Abstract
The recombinant adeno-associated virus (rAAV) vector has been successfully employed in clinical trials for patients with blindness and bleeding diseases as well as neuromuscular disorders. To date, it remains a major challenge to achieve higher transduction efficiency with a lower dose of rAAV vector. Our previous studies have demonstrated that serum proteins are able to directly interact with AAV virions for transduction enhancement. Herein, we explored the effect of the FerA domains, which are derived from ferlin proteins and possess membrane-fusion activity, on AAV transduction. Our results show that FerA domains from dysferlin, myoferlin, and otoferlin proteins are able to directly interact with AAV vectors and enhance AAV transduction in vitro and in mice through either intravenous or intramuscular injections. The enhanced AAV transduction induced by human/mouse FerA domains is achieved in various cell lines and in mice regardless of AAV serotypes. Mechanism studies demonstrated that the FerA domains could effectively enhance the ability of AAV vectors to bind to target cells and cross the vascular barrier. Additionally, FerA domains slow down the blood clearance of AAV. Systemic administration of AAV8/hFIX-FerA complex induced approximate 4-fold more human coagulation factor IX expression and improved hemostasis in hemophilia B mice than that of AAV8/hFIX. Collectively, we show, for the first time, that multiple FerA domains could be tethered on the AAV capsid and enhance widespread tissue distribution in an AAV serotypes-independent manner. This approach therefore holds a promise for future clinical application.
Keywords: adeno-associated virus vector, FerA domain, transduction, hFIX, intramuscular injection, systemic administration
1. Introduction
Recombinant adeno-associated virus (rAAV) vector has been a leading vehicle for clinical application of gene therapies in view of its safety, broad tissue tropism, and sustained effectiveness [1, 2]. Currently, 13 serotypes and numerous AAV variants and mutants have been isolated and studied as gene delivery vehicles [3]. Several AAV serotypes, such as AAV2, AAV8, and AAV9, have been extensively employed in clinical trials and achieved therapeutic effects [4-6]. Nevertheless, emerging concerns about the rAAV limited transduction efficacy and the high vector dose requirement remain crucial barriers for ongoing AAV-based gene therapy in preclinical and clinical settings [7, 8]. Data from AAV-human factor IX (hFIX)-related clinical trials in hemophilia B patients have shown that high vector dose directly correlates with the cellular immune responses to vector capsid [9, 10]. Meanwhile, several studies have demonstrated that AAV capsid or transgene-specific cytotoxic T lymphocytes (CTLs) response and late innate immune activation after long-term AAV transduction hinder the AAV vector transduction efficiency [10-12], which could be strengthened under the condition of high dose of vehicles. In this context, tremendous efforts have been directed to improve the AAV vector transduction efficacy while simultaneously decreasing the vector dose. Several approaches, including but not limited to direct evolution, capsid/genome engineering and polyploid AAV capsids modification, have been undertaken in an effort to produce and impel AAV variants into desired tissues or cell types and achieve therapeutic efficacy [13-17]. Meanwhile, numerous studies with cell surface targeting of AAV vectors by genetic modification of the capsid proteins VP1, VP2, or VP3 have been extensively explored to improve safety and efficacy in preclinical/clinical application [18-20]. Additionally, covalent coupling of targeting ligands to intact AAV particles and magnetically guided AAV delivery system have been also exploited to achieve selective gene transfer in distinct cell types, even in non-permissive cell types [21, 22]. However, the achieved results being implemented in specific mouse strains and other small -animal models cannot always be extrapolated to that of other species such as non-human primates (NHPs) [23-26]. One study has shown that the therapeutic efficacy for liver transduction in large-animal models compared with mouse models is approximately 50-100-fold less efficient [27]. Notably, engineered modification of the AAV capsid may lead to unpredicted structural and tropism changes [28]. Although those risks could be mitigated or eliminated by direct evolution approaches [17], the most mutants developed from direct evolution were isolated from cell lines in vitro or in animal models and have unknown human tropism. Therefore, further exploration of safer and more effective strategies to improve AAV transduction at a lower vector dose, especially in a manner that induces consistent transgene expression without limitation of species and alteration of tissue tropism, is still of critical clinical importance.
It has been demonstrated by multiple studies that serum proteins effectively bind to AAV and affect transduction efficacy via different mechanisms in a serotypes- and species-specific manner [29-35]. For instance, the mouse-derived but not human-derived C-reactive protein can bind to the AAV6 virions [29]. However, both of them did not appreciably interact with either AAV8 or AAV9 [35]. Our recent studies have demonstrated that several human serum proteins are able to directly interact with the specific AAV capsid and increase its transduction [32-34]. Pei, et al. found that the low-density lipoprotein and transferrin could be appropriated by AAV8 virions to enhance the liver transduction by increasing AAV’s ability to bind to target cells, although the enhancement is only limited to hepatocytes after systemic administration [33]. Chai, et al. has demonstrated that serum proteins could directly interact with AAV9 and further enhance AAV9 vascular permeability for global transduction [34]. Nevertheless, it is worthy to note that the effect of serum proteins may have limited application only to specific AAV serotypes [29, 30, 32-35]. In light of the described events, it will undoubtedly require the exploration of novel proteins with the characterization of generally working with multiple AAV serotypes to achieve broader AAV preclinical/clinical application.
Ferlins are a family of type II transmembrane proteins and are characterized by the multiple tandem C2 domains (Ca2+-regulated, phospholipid-binding domains), a centrally positioned FerA domain, and anchored by a C-terminal transmembrane domain [36, 37]. Ferlins are implicated in membrane repair and membrane fusion processes. To date, there are six Fer-1-like genes that form the ferlin family: dysferlin (Fer1L1), otoferlin (Fer1L2), and myoferlin (Fer1L3), as well as three additional yet not well-characterized members Fer1L4, Fer1L5, and Fer1L6 in humans and most mammals [38]. A recent study found that the α-helical ~15-kDa FerA domain possesses Ca2+-dependent membrane-associating activity, and may contribute to the overall membrane fusion activity of ferlin proteins [39]. If the isolated FerA domain can help mediate vesicle fusion, can it then be used to improve the AAV transduction process?
In the present study, we demonstrated for the first time that the isolated FerA domains from the dysferlin, myoferlin, and otoferlin proteins could generally enhance the AAV transduction in vitro and in vivo in a species- and AAV serotypes-independent manner, thus overcoming some of the limitations of the previous AAV adjuvants. Underlying mechanism studies suggest that FerA domains directly bind to AAV, promote the AAV binding activity, delay the blood clearance, and enhance the transcytosis ability of AAV. We applied this approach for treating FIX-deficient hemophilic mice. After systemic administration of self-complementary (sc) AAV8/hFIX vector preincubated with FerA domain, enhanced transgene hFIX expression and hemostasis improvement were achieved.
2. Materials and methods
2.1. Cell lines
Huh7, HeLa, and C2C12 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10 % (v/v) heat inactivated fetal bovine serum (FBS), and antibiotics (penicillin 100-U/mL and streptomycin 100-μg/mL). Human brain endothelial capillary hCMEC/D3 cells (Millipore Sigma, USA), as a model of human blood-brain barrier (BBB), were cultured as previously described [40]. All cells were maintained at 37 °C in an atmosphere of 5 % CO2.
2.2. Expression and Purification of FerA domains
FerA samples were prepared as previously described [39]. Briefly, human dysferlin FerA (hDys-FerA), human GFP-Dys-FerA (GFP-hDys-FerA), human myoferlin FerA (hMyo-FerA) and human or mouse otoferlin FerA (h/mOto-FerA) expression plasmids were transformed into chemically competent BL21 (DE3) cells using kanamycin as the selection antibiotic. One-liter TB (Terrific Broth) cultures with 50-μg/mL kanamycin utilizing IPTG chemical induction were used to grow up large volumes of cells. Cells were lysed using a microfluidizer and purified using affinity, ion exchange, and size-exclusion chromatography. All buffers were made the day of the experiment using fresh Milli-Q water. For Oto-FerA, 5 mM dithiothreitol (DTT) or 1 mM TCEP reducing agents were added to all buffers. Purity was assessed with SDS-PAGE Stain-Free gels (Bio-Rad, USA) for dysferlin and myoferlin. Due to the lack of tryptophan in Oto-FerA, coomassie staining was used for visualization. Protein concentration was measured by OD280 using extinction coefficients for hDys-FerA, GFP-hDys-FerA, and hMyo-FerA while a Bradford assay and reagent (Sigma Aldrich, USA) were used for h/mOto-FerA. FerA domains in a reducing reagent were exchanged with PD-10 desalting column (GE Healthcare Life Sciences, NJ, USA) and dissolved in Dulbecco's phosphate-buffered saline (DPBS). Finally, these proteins were stored at −80 °C in aliquots until ready to use.
