Abstract
Major histocompatibility complex (MHC) molecules play an important role in the susceptibility and/or resistance to many diseases. To gain an insight into the MHC background and to facilitate the experimental use of cynomolgus macaques, the second exon of the MhcMafa-DOB, -DPB1, and -DQB1 genes from 143 cynomolgus macaques were characterized by cloning to sequencing. A total of 16 Mafa-DOB, 16 Mafa-DPB1, and 34 Mafa-DQB1 alleles were identified, which revealed limited, moderate, and marked allelic polymorphism at DOB, DPB1, and DQB1, respectively, in a cohort of cynomolgus macaques of Vietnamese origin. In addition, 16 Mafa-DOB, 5 Mafa-DPB1, and 8 Mafa-DQB1 alleles represented novel sequences that had not been reported in earlier studies. Almost of the sequences detected at the DOB and DQB1 locus in the present study belonged to DOB*01 (100%) and DQB1*06 (62%) lineages, respectively. Interestingly, four, three, and one high-frequency alleles were detected at Mafa-DOB, -DPB1, and -DQB1, respectively, in this monkeys. The alleles with the highest frequency among these monkeys were Mafa-DOB*010102, Mafa-DPB1*13, and Mafa-DQB1*0616, and these were found in 33 (25.6%) of 129 monkeys, 32 (31.37%) of 102 monkeys, and 30 (31%) of 143 monkeys, respectively. The high-frequency alleles may represent high priority targets for additional characterization of immune function. We also carried out evolutionary and population analyses using these sequences to reveal population-specific alleles. This information will not only promote the understanding of MHC diversity and polymorphism in the cynomolgus macaque but will also increase the value of this species as a model for biomedical research.
Keywords: High frequency, Major histocompatibility complex class II, Macaca fascicularis
Introduction
Rhesus macaques have been used as animal models for various human diseases for a long time. With the 1978 ban on exportation of Rhesus macaques from India, researchers have become increasingly interested in an alternative macaque, the cynomolgus macaque, which has a shorter breeding cycle, a docile personality, and requires lower dosages of drugs. The cynomolgus macaque (Macaca fascicularis), also known as the crab-eating monkey or long-tailed macaque, is used mainly in animal models of diabetes, renal transplantation, virological research, SARS, tuberculosis, studies of the pathogenesis of simian immunodeficiency virus (SIV), and pharmacodynamic evaluation (O'Sullivan et al. 1997; Menninger et al. 2002; McAuliffe et al. 2004; Reed et al. 2009). Owing to the need for reliable data on experimental drug reactions provided by animal models, researchers have focused on genes of the immune system of cynomolgus monkeys, in particular, the genes of the major histocompatibility complex (MHC). Molecules of MHC class I and II play an important role in immune regulatory processes by presenting peptides of intracellular or extracellular origin to CD8+ or CD4+ T cells, respectively.
The classical MHC genes of cynomolgus macaques can be divided into MHC-I and MHC-II genes. The MHC-I genes include mainly -A and -B alleles, and MHC-II genes include mainly -DM, -DO, -DP, -DQ, and -DR alleles. DO is a non-classical class II heterodimer that consists of α and β chains, which are encoded by the DOA and DOB genes located in the MHC class II region. The function of MHC-DO is poorly understood; it may act as a negative regulator by binding to HLA-DM and inhibiting the exchange reaction of class II-associated invariant chain peptides for antigenic peptides (Fernandez-Donoso et al. 1970; O'Sullivan et al. 1997). HLA-DPB1 alleles have been demonstrated to be involved in corneal and renal transplantation, myasthenia gravis, multiple sclerosis, Hodgkin’s disease, Beryllium disease, and sarcoidosis. In addition, an MHC-DPB1 allele was found to be involved in the susceptibility to experimental autoimmune encephalomyelitis in rhesus macaques (Slierendregt et al. 1995a, b). Thus, the primate MHC-DPB1 plays a fundamental and important role in the peptide-binding selectivity of the DP antigen (Uda et al. 2005) and is a significant factor in the humoral response (Krebs et al. 2005). It has been reported that HLA-DQB1 alleles have been associated as an increased risk of developing type 1 diabetes (Todd 1990; Todd 1997; Redondo et al. 2001), celiac disease (Murray et al. 2007), multiple sclerosis (Dyment et al. 1997; Schmidt et al. 2007, and narcolepsy (Kadotani et al. 1998).
It is not easy to elucidate the mechanism of naturally occurring immune protection in human immunodeficiency virus (HIV), and most direct supporting data are available from animal models in non-human primates. The SIVmac virus, which was isolated originally from cynomolgus monkeys, is now used frequently for research on vaccines against acquired immune deficiency syndrome (AIDS; Hu 2005). Many reports show that polymorphism of MHC genes in the cynomolgus monkey affects the results obtained with drugs significantly (Walsh et al. 1996; Mothe et al. 2003; Hao and Nei 2005) and is associated with control of viral diseases (Florese et al. 2008). Cynomolgus macaques from Mauritius may be particularly valuable because more than half of these animals share the MHC class I allele combination Mafa-B*430101, Mafa-B*440101, and Mafa-B*460101. The increased sharing of MHC-I allele in cynomolgus macaques of Mauritian origin may reduce the overall number of animals needed to study cellular immune responses in non-human primates dramatically, while simultaneously reducing the confounding effects of genetic heterogeneity in HIV/AIDS research (Krebs et al. 2005).
The MHC class II molecules of Rhesus macaques (Macaca mulatta) have been studied methodically, especially in animals of Indian origin. In contrast to Rhesus macaques, although alleles of the Mafa II class molecules, including Mafa-DPB1 and Mafa-DQB1, have been identified by several groups (Otting et al. 2002; Doxiadis et al. 2006; Blancher et al. 2006; O'Connor et al. 2007), knowledge is limited of the MHC class II genes of the cynomolgus monkey and their degree of polymorphism. In addition, a more detailed study of the Mafa-DPB1 sequence in three different geographical variants of cynomolgus monkeys from south-east Asia demonstrated that polymorphism of MHC-II genes is influenced by species and geography (Sano et al. 2006). It is very rare that cynomolgus macaques from different regions share MHC-II genes (Krebs et al. 2005).
Haplotype screening, which employs multiple markers rather than single genes, would be meaningful in MHC disease association studies because it is well-known that most of the MHC loci are tightly linked and exhibit very little recombination (Dukkipati et al. 2006). The high-frequency alleles may represent high priority targets for additional characterization of immune function (Wiseman et al. 2009). Therefore, to examine whether the MHC class II genes found in a cohort of Vietnamese cynomolgus macaques are common to for other geographical populations of cynomolgus macaques, the Mafa class II region was characterized in the present study by sequencing of the polymorphic exon 2 of the -DOB, -DPB1, and -DQB1 genes.
