Abstract
Peptides are known to contribute to central pattern generator (CPG) flexibility throughout the animal kingdom. However, the role played by receptor diversity/complement in determining this functional flexibility is not clear. The stomatogastric ganglion (STG) of the crab, Cancer borealis, contains CPGs that are models for investigating peptidergic control of rhythmic behavior. Although many Cancer peptides have been identified, their peptide receptors are largely unknown. Thus, the extent to which receptor diversity/complement contributes to modulatory flexibility in this system remains unresolved. Here, a Cancer mixed nervous system transcriptome was used to determine the peptide receptor complement for the crab nervous system as a whole. Receptors for 27 peptide families, including multiple receptors for some groups, were identified. To increase confidence in the predicted sequences, receptors for allatostatin-A, allatostatin-B, and allatostatin-C were cloned, sequenced, and expressed in an insect cell line; as expected, all three receptors trafficked to the cell membrane. RT-PCR was used to determine whether each receptor was expressed in the Cancer STG. Transcripts for 36 of the 46 identified receptors were amplified; these included at least one for each peptide family except RYamide. Finally, two peptides untested on the crab STG were assessed for their influence on its motor outputs. Myosuppressin, for which STG receptors were identified, exhibited clear modulatory effects on the motor patterns of the ganglion, while a native RYamide, for which no STG receptors were found, elicited no consistent modulatory effects. These data support receptor diversity/complement as a major contributor to the functional flexibility of CPGs.
Keywords: Cancer borealis, stomatogastric ganglion (STG), transcriptomics, RT-PCR, electrophysiology
1. Introduction
Neural circuits that control rhythmic movement patterns are generally hard-wired, but are nonetheless capable of producing a wide array of outputs, enabling them to respond appropriately to changing internal and external environmental conditions (e.g., Dickinson et al., 2016; Nusbaum and Blitz, 2012; Nusbaum et al., 2017; Taghert and Nitabach, 2012). Underlying this functional flexibility is a dazzling array of neuromodulators; these include both locally released and hormonally delivered compounds, among which peptides are the largest and most diverse single class (e.g., Christie, 2011; Christie et al., 2010).
The decapod crustacean stomatogastric nervous system (STNS; Fig. 1), which controls the rhythmic movements of the foregut, has long been used as a model for studying the modulation of simple pattern generating networks (e.g., Harris-Warrick et al., 1992; Selverston and Moulins, 1987). Within the STNS is the stomatogastric ganglion (STG), which contains the ~25 neurons that comprise two pattern generating networks: the gastric mill network, which controls the rhythmic chewing movements of the three gastric mill teeth, and the pyloric network, which generates a triphasic output that underlies the rhythmic movements of the pyloric filter (Harris-Warrick et al., 1992; Selverston and Moulins, 1987). Both the gastric mill and pyloric rhythms are highly modulated by a variety of chemical compounds, including peptides (Harris-Warrick et al., 1992; Selverston and Moulins, 1987).
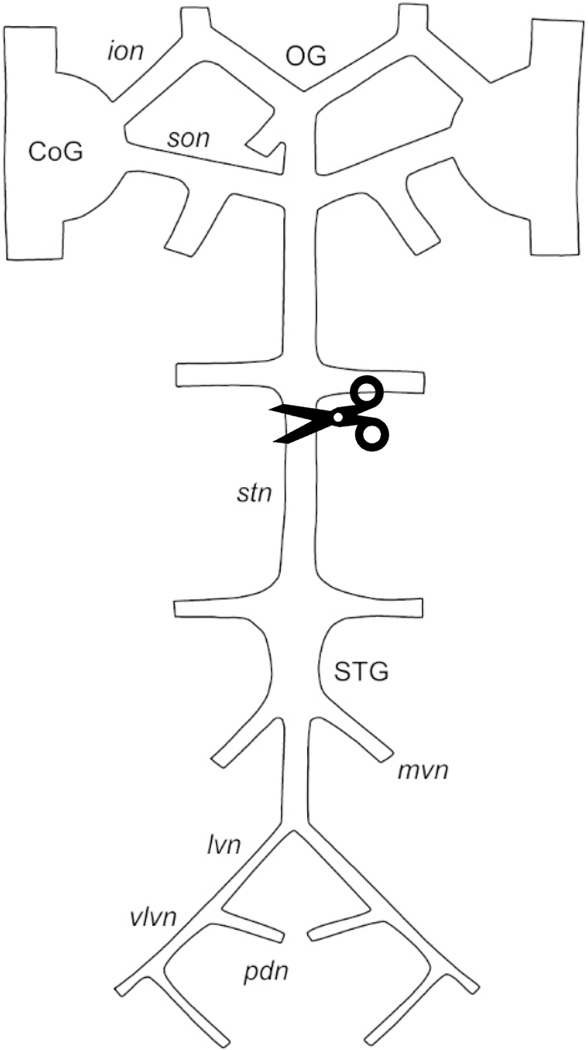
Figure 1.
Schematic diagram of the stomatogastric nervous system (STNS). Scissors indicate the region in which the stomatogastric nerve (stn) was blocked and cut to eliminate input from the anterior ganglia. CoG, commissural ganglion; ion, inferior oesophageal nerve; lvn, lateral ventricular nerve; mvn, medial ventricular nerve; OG, oesophageal ganglion; pdn, pyloric dilator nerve; son, superior oesophageal nerve; STG, stomatogastric ganglion; stn, stomatogastric nerve; vlvn, ventral lateral ventricular nerve.
Previous work on the nervous system of the Jonah crab, Cancer borealis, suggests the presence of ~200 neuropeptides, which includes members of 27 different peptide families (e.g., Christie and Pascual, 2016; Christie et al., 1997; Fu et al., 2005a; Huybrechts et al., 2003; Li et al., 2002, 2003; Stemmler et al., 2007a, 2007b). While members of many of these peptide families have been shown to modulate the gastric mill and/or pyloric networks (e.g., Blitz et al., 2019; Christie et al., 1997; Cruz-Bermúdez et al., 2006; Fu et al., 2007; Li et al., 2002; Ma et al., 2009a; Marder et al., 1986; Saideman et al., 2006, 2007; Swensen and Marder, 2000, 2001; Szabo et al., 2011; Weimann et al., 1993, 1997), the functions of members of other groups remain unknown. Some peptide families consist of a single isoform, e.g., proctolin, whereas others are comprised of a large number of family members, e.g., the A-type allatostatins (AST-As), for which ~30 different isoforms have been identified in C. borealis (e.g., Christie and Pascual, 2016). This raises at least three questions. First, do different isoforms from a common peptide family exert the same or different effects on a given CPG? Several studies have begun to address this question in the STNS and other pattern generating networks in decapods (e.g., Christie et al., 2008; Dickinson et al., 2007, 2015a, 2015b, 2015c, 2018; Ma et al., 2009a; Saideman et al., 2007; Stemmler et al., 2007b; Szabo et al., 2011). Second, are there more receptors for peptides in families that appear to have larger numbers of isoforms, and does the presence of larger numbers of receptors correlate with more variable modulatory responses? The second question remains largely unanswered because the receptors for the vast majority of the peptides identified in C. borealis have not yet been identified (Garcia et al., 2015). In fact, few studies have focused on the large-scale identification/characterization of peptide receptors in any crustacean species (e.g., Buckley et al., 2016; Christie and Yu, 2019; Christie et al., 2013, 2015, 2018a, 2018b). Finally, does the presence or absence of receptors for a peptide family in a CPG provide an accurate means for predicting whether or not a peptide from that family is bioactive on the neural circuits of the pattern generating system in question, e.g., those present in the C. borealis STG?
Here, a transcriptome generated from multiple regions of the C. borealis nervous system, including the STG (BioProject No. PRJNA310325; Northcutt et al., 2016), was used to identify 46 distinct putative receptors for members of 27 peptide families. These data allowed us to compare the number of Cancer peptide precursor genes/peptide isoforms to the number of putative receptor genes within each peptide family. Three receptors were fully cloned, sequenced and expressed in insect cell lines to confirm cell surface trafficking. To determine whether each identified receptor is likely present in the C. borealis STG, and hence might contribute to the modulation of the gastric mill and/or pyloric networks, RNA from the ganglion was isolated and used in RT-PCR to determine the presence/absence of receptor transcript expression. Finally, two peptides that had not previously been tested for bioactivity in C. borealis were selected for physiological testing. We predicted that a peptide for which receptors were present in the STG would modulate the gastric mill and/or pyloric motor patterns, whereas another peptide, which lacks molecular evidence for cognate receptor expression in the ganglion, would not exert consistent modulatory effects on either rhythm.
2. Materials and methods
2.1. Animals
Crabs, C. borealis, were purchased from local (Brunswick/Harpswell, Maine, USA) seafood retailers, and housed in recirculating natural seawater aquaria at 10–12°C; crabs were fed approximately weekly with chopped squid and/or shrimp.
2.2. In silico identification of putative peptide receptors
2.2.1. Database searches
Searches to identify transcripts encoding putative C. borealis peptide receptors for 30 peptide families were conducted using methods modified from a well-established protocol (e.g., Christie and Yu, 2019; Christie et al., 2013, 2015, 2016, 2018a, 2018b). Specifically, the database of the online program tblastn (National Center for Biotechnology Information, Bethesda, MD; http://blast.ncbi.nlm.nih.gov/Blast.cgi) was set to Transcriptome Shotgun Assembly (TSA) and restricted to data from a C. borealis mixed neural tissue transcriptome (BioProject No. PRJNA310325; Northcutt et al., 2016). For the majority of searches, peptide receptors from the fruit fly, Drosophila melanogaster, were used as the query proteins (e.g., Adams et al., 2000). The complete list of peptide receptor families searched for in this study, as well as the specific query proteins used in the BLAST searches, is provided in Table 1.
Table 1.
Cancer borealis (Canbo) peptide receptor-encoding transcripts and their deduced proteins
| Peptide family | Transcript/receptor protein identifications | ||||
|---|---|---|---|---|---|
| Transcript | Deduced protein | ||||
| Accession No. | Length* | Name | Length† | Type | |
| Adipokinetic hormone-corazonin-like peptide (ACP) | GEFB01018628 | 1827 | Canbo-ACPR | 470 | N |
| Allatostatin A (AST-A) | GEFB01012018 | 1522 | Canbo-AST-AR | 454 | F |
| Allatostatin B (AST-B) | GEFB01014490 | 3636 | Canbo-AST-BR | 398 | F |
| Allatostatin C (AST-C) | GEFB01019215 | 3779 | Canbo-AST-CR | 426 | F |
| Allatotropin | None found | - | - | - | - |
| Bursicon (Burs) | GEFB01007449 | 3802 | Canbo-BursR | 1178 | N |
| CCHamide (CCHa) | GEFB01030009 | 2660 | Canbo-CCHaR-I | 409 | F |
| GEFB01036413 | 1541 | Canbo-CCHaR-II | 400 | F | |
| GEFB01015997 | 2011 | Canbo-CCHaR-III | 415 | F | |
| GEFB01038814 | 1333 | Canbo-CCHaR-IV | 235 | C | |
| GEFB01039418 | 634 | Canbo-CCHaR-V | 133 | N | |
| Corazonin (CRZ) | GEFB01026704 | 790 | Canbo-CRZR-I | 243 | N |
| GEFB01031741 | 833 | Canbo-CRZR-II | 277 | I | |
| Crustacean cardioactive peptide (CCAP) | GEFB01008615 | 2904 | Canbo-CCAPR | 390 | N |
| Crustacean hyperglycemic hormone (CHH) | None found | - | - | - | - |
| Diuretic hormone 31 (DH31) | GEFB01018473 | 2168 | Canbo-DH31R | 416 | F |
| Diuretic hormone 44 (DH44) | GEFB01015824 | 1661 | Canbo-DH44R | 391 | C |
| Ecdysis-triggering hormone (ETH) | GEFB01025040 | 2548 | Canbo-ETHR-I | 654 | F |
| GEFB01022057 | 2089 | Canbo-ETHR-II | 472 | F | |
| GEFB01031733 | 1171 | Canbo-ETHR-III | 317 | C | |
| FMRFamide-like peptide (FLP) | GEFB01000837 | 3602 | Canbo-FLPR-I | 463 | F |
| GEFB01020234 | 3110 | Canbo-FLPR-II | 428 | F | |
| Glycoprotein hormone (GPH) | GEFB01005208 | 5862 | Canbo-GPHR | 1605 | F |
| Inotocin | GEFB01030964 | 1528 | Canbo-inotocinR | 363 | N |
| Insulin-like peptide (ILP) | None found | - | - | - | - |
| Leucokinin (LK) | GEFB01016835 | 894 | Canbo-LKR | 298 | I |
| Myosuppressin (MS) | GEFB01024737 | 2151 | Canbo-MSR-I | 338 | F |
| GEFB01027877 | 2615 | Canbo-MSR-II | 408 | F | |
| Neuropeptide F (NPF) | GEFB01022366 | 1605 | Canbo-NPFR-I | 432 | F |
| GEFB01025235 | 1808 | Canbo-NPFR-II | 468 | F | |
| GEFB01030636 | 2699 | Canbo-NPFR-III | 471 | F | |
| GEFB01028428 | 1694 | Canbo-NPFR-IV | 345 | N | |
| Pigment dispersing hormone (PDH) | GEFB01009443 | 1248 | Canbo-PDHR-I | 317 | F |
| GEFB01031257 | 686 | Canbo-PDHR-II | 228 | I | |
| GEFB01037166 | 827 | Canbo-PDHR-III | 212 | N | |
| Proctolin (Proc) | GEFB01004771 | 4715 | Canbo-ProcR | 668 | F |
| Pyrokinin (PK) | GEFB01015867 | 748 | Canbo-PKR-I | 190 | C |
| GEFB01028557 | 1878 | Canbo-PKR-II | 499 | F | |
| Red pigment concentrating hormone (RPCH) | GEFB01027769 | 1493 | Canbo-RPCHR | 278 | N |
| RYamide (RYa) | GEFB01016897 | 1635 | Canbo-RYaR-I | 461 | C |
| GEFB01016602 | 2233 | Canbo-RYaR-II | 447 | C | |
| Short neuropeptide F (sNPF) | GEFB01013521 | 7328 | Canbo-sNPFR | 456 | F |
| SIFamide (SIFa) | GEFB01030224 | 1956 | Canbo-SIFaR-I | 530 | F |
| GEFB01028504 | 1463 | Canbo-SIFaR-II | 487 | I | |
| Sulfakinin (SK) | GEFB01033477 | 2935 | Canbo-SKR | 706 | F |
| Tachykinin-related peptide (TRP) | GEFB01026365 | 2066 | Canbo-TRPR-I | 677 | C |
| GEFB01038657 | 1460 | Canbo-TRPR-II | 318 | N | |
| GEFB01007474 | 3313 | Canbo-TRPR-III | 383 | F | |
| Trissin | GEFB01026709 | 2386 | Canbo-trissinR | 539 | F |
Length in nucleotides.
Length in amino acids.
Protein type abbreviations: F, full-length protein; N, amino-terminal partial protein; I, internal fragment protein; C, carboxyl-terminal partial protein.
Query sequences: ACP, Tribolium castaneum ACP receptor (ABX52400; Hansen et al., 2010); AST-A Drosophila melanogaster allatostatin A receptor 1, isoform B (AAF45884; Adams et al., 2000) and D. melanogaster allatostatin A receptor 2, isoform A (AAF56809; Adams et al., 2000); AST-B, D. melanogaster sex peptide receptor, isoform A (AAF46037; Adams et al., 2000); AST-C, D. melanogaster allatostatin C receptor 1 (AAF49259; Adams et al., 2000) and D. melanogaster allatostatin C receptor 2, isoform B (AAN11677; Adams et al., 2000); allatotropin, Manduca sexta allatotropin receptor (ADX66344; Horodyski et al., 2011); Burs, D. melanogaster rickets (AAF53367; Adams et al., 2000); CCHa, D. melanogaster CCHamide-1 receptor (AAF57819; Adams et al., 2000) and D. melanogaster CCHamide-2 receptor, isoform A (AAF57285; Adams et al., 2000); CZR, D. melanogaster corazonin receptor, isoform A (AAF49928; Adams et al., 2000); CCAP, D. melanogaster crustacean cardioactive peptide receptor (AAF56536; Adams et al., 2000); CHH, Bombyx mori neuropeptide receptor A2 (BAG68400; Yamanaka et al., 2008), B. mori neuropeptide receptor A24 (BAG68423; Yamanaka et al., 2008) and B. mori neuropeptide receptor A34 (BAG68433; Yamanaka et al., 2008); DH31, D. melanogaster diuretic hormone 31 receptor, isoform A (AAN16138; Adams et al., 2000); DH44, D. melanogaster diuretic hormone 44 receptor 1 (AAF58250; Adams et al., 2000) and D. melanogaster diuretic hormone 44 receptor 2, isoform A (AAF58501; Adams et al., 2000); ETH, D. melanogaster ETHR, isoform A (AAF55872; Adams et al., 2000); FLP, D. melanogaster FMRFamide receptor, isoform A (AF47700; Adams et al., 2000); GPH, D. melanogaster leucine-rich repeat-containing G protein-coupled receptor 1, isoform A (AAF55460; Adams et al., 2000); inotocin, T. castaneum arginine vasopressin receptor (ABN79656; Aikins et al., 2008); ILP, D. melanogaster insulin-like receptor, isoform A (AAF55903; Adams et al., 2000); LK, D. melanogaster leucokinin receptor (AAF50775; Adams et al., 2000); MS, D. melanogaster myosuppressin receptor 1, isoform A (AAF47635; Adams et al., 2000) and D. melanogaster myosuppressin receptor 2, isoform A (AAF47633; Adams et al., 2000); NPF, D. melanogaster neuropeptide F receptor, isoform A (AAF51909; Adams et al., 2000); PDH, D. melanogaster pigment-dispersing factor receptor, isoform A (AAF45788; Adams et al., 2000); Proc, D. melanogaster proctolin receptor, isoform A (AAF45980; Adams et al., 2000); PK, D. melanogaster capability receptor, isoform B (AAS65092; Adams et al., 2000), D. melanogaster pyrokinin 1 receptor, isoform D (AAX52950; Adams et al., 2000), D. melanogaster pyrokinin 2 receptor 1 (AAF54930; Adams et al., 2000) and D. melanogaster pyrokinin 2 receptor 2, isoform A (AAF54929; Adams et al., 2000); RPCH, D. melanogaster adipokinetic hormone receptor, isoform A (AAF52426; Adams et al., 2000); RYa, D. melanogaster RYamide receptor, isoform A (AAF56655; Adams et al., 2000); sNPF, D. melanogaster short neuropeptide F receptor, isoform A (AAF49074; Adams et al., 2000); SIFa, D. melanogaster SIFamide receptor, isoform A (AAN13859; Adams et al., 2000); SK, D. melanogaster cholecystokinin-like receptor at 17D3 (AAF48879; Adams et al., 2000) and D. melanogaster cholecystokinin-like receptor at 17D1 (ABW09450; Adams et al., 2000); TRP, D. melanogaster tachykinin-like receptor at 86C, isoform A (AAF54544; Adams et al., 2000) and D. melanogaster tachykinin-like receptor at 99D, isoform A (AAF56979; Adams et al., 2000); trissin, D. melanogaster trissin receptor, isoform B (AAF52294; Adams et al., 2000).
