Abstract
Transmissible gastroenteritis virus (TGEV) is the causative agent of porcine transmissible gastroenteritis, and sensitive detection methods are required for preventing the disease. In this article, reverse transcription-loop-mediated isothermal amplification (RT-LAMP) was developed to detect TGEV. Three pairs of primers targeting the nucleocapsid (N) gene of TGEV were synthesized and used in the RT-LAMP. The optimization, sensitivity, and specificity of the RT-LAMP were evaluated. Our results showed that the RT-LAMP amplified the N gene with high specificity, efficiency, and rapidity at isothermal condition. The optimal reaction condition was achieved at 60°C for 30 min. The RT-LAMP assay was more sensitive than gel-based RT-PCR and PCR. It had a higher sensitivity than enzyme-linked immunosorbent assay (ELISA) using the equal virus templates. In addition, the established RT-LAMP differentiated TGEV from porcine epidemic diarrhea virus, porcine rotavirus, porcine pseudorabies virus, porcine reproductive and respiratory syndrome virus, and avian infectious bronchitis virus. The approach is suitable for detecting TGEV for field diagnostics or in less-equipped laboratories due to its convenience and simplicity.
Keywords: Porcine Epidemic Diarrhea Virus, Infectious Bronchitis Virus, Respiratory Syndrome Virus, Transmissible Gastroenteritis Virus, Avian Infectious Bronchitis Virus
Introduction
Transmissible gastroenteritis virus (TGEV) is the causative agent of transmissible gastroenteritis (TGE) of swine. The disease is a highly contagious disease characterized by vomiting, diarrhea, and dehydration, and its mortality rate in seronegative suckling piglets may reach 100% [4, 16]. Prevalence of TGE often results in considerable economic loss to large swine-breeding units. TGEV belongs to the family Coronaviridae and is an enveloped virus with a positive-stranded RNA genome [32]. TGEV consists of four structural proteins: the spike (S) protein, the integral membrane (M) glycoprotein, the minor envelope (E) protein, and the nucleocapsid (N) protein [16, 23, 32]. The S protein plays a crucial role in the induction of neutralizing antibodies [15, 34], binding to the cellular receptor porcine aminopeptidase N [9, 18, 24], and mediating viral attachment to target cells [1, 10, 22, 31]. The M protein is a trans-membrane protein [35] and is involved in the assembly process of viral nucleocapsid and membrane [29]. The N protein of coronaviruses is phosphorylated protein, interacting with viral genomic RNA forming a helical ribonucleoprotein [19]. The N protein plays crucial roles in viral genome transcription, core formation, and RNA packaging [38]. In addition, the N protein is highly conserved and can be used as a diagnostic target for detecting viral infection.
Loop-mediated isothermal amplification (LAMP) is a novel DNA amplification method. LAMP uses four to six primers that respectively recognize six to eight regions of target DNA, in conjunction with the enzyme Bst polymerase, which has strand-displacement activity. The synchronization DNA synthesis by these primers guarantees the specificity of the method. Meanwhile, the amplification step performed at isothermal conditions leads to the synthesis of a large amount of DNA. LAMP starts when the forward inner primer (FIP) anneals to the complementary region in the target DNA, resulting in the first-strand synthesis. Then, the outer forward primer (F3) hybridizes and displaces the first strand, forming a loop structure at one end. The resulting single-stranded DNA is the template for backward inner primer (BIP)-initiated DNA synthesis and subsequent outer backward (B3)-primed strand-displacement DNA synthesis. The resulting dumbbell-shaped DNA stem-loop structure is the template for subsequent hybridization between one inner primer and the loop and initiates the displacement DNA synthesis. Consequently, the LAMP assay may form the original stem-loop and a new stem-loop that is twice as long and the final products are stem-loop DNAs with several inverted repeats of the target DNA and cauliflower-like structures containing multiple loops [20].
In this study, the N gene of TGEV was selected as a target and using specific primers, we developed an RT-LAMP to detect TGEV and to differentiate it from other viruses. Our data confirm that the RT-LAMP is highly sensitive, convenient, and specific for amplifying the target gene. Importantly, the approach may be used for differentiating TGEV from other viruses.