2.3. AAV vectors production
The detailed construction of the recombinant AAV full particles expressing luciferase, driven by the CBA promoter (AAV/luc), used in the present study was reported previously [41]. In short, the AAV transgene plasmid pTR/CBA-luc, AAV helper plasmid containing AAV Rep and Cap genes (pXR6, pXR8, or pXR9), and Ad helper plasmid pXX-680 were co-transfected into HEK293 cells. HEK293 cells were collected and lysed 48 hours (h) post-transfection. The supernatant was then subjected to CsCl gradient ultra-centrifugation and fractions containing AAV were collected. The scAAV8 vector containing the hFIX cDNA (scAAV8/hFIX) [42] was purchased from UNC Vector Core (Chapel Hill, NC, USA). Finally, viral titer was determined by real-time quantitative PCR (qPCR) using the Light Cycler 480 instrument with SYBR green (Roche, USA) and a pair of primers that were designed to bind to a homologous sequence on the inverted terminal repeat (ITR) region [40]. The purity of all AAV vectors was measured by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) or Western blot with anti-AAV capsid B1 antibody.
2.4. Cell toxicity assay
Huh7 cells were seeded in 96-well plates at a density of about 2.5×104 cells/well and incubated overnight at 37 °C. Triplicate wells were treated with different concentrations of FerA. Cell toxicity assays were performed about 48 h after treatment using Celltiter 96 Aqueous one solution reagent (G3582, Promega, Madison, WI) according to manufacturer's instruction. Cell viability was estimated by assessing the absorbance at 490 nm. All experiments were repeated at least 3 times.
2.5. In vitro transduction assay
At least 4-5 h before AAV/luc transduction, cells were seeded in 48-well plates with a density of about 1×105 cells per well. Different concentrations of FerA (0.05 μg and 0.5 μg) were incubated with AAV/luc at 4 °C for 2 h. After that, cells were transduced with 1×109 vector genomes (vg) of AAV/luc vector or the incubated complex of vector and proteins. After 48 h, the cells were harvested and lysed. Luciferase activity was measured using the Promega Luciferase assay system based on the manufacturer's instructions (Promega, Madison, WI).
2.6. Streptavidin beads-based pull down assay
Biotinylated ligand/protein-based pull down assay was performed with Dynabeads® MyOne™ Streptavidin C1 beads (65001, Thermo Fisher Scientific, USA). For biotinylated ligand-based pull down, 1×1010 vg AAV9 was first incubated with/without 1-2 μg FerA for 2 h at 4 °C. Then the CaptureSelect™ biotin anti-AAV9 conjugate (7103332100, Thermo Fisher Scientific, USA) was added and incubated for 1 h at room temperature (RT). After that, the complexes were isolated with 20 μl pre-washed Dynabeads® MyOne™ Streptavidin C1 beads for another 0.5 h at RT. Incubated complexes of AAV9 with Streptavidin C1 beads, Streptavidin C1 beads with FerA domains, AAV9 and FerA domains with C1 beads, and the biotin anti-AAV9 conjugate and FerA domains with C1 beads were included as controls. For biotinylated protein-based pull down, recombinant fusion protein GFP-hDys-FerA was first biotinylated by incubating GFP-hDys-FerA in 100 mM HEPES buffer (pH 7.3) containing N-hydroxysuccinimide ester (NHS)-water-soluble biotin (SP-1210-50, Vector Laboratories, CA, USA) for 2 h at RT, and then dialyzed according to the manufacturer's instructions. After that, the biotinylated GFP-hDys-FerA was mixed with AAV9 for 2 h at 4 °C. Finally, the Streptavidin C1 beads were added and incubated for 0.5 h at RT. After stringent washing with DPBS four times, the final complex was boiled for 10 minute (min) in elution buffer and separated with 4-20 % SurePAGE™ Bis-Tris gel (Gene Script, NJ, USA). The presence of bound FerA was analyzed by Western blotting with an anti-dysferlin antibody (HPA021945, Sigma Aldrich, USA), anti-myoferlin antibody (ab190264, Abcam, MA, USA), anti-otoferlin antibody (C-12) (sc-271092, Santa Cruz Biotechnology), or B1 mouse antibody. After that, membranes were extensively washed in DPBS containing 0.05 % Tween-20 (DPBST), followed by incubation with horseradish peroxidase-conjugated goat anti-mouse IgG or anti-rabbit IgG (Thermo Fisher Scientific, USA) for 1 h at RT. Membranes were extensively washed in DPBST, and immunoblots were tested by Amersham ECL Western blotting detection kit and visualized with the Amersham imager 600 machine (GE Healthcare Life Sciences, NJ, USA).
2.7. Effect of FerA for AAV9 binding and intracellular trafficking
5×105 Huh7 or HeLa cells in 250 μl sera-free medium were suspended and incubated with 250 μl of medium containing AAV9/luc (10,000 vg/cell) or a complex of AAV9/luc and hDys-FerA previously incubated at 4 °C for 1 h with gentle shaking. For the binding assay, the cells were washed 3 times with cold DPBS to remove unbound particles and collected for total DNA preparation using the QIAamp DNA Blood Mini Kit (Qiagen, USA). For trafficking assays, the washed cells were incubated at 37 °C for another 2 h. Cytoplasmic and nuclear factions were separated using the NE-PER nuclear and cytoplasmic extraction kit (78833, Thermo Fisher Scientific, USA) according to manufacturer protocol. Finally, genome DNA (gDNA) was quantified by qPCR with luciferase primers or ITR primers and reference GAPDH primers [40].
2.8. Transcytosis assay
Transcytosis assay was performed on hCMEC/D3 cell line. Briefly, cells were seeded into the Transwell®-COL collagen-coated membrane inserts (24-Well Permeable Support with 0.4 μm Pore Polycarbonate Membrane and 6.5 mm Inserts, Sigma Aldrich, USA) at a density of 5×104 cells per well in EBM-2 cultured medium. The medium was changed every 2-3 days. After about 2 weeks, the cells were washed with DPBS and cultured in medium and treated with AAV9 alone or the pre-incubated AAV9-0.5 μg hDys-FerA. The medium in the basal chamber was collected at different time points of 0.5 h, 2 h, 4 h, 6 h, and 24 h. Viral titers were calculated by qPCR according to established procedures using primers designed against the ITR region [40]. Meanwhile, the transwell permeability was confirmed using dextran conjugated with FITC (FITC-dextran) according to the manufacturer’s instruction. Briefly, FITC-dextran was added to the apical cell layers and incubated at RT for 0.5 h. After that, cell inserts were removed and the medium remaining in the basolateral chamber was collected and measured for FITC-dextran fluorescence intensity using Perkin Wallac 1420 Victor Microplate Reader with excitation and emission wavelengths of 485 and 530 nm, separately.
2.9. Histopathological and biochemical studies for liver toxicity
To investigate the possible hepatotoxicity, mouse liver fragments in two groups with high doses (AAV9 and AAV9-hDys-FerA cohorts) were stained for hematoxylin and eosin (H&E) by the UNC Histology Research Core. Alanine aminotransaminase (ALT) activity and aspartate aminotransaminase (AST) activity in serum of injected mice by systemic administration were analyzed at different time points using ALT activity assay kit (ab105134, Abcam, Cambridge, MA) and AST activity assay kit (K753, Biovision, CA) according to the manufacturer’s instructions.
2.10. Animal study
C57BL/6 female mice, at 5-6 weeks of age, were purchased from the Jackson Laboratory (Bar Harbor, ME). 5-6 weeks of age FIX knockout (FIX−/−) female mice [43] were bred in house. All mice were randomly divided into groups of 5 animals each and maintained in a specific pathogen-free facility at UNC-Chapel Hill. All animal experimental procedures were approved by the UNC Institutional Animal Care and Use Committee. Mice were administered AAV-CBA/luc or scAAV8-hFIX incubated with FerA via the retro-orbital injection or intramuscular injection. For mice receiving AAV-CBA/luc vectors, following intraperitoneal injection of D-luciferin substrate (Nanolight Pinetop, AZ), luciferase expression was imaged at the indicated time points using a Xenogen IVIS Lumina (Caliper Lifesciences, Waltham, MA). Bioluminescent images were analyzed using Living Image (PerkinElmer, Waltham, MA). For hemophilia B mice, at various time points post injection, blood samples were collected from the retro-orbital plexus into 1:9 parts 3.2 % citrated sodium and stored at −80 °C for later FIX expression and function analysis. The viral titers were tested by qPCR [40].
2.11. Quantitation of luciferase expression in tissues
Animals utilized for imaging studies were sacrificed one-week after imaging work, and the tissues of the heart, liver, kidney, skeletal muscle, and brain were collected, minced, and homogenized in passive lysis buffer. The lysates were centrifuged at 12,000 g for 5 min to remove cellular debris. The supernatant was transferred to 96-well plates for luciferase activity analysis as described above. Total protein concentration in tissue lysates was measured using the Bradford assay (Sigma Aldrich, USA).
2.12. Measurement of AAV genome copy number in the tissues
Different minced tissues, or blood collected in different time points, were treated with Proteinase K and total genome DNA (gDNA) was isolated using the DNeasy blood & tissue kit (Qiagen, USA). The luciferase gene was measured by qPCR assay. The mouse GAPDH gene served as an internal control.
2.13. Detection of hFIX concentration and activity
hFIX protein concentration in the plasma was measured by enzyme-linked immunosorbent assay (ELISA) as previously described [44]. The hFIX-specific one-stage FIX activity assay (hFIX-specific aPTT) was performed using a START 4 Coagulation Analyzer (Diagnostica Stago, Asnières, France) as described previously [43].
