Abstract
The first case of Middle East respiratory syndrome coronavirus (MERS-CoV) was identified in the year 2012, which spread rapidly and increased to more than 2200 in 2018. This highly pathogenic virus with high mortality rate is among one of the major public health concerns. Saudi Arabia remains to be the most affected region with the majority of MERS-CoV cases, and currently, no effective drugs and vaccines are available for prevention and treatment. A large amount of information is now available regarding the virus, its structure, route of transmission and its pathophysiology. Therefore, this review summarizes the current understanding of MERS-CoV's pathogenesis, treatment options and recent scientific advancements in vaccine and other therapeutic developments, and the major steps taken for MERS prevention control.
Keywords: : Arabian Peninsula, coronavirus, global, macrophages, MERS, SARS, Saudi Arabia, therapeutic, vaccine, WHO
Middle East respiratory syndrome coronavirus (MERS-CoV) was identified as a zoonotic virus, whose mode of transmission is from animals to humans. The origin of the virus is believed to be bats, from which it was then transferred to camels. Camels are currently regarded as a major host for MERS-CoV. It has also been identified as the most significant source for human infections [1]. The isolates of MERS-CoV from camels and humans have also been found to more than 99% identical [2]. While camels have been identified as the primary source of MERS-CoV infection through both indirect as well as direct contact, the specific route and role of camels in disease transmission is yet to be identified.
The pathogenic agent of Middle East respiratory syndrome is a new coronavirus which was initially identified from the respiratory content of a patient who was infected, and died, as a result of infection from a mysterious viral disease showing pneumonia like symptoms in Saudi Arabia in 2012 [3]. Initially, a group of healthcare personnel working in a hospital in Jordan contracted a respiratory infection in April 2012, the source of which was not known [4]. Later in June 2012, an elderly businessman with severe pneumonia associated with kidney failure was admitted to a Saudi hospital. The coronavirus detected from his sputum was not known before and for a while it was referred to as human Coronavirus Erasmus Medical Center (hCoV-EMC) [5]. Following that case, another patient from the Middle East with severe respiratory infection, who was transferred to the UK, was found to be infected with the same virus. On analyzing and comparing the samples from the Jordan outbreak with the latter ones, the same virus hCoV-EMC was recognized as causing this new and severe form of respiratory infection. It was later titled as MERS-CoV [6]. Since its first detection to this date, this virus has spread in countries across Middle East and Europe [7]. The highest number of cases has been identified in Saudi Arabia. Infections in European countries have been brought in by travel from the Middle East [7,8].
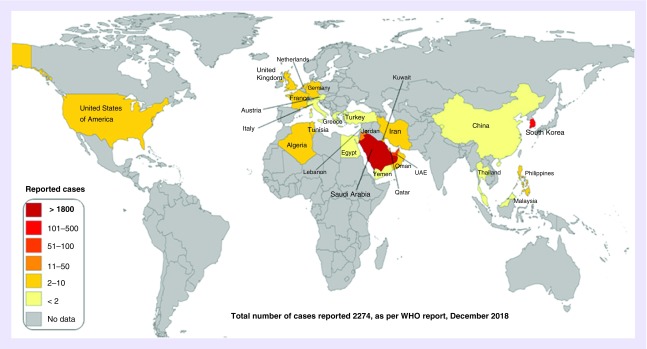
From 2012 through the end of December 2018, the number of confirmed cases of MERS-CoV globally reported to the WHO was 2279 with 806 associated deaths, which corresponds to a fatality rate of approximately 35.36%. Saudi Arabia was on the top of the list of countries with 1901 reports of confirmed cases, including 732 related deaths with a fatality rate of approximately 39% [9]. Due to its high mortality rate (∼36%) [9,10] and pathogenicity, nonavailability of vaccine or any other definite treatment, MERS-CoV is considered a major challenge to global health and presents a pressing need for the research and development of definite therapeutic options and adequate management to prevent its infection [11,12].
The interest in coronaviruses was reignited after the 2002 outbreak of Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) and 2012 outbreak of MERS-CoV. Researchers have tried to understand pandemic potential of MERS-CoV by studying its emergence and ecology. Studies are being carrying out to assess how this virus causes disease so that novel therapeutics and vaccines can be developed. The mode of transmission of MERS-CoV is yet to be established, but it is assumed to have come from bats [13]. Camels found in the Arabian Peninsula may serve as intermediate hosts for human infection [14].
This review will focus on studies trying to elucidate mechanisms by which MERS-CoV escapes host-immune system and causes disease. We will also examine the recent advances in the development of novel therapeutics and vaccines.
Figure 1. . Global map of confirmed MERS-CoV infections, 2012–2018.
Figure modified from [15].
Characteristics of coronavirus & major proteins
Coronaviruses are positive stranded RNA viruses, and while most of these infect animals, particularly bats, a minor number can also cause human diseases [16]. Human coronaviruses can be broadly divided into types, α and β coronavirus [17]. MERS-CoV is from the β-coronavirus family [18]. It has four major surface proteins that help the virus to enter the cells viz. envelope protein (E), spike (S) protein, nucleocapsid (N) protein and membrane (M) protein. The spike (S) protein is a transmembrane glycoprotein made up of S1 and S2 subunits. The S protein is crucial for virus entry through binding and fusion to host cells. The S1 subunit has a receptor binding domain (RBD) that binds with the DPP4 receptor of the host [18,19]. The S2 subunit contains heptad repeats H1 and H2 which forms the main membrane fusion unit [20]. The E protein is required for assembly, intracellular transport and budding of virus [21]. The M protein has its role in viral morphogenesis and assembly [22]. N proteins and S, E and M proteins interact to form complete virus particles [23]. In addition to these structural proteins, MERS-CoV has two large polyproteins called pp1a and pp1ab. These proteins are broken down by proteases to form various nonstructural essential proteins, such as enzymes [24,25]. Recent studies have revealed that these viral structural and nonstructural proteins can be exploited as novel targets for therapeutic purposes [26–28].
