Abstract
Over the last decade, adherent MDCK (Madin Darby canine kidney) and Vero cells have attracted considerable attention for production of cell culture-derived influenza vaccines. While numerous publications deal with the design and the optimization of corresponding upstream processes, one-to-one comparisons of these cell lines under comparable cultivation conditions have largely been neglected. Therefore, a direct comparison of influenza virus production with adherent MDCK and Vero cells in T-flasks, roller bottles, and lab-scale bioreactors was performed in this study. First, virus seeds had to be adapted to Vero cells by multiple passages. Glycan analysis of the hemagglutinin (HA) protein showed that for influenza A/PR/8/34 H1N1, three passages were sufficient to achieve a stable new N-glycan fingerprint, higher yields, and a faster increase to maximum HA titers. Compared to MDCK cells, virus production in serum-free medium with Vero cells was highly sensitive to trypsin concentration. Virus stability at 37 °C for different virus strains showed differences depending on medium, virus strain, and cell line. After careful adjustment of corresponding parameters, comparable productivity was obtained with both host cell lines in small-scale cultivation systems. However, using these cultivation conditions in lab-scale bioreactors (stirred tank, wave bioreactor) resulted in lower productivities for Vero cells.
Keywords: Influenza virus, Vaccine production, TCID50 stability, Trypsin, Glycosylation, Bioreactor
Introduction
With the goal to establish flexible production platforms for high-yield cell culture-derived influenza vaccine manufacturing, cell-line screening has identified on one hand, adherently growing cells such as MDCK (Madin Darby canine kidney) and Vero (African green monkey kidney) cells, and on the other hand, suspension cells such as MDCK 33016, HEK-293, PER.C6, or AGE1.CR® cells (Kessler et al. 1999; Pau et al. 2001; Brands et al. 1999; Kistner et al. 1998; Nakamura and Homma 1981; Sandig and Jordan 2005; Gröner and Vorlop 1997; Le Ru et al. 2010). Each cell line shows its specificities, and its use for vaccine production has advantages but also disadvantages compared to the other cell lines. Based on a thorough understanding of the pros and cons of individual host cells for process performance, properties of other cell lines, specific aspects of virus–host cell interaction and process options can be better evaluated. Overall, a thorough comparison of cell lines might help to develop process optimization strategies, as well as to assess quality differences concerning the virus strains and antigens produced.
Over the last decade, the use of adherent cells for influenza virus manufacturing has attracted much attention (Kistner et al. 1998; Barrett et al. 2009; Tree et al. 2001). In particular, growth and influenza virus replication in adherently growing MDCK and Vero cells has been investigated (Rimmelzwaan et al. 1998; Merten et al. 1999a, b; Nicolson et al. 2005). However, a direct comparison of experimental data for these two cell lines, i.e., an identification of the cell line resulting in optimum process performance, is not straight forward. On one side, cell and virus propagation were investigated in different cultivation systems (static systems, roller bottles, spinner, stirred tank). On the other side, not only different media (serum-containing (SC) as well as serum-free (SF)) but also different microcarriers (Cytodex 1 and 3) were used for large-scale cultures (Merten et al. 1996; Quesney et al. 2001; Kistner et al. 2007; Mendonca and Pereira 1998, 1995). Additional variations resulted from the use of MDCK as well as Vero cell lines from different sources and the use of different influenza virus strains (Genzel and Reichl 2009). And, even when similar cultivation conditions are chosen, differences in productivity as well as product quality are to be expected, as for example, Vero cells are known to be deficient in β-interferon response due to the absence of the β-interferon gene (Mosca 1986; Desmyter et al. 1968). Typically, cell lines from two different hosts will also show variations in the glycosylation machinery, metabolism, enzyme activities, and proteome levels (Ritter et al. 2008; Schwarzer et al. 2009).
Kistner et al. described the use of adherent Vero cells for influenza vaccine production by comparing the production of different virus strains in eggs and Vero cells (Kistner et al. 1998). For scale-up, a 1,200 L stirred tank reactor (STR) was used, and cell concentrations up to 5 × 106 cells/ml were obtained using Cytodex 3 microcarriers. Virus infection was done at 32 °C. Later, Kistner et al. discussed the successful production of an inactivated H5N1 whole virus vaccine with Vero cells (Kistner et al. 2007; Barrett et al. 2009).
It was shown that for most cell culture-derived influenza production, the addition of trypsin at the time of infection resulted in a fast cleavage of hemagglutinin (HA) needed for successful infection. Decreased virus titers (pfu/ml) and slower virus production were described when trypsin was not added to the virus maintenance medium in roller bottles (Tree et al. 2001). Typically, trypsin has to be added several times during influenza production in Vero cells as these cells release a trypsin inhibitor (Kaverin and Webster 1995; Romanova et al. 2003). For MDCK cells, the release of a trypsin inhibitor has also been described recently (Nishiyama et al. 2008). However, influenza virus replication has not been found to be as sensitive to trypsin concentration as in Vero cells, and trypsin is usually added at time of infection only (Tree et al. 2001; Genzel et al. 2004, 2006a, b; Schwartz et al. 2006). Finally, virus adaptation to the producer cell line has often been described for the adaptation from eggs to cells (Lin et al. 1997; Genzel and Reichl 2007; Govorkova et al. 1996). Up to 20 passages were needed to obtain acceptable HA virus titers. However, more typically only three to four passages are needed.
Nevertheless, some viruses were not able to proliferate in some host cell lines (Tobita et al. 1997). Stability of the produced virus as well as influence of multiplicity of infection (moi), trypsin addition and adaptation can result in varying HA titers over the infection time (Rimmelzwaan et al. 1998; Wressnigg et al. 2009; Audsley and Tannock 2005; Youil et al. 2004). Harvest time point might therefore be crucial for the virus titer, but also screening for high titer cannot be done without time profiles of infection.
In the following, we present a thorough analysis of the impact of infection parameters (trypsin concentration and cultivation medium) on human influenza virus production in adherent Vero and MDCK cells. Virus seed was adapted to both cell lines. Differences in virus propagation and data for stability of virus were analyzed for various human influenza strains for small-scale cultivation systems (roller bottle, T-flask) in two media. In addition, HA N-glycan analysis was performed to asses cell-specific differences in HA N-glycosylation patterns. Finally, cultivation conditions resulting in similar virus yields for both cell lines in small-scale cultivations were evaluated in a stirred tank and a wave bioreactor using two different media.
Materials and methods
Cell lines
Adherent MDCK cells (ECACC 84121903) and adherent Vero cells (WHO seed; ECACC 88020401) were either cultivated under serum-containing conditions (GMEM # 22100-093, Gibco) as described by Genzel et al.2004) (referred as SC in the following) or under serum-free conditions (Episerf #10732-022, Gibco) supplemented with glucose (20-mM final concentration, Sigma #G8270), glutamine (2-mM final concentration, Sigma #G3126) and pyruvate (2-mM final concentration, Sigma #P8574) at 37 °C (Genzel et al. 2005). This medium is referred to as SF medium in the following. Cells were cultivated in SF medium at least for three passages before an experiment was done.
Virus
As virus production requires serum-free medium for maximum trypsin activity, cells cultivated in SC medium were washed with phospate-buffered saline (PBS) prior to infection to remove the serum and the medium was changed to ΔSC medium (medium based on SC medium but without serum addition). Cells cultivated in SF medium were directly infected. For all conditions, low levels of porcine trypsin were added to facilitate infection of cells at 37 °C. Therefore, a sterile-filtered stock solution of trypsin (Gibco, #27250-018) was prepared in PBS with 500 units/ml according to the activity given by the supplier. Virus production was followed by titration of viral HA or by tissue culture infectious dose (TCID50) (Govorkova et al. 1995; Genzel et al. 2006a, b). Virus seed from supplier (egg-adapted) was passaged at least three times in MDCK cells to produce a master and working virus seed (from ΔSC medium production, referred to as MDCK cell-derived virus seed) and stored in aliquots at −70 °C. Virus adaptation for production in Vero cells was done by passaging MDCK cell-derived virus seed four times in Vero cells (T-flasks) (Govorkova et al. 1995). MDCK cell-derived virus seeds (abbreviated with ma for MDCK adapted) used were: human influenza A/PR/8/34 H1N1ma supplied by National Institute for Biological Standards and Control (NIBSC code: 06/114) (1.3 × 107 TCID50/ml) and Robert Koch Institute (RKI code: Amp. 3138) (4.0 × 107 TCID50/ml), human influenza A/Wisconsin/67/2005 HGR H3N2ma (NIBSC; NIBSC code: 06/112, prepared from NYMC X-161B, a reassortant with A/PR/8/34 M and NP genes) (3.1 × 107 TCID50/ml), human influenza B/Malaysia/2506/2004ma (NIBSC; NIBSC code: 06/104) (1.3 × 106 TCID50/ml). Vero cell-derived virus seeds (abbreviated with va for Vero adapted) used were: human influenza A/PR/8/34 H1N1va adapted from NIBSC-MDCK cell-derived virus seed (5.6 × 107 TCID50/ml) and from RKI-MDCK cell-derived virus seed (1.1 × 107 TCID50/ml) and human influenza A/Wisconsin/67/2005 H3N2va adapted from MDCK cell-derived virus seed (1.4 × 107 TCID50/ml).