2.14. Neutralization assay
The neutralization antibody (Nab) titers in different groups were measured as previously described [15]. Briefly, Huh7 cells were seeded at a density of 1×105 cells/well. Mouse sera with serial dilutions were incubated with AAV9/luc (1×109 vg/well) for 1 h at 37 °C. The complex was added into cells and incubated for 48 h at 37 °C, and then the luciferase activity in cell lysates was tested. Nab titers were defined as the highest dilution for which luciferase activity was 50 % lower than serum-free controls.
2.15. Statistical analysis
All quantitative data are presented as means ± standard deviation (SD). Data were analyzed by either the two-tailed/sided Student’s t-test or one-way ANOVA, and done using GraphPad Prism Version 6 software. In these figures, *, **, and *** were used to denote p < 0.05, p < 0.01, and p < 0.001, respectively and were considered statistically significant. Graphs are representative of data sets from at least three independent assays.
3. Results
3.1. Mouse otoferlin FerA enhances the transduction of multiple AAV serotypes in vitro without cytotoxicity
Our previous studies have shown that several serum proteins could directly and specifically bind to several AAV serotypes for transduction enhancement [32-34]. To study whether the FerA domains from ferlin proteins have an effect on AAV transduction, we first investigated the effect of the FerA from mouse otoferlin (mOto-FerA) on AAV transduction in vitro. The purity of AAV vectors were confirmed by SDS-PAGE and Western blot, which showed no protein contamination in all the AAV stocks (Fig. S1 A and B). AAV9/luc vectors (10,000 vg/cell) were incubated with mOto-FerA using different molecular ratios (AAV particle to FerA: 1:0, 1:20, 1:200, 1:2000, 1:20,000, and 1:200,000) at 4 °C for 2 h, and then the mixtures were applied onto Huh7 cells. After 48 h, the cell cytotoxicity was tested with MTT assay and no significant cytotoxicity was observed regardless of FerA concentrations (Fig. S2 A). Meanwhile, cell lysate was collected for luciferase analysis. The results showed that the incubated groups at the ratios of 1:20,000 and 1:200,000 significantly enhanced AAV9 transduction. In contrast, AAV9 transduction enhancement was not observed in non-incubated groups with different concentrations of mOto-FerA or incubated groups at the ratios of 1:20, 1:200, and 1:2000 (Fig. S2 B). This result suggests that AAV transduction enhancement requires the direct interaction of AAV virions with a certain amount of FerA molecules. Based on the above results, we further investigated the effect of mOto-FerA with the ratios of 1:20,000 and 1:200,000 on different AAV serotypes transduction in different cell lines. We found that transduction from AAV6, AAV8, and AAV9 was significantly increased in Huh7, hCMEC/D3, HeLa, and C2C12 cell lines (Fig. S2 C, D, E, and F). These results indicate that the mOto-FerA could enhance AAV transduction in a serotype-independent manner.
3.2. Mouse otoferlin FerA enhances the AAV transduction in vivo by systemic or intramuscular administration
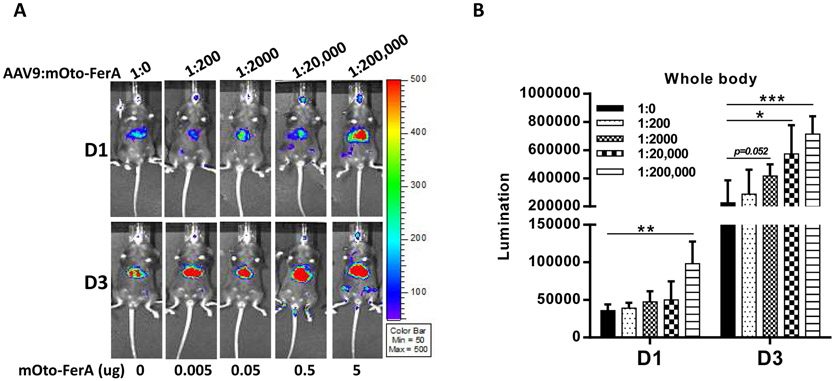
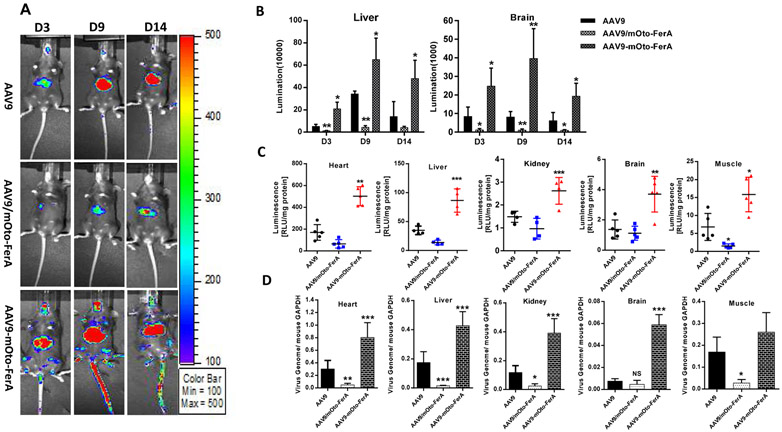
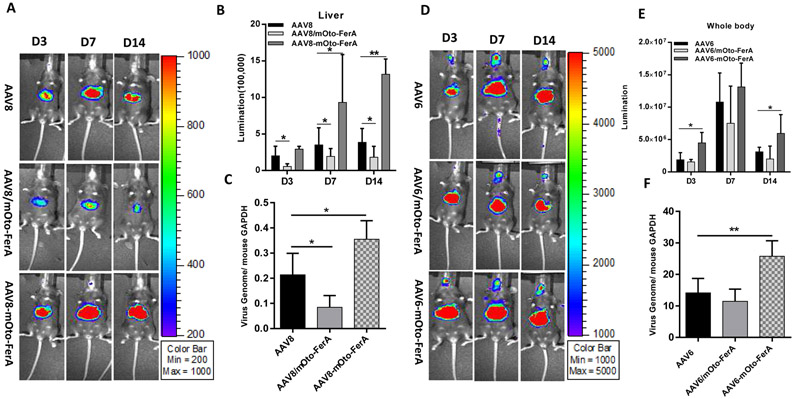
Next, we assessed the effect of mOto-FerA on AAV transduction in vivo. AAV9 vectors preincubated with mOto-FerA at different molecular ratios were first administered in C57BL/6 mice, and the imaging was performed on day 1 and 3. Consistent to the results from cell lines in vitro, significantly increased transduction was achieved in mice with the ratios of 1:20,000 and 200, 000 when compared to mice with the ratios of 1:0, 1:200, and 1:2000 (Fig. 1 A and B). To further confirm whether the AAV transduction enhancement required the direct interaction of AAV virions with FerA domains, three cohorts were designed: AAV9/luc vector (1×1010 vg) was incubated with 5 μg of mOto-FerA domain (AAV-mOto-FerA cohort, the ratio is 1:200, 000) or PBS (AAV cohort) at 4 °C for 2 h and injected into C57BL/6 female mice via the retro-orbital vein. The third cohort was separately injected with 5 μg of mOto-FerA followed by AAV vectors immediately (AAV/mOto-FerA cohort). After 3, 7 or 9, and 14 days, images were taken. The results showed that the AAV9-mOto-FerA cohort significantly outperformed the other two cohorts for transgene expression in the whole body, especially the liver and brain (Fig. 2 A and B), which is consistent with the results observed in the different cell lines in vitro. Similar outcomes were seen on AAV8/luc vector (Fig. 3 A and B). However, the enhancing effect is modest with AAV6/luc vector (Fig. 3 D and E). Interestingly, the non-incubation cohort AAV9/mOto-FerA or AAV8/mOto-FerA had significantly decreased transduction compared to the AAV9 or AAV8 group, respectively.
Fig. 1. Mouse otoferlin FerA domain increased AAV transduction in vivo via systemic administration.
5×1010 vg of AAV9/luc alone or the complex of AAV9/luc incubated with mOto-FerA at multiple molecule ratios (1:0, 1:200, 1:2000, 1:20,000, and 1:200,000 is equal to 0 μg, 0.005 μg, 0.05 μg, 0.5 μg, and 5 μg, respectively) were administered via intravenous injection. Luminescence imaging was performed (A) and the photon signal was measured and calculated (B) on day 1 and day 3. The data of each group represent the average and SD from five mice. *** p < 0.001, ** p < 0.01, and *p < 0.05 compared to control mice with AAV9/luc treatment only.
