Abstract
Cyanovirin-N (CVN) is a promising antiviral candidate that has an extremely low sequence homology with any other known proteins. The efficient and soluble expression of biologically functional recombinant CVN (rCVN) is still an obstacle due to insufficient yield, aggregation, and abnormal modification. Here, we describe an improved approach to preparing native rCVN from Escherichia coli more efficiently. A fusion gene consisting of cvn and sumo (small ubiquitin-related modifier) and a hexahistidine tag was constructed according to the codon bias of the host cell. This small ubiquitin-related modifier (SUMO)-fused CVN is expressed in the cytoplasm of E. coli in a folded and soluble form (>30% of the total soluble protein), yielding 3 to 4 mg of native rCVN from 1 g of wet cells to a purity up to 97.6%. Matrix-assisted laser desorption ionization coupled to time-of-flight mass spectrometry and reverse-phase high-performance liquid chromatographic analysis showed that the purified rCVN was an intact and homogeneous protein with a molecular weight of 11,016.68 Da. Potent antiviral activity of rCVN against herpes simplex virus type 1 and human immunodeficiency virus type 1/IIIB was confirmed in a dose-dependent manner at nanomolar concentrations. Thus, the His-SUMO double-fused CVN provides an efficient approach for the soluble expression of rCVN in the cytoplasm of E. coli, allowing an alternative system to develop bioprocess for the large-scale production of this antiviral candidate.
Keywords: Cyanovirin-N, SUMO, Escherichia coli, Soluble expression, Anti HIV-1
Introduction
Human immunodeficiency virus (HIV) remains a global health problem of unprecedented dimensions. Globally, there were an estimated 33 million people living with HIV (UNAIDS 2008). Cyanovirin-N (CVN), a potent anti-HIV protein shown promising in vivo efficacy recently (Buffa et al. 2009; Tsai et al. 2004), was originally discovered and isolated from the aqueous extract of the cyanobacterium (blue-green alga) Nostoc ellipsosporum (Boyd et al. 1997). CVN can permanently and efficiently inactivate an extensive range of HIV strains as well as prevent cell-to-cell fusion and the transmission of HIV. It acts by binding with high affinity to N-linked high-mannose glycans present on gp120 and gp41 (Dey et al. 2000; Shenoy et al. 2001), thus inhibiting the attachment and fusion of the virus particle to the target cell. In addition, CVN is also active against rhinoviruses, herpes simplex virus type 1 (HSV-1), hepatitis C, Ebola virus, and influenza A and B viruses (Barrientos et al. 2003; Helle et al. 2006; O'Keefe et al. 2003; Smee et al. 2008). Furthermore, CVN is insensitive to denaturants, detergents, organic solvents, and extreme temperatures. The potent HIV-inactivation ability of CVN and its physicochemical properties make this protein suitable for use as a topical anti-HIV drug.
CVN has a unique sequence of 101 amino acid residues (Boyd et al. 1997). Although the two halves (residues 1–50 and 51–101) in the monomeric CVN share a high degree of sequence homology, CVN has an extremely low sequence homology with any other known proteins. It has no homology greater than eight contiguous amino acids or 20% of the total sequence to any other known protein. In addition, it contains two intramolecular disulfide bonds (Bewley et al. 1998; Yang et al. 1999). These properties make the artificial production of this protein very difficult. The expression of recombinant CVN (rCVN) has been attempted in various species including Escherichia coli (Boyd et al. 1997; Colleluori et al. 2005; Mori et al. 1998), Pichia pastoris (Mori et al. 2002), and transgenic Nicotiana tabacum (Sexton et al. 2006), but the efficient expression of biologically functional rCVN is still problematic due to low yield, aggregation, and abnormal modifications. Thus far, E. coli is still considered to be the simplest, most efficient, and least expensive host to produce simple proteins such as rCVN (Liu and Schultz 2006). However, there is a complex reductase system in the cytoplasm of E. coli that renders it difficult to form stable disulfide bonds (Prinz et al. 1997), and these are crucial for the bioactivity of CVN. The periplasm is a more feasible location for the formation of disulfide bonds and protein refolding. But the yield is extremely low (usually less than 10 mg/L) for a heterologous protein produced in the periplasm (Mergulhao et al. 2004). In addition, when expressed in periplasm under the lead of the ompA signal peptide, the final rCVN product was a mixture of intact proteins: Up to 30% of the truncated product was missing the first two N-terminal residues (Mori et al. 1998; Prinz et al. 1997). Alternatively, the expression of CVN as inclusion bodies in the cytoplasm increased the yield to 3.5 mg rCVN/g of wet cell. Nevertheless, expression remains problematic as this method produces a significant fraction of dimeric species consistently, as well as generating uncharacterized side-chain modifications (Colleluori et al. 2005).
Small ubiquitin-related modifier (SUMO) family proteins function as posttranslational modifiers by attaching to other proteins covalently and reversibly (Saitoh and Hinchey 2000). Recently, SUMO, fused at the N-terminus with heterologous proteins, has been found to improve protein folding, enhance expression level, and protect the protein from degradation via its properties as a chaperonin. SUMO-fusion proteins can be specifically cleaved by SUMO protease depending on the 3D structure of the SUMO tag at the fusion site, and the target protein will be released, together with its native N terminus (Malakhov et al. 2004). Recently, a series of SUMO expression vectors have been developed for the efficient production of recombinant proteins in prokaryotic or eukaryotic host cells (Assadi-Porter et al. 2008; Liu et al. 2008; Sun et al. 2008; Ye et al. 2008).
Here, we tested the effect of Saccharomyces cerevisiae SUMO3 on the expression of rCVN in the cytoplasm of E. coli using a construct carrying sumo-fused cvn with a hexahistidine tag. We found that this new system produced soluble and biologically functional rCVN efficiently in the cytoplasm of E. coli, and this could be rapidly purified into intact and homologous native rCVN by two rounds of Ni2+ chelating chromatography intervened with a SUMO protease cleavage step. The low cost and reduced time consumption of this improved system makes it suitable for the large-scale production of rCVN.