Materials and Methods
Cells and Viruses
TGEV strain PUR46-MAD [23], porcine epidemic diarrhea (PEDV) isolate HLJBY, porcine pseudorabies virus (PrV) strain Kaplan [27, 33], porcine rotavirus (PRV), porcine reproductive and respiratory syndrome virus (PRRSV) isolate HH08 [25, 26, 39], and avian infectious bronchitis virus (IBV) strain Beaudette [17] are propagated in susceptible cells.
Primers
Six primers targeting the N gene of TGEV were designed using the Primer Explorer version 3 (http://primerexplorer.jp/lamp3.0.0/index.html). They include an outer pair (F3, B3), an inner pair (FIP, BIP), and a loop pair (F- and B-loop). A pair of primers TGE1 and TGE2 was used for conventional RT-PCR amplifying the N gene. Information regarding the primer names and sequences is provided in Table 1.
Table 1.
Information on the primers used in this study
| Primer name | Sequence 5′-3′ |
|---|---|
| F3 | TGTCAGTTGGGGAGATGA |
| B3 | GCGAGTTTGTCTATTCCAAT |
| FIP | TGGGGTTGAAGAATGAAAGAGGTATTTTTCCAAAACACGTGGTCGTT |
| BIP | CCTCCAACAAGGTTCAAAATTTTGGTTTTCCAATCTGTTGATCCCTGTT |
| F-loop | TCCGACCACGGGAATTGG |
| L-loop | CTTATGTCCGAGAGACTTTGTACC |
| TGE1 (forward) | ATGGCCAACCAGGGACAA |
| TGE2 (reverse) | TTAGTTCGTTACCTCATC |
Virus Propagation and RNA Extraction
Swine testis (ST) cells were cultured in Eagle’s Minimum Essential Medium (EMEM) containing 10% newborn bovine serum (Excell Bio, Shanghai, China) in 6-well plates at 37°C to allow the formation of cell monolayer. The cells were washed with phosphate buffered saline (PBS) buffer, and infected with TGEV (500 μl/well) at a multiplicity of infection (MOI) of 2 at 37°C for 1 h in the absence of serum. After absorption, the serum-free EMEM was then added into the wells (2.5 ml/well), and the culture was maintained at 37°C for 24–36 h.
Total RNAs were extracted from the culture supernatants of TGEV, PEDV, PRV, PRRSV, and IBV using the RNA extraction kit (Keygen Biotech, China), and the genomic DNA of PrV was extracted from virus-infected Vero cell culture using the DNA extraction kit (Omega, Norcross, USA) according to the manufacturer’s instructions. The extracted RNA was subjected to reverse transcription (RT) to synthesize the cDNA using reverse primer TGE2 and a cDNA synthesis kit (HaiGene, China) according to the manufacturer’s instructions.
RT-LAMP Reaction
The reaction mixture contained RNA template (2 μg) inner primers FIP and BIP (2 M/each), outer primers F3 and B3 (0.2 M/each), Loop primers F- and L-loop (0.8 M/each), dNTPs mix (1.4 mM, HaiGene, China), 5 mM of MgSO4, Bst DNA polymerase (8 U, New England Biolabs, USA), 1× ThermoPol buffer consists of 20 mM Tris–HCl, 10 mM KCl, 10 mM(NH4)2SO4, 2 mM MgSO4, and 0.1% Triton X-100 (HaiGene, China), and M-MuLV reverse transcriptase (HaiGene, China). The reaction was performed at a volume of 25 μl at 65°C in a water bath for 1 h followed by stopping at 85°C for 10 min. Sterile water was used as a negative control template. The resulting DNA products were analyzed by separating 5 μl RT-LAMP reaction mixtures in ethidium bromide-stained 2% agarose gel electrophoresis, where the positive reaction mixtures showed a characteristic ladder of multiple bands. The relative quantification of the DNA was performed using the gel documentation system (Uvitec, Cambridge, UK) according to the manufacturer’s instructions.
Optimization of the RT-LAMP Assay
The RT-LAMP reaction mixtures were incubated at 58, 59, 60, 61, or 62°C for 60 min to determine the optimal reaction temperature. Subsequently, the optimal reaction time was determined by performing the RT-LAMP at 60°C for 30, 40, 50, or 60 min. The reactions were terminated by heat inactivation at 80°C for 10 min. The amplified DNA products from the RT-LAMP assays were visualized by agarose gel electrophoresis as above.
