Abstract
Microarrays provide a powerful analytical tool for the simultaneous detection of multiple analytes in a single experiment. The specific affinity reaction of nucleic acids (hybridization) and antibodies towards antigens is the most common bioanalytical method for generating multiplexed quantitative results. Nucleic acid-based analysis is restricted to the detection of cells and viruses. Antibodies are more universal biomolecular receptors that selectively bind small molecules such as pesticides, small toxins, and pharmaceuticals and to biopolymers (e.g. toxins, allergens) and complex biological structures like bacterial cells and viruses. By producing an appropriate antibody, the corresponding antigenic analyte can be detected on a multiplexed immunoanalytical microarray. Food and water analysis along with clinical diagnostics constitute potential application fields for multiplexed analysis. Diverse fluorescence, chemiluminescence, electrochemical, and label-free microarray readout systems have been developed in the last decade. Some of them are constructed as flow-through microarrays by combination with a fluidic system. Microarrays have the potential to become widely accepted as a system for analytical applications, provided that robust and validated results on fully automated platforms are successfully generated. This review gives an overview of the current research on microarrays with the focus on automated systems and quantitative multiplexed applications.
Figure.
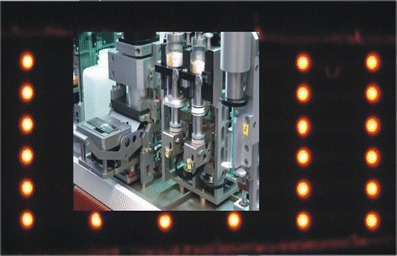
MCR 3: A fully automated chemiluminescence microarray reader for analytical microarrays
Keywords: Analytical microarrays, Biosystems technology, Environmental analysis, Food analysis, Clinical diagnostics
Introduction
Research on microarrays as multianalyte biosystems has generated increased interest in the last decade. The main feature of the microarray technology is the ability to simultaneously detect multiple analytes in one sample by an affinity-binding event at a surface interface. Fifteen years ago, the gene expression analysis of cDNA on microarrays was one of the first applications that successfully detected thousands of labelled target DNA molecules in parallel [1, 2]. Also the first immunoanalytical microarray was described at the same time [3]. In the meantime, a great variety of target analytes capable of interacting selectively with a biomolecular receptor has been adapted to microarrays.
In some cases immunoanalytical microarrays have the potential to replace conventional chromatographic techniques. They are applied if the number of samples is high or analysis by current methods is difficult and/or expensive [4]. Compared to the routinely applied chromatography techniques like GC or HPLC, immunoanalytical techniques are relatively fast and cost-effective [5, 6]. Therefore, microarray platforms have a great potential as monitoring systems for the rapid assessment of water or food samples [7]. Also other disciplines such as clinical diagnostics (especially point-of-care (POC) diagnostics) or forensic science have a demand for highly multiplexed analytical systems. A microarray platform will be accepted as an analytical system provided that it delivers robust and valid results in combination with a routinely used multiplexed application.
An overview of possible applications is given in Table 1. For these applications fully automated platforms are needed. Manually executed process steps would have to be avoided so that untrained personnel can operate an automated microarray platform inside and outside the laboratory. Furthermore, integration of fluidic systems and multiplexing of analytical parameters are key features for the future acceptance of analytical microarrays.
Table 1.
Possible applications for analytical microarrays
| Matrix | Class | Analytes |
|---|---|---|
| Drinking water | Toxins | Microcystin algae toxins |
| Pesticides | Up to 1000 individual pesticides | |
| Microorganisms | E. coli, coliforms, Enterococci, pathogenic bacteria and viruses | |
| Milk | Toxins | Aflatoxin M1, staphylococcal enterotoxins (SEA, SEB, SEC, SED) |
| Microorganisms | Pathogenic bacteria (e.g. C. jejuni; L. enterocolitica) | |
| Pharmaceuticals | e.g. Antibiotics, sulfonamides, prostagens | |
| Cereal products | Toxins | Aflatoxin B1, B2, G1 and G2 |
| Deoxynivalenol | ||
| Ochratoxin A | ||
| Zearalenone | ||
| Fumonisins | ||
| PCB congeners | ||
| Baby food | Pesticides | Up to 1000 individual pesticides |
| Feed | Pharmaceuticals | Diclofenac, clenbuterol, antibiotics |
| Toxins | Mycotoxins | |
| Blood | Inflammation factors | e.g. Cytokines |
| Allergens | IgE Antibodies | |
| Bacteria and viruses | Antibodies against pathogenic bacteria and viruses | |
| Air | Allergens | Pollen proteins |
| Bacteria | e.g. B. anthracis, M. tuberculosis | |
| Viruses | e.g. Influenza, SARS |
In this review the current existing microarray platforms will be discussed with regard to the technical components (fluidics, readout), surface chemistry and multiplexed applications.
Principle of analytical microarrays
All microarrays are based on one general principle—certain (bio)molecular recognition elements are defined on a heterogeneous matrix. Each element is dedicated to an analyte and contains quantitative information. The matrix is a patterned surface where the recognition molecules are immobilized by microprinting or microstructuring processes (Fig. 1a).
Fig. 1.
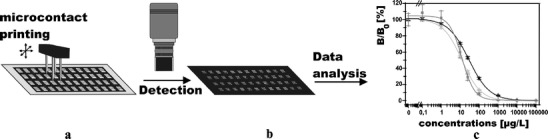
Schematic diagram of production a, imaging b, and data analysis c for a quantitative analytical microarray
An optical matrix on microparticles is also possible by mapping with a chromogenic code. The 2D readout is achieved by scanning or imaging the patterned surface (Fig. 1b), by means of a sensor array, by parallel arrangement of segmented channels, or by parallel detection of two bits of optical information (e.g. two different fluorescence emission wavelengths) encoded in microparticles.
Quantitative results with an analytical microarray are obtained by calibration with standardized analytes (Fig. 1c). Data analysis of an analytical microarray is achieved with software for image processing. Using a segmented microarray on a microtitre plate or with a flat slide with barriers, the multiplexed standard concentrations and the samples are separately placed on the segmented microarray and are simultaneously measured after a washing and incubation step. Microarrays on flat substrates with one flow channel can be calibrated sequentially if they are regenerable. Single-usable microarrays need, for each multi-analyte concentration, a separate microarray and positive and negative control spots. A solution to this problem is a microarray with multiple parallel flow channels using one channel for sample analysis and the other for calibration. Microarrays for screening applications have only positive and negative control spots to ensure that threshold limits for positive responses are maintained and that non-specific adsorption does not create a false-negative signal.
A flow-through microarray platform is a microarray with additional integrated instrumentation for a multiplexed analytical application. It consists of a fluidic system for sample introduction, a reagent supply, a flow cell, a microarray on a substrate, and a detection system. The fluidic system is in some way similar to a flow-injection analysis (FIA) system except for the implemented microarray. The analytical process is carried out automatically with computer-controlled pumps and valves, as depicted in Fig. 2.
Fig. 2.
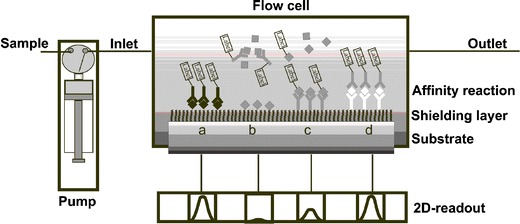
Schematic diagram of set-up of analytical flow-through microarray platforms for quantification of analytes with indirect a, b, direct c, and sandwich d assay formats
Analytical microarrays use the specific affinity reaction of receptor molecules (e.g. proteins, nucleic acids) and analyte or analyte derivatives on the heterogeneous phase. Analyte derivatives are immobilized using the indirect assay format reacting with the receptor molecules from the liquid phase (Fig. 2a,b). The reverse arrangement is the direct assay format by immobilization of receptor molecules as capture molecules (Fig. 2c). Molecules with different binding positions are detected by use of a sandwich assay format (Fig. 2d). The substrate is the ground surface for building up the microarray step-by-step. The top layer is activated in a way that capture molecules or analyte-derivatives are directly immobilized on to the spotted matrix. The quantification of analytes with a microarray is achieved by multiple affinity reactions. The affinity reaction is an equilibrium reaction strongly depending on the concentration of analytes, since the receptor and the other reagents are present at constant concentrations. Small analytes are quantified by an indirect competitive assay format using, preferentially, multiple antibodies. In this case the binding reaction takes place in solution. Only the free antibodies at low analyte concentration react with the immobilized analyte derivatives at the surface. The binding of antibodies at the surface is detected with a microarray readout system such as depicted in Fig. 3 with a chemiluminescence microarray readout principle. Flow-through analytical microarray platforms automatically deliver samples, and reagent solutions and washing buffer, by connection to a fluidic system.
Fig. 3.
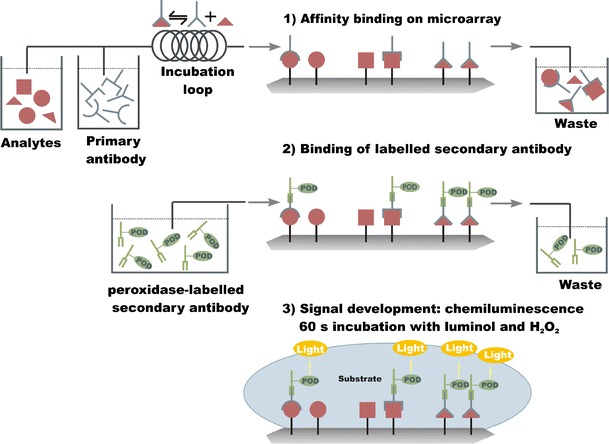
Principle of multiplexed analysis by use of a chemiluminescence microarray with an automated sample and reagent supply
Multiplexed sandwich immunoassay formats quantify proteins (e.g. proteinic toxin targets, cytokines), and bacteria and viruses. The DNA or RNA of cells and viruses can be analysed quantitatively by a sandwich hybridization assay according to sandwich immunoassays. A labelled detection oligonucleotide (ODN) and an immobilized capture ODN as receptor are used for quantifying the native DNA or RNA concentration in a sample.
Semi-quantitative results are obtained with labelled analytes such as target DNA on DNA microarrays for gene expression analysis or antibody profiling of sera on antigen microarrays. Also direct assay formats are used for screening applications by immobilization of the analyte such as ligands and labelled receptors. The types of microarray format and the applications are summarized in Table 2 and explained in detail in the next section.
Table 2.
Types of microarray
| Microarray | Biomolecular receptor | Analyte | Test format | Analytical application |
|---|---|---|---|---|
| Quantitative results | ||||
| Hapten microarray | Antibodies Aptamers | Small toxin targets, pesticides, pharmaceuticals | Indirect immunoassay | Food, forensic, water, process analysis, clinical diagnostics, bioterrorism protection |
| Antibody microarray | Proteinic toxin targets, viruses, bacteria | Sandwich immunoassay | ||
| DNA microarray | DNA, RNA | Bacteria, viruses | Sandwich hybridization assay | |
| Semi-quantitative results | ||||
| Antigen microarray | Antibodies | Cells, allergens cytokines | Direct immunoassay | Drug discovery, clinical diagnostic |
| Protein microarray | Protein receptors | Enzymes, antibodies, receptors | Reversed-phase, functional, and quantitative array | Drug discovery, proteome analysis |
| Carbohydrate microarray | Lectins, antibodies, viruses | Lectins, antibodies, viruses | Ligand binding assay | Clinical diagnostics, drug discovery |
| Peptide microarray | Enzymes, cells | Pharmaceutical inhibitors Protein receptors, cells | Enzyme inhibition assay Ligand binding assay | Drug discovery, Clinical diagnostics |
| Enzyme microarray | Enzymes | Pharmaceuticals inhibitors | Enzyme inhibition assay | Drug discovery |
| Receptor microarray | Membrane receptors, olfactory receptors | Pharmaceuticals | Ligand binding assay | Drug discovery |
| DNA microarray | Nucleic acids | RNA, DNA | PCR amplification and hybridization assay | Food, forensic, water analysis, clinical diagnostics |
| Cell microarray | Cell, tissue | Pharmaceuticals, cDNA transfection, siRNA, miRNA | Immune fluorescence Reporter gene assay | Drug discovery, clinical diagnostics |
Types of analytical microarray format and possible applications
Currently, for most applications with analytical microarrays only proof-of-principle studies are published in the literature. In this chapter, an overview is given over analytical microarray formats in context with a multianalyte application. In the future possible multi-analyte applications have to be validated on automated flow-through microarrays proving the benefit of such a system. With analytical microarrays, quantitative and semiquantitative results are obtained. Quantitative results are important if regulated chemical contaminants are quantified in food or water to monitor the observance of maximum residue limits (MRLs). Other applications are in the analytical field of clinical diagnostics. For example, in a blood sample diverse biomarkers of a disease need to be quantified in parallel. In this section, most of the reviewed analytical applications are with non-automated microarray systems. The aim of the section is to point out the diversity of applications of analytical microarrays developed in the last decade. All applications can be automated on an analytical microarray platform. The section “Analytical microarray platforms” describes the different automated microarray platforms in detail.
Microarrays for quantitative applications
Hapten and antibody microarrays
Antibody-based microarrays are a powerful tool for analytical purposes. Immunoanalytical microarrays are a quantitative analytical technique using antibodies as highly specific biological recognition elements [8, 9]. They can be designed for a variety of analytical applications producing rapid results with low limits of detection (LOD).
The detection antibodies are in direct contact with the sample, without prior sample cleanup. Immobilized haptens in combination with an indirect competitive immunoassay (IA) are the common format for the detection of multiple small molecules (e.g. pesticides, pharmaceuticals, small toxin targets). Haptens provoke an immune response if coupled to a protein by use of their functional group(s). Hapten microarrays use analyte derivatives as immobilized recognition molecules [10]. Antibody microarrays quantify proteins, bacteria, or viruses by using a sandwich immunoassay format.
The need for a certain amount of antibodies for multianalyte applications with analytical microarrays and the cost and time-intensive production of monoclonal antibodies has become crucial [11]. One strategy is the generation of highly specific oligonucleotide or peptide aptamers [12]. Oligonucleotide aptamers were evolutively selected from large libraries (1010–1015 different DNA or RNA molecules) by the SELEX process [13]. An advantage of aptamers is that they can be selected against difficult immunizable analytes such as toxins, viruses, or bacteria [14]. However, only few aptamers with high affinity constants are currently available for analytical applications [15].
Pesticides
A pesticide microarray for the monitoring of food and water has been developed [10, 16]. The restriction is the large number of approximately 1,000 applied pesticides. This quantity exceeds the financial capacity for antibody development. Another complexity is cross-reactivity against closely related compounds. This is still an open problem for multiplexed immunoanalytical pesticide detection.
Pharmaceuticals, hormones and endocrine-disrupting compounds
Pharmaceuticals, hormones, and endocrine-disrupting compounds (EDCs) have become important contaminants in drinking water and food. Pharmaceuticals are introduced by sewage water dispersion or by animals through veterinary use in farming. Approximately 3,000 different pharmaceuticals, for example analgesics, antibiotics, antidiabetic drugs, and ß-blockers, have to be removed from sewage water treatment to protect the environment. Multiplexed analysis with microarrays would be valuable for the online monitoring of possible contaminants.