Materials and methods
Gene splicing by overlap extension
A standard overlap extension was carried out using 20 pmol of each primer, 2 ng of template, 1 U of Pyrobest DNA polymerase (Invitrogen, Carlsbad, CA, USA) and other reagents as per the manufacturer’s instructions. Amplification (25 cycles) consisted of denaturation at 95°C for 1 min, annealing at 53°C for 45 s, and elongation at 72°C for 1 min (Horton et al. 1989).
Construction of the expression plasmid
The DNA sequence encoding SUMO was amplified from the vector, pET-SUMO, using the forward primer (F-SUMO, 5′-CAGCATATGCATCATCATCATC-3′) and the reverse primer (R-SUMO, 5′-CTGGGAGAATTTACCAAGACCACCAATCTGTT CTCTG-3′). The resulting 342-bp fragment was spliced with the encoding sequence of cvn with the primers of F-SUMO and C5 (5′-AGAGGATCCTCATCATTCGTATTT CAGGGTACCGTCGATGTTAGCGATGTGGTC GTC-3′). The open reading frame (ORF) of his6-sumo-cvn (642 bp) was then digested with NdeI and BamHI (Takara, Dalian, China), followed by subcloning into pET-3c (Novagen) to create the corresponding expression plasmid pET-SUMO-CVN, which was then transformed into E. coli DH5α (Invitrogen) to sequence the correct insert. At the same time, another expression plasmid, pET-CVN without SUMO, was also constructed using the same vector in the same site. All fragments of the DNA recombination were purified using agarose gel electrophoresis and the Gel Recovery Kit (Sangon, Shanghai, China).
Protein expression and fractionation
Individual colonies of E. coli BL21(DE3) (Novagen, Madison, WI, USA) harboring the pET-sumo-cvn (or pET-cvn) plasmids were inoculated in Luria–Bertani (LB) medium containing 100 μg/ml ampicillin. For optimal conditions, 0.5% glucose and 1.6 mM MgSO4 were added to the medium. The cells were grown at 37°C, 30°C, or 20°C in a shaking incubator (180 rpm) until the culture reached an OD600 = 0.6–1.0. At this point, 1 or 0.5 mM isopropyl-beta-d-thiogalactopyranoside (IPTG) was added, and the cells were grown for a further 4–24 h after induction. Cells were harvested by centrifugation at 4,000×g for 20 min and lysed in sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer for further analysis.
To determine the solubility of recombinant SUMO-CVN (rCVN), cells from 100 ml of induced culture were freeze-thawed once and thoroughly resuspended in NTA-10 buffer (20 mM Tris-HCl, pH 8.0, 0.5 mol/L NaCl, 10 mM imidazole) at a ratio of 1 g of cells to 5 ml of buffer, followed by the addition of 0.1 g/L lysozyme (Dingguo, China), and incubated at 30°C for 30 min. The whole cell lysate was then centrifuged at 20,000×g for 30 min at 4°C to separate the soluble and insoluble fractions.
Protein purification
A typical procedure for the purification of native rCVN is illustrated in Fig. 4a. The hexahistidine-tagged SUMO-CVN from E. coli BL21(DE3) was initially purified by ion metal affinity chromatography with Ni-NTA agarose (GE Healthcare, USA). Fifty milliliters of the cleared bacterial lysate from 1 L of LB culture was applied to an XK 16/20 column (GE Healthcare, USA) containing 20 ml of Ni-NTA resin that had been equilibrated with ten column volumes of NTA-10 buffer. The resin was then washed with NTA-10 buffer until the OD280 reached a base line. The nonspecifically bound proteins were eluted with 100–150 ml of NTA-40 buffer (20 mM Tris-HCl, pH 8.0, 0.5 M NaCl, 40 mM imidazole). Finally, recombinant hexahistidine-tagged SUMO-CVN was eluted with NTA-200 buffer (20 mM Tris-HCl, pH 8.0, 0.5 mol/L NaCl, 200 mM imidazole).
Fig. 4.
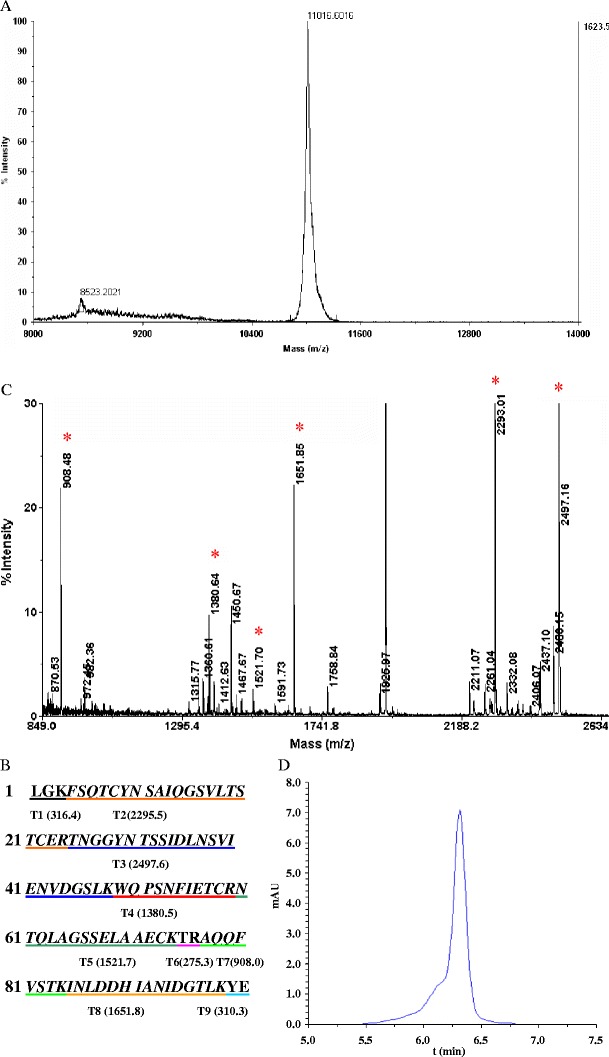
Chemicophysical characterization of the recombinant native CVN. a Molecular weight of rCVN determined by MALDI-TOF MS. b Predicted peptide map of natural CVN. Amino acid sequences underlined with different colors indicate the nine fragments of trypsin-digested CVN. T1 (316.4) indicates that the peptide numbered T1 has a theoretical MW of 316.4 Da. The same labeling scheme applies for T2 through T9. c PMF analysis of rCVN. The rCVN band cut from a 16% Tris-tricine SDS-PAGE gel was digested with trypsin and then analyzed by MALDI-TOF-MS. Mass peaks that match the theoretical MW of fragments deriving from natural CVN are marked with asterisk. d Purity analysis of rCVN by RP-HPLC with C18 column
After being applied to a sephadex G-25 column (GE Healthcare, USA) and washed with buffer A (20 mM Tris-HCl, pH 8.0, 2% Igepal, 1.5 M NaCl, 10 mM dithiothreitol) to remove imidazole, the eluted SUMO-CVN was digested with SUMO protease (1 U enzyme to 10 μg substrate) at 4°C for 1 h (Lee et al. 2008). The Ni-NTA column was used again to remove SUMO, undigested SUMO-CVN, and SUMO protease (in which the hexahistidine tag was also fused to the N-terminal), thus resulting in the native rCVN. The purity of rCVN was analyzed on a 16% reduced Tris-tricine SDS-PAGE gel, and the protein concentration was determined using the Bradford method (Bradford 1976).