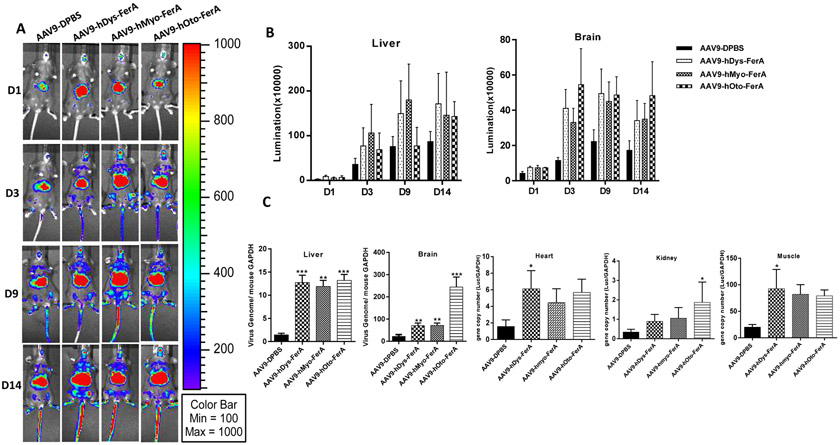
Fig. 2. Direct interaction of mouse otoferlin FerA with AAV9 was required for enhanced transduction in vivo via systemic administration.
Three groups were designed as below: 1×1010 vg AAV9 only (AAV9 cohort), AAV9 vectors and mOto-FerA were injected simultaneously without pre-incubation (AAV9/mOto-FerA cohort), and AAV9 vectors were pre-incubated with 5 μg mOto-FerA for 2 h at 4 °C (AAV9-mOto-FerA cohort). (A) Images were taken for luminescence analysis on days 3, 9, and 14 following the retro-orbital injections of female C57BL/6 mice. (B) The average luciferase signal for liver and brain was calculated. (C) The luciferase expression and (D) gene copy numbers in heart, liver, kidney, brain, and muscle were determined, separately. The data of each group represent the average and SD from five mice. *** p < 0.001, ** p < 0.01, and *p < 0.05 compared to control mice with AAV9/luc treatment only.
Fig. 3. Mouse otoferlin FerA enhanced the transduction of AAV8 and AAV6 vectors in vivo by systemic injection.
1×1010 vg of AAV8/luc or AAV6/luc with or without pre-incubation of mOto-FerA were injected into the 5-6-week-old female C57BL/6 mice via the retro-orbital injection. Imaging was carried out (A and D) and photon signal was calculated (B and E) at day 3, 7, and 14 after vector administration. The viral gene copy numbers (C and F) in liver were determined by qPCR. The data of each group represent the average and SD from five mice. **p <0.01 and *p < 0.05 compared to control mice with AAV/luc treatment only.
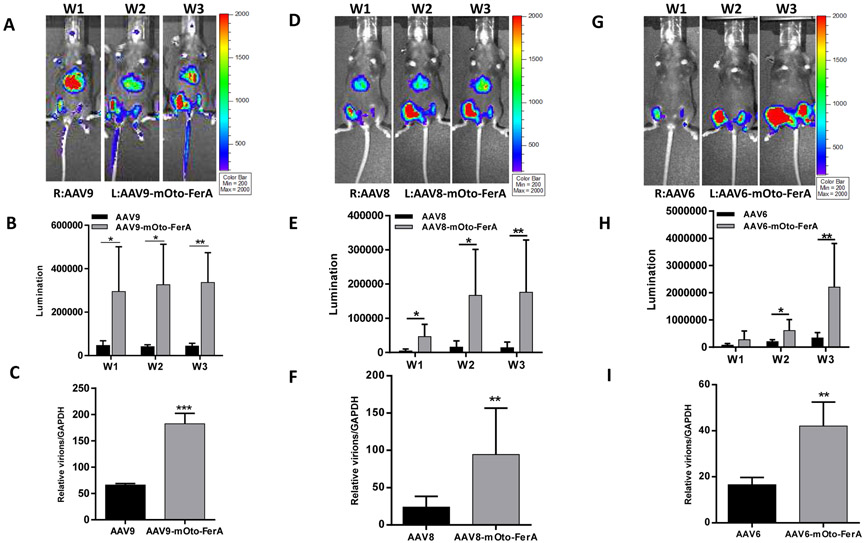
In consideration of the possible effect of FerA domain on the immunological response of AAV-immunized mice, we examined the Nab level to AAV9 in sera collected from individual mice of the different groups 2 and 4 weeks post administration. Similar neutralization profiles were observed among three groups (AAV9, AAV9/mOto-FerA, and AAV9-mOto-FerA) (Fig. S3). At the end of the experiment, we evaluated luciferase protein level and viral genome copy number in different tissues. The results showed that the mOto-FerA generally increased luciferase protein expression and the viral genome number in different tissues, which are in agreement with the imaging results (Fig. 2 C and D, Fig. 3 C and F). Additionally, we administered different AAV vectors (AAV9/luc, AAV8/luc, and AAV6/luc) intramuscularly and found that the mOto-FerA could generally enhance the AAV vectors transduction in the muscle (Fig. 4 A, B, D, E, G, and H). Accordingly, mOto-FerA increased the viral genome number of multiple AAV serotypes in muscle (Fig. 4 C, F, and I). These data further support that mOto-FerA bolsters the AAV vectors transduction in a serotypes-nonspecific manner.
Fig. 4. Mouse otoferlin FerA increased the muscle transduction of AAV.
2×109 vg of AAV9/luc, AAV8/luc, or AAV6/luc particles were first incubated with 1 μg mOto-FerA for 2 h at 4 °C, then administered via intramuscular injection in the left legs (face-up) in 6-week-old female C57BL/6 mice (n = 5). As the internal control, AAV vectors only were injected into the right legs (face-up). Several weeks (W1, W2, and W3) post AAV application, in vivo luminescence imaging was performed (A, D, and G). The photon signal (B, E, and H) and gene copy numbers (C, F, and I) in the leg muscle were measured and calculated. The data of each group represent the average and SD from five mice. *** p < 0.001, **p <0.01, and *p < 0.05 compared to control mice with AAV/luc treatment only. R: right leg, L: left leg.
3.3. Human FerA domains enhances the AAV transduction in vitro
Given that mouse FerA may induce an unwanted immune response when applied in human studies, we then wondered whether human-derived FerA domains were able to exert a similar enhanced effect on AAV transduction to mouse FerA. To this end, human FerA domains derived from dysferlin (hDys-FerA), myoferlin (hMyo-FerA), and otoferlin (hOto-FerA) at the doses of 0.05 μg and 0.5 μg were separately incubated with AAV9 vectors (1×109 vg) at 4 °C for 2 h, then the complex was applied to different cell lines (Huh7, HeLa, and hCMEC/D3). The luciferase value was measured after 48 h treatment. The results showed that the hDys-FerA, hMyo-FerA, and hOto-FerA domains augmented the AAV9 transduction in all tested cell lines (Fig. S4 B, C, and D) under the premise of having no significant cytotoxicity (Fig. S4 A).
3.4. Human FerA domains increase the AAV9 transduction in vivo by systemic or intramuscular administration
To further validate the effect of human FerA domains on AAV transduction in vivo, C57BL/6 mice were randomly grouped and systemically administrated with 1×1010 particles of AAV9/luc alone or the complex of AAV9/luc pre-incubated with 5 μg of hDys-FerA, hMyo-FerA, or hOto-FerA domains. The results showed that, consistent with mOto-FerA (Fig. 2 and 3), all three human FerA domains resulted in vastly and significantly higher level of widespread luciferase expression when compared to control (Fig. 5 A and B). Similarly, higher AAV genomes were detected in tissues of mice receiving human FerA treatment (Fig. 5 C). It is worth noting that the variable gene copy numbers from different human FerA domains were detected in the brain. The AAV genome copy number was much higher in the brain of mice treated with hOto-FerA than that with hDys-FerA and hMyo-FerA. This finding may be partially attributed to specific tissue localization of different ferlin proteins [45]. After direct muscular injection, AAV9 vectors preincubated with FerA domains from three ferlin proteins induced a similar transduction enhancement (Fig. S5 A, B, and C). It is interesting that the hOto-FerA domain has a less pronounced enhancing effect than that of the mOto-FerA domain in liver and muscle by systemic and intramuscular administration, respectively. Further investigation is needed to determine whether the species-specific difference of the FerA domain contributes to it or not.
Fig. 5. Multiple human FerA domains increased the AAV9 transduction by systemic injections.
AAV9/luc vectors were incubated with DPBS or hDys-/hMyo-/hOto-FerA domains for 2 h at 4 °C. The complex was injected into the female C57BL/6 mice by systemic administration. At different time points (D1, 3, 9, 14) post AAV application, images were taken for luminescence analysis (A). The average luciferase signal for liver and brain was calculated (B). The gene copy numbers in liver, brain, heart, kidney, and muscle were separately determined (C). The data of each group represent the average and SD from five mice. *** p < 0.001, ** p < 0.01, and *p < 0.05 compared to control mice with AAV9/luc treatment only.