Sensitivity Analysis of the RT-LAMP
The concentration of TGEV RNA was determined using a ultra-violet photometer (Thermo Scientific NanoDrop 2000) according to the manufacturer’s instructions. Then, the 10-fold serial dilutions of the RNA (76.4 μg/ml) were used as template for RT-LAMP and a conventional RT-PCR. The RT-LAMP system was done as described above.
The RT-PCR was performed using a RT-PCR kit (HaiGene, China). The RT system included 1 μl M-MuLV ReverseTransciptase (200 U/μl, TaKaRa, China), 4 μl 5× RT-PCR buffer, 1 μl dNTP Mixture (10 mM each), 2 μl template, 1 μl Oligo dT, RNase 0.5 μl Inhibitor (40 U/μl), and 11.5 μl sterile water. The RT profile included 30°C for 10 min, 42°C for 60 min, and 95°C for 5 min. The PCR system was comprised of 12.5 μl 2× HG-PCR buffer, 2.5 μl dNTP (2.5 mM), 0.5 μl each primers, 1 μl cDNA template, 0.5 μl super Taq DNA polymerase (HaiGene) and adding of sterile water to a final volume of 25 μl. The PCR parameters included 95°C for 5 min, 30 cycles of 94°C for 36 s, 70°C for 36 s, 72°C for 2 min, and a final extension at 72°C for 5 min. In parallel, the 10-fold serially diluted cDNA (240 μg/ml) of TGEV was subjected to LAMP and conventional PCR. The PCR system and condition were performed as above.
Enzyme-linked immunosorbent assay (ELISA) was performed to compare its sensitivity with RT-LAMP. In brief, purified TGEV particles (2 μg/μl) were serially diluted in carbonate–bicarbonate buffer (15 mM Na2CO3, 35 mM NaHCO3, pH 9.6), and the viruses were coated into ELISA plates (100 μl/well) at 4°C overnight. The next day, the plates were blocked with 5% non-fat dry milk in PBS-0.05% Tween 20 (PBST) at 37°C for 2 h. Subsequently, the wells were incubated with serially diluted anti-TGEV polyclonal antibody (1:800 dilution) or control serum from a non-immunized rabbit at 37°C for 1 h, after complete washing with PBST. The plates were incubated with horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (BOSTER, China 1:5000 diluted in PBST) at 37°C for 1 h. The wells were incubated with o-phenylenediamine dihydrochloride (OPD) substrate for 5 min after complete washing with PBST. The OD492 value was examined using an ELISA reader. The OD492 value of TGEV-containing positive well (P)/the OD492 value of control serum well (N) >2 was regarded as positive reaction of ELISA.
Specificity of the RT-LAMP Assay
To analyze the specificity of the established RT-LAMP assay, TGEV, PEDV, PrV, PRV, PRRSV, and IBV were used as templates and subjected to RT-LAMP as above. The reaction was performed at 60°C for 30 min.
Results
Amplification of N Gene of TGEV by RT-LAMP
The TGEV RNA and specific primers targeting the viral N gene were included in an RT-LAMP performed at 65°C in a water bath for 1 h. The agarose gel electrophoresis analysis indicated that the amplified DNA products showed a characteristic ladder of multiple bands, indicating that the final products were the mixtures of stem-loop DNAs with various stem lengths (Fig. 1). In contrast, the negative control did not show the characteristic bands.
Fig. 1.
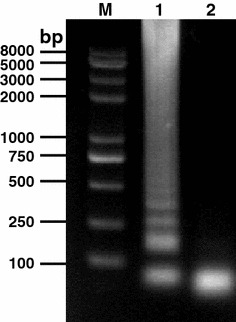
Amplification of N gene of TGEV by RT-LAMP. TGEV RNA was used as template and six primers targeting the viral N gene were used in an RT-LAMP performed at 65°C for 1 h. Lane M DNA marker; lane 1 RT-LAMP product; lane 2 negative control
Optimization of the RT-LAMP Assay
The optimal reaction temperature and time of the RT-LAMP were investigated. As shown in Fig. 2a, the DNA products of the RT-LAMP at different temperatures showed multiple of characteristic ladder bands; however, the intensity of DNAs from the reactions at 60°C was the strongest among the tested reaction temperatures, which was determined as the optimal temperature for RT-LAMP amplifying TGEV N gene. The RT-LAMP was then performed at 60°C at different time points. The subsequent results indicated that the DNA products showed the highest intensity, when the reaction was performed for 30 min (Fig. 2b). The optimal reaction condition of the RT-LAMP for TGEV was 60°C for 30 min.