Materials and methods
Animals
Whole blood samples from 150 unrelated cynomolgus macaques, originally from Vietnam, were provided generously by South China Primates Research Central. Whole blood samples (3–5 ml) withdrawn from each monkey were collected into EDTA vacuum tubes. All the monkeys were clinically normal with no known diseases.
DNA isolation and sequencing of exon 2 of Mafa-DOB, -DPB1, and -DQB1
Genomic DNA was extracted from EDTA blood samples using an Animal Genomics DNA Mini Preparation Kit (NewProbe, China) as per the manufacturer’s instructions. Sequences of exon 2 regions of Mafa-DOB, -DPB1, and -DQB1 were obtained by direct sequencing of polymerase chain reaction (PCR) products according to the following procedures: the PCR amplification was performed in 50 μL reaction mixtures containing 25 μL of 2× Taq Plus PCR Master Mix, 1 μL (10 pm/μL) of each primer, 1 μL of template DNA, and 22 μL of ddH2O. The sequences of the DOB, DPB1, and DQB1 primers are shown in Table 1. In general, amplification was carried out for 3 min at 94°C, 32 cycles of 30 s at 94°C, 30 s at 60°C, and 1 min at 72°C, ending with 3 min at 72°C. The annealing temperature was adjusted on the basis of the T m of the primers. The PCR products were subjected to agarose gel electrophoresis and ethidium bromide staining for visualization.
Table 1.
Primers used to amplify MHC class II alleles
| Locus | Primer name | Sequence (5′ to 3′) | Tm (°C) | Product (bp) |
|---|---|---|---|---|
| DOB | DOB-F | GTAGCATTATTTCCCTTT | 50 | 600 |
| DOB-R | ACAGACAACCGTTTATCC | |||
| DPB1 | DPB1-F1 | CACAGAACTCGGTACTAGGAAA | 52 | 700 |
| DPB1-R1 | CTCAGGAACCTCAAACCC | |||
| DPB1-F2 | CCTGAGTGGGAAGATTTG | 55.5 | 588 | |
| DPB1-R2 | TCTCTCTGCTCCCATCCT | |||
| DQB1 | DQB1-F1 | CACTGGTGAGCGGGAACT | 52 | 700 |
| DQB1-R1 | GGAGGCAAACGCATAAGG | |||
| DQB1-F2 | CCCGCAGAGGATTTCGTG | 62 | 250 | |
| DQB1-R2 | GGCGACGATGCTCACCTC |
Phylogenetic analysis
The sequences of the exon 2 regions of Mafa-DOB, -DPB1, and -DQB1 obtained in the present study were aligned, and the phylogenetic tree was generated, which was done using the neighbor-joining method (Saitou and Nei 1987) and Mega 4.0 software (Tamura et al. 2007). The bootstrap consensus tree inferred from 1,000 replicates is taken to represent the evolutionary history of the taxa analyzed (Felsenstein 1985). Branches corresponding to partitions reproduced in fewer than 50% of bootstrap replicates are collapsed. The evolutionary distances were computed using the Kimura 2-parameter method (Kimura 1980) and are in the units of the number of base substitutions per site. The rate of variation among sites was modeled with a gamma distribution (shape parameter = 1). New alleles were confirmed by three repeats sequencing. The names of new sequences were derived according to the published guidelines, and the Immuno Polymorphism Database of Major Histocompatibility Complex for Non-Human Primates was searched to avoid the same name(s) being assigned to different alleles (Klein et al. 1990; Robinson et al. 2005).
Results and discussion
Allele frequencies of Mafa-DOB, -DPB1, and -DQB1 in Vietnamese cynomolgus macaques
Limited polymorphism at the MHC-DOB locus and extensive polymorphism at the MHC-DPB1 and -DQB1 loci have been reported in the primates tested until now, and most of the variability is confined to exon 2, which encodes a major part of the peptide-binding site.
To analyze the genetic polymorphism and allelic variation in the MhcMafa-DOB gene, which may affect the efficiency of class II restricted antigen presentation and therefore be involved in the susceptibility to MHC associated diseases, 16 Mafa-DOB alleles were identified in this study, none of which had been described previously in rhesus and/or cynomolgus macaques, by direct sequencing of exon 2 of the MhcMafa-DOB gene using blood samples from 129 randomly sampled cynomolgus macaques. These novel sequences were submitted to GenBank and were assigned by the NHP Nomenclature Committee. Their accession numbers are listed in Table 2. All the new sequences are highlighted in italic and boldface type. The novel allele with the highest frequency among these cynomolgus macaques was Mafa-DOB*010102, and it was found in 33 (25.6%) of the 129 monkeys, followed by Mafa-DOB*010401, Mafa-DOB*010103, and Mafa-DOB*010104, which were detected in 20 (15.5%), 17 (13.2%), and 14 (10.9%) of the monkeys, respectively. Five alleles presented only once in these monkeys (Table 2). Only six allelic variations (HLA-DOB*0101101, -*0101102, -*01012, -*01022, -*0104101, and -*0104102) were identified until now. It has been demonstrated that strong linkage disequilibrium exists between HLA-DOB*01022 and HLA-DRB1*1502, with no linkage disequilibrium between the DOA and the DOB genes (Naruse et al. 2002). A new allelic type of DOB*010103 has been described in the Korean population (Gu et al. 2005). So, limited polymorphism in the DOB gene is profitable in the execution of the unique function of its product as a co-chaperone. Therefore, strong selection pressure operates to prevent generic variation in the DOB molecule in its interaction with the DM molecule thus maintaining the specified immunological function of regulating antigen presentation (Naruse et al. 2002; Lith et al. 2002). Ectopic expression of HLA-DO in mouse dendritic cells diminishes MHC class II antigen presentation (Fallas et al. 2004). It has also been demonstrated that a highly significant upregulation of DOA and DOB mRNA occurs in purified malignant cells, when compared with B cells from healthy donors (Souwer et al. 2009). The increased levels of mRNA were not translated into enhanced protein levels but could reflect aberrant transcriptional regulation, which forms a novel and additional prognostic indicator for survival in B cell chronic lymphocytic leukemia (Souwer et al. 2009).
Table 2.