Clinical presentation
The infection caused by MERS-CoV has an average incubation time of 5 days (2–14 days range). The host, in this time period, shows no symptoms of infection. The clinical symptoms of the disease range from mild symptoms of upper respiratory infection like cough, fever and myalgia to severe forms such as pneumonitis, as well as respiratory failure. Patients may also suffer from abdominal pain, appetite loss, nausea, diarrhea, vomiting and other gastrointestinal symptoms. The less commonly occurring symptoms include hemoptysis and diarrhea without any hint of fever [29]. Studies advocate that chronic illnesses, such as chronic heart disease, kidney disease, diabetes and hypertension, increase the risk of MERS-CoV infection and its severity [30], though further proof is required.
MERS-CoV can alter antigen presentation, host immune response and modulate the apoptotic pathways and mitogen-activated protein kinase pathways [7].
Pathogenesis
Before the discovery of MERS-CoV, SARS-CoV was considered the most pathogenic coronavirus. However, the higher pathogenicity of MERS-CoV was apparent by the higher number of deaths caused by this virus. Similar to the virus of SARS, MERS-CoV infects and replicates in the human airway epithelial cells and suppresses the production of interferons [31,32]. However, unlike SARS-CoV, the MERS virus exhibits wider tissue tropism [33,34]. MERS-CoV can also induce pro-inflammatory cytokines but lacks in production of innate antiviral cytokines compared with SARS-CoV. Suggesting MERS-CoV induces delayed pro-inflammatory response and attenuates innate immunity, which suggests that MERS-CoV is more lethal compared with SARS-CoV [35–37]. Primarily the MERS virus interacts with the host DPP4 receptor through its spike (S) protein after entering the respiratory tract. DPP4 receptors are present on the epithelial surface of various human organs such as, the lungs, kidneys, liver, bone marrow, thymus and intestines [38]. The systemic distribution of DPP4 facilitates the dissemination of virus in the human body. Expression of DPP4 on the respiratory tract is mainly on type I and type II pneumocytes, endothelial cells, nonciliated bronchial epithelial cells and a few forms of hematopoietic cells [39,40]. The abundance of the receptor DPP4 is greater on the epithelial cells lining the lower airways and alveoli and lesser on the epithelial surface of upper conducting airways and nasal cavity [41]. Recent findings have suggested that a prior existing pulmonary ailment might increase the chances of such individuals contracting MERS, as chronic pulmonary diseases results in enhanced DPP4 expression [40].
CoV nsp1 is a serious virulence factor, which facilitates the biological actions of MERS-CoV. Studying the nsp1 can advance our understanding of pathogenicity of MERS-CoV and facilitate development of better therapeutics. Host gene expression in infected cells is suppressed by MERS-CoV nsp1, which also promotes virus assembly or budding in in vitro, leading to efficient virus replication, suggesting nsp1 is also critical for MERS-CoV replication and promotes production of virus particles in the host [42]. The severity of MERS-CoV infection is relatively more in patients with co-morbid conditions, such as chronic lung disease, renal failure, diabetes and others with compromised immune systems [9].
The understanding of MERS-CoV pathogenesis has been limited due to nonavailability of patient autopsy or pathological samples from the patients. Our understanding of the disease pathogenesis is based entirely on in vitro studies. Studies were conducted on animal models and human lung cell lines, and the account from a single autopsy [43]. In vitro studies revealed that MERS-CoV can easily replicate in several cultured human cells, as well as, in the differentiated and nondifferentiated human epithelial cells [34,44]. The antigens of MERS-CoV were found on the macrophages in the alveoli of the infected human lung explants, ciliated and nonciliated bronchial epithelial cells, endothelial cells and pneumocytes [33,45]. These findings were in sync with the findings made in the single autopsy where MERS-CoV antigen was detected on pneumocytes, endothelial cells and epithelial cells of the airways and few on macrophages [43,45].
Macrophages are the important phagocytic cells of innate immune system which help remove the pathogenic substances from the body and present their antigens to the T cells. The cytokines as well as chemokines produced by the macrophages help in destroying the pathogens, adjusting the immune system and maintaining tissue homoeostasis [46]. However, in MERS, the virus-infected macrophages contribute considerably to the development of disease symptoms [47]. The infection with MERS-CoV of human epithelial cells induces the release of pro-inflammatory chemokines and cytokines from the monocyte-derived macrophages. It is believed that these chemokines/cytokines cause inflammatory changes and tissue injury through infiltration of immune cells in the lower respiratory tract [47]. The patients suffering from MERS clinically manifest pneumonia, which is progressive in nature and has a large number of macrophages and neutrophils found in the fluids present in the lungs [3,48]. Studies conducted on rhesus macaques have shown the lung tissue infiltration of macrophages and neutrophils on MERS-CoV infection, though their respiratory symptoms were milder as compared with humans [49]. Many scientists believe that this sequestration of immune cells contribute to the development of lymphopenia seen in patients with MERS. The progress in the severity of pneumonia and respiratory dysfunction in the MERS patients is also attributed to cytokine/chemokine induction [30,50]. Zhou et al. found that MERS-CoV can efficiently replicate inside macrophages and, hence, can overcome the host immune system. [47]. Thus, these phagocytes act like reservoirs and means of transportation for these viruses, helping to replicate and disseminate, such as the HIV virus [51].