2.2.2. Identification and vetting of candidate peptide receptors
Candidate C. borealis peptide receptors were predicted and vetted using a workflow developed for identifying a variety of crustacean proteins, including peptide receptors (e.g., Christie and Yu, 2019; Christie et al., 2013, 2015, 2016, 2018a, 2018b). First, nucleotide sequences identified as described above were translated using the Translate tool of ExPASy (http://web.expasy.org/translate/) and assessed for completeness. Receptors reported as full-length contain in-frame codons encoding a start methionine and downstream stop, while those described as partial lack a start methionine (referred to as carboxyl [C]-terminal partial proteins), a stop codon (referred to as amino [N]-terminal partial proteins), or both of these features (referred to as internal fragment proteins). Next, to confirm that each C. borealis receptor is most similar to the D. melanogaster protein used to identify the transcript encoding it, the C. borealis sequence was used as the query in a BLAST search of the annotated D. melanogaster protein dataset present in FlyBase (version FB2017_04; Gramates et al., 2017). For searches using non-Drosophila proteins as the initial queries, a search of the query species’ non-redundant protein dataset was used for the reciprocal BLAST rather than the FlyBase D. melanogaster dataset. The top five arthropod protein hits for each C. borealis sequence were determined by conducting a BLAST search of the non-redundant arthropod proteins curated in NCBI (taxid:6656). Finally, protein structural motifs were predicted for each of the C. borealis receptors using the online program Pfam (version 29.0; http://pfam.xfam.org/; Finn et al., 2016). FlyBase and NCBI non-redundant arthropod protein reciprocal BLAST searches were conducted on or before January 1, 2018. Protein alignments were done using the online program MAFFT version 7 (http://mafft.cbrc.jp/alignment/software/; Katoh and Standley, 2013). To determine amino acid conservation between selected proteins, the sequences in question were aligned using MAFFT, and amino acid identity/similarity subsequently determined using the alignment output. Specifically, percent identity was calculated as the number of identical amino acids divided by the total number of residues in the longest sequence (x100), while amino acid similarity was calculated as the number of identical and similar amino acids divided by the total number of residues in the longest sequence (x100).
2.2.3. Assessment of phylogenetic relationships among deduced receptor proteins
The phylogenetic relationships among the putative C. borealis peptide receptors (sequences shown in Supplemental Figure 1) were inferred from a multiple sequence alignment constructed using default MUSCLE (Edgar, 2004) settings in Geneious v10.1.3. Evolutionary analyses were conducted in MEGA X (Kumar et al., 2018) using the Neighbor-Joining method (Saitou and Nei, 1987). The optimal tree with the sum of branch length = 16.87124773 is shown. The rate variation among sites was modeled with a gamma distribution (shape parameter = 1) across 47 protein sequences. The evolutionary distances were computed using the p-distance method (Nei and Kumar, 2000) and are in the units of the number of amino acid differences per site. All ambiguous positions were removed for each sequence pair, with a total of 1855 positions in the final dataset.
A more refined examination of the phylogenetic relationships between the putative C. borealis AST-A, allatostatin B (AST-B), and allatostatin C (AST-C) receptors with sequences from diverse arthropods annotated as receptors for these peptide families was also conducted; accession numbers of the sequences used are provided in Supplemental Table 1. As before, a multiple sequence alignment was constructed using default MUSCLE settings, but with phylogeny estimated using the Maximum Likelihood method based on the Le and Gascuel model (Le and Gascuel, 2008) implemented in MEGA6 (Tamura et al., 2013). The tree with the highest log likelihood (−13400.7631) is shown. Initial trees for the heuristic search were obtained by applying the Neighbor-Joining method to a matrix of pairwise distances estimated using a Jones-Taylor-Thornton model (Jones et al., 1992). A discrete gamma distribution was used to model evolutionary rate differences among sites (5 categories [+G, parameter = 0.8513]). The analysis involved 51 protein sequences. All positions with less than 95% site coverage were eliminated such that fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position. There were a total of 295 positions in the final dataset.
2.3. In silico searches for transcripts encoding new peptide precursor proteins
Searches of the C. borealis transcriptome for transcripts encoding precursor proteins for several peptide families not searched for previously (Christie and Pascual, 2016), but for which receptor-encoding transcripts were found here, were done using methods described in detail in Christie and Pascual (2016); known lobster, Homarus americanus, precursor proteins (Christie et al., 2017) were used as the query sequences for these tblastn searches. The structures of mature peptides contained within the deduced pre/preprohormones were predicted using a workflow described in detail in Christie and Pascual (2016). In brief, each of the deduced precursor proteins was assessed for the presence of a signal peptide using the online program SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP/; Petersen et al., 2011). Prohormone cleavage sites were identified based on the information presented in Veenstra (2000) and/or by homology to known arthropod pre/preprohormone processing schemes. When present, the sulfation state of tyrosine residues was predicted using the online program Sulfinator (http://www.expasy.org/tools/sulfinator/; Monigatti et al., 2002), while disulfide bonding between cysteine residues was predicted by homology to known peptide isoforms and/or by using the online program DiANNA (http://clavius.bc.edu/~clotelab/DiANNA/; Ferrè and Clote, 2005). Other post-translational modifications were predicted by homology to known arthropod peptide isoforms. All precursor protein alignments were done using MAFFT version 7.
2.4. Confirmational cloning and expression of selected receptors in insect cell lines
2.4.1. Full-length cloning
To extract RNA from the STG, crabs were cold-anaesthetized by packing in ice for 30–60 min, after which the foregut was removed and pinned out in a Sylgard 170 (Dow Corning, Midland, MI, USA)-coated dish filled with cold (~4°C) physiological saline (composition in mM/L: NaCl, 440.0; KCl, 11.0; CaCl2, 13.0; MgCl2, 26.0; Trizma base, 12.0; maleic acid, 1.22; adjusted to pH 7.4–7.5 with NaOH). The STG was manually dissected from the foregut and was placed directly into a sterile 2.0 mL tube containing 3.0 mm triple-pure (molecular biology grade) zirconium beads (Item No. D1032–30; Benchmark Scientific Inc., Edison, NJ, USA), 200 μL of Buffer RA1, and 2 μL TCEP (Nucleospin XS Total RNA isolation kit; Takara Bio Co., Mountain View, CA, USA). Tubes were kept on ice while five STGs were dissected out and added to each tube.
Tissue samples were homogenized using a Model D1030 BeadBug Microtube Bead Homogenizer (Benchmark Scientific Inc.) at 2700 rpm for a total of 3 min, performed in 15-sec intervals, each of which was followed by 15 sec of cooling, during which the samples were placed on ice. Once homogenized, RNA was isolated using the column-based Takara nucleospin XS RNA isolation kit, following the protocol recommended by the manufacturer (Takara Bio Co). The final elution volume was 10 μL; the same 10 μL of RNAse-free water was run through the final column an additional time. RNA quality and quantity were initially assessed with an Agilent 211 Bioanalyzer (Agilent, Santa Clara, CA, USA). All RNA was stored at −80°C until used for cDNA production.
First-strand cDNAs for each of three biological replicates were synthesized from ~35 ng of total RNA using random pentadecamers (IDT, San Diego, CA, USA) and a SuperScript III First-Strand Synthesis System (Life Technologies Corp., Carlsbad, CA, USA). Complete open reading frames (ORFs) corresponding to putative C. borealis AST-A, AST-B, and AST-C receptors (Canbo-AST-AR, Canbo-AST-BR, and Canbo-AST-CR) were amplified from the respective cDNA sets using SapphireAmp Fast PCR Master Mix (Takara Bio Co.) in a 20-μL reaction volume with 0.4 μL cDNA and oligonucleotide primers (Supplemental Table 2) designed to the respective transcriptomic sequences using Primer3 v2.3.7 (Rozen and Skaletsky, 2000) implemented in Geneious v10.1.3 (Biomatters Ltd., Auckland, New Zealand; Kearse et al., 2012). Thermocycler conditions consisted of: 95°C for 2 min, followed by 40 cycles of 95°C for 20 sec, 56°C for 20 sec, and 72°C for 60 sec, with a final extension at 72°C for 5 min. Aliquots from each reaction were electrophoresed on 1.5% agarose gels stained with SYBR Safe (Life Technologies Corp.). Amplimers of the expected sizes were cloned into pCR2.1TOPO TA (Life Technologies Corp.), and the resulting plasmids were sequenced at the Arizona State University DNA Core laboratory (Tempe, AZ, USA).
2.4.2. Receptor expression in insect cell lines and assessments of cellular localization
To observe the cellular localization of the putative C. borealis AST-A, AST-B, and AST-C receptors, insect expression vectors encoding fluorescent chimeras of the three receptors were constructed. The respective chimeric sequences, with the enhanced green fluorescent protein (EGFP) coding sequence fused in-frame to the C-terminal residues of the respective allatostatin receptors, were generated via overlap extension PCR (Wurch et al., 1998). The initial PCR products were generated from sequence-validated plasmid DNA templates with KOD Hot Start DNA polymerase (Toyobo/Novagen, EMD Biosciences, San Diego, CA, USA) using gene-specific and chimeric primers (Supplemental Table 2). Thermocycler conditions consisted of 95°C for 2 min followed by 21 cycles at 95°C for 20 sec, 58°C for 20 sec, and 70°C for 60 sec with a final extension at 70°C for 5 min. Amplimers of the expected sizes were gel excised and purified using an EZNA Gel Extraction kit (Omega Bio-Tek Inc., Norcross, GA, USA). The respective 5’ and 3’ fragments were joined using KOD Hot Start DNA polymerase with gene specific primers (Supplemental Table 2). Thermocycler conditions consisted of 95°C for 2 min followed by 25 cycles at 95°C for 20 sec, 56°C for 20 sec, and 70°C for 90 sec with a final extension at 70°C for 5 min. The resulting PCR products were gel excised, treated with ExTaq DNA polymerase (Clontech) to add 3’A overhangs, and cloned into the pIB/V5-His TOPO TA insect expression vector (Life Technologies). Plasmids were sequenced as before at the Arizona State University DNA Core laboratory.
The cellular localization of heterologously-expressed fluorescent C. borealis AST-A, AST-B and AST-C receptor chimeras was examined in cultured Spodoptera frugiperda Sf9 cells (Allele Biotechnology, San Diego, CA, USA), a cell line derived from pupal ovarian tissue of the fall armyworm (Vaughn et al., 1977). Briefly, Sf9 cells maintained as adherent cultures in Graces supplemented insect culture media (Gibco/Life Technologies) with additional supplementation of 10% fetal bovine serum (Gibco/Life Technologies) were seeded into 35-mm #1.5 glass bottom dishes (Matsunami Glass USA Inc., Bellingham, WA, USA) and allowed to settle for 20 min. Cells were then transfected with 2 μg plasmid DNA for 5 hr using Cellfectin II (Life Technologies). The transfection medium was removed, the cells were washed twice with 1 mL serum-free media, and were then maintained in normal insect media at 28°C. After 48 hr, the transfected cells were washed twice with 1 mL IPL-41 insect media (Life Technologies) before being imaged in 2 mL IPL-41 on a Fluoview FV10i-LIV laser scanning confocal microscope (Olympus, Center Valley, PA, USA) using a 60x phase contrast water-immersion objective (NA 1.2). Images were subsequently processed (tone and contrast) in Adobe Photoshop CS6 v13.0 (Adobe Inc., San Jose, CA, USA).
2.5. RT-PCR assessment of peptide receptor expression in the stomatogastric ganglion
Fragments (~500 base pairs [bp]) of 46 putative C. borealis receptor-encoding transcripts and two transcripts encoding housekeeping genes, i.e., actin (Accession No. GEFB01000224) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Accession No. GEFB01000609), were amplified from the biologically replicated (n=3) STG cDNAs using SapphireAmp Fast PCR Master Mix (Takara Bio Co.) in a 20 μL reaction volume with 0.3 μL cDNA and oligonucleotide primers (Supplemental Table 2). PCR conditions consisted of: 95°C for 2 min, then 35 cycles of 95°C for 20 sec, 56°C for 20 sec, and 72°C for 20 sec, with a final extension at 72°C for 5 min. The resulting PCR products were visualized on 1.5% agarose gels stained with SYBR Safe (Life Technologies) and cloned into pCR2.1TOPO TA (Life Technologies). Multiple clones for each transcript were sequenced at the Arizona State University DNA Core laboratory and compared to the expected transcriptomic sequence. Gel images were generated using an AlphaImager gel documentation system (ProteinSimple, San Jose, CA, USA) and processed in Adobe Photoshop.
2.6. Physiological recordings
To determine whether the peptide myosuppressin (pQDLDHVFLRFamide) and a native C. borealis RYamide isoform (pQGFYSQRYamide), both custom synthesized by GenScript Corporation (Piscataway, NJ, USA), function as modulators of the STNS pattern generating circuits, we examined their effects on the isolated STNS. The foregut was removed from cold-anaesthetized animals, as described above; the entire STNS (Fig. 1), including the paired commissural ganglia (CoGs), the single STG, the single oesophageal ganglion (OG), and connecting and motor nerves, was manually dissected from the foregut and pinned out in a Sylgard 184-lined Petri dish in cold physiological saline. The STG was desheathed to allow peptide access to the neurons and neuropil within the ganglion. Small petroleum jelly wells were made around relevant motor nerves, i.e., the lateral ventricular (lvn), medial ventricular (mvn), pyloric dilator (pdn), and ventral lateral ventricular (vlvn) nerves. Neuronal activity was recorded using bipolar stainless steel electrodes, with one electrode inserted into the petroleum jelly well and the other inserted into the saline bath nearby. Electrical activity was amplified using Model 1700 AC amplifiers (AM Systems, Carlsborg, WA, USA), and Brownlee Precision Instrumentation Amplifiers (Model 210 A; Brownlee Instruments, San Jose, CA, USA). Data were recorded on a computer through a CED Micro 1401 or Power1401 data acquisition interface using Spike2 version 7 software (CED; (Cambridge Electronic Design, Cambridge, UK), with a sampling rate of 10 kHz.
A petroleum jelly wall was built across the petri dish containing the STNS so that the anterior and posterior portions of the system could be separately superfused with saline kept at 10–12°C with a Peltier cooling system (CL-100 bipolar temperature controller and SC-20 solution heater/cooler; Warner Instruments, Hamden, CT, USA) via a peristaltic pump. Flow rate was approximately 2.5 mL/min in each portion of the dish. Peptides were applied through the perfusion system only to the posterior portion of the STNS, where the STG is located. To enable blockage of the passage of action potentials through the stomatogastric nerve (stn), a region of the nerve was desheathed and surrounded with a petroleum jelly well; action potential propagation was blocked by replacing the saline within the well with a solution of isotonic (750 mM) sucrose. To ensure complete block, the stn was subsequently transected within the well.
Electrophysiological data were analyzed using both the functions available within Spike2 and Spike2 scripts available at http://stg.rutgers.edu/Resources.html. Data were analyzed statistically and graphed using Prism 7 (GraphPad Software, San Diego, CA, USA). Measurements were taken and averaged across ten bursts shortly before peptide application and at the peak of the peptide effect, or ~5–10 min into peptide application, when any effects exerted by a peptide are expected to be maximal. Parameters that were measured included overall cycle frequency, burst duration and duty cycle of relevant neurons, i.e., the dorsal gastric (DG) neuron for the gastric mill rhythm and the pyloric dilator (PD), lateral pyloric (LP), pyloric (PY), inferior cardiac (IC), and ventricular dilator (VD) neurons for pyloric motor pattern, as well as the number of spikes per burst in those neurons that occur as single copies or in pairs, i.e., the DG, PD, LP, IC, and VD neurons. We did not determine number of spikes per burst for the PY neurons because there are multiple PY neurons, making an accurate count impossible. For the PD neurons, which occur as a pair, our number of spikes likely underestimates the actual number of spikes to some extent. To determine whether the peptides altered the gastric mill and pyloric rhythms, values of all parameters measured were compared using paired two-tailed t-tests. Only preparations that returned to control values in saline wash after the peptide application were used for analysis. N-values for all experiments refer to individual animals; error bars and values represent standard deviations.