Fig. 2.
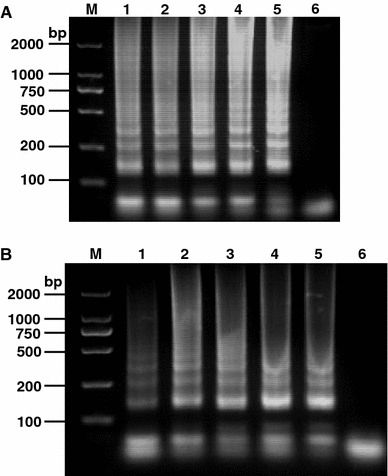
Optimal condition for the RT-LAMP of TGEV N gene. The optimization of the RT-LAMP targeting the TGEV N gene was analyzed by performing the reaction at 58, 59, 60, 61, or 62°C for 1 h. The reaction results are shown in lanes 1–5, respectively (Panel a). The same reaction was performed at 60°C for 20, 30, 40, 50, or 60 min. The RT-LAMP results are shown in lanes 1–5, respectively (Panel b). Lane M DNA marker; lane 6 Negative control
Sensitivity of the RT-LAMP
The sensitivity of the RT-LAMP assay was firstly compared with the conventional RT-PCR amplifying the 10-fold serial dilutions of viral RNA templates. Our results showed that the detection limitation of RT-LAMP was 7.64 × 10−6 μg/ml, in contrast, the RT-PCR has a detection limit of 7.64 × 10−1 μg/ml (Fig. 3). The sensitivity of the LAMP was further confirmed by comparison with PCR using TGEV cDNA (Fig. 4). The former was approximately 10000 times more sensitive than PCR.
Fig. 3.
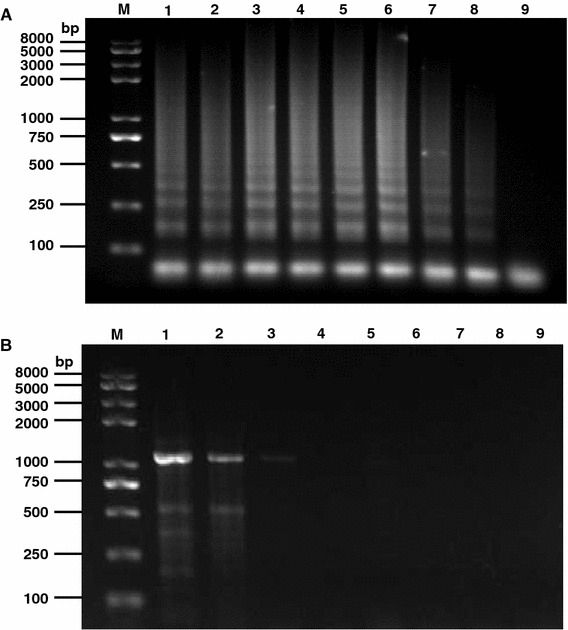
Sensitivity comparison between RT-LAMP and RT-PCR. 10-fold serial dilutions of the TGEV RNA (76.4 μg/ml) were used as template for RT-LAMP and a conventional RT-PCR using specific primers. The results of RT-LAMP and RT-PCR are shown in panels a and b, respectively. Lane M DNA marker; lanes 1–8 are the reaction results from 10-fold serial dilution of RNA templates; lane 9 negative control
Fig. 4.