MALDI-TOF MS analysis
The purified native rCVN was subjected to matrix-assisted laser desorption ionization coupled to time-of-flight mass spectrometry (MALDI-TOF MS), to confirm its peptide mass map and molecular weight (MW; Henzel et al. 2003). Briefly, after separation on the 16% Tris-tricine SDS-PAGE gel, followed by silver staining, the rCVN band was excised and destained with 1% potassium ferricyanide and 1.6% sodium thiosulfate. The protein was then in-gel digested with 20 g/L trypsin at 37°C overnight. The digested peptides were extracted and loaded onto an MTP AnchorChipTM 600/384 TF (Bruker-Daltonik, Bremen, Germany). MALDI-TOF MS analysis was carried out using an UltraflexTM MALDI-TOF mass spectrometer (Bruker-Daltonik). For molecular weight determination, the purified protein was loaded directly onto the palladium plate and analyzed with an UltraflexTM MALDI-TOF mass spectrometer.
RP-HPLC analysis
The purity of the purified rCVN was confirmed using reverse-phase high-performance liquid chromatography (RP-HPLC) in addition to the SDS-PAGE analysis. The sample (10 μL) was loaded onto a 4.6 × 250-mm Zorbax 300 SB-C18 column (Agilent Lifescience, USA) and then eluted with a linear gradient of 30–70% acetonitrile at a flow rate of 1 ml/min in the presence of 0.1% trifluoroacetic acid. The HPLC system (Agilent Lifescience, Model 1100, USA) was configured with a quarter pump and diode array detector.
MTT assay
The activity of rCVN against HSV-1 was determined using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) method (Takeuchi et al. 1991). Briefly, rCVN was twofold serially diluted from 181.55 nM (2 μg/ml) to 11.35 nM (0.125 μg/ml), and then 50 μL of each dilution and HSV-1 (100 TCID50/50 μL; State Key Laboratory of Virology of Wuhan University, China) was added to the monolayer of Vero cells (ATCC, USA) in a 96-well microtiter plate. A cytotoxicity determination was simultaneously carried out in wells with the test samples but without the virus. Concurrently, 25 μg/ml acyclovir was used as a positive control. After incubation at 37°C with 5% CO2 for 48 h, MTT reagent was added for color development. The absorbance of λ 570/630 was measured using a microplate reader (Bio-Rad Laboratories, USA) and the 50% inhibiting concentration (IC50) and 50% cytotoxic concentration (CC50) were calculated.
WST-1 assay
The anti-HIV activity of the recombinant CVN was determined in vitro using the water-soluble tetrazolium salt (WST)-1 method in a P3 laboratory (Boyd et al. 1997; Dueweke et al. 1993). CVN was twofold serially diluted with Rapid Prototyping and Manufacturing Institute 1640 medium (Invitrogen) containing 10% fetal bovine serum. Then 50 μL of each dilution and HIV-1/IIIB (100 TCID50/50 μL, harvested from the culture supernatant of MOLT-4/IIIB cells), were added to MT-4 cells (104 cells/100 μL/well) in 96-well microtiter plate. The final concentration of CVN ranged from 3.52 to 112.5 nM. At the same time, azidothymidine (AZT; Sigma) ranging from 0.98 to 250 nM was used as a positive control. After incubation with 5% CO2 at 37°C for 96 h, WST-1 was added to develop the color. The absorbance of λ 450/650 was determined using a microplate reader (Wako, Japan) to evaluate the activity.
All samples were tested in triplicate, and the reproducibility was confirmed in three separate experiments.
Gel electrophoresis
SDS-PAGE (12%) or 16% tricine SDS-PAGE was carried out under reducing conditions as previously described (Laemmli 1970). The gel was scanned with a densitometer (Bio-Rad, USA) after staining with Coomassie Blue, and the expression level of rCVN was determined.
Results
Construction and soluble expression of SUMO-CVN
An artificial coding sequence for cvn was designed de novo, and nine oligos were synthesized for overlap extension of the coding sequence, according to the codon usage bias of E. coli (Fig. 1a). The sequence of his-sumo was amplified from pET3c-sumo using a primer pair in which a sequence coding for hexahistidine was added to the forward primer. The final product of the multiple overlap extension was cloned into pET-3c at the NdeI and BamHI restriction sites to get the expression plasmid pET-SUMO-CVN, which was then transformed into the expression host, E. coli BL21(DE3) for protein expression.
Fig. 1.