MhcMafa-DOB alleles identified in 129 randomly sampled cynomolgus macaques
| Sequence no. | Accession no. | Allele name | No. of haplotypes | Gene frequency |
|---|---|---|---|---|
| O1 | HM152983 | Mafa-DOB*010101 | 11 | 0.085 |
| O2 | HM152984 | Mafa-DOB*010102 | 33 | 0.256 |
| O3 | HM152985 | Mafa-DOB*010103 | 17 | 0.132 |
| O4 | HM152986 | Mafa-DOB*010104 | 14 | 0.109 |
| O5 | HM152987 | Mafa-DOB*010401 | 20 | 0.155 |
| O6 | HM152988 | Mafa-DOB*010202 | 12 | 0.093 |
| O7 | HM152989 | Mafa-DOB*010301 | 6 | 0.047 |
| O8 | HM152990 | Mafa-DOB*010302 | 1 | 0.008 |
| O9 | HM152991 | Mafa-DOB*010402 | 4 | 0.031 |
| O10 | HM152992 | Mafa-DOB*010105 | 3 | 0.023 |
| O11 | HM152993 | Mafa-DOB*010201 | 1 | 0.008 |
| O12 | HM152994 | Mafa-DOB*010203 | 2 | 0.016 |
| O13 | HM152995 | Mafa-DOB*010303 | 1 | 0.008 |
| O14 | HM152996 | Mafa-DOB*0105 | 1 | 0.008 |
| O15 | HM152997 | Mafa-DOB*010304 | 2 | 0.016 |
| O16 | HM152998 | Mafa-DOB*010204 | 1 | 0.008 |
By direct sequencing of the second exon of the MhcMafa-DPB1 genes using blood samples from 102 randomly sampled cynomolgus macaques, 16 alleles of MhcMafa-DPB1 were identified in this study, of which 11 were identical to alleles described formerly in cynomolgus macaques whose sequences could be retrieved from GenBank. The other five alleles were not documented in the literatures or databases. These novel sequences were submitted to GenBank and were assigned by the NHP Nomenclature Committee. Their accession numbers are listed in Table 3, in which all the new sequences are highlighted in italic and boldface type. The allele with the highest frequency among these cynomolgus macaques was DPB1*13, which was found in 32 (31.37%) of the 102 monkeys. The next most frequent alleles were Mafa-DPB1*21 and Mafa-DPB1*35, which were detected in 14 (13.72%) and 11 (10.77%) of the monkeys. Four alleles presented only once in these monkeys (Table 3). The frequency of the five novel alleles found in this study was less than 4%.
Table 3.
MhcMafa-DPB alleles identified in 102 randomly sampled cynomolgus macaques
| Sequence no. | Accession no. | Allele name | No. of haplotypes | Gene frequency |
|---|---|---|---|---|
| P1 | HM153018 | Mafa-DPB1*35 | 11 | 0.108 |
| P2 | HM153016 | Mafa-DPB1*20 | 6 | 0.059 |
| P3 | HM153013 | Mafa-DPB1*51 | 2 | 0.020 |
| P4 | HM153019 | Mafa-DPB1*21 | 14 | 0.137 |
| P5 | HM153017 | Mafa-DPB1*19 | 1 | 0.010 |
| P6 | HM153015 | Mafa-DPB1*40 | 9 | 0.088 |
| P7 | HM153014 | Mafa-DPB1*50 | 3 | 0.029 |
| P8 | HM371244 | Mafa-DPB1*52 | 4 | 0.039 |
| P9 | HM371245 | Mafa-DPB1*13 | 32 | 0.314 |
| P10 | HM371246 | Mafa-DPB1*53 | 3 | 0.029 |
| P11 | HM371247 | Mafa-DPB1*54 | 1 | 0.010 |
| P12 | HM371248 | Mafa-DPB1*24 | 6 | 0.059 |
| P13 | HM371249 | Mafa-DPB1*55 | 1 | 0.010 |
| P14 | HM371250 | Mafa-DPB1*44 | 2 | 0.020 |
| P15 | HM371251 | Mafa-DPB1*32 | 1 | 0.010 |
| P16 | HM371252 | Mafa-DPB1*17 | 6 | 0.059 |
It has been shown that the most frequent alleles in Vietnam cynomolgus macaques are Mafa-DPB1*13 and -DPB1*35 (Sano et al. 2006), which supports our results above. In contrast to the result from Sano et al. (Sano et al. 2006), a high frequency of the Mafa-DPB1*21 allele was detected in our study. The Mafa-DPB1*21 allele was also observed in Mauritian cynomolgus macaques, but was not at a high frequency (O'Connor et al. 2007). Like HLA-DPB1, the Mafa-DPB1 gene of the cynomolgus macaque also displays moderate polymorphism, and more than 50 alleles have been documented to date (Slierendregt et al. 1995a, b; Otting et al. 1998; Marsh et al. 2005; O'Connor et al. 2007). In cynomolgus macaques, point mutations might play crucial role in generating DPB1 polymorphism, whereas in humans, much of the variability has been produced by frequent exchange of polymorphic sequence motifs (Zangenberg et al. 1995; Bontrop et al. 1999; Doxiadis et al. 2001).
By direct sequencing of the second exon of MhcMafa-DQB1 genes using blood samples from 143 randomly sampled cynomolgus macaques, 34 MhcMafa-DQB1 alleles were identified in this study, of which 26 were identical to alleles described formerly in cynomolgus macaques whose sequences could be retrieved from GenBank. The other eight alleles were not documented in the literatures or databases. These novel sequences were submitted to GenBank and were assigned by the NHP Nomenclature Committee. Their accession numbers are listed in Table 4, in which all the new sequences are highlighted in italic and boldface type. Most of the sequences (62%) observed in this study belong to DQB1*06 lineages (15 alleles), the second most common (27%) belong to DQB1*18 (9 alleles), and the rest (less than 1%) belong to the DQB1*15, DQB1*16, and DQB1*17 lineages. The allele with the highest frequency among these cynomolgus macaques was Mafa-DQB1*0616, which was found in 30 (20.97%) of the 143 monkeys. Eleven alleles presented only once in these monkeys (Table 4). The frequency of the eight novel alleles found in this study was less than 4%, and most of them (seven of eight) were at less than 2%.
Table 4.