3. Results and discussion
3.1. Identification of putative Cancer borealis peptide receptors from a mixed nervous system transcriptome
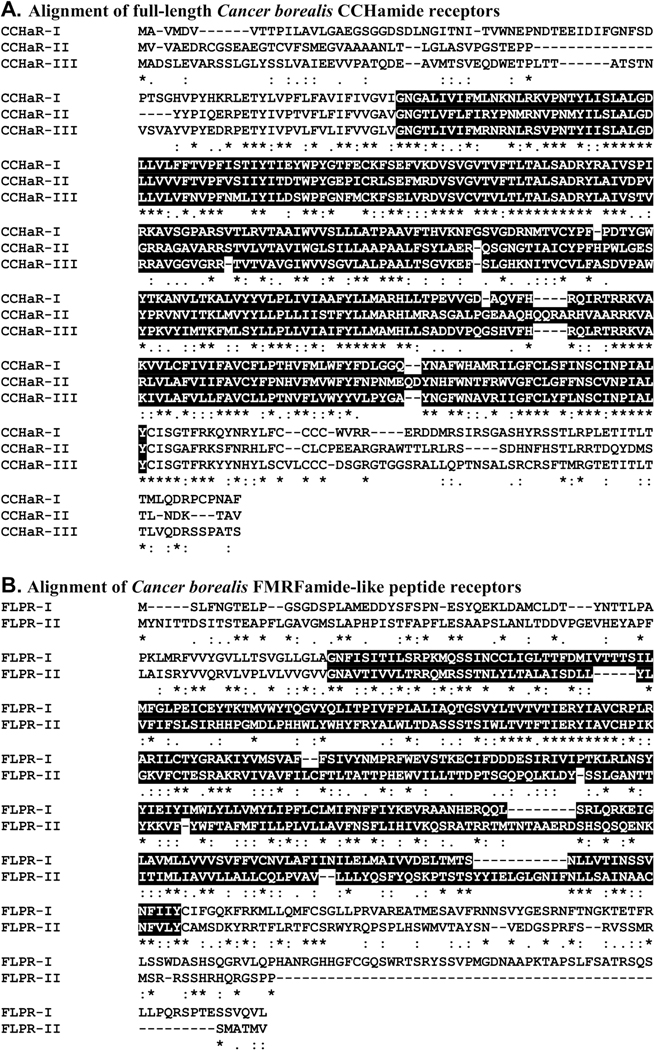
Recently, a mixed nervous system transcriptome was generated for C. borealis and publicly deposited in NCBI under BioProject No. PRJNA310325 (Northcutt et al., 2016). The portions of the nervous system used to produce this assembly were the supraoesophageal ganglion (brain), the complete STNS (Fig. 1; including the STG) and the cardiac ganglion (Northcutt et al., 2016). Here, this assembly was searched for transcripts encoding putative peptide receptors using a well-established workflow. Included in the search were receptors for all of the known crustacean peptide families for which arthropod receptors have been identified (Table 1), i.e., adipokinetic hormone-corazonin-like peptide (ACP), AST-A, AST-B, AST-C, allatotropin, bursicon, CCHamide, corazonin (CRZ), crustacean cardioactive peptide (CCAP), crustacean hyperglycemic hormone (CHH), diuretic hormone 31 (DH31), diuretic hormone 44 (DH44), ecdysis-triggering hormone (ETH), FMRFamide-like peptide (FLP), glycoprotein hormone (GPH), inotocin, insulin-like peptide (ILP), leucokinin (LK), myosuppressin, neuropeptide F (NPF), pigment dispersing hormone (PDH), proctolin, pyrokinin (PK), red pigment concentrating hormone (RPCH), RYamide, short neuropeptide F (sNPF), SIFamide, sulfakinin (SK), tachykinin-related peptide (TRP), and trissin. With the exceptions of allatotropin, CHH, and ILP, transcripts encoding putative receptors for all of the targeted peptide families were identified from the C. borealis mixed nervous system assembly (Table 1). Translation of these transcripts provided evidence for the existence of at least one ACP, one AST-A (Fig. 2), one AST-B (Fig. 3), one AST-C (Fig. 4), one bursicon, five CCHamide (Fig. 5A), two CRZ, one CCAP, one DH31, one DH44, three ETH, two FLP (Fig. 5B), one GPH, one inotocin, one LK, two myosuppressin, four NPF, three PDH, one proctolin, two PK, one RPCH, two RYamide, one sNPF, two SIFamide, one SK, three TRP, and one trissin receptors in the crab nervous system as a whole (Table 1). For the families in which multiple putative receptors were found, alignment of the deduced proteins suggests that each receptor is likely to be the product of a distinct gene; no evidence for receptor diversity arising from alternative splicing was found in the mixed tissue assembly. For example, the three full-length putative C. borealis CCHamide receptor (Canbo-CCHaR) proteins (Fig. 5A) ranged in amino acid identity/similarity from 48%/75% (Canbo-CCHaR-II vs. Canbo-CCHaR-III) to 54%/80% (Canbo-CCHaR-I vs. Canbo-CCHaR-III); the two full-length putative FLP receptors (Fig. 5B) showed 24% identity/56% similarity in amino acid sequence.. The amino acid sequences of all putative receptor proteins identified from the C. borealis mixed nervous system transcriptome are provided in Supplemental Figure 1.
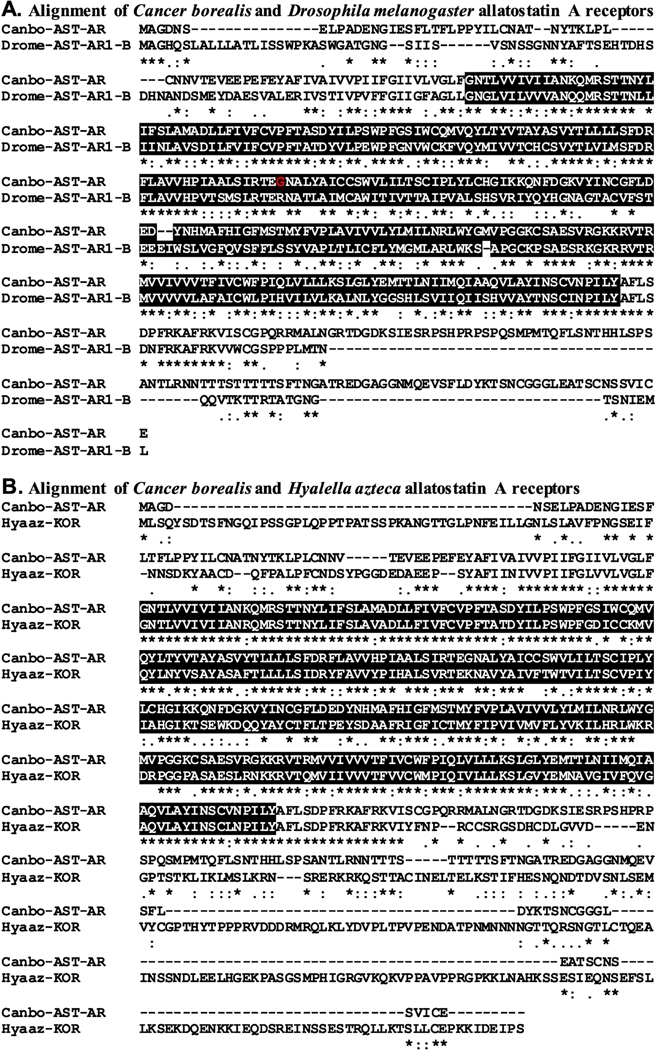
Figure 2.
Alignment of Cancer borealis and related allatostatin A receptors. (A) MAFFT alignment of the putative C. borealis allatostatin A receptor (Canbo-AST-AR; deduced from GEFB01012018) and Drosophila melanogaster allatostatin A receptor 1, isoform B (Drome-AST-AR1-B; Accession No. AAF45884). (B) MAFFT alignment of Canbo-AST-AR and the Hyalella azteca kappa opioid receptor (Hyaaz-KOR; Accession No. XP 018018012). In the line immediately below each sequence grouping, “*” indicates identical amino acid residues, while “:” and “.” denote amino acids that are similar in structure between sequences. In this figure, rhodopsin family seven-transmembrane receptor domains identified by Pfam analyses are highlighted in black. In A, the residue that varies between the transcriptome derived Canbo-AST-AR sequence and that deduced from the cloned transcript MH729782, i.e., Gly172 to Ser, is shown in red font.
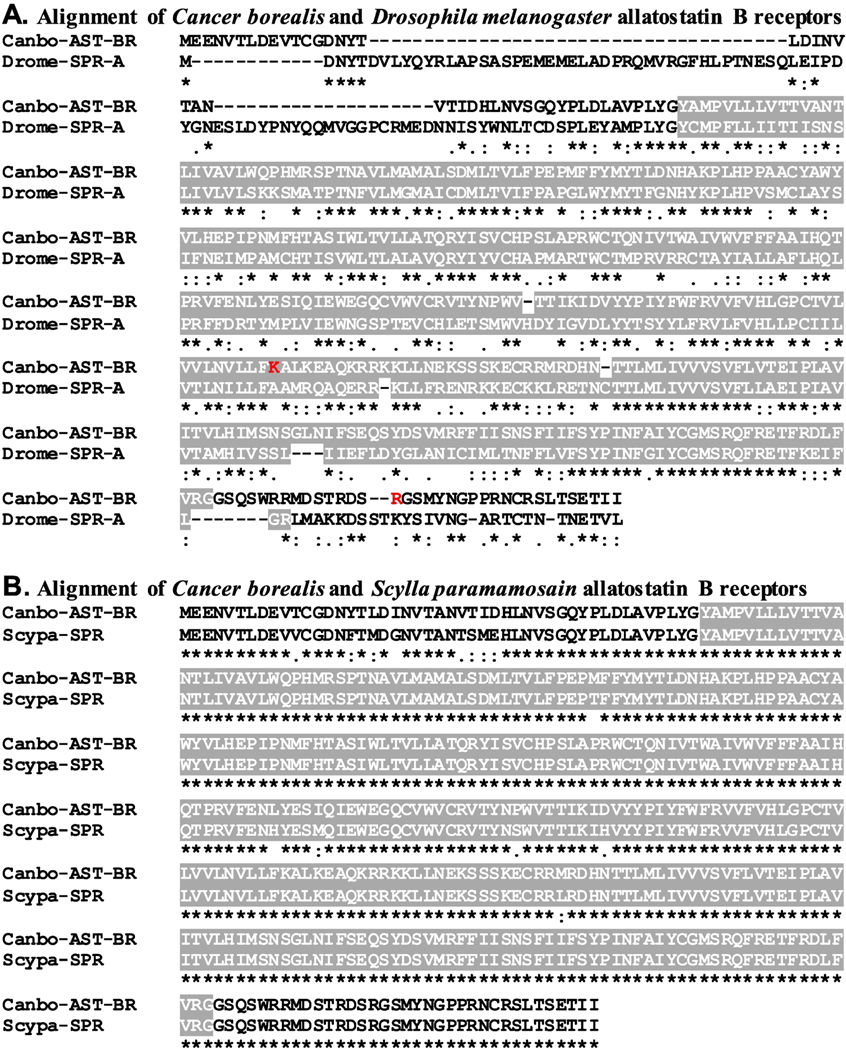
Figure 3.
Alignment of Cancer borealis and related allatostatin B/sex peptide receptors. (A) MAFFT alignment of the putative C. borealis allatostatin B receptor (Canbo-AST-BR; deduced from GEFB01014490) and the Drosophila melanogaster sex peptide receptor (Drome-SPR; Accession No. AAF46037). (B) MAFFT alignment of Canbo-AST-BR and the Scylla paramamosain sex peptide receptor (Scypa-SPR; Accession No. ANF04993). In the line immediately below each sequence grouping, “*” indicates identical amino acid residues, while “:” and “.” denote amino acids that are similar in structure between sequences. In this figure, serpentine receptor class W seven-transmembrane domains identified by Pfam analyses are highlighted in gray. In A, the residues that vary between the transcriptome derived Canbo-AST-BR sequence and that deduced from the cloned transcript MH729783, i.e., Lys250 to Arg and Arg378 to His, are shown in red font.
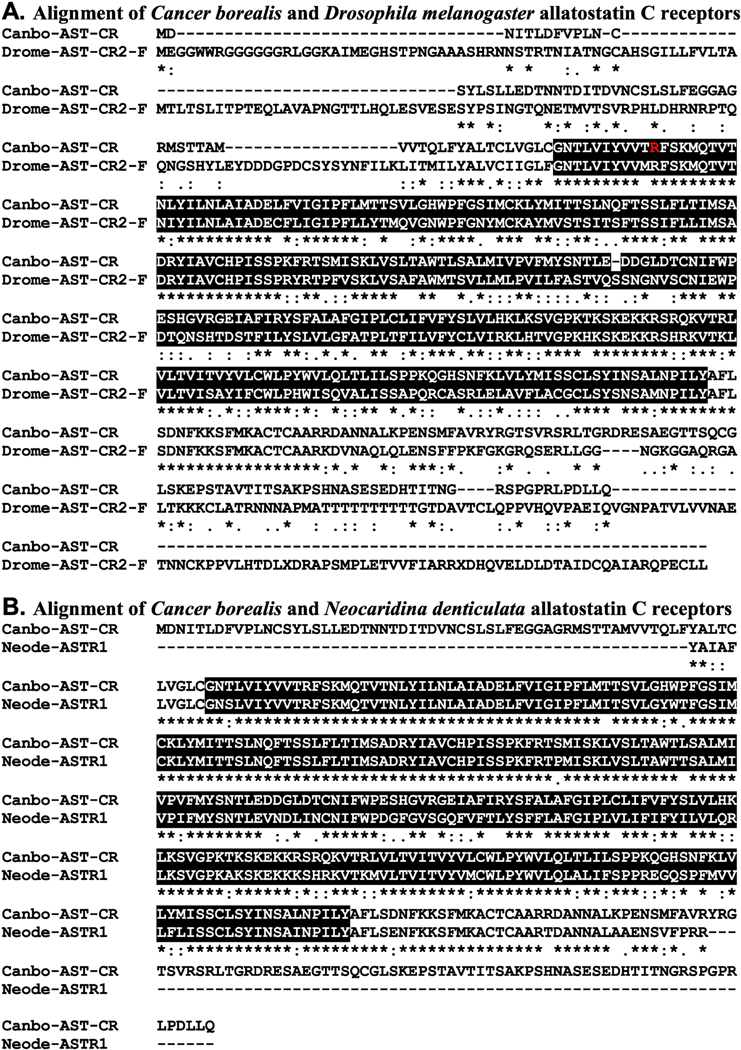
Figure 4.
Alignment of Cancer borealis and related allatostatin C receptors. (A) MAFFT alignment of the putative C. borealis allatostatin C receptor (Canbo-AST-CR; deduced from GEFB01019215) and Drosophila melanogaster allatostatin C receptor 2, isoform F (Drome-AST-CR2-F; Accession No. ALI30485). (B) MAFFT alignment of Canbo-AST-CR and the extant portion of Neocaridina denticulata allatostatin receptor 1 (Neode-ASTR1; Accession No. AIY69136). In the line immediately below each sequence grouping, “*” indicates identical amino acid residues, while “:” and “” denote amino acids that are similar in structure between sequences. In this figure, rhodopsin family seven-transmembrane receptor domains identified by Pfam analyses are highlighted in black. In A, the residue that varies between the transcriptome derived Canbo-AST-CR sequence and that deduced from the cloned transcript MH729784, i.e., Arg76 to Gly, is shown in red font.
Figure 5.
Alignment of Cancer borealis CCHamide and FMRFamide-like peptide receptor proteins putatively derived from separate genes. (A) MAFFT alignment of the putative C. borealis CCHamide receptors I-III (Canbo-CCHaR-I; deduced from GEFB01030009; Canbo-CCHaR-II; deduced from GEFB01036413; Canbo-CCHaR-III; deduced from GEFB01015997).(B) MAFFT alignment of C. borealis FMRFamide-like peptide receptors I and II (Canbo-FLPR-I; deduced from GEFB01000837; Canbo-FLPR-II; deduced from GEFB01020234). In the line immediately below each sequence grouping, “*” indicates identical amino acid residues, while “:” and “.” denote amino acids that are similar in structure between sequences. In this figure, rhodopsin family seven-transmembrane receptor domains identified by Pfam analyses are highlighted in black.