The detection of antibiotics in milk is one of the first applications in food analysis using an automated microarray analyser. A hapten microarray has been developed for quantifying, in parallel, ten of the most important antibiotics in milk [17].
EDCs have become an important eco-toxicological issue. EDCs is a collective term for small molecules interacting with the oestrogen receptor and interfering with the endocrine system of humans and other vertebrates. Natural and synthetic estrogens, phyto-oestrogens, alkylphenols, polychlorinated biphenyls, and pesticides are examples of EDCs. The list is currently open for more chemicals and unknown metabolites. EDCs have to be identified and quantified in waste water-treatment plants at levels as low as parts per trillion [18]. Hapten microarrays for detection of EDCs could be applied as a water monitoring system [19]. Proof-of-principle has been demonstrated with a fully automated AWACSS fluorescence immunosensor [20]. Further developments depend strongly on their market potential. The relevance is increased at the moment by defining MRLs for EDCs at the European legislation level.
Toxins
Toxins are products of microorganisms, fungi, or parasites. They are either small organic molecules or proteins. Toxins can enter the food chain if water, animals, or other food is contaminated with microorganisms and their toxins are accumulated in their habitat. The oral ingestion of contaminated food has toxic effects on human health. Undesired ingredients in food and drinking water have to be avoided. An immunoanalytical platform able to detect microorganisms and toxins would considerably improve the quality and safety of food and drinking water.
Microcystins (MCs) are a family of hepatotoxic cyclic heptapeptides produced by freshwater cyanobacteria. Among more than 80 microcystins identified to date, only a few occur frequently and in high concentrations. Microcystin-LR, which contains leucine (L) and arginine (R), is the most frequently identified microcystin in water, and the most toxic. The WHO has set a provisional threshold limit at 1 μg L−1 for total microcystin-LR (cell-bound and dissolved) [21].
Mycotoxins are produced by fungi and occur in food and feed. The major fungal genera producing mycotoxins include Aspergillus, Fusarium, and Penicillium. They show carcinogenic, mutagenic, immunosuppressive, nephrotoxic, and teratogenic properties. Examples are ochratoxin A, aflatoxins, zearalenone, fumonisins, T-2 toxin, and deoxynivalenol. Regulations for major mycotoxins in food and feed exist, but they differ greatly among countries [22]. The detection limits for mycotoxins on an immunoanalytical platform should be as low as 1 μg L−1. Zheng et al. have reviewed rapid methods for the analysis of mycotoxins [23]. Mycotoxin microarrays are under investigation [24]. No microarray system for routine quantification of toxins in food samples has yet been described in the literature.
In contrast with small organic molecules, protein toxins like staphylococcal enterotoxins (SE) can cause food-borne illnesses. SEs are heat-resistant and are an indication of poor sanitation or postcontamination of processed food, such as dairy and meat products, pasta, instant soups, pastry, etc. The minimum levels of SE causing gastroenteritis in humans are approximately 200 ng per 100 g food. Methods like immunoassays are desired for detecting SE toxins at concentrations at or below 1 μg kg−1 in food. Botulinum neurotoxins (BoNTs), another proteinic toxin target, are produced by Clostridium botulinum and are the most poisonous substances known. The lethal dose for 50% (LD50) of the exposed population is approximately 1 ng kg−1. Cases of botulism are rare, but the potency and ease of distribution of BoNTs have resulted in the listing of botulism as one of the six highest-risk threat agents for bioterrorism. Other extremely toxic protein targets are ricin, cholera toxin, viscumin, or tetanus toxin. Sensitive immunoanalytical microarrays with highly affine antibodies are needed for the rapid monitoring of food and water samples.
Microorganisms
Infectious diseases caused by pathogenic bacteria, viruses, and parasites are the most common widespread health risks associated with drinking water and food. Bacterial food-borne outbreaks account for 91% of total outbreaks [25]. Microorganisms can enter the food chain as a result of poor hygiene, accident, or act of bioterrorism. Conventional methods for detection of microorganisms are time and cost-intensive. However, they are very reliable and are able to identify a single living cell in a 100 mL sample by combining filtration and cultivation. Immunoanalytical and DNA microarrays could have a high impact on the rapid identification of microorganisms, provided that highly specific antibodies are generated and the pre-enrichment steps are integrated within the detection system [26, 27].
Clinical diagnostics
Quantitative antibody microarrays have been developed for clinical diagnostics, although routine clinical use of microarray technology is still in its early stages [28]. The quantitative detection of the prostate-specific antigen (PSA) on a microarray has been shown by Jaras et al. [29]. Cytokines have been quantified in a multiplexed manner in serum or plasma [30, 31]. Antibody microarrays for the detection of cytokines have become robust and stable. Many different antibody microarrays have been offered commercially for the detection and quantification of low-abundance cytokines in serum or plasma [32]. The first commercial fully automated clinical analyser using a 3 × 3 segmented microarray is the Evidence (Randox Laboratories, Northern Ireland). The current range of arrays includes multi-analyte panels for the analysis of free thyroid hormones, tumour markers including PSA, cytokines and growth factors, cardiac markers, and drugs of abuse [28].
Nucleic acid-based microarrays
Nucleic acid-based microarrays are a multiplexed method for detection of bacteria or viruses in food, water, or clinical samples. The analytes are genomic DNA, mRNA, rRNA, or other nucleic acids. Quantitative results are achieved by a sandwich hybridization format, with a capture and a detection oligonucleotide for detection of bacteria [33] and viruses [34]. Preanalytical steps, for example cell lysis and nucleotide extraction, are necessary [35]. Polymerase chain reaction (PCR) enables amplification of a DNA target. These procedures increase sensitivity and selectivity. However, they are time-consuming and not easy to integrate in a fully automated system.
Microarrays for screening applications
A wide range of microarrays has been developed for screening applications and for obtaining semi-quantitative multianalyte results. In contrast with quantitative analytical microarrays using native unlabelled analytes, for screening applications the target analyte is labelled. The types of microarrays depend on the immobilized capture molecule. Since 2000, protein microarrays have been used for high-throughput protein analysis [36, 37]. Three test formats are used: functional, quantitative, and reversed-phase microarrays [38]. Cell lysates are immobilized and react with labelled antibodies by applying the reverse-phase microarray format [39, 40]. Quantitative protein microarrays use a sandwich immunoassay format [41]. Functional protein microarrays have immobilized antigen (cell, protein, or other). In clinical diagnostic applications the functional protein microarray is also called an antigen microarray. They present at each spot a recognition molecule for one specific antibody. With this format allergens have been immobilized for screening of allergen-specific IgE in human serum [42]. Autoantigens have also been spotted to quantify autoantibodies in the sera of patients with autoimmune disease [43]. The immobilization of cells in a microarray format is another application in clinical diagnostics, e.g. for diagnosis of sepsis [44], but also in other fields such as drug screening [45, 46], or screening of successful DNA transfection [47]. Peptide microarrays are applied for epitope mapping, antibody profiling, or drug screening [48–50]. Carbohydrate [51, 52], receptor [53], or enzyme [54] microarrays are also developed for drug discovery. DNA microarrays have not only been developed for gene expression analysis but also for detection of microorganisms and viruses in food and water analysis and for clinical diagnostics. Semi-quantitative results are obtained by using PCR combined with a labelling procedure before the microarrays are analysed [55–59].
Production of microarrays
Immobilization methods
The methodology of surface chemistry is the basic know-how for obtaining reproducible results with analytical microarrays. The key points to consider when selecting an appropriate surface and coating procedure are a low degree of unspecific binding sites and uniform distribution of functional groups on the substrate surface. The reduction of non-specific binding lowers the detection limit of an analytical microarray. The uniformity of coating increases the reproducibility, because of an identical amount of immobilized capture molecules within one microarray and a batch of produced microarrays.
Plastic and nitrocellulose microarrays are used for immobilization of the antibodies. They are bound directly to this monolayer via non-covalent adsorption. The non-covalent immobilization procedure is frequently used because of the simple immobilization procedure and high yield of protein attachment. Antibodies or haptens conjugated to, e.g., bovine serum albumin (BSA) are directly attached to polymer surfaces via hydrophobic or ionic interactions. Polystyrene [60], acrylonitrile-butadiene-styrene (ABS) [61], poly(vinylidene difluoride) (PVDF) [62], and nitrocellulose substrates are very effective for non-covalent antibody attachment. After immobilization, these microarrays have to be blocked intensively in order that the non-specific binding of labelled molecules is reduced. However, high background signals significantly reduce the signal-to-noise ratio of directly adsorptive substrates [63]. Also the desorption of antibodies during washing steps is observed. The only advantage of plastic and nitrocellulose substrates is that no chemical treatment is required. Although, these substrates do not meet the required reproducible results of analytical microarrays.
Glass is the common substrate to build up step-by-step a microarray surface architecture using silane chemistry. The covalent attachment of haptens, antibodies, or other capture molecules is necessary for automated analytical microarrays using flow channels. The desorption of capture molecules has to be prevented. Currently, gold surfaces and plastic microarrays are described less often in the literature, even though covalent coating is as easy as silane chemistry. Self assembled monolayers are formed reproducibly on gold layers with sulfur-containing compounds [64] and can easily be implemented in analytical microarrays [65]. Plastic substrates are activated by photochemistry producing radicals on the surface [66, 67]. With all these strategies a monolayer for covalent attachment is created. This first step serves for activation of the substrate. Glass substrates are generally activated with silanes. They carry terminal head groups like amines, epoxides, thiols or aldehydes which are used for immobilization of proteins [68]. This strategy works also for other molecules containing a terminal amino group such as amine-modified oligonucleotides and haptens. For covalent attachment of proteins, DNA or other molecules, aminosilane-coated glass slides have to be derivatised with a homobifunctional crosslinker. Mercaptosilane-coated glass slides use a heterobifunctional crosslinker. Aldehyde and epoxide-coated slides react directly with terminal amine groups. The silanization process does not produce a high-density monolayer [69, 70]. The free binding sites for non-specific adsorption have to be blocked intensively with proteins such as BSA. The reproducibility and the signal-to-noise ratio is much higher compared with directly immobilized molecules [71].
Microarray slides coated with affinity binding proteins, for example streptavidin, protein A, protein G or concanavalin A, are an alternative strategy to covalently attached capture molecules [72, 73]. These methods are attractive because they need no additional chemical crosslinker.
The copper(I)-catalyzed azide-alkyne cycloaddition is a new innovative coupling chemistry for bioanalytical approaches. It is so far the best example of click chemistry [74–76]. Another coupling strategy is the HyNic chemistry forming a stable hydrazone bond [77, 78].
Hydrogels on microarray substrates have become increasingly important. Hydrogels of aminodextrans [79], carboxydextrans [80], or poly(ethylene glycol) (PEG) [81] are used as a shielding layer on the substrate. They prevent non-specific binding and an additional blocking reagent is not necessary. PEG surfaces, especially, are known to reduce the non-specific binding of proteins [82]. Another advantage of PEG surfaces is the stabilizing effect of immobilized proteins or antibodies on glass substrates [83]. The surface provides a chemical moiety suitable for step-by-step modification. Various methods are available for covalently coupling haptens or capture molecules to surfaces. For instance, the coated surface can be modified to display terminal amine, thiol, or carboxy groups capable of reacting with homobifunctional or heterobifunctional crosslinkers. Homobifunctional crosslinkers have two identical coupling groups such as N-hydroxysuccinimide (NHS) esters, maleimides, epoxides, or aldehydes. A heterobifunctional crosslinker has two different coupling groups. The activated surfaces are suitable for coupling primary amines or thiols. Surfaces coated with PEGs have a monolayer structure. The binding capacity is lower than with hydrogels, generating a 3D structure.
Nanopatterned surfaces with functional nanoparticles is another area of interest with the aim of well organized structures on the surface [84]. Steric hindrance and the binding effectiveness can be adjusted. The steric accessibility is an important factor for formation of the antigen-antibody complex at the surface.
Although many different immobilization methods for microarrays are under investigation, effective methods for characterization of monolayers on microarray substrates are lacking. No analytical method has been established identifying the number of molecules which are covalently bound or only adsorbed per spot. Therefore, the absolute immobilization efficacy is difficult to calculate. An acceptable indirect characterization method is the imaging of fluorescence or chemiluminescence labels on the surfaces. They cannot distinguish between a labelled target that is immobilized covalently or adsorptively. However, the homogeneity and the functionality of the surface can be characterized with, for example, printed antibodies by capturing horseradish peroxidase (HRP) as depicted in Fig. 4 [82]. The additives in the spotting buffer and the production of homogeneous layers and microarray spots are optimized, preventing smeared microarrays and non-uniform spots. Many microarrays are published disregarding quality control of microarrays. Qualitatively excellent microarrays will enforce the acceptance of analytical microarrays.
Fig. 4.
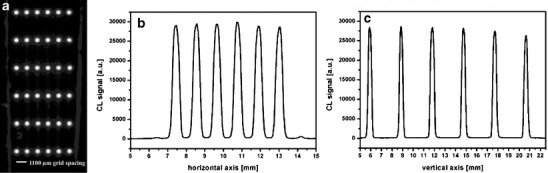
Image of an antibody microarray (anti-HRP antibody) for characterization of surface homogeneity and functionality with HRP as analyte and CL reagent a CL signal in horizontal axis b and vertical axis c (Reprinted with permission from Ref. [82]; Copyright 2007 American Chemical Society)
Patterning
A variety of technologies for patterning of microarrays have been established in the last decade [85]. Diverse micropipetting systems based on ink-jet principles such as thermal [86], piezo-electrically actuated [87], and syringe-solenoid [88] based technologies have been adapted from printing technologies [89]. The newer techniques consist of high-parallel microdispensing devices, such as the TopSpot technology, which acts by using piezo-triggered, short, pneumatic pressure pulses [90, 91]. These non-contact printing systems are used for high-throughput microarray production because of the advantage offered by the very fast printing process. The printing solution is transferred to a reservoir of the pump head and can be delivered to a considerable number of slides as carrier substrate. By using multiple microdispensing units on the printing system the process is made parallel and, therefore, more cost-efficient. The limitation of these non-contact printing devices is the high technical effort required for installing a robot system and the accuracy of microdispensing. The spotting buffer often contains salts and detergents that prevent adsorption on the walls of the microtitre plates used for storage of the printing solution. The disadvantage of added supplementary compounds is that the printing head can become irreversibly clogged. In contrast, the printing process of contact printers is very reproducible and the contact pins have a long lifetime. Solid pins or split pins (contact printing) [2, 92] and microstructured stamps (microcontact printing) [93, 94] have been further developed for diverse microarray formats such as protein, cell, tissue or other microarray formats [95–98]. The printing principle is adapted from stamping or fountain pens. The print head is dipped into the spotting solution and the molecules are transferred to the flat support by contact with the slide. The printing process of all systems is fully automated. For both microprinting technologies regulation of the temperature and humidity is highly essential, since the reproducibility of the printing process depends strongly on maintaining evaporation of the analyte solution on the printing head at a constant rate.
