liberia. PLoS Curr Outbreaks. 10.1371/currents.outbreaks.4d41fe5d6c05e9df30ddce33c66d084c [DOI] [PMC free article] [PubMed]
- Schurz H. Numerical regularization for SDEs: construction of nonnegative solutions. Dyn Syst Appl. 1996;5:323–351. [Google Scholar]
- Tuite AR, Tien J, Eisenberg M, Earn DJ, Ma J, Fisman DN. Cholera epidemic in Haiti, 2010: using a transmission model to explain spatial spread of disease and identify optimal control interventions. Ann Intern Med. 2011;154(9):593–601. doi: 10.7326/0003-4819-154-9-201105030-00334. [DOI] [PubMed] [Google Scholar]
- Ungarala S, Miriyala K, Co TB (2013) On the estimation of time-varying parameters in continuous-time nonlinear systems. In: 10th IFAC international symposium on dynamics and control of process systems the international federation of automatic control, India
- Vajda S, Godfrey KR, Rabitz H. Similarity transformation approach to identifiability analysis of nonlinear compartment models. Math Biosci. 1989;93(2):217–248. doi: 10.1016/0025-5564(89)90024-2. [DOI] [PubMed] [Google Scholar]
- Vynnycky E, White RG. An introduction to infectious disease modelling. Oxford: Oxford University Press; 2010. [Google Scholar]
- West BJ, Bulsara AR, Lindenberg K, Seshadri V, Shuler KE. Stochastic process with non-additive fluctuations: I. Itô versus Stratonovich calculus and the effects of correlations, Physica A. 1979;97:211–233. [Google Scholar]
- Witbooi PJ. An SEIRS epidemic model with stochastic transmission. Adv Differ Equ. 2017;2017:109. [Google Scholar]
- Xu R, Ekiert DC, Krause JC, Hai R, Crowe JE, Jr, Wilson IA. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science. 2010;328:357–360. doi: 10.1126/science.1186430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Sugimoto JD, Halloran ME, Basta NE, Chao DL, Matrajt L, Potter G, Kenah E, Longini IM. The transmissibility and control of pandemic influenza A (H1N1) virus. Science. 2009;326(5953):729–733. doi: 10.1126/science.1177373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Teng Z. On a nonautonomous SEIRS model in epidemiology. Bull Math Biol. 2007;69:2537–2559. doi: 10.1007/s11538-007-9231-z. [DOI] [PubMed] [Google Scholar]