Roller-bottle and T-flasks experiments
Roller-bottle cultures (RBs) (Greiner, 850 cm2) were inoculated with approximately 2.5 × 107 cells (2.9 × 104 cells/cm2) and grown for four days in 250 ml SC or SF medium (caps tightly closed). When fully confluent (1.0–1.4 × 108 cells or 1.2–1.6 × 105 cells/cm2), the cells were infected with moi of 0.001–1 (based on the TCID50 of the virus seed). When grown in SC medium the bottles were washed three times with PBS (without Ca2+/Mg2+) before addition of ΔSC medium. For SF medium, infection was done without washing steps and medium exchange (preliminary experiments showed that the medium sufficiently supported cell growth and virus replication as seen for a different SF medium (Genzel et al. 2006a, b)). T-flask (T75; Greiner, 75 cm2) experiments were carried out accordingly.
Microcarrier culture in 5 L stirred tank bioreactor
Conditions used for the 5 L stirred tank bioreactor were as described by Genzel et al. (2004). The microcarriers (Cytodex 1, GE Healthcare) were added to the medium in a final concentration of 2 g/L (start cell concentration: 2–3 × 105 cells/ml). After four days cultivation the cells were grown confluent. Then human influenza A/Wisconsin/67/2005 HGR H3N2 virus seed and trypsin solution were added. The moi was adjusted to 0.001 based on the TCID50 of the virus seed (details see Table 1) as preliminary experiments had shown that a lower moi resulted in slightly higher HA titers (data not shown). At the end of the virus production step (indicated by an increase of pO2 and empty microcarriers), the cultivation was stopped.
Table 1.
Overview on cultivation conditions for stirred tank and wave bioreactor
| MCS | VCS | MFW | VFW | |
|---|---|---|---|---|
| Reactor | STR | STR | wave | wave |
| Cells | MDCK | Vero | MDCK | Vero |
| Medium | SC/ΔSC | SC/ΔSC | SF | SF |
| Medium lot # | – | – | 371739 | 371739 |
| Gluc (mM) | 28 | 28 | 22 | 22 |
| Gln (mM) | 2 | 2 | 2 | 2 |
| Pyr (mM) | – | – | 2 | 2 |
| Inoculation (× 105 cells/ml) | 2.0 | 3.1 | 2.0 | 2.0 |
| Cell concentration toi (× 106 cells/ml) | 2.0 | 1.6 | 1.2 | 1.0 |
| Trypsin (× 10−6 U/cell) | 10 | 3 | 8.1 | 8.0 |
| Moi | 0.001 | 0.001 | 0.018 | 0.018 |
| Virusa | Wisma | Wisva | A/PR/8-RKIma | A/PR/8-RKIva |
| Max HA | 3.0 | 2.6 | 3.3 | 2.3 |
| Productivity (virions/cell)b | 10,000 | 4,976 | 33,255 | 3,990 |
a Wis, human influenza A/Wisconsin/67/2005 HGR H3N2, and A/PR/8-RKI, human influenza A/PR/8/34 H1N1, supplied by Robert Koch Institute (RKI); ma, MDCK cell-derived virus seed; va, Vero cell-derived virus seed
b Calculated from maximum HA value and cell concentration at toi—HA was converted to virions/ml, assuming the binding of one virus particle per red blood cell with a given concentration of red blood cells of 2 × 107/ml (an HA of 3.0 corresponds to a dilution of 1:1,000; productivity is then: 2 × 107 virions/ml × 1,000 divided by 2 × 106 cells/ml, which equals to 10,000 virions/cell)
Serum-free microcarrier culture in 2 L wave bioreactor
Roller-bottle precultures in SF medium were cultivated as described by Genzel et al. (2006a, b). When fully confluent (1.0–1.4 × 108 cells or 1.2–1.6 × 105 cells/cm2) the cells were washed three times with PBS (3 × 50 ml, without Ca2+/Mg2+) and detached by exposure to trypsin/EDTA solution (10 ml, 2 min, 0.05 g/L trypsin (Gibco); 0.02 g/L EDTA (Merck)). After incubation at 37 °C (MDCK cells: 15 min; Vero cells: 5 min) the reaction was stopped with 20 ml SF medium. The cell suspension was then used to inoculate a 2 L wave bioreactor cellbag (1-L working volume).
Conditions used for the 2 L wave bioreactor were as described by Genzel et al. (2006a, b). For cultivation in a 2 L wave bioreactor (Wave Biotech AG) SF medium (1 L), microcarriers (prepared as described in (Genzel et al. 2006a, b)) (2 g/L) and cells (start cell concentration: 2 × 105 cells/ml) were added to a feed bottle and transferred into the cellbag (CB2L, Wave Biotech AG, LDPE material) (details see Table 1). The reactor was set to a temperature of 37 °C, a platform angle of 7°, a rocking rate of 15 rocks/min and aeration with CO2 mixed with air at 0.1 NL/min (Genzel et al. 2006a, b). The CO2 concentration was set manually as shown in Fig. 6e. For infection, virus (human influenza A/PR/8/34 H1N1 (RKI)) and trypsin were added to 250 ml of fresh SF medium. Afterwards, this medium was transferred into the cellbag when cell concentrations higher than 1 × 106 cells/ml were reached (moi = 0.018) (removal of 250-ml spent medium before infection). Neither pH nor pO2 were controlled.
Fig. 6.
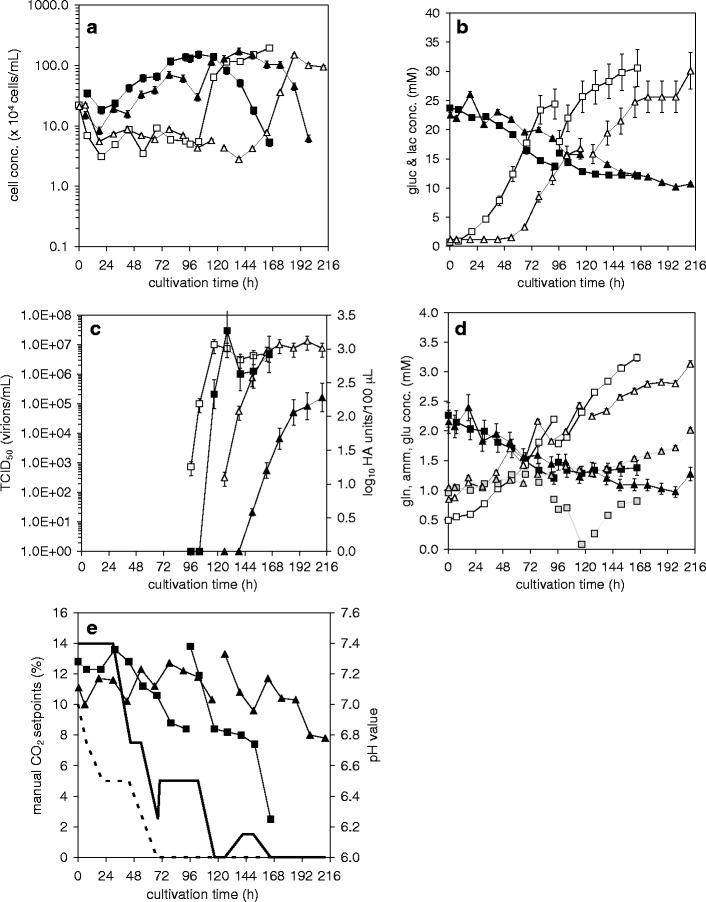
Release and uptake of metabolites, cell concentrations, and virus titers during growth of MDCK and Vero cells on Cytodex 1 microcarriers (2 g/L) in SF medium in a 2 L wave bioreactor (0–96 h and 0–126 h respectively) and subsequent direct influenza A/PR/8/34 (H1N1) (RKI) virus production (cell line adapted viruses) (96–168 h and 126–212 h) (for details see Table 1): MDCK cells (squares) compared to Vero cells (triangles). a cell concentrations: cells on MC (full symbols), cells in suspension (empty symbols); b glucose (full symbols), lactate (empty symbols); c virus titer in log HA units/100 μL (full symbols) and infectious virus titer (TCID50) (empty symbols); d glutamine (full symbols), ammonia (empty symbols), glutamate (grey symbols); e pH value (full symbols) and manual CO2 setpoints (line, Vero; dotted line, MDCK). Relative standard deviations were as given previously ((Lohr et al. 2009)); read-out for the more sensitive HA assay: log10 HA units/100 μL with a relative standard deviation of 9.3%
Sampling and analytics
Sample preparation and storage as well as determination of cell concentration, cell viability, metabolites, and virus titers was carried out as described previously (Genzel and Reichl 2007; Genzel et al. 2004, 2005; Mahy and Kangro 1996). Errors of the different validated assays were as described earlier. For small-scale experiments, given HA titers are expressed as log to the base of 10 with dilution errors of HA: ±0.3 log HA units/100 μL (assay with visual read-out, chicken-erythrocyte solution was set to 2 × 107 cells/ml). For bioreactor cultivations, a more sensitive HA assay was used with a well to well dilution of 1:20.5, and a read-out using a plate reader at 700 nm evaluated after Boltzmann sigmoid fitting (read-out: log10 HA units/100 μL with a relative standard deviation of the method of 9.3%) (Kalbfuss et al. 2008). For TCID50 determination the dilution error was ±0.3 log (Genzel and Reichl 2007). Samples from the wave bioreactor cellbags (6–7 ml) were taken with a syringe through a Luer-Lock-septum in a 12-h interval during cell growth and virus production. Determination of cell concentrations on microcarriers was carried out as described by Genzel and Reichl (2007).