To increase confidence that the proteins identified here as C. borealis peptide receptors are members of the protein families for which they have been named, each sequence was used to search the annotated D. melanogaster proteins in FlyBase and the non-redundant arthropod proteins in NCBI for the most similar sequences. Here, our expectations were that the top hits from each search would be a member of the proposed receptor family or a closely related receptor group. As can be seen from Supplemental Table 3, this was indeed the case for all FlyBase searches. For example, the top FlyBase hit for the putative C. borealis AST-A receptor (Canbo-AST-AR) was an isoform of the D. melanogaster allatostatin A receptor 1 (Fig. 2A and Supplemental Table 3), while the top FlyBase hits for the C. borealis AST-B (Canbo-AST-BR) and AST-C receptors were the D. melanogaster sex peptide receptor (Fig. 3A and Supplemental Table 3), a synonym for the AST-BR, and isoform F of the allatostatin C receptor 2 (Fig. 4A and Supplemental Table 3), respectively. Similarly, the results obtained from searches of the NCBI non-redundant arthropod dataset (Supplemental Table 4) largely support the protein family annotations given to the putative C. borealis peptide receptors reported here. For example, the top non-redundant arthropod protein hit for Canbo-AST-AR was an amphipod, Hyalella azteca, protein annotated as a kappa-type opioid receptor, a synonym for AST-AR (Fig. 2B and Supplemental Table 4), while the top hits in the dataset for Canbo-AST-BR and Canbo-AST-CR were the crab, Scylla paramamosain, sex peptide receptor (Fig. 3B and Supplemental Table 4) and the shrimp, Neocaridina denticulata, allatostatin receptor 1 (Fig. 4B and Supplemental Table 4), respectively. However, for several receptors, e.g., the C. borealis proctolin receptor, there are mismatches, e.g., sex peptide and FMRFamide receptors returned among top NCBI non-redundant arthropod dataset hits, raising uncertainty in the annotations of some receptors.
Structural domain analysis was also conducted on each C. borealis sequence using the online program Pfam; the domains identified by Pfam in each putative Cancer receptor were compared to those identified for the corresponding top FlyBase and NCBI non-redundant protein hit. Our expectation for these analyses was that identical and/or highly similar domain complements would be identified for each of the receptor sets in question, which was the case for essentially all groupings (Pfam results for each C. borealis sequence are provided in Supplemental Table 5). For example, Pfam identified a single rhodopsin family seven-transmembrane receptor domain in Canbo-AST-AR (Fig. 2 and Supplemental Table 5), a domain also predicted by the program for the D. melanogaster allatostatin A receptor 1 (Fig. 2A) and the H. azteca kappa-type opioid receptor (Fig. 2B). Similarly, Pfam identified one serpentine receptor class W seven-transmembrane domain in Canbo-AST-BR (Fig. 3 and Supplemental Table 5), a domain also predicted by Pfam in both the D. melanogaster and S. paramamosain sex peptide receptors (Fig. 3A and 3B, respectively). A single rhodopsin family seven-transmembrane receptor domain was identified in Canbo-AST-CR, the sole domain predicted by the program for both isoform F of the D. melanogaster allatostatin C receptor 2 and N. denticulate allatostatin receptor 1 (Fig. 4A and 4B, respectively). Taken collectively, the structural domain and reciprocal BLAST results obtained for the putative receptors deduced from the C. borealis mixed nervous system transcriptome support the family attributions ascribed to them here.
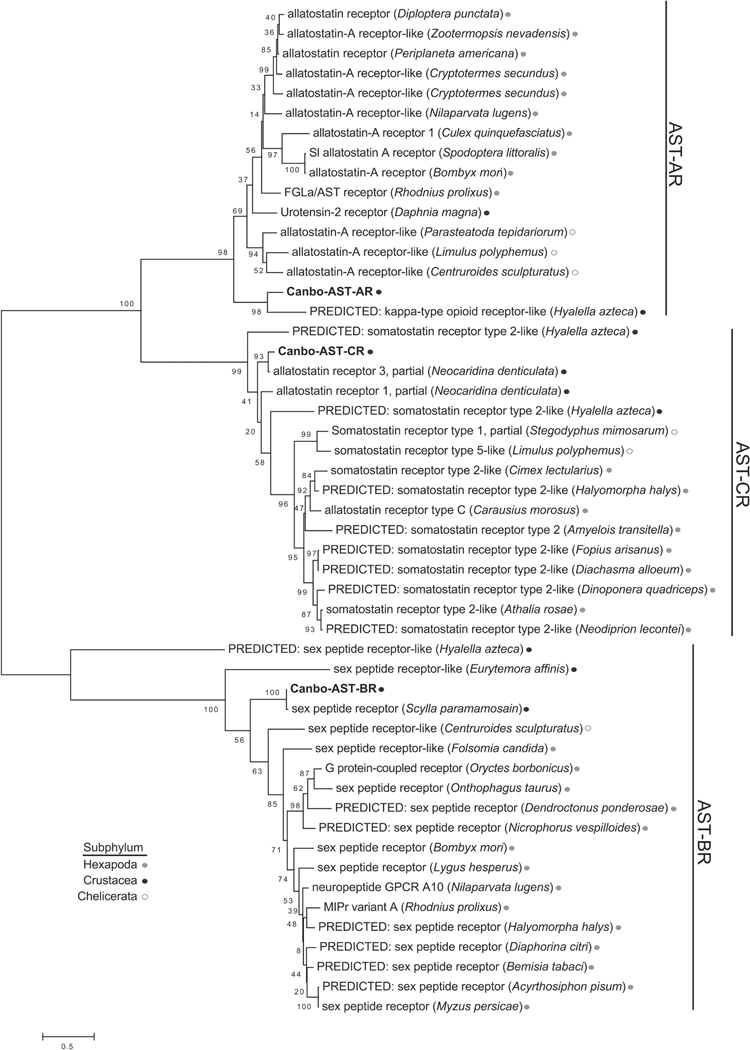
As a final means of vetting the annotations assigned to the C. borealis receptors reported here, phylogenetic relationships among the 46 putative receptor sequences were evaluated (Fig. 6). Consistent with the initial annotations, receptors largely clustered with significant bootstrap support (>70) in sub-type specific clades defined by putative ligand motifs. For example, the four NPF receptors and the sole sNPF receptor, which are predicted to bind ligands containing a C-terminal–RXRFamide motif (where X represents a variable amino acid), clustered together. Similarly, the ETH receptors and Canbo-PKR-II formed a clade characterized by–PRXamide ligands. The presence of Canbo-PKR-I in a CCHaR specific clade, rather than with the other – PRXamide-based receptors, is likely attributable to the relatively limited sequence length of the transcript (190 amino acids), which lacks an amino terminus. Interestingly, FLPR-I, which is predicted to bind –F/YLRFamide ligands, sorted to a more basal sister branch of the clade that includes FLPR-II, suggesting that FLPR-II may have diverged after an initial gene duplication event.
Figure 6.
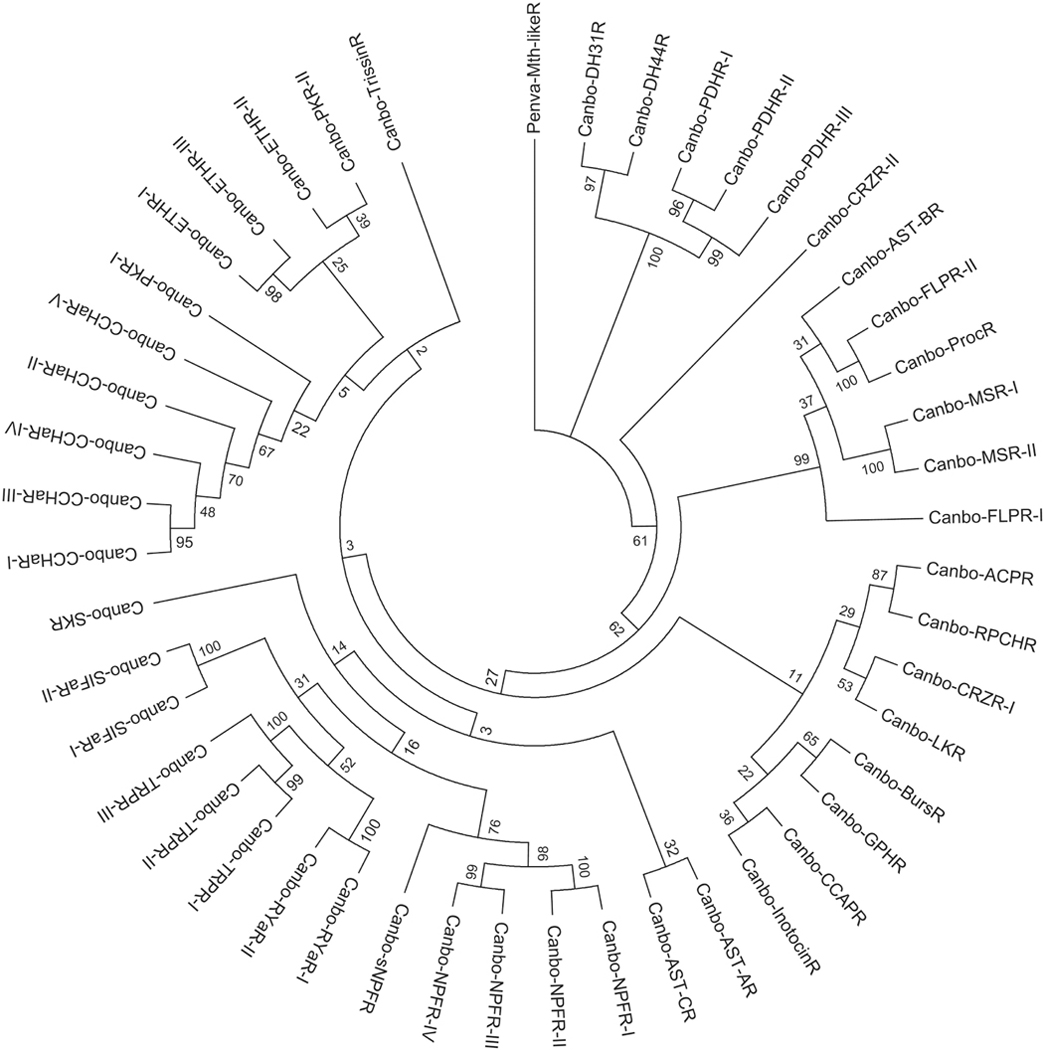
Phylogenetic relationships among putative Cancer borealis peptide receptors. Neighbor-joining tree depicting relationships among the 46 C. borealis peptide receptor sequences identified from mixed neural tissue transcriptomic data. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches (Felsenstein, 1985). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. Abbreviations are as follows: ACPR (adipokinetic hormone-corazonin-like peptide receptor); AST-AR (allatostatin A receptor); AST-BR (allatostatin B receptor); AST-CR (allatostatin C receptor); BursR (bursicon receptor); CCHaR (CCHamide receptor); CRZR (corazonin receptor); CCAPR (crustacean cardioactive peptide receptor); DH31R (diuretic hormone 31 receptor); DH44R (diuretic hormone 44 receptor); ETHR (ecdysis-triggering hormone receptor); FLPR (FMRFamide-like peptide receptor); GPHR (glycoprotein hormone receptor); InotocinR (inotocin receptor); LKR (leucokinin receptor); MSR (myosuppressin receptor); NPFR (neuropeptide F receptor); PDHR (pigment dispersing hormone receptor); ProcR (proctolin receptor); PKR (pyrokinin receptor); RPCHR (red pigment concentrating hormone receptor); RYaR (RYamide receptor); sNPFR (short neuropeptide F receptor); SIFaR (SIFamide receptor); SKR (sulfakinin receptor); TRPR (tachykinin-related peptide receptor); TrissinR (trissin receptor). The tree was rooted using the Penaeus vannamei Methuselah-like 1 GPCR sequence (Penva-Mth-likeR; Accession No. XP 027232612) as an outgroup.
Despite clear C-terminal sequence differences, member of the three allatostatin peptide families, i.e., AST-A (–YXFGLamide), AST-B (–WX6Wamide), and AST-C (–PISCF), were initially identified based on their physiological effects in insects (Tobe and Bendena, 2013). Members of each of these peptide families inhibited juvenile hormone synthesis and/or release from the corpora allata in different groups of insects (Tobe and Bendena, 2013). The discontinuity between ligand structure and function suggests the possibility of some degree of structural overlap at the receptor level. However, the expansion of biological activity ascribed to the allatostatin family of peptides in decapod crustaceans is suggestive of greater ligand-receptor discrimination. At the transcript level, the perception of ligand discrimination is further complicated by the lack of consistent, unified annotations. To assess the utility of the predicted C. borealis receptor repertoire to provide insights into receptor-ligand pairs, we generated a more refined phylogenetic analysis of the allatostatin receptors within a larger evolutionary context representing diverse arthropod species from three subphyla (Hexapoda, Crustacea and Chelicerata) of arthropods. Consistent with the initial C. borealis receptor phylogeny, the three Cancer allatostatin receptors clustered in separate clades with phylogenetic support that suggests a possible common evolutionary ancestor for the AST-A and AST-C receptors (Fig. 7). At the individual receptor level, Canbo-AST-AR aligned with a H. azteca sequence annotated as a kappa-type opioid receptor (Fig. 2B) in a Crustacea-specific grouping. Although the Canbo-AST-B and -C receptors likewise clustered in Crustacea-specific clades, the Canbo-AST-BR clade was composed of Hexapoda sequences largely annotated as sex peptide receptors, which also function as the cognate receptor for the more ancestral myoinhibiting peptides/AST-Bs (Kim et al., 2010; Poels et al., 2010; Yamanaka et al., 2010).
Figure 7.
Phylogenetic analysis of A-, B- and C-type allatostatin receptors from diverse arthropods. Maximum likelihood tree depicting the inferred evolutionary history of the putative Cancer borealis allatostatin receptors with allatostatin-like receptor sequences identified in arthropods from the Hexapoda, Crustacea, and Chelicerata subphyla. The tree with the highest log likelihood is drawn to scale, with branch lengths measured in the number of substitutions per site. The percentage of trees in which the associated taxa clustered together across 1000 replicates is shown next to the branches. Accession numbers for the sequences used for the phylogenetic analysis are provided in Supplemental Table 1.
3.2. Receptor identifications expand the number of known peptidergic signaling system for Cancer borealis
The putative peptide receptors identified here include seven for which no peptide isoforms have been identified in C. borealis. These peptide groups include several for which searches of the Cancer assembly were conducted, but failed to identify precursor-encoding transcripts, i.e., ACP, bursicon, ETH, NPF, and sulfakinin (Christie and Pascual, 2016), as well as two peptide families that have not previously been the subject of searches for precursor proteins in C. borealis, i.e., GPH and trissin. Using known lobster, H. americanus, GPH (both α2 and β5 subunit prehormones) and trissin precursors (Christie et al., 2017), the C. borealis transcriptome was searched for transcripts encoding members of these two peptide families. Transcripts encoding putative precursors for each family were identified. Specifically, for GPH, one transcript encoding a 97 amino acid C-terminal partial GPα2 subunit precursor was identified in the assembly, as was one transcript encoding a 173 amino acid full-length GPβ5 subunit protein. For trissin, one transcript encoding a 206 amino acid full-length preprohormone was found in the C. borealis transcriptome.
Alignments of the Cancer GPH and trissin precursors with their Homarus counterparts are shown in Figure 8. As can be seen from these alignments, the C. borealis GPα2, GPβ5, and trissin isoforms (colored red in the panels shown in Fig. 8) share extensive amino acid identity with those predicted for H. americanus, e.g., the Cancer and Homarus trissin isoforms are identical except for a single substituted position. Although the three peptides are themselves very similar between the two species, there is considerable variation in the signal peptides (colored gray in the panels shown in Fig. 8) for all three proteins, as well as in the precursor-related peptides derived from the trissin preprohormones (colored blue in the panel Fig. 8C). Homology to other predicted crustacean GPH and trissin isoforms (e.g., Christie et al., 2017) suggests extensive disulfide bridging is likely to be present within the C. borealis GPα2 and GPβ5 subunit peptides and in the trissin isoforms. Based on homology to similar glycoprotein hormones, the Cancer GPα2 and GPβ5 subunits are hypothesized to form a classic bioactive glycoprotein cysteine knot-forming heterodimer via non-covalent bonding, with N-linked glycosylation on the GPα2 subunit peptide (e.g., Paluzzi et al., 2014).
Figure 8.

Alignment of Cancer borealis and Homarus americanus glycoprotein hormone and trissin precursor proteins. (A1) MAFFT alignment of the putative C. borealis and H. americanus glycoprotein hormone α2 subunit precursors (Canbo-pre-GPα2; deduced from GEFB01019143; Homam-pre-GPα2; deduced from GFDA01013881). (A2) MAFFT alignment of the putative C. borealis and H. americanus glycoprotein hormone β5 subunit precursors (Canbo-pre-GPβ5; deduced from GEFB01005773; Homam-pre-GPβ5; deduced from GFDA01059529). (B) MAFFT alignment of the putative C. borealis and H. americanus trissin precursors (Canbo-prepro-Tris; deduced from GEFB01013178; Homam-prepro-Tris; deduced from GFDA01095285). In the line immediately below each sequence grouping, “*” indicates identical amino acid residues, while “:” and “” denote amino acids that are similar in structure between sequences. In this figure, signal peptides are shown in gray, while all mono/dibasic cleavage loci are shown in black. For each sequence, the isoform of the peptide for which the precursor is named is shown in red; in B, trissin linker/precursor related peptides shown in blue.