A different method for microarray production on a flat surface is the physically isolated patterning method (PIP) (Fig. 5). This method generates a microarray using six or more flow channels on a flat surface for patterning and analysis [99]. A multi-channel flow cell is placed on the surface of a microarray substrate which has been activated for immobilization [100]. After the patterning process has taken place, the channels are rotated horizontally by 90° relative to the substrate and are now oriented perpendicular to the channel that holds the immobilized binding partner. Thus, a microarray structure is generated manually without complex microdispensing devices.
Fig. 5.
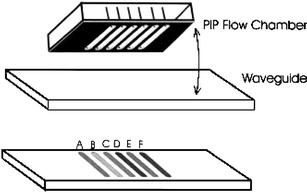
Production of microarrays by a physically isolated patterning (PIP) method (Reprinted from Ref. [99] with permission from Kluwer)
Fluidic systems for automated microarrays
Analytical microarray platforms automatically deliver sample, detection reagents for analysis, washing solutions with a fluidic system. It is essential that the process steps of an assay on a microarray platform are automatically performed with the aim of being simple-to-use and reproducible in operation. Platforms dealing with a segmented microarray on a slide or a multiwell plate can be processed either manually or with a robot pipetting system. Using an automated platform the samples and detecting reagents are dispensed with a pipetting system and the microarray is read separately on a microarray scanner. As each system for processing and readout of the microarray is large and expensive, segmented microarrays are only operated in laboratories and are not suitable for automated monitoring systems outside the laboratory.
The integration of an automated fluidic device containing valves, pumps, a detection system and electronics is the most challenging aspect of platform development. Automated flow-through microarray platforms often use FIA systems for reagent supply. An autosampler is the link between sampling and microarray platform. The autosampler constitutes an effective solution when several samples must be analysed automatically.
Using flow-through microarrays, the microarray is part of a flow cell through which samples and reagents are pumped. In comparison with conventional passive microwell plates, flow-through microarrays present thinner and more constant diffusion layers for the heterogeneous affinity reactions to take place. This substantially reduces the time needed to perform an multianalyte assay. Furthermore, automated fluidic systems have the advantage that the affinity reaction does not have to be in equilibrium as long as flow speed and temperature are reproducible within the analysis. This fact reduces the assay time dramatically. Automated flow-through microarrays allow the analysis of a sample within minutes.
The flow cell of a flow-through microarray and the connection to the fluidic and the readout system are designed in different ways. The fluidic channel on top of the microarray has to be as thin as possible in order to keep the time short for mass transport on to the heterogeneous phase. But, on the other hand, a microfluidic flow channel with heights of a few hundred micrometers increases the flow resistance. This implies that only small flow rates can be achieved or otherwise high pressure in the flow cell is generated. Also the shear rate on the surface is not negligible, because the bound complex can become dissociated [101].
The covering lid seals the flow cell and the support with adhesives or flexible materials. Adhesives or other bonding techniques are used especially for single-usage housings [102]. Flexible polymer sheets enable changing of the microarray substrate. Basically, sealing is enabled by pressing the lid on the flow cell case. If silicone or polytetrafluoroethylene (PTFE) is used as sealing material, high forces on top of the housing are needed, which can lead to breakage of the flow cell or leakage. Another often used flexible material for flow cells is polydimethylsiloxane (PDMS). This elastomeric polymer is known for its ability to mould and reproduce three-dimensional imprinting structures. Other features are its high inertness to biomolecules and the ease of sealing by pressure.
Another point worthy of consideration in order to achieve automation in a flow-through microarray is the choice of pumps and valves. Pumping fluids through the microarray flow cell can be achieved by syringe or peristaltic pumps. It should be kept in mind that buffers contain relatively high salt concentrations and corrosion of metal parts can cause leaks. For this reason pumps and valves have to be changed periodically. The fluidic pathway from the pump to the flow cell is more easily directed with switching valves, but they are more susceptible to leakage. Magnetic valves prevent these problems by squeezing the tubing without coming into direct contact with the buffer solutions.
Centrifugal discs containing a microfluidic system are an alternative solution to the application of pumps. The actuator is a rotating disc, and the solvents and reagents are only driven by centrifugal forces, capillary forces, and directed by hydrophobic barriers [103].
Microarray readout systems
Fluorescence microarray readout systems
Currently, lasers are used for highly sensitive fluorescence microarray readout systems [104]. Microarrays without active fluidic parts are read with a fluorescence laser scanner. The area of a microarray is excited by a laser beam to fluoresce line-by-line. Laser diodes are frequently used in fluorescence microarray scanners because they are inexpensive and compact. Today fluorophores with emission in the far red and NIR range are commercially available. New diode lasers are currently available for excitation in the wavelength range between the UV and the visible. This enables use of new fluorophores such as quantum dots or other fluorescence nanoparticles [105, 106]. Also time-resolved fluorescence is possible for microarray readout systems by using an UV laser [107]. The sensitivity of fluorescence microarrays can be increased with time-resolved fluorescence readout systems. For short decay times background fluorescence and light scattering are no longer detectable after some microseconds. Another sensitive optical microarray readout technique is based on surface plasmon resonance (SPR) combined with fluorescence [108]. Surface plasmons can be excited in noble metals by resonance coupling with a light beam of a laser. The local enhancement of the optical field associated with surface plasmons can then be used to excite surface-bound fluorophores. The distance to the surface should be between 10 and 20 nm, otherwise the fluorophores are quenched or ineffectively excited [109]. Currently, SPR-enhanced fluorescence readout systems can image an area of 3 × 3 spots.
Several evanescent field fluorescence microarray readout systems based on total internal reflection fluorescence (TIRF) have been developed for flow-through and segmented microarrays. One distinguishing feature is the coupling of exciting light into a waveguide to generate an evanescent field along the surface area of the waveguide by total internal reflection. The field strength decays exponentially with distance from the waveguide surface, and its penetration depth into the solution is limited to about a few hundred nanometres. This effect can be utilized to excite only fluorophores located at or near the surface of the waveguide, as depicted in Fig. 6. The emission light generated simultaneously at the surface of the microarray is detected with a charge-coupled device (CCD), a photodiode, or a photomultiplier tube.
Fig. 6.
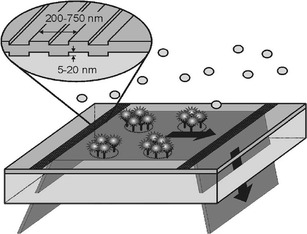
Evanescent field generation for microarrays using diffractive relief gratings (Reprinted from Ref. [155] with permission from Elsevier)
Chemiluminescence microarray readout systems
For multianalytical applications with CL microarrays a close-to-the-surface chemiluminescence reaction is applied. CL microarray readout systems use the effect of light emission caused by a chemical reaction. Every assay format developed as ELISA in microwell platforms is adaptable as a multiplexed assay on CL microarrays. The principle of CL microarrays is based on multiple affinity reactions which are detected in parallel with localized chemiluminescence labels. Enzymes with a short half-life, for example HRP, are required. The light emission takes place very close to the enzyme label by incubation with the CL reagent, e.g. luminol. This reacts with H2O2 in the presence of HRP, with light emission at 425 nm [110]. The CL reagent is pumped into the flow cell and the discrete close-to-the-surface chemiluminescence reaction is imaged. The reaction is stopped after a defined time. Only endpoint measurements are possible because of the washing step between the affinity reaction and the chemiluminescence reaction. CCD or CMOS chips image the CL microarrays. The readout system of CL microarrays can be build up very compactly. Optical components like an exciting light source (laser or lamp), filters and mirrors are not needed. CL microarray instrumentation can even be integrated into a miniaturized handheld system for applications in POC diagnostics and food and water quality monitoring.
Generally, CL microarrays are more sensitive than fluorescence microarrays [111]. One reason for the extremely high sensitivity of CL microarrays is that the background signal is near zero. In contrast with fluorescence, there is no background signal either from the excitation source or from light scattering by the matrix. Moreover, the signal intensity per HRP molecule can be increased using optimised luminol analogues and enhancers [112, 113]. The signal can also be increased by surface-enhanced CL. Metals like gold enhance the CL signal depending on the distance. With a distance between 1 and 10 nm the CL signal is increased by more than one order of magnitude [114].
The limit of detection can be worsened by imperfect surfaces. The enzyme reaction is so sensitive that very small amounts of unspecifically bound enzymes can be detected as a background signal. Small amounts of the enzymes can also remain in a fluidic system with valves and pumps. Surfaces which prevent unspecific binding are necessary, as also are intensive washing steps. Another point is that the sensitivity of the CL depends on the enzyme activity. An enzymatic turnover rate depends on temperature and the concentration of the CL reagent. Both should be constant during the analysis. Automated CL microarray platforms have to be temperature-controlled. This is an important point, especially for robust conditions of analysis over a whole working day.
Electrochemical microarray readout systems
Electrochemical microarray readout systems are based on microelectrode arrays. The multiplexed electrical readout is achieved with interdigitated array microelectrodes (IDA) [115] or complementary metal oxide semiconductor (CMOS) image sensors [116, 117]. CMOS image sensors measure the affinity binding at the microelectrode by cyclic voltammetry [118], differential pulse voltammetry [119], or amperometry [120]. The electrochemical reaction is amplified by enzyme labels such as HRP or alkaline phosphatase (AP), producing redox-active reporter molecules [121, 122].
Currently, two electrochemical microarray instruments based on interdigitated array (IDA) technology are available—the eSensor from Motorola [123] and the eBiochip from Fraunhofer Institute for Silicone Technology (ISIT) [124]. In the literature, the electrochemical readout system from the ISIT is described more in detail and serves as an example for electrochemical readout systems. A multiplexed electrical readout is achieved by interdigitated gold array electrodes. The electrochemical signal is generated by AP converting the electrically inactive p-aminophenyl phosphate (pAPP) into the electrochemical mediator p-aminophenol [125]. The mediator is oxidized at + 350 mV at one finger pair of electrodes and the quinoneimin formed is reduced at −150 mV on the other finger system (Fig. 7).
Fig. 7.
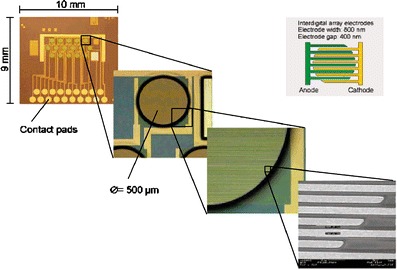
IDA-electrode structure of an electrochemical microarray (Reprinted in part with permission from Ref. [127]; Copyright 2006 American Chemical Society)
The signal is enhanced by redox-cycling, because the anode and the cathode are in close proximity. The electrical signal is detected by a multiplexing 16-channel potentiostat for direct data readout from the chip [126]. A common counter-electrode is also integrated on the microarray. The amperometric measurement is taken versus an external Ag/AgCl reference electrode. The electrochemical microarray carries 16 electrodes as sensor elements. One sensor element contains 204 fingers, each 800 nm wide, and with a 400-nm gap between anode and cathode fingers [127].
Electrochemical microarrays have high potential for cost-effective portable devices for the POC diagnostics and field applications [128]. Semiconductor technology is an established process for manufacturing chips on a production line, and is easily convertible into a cost-effective means of producing electrical biochips based on silicon technology. Restrictions are the quantity of sensor elements on the microelectrode array per chip and inflexibility because changes in chip design are very expensive. Electrochemical microarray readout systems are more applicable to multianalyte applications for low-density microarrays. Currently, the highest density (17,778 electrodes cm−2) is achieved with an electrochemical CMOS image sensor [129].
Label-free microarray readout systems
In recent years, label-free analytical approaches have gone through intense development as optical microarray readout systems [130, 131]. Label-free imaging is based on SPR [132], interferometry [133, 134], reflectometry [135], or ellipsometry [136].
Label-free imaging has not only offered multianalyte detection in real-time, but also provides kinetic information about affinity constants and association/dissociation kinetics. This is important, for example, with protein microarrays for characterization of the functionality of multiple antibodies in parallel [137] or epitope mapping [138]. Another point to keep in mind is that no additional label is needed. Usually, coupling of a fluorescent or enzyme label is performed on relatively high concentrations of proteins. This procedure is an additional expense, and the bioactivity of the protein is often reduced. This is avoided in label-free microarray systems.
SPR is the most prominent label-free technology implemented in automated biosensor platforms for detection of single analytes in biomedical, food, and environmental samples [139]. Multi-analyte SPR biosensors are designed as multi-channel or SPR imaging (SPRi) platforms [140, 141]. SPR spectroscopy based multi-channel biosensors are restricted in the number of channels [142]. SPRi instruments using the Kretschmann configuration are limited in resolution of signal [143] and require all capture probes to be applied at the same density [144]. The first commercialized label-free microarray is the Flex Chip Kinetic Analysis System launched by HTS Biosystems (Hopkinton, USA) and later acquired by Biacore (Uppsala, Sweden), now GE Healthcare, UK. This system has been approved for simultaneous detection of 400 parallel reactions with high resolution using a grating-coupled SPR [145].
In label-free microarray readout systems, it is critical to have a non-fouling sensor interface, especially for analysis of real samples in complex matrices like blood, food, water, or other liquid samples. Currently, label-free readout systems for multiplexed microarray applications in the field of clinical diagnostics, food, and water analysis have to be demonstrated as being as successful as fluorescence, chemiluminescence, or electrochemical microarrays in fully automated platforms.
Analytical microarray platforms
Non-automated microarrays
The direct transfer of DNA microarrays for gene-expression analysis to protein microarrays is achieved with protein microarrays that use quantitative or reversed-phase microarray protocols [146]. Capture molecules are immobilized on quantitative microarrays. The proteome is labelled and incubated with the microarray. In contrast, the other format uses immobilized cell lysates and labelled proteins for screening the functional activity of a protein. There are also microarrays adapted from immunoassay formats in microwell plates. Depending on the readout system they are called fluorescence or chemiluminescence microarray immunoassays (F-MIA or CL-MIA). All these kinds of microarray have one reaction area and a integrated fluidic system is absent. Samples and assay reagents are placed on top of the microarray and are passively incubated. After this incubation step the slides are cleaned and dried. For read out, the microarrays are placed in an appropriate detection system (fluorescence or chemiluminescence microarray reader). Many manual process steps (washing, drying, transfer to reader) are required. The microarrays are used only once. For calibration, positive and negative controls are often included to ensure that threshold limits for positive responses are maintained and that non-specific adsorption does not create a false negative result. This type of analytical microarray is used for screening applications by obtaining semi-quantitative results. If quantitative results are required, each slide is analysed for one multi-analyte concentration. Passive incubation steps dramatically increase the assay time. These platforms are very helpful for protein screening applications in proteome analysis. However, they will not gain acceptance in the field of on-site analysis of food, water, or diagnostics. Some examples of passively incubated microarrays are given as some of these applications could be adapted to flow-through microarrays.