N-Glycan analysis was done according to Schwarzer et al. (2009, 2008). Virus harvest was purified and concentrated by consecutive stepwise g-force gradient centrifugation. Re-dissolved pelleted virus proteins were separated by SDS-PAGE and in-gel-deglycosylation was performed according to Kuster et al. (1997). N-Glycans were extracted from gel pieces and completely dried (Integrated SpeedVac System ISS100 Savant). Subsequently, glycans were fluorescently labeled with 8-amino-1,3,6-pyrenetrisulfonic acid (APTS) (>96.0%, Sigma-Aldrich) by reductive amination (Callewaert et al. 2001). APTS-conjugated N-glycans were then desalted in MultiScreen Deep Well Solvinert Filter Plates (Millipore #MDRL N04) packed with size exclusion chromatography (SEC) material. Separation of the conjugated N-glycans from surplus dye was done with modified methacrylate polymer based Toyopearl HW-40F. N-glycan-containing fractions were identified by scanning with a Typhoon TRIO Variable Mode Imager (GE Healthcare). Capillary gel electrophoresis-laser-induced fluorescence (CGE-LIF) was performed according to Callewaert et al. (2004) on an Applied Biosystems ABI PRISM 3100-Avant genetic analyzer equipped with a 3100-Avant Genetic Analyzer Capillary Array and an effective capillary length of 50 cm with undiluted POP-6 polymer (Applied Biosystems #4316357) as separation matrix. The obtained N-glycan fingerprints were then normalized against an internal standard using the suppliers’ software package.
Results
Options for cell culture-derived influenza vaccine manufacturing using adherent MDCK cells growing on microcarriers were shown in different cultivation systems (Genzel et al. 2004, 2006a, b; Genzel and Reichl 2007) under serum-containing and serum-free conditions. Monitoring of specific analytical data for this process allowed a thorough evaluation of the corresponding production systems (Schwarzer et al. 2009; Vester et al. 2009; Schulze-Horsel et al. 2009; Kalbfuss et al. 2007). As an alternative substrate, an adherent WHO-certified Vero cell line was used (Kaverin and Webster 1995). As a starting point, screening experiments for inoculation conditions, cell growth, and virus production of MDCK and Vero cells were carried out in this study. Comparable conditions were identified for GMEM as SC/ΔSC medium and Episerf as SF medium, leading to acceptable attachment of cells on microcarriers and similar specific cell-growth rates. Preliminary experiments on the influence of moi for Vero cell infections had shown similar results as for MDCK cells. Slightly better virus titers were obtained at lower moi for influenza A Wisconsin/67/2005 HGR H3N2, but for other tested viruses, no influence of moi on virus titer could be seen. The moi had only an effect on the time to reach maximum HA titer. The higher the moi was, the faster virus replication reached maximum HA titer (data not shown). The infection parameters for influenza production in Vero cells (trypsin concentration and virus adaptation) shown in this study, were critical for the following comparison of MDCK against Vero cells in lab-scale cultures (5 L stirred tank and 2 L wave bioreactor). SC/ΔSC and SF medium cultivations were carried out in both reactor types. As an example for stirred tank cultivation, data for SC/ΔSC medium are presented and wave cultivation data represent the results in SF medium (Table 1) (data represent typical cultivations, similar trends have been obtained from further cultivations under the same cultivation conditions, but with other virus strains (adapted and non-adapted)). In the following, the four different cultivations are abbreviated as follows: MDCK in SC/ΔSC medium in stirred tank bioreactor: MCS; Vero in SC/ΔSC medium in stirred tank bioreactor: VCS; MDCK in SF medium in wave bioreactor: MFW; Vero in SF medium in wave bioreactor: VFW.
MDCK cell-derived virus seed adaptation to Vero cells
Preliminary experiments for virus production in Vero cells using influenza virus seed produced in MDCK cells resulted in relatively low HA titers. Furthermore, it took a comparatively long time to reach maximum HA titers. However, after adaptation over at least three passages in Vero cells for both influenza A/PR/8/34 variants and influenza A/Wisconsin/67/2005 HGR, significant improvement was obtained (Figs. 1 and 2). Surprisingly, influenza B/Malaysia/2506/2004 could not be adapted to replication in Vero cells (HA titer below detection limit) even when different adaptation strategies were applied (e.g., changes in temperature from 37 to 33 °C or fast or slow passaging) in ΔSC and SF medium.
Fig. 1.
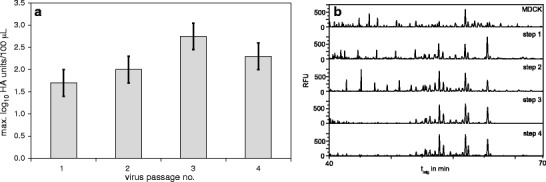
Virus seed adaptation to Vero cells. a Maximum HA titer of each passage during adaptation of human influenza A/PR/8/34 H1N1 supplied by NIBSC from a MDCK cell-derived virus seed to Vero cells in ΔSC medium (dilution error of HA, ±0.3 log10 HA units/100 μL). b Capillary gel electrophoresis—laser-induced fluorescence (CGE-LIF) N-glycan fingerprint of the HA protein fraction from the MDCK cell-derived virus seed (human influenza A/PR/8/34 H1N1ma supplied by NIBSC) and of each adaptation step in Vero cells (Schwarzer et al. 2009) (RFU: relative fluorescence units over migration time (t mig))
Fig. 2.
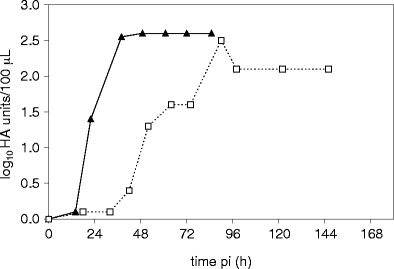
HA titer during infection of Vero cells with MDCK cell-derived virus seed (influenza A/PR/8/34 H1N1ma (NIBSC)) (white square) and adapted Vero cell-derived virus seed from passage four (influenza A/PR/8/34 H1N1va (NIBSC)) (black triangle) (dilution error for the HA assay: ± 0.3 log10 HA units/100 μL)
N-glycan fingerprints of the HA protein fraction from influenza A/PR/8/34 (NIBSC) were monitored during this adaptation process. As expected, N-glycan fingerprints of the produced HA proteins were different for MDCK and Vero cells. After the first adaptation step, the N-glycan fingerprint changed (Fig. 1), and stabilized after adaptation step three indicating that the three steps were sufficient for successful matching this virus to the new host cell. Three adaptation steps were also enough to obtain similar virus yields and infection time profiles in Vero cells compared to MDCK cells.
Influence of trypsin
Vero as well as MDCK cells have been reported to release protease inhibitors that have an effect on the activity of trypsin during infection (Kaverin and Webster 1995; Nishiyama et al. 2008). For the development of production processes using serum-free media, these inhibitors could have a negative effect on virus production when washing and medium exchange steps are omitted before infection. While virus replication in MDCK cells seems to be only marginally effected by protease inhibitors, multiple trypsin additions have been described for Vero cells to overcome trypsin inactivation (Govorkova et al. 1996).
When evaluating different trypsin concentrations added to Vero cells in ΔSC medium in RBs, no effects on time courses of virus replication and maximum HA titers could be seen for the tested concentrations (Fig. 3a). However, in SF medium without washing, an activity of 5.0 × 10−5 U/cell was needed for fast progress of infection to high HA titers (Fig. 3b). An additional washing step before infection (as routinely required in SC/ΔSC medium cultivations) could additionally reduce the time needed for reaching maximum HA titers. Interestingly, multiple trypsin additions did not result in increased HA titers and faster virus replication.
Fig. 3.
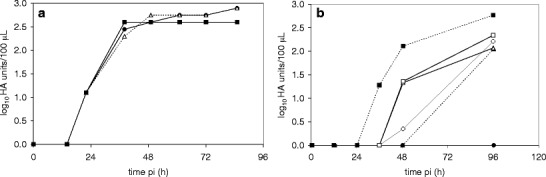
Influence of trypsin activity on influenza virus titer produced in Vero cells. a HA titer for Vero cells in ΔSC medium (RBs) infected with Vero cell-derived virus seed (influenza A/PR/8/34 H1N1va (NIBSC), moi: 0.01) with different trypsin concentrations (washing steps and medium exchange before infection): (black square) 2.0 × 10−5 U/cell, (white triangle) 1.5 × 10−5 U/cell, (black circle) 1.0 × 10−5 U/cell. b HA titer for Vero cells in SF medium (T75s) infected with Vero cell-derived virus seed (influenza A/PR/8/34 H1N1va (RKI), moi: 0.01) with different trypsin concentrations and addition strategies: (black square) 5 × 10−5 U/cell with medium exchange before infection, (white square) 5 × 10−5 U/cell, (black triangle) 3 × 10−5 U/cell, (white triangle) 3 × 10−5 U/cell plus addition of 1 × 10−5 U/cell after 12 and 24 h, (black circle) 1 × 10−5 U/cell with medium exchange before infection, (white circle) 1 × 10−5 U/cell plus addition of 1 × 10−5 U/cell after 12 and 24 h (dilution error for the HA assay: ±0.3 log10 HA units/100 μL)
In microcarrier systems however (STR and wave bioreactor), additional shear forces led to fast cell detachment at these trypsin concentrations and therefore a reduced concentration of 3–10 × 10−6 U/cell had to be used to avoid early cell detachment (see Table 1). Therefore, trypsin addition had to be adapted individually to media (even to different lots) and cultivation systems used for Vero cell infections.