MhcMafa-DQB alleles identified in 143 randomly sampled cynomolgus macaques
| Sequence no. | Accession no. | Allele name | No. of haplotypes | Gene frequency |
|---|---|---|---|---|
| Q1 | HM371224 | Mafa-DQB1*1603 | 1 | 0.007 |
| Q2 | HM371225 | Mafa-DQB1*1503 | 3 | 0.021 |
| Q3 | HM371226 | Mafa-DQB1*1501 | 2 | 0.014 |
| Q4 | HM153006 | Mafa-DQB1*0601 | 4 | 0.028 |
| Q5 | HM153000 | Mafa-DQB1*2401 | 4 | 0.028 |
| Q6 | HM371227 | Mafa-DQB1*1703 | 8 | 0.056 |
| Q7 | HM153008 | Mafa-DQB1*0611 | 2 | 0.014 |
| Q8 | HM371228 | Mafa-DQB1*0622 | 2 | 0.014 |
| Q9 | HM153009 | Mafa-DQB1*1702 | 4 | 0.028 |
| Q10 | HM371229 | Mafa-DQB1*0627 | 1 | 0.007 |
| Q11 | HM371230 | Mafa-DQB1*0623 | 3 | 0.021 |
| Q12 | HM371231 | Mafa-DQB1*1804 | 8 | 0.056 |
| Q13 | HM371232 | Mafa-DQB1*0626 | 7 | 0.049 |
| Q14 | HM153001 | Mafa-DQB1*0619 | 6 | 0.042 |
| Q15 | HM371233 | Mafa-DQB1*0628 | 1 | 0.007 |
| Q16 | HM371234 | Mafa-DQB1*0616 | 30 | 0.210 |
| Q17 | HM371235 | Mafa-DQB1*0614 | 8 | 0.056 |
| Q18 | HM371236 | Mafa-DQB1*0629 | 4 | 0.028 |
| Q19 | HM371237 | Mafa-DQB1*0630 | 6 | 0.042 |
| Q20 | HM371238 | Mafa-DQB1*1818 | 1 | 0.007 |
| Q21 | HM371239 | Mafa-DQB1*1809 | 4 | 0.028 |
| Q22 | HM371240 | Mafa-DQB1*1819 | 1 | 0.007 |
| Q23 | HM371241 | Mafa-DQB1*1817 | 3 | 0.021 |
| Q24 | HM371242 | Mafa-DQB1*1806 | 1 | 0.007 |
| Q25 | HM371243 | Mafa-DQB1*1601 | 1 | 0.007 |
| Q26 | HM152999 | Mafa-DQB1*1816 | 12 | 0.084 |
| Q27 | HM153002 | Mafa-DQB1*0613 | 3 | 0.021 |
| Q28 | HM153003 | Mafa-DQB1*1810 | 4 | 0.028 |
| Q29 | HM153004 | Mafa-DQB1*0610 | 3 | 0.021 |
| Q30 | HM153012 | Mafa-DQB1*170701 | 1 | 0.007 |
| Q31 | HM153005 | Mafa-DQB1*1802 | 2 | 0.014 |
| Q32 | HM153007 | Mafa-DQB1*060702 | 1 | 0.007 |
| Q33 | HM153010 | Mafa-DQB1*170802 | 1 | 0.007 |
| Q34 | HM153011 | Mafa-DQB1*1602 | 1 | 0.007 |
Although the MhcMamu-DQB1*06111 (equivalent to the -DQB1*061101) allele was the most frequent (13%) in 105 randomly sampled Chinese rhesus macaques (Qiu et al. 2008), the allele, which corresponds to MhcMafa-DQB1*0613 in the present study, was at a low frequency (2%) in the 143 monkeys tested. The Mafa-DQB1 polymorphism has been studied earlier (Otting et al. 2002), and only eight of the 34 alleles detected in this study have not been reported previously. Given that the number of different -DQ alleles observed is nearly as high as the number of animals tested, it is likely that cynomolgus macaques display abundant Mafa-DQB1 polymorphism and, when other populations are tested, the levels may reach or even exceed those reported for rhesus macaques (Robinson et al. 2003; Doxiadis et al. 2006).
Amino acid sequences encoded by the MhcMafa-DOB, -DPB1, and -DQB1 genes
Eighty-nine amino acid residues of DOB exon 2 that encode the DO antigen β domain of MHC class II molecules were blasted using 16 Mafa-DOB and six HLA-DOB sequences (Fig. 1). Almost all of these 22 MHC-DOB alleles were conserved except for four different amino acid sequences, located at amino acid positions 50, 58, 68, and 87, respectively. Mafa-DOB*0101 and Mafa-DOB*0104 were the most frequent in the Vietnamese population (Table 2), in which the amino acid positions 58 and 87 were occupied by either Arg and Gly or Ser and Gly (Fig. 1). Of the 89 residue positions, three residues were polymorphic among the Mafa-DOB alleles in contrast to two residues among the HLA-DOB alleles. In addition, two species-specific amino acid residues were identified from the comparison between the Mafa- and HLA-DOB alleles, and of these, one was specific to Mafa-DOB (Fig. 2).
Fig. 1.
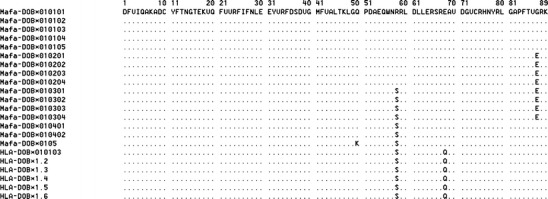
Alignment of the deduced amino acid sequences of the second exon of 16 cynomolgus macaque MhcMafa-DOB alleles and 6 human HLA-DOB alleles
Fig. 2.
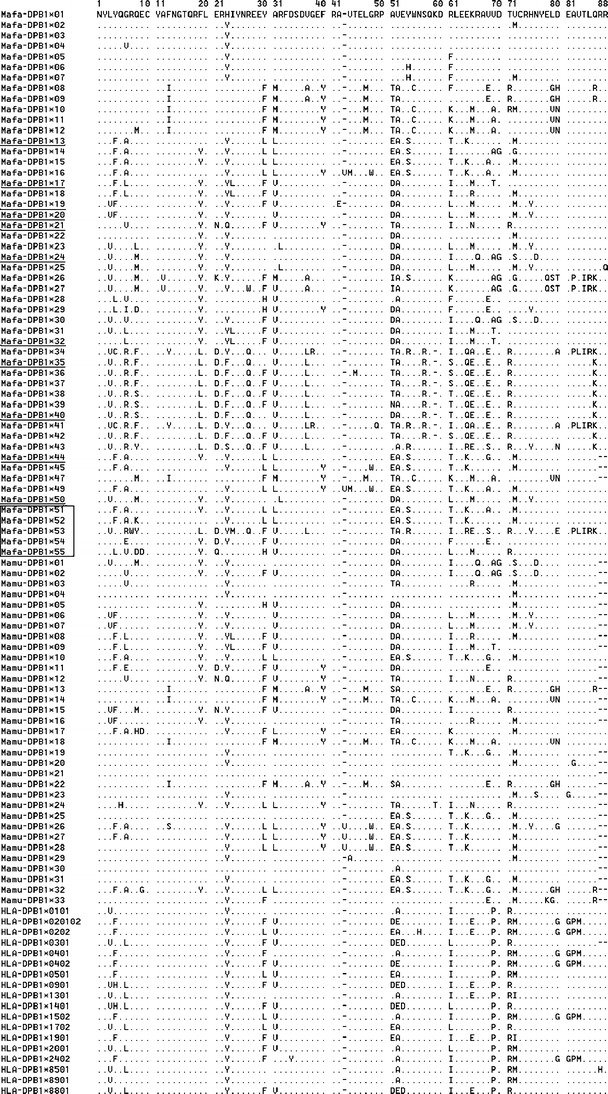
Alignment of the deduced amino acid sequences of the second exon of 52 Mafa-DPB1, 33 Mamu-DPB1, and 18 HLA-DPB1 sequences. Five novel alleles were listed in the middle frame; previously reported alleles also detected in this study were indicated by an underline
Eighty-seven amino acid residues of DPB1 exon 2, which encodes the DP antigen β1 domain, were aligned using 52 Mafa-DPB1, 33 Mamu-DPB1, and 18 HLA-DPB1 sequences (Fig. 2). All 103 MHC-DPB1 alleles had different amino acid sequences. Although all the Mafa-DPB1 alleles were conserved at two cysteine sites (amino acid positions 10 and 72) and did not contain a terminator codon, an amino acid insertion or deletion was seen in nine Mafa-DPB1 alleles (Sano et al. 2006). Mafa-DPB1*16 and Mafa-DPB1*49 contained a methionine residue inserted between amino acid positions 43 and 44, whereas Mafa-DPB1*35, which was the most frequent in the Vietnamese population (Table 2), had a single amino acid residue deleted from position 58 (Fig. 2). Of the 87 residue positions, 54 residues were polymorphic among the Mafa-DPB1 alleles, in contrast to 33 residues among the Mamu-DPB1 alleles (Sano et al. 2006). In addition, 62 species-specific amino acid residues, located at 43 positions, were identified from the comparison between the Mafa- and Mamu-DPB1 alleles, and of these, 59 were specific to Mafa-DPB1 (Fig. 2).