Gehring et al. have designed an antibody-based microarray for detection of E. coli O157:H7 [147, 148]. A sandwich immunoassay format is chosen by immobilization of biotinylated capture antibodies on streptavidin-coated glass slides and by using fluorescein-labelled detection antibodies. The assay time (140 min) on this passive incubation platform is extremely long and the reported LOD of 3 × 106 cells mL−1 for E. coli O157:H7 was rather high.
Rubina et al. demonstrate a F-MIA on protein-gel-based microchips for the toxins ricin, viscumin, SEB, tetanus toxin, diphtheria toxin, and lethal factor of anthrax toxin [149]. The LODs are comparable to those of an ELISA and were in the range of 0.1 and 20 μg L−1 depending on the toxin. The differences between chemiluminescence and fluorescence were compared using direct, indirect, and sandwich assay formats. The LODs of the CL microarray were in the same range as those of the fluorescence microarray. For in-field use the sandwich format was the most practicable and provided the highest sensitivity.
The need for automation became evident when fluorescence microarrays were used as an instrument for biomarker detection on Mars missions [150]. The so-called signs of life detector (SOLID) was built for processing samples and detecting specific biomarkers, for example L. ferrooxidans, A. ferrooxidans, Acidithiobacillus sp., Acidiphilium sp., aspartate, glutathione-S-transferase, thioredoxin, or cortisol via specific antibodies.
A similar concept as the F-MIA platforms is the CL-MIA-based array tube system (ATS) from Clondiag Chip Technologies (Jena, Germany) [151]. A single reagent tube is used for incubation of sample and reagents. The microarray is located on a glass chip forming the bottom of a 1.5 mL plastic tube. Antibodies are deposited on the glass surface. A sandwich format is used to simultaneously detect bacteria, viruses, and toxins. The chemiluminescence signal is generated after an incubation step with streptavidin-poly-HRP. Washing steps and pipetting steps are manually operated. The incubation steps are carried out with a horizontal tube shaker. The detection limit for viruses (TCID50 = 50% tissue culture infection dose) was 6 × 102 to 5 × 106 TCID50 mL−1, for toxins between 0.1 and 0.2 μg L−1, and for bacteria between 5 × 103 and 2 × 106 cfu mL−1. The analytes were detected in 1 h 23 min.
The CombiMatrix oligonucleotide microarray platform has an electrochemical readout system that contains 12,544 individually addressable microelectrodes on a CMOS image sensor [129]. Each working electrode is 44 μm in diameter. Detection is based on the close-to-the-surface reaction of redox-active chemistries such as HRP and the associated reagent tetramethylbenzidine (TMB). CombiMatrix (Mukilteo, USA) has launched the ElectroSense Reader and the ElectroSens 12 K microarray as a platform for genotyping and gene expression analysis. The microarray consist of individual ODN probes synthesized at each microelectrode. Hybridization with biotin-labelled ODN targets is performed in a specially designed chamber called the ElectraSense Hybridization Cap sealed with an O-ring. The captured ODN targets are detected by incubation (45 min) with HRP-avidin conjugate. The LOD of cRNA was 0.75 pmol L−1.
Segmented microarrays
Segmented microarrays are flat slides with a hydrophobic barrier around a subarray. Additionally, a microwell plate with a spotted subarray on the bottom of each well is a segmented microarray. Each subarray contains the same quantity of spotted molecules. A segmented microarray allows for simultaneous calibration and sample measurement in a similar way to an ELISA. Samples and reagents are placed on each segment and are passively incubated. After incubation the slides are cleaned and dried. The readout system for such microarrays is based on fluorescence or chemiluminescence microarray detection. As most systems are not automated, many manually performed assay steps are needed, e.g. drying the microarray surface after washing and incubation, transfer of the microarray to the microarray readout system. These microarrays are difficult to automate and/or need a large amount of additional equipment. Another disadvantage is the long assay time of such a static assay format, because of the increasing diffusion layer of static incubation processes. These facts make segmented microarrays expensive. Some segmented microarray platforms have been commercialized however.
Segmented fluorescence microarrays
A segmented F-MIA with an indirect competitive immunoassay format for pesticide detection was presented by Belleville et al. [152]. Each segment contains spotted hapten derivatives in a submicroarray arrangement. The immunoreaction is performed in each segment by parallel incubation with calibration standards or samples. Hapten derivatives are immobilized via a photolinker on silanized glass substrates. Atrazine and 2,6-dichlorobenzamide (BAM) are simultaneously detected with an assay time of 75 min. The LOD for BAM is 4 ng L−1 and for atrazine 9 ng L−1 [153].
A segmented antibody microarray has been developed for multiplexed toxin detection by using a direct competitive immunoassay format [154]. A 16-chamber polymer tray was fixed to the slide surface. The following six toxins were quantified: cholera toxin β-subunit, diphtheria toxin, anthrax lethal factor, protective antigen, SEB, tetanus C fragment. The LODs were quite high (between 23 and 5,000 μg L−1) even though the toxins are not simultaneously analysed. For toxin analysis a direct assay format is too complicated and expensive, because a multiplexed competitive immunoassay requires each toxin to be labelled and added in relative high concentrations to the sample.
The ZeptoReader is a commercially available platform consisting of a fluorescence segmented microarray. Detection is based on an evanescent field fluorescence microarray readout system. The surface-confined excitation is associated with light guiding in a planar optical waveguide. The excitation light of a laser beam (λ = 635, 532, and 492 nm optional) is coupled to a wave-guiding Ta2O5 film using a diffractive relief grating [155]. To perform an assay a ZeptoPlate with 96 wells was designed. Each well contains a 5 × 5 subarray and has a volume of 15 μL. With this platform a multiplexed antibody microarray was used for detection of three different human cytokines in parallel [156]. The test format was a sandwich immunoassay by immobilization of three different capture antibodies. The fluorescence image was taken after an incubation time of 2 h. The assay was performed without washing steps. The microarray was imaged after a single addition of the pre-incubated interleukin/secondary antibody/Cy5-streptavidin cocktail. IL-2, IL-4, and IL-6 were quantified with a LOD of 15 ng L−1, 10 ng L−1, and 5 ng L−1, respectively.
Segmented chemiluminescence microarrays
Magliulo et al. [157] have presented a rapid multiplexed chemiluminescence immunoassay for detection of E. coli O157:H7, Yersinia enterocolitica, Salmonella typhimurium, and Listeria monocytogenes, four pathogenic bacterial strains that are relevant in food hygiene control. The microarray platform consists of a 96 × 4 multiwell plate with four subwells in each well (Fig. 8).
Fig. 8.
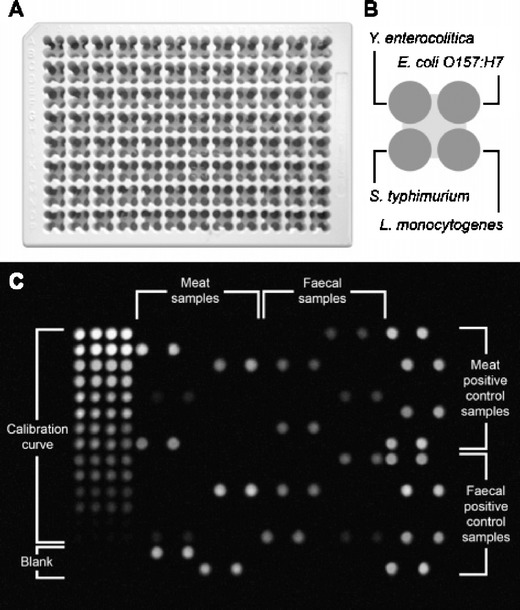
a Image of the microarray platform on a 96 × 4 well microtitre plate. b Position of the immobilized antibodies within each subwell. c CL image of the microtitre plate (Reprinted with permission from Ref. [157]; Copyright 2007 American Chemical Society)
A sandwich CL-EIA was used as test format. Four different bacteria strains could be detected in parallel in each well. The standards for the calibration curve and the samples are simultaneously measured on the same platform, which is an advantage over fluidic microarrays. The multiwell plate platform requires much manual handling in the laboratory. A skilled person has to handle washing and pipetting steps, even though these steps can be automated with pipetting robots and automated plate washers. This CL segmented microarray platform is limited to four analytes. Analytical microarrays with multiplexed approaches are of interest for analysis of up to 10 analytes in parallel.
The first commercially available automated CL microarray platform is the Evidence biochip array analyser from Randox Laboratories (Crumlin, UK). It was developed for the clinical diagnostic market. The platform consists of an automated dispensing station for reagent supply, incubation and washing steps, and sample introduction [158]. The biochip is secured in the base of a plastic well, which is then placed in a carrier holding nine biochips in a 3 × 3 format. Each biochip contains a 5 × 5 array. The capture antibodies are immobilized on an aluminium oxide substrate activated with silanes. Light emission from the CL reactions on the surface of the biochip using HRP-labelled detection antibodies is imaged by a CCD camera. With this platform an antibody microarray has been designed for detection of ten cytokines in parallel in the working range between 1.1 and 2,000 ng L−1. The Evidence analyser enables complete automation of assays at a test frequency of up to 2000 results per hour. This platform is the first fully automated CL microarray based on segmented microarrays. The assay process is automated with a spacious robotic station for dispensing, washing, incubation, and readout. This system is well designed for multi-analyte applications in the field of clinical diagnostics in the laboratory.
Flow-through microarrays
Fluorescence flow-through microarrays
The River Analyser (RIANA) is a multianalyte immunosensor based on total internal reflection fluorescence (TIRF) [159]. A laser beam (λ = 633 nm) is coupled directly to a glass slide via a bevelled end-face and guided through the length of the transducer. A strong evanescent field is produced at each reflection point. Cy5.5-labelled antibodies are used for simultaneous detection of estrone, isoproturon, and atrazine at the surface by means of a multiplexed binding inhibition assay. The analyte derivative was covalently immobilized on aminodextran surfaces. The fluorescence signal is collected with optic fibres and photodiodes for each evanescent spot. An autosampler is used to automatically deliver the samples to the FIA system. The transducer is mounted on the flow cell and sealed with an O-ring. Estrone, isoproturon, and atrazine are detected with LOD of 0.08, 0.05, and 0.16 μg L−1. Total analysis time for a single run is 15 min. In a further development of the fully automated assay procedure four different analytes (atrazine, bisphenol A, estrone, and isoproturone) are detected simultaneously in 12 min with an LOD below 0.020 μg L−1 [160]. The same immunosensor is also used for detection of progesterone in milk [161], and propanil [162]. Additionally, an optimized single-analyte assay procedure has achieved a LOD for estrone of 0.2 ng L−1 [163].
The AWACCS (automated water analyser computer-supported system) immunosensor consists of a fibre pigtailed chip with 32 evanescent field-sensing regions [164]. A semiconductor laser is coupled into an integrated optical (IO)-transducer chip containing a Y-junction splitter structure that generates four parallel waveguides. The fluorescence is picked up with 32 optical fibres transferring the light to 32 photodiodes (Fig. 9).
Fig. 9.
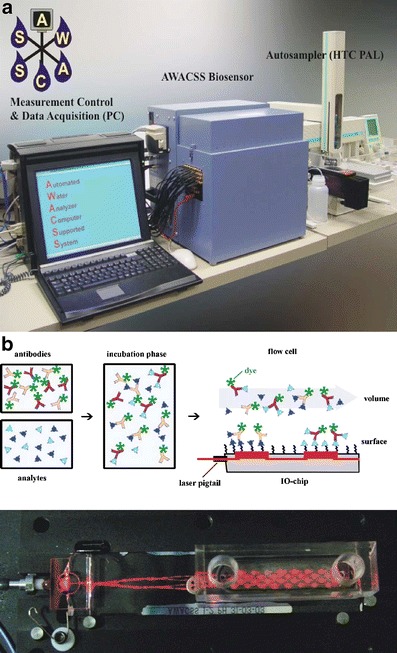
a AWACSS immunosensor set-up. b The fibre-pigtailed chip with 32 evanescent field-sensing regions (Reprinted in part from Ref. [164] with permission from Elsevier)
Six different analytes (atrazine, isoproturon, bisphenol A, estrone, propanil, and sulfamethizole) were calibrated simultaneously with a detection limit for each of them below 0.02 μg L−1 using a multiplexed binding inhibition assay [20]. All important components of the immunoanalytical platform are combined for fully automated immune analysis: immunosensor, the fluidic system, and a software module enable fully automated calibration and determination of analyte concentrations [165].
The NRL array biosensor is a portable flow-through microarray system (Fig. 10). The system consists of a 50-W power supply, a cooled CCD camera, peristaltic pumps, valves, and a multi-channel flow cell. It also contains a 635-nm diode laser equipped with a line generator to spread the laser beam into a fan pattern. The expanded laser beam is launched into the edge of a waveguide (microscope slide) resulting in an evanescent field. A bandpass filter eliminates excitation and scattered light prior to imaging. Some array-based biosensors have the feature of special imaging optics consisting of a two-dimensional array of graded index (GRIN) lenses to provide uniform excitation in the sensing area of the waveguide on to a CCD camera.
Fig. 10.
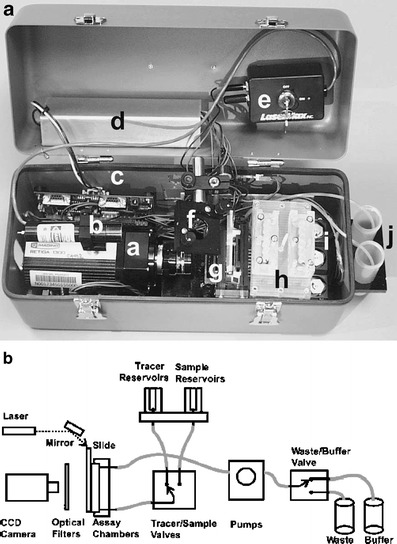
a The portable NRL Array Biosensor. b The fluidic and optic set-up (Reprinted from Ref. [72] with permission from Elsevier)
The multi-channel microarray immunosensor can be calibrated concurrently to the measurement [166] and enables simultaneous detection and quantification of up to nine target analytes [167]. It has been demonstrated that the NRL Array Biosensor can detect small molecular toxins [24, 168, 169], protein toxin targets [170, 171], bacterial and viral targets [172–175] in food and environmental samples. The standard assay time is 15 min. Whole cells have a smaller diffusion coefficient than small organic molecules. As a result, the incubation time needed for the heterogeneous immunoreaction is greater. A 1-h assay time with an increased incubation time by recirculation of sample and tracer solution leads to a tenfold improvement of the detection limit for bacteria [176].
Chemiluminescence flow-through microarrays
A microfluidic CL immunosensor with a direct immunoassay format has been developed for detection of atrazine [177]. The immune reaction between immobilized antibodies and analyte or HRP-tracer was reversible using a mild regeneration buffer containing 0.4 mol L−1 glycine and HCl, pH 2.2. The developed microfluidic CL immunosensor had a total assay time of ~10 min for atrazine, including incubation detection, and regeneration. The IC50 values were in the range of the official European Union requirements for drinking water (0.1 μg L−1). The LOD was 0.8 ng L−1 which is lower than determined with the same antibody on an ELISA microtitre plate. This result indicates that low diffusion times within flat flow cells reduce the IC50 and the LOD values.