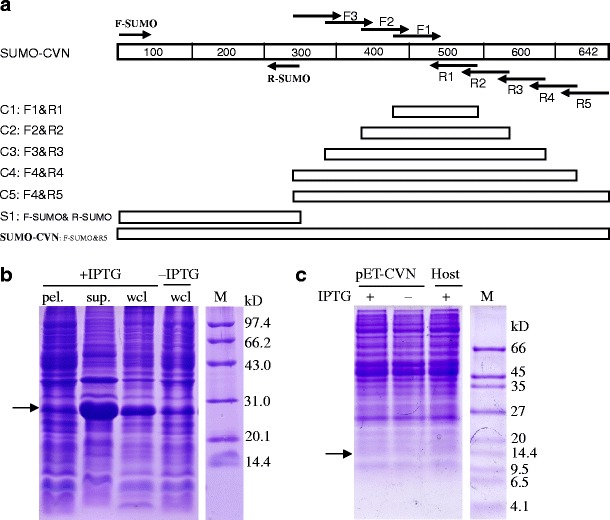
Construction and soluble expression of SUMO-CVN in the cytoplasm of E. coli. a Strategy for the synthesis of hexahistidine-tagged sumo-cvn ORF by multiple overlap extension. Nine primers were designed using the amino acid sequence of natural CVN and the codon bias of E. coli. Elongation of primer pair F1 and R1 resulted in the fragment C1 (97 bp). Taking C1 as a template and using F2 and R2 as primers, the fragment C2 (173 bp) was synthesized. A similar method was used to get the fragments: C3 (241 bp), C4 (298 bp), and C5 (cvn, 337 bp, overlapped with hexahis-sumo by 37 bp). The sequence of hexahis-sumo (342 bp) was amplified from pET3c-sumo using a primer pair in which the sequence corresponding to the hexahistidine tag was added in the forward primer, i.e., F-SUMO. In the final round of overlap extension, C5 and S1 were spliced into hexahis-sumo-cvn (642 bp) by the primer pair of F-sumo and R5. b Expression of SUMO-CVN in E. coli. c Expression of rCVN without SUMO in E. coli. E. coli BL21(DE3) harboring pET-sumo-cvn (b) or pET-cvn (c) was grown in LB medium to exponential phase (OD600 = 0.6) and then induced with 1 mM IPTG for 4 h at 37°C. Cultures harboring an empty vector or without IPTG induction were used as negative controls. Whole cell lysates (wcl) were extracted, fractioned, resolved by SDS-PAGE, and stained with Coomassie Blue. Protein bands with expected sizes are indicated with arrows
After 4 h induction with 1 mM IPTG at 37°C, SUMO-CVN was efficiently expressed in the cytoplasm of E. coli to 17.5% of the total protein (Fig. 1b). Almost all of the target protein was found in the supernatant (up to 32.3% of the total soluble protein), suggesting that the protein was quite soluble. In contrast, expression of rCVN without the SUMO fusion in the same vector and host cell was not detected (Fig. 1c). Similar results were also obtained using a SUMO-negative CVN in pBV220 in various E. coli strains including BL21(DE3), Origami(DE3), JM109, and DH5α (data not shown). pBV220 is an expression vector with a strong promoter that we have successfully used to express hNDPK-A previously (Xiong et al. 2007). We also tested the possibility of expressing SUMO-negative rCVN in the periplasm of E. coli instead of the cytoplasm and still found no visible expression. Thus, we have showed for the first time that SUMO can effectively stabilize and facilitate the soluble expression of rCVN in the cytoplasm of E. coli.
Purification of native rCVN from the SUMO-CVN fusion
Before purification of native rCVN from the SUMO-CVN fusion protein, we first optimized the expression conditions for the target protein in E. coli BL21(DE3) by testing various culture temperatures, induction periods, and IPTG concentrations (Fig. 2). No significant difference in the expression of target protein was found when either 0.5 or 1 mM IPTG was used at induction. For cultures induced at 20°C, 30°C, and 37°C for 4 h, the expression of SUMO-CVN increased gradually with increasing temperature. Longer induction periods up to 24 h further increased the expression level, and the highest expression level was obtained in cultures induced with 0.5 mM IPTG at 20°C for 24 h. At these optimum conditions, the expression level was 28.3 ± 0.56% of the total protein.
Fig. 2.
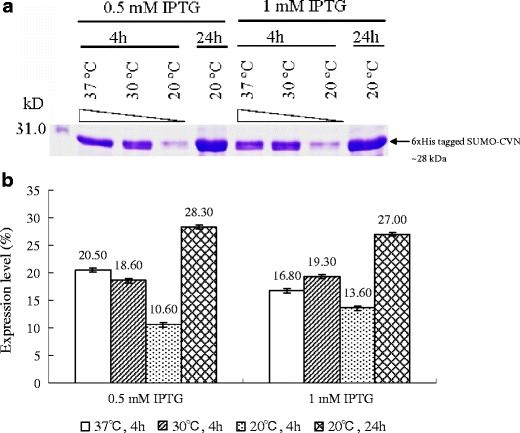
Optimization of the expression of SUMO-CVN in E. coli BL21(DE3) under different induction conditions. Expression of SUMO-CVN was induced at various culture temperatures, induction periods, and IPTG concentrations. Whole cell lysates were resolved in 12% SDS-PAGE gels and stained with Coomassie Blue. a A representative gel image is shown here and the arrow indicates SUMO-CVN, whose apparent molecular weight is approximately 28 kDa. The expression level of SUMO-CVN shown in a was determined by densitometer scanning. b Data are presented as means ± standard deviation (n = 3)
After induction with IPTG under the optimum conditions, the total soluble protein was extracted and applied to the Ni-NTA column. After washing with buffer containing 200 mM imidazole, SUMO-CVN was subsequently eluted with a purity of 73.8% (Fig. 3b) and then digested with SUMO protease to release the native rCVN protein. By running the cleaved proteins through Ni-NTA column again to capture hexahistidine-tagged proteins, native rCVN could be separated from SUMO, undigested SUMO-CVN, and SUMO protease. Approximately 3 to 4 mg of native rCVN could be purified from 1 g of wet cells using this process, to a purity up to 97.6%. This demonstrates that a considerable amount of native rCVN protein could be easily and economically purified with this system, making it suitable for future large-scale production of this protein.