Virus stability at 37 °C
For the newly adapted viruses the time point of harvest to obtain high TCID50, titers had to be tested for the different viruses for virus seed stock preparation. As the stability of virus can clearly influence the virus titer, the stability of two virus variants from human influenza A/PR/8/34 H1N1 (supplied by NIBSC and RKI), produced either in MDCK cells or in Vero cells, was tested. Therefore, virus was incubated in ΔSC medium at 37 °C for 11 days in RBs. From earlier experiments in MDCK cells, it was known that these two variants resulted in different HA and TCID50 titers (Schulze-Horsel et al. 2009).
When following the stability of equine influenza in MDCK cells under similar conditions in RBs, it was seen that the maximum HA titer was relatively constant whereas the TCID50 decreased over time, and after 150 h, no infectious virus could be detected (Genzel et al. 2006a, b). For the two different variants from human influenza A/PR/8/34, surprisingly, only for the NIBSC variant produced in MDCK cells, a similar decrease in the TCID50 was found. After 184 h no more infectious virus could be detected (Fig. 4). For the RKI variant produced in MDCK and Vero cells and for the NIBSC variant produced in Vero cells, clearly, higher stability could be seen. A titer of approximately 104.5 TCID50/ml remained after 11 days. Maximum peak titers were found at 18 h post infection (pi) for TCID50 (for the RKI variant produced in Vero cells: 61 h pi) and between 40–97 h pi for HA (Table 2). As described earlier, for both RKI variants higher TCID50 titers were found (107.9 TCID50/ml) compared to the NIBSC variants (Schulze-Horsel et al. 2009). Surprisingly, the highest TCID50 profile (RKI variant in Vero cells) resulted in a comparatively low HA profile (maximum 2.7 log HA units/100 μL).
Fig. 4.
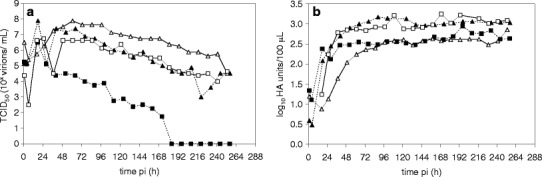
Virus titer during infection of MDCK (RBs) with MDCK cell-derived virus seed (ΔSC medium, moi:1): influenza A/PR/8/34 H1N1ma (NIBSC) (black square), influenza A/PR/8/34 H1N1ma (RKI) (black triangle) and Vero cells (RBs) with Vero cell-derived virus seed (ΔSC medium, moi:1): influenza A/PR/8/34 H1N1va (NIBSC) (white square), influenza A/PR/8/34 H1N1va (RKI) (white triangle). a TCID50, b HA titer (dilution error for the HA assay: ±0.3 log10 HA units/100 μL)
Table 2.
Overview on virus titers and virus stability of different viruses produced in MDCK or Vero cells in ΔSC and SF medium in small-scale cultivations
| Virus | Cells | Medium | moi | max HA value/ time pi a | max TCID50 value/ time pi b | t c1/2 | k dvi |
|---|---|---|---|---|---|---|---|
| A/PR/8/34 H1N1ma (RKI) | MDCK | ΔSC | 1 | 3.2/89 | 7.6 × 107/18 | 19.9 | −0.035 |
| A/PR/8/34 H1N1va (RKI) | Vero | ΔSC | 1 | 2.6/121 | 7.6 × 107/61 | 21.3 | −0.033 |
| A/PR/8/34 H1N1ma (RKI) | MDCK | SF | 0.001 | 2.9/91 | 4.3 × 107/67 | 18.8 | −0.037 |
| A/PR/8/34 H1N1va (RKI) | Vero | SF | 0.001 | 2.7/153 | 1.8 × 108/118 | 52.5 | −0.013 |
| A/PR/8/34 H1N1ma (NIBSC) | MDCK | ΔSC | 1 | 2.6/64 | 3.2 × 106/18 | 12.1 | −0.058 |
| A/PR/8/34 H1N1va (NIBSC) | Vero | ΔSC | 1 | 2.9/61 | 5.6 × 106/26 | 29.1 | −0.024 |
| A/Wis/67/2005 HGR H3N2ma | MDCK | SF | 0.001 | 2.7/67 | 5.6 × 107/67 | 62.4 | −0.011 |
| A/Wis/67/2005 HGR H3N2va | Vero | SF | 0.001 | 3.1/153 | 1.3 × 108/118 | 111.8 | −0.006 |
| B/Mal/2506/2004ma | MDCK | SF | 0.001 | 2.8/91 | 1.8 × 108/43 | 9.6 | −0.072 |
a Maximal HA value reached (log HA units/100 μL) and time point pi for reaching maximal HA (h)
b Maximal TCID50 value reached (virions/ml) and time point pi for reaching maximal TCID50 value (h)
c Half-life time for inactivation of virus at 37 °C in roller bottles in the respective medium (h) (t1/2 = ln2/kvi)
d Specific inactivation rate kvi of virus at 37 °C in roller bottles in the respective medium (h−1) determined from the slope of ln(TCID50) over time during exponential decrease
Finally, virus titers and virus stability were analyzed, for other virus strains were produced with lower moi (moi: 0.001) in MDCK and Vero cells in SF medium. Again, relatively constant HA titers were obtained once maximum HA titer was reached (data not shown). In SF medium, but also in ΔSC medium, MDCK cell-derived virions seemed to be less stable than Vero cell-derived virions (Table 2). Lowest virus stability (k vi = −0.0721 h−1) was found for influenza B/Malaysia/2506/2004ma in SF medium produced with MDCK cells. However, maximum HA titer as well as TCID50 was high. Production of the same virus strain in ΔSC medium in a 5 L STR as well as in static culture showed good HA titers (2.5 log HA units/100 μL), but TCID50 values of only 7.9 × 103 TCID50/ml were reached (data not shown). This indicates poor virus stability.
In SF medium, maximum TCID50 was clearly higher for Vero cell-derived compared to MDCK cell-derived cultivations. An interpretation of the different time points pi when maximal HA titer or TCID50 was obtained is difficult for two reasons: first, a lower moi was used in SF medium, and second, different trypsin concentrations were used for Vero and MDCK cells. Both factors could influence virus replication dynamics. In general, maximum HA titer was typically 30 to 70 h later than maximum TCID50. Only for A/Wisconsin/67/2005ma HGR H3N2 produced in MDCK cells no delay between maximum HA titer and TCID50 could be observed.
Stirred tank cultivation in SC/ΔSC medium
Virus production of influenza A/Wisconsin/67/2005 HGR H3N2 with the MDCK cell-derived seed and the Vero cell-derived seed, respectively, was carried out by cultivating both cell lines to confluency in a 5 L stirred tank bioreactor in SC medium on 2 g/L microcarriers (Cytodex 1) (MCS and VCS, see Table 1). Both cell lines reached 1.0–2.0 × 106 cells/ml after 4 days cultivation from 2.0–3.1 × 105 cells/ml (Fig. 5(b)). Glutamine uptake and ammonia release during cell growth was very similar for both cell lines (detailed MDCK data see (Genzel et al. 2004)), whereas glucose uptake and lactate release was clearly lower for Vero cells (16.9 mM compared to 41.7 mM lactate release) (Fig. 5(d)). While MDCK cells took up glutamate completely at the end of the cell growth phase, Vero cells constantly released glutamate into the medium (up to a concentration of 1.0 mM).
Fig 5.
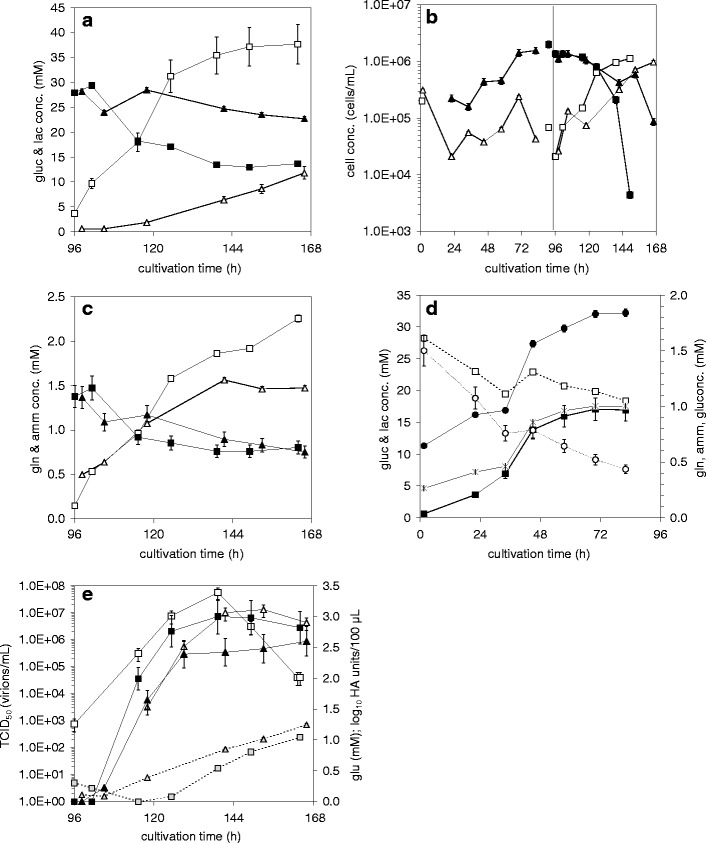
Release and uptake of metabolites, cell concentrations, and virus titers during growth of MDCK and Vero cells on Cytodex 1 microcarriers (2 g/L) in SC/ΔSC medium in a 5 L STR (0–96 h) and subsequent influenza A/Wisconsin/67/2005 HGR H3N2 virus production (cell line adapted viruses) (96–168 h) with washing steps and medium exchange (ΔSC medium) (details see Table 1): MDCK cells (squares) compared to Vero cells (triangles). a Glucose (full symbols), lactate (empty symbols), b cell concentrations: cells on MC (full symbols), cells in suspension (empty symbols), c glutamine (full symbols), ammonia (empty symbols); d metabolite profiles for cell growth of Vero cells only (data for MDCK cells were as in (Genzel et al. 2004): glucose (white square), lactate (black square), glutamine (white circle), ammonia (black circle), glutamate (✱); e) glutamate (grey symbols), virus titer in log10 HA units/100 μL (full symbols), infectious virus titer (TCID50) (empty symbols). Relative standard deviations were as given previously ((Lohr et al. 2009)); read-out for the more sensitive HA assay: log10 HA units/100 μL with a relative standard deviation of 9.3%
During the subsequent virus production phase, Vero cells showed again a lower glucose uptake and lactate release, but a similar glutamine uptake and ammonia release (Fig. 5a, c). The produced HA titer was lower for Vero cells with 2.6 against 3.0 log10 HA units/100 μL for MDCK cells (Fig. 5e). Also, the corresponding TCID50 titer was lower for Vero cells with 1.3 × 107 against 5.6 × 107 infectious virions/ml. This resulted in cell-specific productivities of 10,000 virions/cell against 4,976 virions/cell. Overall, the virus production in the first 24 h was slower in Vero cells than in MDCK cells. However, virus stability seemed to be higher in the Vero production process. For both cell lines, a glutamate release of more than 1 mM could be seen. For Vero cells the time point of increase seemed to correlate to the HA titer. Glutamate release in the MDCK cell process started clearly later ( at 115 h).