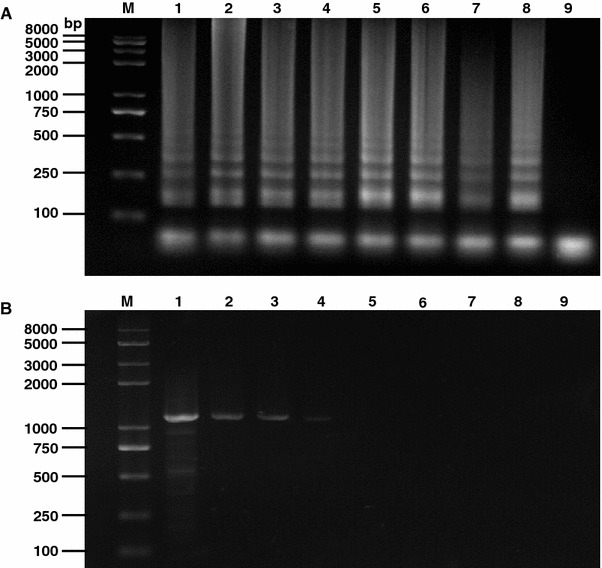
Sensitivity comparison between RT-LAMP and PCR. 10-fold serially diluted cDNA (240 μg/ml) of TGEV was subjected to LAMP and conventional PCR. Their results are shown in panels a and b, respectively. Lane M DNA marker; lanes 1–8 are the reaction results from 10-fold serial dilution of cDNA templates; lane 9 Negative control
The minimal required virus template amount for RT-LAMP and ELISA was analyzed. Our results showed that the minimal limitation for both approaches was 2 × 10−7 μg/μl and 2 × 10−6 μg/μl, respectively, indicating that the former has somewhat higher sensitivity than the latter (Fig. 5).
Fig. 5.
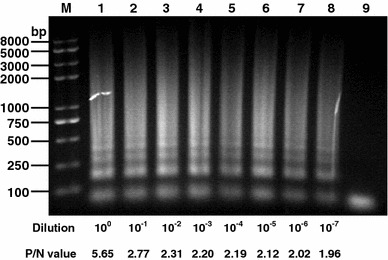
Sensitivity comparison between ELISA and RT-LAMP. Purified TGE virions (2 μg/μl) were serially diluted and used as coating antigen for an indirect ELISA or RT-LAMP. Lane M DNA marker; lanes 1–8 are the RT-LAMP results from 100 to 10−7 diluted virions; lane 9 negative control. The OD492 value of TGEV-containing positive well (P)/the OD492 value of control serum well (N) >2 was regarded as positive reaction in ELISA. The P/N values of different virus dilution are indicated
The RT-LAMP Assay can Differentiate TGEV from Other Viruses
The specificity of the established RT-LAMP assay was analyzed by including several related porcine viruses (i.e., PEDV, PRV, and PrV) and an avian coronavirus, IBV in the reaction. Our result indicated that no positive DNA products of the RT-LAMP assay were observed, when these control viruses were used as templates. In contrast, the positive bands were amplified using TGEV as template (Fig. 6).
Fig. 6.
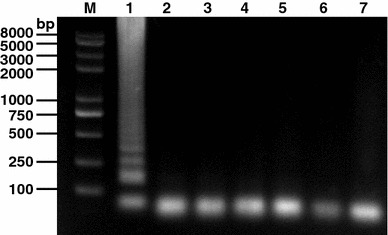
Specificity of the RT-LAMP for TGEV N gene. TGEV and other selected viruses were used as templates and subjected to RT-LAMP performed at 60°C for 30 min. Lane M: DNA marker; lane 1–6 are RT-LAMP results from templates TGEV, PEDV, PRRSV, PRV, PrV, IBV, and negative control (water), respectively
Discussion
The TGE is a severe enteric infection of the pig, and older animals usually recover, but in newborn piglets mortality rates may reach 100% [23, 30]. As a result, TGE prevalence causes enormous economic losses within the pig industry; particularly many related evidences have been reported in China [11–13, 42]. The first case of a TGE outbreak in China was reported in 1956 in Guangdong province [7]. Nowadays, TGE epidemics occur in other parts of mainland China and Taiwan [2, 3, 37, 41], although inactivated or live vaccines against TGEV are used in some regions in China. Establishment of rapid, sensitive, and cost-effective diagnostic assays for detecting TGEV is highly desirable. In addition to conventional virus isolation, there are other diagnostic methods available for TGEV, for example, Multiplex PCR and multiplex RT-PCR, real-time RT-PCR, restriction fragment length polymorphism, microarray hybridization assay, and blocking ELISA [8, 21, 28, 36, 40]. Although these methods are effective, they may require either high-precision instruments or complicated procedures. Therefore, development of simple, sensitive, and cost-effective assays is required for detecting TGEV infection in fields and in less well-equipped laboratories.