The parallel affinity sensor array (PASA) is a CL flow-through microarray platform with an integrated fluidic system [178]. The platform consists of a CCD camera for readout, a fluidic system with pumps for automated reagent supply, and a microarray with spotted analyte derivatives. The first application was a microarray chip for environmental contaminants in water. Triazines, 2,4-D, and TNT could be detected in parallel. The authors also showed that both an indirect and a direct test format is possible for detection of small analyte molecules like pesticides or TNT. The PASA system is also convenient for automatically performed allergy diagnosis. Allergens and recombinant/purified allergens (24 preparations) have been used on the same epoxylated glass surface for screening of allergen-specific IgE. A direct immunoassay format was applied by detecting 24 allergenic proteins in parallel [42].
The same platform is under further development for detection of multiple antibiotics in milk. The antibiotics chip includes ten substances relevant in veterinary medicine. An indirect competitive test format is used. At first, the milk is incubated with a mixture of ten different antibodies. After a short incubation time the immune reactants are passed through the microarray. Free antibodies can bind to the distinct hapten spots [179].
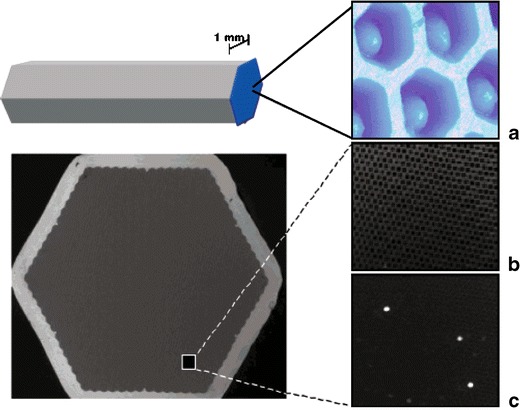
Antibiotics in milk should be detected on-site, so that the driver of a diary van may decide whether the milk is contaminated or not. A new and fully automated detection system (MCR 3) has been constructed for such in-field measurements (Fig. 11).
Fig. 11.

a Schematic drawing of the MCR 3. b Image of the chemiluminescence microarray chip reader (MCR 3; Institute of Hydrochemistry, TU Munich) c The microarray loading unit and the inserted flow-through microarray chip
The platform consists of a more sophisticated fluidic system with rotary valves and pumps. Larger pumps with solutions of primary and secondary antibodies for 50 runs of the regenerated chip are included. All important components of the CL microarray platform are combined for a fully automated multiplexed immune analysis in the field: the microarray, the fluidics, and a software module enable automated calibration and determination of analyte concentrations during a whole working day. The microarray and the reagents are temperature-controlled. The regenerable microarray chip is required to avoid changing the chip during the milk acquisition at the farmer’s house. PEG-ylated chip surfaces are used because of their known durability against chemical treatment. Microspotted antibiotic derivatives are easily coupled to activated PEG surfaces. With the hapten microarray it is possible to determine the five antibiotics sulfamethazine, sulfadiazine, streptomycin, ampicillin, and cloxacillin without any sample-preparation steps; the detection limits are 2.9, 3.3, 2.4, 2,7, and 1.1 μg L−1 [180]. The microarray is designed for parallel analysis of thirteen different antibiotics in fresh milk. Immobilization of the other antibiotics is currently under investigation. Antibiotics are currently detected in 6 min. The whole assay with a regeneration step takes 9.5 min.
Electrochemical flow-through microarrays
The electroanalytical Array Analyser “eMicroLISA” is a fully automated platform with fluidics, rotary valve, reagent reservoirs, and one hand plug and play microarray called ChipStick (Fig. 12).
Fig. 12.

Image of the Array Analyser “eMicroLISA” (Reprinted in part with permission from Ref. [127]; Copyright 2006 American Chemical Society)
The electrochemical microarray is positioned in a polycarbonate cartridge consisting of a flow cell with an external volume of approximately 7 μL. The flow of assay reagents is driven by a peristaltic pump. A six-way rotary valve and two magnetic valves are used to control the different reagents and perform a fully automated assay [127]. The platform is in use for the analysis of viruses or bacteria, either with antibodies or nucleic acids as capture and detection molecules. An rRNA-based assay was established for detecting total rRNA by immobilized capture ODNs and the specific hybridization of supporting ODN, and biotinylated detection ODNs. The detection ODNs bind adjacent to the capture ODNs being in close proximity to the electrode surface. The supporting ODNs enhance the accessibility of the capture and detection ODNs. The automated assay takes 30 min starting with prepared total rRNA. The detection limit of Escherichia coli total rRNA content was 0.5 mg L−1.
Suspension arrays
Suspension arrays analyse the affinity reaction on individual microspheres. With microsphere arrays each array element is prepared in bulk by coupling the appropriate recognition element at the surface of an optically defined microsphere. By optical encoding, micron-sized particles (e.g. polymer particles) can be created to enable highly multiplexed analysis of complex samples [181]. A microsphere array is encoded by one or more intrinsic optical properties or different sizes. The readout of suspension arrays can be performed with a flow cytometer or a fibre-optic readout system.
Suspension microarrays on flow cytometer
The current generation of commercially available microsphere arrays (LabMap System, Luminex Corp, USA) offers a 10 × 10 element array (ten different intensities for each of two fluorescent dyes contained within microspheres) to give a 100-element array. The LabMAP system uses a flow cytometer for analysis of suspension arrays. The flow cytometer provides an encoding method to simultaneously detect analytes by antibody or DNA-based binding assays on microspheres. Two spectrally distinct fluorophores are incorporated in polystyrene microspheres (FlowMetrix Beads). Using defined ratios of these fluorophores, a suspension microarray is created consisting of up to 100 different microsphere sets. The flow cytometer provides two lasers for simultaneous excitation of both fluorophores. A third fluorophore coupled to the reporter molecule quantifies the biomolecular interaction that has occurred at the surface of each microsphere, as depicted in Fig. 13.
Fig. 13.
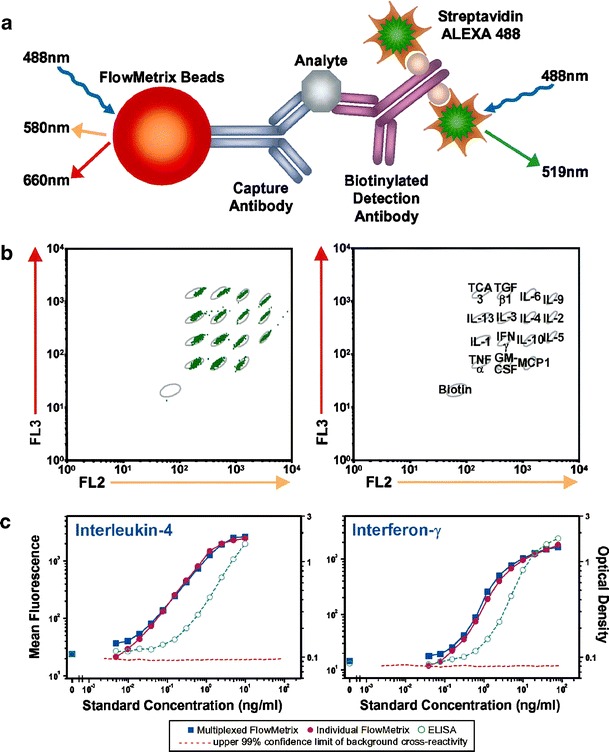
Cytokine analysis by multiplexed FlowMetrix. a Principle of optical encoding. b Fluorescence discrimination of each individual microsphere. c Calibration curves for two different cytokine standards (Reprinted from Ref. [182] with permission from Elsevier)
A suspension of microspheres is individualized by the hydrodynamic focussing effect of a flow cell. High-speed digital signal processing classifies the microspheres according to their spectral properties and quantifies the reaction on the surface. Currently, the assay steps are manually operated. Automated platforms consisting of shakers, autosampler, and dispensing devices are partially applied [183]. For each analyte a defined quantity of microspheres is added to the sample. After mixing and incubating of analytes and microspheres, the detector molecules (e.g. labelled with streptavidin-R-phycoerythrin or streptavidin-Alexa 488) are added. A centrifugation or filtration step is needed after incubation to separate the unbound components. The washed bead suspension is directly read with the flow cytometer.
The Luminex platform is used to quantify 15 human cytokines in a single sample with similar LODs and lower incubation times compared with the ELISA technique [184]. This technology is applied in food analysis to simultaneously detect three different non-milk proteins in milk powders [185]. Moreover bacteria are detected on the Luminex platform with a DNA hybridization assay, and with a sandwich immunoassay format. Amplified target DNA sequences could be detected in 30–40 min with an LOD of 106–107 genome equivalents provided that at least 103 (for E. coli, C. jejuni, and L. monocytogenes) or 105 (S. typhimurium) gene copies were present in a PCR amplification reaction. The sandwich immunoassay was more sensitive (2.5–500 cells mL−1) and the whole assay lasts 3 h [186].
Optical fibre arrays
Optical fibre arrays consist of thousands of individual glass or plastic fibres that are manufactured using an iterative process in which individual fibres are bundled, melted, and pulled through a fibre drawing tower to create arrays of fibres in which all the individual fibres have fused into a single substrate [187].
With this principle a fibre-optic DNA microarray is produced by random distribution of DNA probe-functionalized microspheres (3.1 μm) to microwells of an optical fibre bundle as depicted in Fig. 14. The microwells are created by chemical etching after polishing the fibre. The optical fibre bundles (500 μm diameter) contain approximately 6000 individual microwells [189]. The position of a microsphere is encoded by internal trapping of europium dye in the microsphere’s polymer matrix. The microspheres were functionalized with DNA probes. For detection of target DNA from Salmonella the DNA probes of the microspheres are hybridized and visualized with a Cy-3-labelled secondary probe in a sandwich DNA hybridization assay format. The assay was performed by exposing the microarray to target samples (200 μL) composed of chromosomal DNA from bacterial cell cultures. The chromosomal DNA was released after heat treatment at 90°C for 5 min. After 1 h one could detect 103–104 cfu mL−1 without any additional amplification.
Fig. 14.
Fibre optic DNA microarray platform (Reprinted with permission from Ref. [188]; Copyright 2003 American Chemical Society)
Multiplexed arrays were achieved with an optical bar code of each position-registered microphere. The microspheres contain an internal encoding (Eu3 + dye) and two external fluorescence dyes (Cy5 and TAMRA) at different concentrations. A library of 100 spectroscopically distinguishable microsphere types were prepared using various combinations of the three fluorescence dyes [190]. The fibre-optic microsphere-based array could detect 10 fmol L−1 fluorescein-labelled target DNA of seven different bacteria in a 50-μL sample volume within 30 min of hybridization [191].
Illumina (San Diego, USA) has commercialized a fibre-optic DNA microarray platform by using the intrinsic sequence information of ODN for optical decoding. The fibre optical bundles of the Sentrix Array Matrix can access the wells of a 96-well microtitre plate. In the other format, the Sentrix BioChip, one to several microarrays are arranged on silicon slides. They have created suspension arrays containing 1536 bead types, or more, per bundle that can be assembled into an Array of Arrays capable of performing high-density DNA microarrays for gene expression analysis or pharmacogenomics [192–194].
Conclusions and outlook
An overview of analytical microarray platforms is given in Table 3. Fluorescence, chemiluminescence, and electrochemistry currently dominate as 2D readout systems for analytical platforms. Fluorescence and chemiluminescence microarray platforms, especially, are used for detection of pesticides and pharmaceuticals in an indirect competitive immunoassay format. Protein target toxins, clinical biomarkers, or microorganisms are analysed with a sandwich immunoassay format. The detection methods are optimized for selectivity, sensitivity, and low limits of detection. The assay performance depends now more on the availability of antibodies with high affinity constants and low cross-reactivity. Total assay times are in the range of minutes, which is only possible in fully automated systems under non-equilibrium conditions. Only a few flow-through microarray platforms exist at the moment, consisting of an automated fluidic system for rapid and reproducible multiplexed applications. Automated analytical microarray platforms will compete with applications that completely solve a relevant multi-analyte problem, e.g. the quantification of possible contaminants such as antibiotics in milk, toxins in food, or microorganisms in drinking water. Further example include detection of doping drugs during a sport event, prevention of a bioterrorist attack, or the accurate and early diagnosis of a tumour marker.
Table 3.
Overview of analytical microarray platforms
| Analyte | Assay format | Platform | Assay time | LOD | Ref. |
|---|---|---|---|---|---|
| Fluorescence | |||||
| Estrone | Binding inhibition assay | RIANA | 12 min | 0.019 μg L−1 | [160] |
| Atrazine | 0.002 μg L−1 | ||||
| Isoproturon | 0.016 μg L−1 | ||||
| Bisphenol A | 0.005 μg L−1 | ||||
| Sulfamethizole, atrazine, propanil estrone, bisphenol A, isoproturon | Binding inhibition assay | AWACCS | 15–18 min | Each <0.02 μg L−1 | [164] |
| C. jejun | Sandwich IA | NRL array biosensor | 25 min | 9.7 × 102 cfu mL−1 | [175] |
| S. dysenteria | 4.9 × 104 cfu mL−1 | ||||
| SEB, ricin, cholera toxin, B. anthracis Sterne, B. globigii, F. tularensis LVS, Y. Pestis, F1 antigen, MS coliphage, S. typhimurium | Sandwich IA | NRL array biosensor | 15 min | Toxins: <100 μg L−1 Bacteria: 1 × 103–106 cfu mL−1 Viruses: <109 pfu mL−1 | [176] |
| IL-2 | Sandwich IA | ZeptoReader | 2 h | 0.015 μg L−1 | [156] |
| IL-4 | 0.010 μg L−1 | ||||
| IL-6 | 0.005 μg L−1 | ||||
| E. coli, S. typhimurium L. monocytogenes, C. jejuni | Sandwich IA DNA hybridization | LabMAP | 3 h 30–40 min | 2.5–500 cells mL−1 103–105 genome copies | [186] |
| Salmonella ssp. | DNA sandwich hybridisation assay | Fibre-optic | 1 h | 103–104 cfu mL−1 | [189] |
| Chemiluminescence | |||||
| Atrazine | Indirect competitive IA | PASA | 29 min | 0.04 μg L−1 | [178] |
| Terbutylazine | 0.02 μg L−1 | ||||
| TNT | 0.13 μg L−1 | ||||
| Allergen specific IgE | Direct IA | PASA | 1 h | 0.16–1.9 μg L−1 | [42] |
| E. coli O157:H7, Y. Enterolitica, S. typhimurium, L. monocytogenes | Sandwich IA | CL-MIA | 1 h | Each in the range 104–105 cfu mL−1 | [157] |
| Ten antibiotics | Indirect competitive IA | PASA | 4 min 40 s | 0.12–31.8 μg L−1 | [17] |
| Virus Toxins Bacteria | Sandwich IA | ATS | 1 h 23 min | 6 × 102−5 × 106 TCID50 mL−1 0.1–0.2 μg L−1 5 × 103−2 × 106 cfu mL−1 | [151] |
| Electrochemistry | |||||
| E. coli, P. aerougenosa E. faecali, S. aureus, S. epidermidis | RNA sandwich hybridisation assay | eMicroLISA | 29 min | 0.5 mg L−1 total RNA | [127] |
Automated flow-through microarrays as monitoring systems will become more and more relevant in the future. Regulations for monitoring drinking water and food are enforced in many places. Surface and ground water has to be protected from chemicals in order to guarantee a good water quality [195]. Safe drinking water does not contain contaminants that are harmful to health. The US EPA published a list of regulated contaminants including 53 organic compounds and seven microorganisms, and a list of potential contaminants including bacteria, viruses, pesticides, and toxins [196]. Food safety regulation is important for the consumer who ingests all kinds of foods and is exposed to chemical and microbial contaminants.