Wave cultivation in SF medium
To compare, on one hand, cell line performance in SF medium without a medium exchange before virus infection and, on the other hand, cultivation in a wave bioreactor, serum-free adapted cells (starting concentration: 2 × 105 cells/ml) were grown to confluency on 2 g/L Cytodex 1 microcarriers (1.2 × 106 cells/ml for MDCK; 1.0 × 106 cells/ml for Vero ) (Fig. 6(a)). Vero cells however required about 23 h more to reach the respective cell concentration. Subsequently, the cells of both cellbags were directly infected with the corresponding virus seed (A/PR/8-RKIma and A/PR/8-RKIva). The increase in suspension cells and the decrease in the concentration of cells on microcarriers was clearly faster for MDCK cells after infection. In both cases, the final concentration of suspension cells after detachment corresponded to the concentration of adherent cells at time of infection, i.e., cell lysis rate seemed to be low.
Comparing the required CO2 additions together with the pH profiles, glucose uptake and lactate release (Fig. 6b, e) showed a start of pH decrease for MDCK cells parallel to the lactate release resulting in a possible decrease in CO2 addition. For Vero cells however, clearly, higher CO2 additions were needed at the beginning of the cultivation and pH was more stable compared to the cultivation of MDCK cells. Lactate release started only after 48 h. But this lactate release did not have a similar effect on the pH as for the MDCK cultivation.
For both cell lines, the molar yield coefficients Y lac/gluc were lower during the cell growth phase (MDCK: 2.4; Vero: 1.6) compared to the virus replication phase (MDCK: 3.2; Vero: 2.8). Vero cells showed, during both phases, lower coefficients as MDCK cells. Glutamine uptake and ammonia release for both cells was similar to the stirred tank reactor (Fig. 6d). As in SC/ΔSC medium, Vero cells released glutamate, whereas MDCK cells showed a first slight glutamate release followed by a strong uptake. Again, both cell lines released glutamate during virus production.
Virus production was clearly slower and lower in Vero cells for the total virus yield (HA) (MDCK: 33,255 virions/cell; Vero: 3990 virions/cell). However, for infectious viral particles produced, similar data were obtained for both cell lines (TCID50 1.2 × 107 virions/ml, Fig. 6c). When calculating the ratio of infectious to total virions, the Vero cultivation resulted in 3 × 10−3, whereas the MDCK cultivation led to 3 × 10−4 (corresponding to experimental data obtained for the roller bottle cultivations (see Table 2)). Delay between increase in TCID50 and HA titer was clearly bigger for Vero cells than for MDCK cells. For both cell lines increase in TCID50 was before HA titer increase.
Discussion
Virus seed adaptation
Virus production in Vero cells was significantly improved with respect to maximum HA titer as well as the time period to reach maximum HA titer by adapting MDCK cell-derived virus seed to Vero cells. N-Glycan fingerprints and maximum HA titers for each virus passage indicated that three to four passages were sufficient for the adaptation process. However, success of the adaptation seems to depend strongly on virus strains as influenza B/Malaysia/2506/2004, as well as B/Florida/4/2006 (NIBSC code: 08/138) and A/WSN/33 H1N1 (kind gift by G. Kochs, University Freiburg, Germany) could not be adapted to replication in Vero cells (data not shown).
For other viruses such as plum virus or rinderpest virus, such adaptations were also described, resulting typically in an improved in vitro virus production by passaging virus isolates (Wallis et al. 2007; Ishii et al. 1986). Govorkova et al. described a 26-fold increase in infectious virus titer after 20 passages at limiting dilution of influenza A/England/1/53 in Vero cells (Govorkova et al. 1995). Here, RNA sequence analysis for the HA1 protein showed no changes due to the adaptation process, thus indicating that there is no selection of viruses with antigenic changes in the HA. Additionally, RNA sequence analysis of cold-adapted live-attenuated influenza viruses used as vaccine in children showed that mutations related to attenuation did not change 8 days after vaccination (Klimov et al. 1995). Similar improvements in virus yield as reported by Govorkova et al. (1995) were also described for MDCK cells comparing titers from passage two and five for different viral samples of H1N1 and H3N2 virus subtypes (Tsai and Trusheim 2008).
The exact causes for the changes in the HA N-glycan fingerprints during this adaptation process are not yet clear. Previous findings comparing N-glycan fingerprints from different viruses grown in various cell lines showed specific HA N-glycan fingerprints for each virus–host cell line combination (Schwarzer et al. 2008, 2009). This is in agreement with the differences in HA fingerprints from MDCK and Vero cell-derived A/PR/8/34 H1N1 virus observed in this study. Changes in glycosylation between influenza B viruses grown in eggs or MDCK cells were described in detail by Chen et al. using Western blotting and sequence analysis of the HA1 protein (Chen et al. 2008). For influenza A H3N2 the glycosylation patterns resulting from production in Vero or MDCK cells were compared by Romanova et al. by treatment with glycosidases and Western blotting (Romanova et al. 2003). It was shown that H3N2—but not H1N1—viruses adapted to Vero cells could no longer infect chicken eggs or agglutinate chicken erythrocytes. However, in our study, no problem with HA determination was noticed for samples from the VCS cultivation using A/Wisconsin/67/2005 HGR H3N2. The authors concluded that changes in HA N-glycosylation are most likely linked to changes in the amino acid sequence. Changes of single amino acids were also found for the adaptation of coronaviruses to Vero cells at different temperatures (Shen et al. 2004). Overall, it seems that the interrelation between HA N-glycosylation pattern, RNA, and amino acid sequence has not yet been clearly understood and further studies in this respect should be performed. Based on our findings we assume that the changes in HA N-glycan fingerprints are mainly dependent on the host cell line.
In contrast to the difficulties in adapting influenza B viruses to Vero cells reported here, Wressnigg et al. described successful production of a B/Malaysia/2506/2004-like virus strain (B/Vienna/33/06) in SFM at 33 °C after adaptation (Wressnigg et al. 2009). As Vero cells from different origin (ATCC CCL-81) and different virus strains were used, no clear explanations for this discrepancy to the results reported here can be given. Also, a decrease in incubation temperature to 33 °C did not improve virus yields for the tested viruses described. Studies by Tobita et al. and Nakamura and Homma discuss that Vero cells seem to generally produce influenza B viruses only, poorly, compared to MDCK cells (Nakamura and Homma 1981; Tobita et al. 1997). However, no virus adaptation of the virus seed to Vero cells was described.
In total, various changes can result from passaging of viruses in different cell lines indicating a need for considering specific aspects of virus–host cell interaction (virus dynamics, maximum titers, glycosylation) for process optimization as well as for assessment of antigen quality of vaccine candidates.
Influence of trypsin
Compared to MDCK cells, successful virus production in Vero cells not only depends on virus adaptation but also on the strategy of trypsin addition. MDCK cells, but in particular Vero cells, release protease inhibitors into the cell culture medium affecting the activity of trypsin needed for virus infection (Kaverin and Webster 1995; Nishiyama et al. 2008). Washing and medium exchange steps before infection could clearly reduce the activity of trypsin inhibitors, potentially counteracting successful virus replication. However, for a vaccine production process, these steps mean additional medium costs and increased contamination risks. On the other hand, too high trypsin activities in the virus production medium could result in cell detachment. Therefore, as was seen in this study, optimizing trypsin activity is essential. In particular, batch to batch variations of trypsin, media, and differences in shear stress of cultivation systems need to be considered. Increase in trypsin activity resulted in an earlier increase in HA titer in SF medium, while too low concentrations prevented successful virus replication. In SC medium, however, all trypsin concentrations tested resulted in similar HA profiles. Multiple additions of trypsin as described by Govorkova et al. (1995, 1996), did not result in improvement of HA titers. As this method would complicate large-scale production processes, a single addition of trypsin at time of infection should be preferred.