Infection of epithelial cells with the MERS virus induce slow, but significant, IFN type I and II responses [36]. Macrophages release pro-inflammatory chemokines and cytokines such as IL-1β, IL-6 and IL-8 upon MERS-CoV infection [37]. Similarly, MERS-CoV infection in blood monocyte-derived macrophages and dendritic cells leads to the release of chemokines and cytokines, for example, IL-2, IL-3, CCL-2, CCL-3 and RANTES [37,48]. Infection of activated T cells induces apoptosis via different pathways which may also explain the occurrence of lymphopenia [52]. With the present available knowledge, it is difficult to describe the exact pathogenesis of MERS-CoV, but it seems that viral replication in the macrophages results in extreme cytotoxicity and triggers the induction of pro-inflammatory chemicals which may lead to MERS-associated complications.
A small number of epithelial, pneumocytes, lymphoid aggregates and inflammatory cells of submucosal glands were positive for MERS spike protein in nonhuman primates (NHPs). Whereas, MERS spike antigen were positive in epithelial layers of submucosal bronchial glands, lungs of NHPs and in some cells in BALTs [53]. Dual Immunohistochemistry method was applied using monoclonal antibody against MERS spike protein and CD26, the staining showed that MERS was found in CD26 positive cells but were negative in NHPs [53].
Two NHPs, the common marmoset and rhesus macaque model were established for MERS-CoV infection by three different research groups [49,54–59]. The clinical symptoms included respiratory disease which was mild in nature and could be diagnosed with radio imaging and computed tomography [54,56,59] and respiratory disease of severe nature showing fatal clinical symptoms requiring euthanasia [54,59]. Exposure to the MERS-0 infectious clone (icMERS-0) strain in Rhesus monkeys is reported to cause respiratory disease, wherein virus antigen has been detected. Respiratory disease in such cases was found to be transient and mild in nature, which resolved by 30 days from infection. Pulmonary disease was also found to be mild upon MERS-CoV infection in NHPs. Earlier studies found ocular, intratracheal or intranasal exposure of MERS-EMC isolate to cause lethal disease. However, it has been argued that lethality was due to manipulations of marmoset, which have higher sensitivity than macaque species [54,57]. Cases of mild to moderate symptoms even after higher viral titer have also been reported in other studies [56]. Small animals, such as hamsters and mice, have been found to be resistant to MERS-CoV infection and development of models of severe respiratory disease has been a challenge [60].
Therapeutics against MERS-CoV infection
The identification of highly pathogenic MERS-CoV suggests that coronaviruses present an incessant and long-term hazard to humans. Development of effective therapeutic and prophylactic agents to contain their infections is an urgent need, and yet currently no antiviral treatments against MERS-CoV are available. MERS-CoV is known to interact with host cell surface receptor DPP4 or CD26 with its spike S protein, which subsequently leads to its entry in the host cell [61]. Our knowledge regarding the exact mechanisms that follow thereafter is still limited and requires more research. Due to lack of definite treatment, supportive therapy remains the only solution. Present development consists of the previous experiences of other coronavirus diseases like SARS-CoV, which have been studied in in vitro and in vivo models. Tremendous efforts are being made in this direction and various antiviral and related therapies have been identified. Use of humanized monoclonal antibodies, convalescent plasma and therapeutic peptides has been shown efficacious. Repurposing of the currently available drugs is also being tried to extend their efficacy against MERS-CoV. Some potential options are discussed in detail below.
Repurposing of drugs
The concept of using clinically available drugs to treat new diseases is known as repurposing. In this method, a new target profile is created for the existing drugs through screening of large molecular databases. Owing to advances in computational approaches and development of effective antiviral agents, repurposing has become faster. Using high-throughput screening, researchers have been able to screen large libraries of drugs against novel targets and evaluate their antiviral activity in vitro [62,63]. Some of repurposed drugs have shown potent antiviral activity against MERS-CoV, for example, ribavirin, nitazoxanide and hexachloropene [64]. Some other studies have repurposed the drugs by using a combination of two or more existing drugs. Wilde et al. combined alisporivir with ribavirin for enhanced antiviral activity against MERS and SARs-CoV [65]. In another study on MERS-CoV infected marmosets, hybrid of ritonavir/lopinavir and IFN-β1b had positive effect [66].
Convalescent plasma
Convalescent plasma therapy utilizes plasma or whole blood from people who have been infected with viral diseases and recovered. This therapy has been used during outbreaks when no particular medicines or vaccines are available for treatment [67]. The use of convalescent plasma has been indicated to be an efficient therapeutic strategy for diseases like MERS-CoV [68]. Studies have been carried out to confirm its feasibility and safety in treating MERS; however, the data are insufficient. There are a few drawbacks of this therapy, as often large-scale screening is required to obtain sufficient amount of antibodies from potential donors [69].
Monoclonal & polyclonal antibodies
Monoclonal antibodies have been commonly used in the diagnosis of various diseases. Therapeutics based on monoclonal antibodies have been used successfully in the therapy of various diseases [70,71]. The potential of this approach was acknowledged against coronaviruses during the SARS outbreak [72,73]. And when the other deadly coronavirus attacked humans, this previous knowledge helped greatly in the improving the response against the risk of MERS-CoV.