Responsibility for ensuring that food remains in prime condition is shared by everyone involved in food production and handling, including primary producers, food manufacturers, establishments which serve food, and consumers [197]. Strict legal examinations are enforced across Europe to ensure high levels of safety, hygiene, and quality in commercial processing and food handling, as described in EU directive 93/43/EEC [198].
The advantage of microarray platforms as a rapid and cost-effective system has been addressed. In a central laboratory, chromatography techniques and/or mass spectrometry are established. They will persist in the future, as long as further confirmation in food and water analysis or clinical diagnostics, with independent techniques, is needed to exclude false-positive results. Nevertheless, immunochemical methods are seen as one type of screening technique for water and food monitoring [199]. In clinical diagnostics immunoassays are accepted. The automated systems analyse single analyte in blood samples. Multi-analyte detection would reduce the costs of analysis. However, the different antigen/antibody complexes have to be compatible with a single microarray platform. Nowadays, antibodies are available for many analytes, although most of them are not applied to analytical microarrays. A reliable microarray platform needs validated reagents including DNA or antibodies and standard analytes. Furthermore, the characterization and validation of the immobilized capture molecules and the coating have to be improved.
The combination and integration of sample enrichment techniques with an analytical microarray platform is needed as zero-tolerance is adopted for hygienically safe food and drinking water. The combination of immunomagnetic separation and electrochemiluminescence detection for Cryptosporidia in water is one example [200]. Enrichment steps prior to identification of microorganisms on an immunoanalytical platform are still faster than PCR methods or culture techniques.
Additional demands for microarray platforms are addressed in this review. Some of the discussed instruments have passed first field tests. The next step for fully automated microarray platforms is implementation of an intelligent software controlling the fluidics automatically during the analysis and the data processing afterwards. Consequently, little training will be required for a walk-away system. The software should recognize analytical problems or errors of the fluidics automatically and should generate a warning note therefore. These tools would improve the reliability of data generated from automated analytical microarrays.
The claim to have a portable system for monitoring units is not fulfilled at the moment. As they also need pumps and valves, they are transportable but not really hand-held. Handheld analytical microarray platforms for rapid food and water control are a task for future research and development. Integrated systems consisting of microfluidics and microoptical components will reduce volume and weight. The reagents will be stored on the platform in lyophilized form and are activated before the measurement starts. Currently, the fluidics of miniaturized platforms lack robustness and reproducibility. Intensive interdisciplinary research is essential for success in the future. The development of automated analytical microarray platforms should focus even more on engineering aspects, e.g. optimization of user interfaces and sample handling.
Acknowledgement
The authors like to thank the German Research Foundation, the Federal Ministry of Education and Research, the Federal Ministry of Defence, and the Consortium of Industrial Research Organisation (AiF “Otto von Guericke”) for continuous financial support in the field of microarray development.
References
- 1.Fodor SPA, Read JL, Pirrung MC, Stryer L, Tsai LA, Solas D. Science. 1991;251:767–773. doi: 10.1126/science.1990438. [DOI] [PubMed] [Google Scholar]
- 2.Schena M, Shalon D, Davis RW, Brown PO. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 3.Ekins R, Chu F. Ann Biol Clin. 1992;50:337–353. [PubMed] [Google Scholar]
- 4.Gabaldon JA, Maquieira A, Puchades R. Crit Rev Food Sci Nut. 1999;39:519–538. doi: 10.1080/10408699991279277. [DOI] [PubMed] [Google Scholar]
- 5.Hage DS. Anal Chem. 1999;71:294R–304R. doi: 10.1021/a1999901+. [DOI] [PubMed] [Google Scholar]
- 6.Miller JN, Niessner R, Knopp D. Ullmann’s Biotechnol Biochem Eng. 2007;2:585–612. [Google Scholar]
- 7.Rodriguez-Mozaz S, Lopez de Alda MJ, Barcelo D. Anal Bioanal Chem. 2006;386:1025–1041. doi: 10.1007/s00216-006-0574-3. [DOI] [PubMed] [Google Scholar]
- 8.Ekins R, Chu F. J Int Fed Clin Chem. 1997;9:100–109. [PubMed] [Google Scholar]
- 9.Ekins RP. Clin Chem. 1998;44:2015–2030. [PubMed] [Google Scholar]
- 10.Schütz AJ, Winklmair M, Weller MG, Niessner R. Fres J Anal Chem. 1999;363:625–631. [Google Scholar]
- 11.Templin MF, Stoll D, Schrenk M, Traub PC, Vöhringer CF, Joos TO. Trends Biotechnol. 2002;20:160–166. doi: 10.1016/s0167-7799(01)01910-2. [DOI] [PubMed] [Google Scholar]
- 12.Cho EJ, Collett JR, Szafranska AE, Ellington AD. Anal Chim Acta. 2006;564:82–90. doi: 10.1016/j.aca.2005.12.038. [DOI] [PubMed] [Google Scholar]
- 13.Ellington AD, Szostak JW. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 14.Tombelli S, Minunni A, Mascini A. Biosens Bioelectron. 2005;20:2424–2434. doi: 10.1016/j.bios.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 15.Lee M, Walt DR. Anal Biochem. 2000;282:142–146. doi: 10.1006/abio.2000.4595. [DOI] [PubMed] [Google Scholar]
- 16.Belleville E, Dufva M, Aamand J, Bruun L, Christensen CBV. Biotechniques. 2003;35:1044–1051. doi: 10.2144/03355rr04. [DOI] [PubMed] [Google Scholar]
- 17.Knecht BG, Strasser A, Dietrich R, Märtlbauer E, Niessner R, Weller MG. Anal Chem. 2004;76:646–654. doi: 10.1021/ac035028i. [DOI] [PubMed] [Google Scholar]
- 18.Roda A, Mirasoli M, Michelini E, Magliulo M, Simoni P, Guardigli M, Curini R, Sergi M, Marino A. Anal Bioanal Chem. 2006;385:742–752. doi: 10.1007/s00216-006-0473-7. [DOI] [PubMed] [Google Scholar]
- 19.Proll G, Tschmelak J, Gauglitz G. Anal Bioanal Chem. 2005;382:61–63. doi: 10.1007/s00216-004-2897-2. [DOI] [PubMed] [Google Scholar]
- 20.Tschmelak J, Proll G, Riedt J, Kaiser J, Kraemmer P, Barzaga L, Wilkinson JS, Hua P, Hole JP, Nudd R, Jackson M, Abuknesha R, Barcelo D, Rodriguez-Mozaz S, Lopez de Alda MJ, Sacher F, Stien J, Slobodnik J, Oswald P, Kozmenko H, Korenkova E, Tothova L, Krascsenits Z, Gauglitz G. Biosens Bioelectron. 2005;20:1509–1519. doi: 10.1016/j.bios.2004.07.033. [DOI] [PubMed] [Google Scholar]
- 21.WHO . Guidelines for drinking water quality. 3. Geneva: WHO; 2004. [Google Scholar]
- 22.Van Egmond HP, Jonker MA (2004) Rome: Food and Agriculture Organization of the United Nations
- 23.Zheng MZ, Richard JL, Binder J. Mycopathologia. 2006;161:261–273. doi: 10.1007/s11046-006-0215-6. [DOI] [PubMed] [Google Scholar]
- 24.Ngundi MM, Qadri SA, Wallace EV, Moore MH, Lassman ME, Shriver-Lake LC, Ligler FS, Taitt CR. Environ Sci Technol. 2006;40:2352–2356. doi: 10.1021/es052396q. [DOI] [PubMed] [Google Scholar]
- 25.Ivnitski D, Abdel-Hamid I, Atanasov P, Wilkins E. Biosens Bioelectron. 1999;14:599–624. doi: 10.1016/s0956-5663(99)00004-4. [DOI] [PubMed] [Google Scholar]
- 26.Iqbal SS, Mayo MW, Bruno JG, Bronk BV, Batt CA, Chambers JP. Biosens Bioelectron. 2000;15:549–578. doi: 10.1016/s0956-5663(00)00108-1. [DOI] [PubMed] [Google Scholar]
- 27.Lim DV, Simpson JM, Kearns EA, Kramer MF. Clin Microbiol Rev. 2005;18:583–607. doi: 10.1128/CMR.18.4.583-607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kricka LJ, Master SR, Joos TO, Fortina P. Ann Clin Biochem. 2006;43:457–467. doi: 10.1258/000456306778904731. [DOI] [PubMed] [Google Scholar]
- 29.Jaras K, Ressine A, Nilsson E, Malm J, Marko-Vargo G, Lilja H, Laurel T. Anal Chem. 2007;79:5817–5825. doi: 10.1021/ac0709955. [DOI] [PubMed] [Google Scholar]
- 30.Huang RP, Huang R, Fan Y, Lin Y. Anal Biochem. 2001;294:55–62. doi: 10.1006/abio.2001.5156. [DOI] [PubMed] [Google Scholar]
- 31.Schweitzer B, Roberts S, Grimwade B, Shao W, Wang M, Fu Q, Shu Q, Laroche I, Zhou Z, Tchernev VT, Christiansen J, Velleca M, Kingsmore SF. Nat Biotechnol. 2002;20:359–365. doi: 10.1038/nbt0402-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pollard HB, Srivastava M, Eidelman O, Jozwik C, Rothwell SW, Mueller GP, Jacobowitz DM, Darling T, Guggino WB, Wright J, Zeitlin PL, Paweletz CP. Proteomics Clin Appl. 2007;1:934–952. doi: 10.1002/prca.200700154. [DOI] [PubMed] [Google Scholar]
- 33.Bavykin SG, Akowski JP, Zakhariev VM, Barsky VE, Perov AN, Mirzabekov AD. Appl Environ Microbiol. 2001;67:922–928. doi: 10.1128/AEM.67.2.922-928.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dankbar DM, Dawson ED, Mehlmann M, Moore CL, Smagala JA, Shaw MW, Cox NJ, Kuchta RD, Rowlen KL. Anal Chem. 2007;79:2084–2090. doi: 10.1021/ac061960s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barken KB, Haagensen JJ, Tolker-Nielsen T. Clin Chim Acta. 2007;384:1–11. doi: 10.1016/j.cca.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 36.MacBeath G, Schreiber L. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 37.Haab BB, Dunham MJ, Brown PO. Genome Biol. 2001;2:1–13. doi: 10.1186/gb-2001-2-2-research0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Griffiths J. Anal Chem. 2007;79:8833–8837. [Google Scholar]
- 39.Paweletz CP, Charbanneau L, Bichsel VE, Simone NL, Chen R, Gillespie JW, Emmert-Buck MR, Roth MJ, Petricoin IE, Liotta LA. Oncogene. 2001;20:1981–1989. doi: 10.1038/sj.onc.1204265. [DOI] [PubMed] [Google Scholar]
- 40.Lueking A, Horn M, Eickhoff H, Büssow K, Lehrach H, Walter G. Anal Biochem. 1999;270:103–111. doi: 10.1006/abio.1999.4063. [DOI] [PubMed] [Google Scholar]
- 41.Nielsen UB, Geierstange BH. J Immunol Methods. 2004;290:107–120. doi: 10.1016/j.jim.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 42.Fall BI, Eberlein-König B, Behrendt H, Niessner R, Ring J, Weller MG. Anal Chem. 2003;75:556–562. doi: 10.1021/ac026016k. [DOI] [PubMed] [Google Scholar]
- 43.Joos TO, Schrenk M, Höpfl P, Kröger K, Chowdhury U, Stoll D, Schörner D, Dürr M, Herick K, Rupp S, Sohn K, Hämmerle H. Electrophoresis. 2000;21:2641–2650. doi: 10.1002/1522-2683(20000701)21:13<2641::AID-ELPS2641>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 44.Thirumalapura NR, Ramachandra A, Morton RJ, Malayer JR. J Immunol Methods. 2006;309:48–54. doi: 10.1016/j.jim.2005.11.016. [DOI] [PubMed] [Google Scholar]
- 45.Xu CW. Genome Res. 2002;12:482–486. doi: 10.1101/gr.213002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Castel D, Pitaval A, Debily MA, Gidrol X. Drug Disc Today. 2006;11:616–622. doi: 10.1016/j.drudis.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 47.Ziauddin J, Sabatini DM. Nature. 2001;411:107–110. doi: 10.1038/35075114. [DOI] [PubMed] [Google Scholar]
- 48.Poetz O, Ostendorp R, Brocks B, Schwenk JM, Stoll D, Joos TO, Templin MF. Proteomics. 2005;5:2402–2411. doi: 10.1002/pmic.200401299. [DOI] [PubMed] [Google Scholar]