Fig. 3.
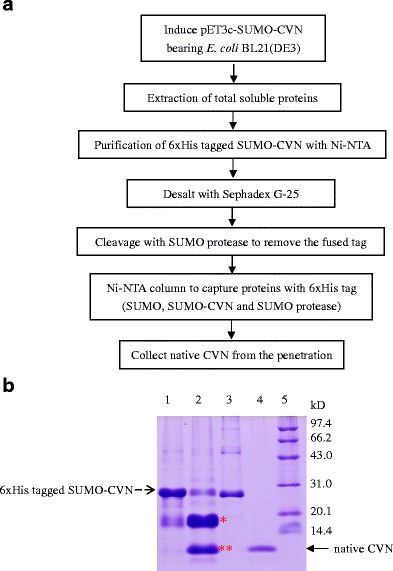
Purification of native CVN from the hexahistidine-tagged SUMO-CVN. a Flow chart of the purification process. Native rCVN was purified by two rounds of Ni2+ chelating chromatography, intervened with one round of hexahistidine-tagged SUMO protease cleavage. b SDS-PAGE analysis of purified rCVN. pET-sumo-cvn bearing E. coli BL21(DE3) was cultured in medium containing 0.5 mM IPTG at 20°C for 24 h. Fractions from the purification were collected and resolved using SDS-PAGE. Lane 1 eluted fraction of the first Ni2+-chelating chromatography; lane 2 SUMO protease cleaved mixture (single asterisk indicates the released hexahistidine-tagged SUMO, double asterisks indicates the rCVN); lane 3 hexahistidine-tagged SUMO protease; lane 4 the eluted fraction of the second Ni2+-chelating chromatography; lane 5 low-molecular-weight protein markers
Chemicophysical characterization of the recombinant native CVN
In addition to the formation of inclusion bodies, other unresolved problems for the expression of rCVN in E. coli include N-terminal deletion, dimerization, and side-chain modification, resulting in different apparent MWs of the final recombinant product (Mori et al. 1998). To confirm the structural integrity of rCVN expressed from our system, we determined the MW of the purified rCVN using MALDI-TOF MS. It was found that the MW of the rCVN was 11,016.68 Da, in agreement with the theoretical MW for natural CVN (Fig. 4a). There are eight known trypsin cleavage sites in natural CVN, making it possible to confirm the sequence identity of the rCVN product. We determined the peptide profile of rCVN by peptide mass fingerprint (PMF) analysis. The predicted cleavage map of natural CVN is shown in Fig. 4b. From MALDI-TOF/TOF MS spectra of in-gel digested rCVN (Fig. 4c), six peptide fragments with molecular weights comparative to their natural counterpart were identified. Database interpretation also confirmed the identity of the rCVN. Purity is another important characteristic for a recombinant drug candidate. In addition to SDS-PAGE analysis (Fig. 3b), RP-HPLC was also employed to determine the purity of the final product. Figure 4d showed that the elution profile of rCVN in a C18 column is a smooth Gaussian curve without any shoulder peak. Taken together, these results provide evidence that the SUMO expression system we used is able to produce high-quality rCVN.
Antiviral activity of recombinant native CVN
Finally, we examined the biological activity of purified rCVN, using the RNA virus HIV-1/III B and the DNA virus HSV-1 as models to evaluate the antiviral activity of rCVN. A WST-1 assay revealed that native rCVN possesses significant antiviral activity. For HIV-1/IIIB, the IC50 and CC50 were 22.35 ± 3.74 and 164.31 ± 5.90 nM, respectively (Table 1). At concentrations of 56.25 and 28.13 nM, the inhibition rates of rCVN were 71.2% and 62.8%, respectively. The control, AZT, a well-known anti-HIV compound, exhibited 64.62% inhibition of the virus at a concentration of 62.5 nM (Fig. 5). The IC50 of rCVN for HSV-1 was 28.14 ± 2.72 nM, and the CC50 was 190.63 ± 9.07 nM (Table 1). The above results indicate that purified rCVN is functionally active. In addition, SUMO-CVN also exhibited activity against HSV-1 with an IC50 of 31.37 ± 2.15 nM, comparable to that of native rCVN. This result serves as evidence that SUMO-CVN is produced in the cytoplasm as the fully folded and biologically functional form, providing an alternative source of the protein for practical applications.
Table 1.
Summary of IC50 and CC50 of native rCVN against HIV-1/IIIB and HSV-1
| Viral strains | Tested samples | IC50 (nM) | CC50 (nM) |
|---|---|---|---|
| HIV-1/III B | rCVN | 22.35 ± 3.74 | 164.31 ± 5.90 |
| AZTa | 35.88 ± 4.30 | >2,500 | |
| HSV-1 | rCVN | 28.14 ± 2.72 | 190.63 ± 9.07 |
| SUMO-CVN | 31.37 ± 2.15 | ND |
IC50 50% inhibitory concentration, CC50 50% cytotoxic concentration, ND not done, AZT azidothymidine
aAZT was used as a positive control for the anti-HIV-1/IIIB assay
Fig. 5.
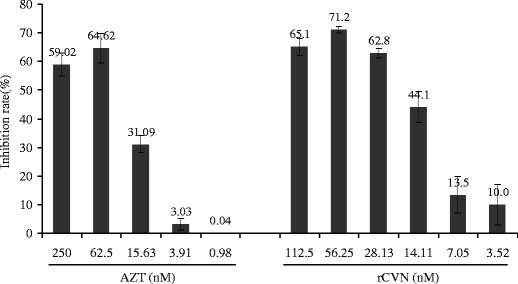
Inhibition effect of rCVN on HIV-1/IIIB. The anti-HIV-1/IIB activity of rCVN was evaluated by WST-1 assay. Data are presented as means ± standard deviation from three independent experiments in which all of the samples were tested in triplicate
Discussion
Large-scale production of rCVN as a potential antiviral drug using genetically engineered microorganisms is required to provide a ready source of material for practical application (Fischetti et al. 2009; Sexton et al. 2006). However, the sequence properties of CVN make it difficult to be produced in heterologous hosts. The expression of rCVN in transgenic plants is time-consuming, with low yields, undeveloped downstream processes, and potential posttranslational modifications (Sexton et al. 2006). Another choice is to produce rCVN in a different eukaryotic system, P. pastoris. However, it seems difficult to obtain native rCVN using this system because of high-mannose N-glycosylation at position N30 and the dimerization of the final product mediated by P51 in the hinge region, both of which result in a loss of antiviral activity of rCVN (Mori et al. 2002). Although E. coli is so far the best-studied host, considerable problems still exist in its practical use for the expression of rCVN. Low protein yield remains a major issue. In addition, rCVN as expressed in E. coli tends to aggregate into nonfunctional forms, which have been attributed to the lack of stable disulfide bond formation and proper folding.