Initial studies suggested that MERS-CoV RBD domain in S1 glycoprotein represents a suitable target for the development of neutralizing monoclonal antibodies [62,74–76]. Hence, the idea to develop neutralizing mouse mAbs as a strategy to prevent the entry of MERS-CoV into host cells was led by Du et al. [75]. mAbs were made by vaccinating mice with IgG1 Fc to which recombinant MERS-CoV S1 was fused [75]. Consequently, Mersmab1, the most effective murine mAb was developed that targeted MERS-CoV RBD, and successfully neutralized MERS-CoV infection in Vero E6, Huh-7 and Calu-3 cells [76]. These studies indicated the potential of humanized mAbs as efficient curative agents against infection caused by MERS-CoV. Around April 2014, three independent studies first reported to have developed complete humanized MERS-CoV neutralizing mAbs [77,78]. All these humanized mAbs specifically targeted the MERS-CoV RBD glycoprotein. The efficacy of human mAbs against MERS-CoV infection was first exhibited by Qiu et al. [79]. They were able to completely treat MERS-CoV infection of lethal nature with a single dose of humanized mAbs in the hDPP4 transgenic mouse [79].
Development of humanized monoclonal antibodies against emerging viral diseases requires a significant production cost and poses as a major drawback. To overcome this obstacle and to reduce the cost, extremely powerful neutralizing mAbs which can be given in smaller doses without compromising the efficacy is needed. One strategy to achieve this could be to design humanized mAbs with exceptional binding attraction and targeting them against the potential targets, such as RBD.
Peptides as potential antiviral therapeutic agents
Recently, interest has been generated in the development of peptide therapeutics as potential drug targets for different pathogens. Currently, more than 140 peptide therapeutics are lined up for clinical evaluation presenting peptide research as a component of pharmaceutical research [80]. As compared with chemical drugs, peptide drugs show high specificity toward the target as well as little side effect and drug tolerance [19]. Many peptide therapeutics have shown positive results against viral infections, for instance RVFV-6 peptide against rift valley fever virus and Kn2-7 peptide derived from scorpion venom against HIV-1 [81–83]. Antimicrobial peptides are cited as potential novel antiviral therapeutics against coronaviruses [84]. Antimicrobial peptides are produced by host immune system upon initial exposure to pathogens. These are gene encoded and positively charged peptides that are selectively toxic against their targets [85,86]. This selectivity is due to their positive charge which attacks the negatively charged membrane bilayer of microbes [87]. The antimicrobial peptides block the receptors present on the host cell's surface, which in turn inhibit different steps of viral fusion and replication causing virolysis and activation of host's adaptive immune response [88].
Future perspective
Since the emergence of MERS-CoV, research has greatly enhanced our knowledge of the pathogenesis caused by it and other contemporary coronavirus, such as SARS-CoV. The efforts toward development of vaccines against this deadly virus have also been continuously increasing leading to the emergence of promising interventions. The two viruses, in other words, SARS and MERS have some common challenges in the development of an efficacious vaccine. As evident from reports, the aged population is more vulnerable to MERS-CoV. The lesson learnt from preclinical studies of SARS-CoV suggests that vaccines fail to protect aged animals, while being effective in young ones. Similarly, in clinical settings, the risk of mortality is even higher in individuals with chronic conditions or an immunocompromised state. An effective vaccine, thus, should offer universal protection, including the vulnerable populations. To do so, there is a need to evaluate the promising vaccines in the comorbid chronic conditions and the immunocompromised rodent models. Since the virus can comfortably replicate in macrophages, the risk of vaccine-derived immunopathology cannot be negated and must be considered using heterologous challenge models. Further, balance must be struck between protection and excessive immune activation while evaluating a successful vaccine candidate. The vaccine development against MERS-CoV is mostly influenced by the SARS-CoV. However, to aid the development of better vaccines, there is need to further enhance the knowledge of pathology of MERS-CoV and outline critical differences between SARS and MERS-CoV. mAbs offers a window of opportunity, as a few candidates have shown potency in in vitro testing. However, care needs to be taken in humanizing mAbs so as to minimize the antimouse antibody response. Epitopes identified from mouse neutralizing mAbs can be neutralized for humanizing MERS-CoV. mAbs targeting RBD are reported to have higher potency than therapies directed against other S protein regions of MERS-CoV, as these could recognize critical residues for DPP4 binding. However, changes in such critical residues may render these mAbs ineffective and lead to development of escape mutant strains of the virus. As discussed, a few promising peptides have also been developed against MERS-CoV. The antiviral activity, stability and solubility of these peptides can be further improved similar to the peptides developed against HIV. This will lead to the development of optimized next generation peptides having better inhibitory action directed against MERS-CoV. Alternatively, as a novel approach, the peptide inhibitors can be combined with mAbs (e.g., RBD specific). The combination can be evaluated for the synergistic effect against divergent and resistant strains. Studies are also needed to explore novel delivery technologies and optimizing vaccine immunity with a suitable combination of adjuvants. In summary, there have been encouraging results in the area of the development of MERS-CoV vaccines in preclinical settings. However, there are challenges related to efficacy, safety and drug delivery that need further consideration before proceeding to clinical trials. Focused research in this direction can help reduce the disease burden caused by MERS-CoV and prevent outbreaks, especially among the aged and immunocompromised population.