- 49.Houseman BT, Huh JH, Kron SJ, Mrksich M. Nat Biotechnol. 2002;20:270–274. doi: 10.1038/nbt0302-270. [DOI] [PubMed] [Google Scholar]
- 50.Frank R. J Immunol Methods. 2002;267:13–26. doi: 10.1016/s0022-1759(02)00137-0. [DOI] [PubMed] [Google Scholar]
- 51.Wang DN, Liu SY, Trumme BJ, Deng C, Wang AL. Nat Biotechnol. 2002;20:275–281. doi: 10.1038/nbt0302-275. [DOI] [PubMed] [Google Scholar]
- 52.Blixt O, Head S, Mondala T, Scanlan C, Huflejt ME, Alvarez R, Bryan MC, Fazio F, Calarese D, Stevens J, Razi N Stevens DJ, Skehel JJ, van Die I, Burton DR, Wilson IA, Cummings R, Bovin N, Wong CH, Paulson JC. Proc Nat Acad Sci. 2004;101:17033–17038. doi: 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fang Y, Lahiri J, Picard L. Drug Disc Today. 2003;8:755–761. doi: 10.1016/s1359-6446(03)02779-x. [DOI] [PubMed] [Google Scholar]
- 54.Funeriu DP, Eppinger J, Denizot L, Miyake M, Miyake J. Nat Biotechnol. 2005;23:622–526. doi: 10.1038/nbt1090. [DOI] [PubMed] [Google Scholar]
- 55.Lemarchand K, Masson L, Brousseau R. Crit Rev Microbiol. 2004;30:145–172. doi: 10.1080/10408410490435142. [DOI] [PubMed] [Google Scholar]
- 56.Lin B, Wang Z, Vora GJ, Thornton JA, Schnur JM, Thach DC, Blaney KM, Ligler AG, Malanoski AP, Santiago J, Walter EA, Agan BK, Metzgar D, Seto D, Daum LT, Kruzelock R, Rowley RK, Hanson EH, Tibbetts C, Stenger DA. Genome Res. 2006;16:527–535. doi: 10.1101/gr.4337206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kostrzynska M, Bachand A. Can J Microbiol. 2006;52:1–8. doi: 10.1139/w05-105. [DOI] [PubMed] [Google Scholar]
- 58.Maynard C, Berthiaume F, Lemarchand K, Harel J, Payment P, Bayardelle P, Masson L, Brousseau R. App Environ Microbiol. 2005;71:8548–8557. doi: 10.1128/AEM.71.12.8548-8557.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Loy A, Bodrossy L. Clin Chim Acta. 2006;363:106–119. doi: 10.1016/j.cccn.2005.05.041. [DOI] [PubMed] [Google Scholar]
- 60.Feng Y, Ke X, Ma R, Chen Y, Hu G, Liu F. Clin Chem. 2004;50:416–422. doi: 10.1373/clinchem.2003.023994. [DOI] [PubMed] [Google Scholar]
- 61.Seidel M, Gauglitz G. Trends Anal Chem. 2003;22:385–394. [Google Scholar]
- 62.Holt LJ, Büssow K, Walter G, Tomlinson IM. Nucleic Acids Res. 2000;28:E72. doi: 10.1093/nar/28.15.e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kusnezow W, Jacob A, Walijew A, Diehl F, Hoheisel JD. Proteomics. 2003;3:254–264. doi: 10.1002/pmic.200390038. [DOI] [PubMed] [Google Scholar]
- 64.Ferretti S, Paynter S, Russell DA, Sapsford KE. Trends Anal Chem. 2000;19:530–540. [Google Scholar]
- 65.Seidel M, Dankbar D, Gauglitz G. Anal Bioanal Chem. 2004;379:904–912. doi: 10.1007/s00216-004-2651-9. [DOI] [PubMed] [Google Scholar]
- 66.Naumann CA, Prucker O, Lehmann T, Knoll W, Frank CW. Biomacromolecules. 2002;3:27–35. doi: 10.1021/bm0100211. [DOI] [PubMed] [Google Scholar]
- 67.Dankbar D, Gauglitz G. Anal Bioanal Chem. 2006;386:1964–1976. doi: 10.1007/s00216-006-0871-x. [DOI] [PubMed] [Google Scholar]
- 68.Angenendt P. Drug Disc Today. 2005;10:503–511. doi: 10.1016/S1359-6446(05)03392-1. [DOI] [PubMed] [Google Scholar]
- 69.Kamisetty NK, Pack SP, Nonogawa M, Devarayapalli KC, Kodaki T, Makino K. Anal Bioanal Chem. 2006;286:1649–1655. doi: 10.1007/s00216-006-0741-6. [DOI] [PubMed] [Google Scholar]
- 70.Wayment JR, Harris JM. Anal Chem. 2006;78:7841–7841. doi: 10.1021/ac061392g. [DOI] [PubMed] [Google Scholar]
- 71.Angenendt P, Glökler J, Sobek J, Lehrach H, Cahill DJ. J Chromatogr A. 2003;1009:97–104. doi: 10.1016/s0021-9673(03)00769-6. [DOI] [PubMed] [Google Scholar]
- 72.Golden JP, Taitt CR, Shriver-Lake LC, Shubin YA, Ligler FS. Talanta. 2005;56:1078–1085. doi: 10.1016/j.talanta.2004.03.072. [DOI] [PubMed] [Google Scholar]
- 73.Taitt CR, Anderson GP, Ligler FS. Biosens Bioelectron. 2005;20:2470–2487. doi: 10.1016/j.bios.2004.10.026. [DOI] [PubMed] [Google Scholar]
- 74.Chevolot Y, Bouillon C, Vial S, Morvan F, Meyer A, Cloarec JP, Jochum A, Praly JP, Vasseur JJ, Souteyrand E. Angew Chem Int Ed. 2007;46:2398–2402. doi: 10.1002/anie.200604955. [DOI] [PubMed] [Google Scholar]
- 75.Hawker CJ, Fokin VV, Finn MG, Sharpless KB. Aust J Chem. 2007;60:381–383. [Google Scholar]
- 76.Gallant ND, Lavery KA, Amis EJ, Becker ML. Adv Mater. 2007;19:965–969. [Google Scholar]
- 77.Bailey RC, Kwong GA, Radu CG, Witte ON, Health JR. J Am Chem Soc. 2007;129:1959–1967. doi: 10.1021/ja065930i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Buhl A, Metzger JH, Heegaard HH, von Landenberg P, Fleck M, Luppa PB. Clin Chem. 2007;53:334–341. doi: 10.1373/clinchem.2006.077339. [DOI] [PubMed] [Google Scholar]
- 79.Jung A, Stemmler I, Brecht A, Gauglitz G. Fres J Anal Chem. 2001;371:128–136. doi: 10.1007/s002160101001. [DOI] [PubMed] [Google Scholar]
- 80.Masson JF, Battaglia TM, Davidson MJ, Kim YC, Prakash AMC, Beaudoin S, Booksh KS. Talanta. 2005;67:918–925. doi: 10.1016/j.talanta.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 81.Piehler J, Brecht A, Valiokas R, Liedberg B, Gauglitz G. Biosens Bioelectron. 2000;15:473–481. doi: 10.1016/s0956-5663(00)00104-4. [DOI] [PubMed] [Google Scholar]
- 82.Wolter A, Niessner R, Seidel M. Anal Chem. 2007;79:4529–4537. doi: 10.1021/ac070243a. [DOI] [PubMed] [Google Scholar]
- 83.Lee CS, Kim BG. Biotechnol Lett. 2002;24:839–844. [Google Scholar]
- 84.Marquette CA, Cretich M, Blum LJ, Chiari M. Talanta. 2007;71:1312–1318. doi: 10.1016/j.talanta.2006.06.039. [DOI] [PubMed] [Google Scholar]
- 85.Schena M, Heller RA, Theriault TP, Konrad K, Lachenmeier E, Davis RW. Trends Biotechnol. 1998;16:301–306. doi: 10.1016/s0167-7799(98)01219-0. [DOI] [PubMed] [Google Scholar]
- 86.Okomoto T, Suzuki N, Yakamoto N. Nature Biotechnol. 2000;18:438–441. doi: 10.1038/74507. [DOI] [PubMed] [Google Scholar]
- 87.Schober A, Schienhorst GA, Döring M, Linemann BF. Biotechniques. 1993;15:324–329. [PubMed] [Google Scholar]
- 88.Lemmo AV, Rose DJ, Tisone TC. Curr Opinion Biotechnol. 1998;9:615–617. doi: 10.1016/s0958-1669(98)80139-0. [DOI] [PubMed] [Google Scholar]
- 89.Rose D. Drug Disc Today. 1999;4:411–419. doi: 10.1016/s1359-6446(99)01388-4. [DOI] [PubMed] [Google Scholar]
- 90.Ducree J, Gruhler H, Hey H, Muller N, Bekesi S, Freygang M, Sandmaier H, Zengerle R (2000) Ann Int Conf Micro Electro Mech Systems, Proc, 13th, Miyazaki, Japan, Jan 23–27, 2000:317–322
- 91.Gutmann O, Kuehlewein R, Reinbold S, Niekrawietz R, Steinert CP, de Heij B, Zengerle R, Daub M. Lab Chip. 2005;5:675–681. doi: 10.1039/b418765b. [DOI] [PubMed] [Google Scholar]
- 92.Shalon D, Smith SJ, Brown PO. Gen Res. 1996;6:639–645. doi: 10.1101/gr.6.7.639. [DOI] [PubMed] [Google Scholar]
- 93.Kane RS, Takayama S, Ostuni E, Ingber DE, Whitesides GM. Biomaterials. 1999;20:2363–2371. doi: 10.1016/s0142-9612(99)00165-9. [DOI] [PubMed] [Google Scholar]
- 94.Bernard A, Delamarche E, Schmid H, Michel B, Bossard HR, Biebuyck H. Langmuir. 1998;14:2225–2229. [Google Scholar]
- 95.Angenendt P, Glökler J, Konthur Z, Lehrach H, Cahill DJ. Anal Chem. 2003;75:4368–4372. doi: 10.1021/ac034260l. [DOI] [PubMed] [Google Scholar]
- 96.Tehan EC, Higbee DJ, Wood TD, Bright FV. Anal Chem. 2007;79:5429–5434. doi: 10.1021/ac070533r. [DOI] [PubMed] [Google Scholar]
- 97.Tseng FG, Lin SC, Huang HM, Huang CY, Chieng CC. Sens Actuators B. 2002;83:22–29. [Google Scholar]
- 98.Rozkiewicz DI, Brugmann W, Kerkhoven RM, Ravoo BJ, Reinhoudt DN. J Am Chem Soc. 2007;129:11593–11599. doi: 10.1021/ja073574d. [DOI] [PubMed] [Google Scholar]
- 99.Feldstein MJ, Golden JP, Rowe CA, MacCraith BD, Ligler FS. J Biomed Microdev. 1999;1:139–153. doi: 10.1023/A:1009900608757. [DOI] [PubMed] [Google Scholar]
- 100.Rowe CA, Tender LM, Feldstein MJ, Golden JP Scruggs SB, MacCraith BD, Cras JJ, Ligler FS. Anal Chem. 1999;71:3846–3852. doi: 10.1021/ac981425v. [DOI] [PubMed] [Google Scholar]
- 101.Benn JA, Hu J, Hogan BJ, Fry RC, Samson LD, Thorsen T. Anal Biochem. 2006;348:284–293. doi: 10.1016/j.ab.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 102.Meusel PR, Grawe F, Katerkamp A, Cammann K, Börchers T. Fresenius J Anal Chem. 2001;371:120–127. doi: 10.1007/s002160101006. [DOI] [PubMed] [Google Scholar]
- 103.Madou MJ, Lee LJ, Daunert S, Lai S, Shih CH. Biomed Microdevices. 2001;3:245–254. [Google Scholar]
- 104.Yu F, Yao D, Knoll W. Anal Chem. 2003;75:2610–2617. doi: 10.1021/ac026161y. [DOI] [PubMed] [Google Scholar]
- 105.Nickova M, Dosev D, Gee SJ, Hammock BD, Kennedy M. Anal Chem. 2005;77:6864–6873. doi: 10.1021/ac050826p. [DOI] [PubMed] [Google Scholar]
- 106.Smith AM, Dave S, Nie SM, True L, Gao XH. Exp Rev Mol Diagn. 2006;5:231–244. doi: 10.1586/14737159.6.2.231. [DOI] [PubMed] [Google Scholar]
- 107.Schäferling M, Nagl S. Anal Bioanal Chem. 2006;385:500–517. doi: 10.1007/s00216-006-0317-5. [DOI] [PubMed] [Google Scholar]
- 108.Liebermann T, Knoll W. Langmuir. 2003;19:1567–1572. [Google Scholar]
- 109.Yu F, Yao DF, Knoll W. Anal Chem. 2003;75:2610–2617. doi: 10.1021/ac026161y. [DOI] [PubMed] [Google Scholar]
- 110.Marquette CA, Blum LJ. Anal Bioanal Chem. 2006;385:546–554. doi: 10.1007/s00216-006-0439-9. [DOI] [PubMed] [Google Scholar]
- 111.Roda A, Guardigli M, Michelini E, Mirasoli M, Pasini P. Anal Chem. 2003;75:462A–470A. [PubMed] [Google Scholar]
- 112.Creton R, Jaffe LF. Biotechniques. 2001;31:1098–1105. doi: 10.2144/01315rv01. [DOI] [PubMed] [Google Scholar]
- 113.Chouhan RS, Vivek Babu K, Kumar MA, Neeta NS, Thakur MS, Amitha BE, Pasha A, Karanth NGK, Karanth NG. Biosens Bioelectron. 2006;21:1264–1271. doi: 10.1016/j.bios.2005.05.018. [DOI] [PubMed] [Google Scholar]
- 114.Lu G, Shem H, Cheng B, Chen Z, Marquette CA, Blum LJ, Tillement O, Roux S, Ledoux G, Ou M, Perriat P. Appl Phys Lett. 2006;89:223128–1–3. [Google Scholar]
- 115.Niwa O. Electroanalysis. 1995;7:606–613. [Google Scholar]
- 116.Schienle M, Paulus C, Frey A, Hofmann F, Holzapfl B, Schindler-Bauer P, Thewes R (2004) IEEE J Solid-State Circuits 39
- 117.Dill K, Montgomery DD, Ghindilis AL, Schwarzkopf KR, Ragsdale SR, Oleinikov AV. Biosens Bioelectron. 2004;20:736–742. doi: 10.1016/j.bios.2004.06.049. [DOI] [PubMed] [Google Scholar]
- 118.Tokuda T, Tanaka K, Matsuo M, Kagawa K, Nunoshita M, Ohta J. Sens Actuators A. 2007;135:315–322. [Google Scholar]
- 119.Hashimoto K, Ito K, Ishimori Y. Sens Actuators B. 1998;46:220–225. [Google Scholar]
- 120.Ayers S, Gillis KD, Lindau M, Minch BA. IEEE Trans Circuits Systems. 2007;54:736–744. doi: 10.1109/TCSI.2006.888777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Drummond TG, Hill MG, Barton JK. Nat Biotechnol. 2003;10:1192–1199. doi: 10.1038/nbt873. [DOI] [PubMed] [Google Scholar]
- 122.Marquette CA, Blum LJ. Biosens Bioelectron. 2006;21:1424–1433. doi: 10.1016/j.bios.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 123.Liu RH, Yang J, Lenigk R, Bonanno J, Grodzinski P. Anal Chem. 2004;76:1824–1831. doi: 10.1021/ac0353029. [DOI] [PubMed] [Google Scholar]
- 124.Hintsche R, Elsholz B, Piechotta R, Woerl R, Scgmueller CCJ, Albers J, Dharuman V, Nebling E, Hanisch A, Blohm L, Hofmann F, Holzapfl B, Frey A, Paulus C, Schienle M, Thewes R. In: Electrochemistry of nucleic acids and proteins. Palacek E, Scheller F, Wang J, editors. Amsterdam: Elsevier; 2005. pp. 247–279. [Google Scholar]