The second exon in the MHC-DQB1 sequences encodes the β1 domain of MHC class II molecules, which contributes a major part to the peptide-binding domain that has a high degree of polymorphism. The amino acid sequence variations of MhcMafa-DQB1 identified so far (48 amino acid sequences) are shown in Fig. 3. The differences between the alleles and the consensus sequences range from 7 to 26 amino acid positions, averaging approximately 16 amino acid positions. There are 47 amino acid positions with codons for more than one amino acid residue, among which are three positions (26:7, 28:5, 57:5) with codons for five to seven amino acid residues. Similarly, there are 47 amino acid positions with codons for more than one amino acid residue, among which are four positions (26:8, 28:6, 57:6, 86:5) with codons for five to eight amino acid residues, in Mamu-MHC-DQB1 (Qiu et al. 2008). Among the amino acid positions classified as participating in pockets, based on HLA-DR and DQ structure (9, 11, 13, 28, 47, 57, 61, 67, 70, 71, 74, 74, 85, 86, 89, and 90; Diaz et al. 2000; Siebold et al. 2004; Ettinger et al. 2006), most are with codons for two to five amino acid residues. Among the eight novel sequences (under the frame in Fig. 3), 36 amino acid positions with variable numbers of codons were found; for the 14 sequences not observed in this study (above the frame in Fig. 3), 34 such positions were found.
Fig. 3.
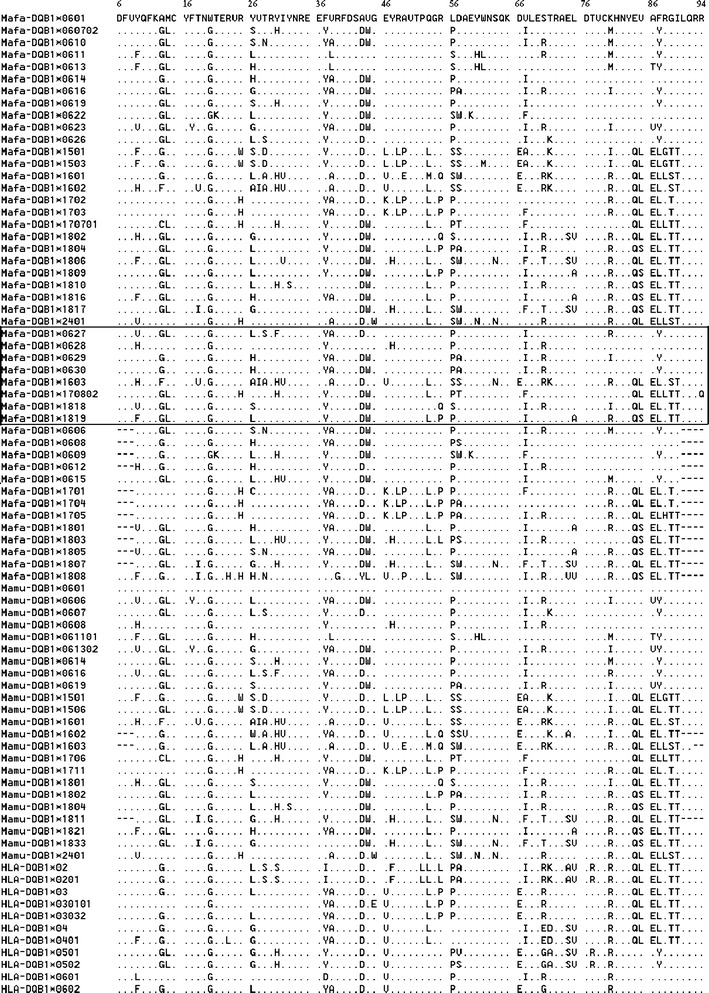
Alignment of the deduced amino acid sequences of the second exon of 47 Mafa-DQB1, 23 Mamu-DQB1, and 11 HLA-DQB1 sequences. Eight novel alleles were listed in the middle frame, 26 sequences common to the earlier studies were listed in the over frame, and other alleles not detected in this study were listed under the frame
Phylogenetic tree of the MhcMafa-DOB, -DPB1, and -DQB1 genes
A phylogenetic tree was created using the novel MhcMafa-DOB gene sequences obtained in this study and those (1 BoLA-DOB, 1 HLA-DOB sequences) retrieved from GenBank. As evident in the phylogenetic tree (Fig. 4), all the novel sequences identified in this study grouped into three DOB allele lineages, including group 1 (Mafa-DOB*0101 and -DOB*0102), group 2 (Mafa-DOB*0103 and -DOB*0104), and one other (Mafa-DOB*0105), whereas the HLA-DOB alleles belonged to a separate group.
Fig. 4.
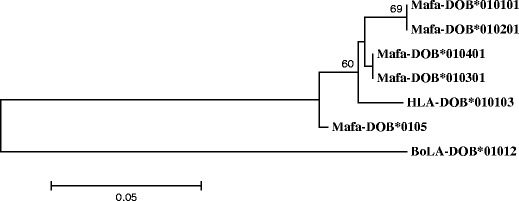
Phylogenetic tree of 5 Mafa-DOB, 1 HLA-DOB, and 1 BoLA-DOB amino acid sequences
A phylogenetic tree constructed from a neighbor-joining analysis of 105 MHC-DPB1 exon 2 sequences using 54 Mafa-DPB1, 33 Mamu-DPB1, and 18 HLA-DPB1 allelic sequences is shown in Fig. 5. The general appearance of the tree is similar to a previously reported tree for the MHC class II polymorphisms in Mafa-DPB1 and other primates (Bontrop et al. 1999; Sano et al. 2006). The 13 Mafa-DPB1 alleles and all HLA-DPB1 alleles were included in the separated group allele clusters. Of the 54 Mafa-DPB1 alleles, 41 alleles were related closely to Mamu-DPB1 alleles, suggesting the possibility of interspecies inheritance. In particular, in our present study Mafa-DPB1*21, -DPB1*18, and -DPB1*24 were matched perfectly with Mamu-DPB1*12, -DPB1*08, and -DPB1*01, respectively, whereas Mafa-DPB1*02, -DPB1*18, -DPB1*20, and -DPB1*24 were matched perfectly with Mamu-DPB1*04, -DPB1*08, -DPB1*06, and -DPB1*01, respectively (Sano et al. 2006).