Footnotes
Financial & competing interests disclosure
This work was supported by King Abdulaziz City for Science and Technology and Deanship of Scientific Research, King Abdulaziz University. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Medical writing support was provided by Scidra Consulting, INC and was funded by the authors.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Zhou J, Li C, Zhao G, et al. Human intestinal tract serves as an alternative infection route for Middle East respiratory syndrome coronavirus. Sci. Adv. 2017;3(11):eaao4966. doi: 10.1126/sciadv.aao4966. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Direct intragastric inoculation of Middle East respiratory syndrome coronavirus (MERS-CoV) caused a lethal infection in human dipeptidyl peptidase 4 transgenic mice.
- 2.Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17(3):181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Severe acute respiratory syndrome coronavirus (SARS-CoV) and MERS-CoV are two highly transmissible and pathogenic viruses that emerged in humans at the beginning of the 21st century.
- 3.Zaki AM, Van Boheemen S, Bestebroer TM, et al. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367(9):1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 4.Al-Abdallat MM, Payne DC, Alqasrawi S, et al. Hospital-associated outbreak of Middle East respiratory syndrome coronavirus: a serologic, epidemiologic, and clinical description. Clin. Infect. Dis. 2014;59(9):1225–1233. doi: 10.1093/cid/ciu359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shapiro M, London B, Nigri D, et al. Middle east respiratory syndrome coronavirus: review of the current situation in the world. Disaster Mil. Med. 2016;2:9. doi: 10.1186/s40696-016-0019-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Groot RJ, Baker SC, Baric RS, et al. Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus Study Group. J. Virol. 2013;87(14):7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coleman CM, Frieman MB. Emergence of the Middle East respiratory syndrome coronavirus. PLoS Pathogens. 2013;9(9):e1003595. doi: 10.1371/journal.ppat.1003595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldstein SA, Weiss SR. origins and pathogenesis of Middle East respiratory syndrome-associated coronavirus: recent advances. F1000Res. 2017;6(F1000 Faculty Rev):1628. doi: 10.12688/f1000research.11827.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Middle East respiratory syndrome coronavirus (MERS-CoV)-Saudi Arabia. WHO. 2018 www.who.int/csr/don/28-december-2018-mers-saudi-arabia/en/ [Google Scholar]; • Laboratory-confirmed MERS-CoV cases reported globally.
- 10.Fehr AR, Channapannavar R, Perlman S. Middle East respiratory syndrome (MERS): emergence of a pathogenic human coronavirus. Annu. Rev. Med. 2017;68:387–399. doi: 10.1146/annurev-med-051215-031152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu L, Liu Q, Du L, et al. Middle East respiratory syndrome coronavirus (MERS-CoV): challenges in identifying its source and controlling its spread. Microbes Infect. 2013;15(8–9):625–629. doi: 10.1016/j.micinf.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hotez PJ, Bottazzi ME, Tseng CT, et al. Calling for rapid development of a safe and effective MERS vaccine. Microbes Infect. 2014;16(7):529–531. doi: 10.1016/j.micinf.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chowell G, Blumberg S, Simonsen L, et al. Synthesizing data and models for the spread of MERS-CoV, 2013: key role of index cases and hospital transmission. Epidemics. 2014;9:40–51. doi: 10.1016/j.epidem.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ge X-Y, Li J-L, Yang X-L, et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503(7477):535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]; • MERS-CoV are far more closely related to SARS-CoV than any previously identified bat coronaviruses.
- 15.Durai P, Batool M, Shah M, et al. Middle East respiratory syndrome coronavirus: transmission, virology and therapeutic targeting to aid in outbreak control. Exp. Mol. Med. 2015;47:e181. doi: 10.1038/emm.2015.76. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Human-to-human ‘superspreading’ of MERS-CoV.
- 16.Mohd HA, Al-Tawfiq JA, Memish ZA. Middle East respiratory syndrome coronavirus (MERS-CoV) origin and animal reservoir. Virol. J. 2016;13:87. doi: 10.1186/s12985-016-0544-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan JF, Lau SK, To KK, et al. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin. Microbiol. Rev. 2015;28(2):465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zumla A, Hui DS, Perlman S. Middle East respiratory syndrome. Lancet. 2015;386(9997):995–1007. doi: 10.1016/S0140-6736(15)60454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mustafa S, Balkhy H, Gabere MN. Current treatment options and the role of peptides as potential therapeutic components for Middle East respiratory syndrome (MERS): a review. J. Infect. Public Health. 2018;11(1):9–17. doi: 10.1016/j.jiph.2017.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xia S, Liu Q, Wang Q, et al. Middle East respiratory syndrome coronavirus (MERS-CoV) entry inhibitors targeting spike protein. Virus Res. 2014;194:200–210. doi: 10.1016/j.virusres.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Surya W, Li Y, Verdià-Bàguena C, et al. MERS coronavirus envelope protein has a single transmembrane domain that forms pentamericion channels. Virus Res. 2015;201:61–66. doi: 10.1016/j.virusres.2015.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J, Sun Y, Qi J, et al. The membrane protein of severe acute respiratory syndrome coronavirus acts as a dominant immunogen revealed by a clustering region of novel functionally and structurally defined cytotoxic T-lymphocyte epitopes. J. Infect. Dis. 2010;202(8):1171–1180. doi: 10.1086/656315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Haan CAM, Rottier PJM. Adv Virus Res. 2005. Molecular interactions in the assembly of coronaviruses; pp. 165–230.www.sciencedirect.com/bookseries/advances-in-virus-research [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cho CC, Lin MH, Chuang CY, et al. Macro domain from Middle East respiratory syndrome coronavirus (MERS-CoV) is an efficient ADP-ribose binding module: crystal structure and biochemical studies. J. Biol. Chem. 2016;291(10):4894–4902. doi: 10.1074/jbc.M115.700542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zumla A, Chan JFW, Azhar EI, et al. Coronaviruses drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15(5):327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]; • SARS and MERS poses major challenges to clinical management.