3.5. FerA significantly improved hemostasis in hemophilia B mice after systemic injection of scAAV8/hFIX vector
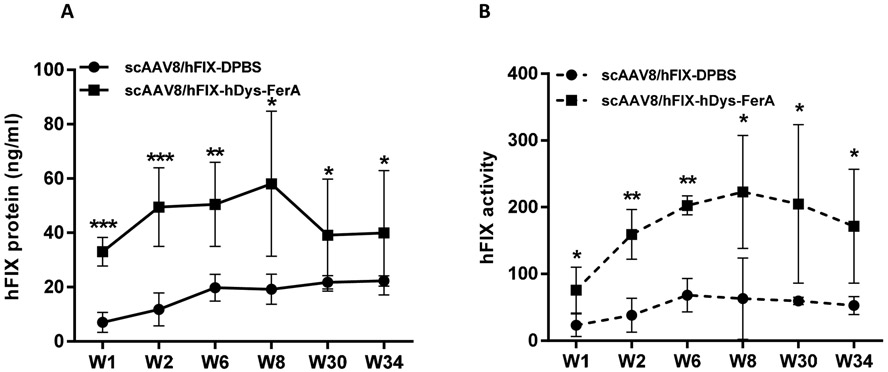
The aforementioned findings demonstrated that the FerA generally increases AAV transduction, which suggests similar transduction efficacy could be achieved with a lower dose of AAV vector coupled with the FerA domain. To apply this finding to treat a disease, we injected scAAV8/hFIX or the complex scAAV8/hFIX-hDys-FerA into adult female FIX−/− mice via the retro-orbital injection at a dose of 1×1010 vg per mouse. At different time points (week 1, 2, 6, 8, 30, and 34) after vector injection, we collected the plasma and tested the FIX expression and coagulation function. As predicted, FerA endowed a 4-fold higher hFIX protein expression and a better hemostasis improvement in the hemophilic mice when compared to mice treated with scAAV8/hFIX (Fig. 6 A and B). Notably, long-term improved transduction efficacy by the FerA domain was continuously observed at weeks 30 and 34.
Fig. 6. FerA significantly improved hFIX expression and hemostasis in FIX−/− hemophilic mice.
1×1010 vg of pre-incubated scAAV8-hFIX vectors with DPBS or hDys-FerA domain were administered in FIX −/− mice via the retro-orbital injection. At 1, 2, 6, 8, 30, and 34 weeks after vector administration, plasma from FIX−/− mice were collected and isolated. (A) Total expressed hFIX protein in mouse plasma was determined by an ELISA assay. (B) Circulating functional hFIX activity was determined by an in vitro aPTT assay. The data of each group represent the average and SD from five mice. *** p < 0.001, **p <0.01, and *p < 0.05 compared to control mice with scAAV8/hFIX treatment only.
3.6. Diverse FerA domains directly interact with AAV9
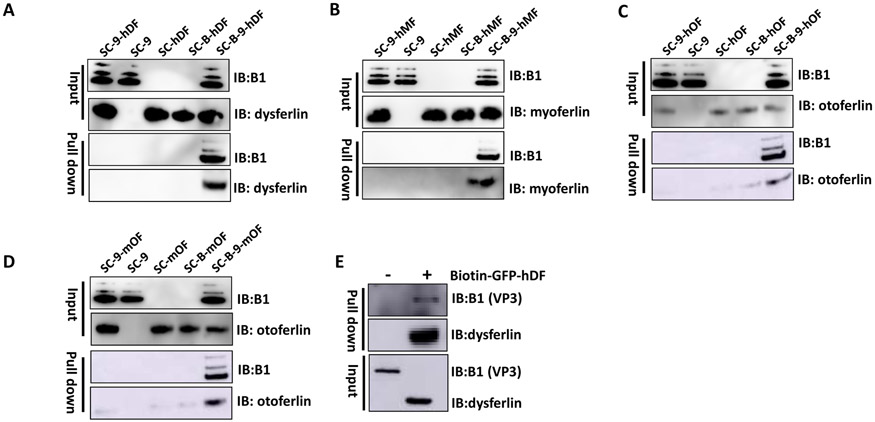
Next, we moved forward to explore the potential mechanism of which diverse FerA domains enhance multiple AAV serotypes transduction in vitro and in vivo. Our recent studies have reported that the serum proteins or the BBB shuttle peptides are able to directly interact with AAV virions and increase their transduction [32, 33, 40]. The data from non-incubation of AAV virions with mOto-FerA have suggested that direct interaction of AAV virions with FerA is required for transduction enhancement in cell lines and in mice. To more definitely substantiate the observation, we conducted a series of Streptavidin C1 beads-based pull down assays. The CaptureSelect™ biotin anti-AAV9 conjugate was added into the complex of pre-incubated AAV9 and hDys-, hMyo-, hOto-, or mOto-FerA domain for 1 h at RT, then Streptavidin C1 beads were put in for another 0.5 h at RT. Single or combined incubation of AAV9 or FerA with C1 beads were included as controls. Potential AAV9-bound FerA was detected by Western blotting with corresponding anti-FerA antibodies. The results showed that FerA domains were able to bind to the AAV9 effectively (Fig. 7 A, B, C, and D). It is also worth noting that the negatively-charged m/hOto-FerA domains slightly and inconspicuously bound to C1 beads, even with a stringent washing step during the pull-down assays (Fig. 7 C and D). The non-specific binding could be attributable to the propensity of Streptavidin C1 beads to non-specifically bind negatively charged molecules, according to the manufacturer’s product information. On the other hand, AAV9 particles could also be pulled down by the biotin-labeled GFP-hDys-FerA domain (Fig. 7 E). These results provide an alternative explanation for the in vivo observation noted above that the non-incubated group of mOto-FerA and AAV9 did not increase transduction, but on the contrary, significantly repressed the AAV9 transduction efficiency.
Fig. 7. FerA domains directly interacted with AAV9.
Purified AAV9 was first incubated with hDys-FerA (A), hMyo-FerA (B), hOto-FerA (C), or mOto-FerA domain (D) for 2 h at 4 °C, then CaptureSelect™ biotin anti-AAV9 conjugate was added into the complex for another 1 h at RT, followed by Streptavidin C1 beads-based pull down assays and immunoblotting analysis with indicated ferlin antibodies. Meanwhile, the incubated complexes of AAV9 with Streptavidin C1 beads, Streptavidin C1 beads with FerA domains, AAV9 and FerA domains with C1 beads, and the biotin anti-AAV9 conjugate and FerA domains with C1 beads were separately included as controls. sc: streptavidin C1 beads, 9: AAV9, B: biotin anti-AAV9 conjugate, hDF: hDys-FerA, hMF: hMyo-FerA, hOF: hOto-FerA, mOF: mOto-FerA. IB: immunoblot. (E) Immunoblot using B1 and dysferlin antibodies of pull down assay in which biotin-GFP-hDys-FerA protein was incubated with AAV9, followed by addition of Streptavidin C1 beads for 0.5 h at RT. IB: immunoblot.
3.7. FerA increases the AAV9 binding and trafficking ability to target cells
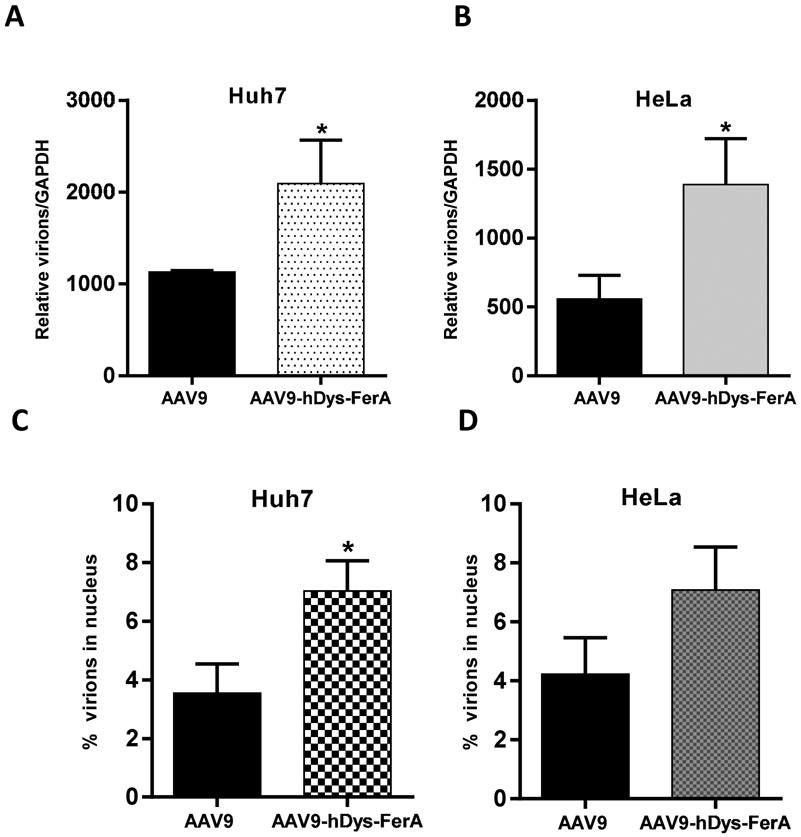
FerA domains from different ferlins are characterized by their membrane associating activity, even though the three FerA domains share rather low sequence similarity [36]. It has been known that membrane fusion proteins induce the joining of phospholipid bilayers for purposes such as endo- or exocytic trafficking. Such proteins are commonly employed by enveloped viruses for cellular entry. Different FerA domains could diversely bind to the phospholipid-containing liposomes in a Ca2+-dependent manner [39]. We hypothesized that FerA might affect the AAV binding ability to target cells. To examine this quantitatively, we performed an AAV9 virion-binding assay in Huh7 and HeLa cells. After AAV9/luc only or a pre-incubated AAV9-hDys-FerA complex was allowed to attach during incubation at 4 °C for 1 h, the unbound vectors were washed completely and gDNA was extracted. Finally, the viral gene copy numbers were determined by qPCR. The results showed that higher gene copy numbers were detected in the AAV9-hDys-FerA complex group (Fig. 8 A and B), which suggested that the FerA dramatically expedited the binding of AAV virions to the cell surface. Meanwhile, we also investigated the intracellular process after AAV binding to the cell surface via trafficking assay. The higher percentage of AAV vectors were detected in the nucleus compared to the total binding gene copy number in cells transduced with AAV pre-incubated with FerA (Fig. 8 C and D), which means that the FerA domain could accelerate AAV particles release from the endosomal compartment and entry into nucleus.