The RT-LAMP developed in this study is a useful method for detecting TGEV, since the approach owns numerous advantages such as simplicity, rapidity, and inexpensiveness. Particularly, the assay can be performed at isothermal conditions using a conventional water bath or heat block. In general, the LAMP reaction was performed under isothermal conditions (60–65°C). In this study, we carried out the RT-LAMP at 65°C, which gave rise to the positive results; however, the density of the RT-LAMP product was not very significant. Therefore, we optimized the reaction conditions of the RT-LAMP by performing the test at different temperature and time to have the highest efficiency. Based on the optimal reaction condition, the sensitivity of the RT-LAMP was compared with that of PCR and RT-PCR. Our data indicated that the sensitivity of the RT-LAMP specific for TGEV was more than 10000 folds than the other methods. Using the inactivated TGEV as coating antigen or template, we observed the higher sensitivity of RT-LAMP, compared with conventional sensitive ELISA, confirming the sensitivity of the N gene-based RT-LAMP.
The TGEV and PEDV belong to the Group I coronaviruses, order Nidovirus, and they are closely related [5]. IBV and PRRSV belong to the Group III coronavirus and arterivirus, respectively; however, both viruses also belong to the order Nidovirus [6, 25]. The structural similarity between the N proteins of IBV and PRRSV implies that members of the Coronaviridae and Arteriviridae families share a mechanism of filamentous nucleocapsid formation, with suitable alterations necessary to interact specifically with their respective genomes [14, 26]. Although PrV and PRV are members of the families Herpesviridae and Reoviridae, respectively; they may cause co-infection in pig with TGEV, PEDV, or PRRSV. Therefore, we selected these related animal viruses as control templates to evaluate the utility of the RT-LAMP for TGEV. Our results showed that the RT-LAMP is very specific for TGEV and can be used for differentiation diagnosis. Taken together, our data demonstrate that the developed RT-LAMP assay is simple, sensitive, specific and can be applied in discriminating ELISA for distinguishing TGEV from other viruses.
Acknowledgments
National Natural Science Foundation of China (30700590; 30972195) and Funding supported by Program for New Century Excellent Talents in Heilongjiang Provincial University (1155–NCET–005) are acknowledged.
References
- 1.Antón IM, González S, Bullido MJ, Corsín M, Risco C, Langeveld JP, Enjuanes L. Cooperation between transmissible gastroenteritis coronavirus (TGEV) structural proteins in the in vitro induction of virus-specific antibodies. Virus Res. 1996;46:111–124. doi: 10.1016/S0168-1702(96)01390-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ao Z, Zhao Y, Lin S. Diagnosis and therapy of a case of transmissible gastroenteritis. Livest Poult Ind. 2006;191:50–51. [Google Scholar]
- 3.Bai W. Report on the diagnosis and therapy of porcine transmissible gastroenteritis virus. Fujian J Anim Husb Vet. 2005;27:34. [Google Scholar]
- 4.Bohl EH (1981) Enteric viral infections as related to diarrhea in swine. Proceedings of the 3rd international symposium on neonatal diarrhea. Veterinary Infectious Diseases Organization, Saskatoon, Saskatchewan, Canada, pp 1–9
- 5.Bridgen A, Duarte M, Tobler K, Laude H, Ackermann M. Sequence determination of the nucleocapsid protein gene of the porcine epidemic diarrhea virus confirms that this virus is a coronavirus related to human coronavirus 229E and porcine transmissible gastroenteritis virus. J Gen Virol. 1993;74:1795–1804. doi: 10.1099/0022-1317-74-9-1795. [DOI] [PubMed] [Google Scholar]
- 6.Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch Virol. 1997;142:629–633. [PubMed] [Google Scholar]
- 7.Chen X, Liu J. Porcine transmissible gastroenteritis. Fujian J Anim Husb Vet. 2000;22:36–37. [Google Scholar]