- 26.St John SE, Mesecar AD. Design, synthesis, and development of broad spectrum coronaviral 3C-like protease inhibitors to target emerging human pathogens: a phylochemical approach. FASEB J. 2016;30(1 Suppl.):842.4. [Google Scholar]
- 27.Tomar S, Johnston ML, St John SE, et al. Ligand-induced dimerization of Middle East respiratory syndrome (MERS) coronavirus nsp5 Protease (3CL [pro]): implications for nsp5 regulation and the development of antivirals. J. Biol. Chem. 2015;290(32):19403–19422. doi: 10.1074/jbc.M115.651463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang F, Chen C, Tan W, et al. Structure of main protease from human coronavirus NL63: insights for wide spectrum anti-coronavirus drug design. Sci. Rep. 2016;6:22677. doi: 10.1038/srep22677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Middle East Respiratory Syndrome (MERS)/symptoms and complications. Centers for Disease Control and Prevention website. 2015 www.cdc.gov/coronavirus/mers/about/symptoms.html [Google Scholar]
- 30.Drosten C, Seilmaier M, Corman VM, et al. Clinical features and virological analysis of a case of Middle East respiratory syndrome coronavirus infection. Lancet Infect. Dis. 2013;13(9):745–751. doi: 10.1016/S1473-3099(13)70154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kindler E, Jonsdottir HR, Muth D, et al. Efficient replication of the novel human betacoronavirus EMC on primary human epithelium highlights its zoonotic potential. MBio. 2013;4(1):e00611–e00612. doi: 10.1128/mBio.00611-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan JF, Chan KH, Choi GK, et al. Differential cell line susceptibility to the emerging novel human betacoronavirus 2c EMC/2012: implications for disease pathogenesis and clinical manifestation. J. Infect. Dis. 2013;207(11):1743–1752. doi: 10.1093/infdis/jit123. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Implications for the diagnosis, pathogenesis and transmission.
- 33.Chan RW, Chan MC, Agnihothram S, et al. Tropism and innate immune responses of the novel human betacoronavirus lineage C virus in human ex vivo respiratory organ cultures. J. Virol. 2013;87(12):6604–6614. doi: 10.1128/JVI.00009-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zielecki F, Weber M, Eickmann M, et al. Human cell tropism and innate immune system interactions of human respiratory coronavirus EMC compared to those of severe acute respiratory syndrome coronavirus. J. Virol. 2013;87(9):5300–5304. doi: 10.1128/JVI.03496-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yin Y, Wunderink RG. MERS, SARS and other coronaviruses as causes of pneumonia. Respirology. 2018;23(2):130–137. doi: 10.1111/resp.13196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Menachery VD, Eisfeld AJ, Schafer A, et al. Pathogenic influenza viruses and coronaviruses utilize similar and contrasting approaches to control interferon-stimulated gene responses. MBio. 2014;5(3):e01174–14. doi: 10.1128/mBio.01174-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lau SK, Lau CC, Chan KH, et al. Delayed induction of proinflammatory cytokines and suppression of innate antiviral response by the novel Middle East respiratory syndrome coronavirus: implications for pathogenesis and treatment. J. Gen. Virol. 2013;94(pt 12):2679–2690. doi: 10.1099/vir.0.055533-0. [DOI] [PubMed] [Google Scholar]
- 38.Boonacker E, Van Noorden CJ. The multifunctional or moonlighting protein CD26/DPPIV. Eur. J. Cell Biol. 2003;82(2):53–73. doi: 10.1078/0171-9335-00302. [DOI] [PubMed] [Google Scholar]
- 39.Raj VS, Mou H, Smits SL, et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495(7440):251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyerholz DK, Lambertz AM, McCray PB., Jr Dipeptidyl peptidase 4 distribution in the human respiratory tract: implications for the Middle East respiratory syndrome. Am. J. Pathol. 2016;186(1):78–86. doi: 10.1016/j.ajpath.2015.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Widagdo W, Raj VS, Schipper D, et al. Differential expression of the Middle East respiratory syndrome coronavirus receptor in the upper respiratory tracts of humans and dromedary camels. J. Virol. 2016;90(9):4838–4842. doi: 10.1128/JVI.02994-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakagawa K, Narayanan K, Wada M, et al. The endonucleolytic RNA cleavage function of nsp1 of Middle East respiratory syndrome coronavirus promotes the production of infectious virus particles in specific human cell lines. J. Virol. 2018;92(21):e01157–18. doi: 10.1128/JVI.01157-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ng DL, Al Hosani F, Keating MK, et al. Clinicopathologic, immunohistochemical, and ultrastructural findings of a fatal case of Middle East respiratory syndrome coronavirus infection in the United Arab Emirates, April 2014. Am. J. Pathol. 2016;186(3):652–658. doi: 10.1016/j.ajpath.2015.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dijkman R, Jebbink MF, Koekkoek SM, et al. Isolation and characterization of current human coronavirus strains in primary human epithelial cell cultures reveal differences in target cell tropism. J. Virol. 2013;87(11):6081–6090. doi: 10.1128/JVI.03368-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hocke AC, Becher A, Knepper J, et al. Emerging human middle East respiratory syndrome coronavirus causes widespread infection and alveolar damage in human lungs. Am. J. Respir. Crit. Care Med. 2013;188(7):882–886. doi: 10.1164/rccm.201305-0954LE. [DOI] [PubMed] [Google Scholar]; •• MERSCoV was detected in urine samples of a patient.