In an attempt to solve the above problems, we fused CVN with yeast SUMO3, preceded by a hexahistidine tag, and we studied the efficiency of this system at expressing rCVN in E. coli. SUMO is a common protein in eukaryotic cells, and it regulates the activities and cellular localizations of target proteins through a modification process similar to ubiquitination (Butt et al. 2005; Kim et al. 2002). Several proteins that are difficult to express in conventional systems, such as SARS-CoV structure proteins (Zuo et al. 2005), metalloprotease (MMP13) (Marblestone et al. 2006), brazzein (Assadi-Porter et al. 2008), urodilatin (Sun et al. 2008), and antibody fragment (Ye et al. 2008) have been successfully expressed in E. coli using a SUMO fusion strategy. Our data showed that a SUMO-CVN fusion construct was efficiently expressed in a soluble form in the cytoplasm of E. coli, supporting the idea that SUMO facilitates the soluble expression of foreign (heterologous) proteins in E. coli. As SUMO is a fusion label, the mechanism of the yield enhancement and solubility improvement is still unclear. Several studies implicate that it may function as a chaperonin or act as a core for the folding of recombinant proteins in E. coli (Butt et al. 2005).
As there is no specific affinity method available, native CVN is usually isolated by general chromatography techniques such as RP-HPLC and anion exchange. Ni2+ affinity chromatography has also been applied to rapidly purify rCVN produced in P. pastoris (Mori et al. 2002), but the final product is His-tagged rather than native rCVN. It is possible to release native rCVN from SUMO-CVN by cleaving with SUMO protease at the site of the fusion. An advantage of our system is that a hexahistidine tag was fused at the N-terminal of SUMO so that the SUMO-CVN could be rapidly purified from the total protein extraction by simply running it through a Ni-NTA column. After digestion with SUMO protease, the released native rCVN could be subsequently purified using the same column to over 97% purity. With this purification process, we obtained a final yield of about 3–4 mg/g of wet cells. The amount is considerable as the protein was induced in E. coli in a conventional flask-shaking culture. E. coli has been cultured to densities greater than 150 g DCW/L in optimized conditions (Lee 1996; Xia et al. 2008). In an ordinary fermentor culture, the biomass will be over 50 g WCW/L, a level that is usually reached in our laboratory (Xiong et al. 2007). The yield of rCVN from our SUMO-CVN system in a fermentor fed-batch culture is therefore expected to be over 150 mg/L. Efforts are now being made to further improve the protein yield of native rCVN using our system.
The expression of soluble SUMO-CVN is a major advantage of our expression system. The highest yield of rCVN reported thus far is 140 mg/L, which has been achieved using a fermentor culture (Colleluori et al. 2005). However, the protein was expressed as inclusion bodies, which dramatically decreased protein quality. Other problems reported for rCVN expression in E. coli include N-terminal deletion, dimerization, and side-chain modification. MW determination, PMF analysis, RP-HPLC chromatography, and SDS-PAGE demonstrated that the purified rCVN derived from our system was intact, homogeneous, and monomeric. Another advantage of this system is that the intermediate recombinant product, SUMO-CVN, was also biologically functional and had an antiviral activity against HSV that was comparable to native rCVN. This result demonstrates that the SUMO-CVN produced in the cytoplasm of E. coli is fully folded and in its functional form, providing an alternative source for practical applications.
Comparing the IC50 and CC50 of rCVN against HIV-1 with that of the natural one (Boyd et al. 1997), it looks like that the rCVN is not as potent as the natural product because the IC50 reported here is significant higher than the previous report (0.4 nM). This might have resulted from the different bioassay parameters such as the host cell, the origination of virus and its titer in the detection system, or from the trace impurity in the recombinant product. The data of commercialized AZT also displayed higher IC50 than the previous report (6 nm; Mitsuya et al. 1985), suggesting a system difference in our bioassay method.
CVN is a promising candidate for a topical HIV microbicide. A 5-mg dose, which proved to be effective in macaque studies, requires a production capacity of 5,000 kg a year to supply just ten million women twice a week (Shattock and Moore 2003). This scale of protein production may be achievable only by expression system that could produce rCVN with high productivity and low cost. Here, we propose an improved approach that efficiently expresses rCVN in the cytoplasm of E. coli. By fusing rCVN with SUMO, it can be efficiently produced in the cytoplasm of E. coli at a level of >30% of the total soluble protein. With the addition of a hexahistidine tag fused at the N-terminal of SUMO, intact and native rCVN can be rapidly purified in a soluble and biologically active form. Further improvement is still required to eventually bring this promising antiviral candidate from bench to clinical application.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (30873082, U0632010), the Ministry of Science and Technology of China (2008BAI63B05), the Program for New Century Excellent Talents in University (NCET-07-0376), and the 211 Project of Jinan University. The anti-HIV bioactivity determination was supported by Tokyo Biochemistry Research Foundation of Japan (TBRF-RF-08-52). We are most grateful to Dr. Jun Fan of Kyoto University for helpful discussions and revision of the manuscript.
Conflict of interest
The authors have no conflicts of interest.