Virus stability
When testing the virus stability of different MDCK and Vero cell-derived virus seeds, stable HA titers where obtained at least until 250 h pi at 37 °C. However, virus strains clearly differed in stability of TCID50. Especially MDCK cell-derived A/PR/8/34 H1N1ma (NIBSC) in ΔSC medium showed low stability. No infectious virions were detected 184 h pi, which was also seen for equine influenza in MDCK cells at 150 h pi (Genzel et al. 2006a, b). All other virus strains tested in this study showed better stabilities. In SF medium with lower moi A/Wisconsin/67/2005va HGR produced in Vero cells was most stable (Table 2, t 1/2 = 112 h). For different influenza B viruses in SFM at 33 °C (moi: 0.01), Wressnigg et al. showed TCID50 data up to 96 h pi (Wressnigg et al. 2009). Here, all virus strains had a stable titer for the time frame shown, once the maximum titer of about 107–108 virions/ml was reached. For influenza A/Neth/18/94 produced in MDCK cells Rimmelzwaan et al. also showed a decrease of TCID50 and a stable HA titer over a time period of 144 h (Rimmelzwaan et al. 1998). In contrast, Schwartz et al. described for the production of A/Vietnam/1203/2004 (H5N1) no decrease in TCID50 until 144 h pi (Schwartz et al. 2006). It thus seems that adaptation of the virus seed to the host cell line as well as the virus strain has an influence on virus stability and has to be characterized to determine the optimum harvest time especially for seed virus preparation or manufacturing of live-attenuated vaccines.
Cell growth of lab-scale cultivations
Vero cells have been used since many years for the production of viral vaccines. The spreading of Vero cells onto Cytodex 1 microcarriers (2 g/L) was observed in detail by Mendonça et al. (1999). For inoculation they found an optimum cells-to-microcarrier ratio of eight cells per microcarrier leading to 1.6 × 106 cells/ml after 7 days of cell growth in spinner flasks in SC medium. Cultivations in a 3.7 L stirred tank bioreactor resulted in 0.8 × 106 cells/ml after 9 days in batch, and 2.1 × 106 cells/ml after 12 days in perfusion mode (Mendonca and Pereira 1998). Cultivation of Vero cells on Cytodex 1 at 3 g/L was shown by Quesney et al. (2003). From a 4 × 105 cells/ml starting concentration, 1.8 × 106 cells/ml were obtained after 5 days. A study in spinner flasks showed that the optimal pH for attachment of Vero cells onto Cytodex 1 microcarriers was pH 7.1, and the use of serum-supported cell growth but not cell attachment (Ng et al. 1996). Furthermore, continuous stirring at 40 rpm during the attachment phase was favored compared to intermittent stirring. For production of Japanese encephalitis virus in Vero cells using 3 g/L Cytodex 1 in a spinner flask resulted in a maximum cell concentration of 1.8 × 106 cells/ml after 6 days in SF medium (Toriniwa and Komiya 2007). The production of rabies virus with Vero cells was shown in VPSFM medium using Cytodex 1 microcarriers (3 g/L) in a 2 L stirred tank bioreactor by Rourou et al. (2007). Here, a maximum cell concentration of 2.6 × 106 cells/ml was reached after 3 days. Finally, Merten et al. compared MDCK and Vero cell growth for influenza infection in the SF medium MDSS2N (Merten et al. 1999a, b). For Vero cells a maximum concentration of 3.1 × 106 cells/ml was obtained after 5 days (Cytodex 1 microcarriers, 2.5 g/L).
Experimental data obtained in this study in SC medium as well as in SF medium in both bioreactors (Cytodex 1 microcarriers, 2 g/L) are thus comparable to reported results for Vero cells. In particular, maximum cell concentrations of 1–2 × 106 cells/ml after 4 to 5 days with 2 g/L microcarriers were reached. Thus, the chosen process conditions resulted in similar cell growth for MDCK and Vero cells.
Virus production in lab-scale cultivations
From preliminary experiments in small-scale T-flasks and roller bottles, optimal conditions for the direct comparison of MDCK and Vero derived influenza vaccine production at a larger scale could be identified. At small-scale, Vero cells produced similar or even higher virus titers than MDCK cells. But also conditions for lower titers were possible. Especially, medium and virus strain had a strong influence on the possible titers. For the production of A/Wisconsin/67/2005 HGR in SF medium, Vero cells showed higher virus titers than MDCK cells (HA as well as TCID50) (Table 2). On the other hand, for the cultivation in 5 L stirred tank bioreactor in SC medium, Vero cells produced lower virus titers than MDCK cells. Nevertheless, titers above 2.6 log10 HA units/100 μL are very reasonable. For the production of two different H5N1 influenza viruses in 30 or 100 L scale bioreactors, Kistner et al. used the Vero ATCC CCL81 cell line and virus adapted over four passages (Kistner et al. 2007). With a moi of 0.001 and infection at 32 °C, HA titers of 2.7–3.0 log10 HA units/100 μL were obtained. Earlier, Kistner et al. showed for different virus strains a comparison between Vero-derived and egg-derived virus production. HA titers of 2.1–2.4 log10 HA units/100 μL were typically obtained for Vero cells and all cultivations shown from a 1,200 L bioreactor reached HA titers of 2.4 log10 HA units/100 μL (Kistner et al. 1998).
In this study, during virus replication phase in both bioreactors (STR and wave), the concentration of cells on microcarriers decreased faster for MDCK cells than for Vero cells. Correspondingly, the respective concentration of MDCK suspension cells increased earlier indicating faster induction of virus-induced apoptosis in these cells.
Metabolism in lab-scale cultivations
Vero and MDCK cells cultivated in both media and both reactor types showed a similar glutamine uptake and ammonia release, but a slightly lower glucose uptake and respective lactate release for Vero cells. A clear difference was noticed for glutamate. Vero cells released glutamate constantly, whereas MDCK cells took up glutamate completely towards the end of the cell growth phase. Mendonca et al. described the metabolism of Vero cells in Leibovitz medium supplemented with 5% FCS and 2 mM glutamine in a stirred tank bioreactor cultivation (Mendonca and Pereira 1998). Cell concentrations of 0.8 × 106 cells/ml were reached in batch mode after 8 days of cultivation. They observed an uptake of lactate when galactose was used as a carbon source. Glutamine uptake and ammonia release was similar but changes in glutamate concentrations were not reported. The effect of ammonia and glutamine/glutamate on growth of Vero cells in DMEM medium was analyzed in detail by Huang et al. (2006). During growth in medium without glutamine but containing glutamate and ammonia, ammonia was slightly taken up. No ammonia release was seen during growth in medium containing only glutamate or glutamate and asparagine. Only for growth in glutamine medium, ammonia release was found, which was similar to the release observed in this study. For the glutamate-containing media Huang et al. also described an uptake of glutamate similar to what was found for MDCK cells in SC medium in this study. This medium, however, did not contain glutamine. It seems that Vero cells use glutamate to produce glutamine via the enzyme glutamine synthetase. However, if glutamine is added to the medium, glutamine is taken up preferably while glutamine synthetase activity is reduced. In this case, even a release of glutamate is possible. Work is in progress to use specific enzyme actvity assays (Janke et al. 2010) to characterize cultivations in different media, ie., media which do not contain glutamine. Details on metabolism of VPSFM/William’s E medium in a stirred tank bioreactor performed under similar conditions as described in this study were also given by Quesney et al. (Quesney et al. 2003). Compared to our results, glutamine uptake was clearly higher in VPSFM/William’s E medium leading to a complete uptake after 4 days. Furthermore, also a release of glutamate was observed, which was explained by the authors as a transformation of glutamine to glutamate. Vero cells were also cultivated in VPSFM medium (4 mM gln) by Rourou et al. for rabies virus propagation (Rourou et al. 2007). Although lower glucose starting concentrations were used, higher lactate levels were reached compared to both lab-scale cultivations in this study. In contrast to glucose, higher glutamine starting concentrations were used by Rourou et al., but lower ammonia release was observed. No glutamate data was given.
Previous cultivation data for MDCK cells in a different serum-free medium in a wave bioreactor under similar conditions (Genzel et al. 2006a, b) and in SC/ΔSC medium in a stirred tank bioreactor (Genzel et al. 2004) showed very similar glucose uptake and lactate release as well as glutamine uptake and ammonia release. Only the uptake of glutamate in both reactors and media was different. In the previously described serum-free medium glutamate was slightly released (Genzel et al. 2006a, b). How glutamine metabolism and glutamate uptake or release is regulated in cells still needs further clarification. A possible approach would be a study monitoring intracellular metabolite concentrations and specific enzyme activities (Ritter et al. 2008) together with the concentration of extracellular substrates and growth inhibitors.
MDCK or Vero cells for influenza vaccine production?
On one hand, comprehensive experimental data on Vero cell lines used for production of polio and rabies vaccines but also influenza virus can be found. On the other hand MDCK cells have been shown to be equally good host cells for influenza vaccine manufacturing. However, as studies were typically performed using different media, cultivation vessels, cultivation conditions, and virus strains, a direct comparison of both host cell lines was difficult.
Data presented here now allow a better assessment of results obtained for influenza virus production in MDCK and Vero cells, emphasizing the influence of virus adaptation, trypsin addition, medium selection, and time of harvest. Similar virus titers can be obtained for Vero and MDCK cells when conditions are optimized in small-scale experiments. In bioreactor cultivations however, MDCK cells seem to be favorable. Additionally, not all virus strains seem to be equally well adaptable to the different cell lines. Furthermore, differences in resulting HA N-glycan fingerprints might be relevant for immunogenicity of vaccines. Therefore, having alternative host cells seems to be essential for worldwide vaccine production, especially in case of a pandemic.