- 46.Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat. Rev. Immunol. 2011;11(11):723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou J, Chu H, Li C, et al. Active replication of MERS-CoV and aberrant induction of inflammatory cytokines and chemokines in human macrophages: implications for pathogenesis. J. Infect. Dis. 2014;209(9):1331–1342. doi: 10.1093/infdis/jit504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guery B, Poissy J, el Mansouf L, et al. Clinical features and viral diagnosis of two cases of infection with Middle East respiratory syndrome coronavirus: a report of nosocomial transmission. Lancet. 2013;381(9885):2265–2272. doi: 10.1016/S0140-6736(13)60982-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Munster VJ, de Wit E, Feldmann H. Pneumonia from human coronavirus in a macaque model. N. Engl. J. Med. 2013;368(16):1560–1562. doi: 10.1056/NEJMc1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Memish ZA, Zumla AI, Al-Hakeem RF, et al. Family cluster of Middle East respiratory syndrome coronavirus infections. N. Engl. J. Med. 2013;368:2487–2494. doi: 10.1056/NEJMoa1303729. [DOI] [PubMed] [Google Scholar]
- 51.Orenstein JM. The macrophage in HIV infection. Immunobiology. 2001;204:598–602. doi: 10.1078/0171-2985-00098. [DOI] [PubMed] [Google Scholar]
- 52.Chu H, Zhou J, Wong BH, et al. Middle East respiratory syndrome coronavirus efficiently infects human primary T lymphocytes and activates the extrinsic and intrinsic apoptosis pathways. J. Infect. Dis. 2016;213(6):904–914. doi: 10.1093/infdis/jiv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cockrell AS, Johnson JC, Moore IN, et al. A spike modified Middle East respiratory syndrome coronavirus (MERS-CoV) infectious clone elicits mild respiratory disease in infected rhesus macaques. Sci. Rep. 2018;8(1):10727. doi: 10.1038/s41598-018-28900-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chan JF, Yao Y, Yeung ML, et al. Treatment with lopinavir/ritonavir or interferon- β1b improves outcome of MERS-CoV infection in a nonhuman primate model of common marmoset. J. Infect. Dis. 2015;212(12):1904–1913. doi: 10.1093/infdis/jiv392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Johnson RF, Bagci U, Keith L, et al. 3B11-N, a monoclonal antibody against MERS-CoV, reduces lung pathology in rhesus monkeys following intratracheal inoculation of MERS-CoV Jordan-n3/2012. Virology. 2016;490:49–58. doi: 10.1016/j.virol.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson RF, Via LE, Kumar MR, et al. Intratracheal exposure of common marmosets to MERS-CoV Jordan-n3/2012 or MERS-CoV EMC/2012 isolates does not result in lethal disease. Virology. 2015;485:422–430. doi: 10.1016/j.virol.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yao Y, Bao L, Deng W, et al. An animal model of MERS produced by infection of rhesus macaques with MERS coronavirus. J. Infect. Dis. 2014;209(2):236–242. doi: 10.1093/infdis/jit590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.de Wit E, Rasmussen AL, Falzarano D, et al. Middle East respiratory syndrome coronavirus (MERS-CoV) causes transient lower respiratory tract infection in rhesus macaques. Proc. Natl Acad. Sci. USA. 2013;110(41):16598–603. doi: 10.1073/pnas.1310744110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Falzarano D, de Wit E, Feldmann F, et al. Infection with MERS-CoV causes lethal pneumonia in the common marmoset. PLoS Pathog. 2014;10(8):e1004250. doi: 10.1371/journal.ppat.1004250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cockrell AS, Yount BL, Scobey T, et al. A mouse model for MERS coronavirus-induced acute respiratory distress syndrome. Nat. Microbiol. 2016;2:16226. doi: 10.1038/nmicrobiol.2016.226. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Small-animal models traditionally used to investigate viral pathogenesis.