References
- Assadi-Porter FM, Patry S, Markley JL. Efficient and rapid protein expression and purification of small high disulfide containing sweet protein brazzein in E-coli. Protein Expr Purif. 2008;58:263–268. doi: 10.1016/j.pep.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos LG, O'Keefe BR, Bray M, Anthony S, Gronenborn AM, Boyd MR. Cyanovirin-N binds to the viral surface glycoprotein, GP(1, 2) and inhibits infectivity of Ebola virus. Antiviral Res. 2003;58:47–56. doi: 10.1016/S0166-3542(02)00183-3. [DOI] [PubMed] [Google Scholar]
- Bewley CA, Gustafson KR, Boyd MR, Covell DG, Bax A, Clore GM, Gronenborn AM. Solution structure of cyanovirin-N, a potent HIV-inactivating protein. Nat Struct Biol. 1998;5:571–578. doi: 10.1038/828. [DOI] [PubMed] [Google Scholar]
- Boyd MR, Gustafson KR, McMahon JB, Shoemaker RH, O'Keefe BR, Mori T, Gulakowski RJ, Wu L, Rivera MI, Laurencot CM, Currens MJ, Cardellina JH, 2nd, Buckheit RW, Jr, Nara PL, Pannell LK, Sowder RC, 2nd, Henderson LE. Discovery of cyanovirin-N, a novel human immunodeficiency virus-inactivating protein that binds viral surface envelope glycoprotein gp120: potential applications to microbicide development. Antimicrob Agents Chemother. 1997;41:1521–1530. doi: 10.1128/aac.41.7.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Buffa V, Stieh D, Mamhood N, Hu Q, Fletcher P, Shattock RJ. Cyanovirin-N potently inhibits human immunodeficiency virus type 1 infection in cellular and cervical explant models. J Gen Virol. 2009;90:234–243. doi: 10.1099/vir.0.004358-0. [DOI] [PubMed] [Google Scholar]
- Butt TR, Edavettal SC, Hall JP, Mattern MR. SUMO fusion technology for difficult-to-express proteins. Protein Expr Purif. 2005;43:1–9. doi: 10.1016/j.pep.2005.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colleluori DM, Tien D, Kang F, Pagliei T, Kuss R, McCormick T, Watson K, McFadden K, Chaiken I, Buckheit RW, Jr, Romano JW. Expression, purification, and characterization of recombinant cyanovirin-N for vaginal anti-HIV microbicide development. Protein Expr Purif. 2005;39:229–236. doi: 10.1016/j.pep.2004.10.009. [DOI] [PubMed] [Google Scholar]
- Dey B, Lerner DL, Lusso P, Boyd MR, Elder JH, Berger EA. Multiple antiviral activities of cyanovirin-N: blocking of human immunodeficiency virus type 1 gp120 interaction with CD4 and coreceptor and inhibition of diverse enveloped viruses. J Virol. 2000;74:4562–4569. doi: 10.1128/JVI.74.10.4562-4569.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dueweke TJ, Poppe SM, Romero DL, Swaney SM, So AG, Downey KM, Althaus IW, Reusser F, Busso M, Resnick L, et al. U-90152, a potent inhibitor of human immunodeficiency virus type 1 replication. Antimicrob Agents Chemother. 1993;37:1127–1131. doi: 10.1128/aac.37.5.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischetti L, Barry SM, Hope TJ, Shattock RJ. HIV-1 infection of human penile explant tissue and protection by candidate microbicides. Aids. 2009;23:319–328. doi: 10.1097/QAD.0b013e328321b778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helle F, Wychowski C, Vu-Dac N, Gustafson KR, Voisset C, Dubuisson J. Cyanovirin-N inhibits hepatitis C virus entry by binding to envelope protein glycans. J Biol Chem. 2006;281:25177–25183. doi: 10.1074/jbc.M602431200. [DOI] [PubMed] [Google Scholar]
- Henzel WJ, Watanabe C, Stults JT. Protein identification: the origins of peptide mass fingerprinting. J Am Soc Mass Spectrom. 2003;14:931–942. doi: 10.1016/S1044-0305(03)00214-9. [DOI] [PubMed] [Google Scholar]
- Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- Kim KI, Baek SH, Chung CH. Versatile protein tag, SUMO: its enzymology and biological function. J Cell Physiol. 2002;191:257–268. doi: 10.1002/jcp.10100. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee SY. High cell-density culture of Escherichia coli. Trends Biotechnol. 1996;14:98–105. doi: 10.1016/0167-7799(96)80930-9. [DOI] [PubMed] [Google Scholar]
- Lee CD, Sun HC, Hu SM, Chiu CF, Homhuan A, Liang SM, Leng CH, Wang TF. An improved SUMO fusion protein system for effective production of native proteins. Protein Sci. 2008;17:1241–1248. doi: 10.1110/ps.035188.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CC, Schultz PG. Recombinant expression of selectively sulfated proteins in Escherichia coli. Nat Biotechnol. 2006;24:1436–1440. doi: 10.1038/nbt1254. [DOI] [PubMed] [Google Scholar]
- Liu L, Spurrier J, Butt TR, Strickler JE. Enhanced protein expression in the baculovirus/insect cell system using engineered SUMO fusions. Protein Expr Purif. 2008;62:21–28. doi: 10.1016/j.pep.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malakhov MP, Mattern MR, Malakhova OA, Drinker M, Weeks SD, Butt TR. SUMO fusions and SUMO-specific protease for efficient expression and purification of proteins. J Struct Funct Genomics. 2004;5:75–86. doi: 10.1023/B:JSFG.0000029237.70316.52. [DOI] [PubMed] [Google Scholar]
- Marblestone JG, Edavettal SC, Lim Y, Lim P, Zuo X, Butt TR. Comparison of SUMO fusion technology with traditional gene fusion systems: enhanced expression and solubility with SUMO. Protein Sci. 2006;15:182–189. doi: 10.1110/ps.051812706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergulhao FJ, Taipa MA, Cabral JM, Monteiro GA. Evaluation of bottlenecks in proinsulin secretion by Escherichia coli. J Biotechnol. 2004;109:31–43. doi: 10.1016/j.jbiotec.2003.10.024. [DOI] [PubMed] [Google Scholar]