Acknowledgment
The authors thank N. Wynserski, C. Best, S. König and I. Behrendt for their excellent technical assistance. The authors would like to express their gratitude to V. Lohr and J. Rödig for critical reading of this manuscript.
References
- Audsley JM, Tannock GA. The growth of attenuated influenza vaccine donor strains in continuous cell lines. J Virol Methods. 2005;123(2):187–193. doi: 10.1016/j.jviromet.2004.09.015. [DOI] [PubMed] [Google Scholar]
- Barrett PN, Mundt W, Kistner O, Howard MK. Vero cell platform in vaccine production: moving towards cell culture-based viral vaccines. Expert Rev Vaccines. 2009;8(5):607–618. doi: 10.1586/erv.09.19. [DOI] [PubMed] [Google Scholar]
- Brands R, Visser J, Medema J, Palache AM, van Scharrenburg GJM. Influvac: a safe Madin Darby Canine Kidney (MDCK) cell culture-based influenza vaccine. In: Brown F, Robertson JS, Schild GC, Wood JM, editors. Inactivated influenza vaccines prepared in cell culture. Basel: Karger; 1999. pp. 93–100. [PubMed] [Google Scholar]
- Callewaert N, Van Hecke A, Schollen E, Matthijs G, Contreras R. Carbohydrate electrophoresis on the DNA-sequencer: technology development and first applications. Glycobiology. 2001;11(10):922–923. [Google Scholar]
- Callewaert N, Van Vlierberghe H, Van Hecke A, Laroy W, Delanghe J, Contreras R. Noninvasive diagnosis of liver cirrhosis using DNA sequencer-based total serum protein glycomics. Nat Med. 2004;10(4):429–434. doi: 10.1038/nm1006. [DOI] [PubMed] [Google Scholar]
- Chen Z, Aspelund A, Jin H. Stabilizing the glycosylation pattern of influenza B hemagglutinin following adaptation to growth in eggs. Vaccine. 2008;26(3):361–371. doi: 10.1016/j.vaccine.2007.11.013. [DOI] [PubMed] [Google Scholar]
- Desmyter J, Melnick JL, Rawls WE. Defectiveness of interferon production and of rubella virus interference in a line of African green monkey kidney cells (Vero) J Virol. 1968;2(10):955–961. doi: 10.1128/jvi.2.10.955-961.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genzel Y, Reichl U. Vaccine production-state of the art and future needs in upstream processing. In: Pörtner R, editor. Methods in biotechnology: animal cell biotechnology—methods and protocols. Totowa: Humana Press Inc.; 2007. pp. 457–473. [Google Scholar]
- Genzel Y, Reichl U. Continuous cell lines as a production system for influenza vaccines. Expert Rev Vaccines. 2009;8(12):1681–1692. doi: 10.1586/erv.09.128. [DOI] [PubMed] [Google Scholar]
- Genzel Y, Behrendt I, Konig S, Sann H, Reichl U. Metabolism of MDCK cells during cell growth and influenza virus production in large-scale microcarrier culture. Vaccine. 2004;22(17– 18):2202–2208. doi: 10.1016/j.vaccine.2003.11.041. [DOI] [PubMed] [Google Scholar]
- Genzel Y, Ritter JB, Konig S, Alt R, Reichl U. Substitution of glutamine by pyruvate to reduce ammonia formation and growth inhibition of mammalian cells. Biotechnol Prog. 2005;21(1):58–69. doi: 10.1021/bp049827d. [DOI] [PubMed] [Google Scholar]
- Genzel Y, Fischer M, Reichl U. Serum-free influenza virus production avoiding washing steps and medium exchange in large-scale microcarrier culture. Vaccine. 2006;24(16):3261–3272. doi: 10.1016/j.vaccine.2006.01.019. [DOI] [PubMed] [Google Scholar]
- Genzel Y, Olmer RM, Schafer B, Reichl U. Wave microcarrier cultivation of MDCK cells for influenza virus production in serum containing and serum-free media. Vaccine. 2006;24(35–36):6074–6087. doi: 10.1016/j.vaccine.2006.05.023. [DOI] [PubMed] [Google Scholar]
- Govorkova EA, Kaverin NV, Gubareva LV, Meignier B, Webster RG. Replication of influenza A viruses in a green monkey kidney continuous cell line (Vero) J Infect Dis. 1995;172(1):250–253. doi: 10.1093/infdis/172.1.250. [DOI] [PubMed] [Google Scholar]
- Govorkova EA, Murti G, Meignier B, deTaisne C, Webster RG. African green monkey kidney (Vero) cells provide an alternative host cell system for influenza A and B viruses. J Virol. 1996;70(8):5519–5524. doi: 10.1128/jvi.70.8.5519-5524.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gröner A, Vorlop J (1997) Animal cells and processes for the replication of influenza viruses patent WO97/37000
- Huang HY, Yi XP, Zhang YX. Improvement of Vero cell growth in glutamate-based culture by supplementing ammoniagenic compounds. Process Biochem. 2006;41(12):2386–2392. doi: 10.1016/j.procbio.2006.06.018. [DOI] [Google Scholar]
- Ishii H, Yoshikawa Y, Yamanouchi K. Adaptation of the lapinized rinderpest virus to invitro growth and attenuation of its virulence in rabbits. J Gen Virol. 1986;67:275–280. doi: 10.1099/0022-1317-67-2-275. [DOI] [PubMed] [Google Scholar]
- Janke R, Genzel Y, Wahl AS, Reichl U. Measurement of key metabolic enzyme activities in mammalian cells using rapid and sensitive microplate-based assays. Biotechnol Bioeng. 2010 doi: 10.1002/bit.22817. [DOI] [PubMed] [Google Scholar]
- Kalbfuss B, Genzel Y, Wolff M, Zimmermann A, Morenweiser R, Reichl U. Harvesting and concentration of human influenza A virus produced in serum-free mammalian cell culture for the production of vaccines. Biotechnol Bioeng. 2007;97(1):73–85. doi: 10.1002/bit.21139. [DOI] [PubMed] [Google Scholar]
- Kalbfuss B, Knochlein A, Krober T, Reichl U. Monitoring influenza virus content in vaccine production: precise assays for the quantitation of hemagglutination and neuraminidase activity. Biologicals. 2008;36(3):145–161. doi: 10.1016/j.biologicals.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Kaverin NV, Webster RG. Impairment of multicycle influenza-virus growth in vero (WHO) cells by loss of trypsin activity. J Virol. 1995;69(4):2700–2703. doi: 10.1128/jvi.69.4.2700-2703.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler N, Thomas-Roche G, Gerentes L, Aymard M. Suitability of MDCK cells grown in serum-free medium for influenza virus production. In: Brown F, Robertson JS, Schild GC, Wood JM, editors. Inactivated influenza vaccines prepared in cell culture. Karger, Basel: Dev Biol Stand; 1999. pp. 13–21. [PubMed] [Google Scholar]
- Kistner O, Barrett PN, Mundt W, Reiter M, Schober-Bendixen S, Dorner F. Development of a mammalian cell (Vero) derived candidate influenza virus vaccine. Vaccine. 1998;16(9–10):960–968. doi: 10.1016/S0264-410X(97)00301-0. [DOI] [PubMed] [Google Scholar]
- Kistner O, Howard MK, Spruth M, Wodal W, Bruhl P, Gerencer M, Crowe BA, Savidis-Dacho H, Livey I, Reiter M, Mayerhofer I, Tauer C, Grillberger L, Mundt W, Falkner FG, Barrett PN. Cell culture (Vero) derived whole virus (H5N1) vaccine based on wild-type virus strain induces cross-protective immune responses. Vaccine. 2007;25(32):6028–6036. doi: 10.1016/j.vaccine.2007.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimov AI, Egorov AY, Gushchina MI, Medvedeva TE, Gamble WC, Rudenko LG, Alexandrova GI, Cox NJ. Genetic stability of cold-adapted A/Leningrad/134/47/57 (H2N2) influenza virus: sequence analysis of live cold-adapted reassortant vaccine strains before and after replication in children. J Gen Virol. 1995;76(Pt 6):1521–1525. doi: 10.1099/0022-1317-76-6-1521. [DOI] [PubMed] [Google Scholar]
- Kuster B, Wheeler SF, Hunter AP, Dwek RA, Harvey DJ. Sequencing of N-linked oligosaccharides directly from protein gels: in-gel deglycosylation followed by matrix-assisted laser desorption/ionization mass spectrometry and normal-phase high-performance liquid chromatography. Anal Biochem. 1997;250(1):82–101. doi: 10.1006/abio.1997.2199. [DOI] [PubMed] [Google Scholar]
- Le Ru A, Jacob D, Transfiguracion J, Ansorge S, Henry O, Kamen AA. Scalable production of influenza virus in HEK-293 cells for efficient vaccine manufacturing. Vaccine. 2010;28(21):3661–3671. doi: 10.1016/j.vaccine.2010.03.029. [DOI] [PubMed] [Google Scholar]
- Lin YP, Wharton SA, Martin J, Skehel JJ, Wiley DC, Steinhauer DA. Adaptation of egg-grown and transfectant influenza viruses for growth in mammalian cells: selection of hemagglutinin mutants with elevated pH of membrane fusion. Virology. 1997;233(2):402–410. doi: 10.1006/viro.1997.8626. [DOI] [PubMed] [Google Scholar]
- Lohr V, Rath A, Genzel Y, Jordan I, Sandig V, Reichl U. New avian suspension cell lines provide production of influenza virus and MVA in serum-free media: studies on growth, metabolism and virus propagation. Vaccine. 2009;27(36):4975–4982. doi: 10.1016/j.vaccine.2009.05.083. [DOI] [PubMed] [Google Scholar]