Fig. 5.
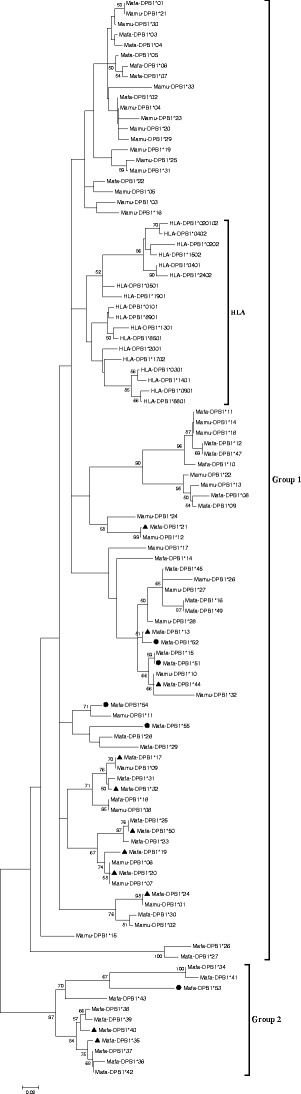
Phylogenetic tree of 52 Mafa-DPB1, 33 Mamu-DPB1, and 18 HLA-DPB1 amino acid sequences. Novel alleles identified in this study were shown with a solid round spot; previously reported alleles also detected in this study were shown with a solid triangle
A phylogenetic tree was created using 34 MhcMafa-DQB1 gene sequences obtained in this study and those (14 Mafa-DQB1, 23 Mamu-DQB1, and 11 HLA-DQB1 sequences) retrieved from GenBank. The relationships among the sequences of exon 2 from this study and those from other studies are shown in Fig. 6. As evident in the phylogenetic tree, novel sequences (shown with a solid round spot) identified in this study grouped into the DQB1 allele lineages DQB1*06 (4), *16 (1), *17 (1), and *18 (2). They tend to cluster with sequences that are common to the earlier studies of Mamu-DQB1 (shown with a solid triangle), rather than clustering with sequences detected only in the earlier reports on Mafa-DQB1 (shown in black), with the exception of DQB1*0629 and DQB1*0630, which clustered together. In addition, the high-frequency DQB1*0616 allele identified in the present study clustered with DQB1*0629 and DQB1*0630. All major MhcMafa-DQB1 lineages that have been reported previously were detected (DQB1*06, *15, *16, *17, *18, *24).
Fig. 6.
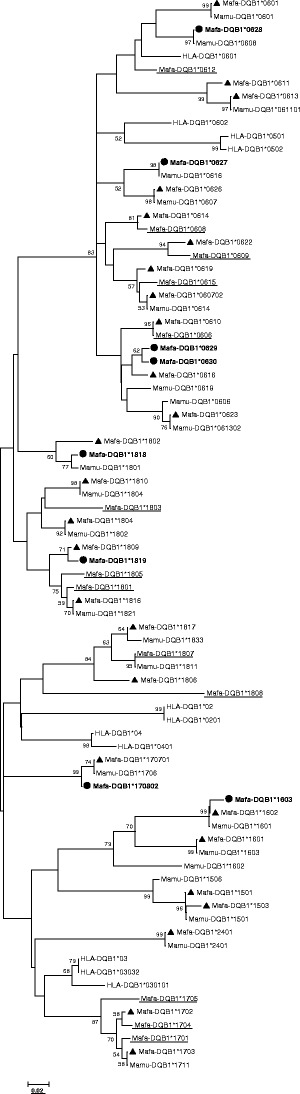
Phylogenetic tree of 47 Mafa-DQB1, 23 Mamu-DQB1, and 11HLA-DQB1 amino acid sequences. Novel alleles identified in this study were shown with a solid round spot and boldface, previously reported alleles also detected in this study were shown with a solid triangle; alleles not detected in this study were indicated by underline
In conclusion, the Mafa-DOB, -DPB1, and -DQB1 alleles detected in this manuscript are mostly specific for a given geographic area, and only a small number of alleles appears to be shared with other populations, providing an important addition to the limited immunogenetic information available for Vietnamese cynomolgus macaques. This suggests the fast evolution of Mafa-DOB, -DPB1, and -DQB1 alleles due to adaptation to new environments. The high-frequency alleles among Vietnamese population, Mafa-DOB*010102, Mafa-DPB1*13, and Mafa-DQB1*0616, may represent high priority targets for additional characterization of immune function. Characterization of shared and unique MHC class II DNA sequences may be vital for disease research and may help better elucidate the biogeography of non-human primates.
Acknowledgment
We thank the Primate Research Center of South China for providing blood samples from animals. We thank Dr. Natasja de Groot, Dr. Nel Otting, and IMGT Non-human Primate Nomenclature Committee for naming the Mafa-DOB, -DPB1, and -DQB1 sequences. We wish to thank Dr. Xiao-hui Liu and David Cushley for editing the manuscript. This project was supported by the National Basic Research Program of China (2006CB701500 and 2007CB512402), the Fundamental Research Funds for the Central Universities of South China University of Technology (2009ZM0105) and Science and Technology Planning Project of Guangdong Province, China (2010B060200007).
Contributor Information
Fei Ling, Email: fling@scut.edu.cn.
Xiao-ning Wang, FAX: +86-020-39380601, Email: xnwang@scut.edu.cn.