- 125.Los M, Los JM, Blohm L, Spillner E, Grunwald T, Albers J, Hintsche R, Wegrzyn G. Lett Appl Microbiol. 2005;40:479–485. doi: 10.1111/j.1472-765X.2005.01706.x. [DOI] [PubMed] [Google Scholar]
- 126.Hintsche R, Albers J, Bernt H, Eder A. Electroanalysis. 2000;12:660–665. [Google Scholar]
- 127.Elsholz B, Wörl R, Blohm L, Albers J, Feucht H, Grunewald T, Jürgen B, Schweder T, Hintsche R. Anal Chem. 2006;78:4794–4802. doi: 10.1021/ac0600914. [DOI] [PubMed] [Google Scholar]
- 128.Albers J, Grunwald T, Nebling E, Piechotta G, Hintsche R. Anal Bioanal Chem. 2003;37:521–527. doi: 10.1007/s00216-003-2192-7. [DOI] [PubMed] [Google Scholar]
- 129.Ghindilis AL, Smith MW, Schwarzkopf KR, Roth KM, Peyvan K, Munro SB, Lodes MJ, Stöver AG, Bernards K, Dill K, McShea A. Biosens Bioelectron. 2007;22:1853–1860. doi: 10.1016/j.bios.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 130.Yu X, Xu D, Cheng Q. Proteomics. 2006;6:5493–5503. doi: 10.1002/pmic.200600216. [DOI] [PubMed] [Google Scholar]
- 131.Bally M, Halter M, Vörös J, Grandin HM. Surf Interface Anal. 2006;38:1442–1458. [Google Scholar]
- 132.Berger CEH, Beumer TAM, Kooyman RPH, Greve J. Anal Chem. 1998;70:703–706. [Google Scholar]
- 133.Gauglitz G. Anal Bioanal Chem. 2005;381:141–155. doi: 10.1007/s00216-004-2895-4. [DOI] [PubMed] [Google Scholar]
- 134.Nikitin PI, Gorshov BG, Nikitin EP, Ksenevich TI. Sens Actuators B. 2005;111:500–504. [Google Scholar]
- 135.Gao T, Rothberg LJ. Anal Chem. 2007;79:7589–7595. doi: 10.1021/ac071082d. [DOI] [PubMed] [Google Scholar]
- 136.Venkatasubbarao S, Beaudry N, Zhao YM, Chipman R (2006) J Biomed Optics 11:Art. No 014028 [DOI] [PubMed]
- 137.Wassaf D, Kuang G, Kopacz K, Wu QL, Nguyen Q, Toews M, Cosic J, Jacques J, Wiltshire S, Lambert J, Pazmany CC, Hogan S, Ladner RC, Nixon AE, Sexton DJ. Anal Biochem. 2006;351:241–253. doi: 10.1016/j.ab.2006.01.043. [DOI] [PubMed] [Google Scholar]
- 138.Kröger K, Bauer J, Fleckenstein B, Rademann J, Jung G, Gauglitz G. Biosens Bioelectron. 2002;17:937–944. doi: 10.1016/s0956-5663(02)00085-4. [DOI] [PubMed] [Google Scholar]
- 139.Shankaran DR, Gobi KV, Miura N. Sens Actuators B. 2007;121:158–177. [Google Scholar]
- 140.Homola J, Vaisocherova J, Dostalek J, Piliarik M. Methods. 2005;37:26–36. doi: 10.1016/j.ymeth.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 141.Rich RL, Myszka (2007) Anal Biochem 361:1–6 [DOI] [PubMed]
- 142.Mauriz E, Calle E, Manclus JJ, Montoya A, Lechuga LM. Anal Bioanal Chem. 2007;387:1449–1458. doi: 10.1007/s00216-006-0800-z. [DOI] [PubMed] [Google Scholar]
- 143.Lee KH, Su YD, Chen SJ, Tseng FG, Lee GB. Biosens Bioelectron. 2007;23:466–472. doi: 10.1016/j.bios.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 144.Li PY, Lin B, Gerstenmaier J, Cunningham BT. Sens Actuators B: 2004;99:6–13. [Google Scholar]
- 145.Unfricht DW, Colpitts SL, Fernandez SM, Lynes MA. Proteomics. 2005;5:4432–4442. doi: 10.1002/pmic.200401314. [DOI] [PubMed] [Google Scholar]
- 146.Jona G, Snyder M. Curr Opin Mol Ther. 2003;5:271–277. [PubMed] [Google Scholar]
- 147.Gehring AG, Albin DM, Bhunia AK, Reed SA, Tu SI, Uknalis J. Anal Chem. 2006;78:6601–6607. doi: 10.1021/ac0608467. [DOI] [PubMed] [Google Scholar]
- 148.Gehring AG, Albin DM. J Rapid Methods Autom Microbiol. 2007;15:49–66. [Google Scholar]
- 149.Rubina AY, Dyukova VI, Dementieva EI, Stomakhin AA, Nesmeyanov VA, Grishin EV, Zasedatelelev AS. Anal Biochem. 2005;340:317–329. doi: 10.1016/j.ab.2005.01.042. [DOI] [PubMed] [Google Scholar]
- 150.Parro V, Rodriguez-Manfredi JA, Briones C, Compostizo C, Herrero PL, Vez E, Sebastian E, Moreno-Paz, Garcia-Villadangos M, Fernandez-Calvo P, Gonzales-Toril E, Perez-Mercader J, Fernandez-Remolar, Gomez-Elvira J (2005) Planet Space Sci, 53:729–737
- 151.Huelseweh B, Ehricht R, Marschall HJ. Proteomics. 2006;6:2972–2981. doi: 10.1002/pmic.200500721. [DOI] [PubMed] [Google Scholar]
- 152.Belleville E, Dufva M, Aamand J, Bruun L, Christensen CBV. Biotechniques. 2003;35:1044–1051. doi: 10.2144/03355rr04. [DOI] [PubMed] [Google Scholar]
- 153.Belleville E, Dufva M, Aamand J, Bruun L, Clausen L, Christensen CBV. J Immunol Methods. 2004;286:219–229. doi: 10.1016/j.jim.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 154.Rucker VC, Havenstrite KL, Herr AE. Anal Biochem. 2005;339:262–270. doi: 10.1016/j.ab.2005.01.030. [DOI] [PubMed] [Google Scholar]
- 155.Duveneck GL, Abel AP, Bopp MA, Kresbach GM, Ehrat M. Anal Chim Acta. 2002;469:49–61. [Google Scholar]
- 156.Pawlac M, Schick E, Bopp MA, Schneider MJ, Oroszlan P, Ehrat M. Proteomics. 2002;2:383–393. doi: 10.1002/1615-9861(200204)2:4<383::AID-PROT383>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 157.Magliulo M, Simoni P, Guardigli M, Michelini E, Luciani M, Lelli R, Roda A. J Agric Food Chem. 2007;55:4933–4939. doi: 10.1021/jf063600b. [DOI] [PubMed] [Google Scholar]
- 158.Fitzgerald SP, Lamont JV, McConnell RI, Benchikh EO. Clin Chem. 2005;51:1165–1176. doi: 10.1373/clinchem.2005.049429. [DOI] [PubMed] [Google Scholar]
- 159.Rodriguez-Mozaz S, Reder S, Lopez de Alda M, Gauglitz G, Barcelo D. Biosens Bioelectron. 2004;19:633–640. doi: 10.1016/s0956-5663(03)00255-0. [DOI] [PubMed] [Google Scholar]
- 160.Tschmelak J, Proll G, Gauglitz G. Biosens Bioelectron. 2004;20:743–752. doi: 10.1016/j.bios.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 161.Käppel ND, Pröll F, Gauglitz G. Biosens Bioelectron. 2007;22:2295–2300. doi: 10.1016/j.bios.2006.11.030. [DOI] [PubMed] [Google Scholar]
- 162.Tschmelak J, Proll G, Gauglitz G. Anal Bioanal Chem. 2004;379:1004–1012. doi: 10.1007/s00216-004-2695-x. [DOI] [PubMed] [Google Scholar]
- 163.Tschmelak J, Proll G, Gaulitz G. Talanta. 2005;65:313–323. doi: 10.1016/j.talanta.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 164.Tschmelak J, Proll G, Riedt J, Kaiser J, Kraemmer P, Barzaga L, Wilkinson JS, Hua P, Hole JP, Nudd R, Jackson M, Abuknesha R, Barcelo D, Rodriguez-Mozaz S, Lopez de Alda MJ, Sacher F, Stien J, Slobodnik J, Oswald P, Kozmenko H, Korenkova E, Tothova L, Krascsenits Z, Gauglitz G. Biosens Bioelectron. 2005;20:1499–1508. doi: 10.1016/j.bios.2004.07.032. [DOI] [PubMed] [Google Scholar]
- 165.Proll G, Tschmelak J, Gauglitz G. Anal Bioanal Chem. 2005;381:61–63. doi: 10.1007/s00216-004-2897-2. [DOI] [PubMed] [Google Scholar]
- 166.Ligler FS, Sapsford KE, Golden JP, Shriver-Lake LC, Taitt CR, Dyer MA, Barone S, Myatt CJ. Anal Sci. 2007;23:5–10. doi: 10.2116/analsci.23.5. [DOI] [PubMed] [Google Scholar]
- 167.Taitt CR, Anderson GP, Lingerfelt BM, Feldstein MJ, Ligler FS. Anal Chem. 2002;74:6114–6120. doi: 10.1021/ac0260185. [DOI] [PubMed] [Google Scholar]
- 168.Shriver-Lake LC, Ligler FS. IEEE Sensors J. 2005;5:751–756. [Google Scholar]
- 169.Sapsford KE, Taitt CR, Fertig S, Moore MH, Lassman ME, Maragos CA. Biosens Bioelectron. 2006;21:2298–2305. doi: 10.1016/j.bios.2005.10.021. [DOI] [PubMed] [Google Scholar]
- 170.Sapsford KE, Taitt CR, Loo N, Ligler FS. Appl Environ Microbiol. 2005;71:5590–5592. doi: 10.1128/AEM.71.9.5590-5592.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Rowe CA, Scruggs SB, Feldstein MJ, Golden JP, Ligler FS. Anal Chem. 1999;71:433–439. doi: 10.1021/ac980798t. [DOI] [PubMed] [Google Scholar]
- 172.Rowe-Taitt CA, Hazzard JW, Hofman KE, Cras JJ, Golden JP, Ligler FS. Biosens Bioelectron. 2000;15:579–589. doi: 10.1016/s0956-5663(00)00122-6. [DOI] [PubMed] [Google Scholar]
- 173.Rowe-Taitt CA, Golden JP, Feldstein MJ, Cras JJ, Hofman KE, Ligler FS. Biosens Bioelectron. 2000;14:785–794. doi: 10.1016/s0956-5663(99)00052-4. [DOI] [PubMed] [Google Scholar]
- 174.Rowe CA, Tender LM, Feldstein MJ, Golden JP, Scruggs SB, MacCraith BD, Cras JJ, Ligler FS. Anal Chem. 1999;71:3846–3852. doi: 10.1021/ac981425v. [DOI] [PubMed] [Google Scholar]
- 175.Sapsford KE, Rasooly A, Taitt CR, Ligler FS. Anal Chem. 2004;76:433–440. doi: 10.1021/ac035122z. [DOI] [PubMed] [Google Scholar]
- 176.Taitt CR, Shubin YS, Angel R, Ligler FS. Appl Environ Microbiol. 2004;70:152–158. doi: 10.1128/AEM.70.1.152-158.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Yakovleva J, Davidsson R, Lobanova A, Bengtsson M, Eremin S, Laurell T, Emneus J. Anal Chem. 2002;74:2994–3004. doi: 10.1021/ac015645b. [DOI] [PubMed] [Google Scholar]
- 178.Weller MG, Schuetz AJ, Winklmair M, Niessner R. Anal Chim Acta. 1999;393:29–41. [Google Scholar]
- 179.Knecht BG, Strasser A, Dietrich R, Märtlbauer E, Niessner R, Weller MG. Anal Chem. 2004;76:646–654. doi: 10.1021/ac035028i. [DOI] [PubMed] [Google Scholar]
- 180.Kloth K, Niessner R, Seidel M (2007) unpublished
- 181.Nolan JP, Sklar LA. Trends Biotechnol. 2002;20:9–12. doi: 10.1016/s0167-7799(01)01844-3. [DOI] [PubMed] [Google Scholar]
- 182.Vignali DAA. J Immunol Methods. 2000;243:243–255. doi: 10.1016/s0022-1759(00)00238-6. [DOI] [PubMed] [Google Scholar]
- 183.Dias D, Vam Doren J, Schlottmann S, Kelly S, Puchalski D, Ruiz W, Boerckel P, Kessler J, Antonello JM, Green T, Brown M, Smith J, Chirmule N, Barr E, Jansen KU, Esser MT. Clin Diagn Lab Immunol. 2005;12:959–969. doi: 10.1128/CDLI.12.8.959-969.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.De Jager W, te Velthius H, Prakken BJ, Kuis W, Rijkers GT. Clin Diagn Lab Immunol. 2003;10:133–139. doi: 10.1128/CDLI.10.1.133-139.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Haasnoot W, Du Pre JG. J Agric Food Chem. 2007;55:3771–3777. doi: 10.1021/jf063281o. [DOI] [PubMed] [Google Scholar]
- 186.Dunbar SA, Vander Zee CA, Oliver KG, Karem KL, Jacobson JW. J Microbiol Methods. 2003;53:245–252. doi: 10.1016/s0167-7012(03)00028-9. [DOI] [PubMed] [Google Scholar]
- 187.Walt DR. Biotechniques. 2006;41:529–535. doi: 10.2144/000112303. [DOI] [PubMed] [Google Scholar]
- 188.Epstein JR, Ferguson JA, Lee KH, Walt DR. J Am Chem Soc. 2003;125:13753–13759. doi: 10.1021/ja0365577. [DOI] [PubMed] [Google Scholar]
- 189.Ahn S, Walt DR. Anal Chem. 2005;77:5041–5047. doi: 10.1021/ac0505270. [DOI] [PubMed] [Google Scholar]
- 190.Ferguson JA, Steemers FJ, Walt DR. Anal Chem. 2000;72:5618–5624. doi: 10.1021/ac0008284. [DOI] [PubMed] [Google Scholar]
- 191.Song L, Ahn S, Walt DR. Anal Chem. 2006;78:1023–1033. doi: 10.1021/ac051417w. [DOI] [PubMed] [Google Scholar]
- 192.Burnes M, Freudenberg J, Thompson S, Aronow B, Pavlidis P. Nucleic Acid Res. 2005;33:5914–5923. doi: 10.1093/nar/gki890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Steemers FJ, Gunderson KL. Pharmacogen. 2005;6:777–782. doi: 10.2217/14622416.6.7.777. [DOI] [PubMed] [Google Scholar]
- 194.Fan JB, Hu SX, Craumer WC, Barker DL. Biotechniques. 2005;39:583–588. [PubMed] [Google Scholar]
- 195.European Commission. Commission Directive Off J L. 2000/60/EC;327:1–72. [Google Scholar]
- 196.Richardson SA. Anal Chem. 2007;79:4295–4324. doi: 10.1021/ac070719q. [DOI] [PubMed] [Google Scholar]
- 197.European Commission. Regulation 2002/178/EC Off J L 31:1–24
- 198.European Commission. Council Regulation 315/93/EEC off J L37:1–3
- 199.Krämer PM, Martens D, Forster S, Ipolyi I, Brunori C, Morabito R. Anal Bioanal Chem. 2007;287:1435–1448. doi: 10.1007/s00216-006-0793-7. [DOI] [PubMed] [Google Scholar]
- 200.Kuczynska E, Boyer DG, Shelton DR. J Microbiol Methods. 2003;53:17–26. doi: 10.1016/s0167-7012(02)00211-7. [DOI] [PubMed] [Google Scholar]