- 61.Pascal KE, Coleman CM, Mujica AO, et al. Pre- and post-exposure efficacy of fully human antibodies against Spike protein in a novel humanized mouse model of MERS-CoV infection. Proc. Natl Acad. Sci. USA. 2015;112(28):8738–8743. doi: 10.1073/pnas.1510830112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adedeji AO, Sarafianos SG. Antiviral drugs specific for coronaviruses in pre-clinical development. Curr. Opin. Virol. 2014;8:45–53. doi: 10.1016/j.coviro.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.de Wilde AH, Jochmans D, Posthuma CC, et al. Screening of an FDA-approved compound library identifies four small-molecule inhibitors of Middle East respiratory syndrome coronavirus replication in cell culture. Antimicrob. Agents Chemother. 2014;58(8):4875–4884. doi: 10.1128/AAC.03011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cao J, Forrest JC, Zhang X. A screen of the NIH clinical collection small molecule library identifies potential anti-coronavirus drugs. Antiviral Res. 2015;114:1–10. doi: 10.1016/j.antiviral.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.de Wilde AH, Falzarano D, Zevenhoven-Dobbe JC, et al. Alisporivir inhibits MERS- and SARS-coronavirus replication in cell culture, but not SARS-coronavirus infection in a mouse model. Virus Res. 2017;228:7–13. doi: 10.1016/j.virusres.2016.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marano G, Vaglio S, Pupella S, et al. Convalescent plasma: new evidence for an old therapeutic tool? Blood Transfus. 2016;14(2):152–157. doi: 10.2450/2015.0131-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Arabi Y, Balkhy H, Hajeer AH, et al. Feasibility, safety, clinical, and laboratory effects of convalescent plasma therapy for patients with Middle East respiratory syndrome coronavirus infection: a study protocol. Springer Plus. 2016;4(709) doi: 10.1186/s40064-015-1490-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chong YP, Song JY, Seo YB, et al. Rapid response T. Antiviral treatment guidelines for Middle East respiratory syndrome. Infect. Chemo. Ther. 2015;47(3):212–222. doi: 10.3947/ic.2015.47.3.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dimitrov DS. Therapeutic proteins. Methods Mol. Biol. 2012;899:1–26. doi: 10.1007/978-1-61779-921-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carter PJ. Potent antibody therapeutics by design. Nat. Rev. Immunol. 2006;6(5):343–357. doi: 10.1038/nri1837. [DOI] [PubMed] [Google Scholar]
- 71.Meulen J, Bakker AB, van den Brink EN, et al. Human monoclonal antibody as prophylaxis for SARS coronavirus infection in ferrets. Lancet. 2004;363(9427):2139–2141. doi: 10.1016/S0140-6736(04)16506-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.ter Meulen J, van den Brink EN, Poon LL, et al. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 2006;3(7):e237. doi: 10.1371/journal.pmed.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Du L, Kou Z, Ma C, et al. A truncated receptor-binding domain of MERS-CoV spike protein potently inhibits MERS-CoV infection and induces strong neutralizing antibody responses: implication for developing therapeutics and vaccines. PLoS ONE. 2013;8(12):e81587. doi: 10.1371/journal.pone.0081587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mou H, Raj VS, van Kuppeveld FJ, et al. The receptor binding domain of the new Middle East respiratory syndrome coronavirus maps to a 231-residue region in the spike protein that efficiently elicits neutralizing antibodies. J. Virol. 2013;87(16):9379–9383. doi: 10.1128/JVI.01277-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Du L, Zhao G, Yang Y, et al. A conformation-dependent neutralizing monoclonal antibody specifically targeting receptor-binding domain in Middle East respiratory syndrome coronavirus spike protein. J. Virol. 2014;88(12):7045–7053. doi: 10.1128/JVI.00433-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ying T, Du L, Ju TW, et al. Exceptionally potent neutralization of Middle East respiratory syndrome coronavirus by human monoclonal antibodies. J. Virol. 2014;88(14):7796–7805. doi: 10.1128/JVI.00912-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jiang L, Wang N, Zuo T, et al. Potent neutralization of MERS-CoV by human neutralizing monoclonal antibodies to the viral spike glycoprotein. Sci. Transl. Med. 2014;6(234):234ra59. doi: 10.1126/scitranslmed.3008140. [DOI] [PubMed] [Google Scholar]
- 78.Tang XC, Agnihothram SS, Jiao Y, et al. Identification of human neutralizing antibodies against MERS-CoV and their role in virus adaptive evolution. Proc. Natl Acad. Sci. USA. 2014;111(19):E2018–26. doi: 10.1073/pnas.1402074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Qiu H, Sun S, Xiao H, et al. Single-dose treatment with a humanized neutralizing antibody affords full protection of a human transgenic mouse model from lethal Middle East respiratory syndrome-coronavirus infection. Antiviral Res. 2016;132:141–148. doi: 10.1016/j.antiviral.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fosgerau K, Hoffmann T. Peptide therapeutics: current status and future directions. Drug Discov. Today. 2015;20(1):122–128. doi: 10.1016/j.drudis.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 81.Ali MM, Karasneh GA, Jarding MJ, et al. A 3-O-sulfated heparansulfate binding peptide preferentially targets herpes simplex virus 2-infectedcells. J. Virol. 2012;86(12):6434–6443. doi: 10.1128/JVI.00433-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tincho MB, Gabere MN, Pretorius A. In silico identification and molecular validation of putative antimicrobial peptides for HIV therapy. J. AIDS Clin. Res. 2016;7(606) [Google Scholar]
- 83.Melnik LI, Garry RF, Morris CA. Peptide inhibition of human cytomegalovirus infection. Virol. J. 2011;8(1):76. doi: 10.1186/1743-422X-8-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Qureshi A, Thakur N, Tandon H, et al. AVPdb: a database of experimentally validated antiviral peptides targeting medically important viruses. Nucleic Acids Res. 2014;42:D1147–D1153. doi: 10.1093/nar/gkt1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fjell CD, Hancock REW, Cherkasov A. AMPer: a database and an automated discovery tool for antimicrobial peptides. Bioinformatics. 2007;23(9):1148–1155. doi: 10.1093/bioinformatics/btm068. [DOI] [PubMed] [Google Scholar]
- 86.Fjell CD, Hiss JA, Hancock REW, et al. Designing antimicrobial pep-tides: form follows function. Nat. Rev. Drug Discov. 2012;11(1):37–51. doi: 10.1038/nrd3591. [DOI] [PubMed] [Google Scholar]
- 87.Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 2003;3(9):710–720. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- 88.Harder J, Schröder JM. In: Antimicrobial Peptides: Role in Human Health and Disease. Harder J, Schröder JM, editors. Springer International Publishing; 2015. [Google Scholar]