- Mahy BWJ, Kangro HO. Virology methods manual. London: Academic; 1996. [Google Scholar]
- Mendonca RZ, Pereira CA. High-density vero cell-culture on microcarriers in a cell bioreactor. Bioprocess Eng. 1995;12(6):279–282. doi: 10.1007/BF00369503. [DOI] [Google Scholar]
- Mendonca RZ, Pereira CA. Cell metabolism and medium perfusion in VERO cell cultures on microcarriers in a bioreactor. Bioprocess Eng. 1998;18(3):213–218. doi: 10.1007/s004490050433. [DOI] [Google Scholar]
- Mendonca RZ, Prado JCM, Pereira CA. Attachment, spreading and growth of VERO cells on microcarriers for the optimization of large scale cultures. Bioprocess Eng. 1999;20(6):565–571. doi: 10.1007/s004490050630. [DOI] [Google Scholar]
- Merten OW, Hannoun C, Manuguerra JC, Ventre F, Petres S. Production of influenza virus in cell cultures for vaccine preparation. In: Cohen S, Shafferman A, editors. Novel strategies in design and production of vaccines. New York: Plenum Press; 1996. pp. 141–151. [DOI] [PubMed] [Google Scholar]
- Merten OW, Manuguerra JC, Hannoun C, van der Werf S. Production of influenza virus in serum-free mammalian cell cultures. Dev Biol Stand. 1999;98:23–37. [PubMed] [Google Scholar]
- Merten OW, Kallel H, Manuguerra JC, Tardy-Panit M, Crainic R, Delpeyroux F, Van der Werf S, Perrin P. The new medium MDSS2N, free of any animal protein supports cell growth and production of various viruses. Cytotechnology. 1999;30(1–3):191–201. doi: 10.1023/A:1008021317639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosca JD (1986) Virus-induced stabilization of a intrinsically unstable messenger-Rna. J Cell Biochem: 176–176
- Nakamura K, Homma M. Protein synthesis in Vero cells abortively infected with influenza B virus. J Gen Virol. 1981;56(Pt 1):199–202. doi: 10.1099/0022-1317-56-1-199. [DOI] [PubMed] [Google Scholar]
- Ng YC, Berry JM, Butler M. Optimization of physical parameters for cell attachment and growth on macroporous microcarriers. Biotechnol Bioeng. 1996;50(6):627–635. doi: 10.1002/(SICI)1097-0290(19960620)50:6<627::AID-BIT3>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Nicolson C, Major D, Wood JM, Robertson JS. Generation of influenza vaccine viruses on Vero cells by reverse genetics: an H5N1 candidate vaccine strain produced under a quality system. Vaccine. 2005;23(22):2943–2952. doi: 10.1016/j.vaccine.2004.08.054. [DOI] [PubMed] [Google Scholar]
- Nishiyama K, Sugawara K, Nouchi T, Kawano N, Soejima K, Abe S, Mizokami H. Purification and cDNA cloning of a novel protease inhibitor secreted into culture supernatant by MDCK cells. Biologicals. 2008;36(2):122–133. doi: 10.1016/j.biologicals.2007.07.004. [DOI] [PubMed] [Google Scholar]
- Pau MG, Ophorst C, Koldijk MH, Schouten G, Mehtali M, Uytdehaag F. The human cell line PER.C6 provides a new manufacturing system for the production of influenza vaccines. Vaccine. 2001;19(17–19):2716–2721. doi: 10.1016/S0264-410X(00)00508-9. [DOI] [PubMed] [Google Scholar]
- Quesney S, Marvel J, Marc A, Gerdil C, Meignier B. Characterization of Vero cell growth and death in bioreactor with serum-containing and serum-free media. Cytotechnology. 2001;35(2):115–125. doi: 10.1023/A:1017589526145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesney S, Marc A, Gerdil C, Gimenez C, Marvel J, Richard Y, Meignier B. Kinetics and metabolic specificities of Vero cells in bioreactor cultures with serum-free medium. Cytotechnology. 2003;42(1):1–11. doi: 10.1023/A:1026185615650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimmelzwaan GF, Baars M, Claas EC, Osterhaus AD. Comparison of RNA hybridization, hemagglutination assay, titration of infectious virus and immunofluorescence as methods for monitoring influenza virus replication in vitro. J Virol Methods. 1998;74(1):57–66. doi: 10.1016/S0166-0934(98)00071-8. [DOI] [PubMed] [Google Scholar]
- Ritter JB, Genzel Y, Reichl U. Simultaneous extraction of several metabolites of energy metabolism and related substances in mammalian cells: optimization using experimental design. Anal Biochem. 2008;373(2):349–369. doi: 10.1016/j.ab.2007.10.037. [DOI] [PubMed] [Google Scholar]
- Romanova J, Katinger D, Ferko B, Voglauer R, Mochalova L, Bovin N, Lim W, Katinger H, Egorov A. Distinct host range of influenza H3N2 virus isolates in Vero and MDCK cells is determined by cell specific glycosylation pattern. Virology. 2003;307(1):90–97. doi: 10.1016/S0042-6822(02)00064-8. [DOI] [PubMed] [Google Scholar]
- Rourou S, van der Ark A, van der Velden T, Kallel H. A microcarrier cell culture process for propagating rabies virus in Vero cells grown in a stirred bioreactor under fully animal component free conditions. Vaccine. 2007;25(19):3879–3889. doi: 10.1016/j.vaccine.2007.01.086. [DOI] [PubMed] [Google Scholar]
- Sandig V, Jordan I (2005) Immortalized avian cell lines for virus production patent WO2005/042728A2
- Schulze-Horsel J, Schulze M, Agalaridis G, Genzel Y, Reichl U. Infection dynamics and virus-induced apoptosis in cell culture-based influenza vaccine production-flow cytometry and mathematical modeling. Vaccine. 2009;27(20):2712–2722. doi: 10.1016/j.vaccine.2009.02.027. [DOI] [PubMed] [Google Scholar]
- Schwartz R, Berry JM, Subramanian A, Shi X (2006) Non-tumorigenic MDCK cell line for propagating viruses patent WO2006/071563A2
- Schwarzer J, Rapp E, Reichl U. N-glycan analysis by CGE-LIF: profiling influenza A virus hemagglutinin N-glycosylation during vaccine production. Electrophoresis. 2008;29(20):4203–4214. doi: 10.1002/elps.200800042. [DOI] [PubMed] [Google Scholar]
- Schwarzer J, Rapp E, Hennig R, Genzel Y, Jordan I, Sandig V, Reichl U. Glycan analysis in cell culture-based influenza vaccine production: Influence of host cell line and virus strain on the glycosylation pattern of viral hemagglutinin. Vaccine. 2009;27(32):4325–4336. doi: 10.1016/j.vaccine.2009.04.076. [DOI] [PubMed] [Google Scholar]
- Shen S, Law YC, Liu DX. A single amino acid mutation in the spike protein of coronavirus infectious bronchitis virus hampers its maturation and incorporation into virions at the nonpermissive temperature. Virology. 2004;326(2):288–298. doi: 10.1016/j.virol.2004.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobita K, Tanaka T, Hayase Y. Rescue of a viral gene from VERO cells latently infected with influenza virus B/Lee/40. Virology. 1997;236(1):130–136. doi: 10.1006/viro.1997.8716. [DOI] [PubMed] [Google Scholar]
- Toriniwa H, Komiya T. Japanese encephalitis virus production in Vero cells with serum-free medium using a novel oscillating bioreactor. Biologicals. 2007;35(4):221–226. doi: 10.1016/j.biologicals.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Tree JA, Richardson C, Fooks AR, Clegg JC, Looby D. Comparison of large-scale mammalian cell culture systems with egg culture for the production of influenza virus A vaccine strains. Vaccine. 2001;19(25–26):3444–3450. doi: 10.1016/S0264-410X(01)00053-6. [DOI] [PubMed] [Google Scholar]
- Tsai TF, Trusheim H (2008) Making influenza virus vaccines without using eggs patent WO2008032219 (A2)
- Vester D, Rapp E, Gade D, Genzel Y, Reichl U. Quantitative analysis of cellular proteome alterations in human influenza A virus-infected mammalian cell lines. Proteomics. 2009;9(12):3316–3327. doi: 10.1002/pmic.200800893. [DOI] [PubMed] [Google Scholar]
- Wallis CM, Stone AL, Sherman DJ, Damsteegt VD, Gildow FE, Schneider WL. Adaptation of plum pox virus to a herbaceous host (Pisum sativum) following serial passages. J Gen Virol. 2007;88:2839–2845. doi: 10.1099/vir.0.82814-0. [DOI] [PubMed] [Google Scholar]
- Wressnigg N, Shurygina AP, Wolff T, Redlberger-Fritz M, Popow-Kraupp T, Muster T, Egorov A, Kittel C. Influenza B mutant viruses with truncated NS1 proteins grow efficiently in Vero cells and are immunogenic in mice. J Gen Virol. 2009;90:366–374. doi: 10.1099/vir.0.006122-0. [DOI] [PubMed] [Google Scholar]
- Youil R, Su Q, Toner TJ, Szymkowiak C, Kwan WS, Rubin B, Petrukhin L, Kiseleva I, Shaw AR, DiStefano D. Comparative study of influenza virus replication in Vero and MDCK cell lines. J Virol Methods. 2004;120(1):23–31. doi: 10.1016/j.jviromet.2004.03.011. [DOI] [PubMed] [Google Scholar]