One peptide group for which there is only one report of an isoform from a member of the genus Cancer is allatotropin. Specifically, the peptide GFKNVEMMTARGFamide, which contains the C-terminal sequence –TARGFamide, the hallmark of the allatotropin family, was identified via mass spectrometry from the crab, Cancer productus, sinus gland (Fu et al., 2005b), a major neuroendocrine organ in decapod species (e.g., Christie, 2011). Although it has been searched for in a number of decapod transcriptomes, including the C. borealis assembly investigated here (Christie and Pascual, 2016), no transcripts encoding allatotropin precursors have been identified from any member of the Decapoda (e.g., Christie 2016a, 2016b, Christie and Chi, 2015; Christie and Pascual, 2016; Christie et al., 2015, 2017), nor have any in silico searches revealed transcripts for putative allatotropin receptors in any decapod (e.g., Christie and Yu, 2019; Christie et al., 2015). Moreover, while mass spectral studies on a diverse array of decapods have been conducted (e.g., Hui et al., 2012, 2013; Ma et al., 2008, 2009b, 2010; Ye et al., 2015), there have been no reports of allatotropin isoforms in any decapod species other than the one report from C. productus (Fu et al., 2005b). The lack of in silico allatotropin precursor/receptor identifications, the lack of additional mass spectral support for allatotropin isoforms in decapods, and the fact that the original identification from C. productus was via accurate mass matching (to an insect allatotropin mass) and not de novo sequencing (Fu et al., 2005b), leads us to suspect that the original identification of GFKNVEMMTARGFamide in the C. productus sinus gland was a misidentification, and that members of the Decapoda, including C. borealis, likely lack an allatotropin signaling system.
3.2. Correlations between receptor gene and peptide precursor gene/peptide isoform diversity
To date, ~200 neuropeptides have been identified from the C. borealis nervous system via a combination of biochemistry, mass spectrometry and/or in silico transcriptome mining (e.g., Christie and Pascual, 2016; Christie et al., 1997; Fu et al., 2005a; Huybrechts et al., 2003; Li et al., 2002, 2003; Stemmler et al., 2007a, 2007b). Previously reported peptides include members of 27 different families (AST-A, AST-B, AST-C, CCHamide, CRZ, CCAP, CHH, DH31, DH44, FLP, GSEFLamide, HIGSLYRamide, inotocin, ILP, LK, myosuppressin, neuroparsin, orcokinin, orcomyotropin, PDH, proctolin, PK, RPCH, RYamide, sNPF, SIFamide, and TRP), as well as a large number of precursor-related peptides. Only one previous study has examined the peptide precursors in a C. borealis transcriptome (Christie and Pascual, 2016). This study used the same transcriptome mined here for putative peptide receptors, which enabled us to compare the number of genes encoding peptide precursors to the number of receptor genes for each peptide family (Table 2). A one-for-one correspondence between precursor and receptor genes was found for CCAP, DH31, DH44, inotocin, RPCH, and sNPF, i.e., one precursor and one receptor gene for each family (Table 2). In contrast, while a single precursor gene was identified for both PK and SIFamide, two receptor sequences were identified for each family (Table 2).
Table 2.
Comparisons of the number of putative peptide receptor genes with the number of putative peptide precursor genes/peptide isoforms for Cancer borealis
| Peptide family | Number of receptor genes | Number of peptide precursor genes | Number of peptide isoforms |
|---|---|---|---|
| Adipokinetic hormone-corazonin-like peptide | 1 | NI | 1e |
| Allatostatin A | 1 | 1 | 30b |
| Allatostatin B | 1 | 1 | 8b |
| Allatostatin C | 1 | 2 | 2c |
| Bursicon | 1 | NI | 1e† |
| CCHamide | 5 | 1 | 1c |
| Corazonin | 2 | NI | 1d |
| Crustacean cardioactive peptide | 1 | 1 | 1a |
| Crustacean hyperglycemic hormone | NI | 1 | 1c |
| Diuretic hormone 31 | 1 | 1 | 1a |
| Diuretic hormone 44 | 1 | 1 | 1a |
| Ecdysis-triggering hormone | 3 | NI | ?f |
| FMRFamide-like peptide | 2 | 1 | 9b |
| Glycoprotein hormone | 1 | 2a | 1a† |
| GSEFLamide | RU | 1 | 2b |
| HIGSLYRamide | RU | 1 | 5b |
| Inotocin | 1 | 1 | 1a |
| Insulin-like peptide | NI | 1 | 1a* |
| Leucokinin | 1 | 1 | 2b |
| Myosuppressin | 2 | NI | 1d |
| Neuroparsin | RU | 4 | 4a |
| Neuropeptide F | 4 | NI | 3e |
| Orcokinin | RU | NI | 4e |
| Orcomyotropin | RU | NI | 1d |
| Pigment dispersing hormone | 3 | 1 | 1c |
| Proctolin | 1 | NI | 1d |
| Pyrokinin | 1 | 1 | 10a |
| Red pigment concentrating hormone | 1 | 1 | 1a |
| RYamide | 2 | NI | 3d |
| Short neuropeptide F | 1 | 1 | 3a |
| SIFamide | 2 | 1 | 1a |
| Sulfakinin | 1 | NI | 2e |
| Tachykinin-related peptide | 3 | NI | 2d |
| Trissin | 1 | 1 | 1a |
Abbreviations: NI, none identified (searched for in the C. borealis transcriptome, but no transcript encoding the protein in question identified); RU, receptor(s) unknown.
Isoform diversity based on the number of isoforms present in full-length pre/preprohormones deduced from C. borealis transcriptomic data (Christie and Pascual, 2016) and where the number of genes identified is likely complete.
Isoform diversity based on the number of isoforms present in partial pre/preprohormones deduced from C. borealis transcriptomic data (Christie and Pascual, 2016) and where the number of genes identified is likely complete; number of isoforms reported may be an underestimate.
Isoform diversity based on the number of isoforms present in full-length and/or partial pre/preprohormones deduced from C. borealis transcriptomic data (Christie and Pascual, 2016) but where the number of genes identified is likely incomplete based on data from other decapod species (e.g., Christie and Yu, 2019; Christie et al., 2017); number of isoforms reported is likely to be an underestimate.
Isoform diversity based on the number of isoforms detected via mass spectrometry and/or other methods in C. borealis and where the reported number is likely complete (e.g., Fu et al., 2005a; Huybrechts et al., 2003; Li et al., 2002, 2003; Stemmler et al., 2007a, 2007b).
No isoform diversity data from C. borealis; the number of isoforms reported is a prediction based on isoform conservation in decapods for which members of the family have been identified (e.g., Christie and Yu, 2019; Christie et al., 2017).
No authentic isoforms of ecdysis-triggering hormone have been identified in any member of the Decapoda and thus there is no ability at this time to predict isoform diversity for this peptide family.
Mature bioactive hormone consists of a heterodimer with each peptide subunit derived from separate genes.
Mature bioactive hormone consists of a heterodimer with each peptide subunit derived from the same gene.
In addition, the previous study identified full-length preprohormones for ten of the peptide families examined, i.e., CCAP, DH31, DH44, inotocin, ILP, neuroparsin, PK, RPCH, sNPF, and SIFamide (Christie and Pascual, 2016); these likely represent the complete set of pre/preprohormones for those peptide families. Although no ILP or neuroparsin receptors were identified from the mixed nervous system transcriptome examined here, receptors for members of the other eight families were identified. Using these data, we compared the number of distinct isoforms of a peptide group to the number of receptor genes for the family (Table 2). Comparison of peptide isoform diversity to receptor gene number showed a one-for-one correspondence for CCAP, DH31, DH44, inotocin, and RPCH, with one peptide isoform and one receptor present for each group (Table 2). In contrast, mismatches were seen for the other families, i.e., one SIFamide isoform and two putative SIFamide receptors, three distinct isoforms of sNPF with a single sNPF receptor, and ten distinct PKs with two putative receptors for members of that family (Table 2). Although only partial precursors were found for other families (Christie and Pascual, 2016), clear mismatches are likely to exist between their peptide isoform diversity and receptor gene number (Table 2). For example, while single precursor and receptor genes likely exist in C. borealis for both the AST-A and AST-B families, at least 30 and eight distinct peptide isoforms, respectively, are predicted to exist in the crab (Table 2). Similarly, although biochemical and/or mass spectral analyses have shown that C. borealis has a single myosuppressin isoform and two TRP isoforms (e.g., Christie et al., 1997; Stemmler et al., 2007a, 2007b), there appear to be two and three receptors for these peptide families, respectively, in Cancer (Table 2). Thus, there do not appear to be any hard and fast rules between either peptide precursor and receptor gene number or between peptide isoform diversity and peptide receptor gene number in C. borealis.
Although there is no clear correspondence between peptide gene/isoform diversity and receptor gene number, the fact that there appear to be more receptor genes for some peptide groups than there are distinct peptide isoforms in the family provides a potential mechanism for increasing the functional flexibility for some peptide groups. Peptide families for which this seems likely in C. borealis include CCHamide (five putative receptors for likely two peptides), CRZ (two receptors for one peptide), myosuppressin (two receptors for one peptide), SIFamide (two receptors for one peptide), and TRP (three receptors for two peptides). Moreover, if receptors are differentially selective for specific isoforms of a given family, the presence of multiple receptors in C. borealis could expand the functional flexibility for members of the CCHamide, FLP, NPF, PDH, PK, RYamide, and TRP families, as each has, or is likely to have (based on data from other decapods [e.g., Christie and Yu, 2019; Christie et al., 2015, 2017]), multiple peptide isoforms. Thus, taken collectively, receptor complement may provide a means for expanding the functional flexibility of nearly half of the peptide families thus far identified in C. borealis.
3.3. Full-length cloning and expression of AST-A, AST-B, and AST-C receptors
3.3.1. Full-length cloning
To determine whether the receptor sequences predicted from the mixed tissue transcriptome reflected the sequences of receptors in specific tissues, and therefore could provide insights into neuromodulation in those portions of the nervous system, we selected three receptors, those for AST-A, AST-B, and AST-C, to examine in more detail. STG cDNAs were used as templates to amplify the full-length AST-A, AST-B and AST-C receptor ORFs identified in the transcriptomic data. RT-PCR products of the expected sizes (Canbo-AST-AR, 1365 bp; Canbo-AST-BR, 1197 bp; Canbo-AST-CR, 1281bp) were generated for each receptor; nucleotide sequence identity relative to the transcriptomic sequence was >99% for all three receptors. There were, however, small discrepancies in the nucleotide sequences of the cloned products relative to those identified from the mixed nervous system assembly that resulted in amino acid variations in their deduced proteins. Specifically, the AST-AR deduced from the cloned sequence had a single conserved amino acid substitution vs. that deduced from transcriptomic data (Gly172 to Ser). The cloned vs. transcriptome-derived AST-BRs had two conserved substitutions (Lys250 to Arg and Arg378 to His). The cloned AST-CR differed from the transcriptome-predicted AST-CR by a single non-conserved substitution (Arg76 to Gly); these substituted residues are noted in red font in Figures 2–4. It is possible that these differences represent individual or population-specific differences in the crabs used as the initial sources of RNA, as those used for transcriptome development were obtained from a supplier in Gloucester, Massachusetts (Northcutt et al., 2016), whereas those used for cloning were from suppliers in the mid-coast area of Maine (see Materials and methods). No evidence for alternatively spliced transcripts was observed during cloning. Consensus sequences for the respective cloned transcripts have been deposited with GenBank under Accession Nos. MH729782 (Canbo-AST-AR), MH729783 (Canbo-AST-BR), and MH729784 (Canbo-AST-CR).
3.3.2. Cell surface trafficking of AST-A, AST-B, and AST-C receptor fluorescent chimeras
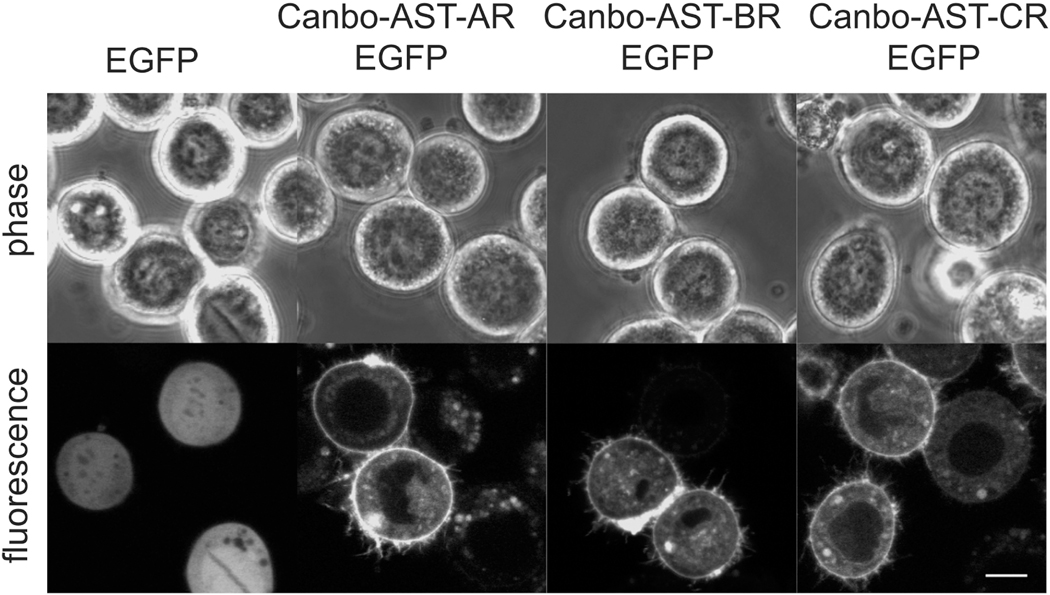
Peptide receptors relay information from extracellular peptide signals to various intracellular pathways. We thus sought to examine the potential for Canbo-AST-AR, Canbo-AST-BR and Canbo-AST-CR to function in signal transduction by assessing their cellular localization. Plasmids encoding each of the receptors, tagged at their respective C-termini with the fluorescent protein EGFP, were transiently expressed in cultured Sf9 insect cells, and localization was assessed using confocal microscopy. In cells expressing EGFP alone, fluorescence was completely intracellular (Fig. 9). In contrast, fluorescence in cells expressing each chimeric receptor was predominantly localized at the cell surface (Fig. 9), indicating that each receptor had undergone typical plasma membrane trafficking and would thus be accessible to activation by an extracellular ligand, as would be necessary for a functional peptide receptor.
Figure 9.
Cell surface localization of Cancer borealis allatostatin A (AST-AR), allatostatin B (AST-BR) and allatostatin C (AST-CR) receptors. Fluorescent (enhanced green fluorescent protein [EGFP]) chimeras of the three receptors (A) AST-AR-EGFP, (B) AST-BR-EGFP and (C) AST-CR-EGFP were transiently expressed in cultured Sf9 cells. EGFP-associated fluorescence for all three receptor constructs was observed at the cell surface. Corresponding phase contrast images are shown. Images are representative of at least two independent transfections. Scale bar = 10 μm.
3.4. RT-PCR profile of peptide receptor expression in the stomatogastric ganglion
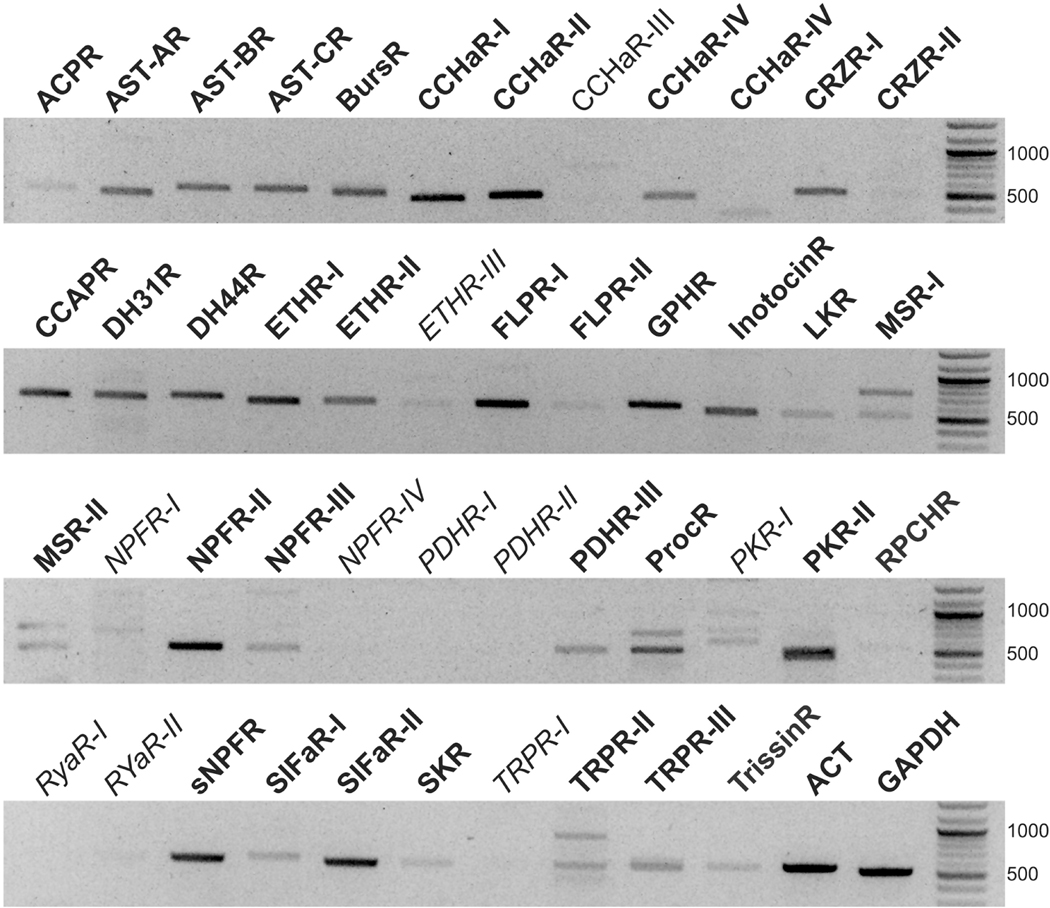
To determine which of the peptide receptors identified from the C. borealis mixed nervous system transcriptome are part of the expressed crab STG receptor repertoire, ~500 bp fragments of the 46 identified peptide receptor transcripts were PCR amplified, sequenced, and compared to their respective transcriptome-derived sequences. Sequence-validated PCR products were generated for 36 of the transcripts (Fig. 10). Although amplicons were visible for Canbo-CCHaR-III, Canbo-ETHR-III, Canbo-NPFR-1, Canbo-PKR-1, and Canbo-RYaR-II (Fig. 10), the product sizes differed from the expected, and sequence analysis of multiple clones indicated that the products were the result of non-specific amplification. No amplicons were detected using primer sets designed to Canbo-NPFR-V, Canbo-PDHR-I, Canbo-PDHR-II, Canbo-RYaR-I, or Canbo-TRPR-I (Fig. 10).