References
- Blancher A, Tisseyre P, Dutaur M, Apoil P, Maurer C, Quesniaux V, Raulf F, Bigaud M, Abbal M. Study of cynomolgus monkey (Macaca fascicularis) MhcDRB (Mafa-DRB) polymorphism in two populations. Immunogenetics. 2006;58:269–282. doi: 10.1007/s00251-006-0102-9. [DOI] [PubMed] [Google Scholar]
- Bontrop RE, Otting N, de Groot NG, Doxiadis GG. Major histocompatibility complex class II polymorphisms in primates. Immunol Rev. 1999;167:339–350. doi: 10.1111/j.1600-065X.1999.tb01403.x. [DOI] [PubMed] [Google Scholar]
- Diaz D, Naegeli M, Rodriguez R, Nino-Vasquez JJ, Moreno A, Patarroyo ME, Pluschke G, Daubenberger GA. Sequence and diversity of Mhc DQA and DQB genes of the owl monkey Aotus nancymaae. Immunogenetics. 2000;51:528–537. doi: 10.1007/s002510000189. [DOI] [PubMed] [Google Scholar]
- Doxiadis GG, Otting N, Groot NG, Bontrop RE. Differential evolutionary MHC class II strategies in humans and rhesus macaques: relevance for biomedical studies. Immunol Rev. 2001;183:76–85. doi: 10.1034/j.1600-065x.2001.1830106.x. [DOI] [PubMed] [Google Scholar]
- Doxiadis GG, Rouweler AJ, de Groot NG, Louwerse A, Otting N, Verschoor EJ, Bontrop RE. Extensive sharing of MHC class II alleles between rhesus and cynomolgus macaques. Immunogenetics. 2006;58:259–268. doi: 10.1007/s00251-006-0083-8. [DOI] [PubMed] [Google Scholar]
- Dukkipati VS, Blair HT, Garrick DJ, Murray A. ‘Ovar-Mhc’—ovine major histocompatibility complex: structure and gene polymorphisms. Genet Mol Res. 2006;5:581–608. [PubMed] [Google Scholar]
- Dyment DA, Sadovnick AD, Ebers GC, Sadnovich AD. Genetics of multiple sclerosis. Hum Mol Genet. 1997;6:1693–1698. doi: 10.1093/hmg/6.10.1693. [DOI] [PubMed] [Google Scholar]
- Ettinger RA, Papadopoulos GK, Moustakas AK, Nepom GT, Kwok WW. Allelic variation in key peptide-binding pockets discriminates between closely related diabetes-protective and diabetes-susceptible HLA-DQB1*06 alleles. J Immunol. 2006;176:1988–1998. doi: 10.4049/jimmunol.176.3.1988. [DOI] [PubMed] [Google Scholar]
- Fallas JL, Tobin HM, Lou O, Guo D, Sant'Angelo DB, Denzin LK. Ectopic expression of HLA-DO in mouse dendritic cells diminishes MHC class II antigen presentation. J Immunol. 2004;173:1549–1560. doi: 10.4049/jimmunol.173.3.1549. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- Fernandez-Donoso R, Lindsten J, Norrby E. The chromosomes of the cynomolgus macaque (Macaca fascicularis) Hereditas. 1970;65:269–275. doi: 10.1111/j.1601-5223.1970.tb02325.x. [DOI] [PubMed] [Google Scholar]
- Florese RH, Wiseman RW, Venzon D, Karl JA, Demberg T, Larsen K, Flanary L, Kalyanaraman VS, Pal R, Titti F, Patterson LJ, Heath MJ, O'Connor DH, Cafaro A, Ensoli B, Robert-Guroff M. Comparative study of Tat vaccine regimens in Mauritian cynomolgus and Indian rhesus macaques: influence of Mauritian MHC haplotypeson susceptibility/resistance to SHIV(89.6P)infection. Vaccine. 2008;26:3312–3321. doi: 10.1016/j.vaccine.2008.03.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Lee KJ, Oh B. Identification of a novel HLA-DOB-allele, DOB*010103, by sequence-based typing in the Korean population. Tissue Antigens. 2005;65:287–288. doi: 10.1111/j.1399-0039.2005.00375.x. [DOI] [PubMed] [Google Scholar]
- Hao L, Nei M. Rapid expansion of killer cell immunoglobulin-like receptor genes in primates and their coevolution with MHC class I genes. Gene. 2005;347:149–159. doi: 10.1016/j.gene.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Hu SL. Non-human primate models for AIDS vaccine research. Curr Drug Targets Infect Disord. 2005;5:193–201. doi: 10.2174/1568005054201508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadotani H, Faraco J, Mignot E. Genetic studies in the sleep disorder narcolepsy. Genome Res. 1998;8:427–434. doi: 10.1101/gr.8.5.427. [DOI] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Klein J, Bontrop RE, Dawkins RL, Erlich HA, Gyllensten UB, Heise ER, Jones PP, Parham P, Wakeland EK, Watkins DI. Nomenclature for the major histocompatibility complexes of different species: a proposal. Immunogenetics. 1990;31:217–219. doi: 10.1007/BF00204890. [DOI] [PubMed] [Google Scholar]
- Krebs KC, Jin ZJ, Rudersdorf R, Hughes AL, O’Connor DH. Unusually high frequency MHC class I alleles in Mauritian origin cynomolgus macaques. J Immunol. 2005;175:5230–5239. doi: 10.4049/jimmunol.175.8.5230. [DOI] [PubMed] [Google Scholar]
- Lith M, Ham W, Neefjes J. Novel polymorphisms in HLA-DOA and HLA-DOB in B-cell malignancies. Immunogenetics. 2002;54:591–595. doi: 10.1007/s00251-002-0500-6. [DOI] [PubMed] [Google Scholar]
- Menninger K, Wieczorek G, Riesen S, Kunkler A, Audet M, Blancher A, Schuurman HJ, Quesniaux V, Bigaud M. The origin of cynomolgus monkey affects the outcome of kidney allografts under Neoral immunosuppression. Transplant Proc. 2002;34:2887–2888. doi: 10.1016/S0041-1345(02)03547-9. [DOI] [PubMed] [Google Scholar]
- Marsh SG, Albert ED, Bodmer WF, Bontrop RE, Dupont B, Erlich HA, Geraghty DE, Hansen JA, Hurley CK, Mach B, Mayr WR, Parham P, Petersdorf EW, Sasazuki T, Schreuder GM, Strominger JL, Svejgaard A, Terasaki PI, Trowsdale J. Nomenclature for factors of the HLA system, 2004. Hum Immunol. 2005;66:571–636. doi: 10.1016/j.humimm.2005.02.002. [DOI] [PubMed] [Google Scholar]
- McAuliffe J, Vogel L, Roberts A, Fahle G, Fischer S, Shieh WJ, Butler E, Zaki S, St Claire M, Murphy B, Subbarao K. Replication of SARS coronavirus administered into the respiratory tract of African Green, rhesus and cynomolgus monkeys. Virology. 2004;330:8–15. doi: 10.1016/j.virol.2004.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mothe BR, Weinfurter J, Wang CX, Rehrauer W, Wilson N, Allen TM, Allison DB, Watkins DI. Expression of the major histocompatibility complex class I molecule Mamu-A*01 is associated with control of simian immunodeficiency virus SIVmac239 replication. J Virol. 2003;77:2736–2740. doi: 10.1128/JVI.77.4.2736-2740.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JA, Moore SB, Van Dyke CT, Lahr BD, Dierkhising RA, Zinsmeister AR, Melton LJ, 3rd, Kroning CM, El-Yousseff M, Czaja AJ. HLA DQ gene dosage and risk and severity of celiac disease. Clin Gastroenterol Hepatol. 2007;5:1406–1412. doi: 10.1016/j.cgh.2007.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naruse TK, Kawata H, Inoko H, Isshiki G, Yamano K, Hino M, Tatsumi N. The HLA-DOB gene displays limited polymorphism with only one amino acid substitution. Tissue Antigens. 2002;59:512–519. doi: 10.1034/j.1399-0039.2002.590608.x. [DOI] [PubMed] [Google Scholar]