- Mitsuya H, Weinhold KJ, Furman PA, St Clair MH, Lehrman SN, Gallo RC, Bolognesi D, Barry DW, Broder S. 3'-Azido-3'-deoxythymidine (BW A509U): an antiviral agent that inhibits the infectivity and cytopathic effect of human T-lymphotropic virus type III/lymphadenopathy-associated virus in vitro. Proc Natl Acad Sci U S A. 1985;82:7096–7100. doi: 10.1073/pnas.82.20.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Gustafson KR, Pannell LK, Shoemaker RH, Wu L, McMahon JB, Boyd MR. Recombinant production of cyanovirin-N, a potent human immunodeficiency virus-inactivating protein derived from a cultured cyanobacterium. Protein Expr Purif. 1998;12:151–158. doi: 10.1006/prep.1997.0838. [DOI] [PubMed] [Google Scholar]
- Mori T, Barrientos LG, Han Z, Gronenborn AM, Turpin JA, Boyd MR. Functional homologs of cyanovirin-N amenable to mass production in prokaryotic and eukaryotic hosts. Protein Expr Purif. 2002;26:42–49. doi: 10.1016/S1046-5928(02)00513-2. [DOI] [PubMed] [Google Scholar]
- O'Keefe BR, Smee DF, Turpin JA, Saucedo CJ, Gustafson KR, Mori T, Blakeslee D, Buckheit R, Boyd MR. Potent anti-influenza activity of cyanovirin-N and interactions with viral hemagglutinin. Antimicrob Agents Chemother. 2003;47:2518–2525. doi: 10.1128/AAC.47.8.2518-2525.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz WA, Aslund F, Holmgren A, Beckwith J. The role of the thioredoxin and glutaredoxin pathways in reducing protein disulfide bonds in the Escherichia coli cytoplasm. J Biol Chem. 1997;272:15661–15667. doi: 10.1074/jbc.272.25.15661. [DOI] [PubMed] [Google Scholar]
- Saitoh H, Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem. 2000;275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- Sexton A, Drake PM, Mahmood N, Harman SJ, Shattock RJ, Ma JK. Transgenic plant production of cyanovirin-N, an HIV microbicide. FASEB J. 2006;20:356–358. doi: 10.1096/fj.05-4742fje. [DOI] [PubMed] [Google Scholar]
- Shattock RJ, Moore JP. Inhibiting sexual transmission of HIV-1 infection. Nat Rev Microbiol. 2003;1:25–34. doi: 10.1038/nrmicro729. [DOI] [PubMed] [Google Scholar]
- Shenoy SR, O'Keefe BR, Bolmstedt AJ, Cartner LK, Boyd MR. Selective interactions of the human immunodeficiency virus-inactivating protein cyanovirin-N with high-mannose oligosaccharides on gp120 and other glycoproteins. J Pharmacol Exp Ther. 2001;297:704–710. [PubMed] [Google Scholar]
- Smee DF, Bailey KW, Wong MH, O'Keefe BR, Gustafson KR, Mishin VP, Gubareva LV. Treatment of influenza A (H1N1) virus infections in mice and ferrets with cyanovirin-N. Antiviral Res. 2008;80:266–271. doi: 10.1016/j.antiviral.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Xia Z, Bi F, Liu JN. Expression and purification of human urodilatin by small ubiquitin-related modifier fusion in Escherichia coli. Appl Microbiol Biotechnol. 2008;78:495–502. doi: 10.1007/s00253-007-1330-0. [DOI] [PubMed] [Google Scholar]
- Takeuchi H, Baba M, Shigeta S. An application of tetrazolium (MTT) colorimetric assay for the screening of anti-herpes simplex virus compounds. J Virol Methods. 1991;33:61–71. doi: 10.1016/0166-0934(91)90008-N. [DOI] [PubMed] [Google Scholar]
- Tsai CC, Emau P, Jiang Y, Agy MB, Shattock RJ, Schmidt A, Morton WR, Gustafson KR, Boyd MR. Cyanovirin-N inhibits AIDS virus infections in vaginal transmission models. AIDS Res Hum Retroviruses. 2004;20:11–18. doi: 10.1089/088922204322749459. [DOI] [PubMed] [Google Scholar]
- UNAIDS (2008) Report on the global AIDS epidemic 2008. In: Unaids.org. http://www.unaids.org/en/KnowledgeCentre/HIVData/GlobalReport/2008/2008_Global_report.asp. Accessed 6 Nov 2008
- Xia XX, Han MJ, Lee SY, Yoo JS. Comparison of the extracellular proteomes of Escherichia coli B and K-12 strains during high cell density cultivation. Proteomics. 2008;8:2089–2103. doi: 10.1002/pmic.200700826. [DOI] [PubMed] [Google Scholar]
- Xiong S, Qian CW, Guo CW, Huang L, Liu QY, Zhang MY, Wang YF. Efficient purification of recombinant human NDPK-A in pilot-scale. Chinese J Biotechnol. 2007;23:508–513. [PubMed] [Google Scholar]
- Yang F, Bewley CA, Louis JM, Gustafson KR, Boyd MR, Gronenborn AM, Clore GM, Wlodawer A. Crystal structure of cyanovirin-N, a potent HIV-inactivating protein, shows unexpected domain swapping. J Mol Biol. 1999;288:403–412. doi: 10.1006/jmbi.1999.2693. [DOI] [PubMed] [Google Scholar]
- Ye T, Lin Z, Lei H. High-level expression and characterization of an anti-VEGF165 single-chain variable fragment (scFv) by small ubiquitin-related modifier fusion in Escherichia coli. Appl Microbiol Biotechnol. 2008;81:311–317. doi: 10.1007/s00253-008-1655-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X, Li S, Hall J, Mattern MR, Tran H, Shoo J, Tan R, Weiss SR, Butt TR. Enhanced expression and purification of membrane proteins by SUMO fusion in Escherichia coli. J Struct Funct Genomics. 2005;6:103–111. doi: 10.1007/s10969-005-2664-4. [DOI] [PMC free article] [PubMed] [Google Scholar]