Figure 10.
RT-PCR profiling of Cancer borealis receptor transcripts in the STG. Fragments (~ 500 base pair) of the putative C. borealis receptors were amplified from three STG biological replicates with data shown representative. Similar sized fragments of the C. borealis housekeeping genes, actin (ACT) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), were likewise amplified. For greater band clarity, negative gel images are shown. PCR products that were sequence validated are indicated in bold font, non-specific products and transcripts that were not amplified are indicated in italics. Unless otherwise indicated, the abbreviations are the same as those in Figure 6.
Based on the RT-PCR profiling data, the C. borealis STG is predicted to have one or more receptors for ACP, AST-A, AST-B, AST-C, bursicon, CCHamide, corazonin, CCAP, DH31, DH44, ETH, FLP, GPH, inotocin, leucokinin, myosuppressin, NPF, PDH, proctolin, pyrokinin, RPCH, sNPF, SIFamide, sulfakinin, TRP, and trissin (Fig. 10). Although the effects of ACP, bursicon, CCHamide, corazonin, DH31, DH44, ETH, GPH, inotocin, myosuppressin, NPF, sNPF, sulfakinin, and trissin on the neural networks within the STG are currently unknown, each of the other peptide groups for which putative receptors were found in the STG has been shown to modulate the gastric mill and/or pyloric rhythms. Specifically, members of the AST-A, AST-B, and AST-C families have all been shown to have inhibitory effects on the pyloric motor pattern (e.g., Fu et al., 2007; Ma et al., 2009a; Skiebe and Schneider, 1994; Szabo et al., 2011), while isoforms of CCAP, FLP, leucokinin, proctolin, pyrokinin, RPCH, SIFamide and TRP have been shown to enhance aspects of the pyloric rhythm (e.g., Blitz et al., 2019; Christie et al., 1997; Cruz-Bermúdez et al., 2006; Marder et al., 1986; Nusbaum and Marder, 1988; Saideman et al., 2006, 2007; Swensen and Marder, 2000, 2001;Weimann et al., 1993, 1997); pyrokinin and SIFamide have been shown to activate gastric mill rhythms as well (Blitz et al., 2019; Saideman et al., 2006). Thus, there is a strong correlation between the detection of a putative receptor for a peptide family in the C. borealis STG and the ability of peptide family members to modulate the motor patterns produced by the neural circuits of the ganglion (Table 3).
Table 3.
Correlation between receptor presence in the Cancer borealis stomatogastric ganglion (STG) and peptide bioactivity
| Peptide family | Demonstration of bioactivity, or lack thereof, on the STG (example reference) | RT-PCR detection of putative receptor(s) in the STG | Correlation between bioactivity/lack of bioactivity and receptor detection |
|---|---|---|---|
| Adipokinetic hormone-corazonin-like peptide | Not tested | Yes | - |
| Allatostatin A | Bioactive (Skiebe and Schneider, 1994) | Yes | Yes |
| Allatostatin B | Bioactive (Fu et al., 2007) | Yes | Yes |
| Allatostatin C | Bioactive (Ma et al., 2009a) | Yes | Yes |
| Bursicon | Not tested | Yes | - |
| CCHamide | Not tested | Yes | - |
| Corazonin | Not tested | Yes | - |
| Crustacean cardioactive peptide | Bioactive (Weimann et al., 1997) | Yes | Yes |
| Diuretic hormone 31 | Not tested | Yes | - |
| Diuretic hormone 44 | Not tested | Yes | - |
| Ecdysis-triggering hormone | Not tested | Yes | - |
| FMRFamide-like peptide | Bioactive (Weimann et al., 1993) | Yes | Yes |
| Glycoprotein hormone | Not tested | Yes | - |
| Inotocin | Not tested | Yes | - |
| Leucokinin | Bioactive (Saideman et al., 2006) | Yes | Yes |
| Myosuppressin | Bioactive (This study) | Yes | Yes |
| Neuropeptide F | Not tested | Yes | - |
| Pigment dispersing hormone | Not tested | Yes | - |
| Proctolin | Bioactive (Marder et al., 1986) | Yes | Yes |
| Pyrokinin | Bioactive (Saideman et al., 2007) | Yes | Yes |
| Red pigment concentrating hormone | Bioactive (Nusbaum and Marder, 1988) | Yes | Yes |
| RYamide | Inactive (This study) | No | Yes |
| Short neuropeptide F | Not tested | Yes | - |
| SIFamide | Bioactive (Blitz et al., 2019) | Yes | Yes |
| Sulfakinin | Not tested | Yes | - |
| Tachykinin-related peptide | Bioactive (Christie et al., 1997) | Yes | Yes |
| Trissin | Not tested | Yes | - |
3.5. Assessment of the modulatory actions of myosuppressin and RYamide on the Cancer borealis stomatogastric ganglion
As noted above, RT-PCR showed that most of the receptors identified from the C. borealis mixed nervous system transcriptome are likely present in the STG of this species (Fig. 10). Numerous previous studies have examined the modulatory effects of members of the peptide families for which we found evidence that putative receptors are present in the crab STG; all of these peptides were shown to exert clear and consistent modulatory effects on the gastric mill and/or pyloric motor patterns (e.g., Blitz et al., 2019; Christie et al., 1997; Cruz-Bermúdez et al., 2006; Fu et al., 2007; Ma et al., 2009a; Marder et al., 1986; Saideman et al., 2006, 2007; Swensen and Marder, 2000, 2001; Szabo et al., 2011; Weimann et al., 1993, 1997). This leads to the hypothesis that for members of peptide groups untested in C. borealis, the detection of putative receptors in the STG indicates a high likelihood that they will be bioactive on the gastric mill and/or pyloric circuits; conversely, a lack of detection of putative receptors for a peptide family in the ganglion suggests that members of the group are unlikely to serve as modulators of the STG pattern generators.
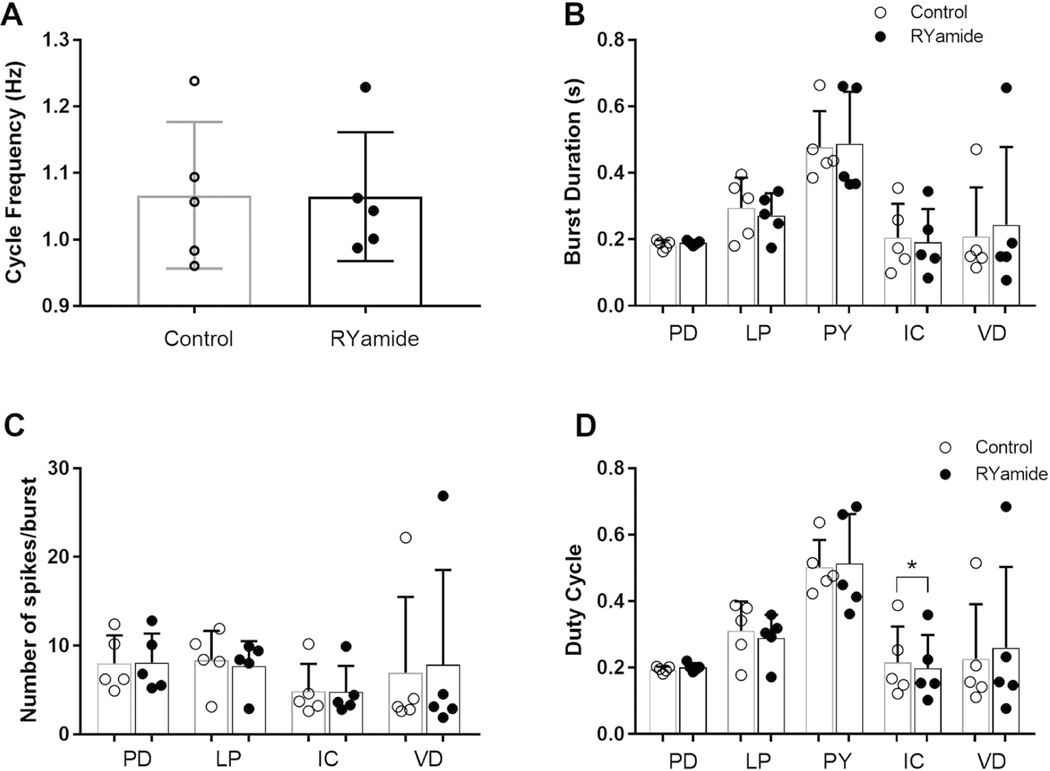
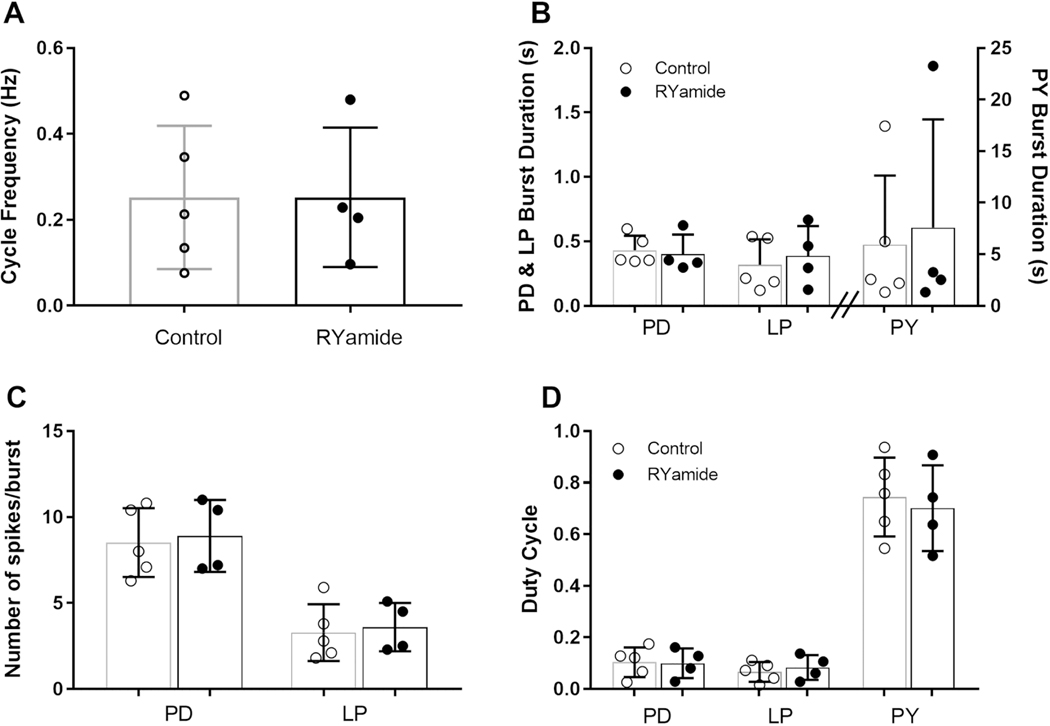
One peptide for which putative receptors were confirmed by RT-PCR as present in the STG of C. borealis, but whose effects on the crab stomatogastric networks have not yet been assessed, is myosuppressin. This peptide has been shown to exert modulatory effects on both the gastric and pyloric networks in the lobster, H. americanus (Kwiatkowski et al., 2013). In the lobster, myosuppressin not only activates neurons in each of the motor patterns, but also increases the interactions between the gastric mill and pyloric rhythms (Kwiatkowski et al., 2013). Based on the identification of two putative myosuppressin receptors in the C. borealis STG, i.e., Canbo-MSR-I and II, we predicted that myosuppressin would modulate the gastric mill and/or pyloric rhythms of the crab, likely in a manner similar to that reported previously for the lobster. In contrast, while two RYamide receptors were identified from the crab mixed nervous system assembly, i.e., Canbo-RYaR-I and II, neither appears to be present in the C. borealis STG; here, we predicted that RYamide would not exert consistent modulatory effects on the motor outputs of the ganglion.
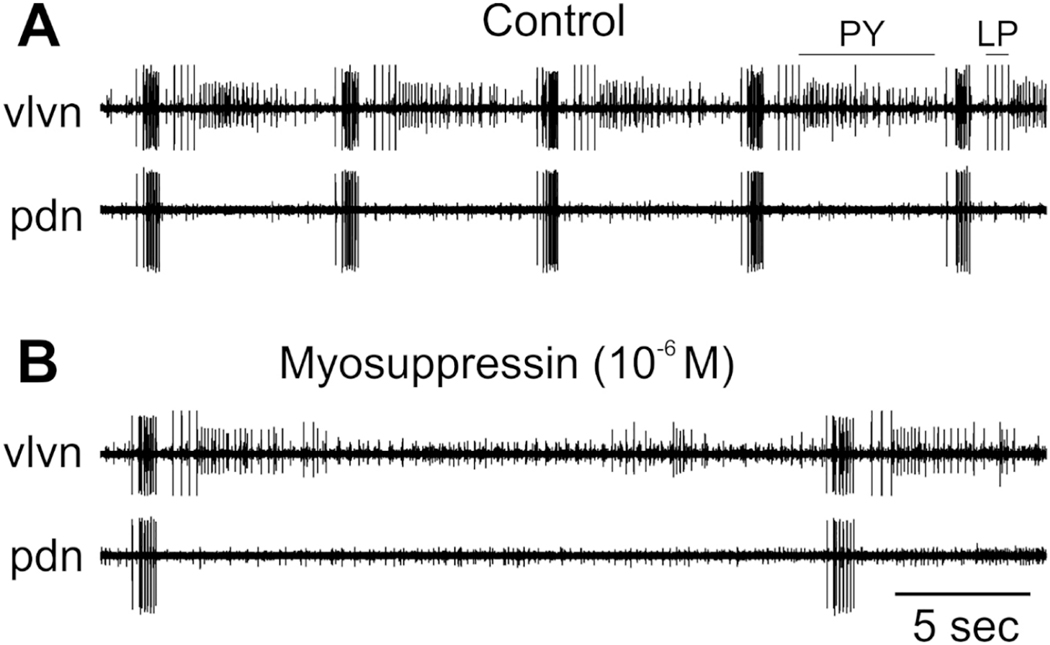
To assess the effects of myosuppressin on both the gastric and pyloric networks of C. borealis, we monitored the gastric mill rhythm using the activity of the DG neuron, and we assessed effects on the pyloric rhythm by recording the activity of all five of the neurons that commonly participate in the motor pattern, i.e., the PD, LP, PY, VD, and IC neurons. Interestingly, although myosuppressin did modulate the motor output of the STG in C. borealis, its effects differed markedly from those previously reported in H. americanus (Kwiatkowski et al., 2013). Specifically, in experiments in which the single input nerve to the STG, the stn, was intact, so that the gastric mill pattern was active, myosuppressin had no significant effects on any of the gastric mill parameters measured. These included gastric cycle period, DG burst duration, DG duty cycle, and spike frequency within DG bursts (paired t-tests between control and myosuppressin, p> 0.05 for all parameters, n=3). When the stn was blocked, no gastric activity was recorded; myosuppressin had no effects on these preparations. This is not surprising as just two peptides, pyrokinin (Saideman et al., 2007) and SIFamide (Blitz et al., 2019), have been shown to activate gastric mill activity in such stn-blocked preparations, in which all other modulatory inputs are absent.