Fig. 8. FerA increased the AAV9 binding and trafficking ability to target cells in vitro.
The effect of FerA on the ability of AAV9 binding and trafficking to cells was assessed in Huh7 (A and C) and HeLa (B and D) cells. For the binding assay, the complex of AAV9 with DPBS or hDys-FerA domain was incubated with suspended cells for 1 h at 4 °C. After that, cells were washed by centrifugation to get rid of unbound viruses. Finally, gDNA from virus bound to cells was extracted. The AAV9 genome copy number was measured and normalized to GAPDH. For the trafficking assay, the cells were stringently washed 3 times and incubated at 37 °C for another 2 h after the binding process. The percentage of viral gene copy number in the nucleus of total bound viruses in Huh7 (C) or Hela (D) cells were calculated. All treatments were performed in triplicate. Asterisks indicate statistical significance compared to the AAV9/luc alone treatment group (*p < 0.05).
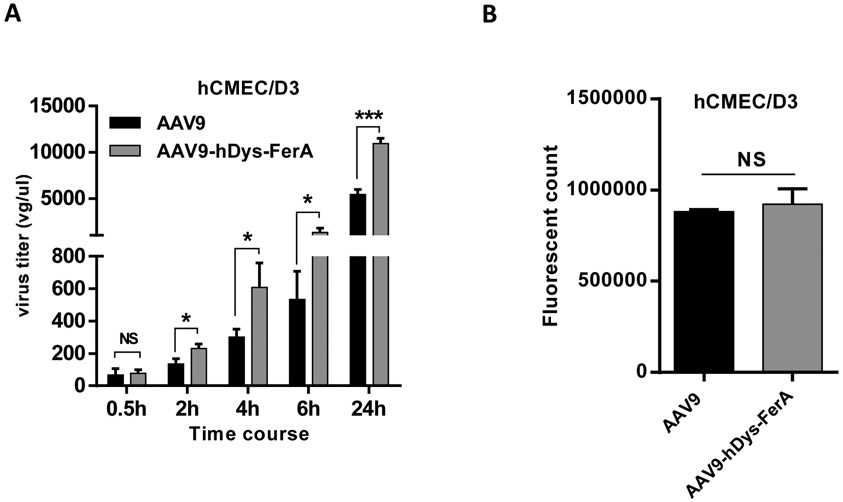
3.8. hDys-FerA enhances the transcytosis ability of AAV9
It has been demonstrated that enhanced global transduction was observed after systemic administration of AAV9 pre-incubated with FerA domains as described above (Fig. 2, 3, and 5). These results suggest that FerA has the potential to increase the ability of AAV virions to cross the blood vascular barrier. To assess the effect of FerA on the ability of AAV9 to cross the endothelial cell layer, we carried out an in vitro endothelial cell permeability analysis in the brain microvascular endothelial cell model hCMEC/D3 [46]. Two groups were designed as follows: AAV9 vector incubated with DPBS (AAV9 cohort) and AAV9 vector incubated with hDys-FerA (AAV9-hDys-FerA cohort). Increased cell permeability was observed at different time points, especially the 24 h, in the AAV9-hDys-FerA cohort groups (Fig. 9 A). The integrity of the endothelial monolayers was confirmed by FITC dextran fluorescence intensity analysis (Fig. 9 B). These results indicate that FerA boosts AAV9 ability to penetrate the BBB via transcytosis, which further explains the observations of enhanced AAV transduction in the mouse brain in vivo (Fig. 2 and 5).
Fig. 9. hDys-FerA enhanced the transcytosis ability of AAV9 in hCMEC/D3 cells line.
(A) hCMEC/D3 cells were cultured in a monolayer and incubated with either AAV9 or AAV9-hDys-FerA complex. The media in the basal chamber was collected at different indicated time points and the virus genome copy number was determined by qPCR. At the same time, the permeability assay for dextran conjugated with FITC (FITC-dextran) was also performed to check membrane integrity (B). In brief, at the indicated time points, FITC-dextran was added to the cells seeded on the collagen-coated cell inserts and incubated at RT for 0.5 h. Following incubation, inserts were removed and the medium remaining in the basal chamber was collected and analyzed for FITC-dextran fluorescence intensity using a microplate reader (BioTek Instruments Inc., Winooski, VT) with excitation and emission wavelengths of 485 nm and 530 nm. All treatments were performed in triplicate. ns p >0.05, ***p < 0.001 and *p < 0.05 compared to cells with AAV9/luc treatment only.
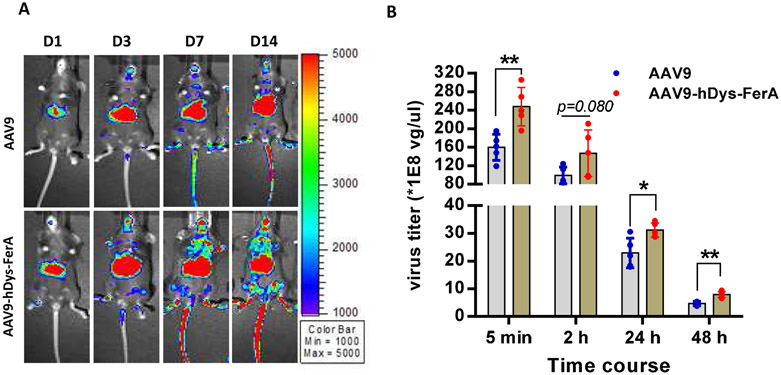
3.9. FerA decreases the AAV9 clearance in the blood without liver cytotoxicity
Slow clearance of AAV9 vector has been suggested to be associated with AAV9 global transduction [47]. Therefore, we examined whether FerA can influence AAV virions clearance in the blood after systemic administration. C57BL/6 female mice were intravenously administered 1×1011 vg per mouse of AAV9/luc or complex of vector pre-incubated with hDys-FerA (AAV9-hDys-FerA). Blood was collected at indicated time points (5 min, 2 h, 24 h, and 48 h) and the AAV genome copy number in the plasma was detected via qPCR. Consistent with the findings above, the hDys-FerA robustly enhanced the AAV9 global transduction (Fig. 10 A). The mice injected with the AAV9-hDys-FerA exhibited higher AAV virions in circulation at both early (5 min) and later time points (48 h) after systemic administration than mice treated with AAV9 alone (Fig. 10 B). This observation suggests that slow clearance of the AAV9 vector may increase the ability of AAV9 to cross the blood vascular barrier and enhance transduction in peripheral tissues after systemic administration. We also investigated the potential of liver cytotoxicity after AAV transduction by the detection of ALT and AST enzyme levels in the circulating blood and H&E staining. There was no obvious increase of the ALT and AST levels in the plasma after systemic administration (Fig. S6 A and B). In addition, no pathological change was found in mouse livers of two groups via H&E staining (Fig. S6 C).
Fig. 10. FerA decreased the AAV9 clearance in the blood.
(A) Mice were administrated with the incubated complex of 1×1011 vg AAV9/luc and 5 μg hDys-FerA via the retro-orbital injection. Luciferase expression was measured on days 1, 3, 7, and 14. (B) Blood was also collected from the retro-orbital plexus at different time points (5 min, 2 h, 24 h, and 48 h) after injection, and the viral titers were tested by qPCR. The data represent the average and SD from five mice. *p < 0.05 and **p <0.01 compared to the AAV9/luc alone treatment group.
4. Discussion
In the current study, we demonstrated that isolated FerA domains derived from different ferlin proteins were able to serve as “native molecules” and augment divergent AAV serotypes transduction by systemic or intramuscular administration. Mechanism researches showed that the FerA domain could directly interact with AAV, increase AAV’s binding activity on target cells, and its transcytosis ability. Meanwhile, it could delay the clearance of AAV particles from circulation to enhance AAV global transduction.
Currently, ongoing AAV-based gene delivery faces several major obstacles including the pre-existing neutralization antibody, the cellular immune response to AAV capsids and transgenes, and limited transgene expression. Numerous efforts have been focused on the genetic modification of AAV capsids and screenings of AAV variants to increase AAV transduction. However, these strategies may not be optimal when transferring to clinical trials because of unexpected tropism and toxicity [8, 26]. Exploration of “small molecules”, such as peptides, oligoes, or small proteins, are good candidates for AAV application considering their relatively low immunogenicity and cytotoxicity. Many studies have verified its feasibility. Previous studies including ours have shown that several serum proteins are able to directly interact with different AAV serotypes and enhance AAV transduction [29, 30, 32-34]. Herein, we found that FerA domains from four ferlin proteins of different species were capable of direct interaction with AAV and enhanced corresponding transduction. Importantly, due to the physical contact between FerA domains and AAV particles, the results could translate from mouse to larger species, even human beings, which suggests these human-derived “small proteins” could be immediately applied to patients who need a systemic administration of AAV vectors with lower anticipated immune responses. Therefore, it could represent a promising strategy for exploring natural and human-derived molecules to enhance the AAV transduction efficacy.