- 8.Chen Q, Li J, Deng Z, Xiong W, Wang Q, Hu YQ. Comprehensive detection and identification of seven animal coronaviruses and human respiratory coronavirus 229E with a microarray hybridization assay. Intervirology. 2010;53:95–104. doi: 10.1159/000264199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delmas B, Gelfi J, L’Haridon R, Vogel LK, Sjostrom H, Noren O, Laude H. Aminopeptidase N is a major receptor for the entero-pathogenic coronavirus TGEV. Nature. 1992;357:417–420. doi: 10.1038/357417a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duarte M, Laude H. Sequence of the spike protein of the porcine epidemic diarrhoea virus. J Gen Virol. 1994;75:1195–1200. doi: 10.1099/0022-1317-75-5-1195. [DOI] [PubMed] [Google Scholar]
- 11.Fang D. Taiwan’s preventive and therapy experience of porcine transmissible gastroenteritis. Taiwan Agri Res. 1986;2:23. [Google Scholar]
- 12.He S. A preliminary report on the prevalence, diagnosis and therapy of porcine transmissible gastroenteritis. Anim Husb Vet Med. 1985;5:48. [Google Scholar]
- 13.Huang S. Diagnosis and therapy report of porcine transmissible gastroenteritis virus. Guangxi J Anim Husb Vet Med. 2001;17:34–35. [Google Scholar]
- 14.Jayaram H, Fan H, Bowman BR, Ooi A, Jayaram J, Collisson EW, Lescar J, Prasad BV. X-Ray structures of the N- and C-terminal domains of a coronavirus nucleocapsid protein: implications for nucleocapsid formation. J Virol. 2006;80:6612–6620. doi: 10.1128/JVI.00157-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiménez G, Correa I, Melgosa MP, Bullido MJ, Enjuanes L. Critical epitopes in transmissible gastroenteritis virus neutralization. J Virol. 1986;60:131–139. doi: 10.1128/jvi.60.1.131-139.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laude H, Reeth KV, Pensaert M. Porcine respiratory coronavirus: molecular features and virus-host interactions. Vet Res. 1993;24:125–150. [PubMed] [Google Scholar]
- 17.Li J, Yin J, Sui X, Li G, Ren X. Comparative analysis on the effect of glycyrrhizin diammonium and lithium chloride on infectious bronchitis virus infection in vitro. Avian Pathol. 2009;38:215–221. doi: 10.1080/03079450902912184. [DOI] [PubMed] [Google Scholar]
- 18.Liu B, Li G, Sui X, Yin J, Wang H, Ren X. Expression and functional analysis of porcine aminopeptidase N produced in prokaryotic expression system. J Biotechnol. 2009;141:91–96. doi: 10.1016/j.jbiotec.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macnaughton MR, Madge MH. The genome of human coronavirus strain 229E. J Gen Virol. 1978;39:497–504. doi: 10.1099/0022-1317-39-3-497. [DOI] [PubMed] [Google Scholar]
- 20.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:e63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogawa H, Taira O, Hirai T, Takeuchi H, Nagao A, Ishikawa Y, Tuchiya K, Nunoya T, Ueda S. Multiplex PCR and multiplex RT-PCR for inclusive detection of major swine DNA and RNA viruses in pigs with multiple infections. J Virol Methods. 2009;160:210–214. doi: 10.1016/j.jviromet.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 22.Ren X, Glende J, Al-Falah M, de Vries V, Schwegmann-Wessels C, Qu X, Tan L, Tschernig T, Deng H, Naim HY, Herrler G. Analysis of ACE2 in polarized epithelial cells: surface expression and function as receptor for severe acute respiratory syndrome-associated coronavirus. J Gen Virol. 2006;87:1691–1695. doi: 10.1099/vir.0.81749-0. [DOI] [PubMed] [Google Scholar]
- 23.Ren X, Glende J, Yin J, Schwegmann-Wessels C, Herrler G. Importance of cholesterol for infection of cells by transmissible gastroenteritis virus. Virus Res. 2008;137:220–224. doi: 10.1016/j.virusres.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ren X, Li G, Liu B. Binding characterization of determinants in porcine aminopeptidease N, the cellular receptor for transmissible gastroenteritis virus. J Biotechnol. 2010;150(1):202–206. doi: 10.1016/j.jbiotec.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ren X, Wang M, Yin J, Ren Y, Li G. Heterologous expression of fused genes encoding the glycoprotein 5 from PRRSV: a way for producing functional protein in prokaryotic microorganism. J Biotechnol. 2010;147:130–135. doi: 10.1016/j.jbiotec.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ren X, Wang M, Yin J, Li G. Phages harboring specific peptides to N protein of PRRSV distinguish the virus from other viruses. J Clin Microbiol. 2010;48:1875–1881. doi: 10.1128/JCM.01707-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ren X, Yin J, Li G, Herrler G (2010) Cholesterol dependence of pseudorabies herpesvirus entry. Curr Microbiol. doi:10.1007/s00284-010-9700-8 [DOI] [PMC free article] [PubMed]