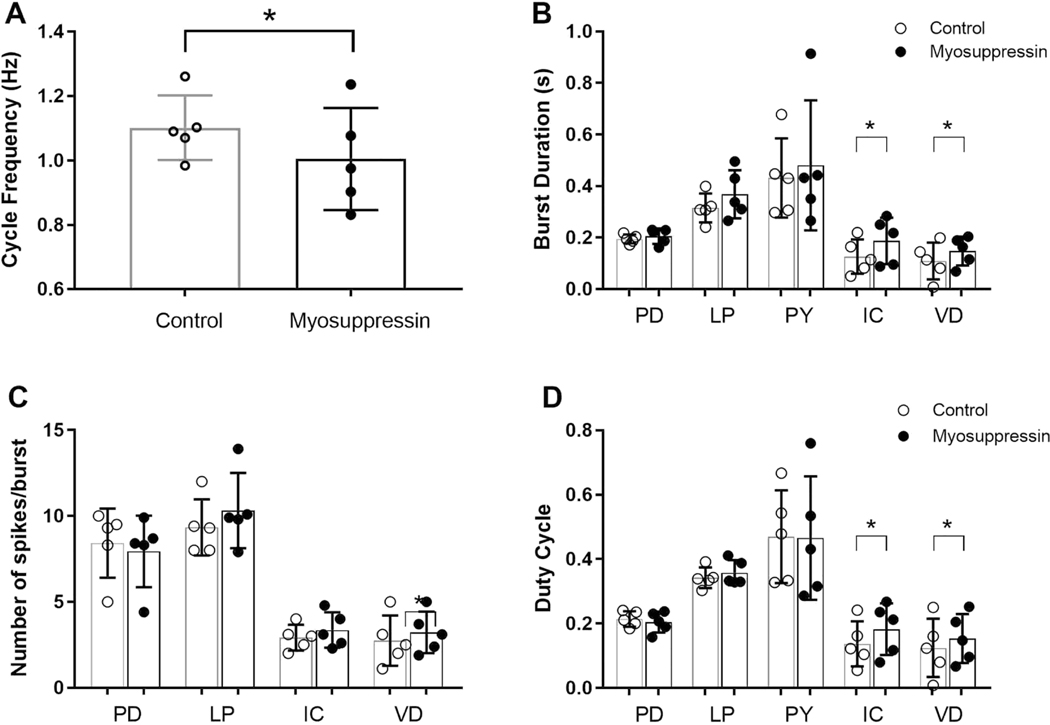
In contrast to its effects on the gastric mill pattern, myosuppressin exerted clear inhibitory effects on the pyloric pattern. When the stn was intact, so that projection neurons originating in the CoGs and OG (e.g., Coleman et al., 1992) were able to influence neurons in the STG, the pyloric pattern was strongly active. Under these conditions, myosuppressin weakly inhibited pyloric activity; notably, pyloric cycle frequency decreased significantly (Fig. 11A; paired t-test, p = 0.041, n = 5). Other burst characteristics of the PD, LP, and PY neurons, which form the core of the triphasic pattern, were not significantly changed (Fig. 11B, D). However, the burst duration and duty cycle of both the IC and VD neurons was increased in concert with the longer cycle period (Fig. 11B, D; paired t-tests, p < 0.05, n=5). Spike frequency within the bursts did not change for any of the neurons for which it was recorded (PD, LP, IC, VD; Fig. 11C; paired t-tests, p > 0.2 for all neurons, n=5). Similarly, the phases at which neurons began to fire or terminated bursts were not altered by myosuppressin (paired t-tests, p > 0.12 for all phases, n=5; data not shown).
Figure 11.
Myosuppressin, superfused at 10−6 M over the STG, exerted modest modulatory effects on the pyloric motor pattern when the single input nerve to the STG, the stn, which carries other inputs to the STG, was intact. (A) Myosuppressin elicited a decrease in pyloric cycle frequency. (B) Burst duration in the PD, LP and PY neurons was not altered by myosuppressin, but burst duration in both the IC and VD neurons increased in duration. (C) Number of spikes per burst remained unchanged by myosuppressin in all neurons (PD, LP, IC, VD). (D) Myosuppressin did not alter duty cycle in the PD, LP or PY neurons, but elicited an increase in this parameter in the IC and VD neurons. * indicates significant differences, paired t-tests, two-tailed, p <0.05. Error bars indicate standard deviations.
The effects of myosuppressin on the pyloric pattern when the stn was blocked were much more dramatic. Under these conditions, cycle frequency in control saline was relatively low (mean 0.22 ± 0.06 Hz with the stn blocked compared to 1.10 ± 0.10 Hz when it was intact), and the IC and VD neurons were not active. Myosuppressin (10−6 M) completely eliminated all pyloric activity in four of the five stn-blocked preparations in which it was tested. In the remaining preparation, activity was suppressed, with the pyloric cycle frequency decreasing from 0.13 Hz to 0.03 Hz, with concomitant decreases in the activity of all pyloric neurons (Fig. 12).
Figure 12.
Myosuppressin superfused over the STG when the stn was blocked resulted in a suppression of pyloric activity. (A) In control saline, regular bursts of action potentials in the PY, LP and PY neurons were recorded, with a cycle period of ~6 sec. (B) In the presence of 10−6 M myosuppressin, firing in all three neuron types continued, but with a much longer cycle period.
Unlike myosuppressin, RYamide did not elicit clear or consistent effects on either the pyloric or the gastric mill motor pattern. When the stn was intact, neither gastric mill cycle frequency nor any of the DG bursting parameters was altered by superfusion with 10−6 M RYamide (data not shown). Similarly, RYamide elicited virtually no effects on the pyloric pattern in the stn-intact preparation, in which the pyloric rhythm was strongly active (Fig. 13). The only statistically significant difference between any bursting parameters was a slight decrease in IC duty cycle (paired t-test, p = 0.0457, n=5; Fig. 13D); however, this change would likely be physiologically insignificant since the average change was a decrease of less than 0.02 (i.e., from 0.22 ± 0.11 to 0.20 ± 0.10). All other parameters, including cycle frequency, burst duration, duty cycle, relative timing of bursts (i.e., the phases at which bursts started and ended), and spike frequency within bursts of all other pyloric neurons remained unchanged (paired t-tests, p > 0.05 for all parameters, n=5; Fig. 13).
Figure 13.
RYamide, superfused over the STG at a concentration of 10−6 M, did not alter any of the measured parameters of the pyloric network when the pattern was active and the stn was intact. (A) Pyloric cycle frequency was unchanged by RYamide superfusion. (B) Burst duration in the PD, LP, PY, IC, and VD neurons was unchanged by RYamide superfusion. (C) Number of spikes per burst did not change in the PD, LP, IC or VD neurons in the presence of RYamide. (D) Duty cycle of the PD, LP, PY, IC, and VD neurons did not change in the presence of RYamide. Error bars indicate standard deviations.
In contrast to the responses to myosuppressin, which were much more evident when the stn was blocked, RYamide elicited no effects on the pyloric pattern even when the stn had been blocked to remove the influence of other neuromodulators as well as that of projection neurons from the anterior ganglia (CoGs and OG) (Fig. 14; paired t-tests, p > 0.098 for all parameters). These data are consistent with our predictions; the absence of detectable transcripts for the RYamide receptors in the samples of RNA we isolated from the STG suggested that this peptide would not exert consistent modulatory effects on the networks within the ganglion.
Figure 14.
Although the pyloric pattern as a whole was less active when the stn was blocked than when it was intact (compare with Fig. 13), RYamide (10−6 M) did not exert any significant effects on the pyloric pattern in these preparations. The IC and VD neurons were not active in these preparations and were not significantly activated by RYamide. (A) Cycle frequency remained unchanged in the presence of RYamide. (B) Duty cycle in the PD, LP, and PY neurons was not altered by RYamide. (C) Number of spikes per burst remained unchanged in the PD and LP neurons during RYamide superfusion. (D) RYamide failed to alter duty cycle in the PD, LP or PY neurons. Error bars indicate standard deviations.
While there were no consistent effects of RYamide on either the gastric or pyloric patterns in C. borealis, the peptide did appear to elicit individual-specific effects in two of the five stn-blocked preparations tested. In one case, the VD neuron appeared to be activated by RYamide superfusion; in the other, the ongoing weak bursting in the PD and LP neurons appeared to be suppressed by RYamide superfusion. Neither of these effects was seen in any of the other preparations. Any of a number of factors might explain these effects. For example, the peptide, when present at a concentration of 10−6 M, might activate other peptide receptors in the ganglion (e.g., Canbo-Homam-FLPR-I and II, Canbo-NPFR-II and III, Canbo-sNPFR, or even the two Cancer myosuppressin receptors, all of whose putative ligands end in –RFamide), thus exerting effects through non-specific binding. Alternatively, RYamide receptors could be present in the terminals of the projection neurons within the STG. Because their cell bodies are located in the CoGs and/or OG (Coleman et al., 1992), functional RYamide receptors could be present on terminals within the ganglion even though the encoding transcripts might not be detectable. Previous studies have shown that the activity of these terminals can be altered by interactions within the ganglion and that these terminals can exert local effects (e.g., Coleman and Nusbaum, 1994). Thus, it is possible that such terminals are mediating the effects of RYamide within the STG. However, the sporadic and inconsistent nature of the RYamide effects on the STG networks argues against this explanation. Finally, although we detected no RYamide receptor transcripts in any of the biological replicates that we examined (three STG RNA samples representing 15 crabs in total with five individuals per sample), it is possible that RYamide receptors are conditionally expressed. Thus, the crabs that appeared to respond to RYamide might have been in a state that led to the expression of RYamide receptors in particular neurons, whereas the other crabs were not in states that lead to this putative transcriptional activation.
Given the myosuppressin and RYamide physiological results, we predict that a number of other peptide groups not yet tested on the C. borealis STG will ultimately be shown to function as modulators of the gastric mill and/or pyloric motor patterns. These peptide families include ACP, bursicon, CCHamide, corazonin, DH31, DH44, ETH, GPH, inotocin, NPF, PDH, sNPF, sulfakinin and trissin, for which one or more receptor for each group is predicted to exist in the ganglion. For sulfakinin, modulation of the C. borealis STG is likely to be both local and hormonal, as cholecystokinin-like immunoreactivity (a proxy for sulfakinin) is present in the neuropil of the ganglion and in release terminals in the neuroendocrine pericardial organ (Christie et al., 1995a, 1995b). Immunohistochemical and mass spectrometric data suggest that corazonin, PDH and sNPF may function, at least in part, as hormonally delivered modulators of the C. borealis gastric mill and/or pyloric neural networks (e.g., Chen et al., 2009; Christie et al., 1995b; Li et al., 2003). As additional physiological investigations are conducted, it will be interesting to see to what extent this hypothesis is borne out.
4. Summary and conclusions
Here, a C. borealis mixed nervous system transcriptome was searched for transcripts encoding putative peptide receptors, with 46 distinct receptors encompassing 27 peptide families identified via this analysis. The identified receptors included one ACP, one AST-A, one AST-B, one AST-C, one bursicon, five CCHamide, two CRZ, one CCAP, one DH31, one DH44, three ETH, two FLP, one GPH, one inotocin, one LK, two myosuppressin, four NPF, three PDH, one proctolin, two PK, one RPCH, two RYamide, one sNPF, two SIFamide, one SK, three TRP, and one trissin receptor. The AST-A, AST-B, and AST-C receptors were cloned, sequenced and expressed in an insect cell line; each receptor trafficked to the cell surface as expected for functional receptors. Profiling of the C. borealis STG for evidence of the 46 receptors suggested that 36 are likely expressed in the ganglion. These include at least one for all but one peptide family for which receptor proteins were identified; no receptor for RYamide appears to be present in the STG. Based on the profiling data, two peptides untested for modulatory actions on the C. borealis STG were assessed for bioactivity on the ganglion. Myosuppressin, for which receptors are likely present in the ganglion, exhibited clear modulatory effects, whereas a native RYamide isoform, for which no receptor appears to be present in the STG, elicited no consistent modulatory effects. Taken collectively, the data presented here support the hypothesis that receptor complement/diversity likely plays an important role in providing functional flexibility to the CPGs the C. borealis STG, and, in all likelihood, to the CPGs of animals generally.
Supplementary Material
Highlights.
Transcriptomics was used to identify Cancer borealis neuronal peptide receptors
46 putative receptors encompassing 27 peptide families were identified
36 of the 46 receptors appear to be expressed in the stomatogastric ganglion
Myosuppressin (receptors +) but not RYamide (receptors -) modulates STG output
Receptor diversity likely contributes significantly to STG functional flexibility
Acknowledgements
The authors thank Micah Pascual and Andy Yu to help with early searches of the Cancer transcriptome for peptide receptor-encoding transcripts. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture; the USDA is an equal opportunity provider and employer.
Funding
This study was funded by the National Science Foundation (IOS-1353023 and IOS-1354567), the National Institutes of Health (5P20RR016463–12 and 8P20GM103423–12), base CRIS funding (Project #2020–22620-022–00D), the Cades, Paller Family and Henry L. and Grace Doherty Charitable Foundations, and the Grua-O’Connell Fund of Bowdoin College.
Footnotes
Conflict of Interest Statement
The authors, Patsy S. Dickinson, J. Joe Hull, Alexandra Miller, Emily R. Oleisky and Andrew E. Christie, of the manuscript entitled “To what extent may peptide receptor gene diversity/complement contribute to functional flexibility in a simple pattern-generating neural network?” that has been submitted to Comp Biochem Physiol D have no conflicts of interest to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, George RA, Lewis SE, Richards S, Ashburner M, Henderson SN, Sutton GG, Wortman JR, Yandell MD, Zhang Q, Chen LX, Brandon RC, Rogers YH, Blazej RG, Champe M, Pfeiffer BD, Wan KH, Doyle C, Baxter EG, Helt G, Nelson CR, Gabor GL, Abril JF, Agbayani A, An HJ, Andrews-Pfannkoch C, Baldwin D, Ballew RM, Basu A, Baxendale J, Bayraktaroglu L, Beasley EM, Beeson KY, Benos PV, Berman BP, Bhandari D, Bolshakov S, Borkova D, Botchan MR, Bouck J, Brokstein P, Brottier P, Burtis KC, Busam DA, Butler H, Cadieu E, Center A, Chandra I, Cherry JM, Cawley S, Dahlke C, Davenport LB, Davies P, de Pablos B, Delcher A, Deng Z, Mays AD, Dew I, Dietz SM, Dodson K, Doup LE, Downes M, Dugan-Rocha S, Dunkov BC, Dunn P, Durbin KJ, Evangelista CC, Ferraz C, Ferriera S, Fleischmann W, Fosler C, Gabrielian AE, Garg NS, Gelbart WM, Glasser K, Glodek A, Gong F, Gorrell JH, Gu Z, Guan P, Harris M, Harris NL, Harvey D, Heiman TJ, Hernandez JR, Houck J, Hostin D, Houston KA, Howland TJ, Wei MH, Ibegwam C, Jalali M, Kalush F, Karpen GH, Ke Z, Kennison JA, Ketchum KA, Kimmel BE, Kodira CD, Kraft C, Kravitz S, Kulp D, Lai Z, Lasko P, Lei Y, Levitsky AA, Li J, Li Z, Liang Y, Lin X, Liu X, Mattei B, McIntosh TC, McLeod MP, McPherson D, Merkulov G, Milshina NV, Mobarry C, Morris J, Moshrefi, , Mount SM, Moy M, Murphy B, Murphy L, Muzny DM, Nelson DL, Nelson DR, Nelson KA, Nixon K, Nusskern DR, Pacleb JM, Palazzolo M, Pittman GS, Pan S, Pollard J, Puri V, Reese MG, Reinert K, Remington K, Saunders RD, Scheeler F, Shen H, Shue BC, Sidén-Kiamos I, Simpson M, Skupski MP, Smith T, Spier E, Spradling AC, Stapleton M, Strong R, Sun E, Svirskas R, Tector C, Turner R, Venter, , Wang AH, Wang X, Wang ZY, Wassarman DA, Weinstock GM, Weissenbach J, Williams SM, Woodage T, Worley KC, Wu D, Yang S, Yao QA, Ye J, Yeh RF, Zaveri JS, Zhan M, Zhang G, Zhao Q, Zheng L, Zheng XH, Zhong FN, Zhong W, Zhou X, Zhu S, Zhu X, Smith HO, Gibbs RA, Myers EW, Rubin GM, Venter JC, 2000. The genome sequence of Drosophila melanogaster. Science 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- Aikins MJ, Schooley DA, Begum K, Detheux M, Beeman RW, Park Y, 2008. Vasopressin-like peptide and its receptor function in an indirect diuretic signaling pathway in the red flour beetle. Insect Biochem. Mol. Biol. 38, 740–748. [DOI] [PubMed] [Google Scholar]
- Blitz DM, Christie AE, Cook AP, Dickinson PS, Nusbaum MP, 2019. Similarities and differences in circuit responses to applied Gly1-SIFamide and peptidergic (Gly1-SIFamide) neuron stimulation. J. Neurophysiol. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley SJ, Fitzgibbon QP, Smith GG, Ventura T, 2016. In silico prediction of the G-protein coupled receptors expressed during the metamorphic molt of Sagmariasus verreauxi (Crustacea: Decapoda) by mining transcriptomic data: RNA-seq to repertoire. Gen. Comp. Endocrinol. 228, 111–127. [DOI] [PubMed] [Google Scholar]
- Chen R, Ma M, Hui L, Zhang J, Li L, 2009. Measurement of neuropeptides in crustacean hemolymph via MALDI mass spectrometry. J. Am. Soc. Mass. Spectrom. 20, 708–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie AE, 2016a. Prediction of Scylla olivacea (Crustacea; Brachyura) peptide hormones using publicly accessible transcriptome shotgun assembly (TSA) sequences. Gen. Comp. Endocrinol. 230–231, 1–16. [DOI] [PubMed] [Google Scholar]
- Christie AE, 2016b. Expansion of the neuropeptidome of the globally invasive marine crab Carcinus maenas. Gen. Comp. Endocrinol. 235, 150–169. [DOI] [PubMed] [Google Scholar]
- Christie AE, 2011. Crustacean neuroendocrine systems and their signaling agents. Cell Tissue Res. 345, 41–67. [DOI] [PubMed] [Google Scholar]
- Christie AE, Baldwin D, Turrigiano G, Graubard K, Marder E, 1995a. Immunocytochemical localization of multiple cholecystokinin-like peptides in the stomatogastric nervous system of the crab Cancer borealis. J. Exp. Biol. 198, 263–271. [DOI] [PubMed] [Google Scholar]