Different ferlin proteins have diverse tissue expression. For example, otoferlin is expressed in a wide range of tissues with high expression in the brain, vestibular system, and cochlea of the ear [37, 45]. It is interesting that higher brain transduction with Oto-FerA domain treatment was observed compared to that with the other two FerA domains (Fig. 5 C). One possible explanation is that the Oto-FerA domain being bound to AAV9 might facilitate AAV’s entry into the brain by increasing AAV’s ability to cross the BBB as demonstrated in the transcytosis assay (Fig. 9 A). Myoferlin shows ubiquitous expression, with high expression in developing skeletal muscle, cardiac muscle, and in the placenta [48]. Dysferlin is also ubiquitously expressed, with high expression in skeletal muscle, heart, and brain [49], implicating a role in surface membrane repair in muscle [50]. Herein, diverse FerA domains derived from different ferlin proteins could universally render an improved transduction in multiple AAV serotypes. Given the disparate distribution of ferlin proteins among various tissues and the variety of net surface charges among the different FerA domains, it merits further investigation of the affinity of various FerA domains with different AAV serotypes in the future.
The membrane fusion traits are commonly characterized by enveloped viruses for target cell binding and cellular entry [51]. Nevertheless, no consensus about the fusion mechanisms is elucidated. For the non-enveloped AAV infectious cycle, binding to the various attachment factors, such as distinct glycans and receptors, on the cell surface is the primary and crucial step for effective transduction [52, 53]. Our data showed that the FerA domains could directly interact with AAV particles and increase their binding to targeted cells, which indicates that the direct interaction of FerA domains with AAV virions for high target cell binding capacity play a role in AAV transduction enhancement.
One notable phenomenon is that the groups with non-incubated mOto-FerA and AAV9 or AAV8 could not increase the transduction, but significantly inhibited the AAV transduction after systemic administration (Fig. 2 and Fig. 3 B), which is in line with our recent finding that non-incubated THR peptide and AAV8 lead to decreased AAV8 transduction [40]. However, the concrete mechanisms leading to lower transduction from the simultaneous administration of mOto-FerA domain and AAV vectors is unknown. Several possibilities might explain the above observation. 1) The FerA domains bind to target cell surface and immediately induce some cellular response, which will block AAV virions to bind on target cells or later intracellular trafficking since AAV vector transduction is a slow process. 2) The excessive and dissociative FerA quickly and competitively occupies the AAV9 or AAV8 binding sites, leading to less AAV transduction. 3) Another possibility is that liver cells interacting with extra FerA domains may influence their binding ability to AAV virions via unknown mechanisms. These hypotheses need further investigation.
Safety remains the utmost important concern for AAV-mediated gene therapy in preclinical and clinical trials. Recent study showed that high-dose intravenous administration of AAV led to extreme toxicity affecting the liver and motor neurons, with symptoms appearing within five days of treatment in nonhuman primates and piglets [8]. FerA domains are naturally occurring intrinsic molecules in the human body, so, their use as accessory components for AAV transduction will exert a minimal impact on any immune response, especially a CTL response. Moreover, the solubility of isolated and purified FerA domain would likely not form complex aggregates, which would benefit its future manufacture and application.
5. Conclusion
In summary, we provide a proof of concept experiment that the FerA domains from different ferlin proteins could be leveraged as an “assistant cargo” to augment gene delivery of multiple AAV serotypes without altering inherent tissue tropism. Further deciphering the structural features of how the FerA domain interacts with AAV virions may assist in the design of more efficient AAV vectors for future clinical application.
Supplementary Material
Highlight.
FerA domains from diverse ferlin proteins enhance AAV transduction
FerA domains augment AAV transduction in AAV serotypes-independent manner
FerA domains directly interact with AAV virion
FerA domains fuel AAV binding, trafficking, and traversing ability
FerA domains slow down the blood clearance of AAV
ScAAV8/hFIX-FerA injection improves hemostasis in FIX−/− hemophilia B mice
Acknowledgments
We would like to thank Ellie Frost and Taylor Ralph for their great technical assistance. We acknowledge the UNC Biomedical Research Imaging Center (BRIC) Small Animal Imaging (SAI) facility for assistance with mouse imaging and UNC Histology Research Core Facility for histological services. This work was supported by National Institutes of Health Grants R01AI117408, R01HL144661, R01HL125749 (to C.L.), R01AR063634 (to R.B.S), P30-CA016086-35-37, and U54-CA151652-01-04 (to the BRIC SAI facility).
Footnotes
Conflicts of interest
Chengwen Li and Roger Bryan Sutton are cofounders of Bedrock Therapeutics, Inc. Chengwen Li has licensed patents by UNC and received royalties from Bedrock Therapeutics and Asklepios BioPharmaceutical, Inc.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Maguire AM, Simonelli F, Pierce EA, Pugh EN Jr., Mingozzi F, Bennicelli J, et al. Safety and efficacy of gene transfer for Leber's congenital amaurosis. The New England journal of medicine. 2008;358:2240–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Bainbridge JWB, Mehat MS, Sundaram V, Robbie SJ, Barker SE, Ripamonti C, et al. Long-Term Effect of Gene Therapy on Leber’s Congenital Amaurosis. New England Journal of Medicine. 2015;372:1887–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lisowski L, Tay SS, Alexander IE. Adeno-associated virus serotypes for gene therapeutics. Current opinion in pharmacology. 2015;24:59–67. [DOI] [PubMed] [Google Scholar]
- [4].Nathwani AC, Rosales C, McIntosh J, Rastegarlari G, Nathwani D, Raj D, et al. Long-term Safety and Efficacy Following Systemic Administration of a Self-complementary AAV Vector Encoding Human FIX Pseudotyped With Serotype 5 and 8 Capsid Proteins. Molecular Therapy. 2011;19:876–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bowles DE, McPhee SWJ, Li C, Gray SJ, Samulski JJ, Camp AS, et al. Phase 1 Gene Therapy for Duchenne Muscular Dystrophy Using a Translational Optimized AAV Vector. Molecular Therapy. 2012;20:443–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hudry E, Vandenberghe LH. Therapeutic AAV Gene Transfer to the Nervous System: A Clinical Reality. Neuron. 2019;101:839–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Colella P, Ronzitti G, Mingozzi F. Emerging Issues in AAV-Mediated In Vivo Gene Therapy. Molecular therapy Methods & clinical development. 2018;8:87–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hinderer C, Katz N, Buza EL, Dyer C, Goode T, Bell P, et al. Severe Toxicity in Nonhuman Primates and Piglets Following High-Dose Intravenous Administration of an Adeno-Associated Virus Vector Expressing Human SMN. Human gene therapy. 2018;29:285–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Nathwani AC, Reiss UM, Tuddenham EG, Rosales C, Chowdary P, McIntosh J, et al. Long-term safety and efficacy of factor IX gene therapy in hemophilia B. The New England journal of medicine. 2014;371:1994–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Manno CS, Pierce GF, Arruda VR, Glader B, Ragni M, Rasko JJ, et al. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nature medicine. 2006;12:342–7. [DOI] [PubMed] [Google Scholar]
- [11].Siders WM, Shields J, Kaplan J, Lukason M, Woodworth L, Wadsworth S, et al. Cytotoxic T lymphocyte responses to transgene product, not adeno-associated viral capsid protein, limit transgene expression in mice. Human gene therapy. 2009;20:11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Shao W, Earley LF, Chai Z, Chen X, Sun J, He T, et al. Double-stranded RNA innate immune response activation from long-term adeno-associated virus vector transduction. JCI insight. 2018;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Muller OJ, Kaul F, Weitzman MD, Pasqualini R, Arap W, Kleinschmidt JA, et al. Random peptide libraries displayed on adeno-associated virus to select for targeted gene therapy vectors. Nature biotechnology. 2003;21:1040–6. [DOI] [PubMed] [Google Scholar]
- [14].Deverman BE, Pravdo PL, Simpson BP, Kumar SR, Chan KY, Banerjee A, et al. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nature biotechnology. 2016;34:204–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Chai Z, Sun JJ, Rigsbee KM, Wang M, Samulski RJ, Li CW. Application of polyploid adeno-associated virus vectors for transduction enhancement and neutralizing antibody evasion. Journal of Controlled Release. 2017;262:348–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Asokan A, Conway JC, Phillips JL, Li C, Hegge J, Sinnott R, et al. Reengineering a receptor footprint of adeno-associated virus enables selective and systemic gene transfer to muscle. Nature biotechnology. 2010;28:79–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bartel MA, Weinstein JR, Schaffer DV. Directed evolution of novel adeno-associated viruses for therapeutic gene delivery. Gene therapy. 2012;19:694–700. [DOI] [PubMed] [Google Scholar]