- 28.Rodák L, Smíd B, Nevoránková Z, Valícek L, Smítalová R. Use of monoclonal antibodies in blocking ELISA detection of transmissible gastroenteritis virus in faeces of piglets. J Vet Med B Infect Dis Vet Public Health. 2005;52:105–111. doi: 10.1111/j.1439-0450.2005.00829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rottier PJM. The coronavirus membrane protein. In: Siddel SG, editor. The Coronaviridae. New York: Plenum Press; 1995. pp. 115–139. [Google Scholar]
- 30.Schwegmann-Wessels C, Zimmer G, Laude H, Enjuanes L, Herrler G. Binding of transmissible gastroenteritis coronavirus to cell surface sialoglycoproteins. J Virol. 2002;76:6037–6043. doi: 10.1128/JVI.76.12.6037-6043.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwegmann-wessels C, Glende J, Ren X, Qu X, Deng H, Enjuanes L, Herrler G. Comparison of vesicular stomatitis virus pseudotyped with the S proteins from a porcine and a human coronavirus. J Gen Virol. 2009;90:1724–1729. doi: 10.1099/vir.0.009704-0. [DOI] [PubMed] [Google Scholar]
- 32.Spaan W, Cavanagh D, Horzinek C. Coronaviruses: structure and genome expression. J Gen Virol. 1988;69:2939–2952. doi: 10.1099/0022-1317-69-12-2939. [DOI] [PubMed] [Google Scholar]
- 33.Sui X, Yin J, Ren X. Antiviral effect of diammonium glycyrrhizinate and lithium chloride on cell infection by pseudorabies herpesvirus. Antiviral Res. 2010;85:346–353. doi: 10.1016/j.antiviral.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sune C, Jimenez G, Correa I, Bullido MJ, Gebauer F, Smerdou C, Enjuanes L. Mechanisms of transmissible gastroenteritis coronavirus neutralization. Virology. 1990;177:559–569. doi: 10.1016/0042-6822(90)90521-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Utiger A, Tobler K, Bridgen A, Suter M, Singh M, Ackermann M. Identification of proteins specified by porcine epidemic diarrhea virus. Adv Exp Med Biol. 1995;380:87–90. doi: 10.1007/978-1-4615-1899-0_46. [DOI] [PubMed] [Google Scholar]
- 36.Vemulapalli R, Gulani J, Santrich C. A real-time TaqMan RT-PCR assay with an internal amplification control for rapid detection of transmissible gastroenteritis virus in swine fecal samples. J Virol Methods. 2009;162:231–235. doi: 10.1016/j.jviromet.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang S, Gao J. Diagnosis and prevention of a case of porcine transmissible gastroenteritis. Hebei Anim Husb Vet Med. 2003;19:38. [Google Scholar]
- 38.Wang Y, Zhang X. The nucleocapsid protein of coronavirus mouse. Nuclear ribonucleoprotein a1 in vitro and in vivo. Virology. 1999;265:96–109. doi: 10.1006/viro.1999.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang M, Li G, Yin J, Ren X. Phylogenetic characterization of genes encoding for glycoprotein 5 and membrane protein of PRRSV isolate HH08. J Vet Sci. 2009;10:309–315. doi: 10.4142/jvs.2009.10.4.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang C, Chen J, Shi H, Qiu HJ, Xue F, Liu S, Liu C, Zhu Y, Almazán F, Enjuanes L, Feng L. Rapid differentiation of vaccine strain and Chinese field strains of transmissible gastroenteritis virus by restriction fragment length polymorphism of the N gene. Virus Genes. 2010;41:47–58. doi: 10.1007/s11262-010-0481-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei Y. Diagnosis and therapy report of a case of porcine transmissible gastroenteritis. Shanghai J Anim Husb Vet Med. 2001;4:37. [Google Scholar]
- 42.Yi S. Control of porcine transmissible gastroenteritis virus. Guangdong J Anim Vet Sci. 1997;22:48. [Google Scholar]