- O'Connor SL, Blasky AJ, Pendley CJ, Becker EA, Wiseman RW, Karl JA, Hughes AL, O’Connor DH. Comprehensive characterization of MHC class II haplotypes in Mauritian cynomolgus macaques. Immunogenetics. 2007;59:449–462. doi: 10.1007/s00251-007-0209-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan MG, Anderson DK, Goodrich JA, Tulli H, Green SW, Young NS, Brown KE. Experimental infection of cynomolgus monkeys with simian parvovirus. J Virol. 1997;71:4517–4521. doi: 10.1128/jvi.71.6.4517-4521.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otting N, de Groot NG, Doxiadis GG, Bontrop RE. Extensive Mhc-DQB variation in humans and non-human primate species. Immunogenetics. 2002;54:230–239. doi: 10.1007/s00251-002-0461-9. [DOI] [PubMed] [Google Scholar]
- Otting N, Doxiadis GG, Versluis L, Versluisb L, Groota N, Anholtsc J, Verduinc W, Rozemullerb E, Claasc F, Tilanusb MJ, Bontropa R. Characterization and distribution of Mhc-DPB1 alleles in chimpanzee and rhesus macaque populations. Hum Immunol. 1998;59:656–664. doi: 10.1016/S0198-8859(98)00070-6. [DOI] [PubMed] [Google Scholar]
- Qiu CL, Zhao H, Yang GB, Liu Q, Shao Y. Flow cytometric characterization of T lymphocyte subsets in the peripheral blood of Chinese rhesus macaques: normal range, age- and sex-related differences. Vet Immunol Immunopathol. 2008;124:313–321. doi: 10.1016/j.vetimm.2008.04.005. [DOI] [PubMed] [Google Scholar]
- Reed SG, Coler RN, Dalemans W, Tan EV, DeLa Cruz EC, Basaraba RJ, Orme IM, Skeiky YA, Alderson MR, Cowgill KD, Prieels JP, Abalos RM, Dubois MC, Cohen J, Mettens P, Lobet Y. Defined tuberculosis vaccine, Mtb72F/AS02A, evidence of protection in cynomolgus monkeys. Proc Natl Acad Sci USA. 2009;106:2301–2306. doi: 10.1073/pnas.0712077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redondo MJ, Fain PR, Eisenbarth GS. Genetics of type 1A diabetes. Recent Prog Horm Res. 2001;56:69–89. doi: 10.1210/rp.56.1.69. [DOI] [PubMed] [Google Scholar]
- Robinson J, Waller MJ, Parham P, de Groot N, Bontrop R, Kennedy LJ, Stoehr P, Marsh SG. IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex. Nucleic Acids Res. 2003;31:311–314. doi: 10.1093/nar/gkg070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J, Waller MJ, Stoehr P, Marsh SG. IPD—the immunopolymorphism database. Nucleic Acids Res. 2005;33:D523. doi: 10.1093/nar/gki032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sano K, Shiina T, Kohara S. Novel cynomolgus macaque MHC-DPB1 polymorphisms inthree South-East Asian populations. Tissue Antigens. 2006;67:297–306. doi: 10.1111/j.1399-0039.2006.00577.x. [DOI] [PubMed] [Google Scholar]
- Schmidt H, Williamson D, Ashley-Koch A. HLA-DR15 haplotype and multiple sclerosis: a HuGE review. Am J Epidemiol. 2007;165:1097–1109. doi: 10.1093/aje/kwk118. [DOI] [PubMed] [Google Scholar]
- Siebold C, Hansen BE, Wyer JR, Harlos K, Esnouf RE, Svejgaard A, Bell JI, Strominger JL, Jones EY, Fugger L. Crystal structure of HLA-DQ0602 that protects against type 1 diabetes and confers strong susceptibility to narcolepsy. Proc Natl Acad Sci USA. 2004;101:1999–2004. doi: 10.1073/pnas.0308458100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slierendregt BL, Otting N, Kenter M, Bontrop RE. Allelic diversity at the Mhc-DP locus in rhesus macaques (Macaca mulatta) Immunogenetics. 1995;41:29–37. doi: 10.1007/BF00188429. [DOI] [PubMed] [Google Scholar]
- Slierendregt BL, Hall M, Bert’t H, Otting N, Anholts J, Verduin W, Claas F, Jonker M, Lanchbury JS, Bontrop RE. Identification of an Mhc-DPB1 allele involved in susceptibility to experimental autoimmune encephalomyelitis in rhesus macaques. Int Immunol. 1995;7:1671–1679. doi: 10.1093/intimm/7.10.1671. [DOI] [PubMed] [Google Scholar]
- Souwer Y, Chamuleau ME, van de Loosdrecht AA, Tolosa E, Jorritsma T, Muris JJ, Dinnissen-van Poppel MJ, Snel SN, van de Corput L, Ossenkoppele GJ, Meijer CJ, Neefjes JJ, Marieke van Ham S. Detection of aberrant transcription of major histocompatibility complex class II antigen presentation genes in chronic lymphocytic leukaemia identifies HLA-DOA mRNA as a prognostic factor for survival. Br J Haematol. 2009;145:334–343. doi: 10.1111/j.1365-2141.2009.07625.x. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Todd JA. Genetic control of autoimmunity in type 1 diabetes. Immunol Today. 1990;11:122–129. doi: 10.1016/0167-5699(90)90049-F. [DOI] [PubMed] [Google Scholar]
- Todd JA. Genetics of type 1 diabetes. Pathol Biol. 1997;45:219–227. [PubMed] [Google Scholar]
- Uda A, Terao K, Tanabayashi K, Fujita O, Hotta A, Terao K, Yamada A. Identification of the MHC class I B locus in cynomolgus monkeys. Immunogenetics. 2005;57:189–197. doi: 10.1007/s00251-005-0782-6. [DOI] [PubMed] [Google Scholar]
- Walsh GP, Tan EV, Dela Cruz EC, Abalos RM, Villahermosa LG, Young LJ, Cellona RV, Narareno JB, Horwitz MA. The Philippine cynomolgus monkey (Macaca fasicularis) provides a new nonhuman primate model of tuberculosis that resembles human disease. Nat Med. 1996;2:430–436. doi: 10.1038/nm0496-430. [DOI] [PubMed] [Google Scholar]
- Wiseman RW, Karl JA, Bimber BN, O'Leary CE, Lank SM, Tuscher JJ, Detmer AM, Bouffard P, Levenkova N, Turcotte CL, Jr, ES WC, Harkins T, O'Connor DH. Major histocompatibility complex genotyping with massively parallel pyrosequencing. Nat Med. 2009;15:1322–1326. doi: 10.1038/nm.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zangenberg G, Huang MM, Arnheim N, Erlich H. New HLA-DPBI alleles generated by interallelic gene conversion detected by analysis of sperm. Nat Genet. 1995;10:407–414. doi: 10.1038/ng0895-407. [DOI] [PubMed] [Google Scholar]