- [18].Yang Q, Mamounas M, Yu G, Kennedy S, Leaker B, Merson J, et al. Development of novel cell surface CD34-targeted recombinant adenoassociated virus vectors for gene therapy. Human gene therapy. 1998;9:1929–37. [DOI] [PubMed] [Google Scholar]
- [19].Munch RC, Janicki H, Volker I, Rasbach A, Hallek M, Buning H, et al. Displaying high-affinity ligands on adeno-associated viral vectors enables tumor cell-specific and safe gene transfer. Molecular therapy : the journal of the American Society of Gene Therapy. 2013;21:109–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Büning H, Srivastava A. Capsid Modifications for Targeting and Improving the Efficacy of AAV Vectors. Molecular therapy Methods & clinical development. 2019;12:248–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kim E, Oh JS, Ahn IS, Park KI, Jang JH. Magnetically enhanced adeno-associated viral vector delivery for human neural stem cell infection. Biomaterials. 2011;32:8654–62. [DOI] [PubMed] [Google Scholar]
- [22].Muik A, Reul J, Friedel T, Muth A, Hartmann KP, Schneider IC, et al. Covalent coupling of high-affinity ligands to the surface of viral vector particles by protein trans-splicing mediates cell type-specific gene transfer. Biomaterials. 2017;144:84–94. [DOI] [PubMed] [Google Scholar]
- [23].Hordeaux J, Wang Q, Katz N, Buza EL, Bell P, Wilson JM. The Neurotropic Properties of AAV-PHP.B Are Limited to C57BL/6J Mice. Molecular therapy : the journal of the American Society of Gene Therapy. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Matsuzaki Y, Konno A, Mochizuki R, Shinohara Y, Nitta K, Okada Y, et al. Intravenous administration of the adeno-associated virus-PHP.B capsid fails to upregulate transduction efficiency in the marmoset brain. Neuroscience letters. 2017. [DOI] [PubMed]
- [25].Matsuzaki Y, Tanaka M, Hakoda S, Masuda T, Miyata R, Konno A, et al. Neurotropic Properties of AAV-PHP.B Are Shared among Diverse Inbred Strains of Mice. Molecular Therapy. 2019. [DOI] [PMC free article] [PubMed]
- [26].Liguore WA, Domire JS, Button D, Wang Y, Dufour BD, Srinivasan S, et al. AAV-PHP.B Administration Results in a Differential Pattern of CNS Biodistribution in Non-human Primates Compared with Mice. Molecular therapy : the journal of the American Society of Gene Therapy. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hurlbut GD, Ziegler RJ, Nietupski JB, Foley JW, Woodworth LA, Meyers E, et al. Preexisting immunity and low expression in primates highlight translational challenges for liver-directed AAV8-mediated gene therapy. Molecular therapy : the journal of the American Society of Gene Therapy. 2010;18:1983–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Michelfelder S, Varadi K, Raupp C, Hunger A, Korbelin J, Pahrmann C, et al. Peptide Ligands Incorporated into the Threefold Spike Capsid Domain to Re-Direct Gene Transduction of AAV8 and AAV9 In Vivo. PloS one. 2011;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Denard J, Beley C, Kotin R, Lai-Kuen R, Blot S, Leh H, et al. Human galectin 3 binding protein interacts with recombinant adeno-associated virus type 6. Journal of virology. 2012;86:6620–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Denard J, Rouillon J, Leger T, Garcia C, Lambert MP, Griffith G, et al. AAV-8 and AAV-9 Vectors Cooperate with Serum Proteins Differently Than AAV-1 and AAV-6. Molecular therapy Methods & clinical development. 2018;10:291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Rapti K, Louis-Jeune V, Kohlbrenner E, Ishikawa K, Ladage D, Zolotukhin S, et al. Neutralizing Antibodies Against AAV Serotypes 1, 2, 6, and 9 in Sera of Commonly Used Animal Models. Molecular Therapy. 2012;20:73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wang M, Sun J, Crosby A, Woodard K, Hirsch ML, Samulski RJ, et al. Direct interaction of human serum proteins with AAV virions to enhance AAV transduction: immediate impact on clinical applications. Gene therapy. 2017;24:49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Pei X, He T, Hall NE, Gerber D, Samulski RJ, Li C. AAV8 virions hijack serum proteins to increase hepatocyte binding for transduction enhancement. Virology. 2018;518:95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Chai Z, Zhang X, Rigsbee KM, Wang M, Samulski RJ, Li C. Cryoprecipitate augments the global transduction of the adeno-associated virus serotype 9 after a systemic administration. Journal of controlled release : official journal of the Controlled Release Society. 2018;286:415–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Denard J, Marolleau B, Jenny C, Rao TN, Fehling HJ, Voit T, et al. C-Reactive Protein (CRP) Is Essential for Efficient Systemic Transduction of Recombinant Adeno-Associated Virus Vector 1 (rAAV-1) and rAAV-6 in Mice. Journal of virology. 2013;87:10784–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Posey AD Jr., Demonbreun A, McNally EM. Ferlin proteins in myoblast fusion and muscle growth. Current topics in developmental biology. 2011;96:203–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lek A, Evesson FJ, Sutton RB, North KN, Cooper ST. Ferlins: Regulators of Vesicle Fusion for Auditory Neurotransmission, Receptor Trafficking and Membrane Repair. Traffic. 2012;13:185–94. [DOI] [PubMed] [Google Scholar]
- [38].Therrien C, Dodig D, Karpati G, Sinnreich M. Mutation impact on dysferlin inferred from database analysis and computer-based structural predictions. Journal of the neurological sciences. 2006;250:71–8. [DOI] [PubMed] [Google Scholar]
- [39].Harsini FM, Chebrolu S, Fuson KL, White MA, Rice AM, Sutton RB. FerA is a Membrane-Associating Four-Helix Bundle Domain in the Ferlin Family of Membrane-Fusion Proteins. Scientific reports. 2018;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zhang X, He T, Chai Z, Samulski RJ, Li C. Blood-brain barrier shuttle peptides enhance AAV transduction in the brain after systemic administration. Biomaterials. 2018;176:71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Grieger JC, Choi VW, Samulski RJ. Production and characterization of adeno-associated viral vectors. Nature protocols. 2006;1:1412–28. [DOI] [PubMed] [Google Scholar]
- [42].Wu Z, Sun J, Zhang T, Yin C, Yin F, Van Dyke T, et al. Optimization of self-complementary AAV vectors for liver-directed expression results in sustained correction of hemophilia B at low vector dose. Molecular therapy : the journal of the American Society of Gene Therapy. 2008;16:280–9. [DOI] [PubMed] [Google Scholar]
- [43].Jin DY, Zhang TP, Gui T, Stafford DW, Monahan PE. Creation of a mouse expressing defective human factor IX. Blood. 2004;104:1733–9. [DOI] [PubMed] [Google Scholar]
- [44].Wu Z, Sun J, Zhang T, Yin C, Yin F, Van Dyke T, et al. Optimization of Self-complementary AAV Vectors for Liver-directed Expression Results in Sustained Correction of Hemophilia B at Low Vector Dose. Mol Ther. 2008;16:280–9. [DOI] [PubMed] [Google Scholar]
- [45].Yasunaga S, Grati M, Chardenoux S, Smith TN, Friedman TB, Lalwani AK, et al. OTOF encodes multiple long and short isoforms: Genetic evidence that the long ones underlie recessive deafness DFNB9. Am J Hum Genet. 2000;67:591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Weksler B, Romero IA, Couraud PO. The hCMEC/D3 cell line as a model of the human blood brain barrier. Fluids and barriers of the CNS. 2013;10:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kotchey NM, Adachi K, Zahid M, Inagaki K, Charan R, Parker RS, et al. A potential role of distinctively delayed blood clearance of recombinant adeno-associated virus serotype 9 in robust cardiac transduction. Molecular therapy : the journal of the American Society of Gene Therapy. 2011;19:1079–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Davis DB, Delmonte AJ, Ly CT, McNally EM. Myoferlin, a candidate gene and potential modifier of muscular dystrophy. Hum Mol Genet. 2000;9:217–26. [DOI] [PubMed] [Google Scholar]
- [49].Anderson LVB, Davison K, Moss JA, Young C, Cullen MJ, Walsh J, et al. Dysferlin is a plasma membrane protein and is expressed early in human development. Hum Mol Genet. 1999;8:855–61. [DOI] [PubMed] [Google Scholar]
- [50].Doherty KR, McNally EM. Repairing the tears: dysferlin in muscle membrane repair. Trends Mol Med. 2003;9:327–30. [DOI] [PubMed] [Google Scholar]
- [51].Harrison SC. Viral membrane fusion. Virology. 2015;479–480:498–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Huang L- Y, Halder S, Agbandje-McKenna M. Parvovirus glycan interactions. Current Opinion in Virology. 2014;7:108–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Pillay S, Meyer NL, Puschnik AS, Davulcu O, Diep J, Ishikawa Y, et al. An essential receptor for adeno-associated virus infection. Nature. 2016;530:108–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.