- Christie AE, Cashman CR, Stevens JS, Smith CM, Beale KM, Stemmler EA, Greenwood SJ, Towle DW, Dickinson PS, 2008. Identification and cardiotropic actions of brain/gut-derived tachykinin-related peptides (TRPs) from the American lobster Homarus americanus. Peptides 29, 1909–1918. [DOI] [PubMed] [Google Scholar]
- Christie AE, Chi M, 2015. Prediction of the neuropeptidomes of members of the Astacidea (Crustacea, Decapoda) using publicly accessible transcriptome shotgun assembly (TSA) sequence data. Gen. Comp. Endocrinol. 224, 38–60. [DOI] [PubMed] [Google Scholar]
- Christie AE, Chi M, Lameyer TJ, Pascual MG, Shea DN, Stanhope ME, Schulz DJ, Dickinson PS, 2015. Neuropeptidergic signaling in the American Lobster Homarus americanus: new insights from high-throughput nucleotide sequencing. PLoS One 10, e0145964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie AE, Lundquist CT, Nassel DR, Nusbaum MP, 1997. Two novel tachykinin-related peptides from the nervous system of the crab Cancer borealis. J. Exp. Biol. 200, 2279–2294. [DOI] [PubMed] [Google Scholar]
- Christie AE, Pascual MG, 2016. Peptidergic signaling in the crab Cancer borealis: Tapping the power of transcriptomics for neuropeptidome expansion. Gen. Comp. Endocrinol. 237, 53–67. [DOI] [PubMed] [Google Scholar]
- Christie AE, Pascual MG, Yu A, 2018a. Peptidergic signaling in the tadpole shrimp Triops newberryi: A potential model for investigating the roles played by peptide paracrines/hormones in adaptation to environmental change. Mar. Genomics 39, 45–63. [DOI] [PubMed] [Google Scholar]
- Christie AE, Roncalli V, Cieslak MC, Pascual MG, Yu A, Lameyer TJ, Stanhope ME, Dickinson PS, 2017. Prediction of a neuropeptidome for the eyestalk ganglia of the lobster Homarus americanus using a tissue-specific de novo assembled transcriptome. Gen. Comp. Endocrinol. 243, 96–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie AE, Roncalli V, Lenz PH, 2016. Diversity of insulin-like peptide signaling system proteins in Calanus finmarchicus (Crustacea; Copepoda) - Possible contributors to seasonal preadult diapause. Gen. Comp. Endocrinol. 236, 157–173. [DOI] [PubMed] [Google Scholar]
- Christie AE, Roncalli V, Wu LS, Ganote CL, Doak T, Lenz PH, 2013. Peptidergic signaling in Calanus finmarchicus (Crustacea, Copepoda): in silico identification of putative peptide hormones and their receptors using a de novo assembled transcriptome. Gen. Comp. Endocrinol. 187, 117–35. [DOI] [PubMed] [Google Scholar]
- Christie AE, Skiebe P, Marder E, 1995b. Matrix of neuromodulators in neurosecretory structures of the crab Cancer borealis. J. Exp. Biol. 198, 2431–2439. [DOI] [PubMed] [Google Scholar]
- Christie AE, Stemmler EA, Dickinson PS, 2010. Crustacean neuropeptides. Cell. Mol. Life Sci. 67, 4135–4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie AE, Yu A, 2019. Identification of peptide hormones and their cognate receptors in Jasus edwardsii – a potential resource for the development of new aquaculture management strategies for rock/spiny lobsters. Aquaculture. In press. [Google Scholar]
- Coleman MJ, Nusbaum MP, 1994. Functional consequences of compartmentalization of synaptic input. J. Neurosci. 14, 6544–6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman MJ, Nusbaum MP, Cournil I, Claiborne BJ, 1992. Distribution of modulatory inputs to the stomatogastric ganglion of the crab, Cancer borealis. J. Comp. Neurol. 325, 581–594. [DOI] [PubMed] [Google Scholar]
- Cruz-Bermúdez ND, Fu Q, Kutz-Naber KK, Christie AE, Li L, Marder E, 2006. Mass spectrometric characterization and physiological actions of GAHKNYLRFamide, a novel FMRFamide-like peptide from crabs of the genus Cancer. J. Neurochem. 97, 784–799. [DOI] [PubMed] [Google Scholar]
- Dickinson PS, Armstrong MK, Dickinson ES, Fernandez R, Miller A, Pong S, Powers BW, Pupo-Wiss A, Stanhope ME, Walsh PJ, Wiwatpanit T, Christie AE, 2018. Three members of a peptide family are differentially distributed and elicit differential state-dependent responses in a pattern generator-effector system. J. Neurophysiol. 119, 1767–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson PS, Calkins A, Stevens JS, 2015a. Related neuropeptides use different balances of unitary mechanisms to modulate the cardiac neuromuscular system in the American lobster, Homarus americanus. J. Neurophysiol. 113, 856–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson PS, Kurland SC, Qu X, Parker BO, Sreekrishnan A, Kwiatkowski MA, Williams AH, Ysasi AB, Christie AE, 2015c. Distinct or shared actions of peptide family isoforms: II. Multiple pyrokinins exert similar effects in the lobster stomatogastric nervous system. J. Exp. Biol. 218, 2905–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson PS, Qu X, Stanhope ME, 2016. Neuropeptide modulation of pattern-generating systems in crustaceans: comparative studies and approaches. Curr. Opin. Neurobiol. 41, 149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson PS, Sreekrishnan A, Kwiatkowski MA, Christie AE, 2015b. Distinct or shared actions of peptide family isoforms: I. Peptide-specific actions of pyrokinins in the lobster cardiac neuromuscular system. J. Exp. Biol. 218, 2892–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson PS, Stevens JS, Rus S, Brennan HR, Goiney CC, Smith CM, Li L, Towle DW, Christie AE, 2007. Identification and cardiotropic actions of sulfakinin peptides in the American lobster Homarus americanus. J. Exp. Biol. 210, 2278–2289. [DOI] [PubMed] [Google Scholar]
- Edgar RC, 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J, 1985. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39, 783–791. [DOI] [PubMed] [Google Scholar]
- Ferré F, Clote P, 2005. DiANNA: a web server for disulfide connectivity prediction. Nucleic Acids Res. 33, W230–W232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A, 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Q, Goy MF, Li L, 2005a. Identification of neuropeptides from the decapod crustacean sinus glands using nanoscale liquid chromatography tandem mass spectrometry. Biochem. Biophys. Res. Commun. 337, 765–778. [DOI] [PubMed] [Google Scholar]
- Fu Q, Kutz KK, Schmidt JJ, Hsu YW, Messinger DI, Cain SD, de la Iglesia HO, Christie AE, Li L, 2005b. Hormone complement of the Cancer productus sinus gland and pericardial organ: an anatomical and mass spectrometric investigation. J. Comp. Neurol. 493, 607–626. [DOI] [PubMed] [Google Scholar]
- Fu Q, Tang LS, Marder E, Li L, 2007. Mass spectrometric characterization and physiological actions of VPNDWAHFRGSWamide, a novel B type allatostatin in the crab, Cancer borealis. J. Neurochem. 101, 1099–1107. [DOI] [PubMed] [Google Scholar]
- Garcia VJ, Daur N, Temporal S, Schulz DJ, Bucher D, 2015. Neuropeptide receptor transcript expression levels and magnitude of ionic current responses show cell type-specific differences in a small motor circuit. J. Neurosci. 35, 6786–6800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramates LS, Marygold SJ, Santos GD, Urbano JM, Antonazzo G, Matthews BB, Rey AJ, Tabone CJ, Crosby MA, Emmert DB, Falls K, Goodman JL, Hu Y, Ponting L, Schroeder AJ, Strelets VB, Thurmond J, Zhou P, the FlyBase Consortium, 2017. FlyBase at 25: looking to the future. Nucleic Acids Res. 45, D663–D671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KK, Stafflinger E, Schneider M, Hauser F, Cazzamali G, Williamson M, Kollmann M, Schachtner J, Grimmelikhuijzen CJ, 2010. Discovery of a novel insect neuropeptide signaling system closely related to the insect adipokinetic hormone and corazonin hormonal systems. J. Biol. Chem. 285, 10736–10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris-Warrick RM, Marder E, Selverston AI, Moulins M, 1992. Dynamic biological networks: the stomatogastric nervous system. MIT press; Cambridge. [Google Scholar]
- Hauser F, Williamson M, Cazzamali G, Grimmelikhuijzen CJ, 2006. Identifying neuropeptide and protein hormone receptors in Drosophila melanogaster by exploiting genomic data. Brief. Funct. Genomic. Proteomic. 4, 321–330. [DOI] [PubMed] [Google Scholar]
- Horodyski FM, Verlinden H, Filkin N, Vandersmissen HP, Fleury C, Reynolds SE, Kai ZP, Broeck JV, 2011. Isolation and functional characterization of an allatotropin receptor from Manduca sexta. Insect Biochem. Mol. Biol. 41, 804–814. [DOI] [PubMed] [Google Scholar]
- Hui L, D’Andrea BT, Jia C, Liang Z, Christie AE, Li L, 2013. Mass spectrometric characterization of the neuropeptidome of the ghost crab Ocypode ceratophthalma (Brachyura, Ocypodidae). Gen. Comp. Endocrinol. 184, 22–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui L, Xiang F, Zhang Y, Li L, 2012. Mass spectrometric elucidation of the neuropeptidome of a crustacean neuroendocrine organ. Peptides 36, 230–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM, 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8, 275–282. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM, 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A, 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Bartalska K, Audsley N, Yamanaka N, Yapici N, Lee JY, Kim YC, Markovic M, Isaac E, Tanaka Y, Dickson BJ, 2010. MIPs are ancestral ligands for the sex peptide receptor. Proc. Natl. Acad. Sci. USA. 107, 6520–6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K, 2018. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski MA, Gabranski ER, Huber KE, Chapline MC, Christie AE, Dickinson PS, 2013. Coordination of distinct but interacting rhythmic motor programs by a modulatory projection neuron using different co-transmitters in different ganglia. J. Exp. Biol. 216, 1827–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le SQ, Gascuel O, 2008. An improved general amino acid replacement matrix. Mol. Biol. Evol. 25, 1307–1320. [DOI] [PubMed] [Google Scholar]
- Li L, Kelley WP, Billimoria CP, Christie AE, Pulver SR, Sweedler JV, Marder E, 2003. Mass spectrometric investigation of the neuropeptide complement and release in the pericardial organs of the crab, Cancer borealis. J Neurochem. 87, 642–656. [DOI] [PubMed] [Google Scholar]
- Li L, Pulver SR, Kelley WP, Thirumalai V, Sweedler JV, Marder E, 2002. Orcokinin peptides in developing and adult crustacean stomatogastric nervous systems and pericardial organs. J. Comp. Neurol. 444, 227–444. [DOI] [PubMed] [Google Scholar]
- Ma M, Bors EK, Dickinson ES, Kwiatkowski MA, Sousa GL, Henry RP, Smith CM, Towle DW, Christie AE, Li L, 2009b. Characterization of the Carcinus maenas neuropeptidome by mass spectrometry and functional genomics. Gen. Comp. Endocrinol. 161, 320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M, Chen R, Sousa GL, Bors EK, Kwiatkowski MA, Goiney CC, Goy MF, Christie AE, Li L, 2008. Mass spectral characterization of peptide transmitters/hormones in the nervous system and neuroendocrine organs of the American lobster Homarus americanus. Gen. Comp. Endocrinol. 156, 395–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M, Gard AL, Xiang F, Wang J, Davoodian N, Lenz PH, Malecha SR, Christie AE, Li L, 2010. Combining in silico transcriptome mining and biological mass spectrometry for neuropeptide discovery in the Pacific white shrimp Litopenaeus vannamei. Peptides 31, 27–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M, Szabo TM, Jia C, Marder E, Li L, 2009a. Mass spectrometric characterization and physiological actions of novel crustacean C-type allatostatins. Peptides 30, 1660–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marder E, Hooper SL, Siwicki KK, 1986. Modulatory action and distribution of the neuropeptide proctolin in the crustacean stomatogastric nervous system. J. Comp. Neurol. 243, 454–467. [DOI] [PubMed] [Google Scholar]
- Monigatti F, Gasteiger E, Bairoch A, Jung E, 2002. The Sulfinator: predicting tyrosine sulfation sites in protein sequences. Bioinformatics 18, 769–770. [DOI] [PubMed] [Google Scholar]
- Nei M, Kumar S, 2000. Molecular Evolution and Phylogenetics. Oxford University Press, New York. [Google Scholar]
- Nusbaum MP, Blitz DM, 2012. Neuropeptide modulation of microcircuits. Curr. Opin. Neurobiol. 22, 592–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusbaum MP, Blitz DM, Marder E, 2017. Functional consequences of neuropeptide and small-molecule co-transmission. Nat. Rev. Neurosci. 18, 389–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusbaum MP, Marder E, 1988. A neuronal role for a crustacean red pigment concentrating hormone-like peptide: neuromodulation of the pyloric rhythm in the crab, Cancer borealis. J. Exp. Biol. 135, 165–181. [Google Scholar]
- Paluzzi JP, Vanderveken M, O’Donnell MJ, 2014. The heterodimeric glycoprotein hormone, GPΑ2/GPB5, regulates ion transport across the hindgut of the adult mosquito, Aedes aegypti. PLoS One 9, e86386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H, 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8,785–786. [DOI] [PubMed] [Google Scholar]
- Poels J, Van Loy T, Vandersmissen HP, Van Hiel B, Van Soest S, Nachman RJ, Vanden Broeck J, 2010. Myoinhibiting peptides are the ancestral ligands of the promiscuous Drosophila sex peptide receptor. Cell. Mol. Life Sci. 67, 3511–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H, 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132, 365–386. [DOI] [PubMed] [Google Scholar]
- Saideman SR, Christie AE, Torfs P, Huybrechts J, Schoofs L, Nusbaum MP, 2006. Actions of kinin peptides in the stomatogastric ganglion of the crab Cancer borealis. J. Exp. Biol. 209, 3664–3676. [DOI] [PubMed] [Google Scholar]
- Saideman SR, Ma M, Kutz-Naber KK, Cook A, Torfs P, Schoofs L, Li L, Nusbaum MP, 2007. Modulation of rhythmic motor activity by pyrokinin peptides. J. Neurophysiol. 97, 579–595. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M, 1987. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. [DOI] [PubMed] [Google Scholar]
- Selverston AI, Moulins M, 1987. The Crustacean Stomatogastric System. Springer; Berlin Heidelberg. [Google Scholar]
- Skiebe P, Schneider H, 1994. Allatostatin peptides in the crab stomatogastric nervous system: inhibition of the pyloric motor pattern and distribution of allatostatin-like immunoreactivity. J. Exp. Biol. 194, 195–208. [DOI] [PubMed] [Google Scholar]
- Stemmler EA, Cashman CR, Messinger DI, Gardner NP, Dickinson PS, Christie AE, 2007a. High-mass-resolution direct-tissue MALDI-FTMS reveals broad conservation of three neuropeptides (APSGFLGMRamide, GYRKPPFNGSIFamide and pQDLDHVFLRFamide) across members of seven decapod crustacean infraorders. Peptides 28, 2104–2115. [DOI] [PubMed] [Google Scholar]
- Stemmler EA, Peguero B, Bruns EA, Dickinson PS, Christie AE, 2007b. Identification, physiological actions, and distribution of TPSGFLGMRamide: a novel tachykinin-related peptide from the midgut and stomatogastric nervous system of Cancer crabs. J. Neurochem. 101, 1351–1366. [DOI] [PubMed] [Google Scholar]
- Swensen AM, Marder E, 2000. Multiple peptides converge to activate the same voltage-dependent current in a central pattern-generating circuit. J. Neurosci. 20, 6752–6759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swensen AM, Marder E, 2001. Modulators with convergent cellular actions elicit distinct circuit outputs. J. Neurosci. 21, 4050–4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo TM, Chen R, Goeritz ML, Maloney RT, Tang LS, Li L, Marder E, 2011. Distribution and physiological effects of B-type allatostatins (myoinhibitory peptides, MIPs) in the stomatogastric nervous system of the crab Cancer borealis. J. Comp. Neurol. 519, 2658–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghert PH, Nitabach MN, 2012. Peptide neuromodulation in invertebrate model systems. Neuron 76, 82–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S, 2013. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 30, 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobe SS, Bendena WG, 2013. Allatostatins. In Handbook of Biologically Active Peptides (Second Edition), 191–196. [Google Scholar]
- Vaughn JL, Goodwin RH, Tompkins GJ, McCawley P, 1977. The establishment of two cell lines from the insect Spodoptera frugiperda (Lepidoptera; Noctuidae). In Vitro 13, 213–217. [DOI] [PubMed] [Google Scholar]
- Veenstra JA, 2000. Mono- and dibasic proteolytic cleavage sites in insect neuroendocrine peptide precursors. Arch. Insect. Biochem. Physiol. 43, 49–63. [DOI] [PubMed] [Google Scholar]
- Weimann JM, Marder E, Evans B, Calabrese RL, 1993. The effects of SDRNFLRFamide and TNRNFLRFamide on the motor patterns of the stomatogastric ganglion of the crab Cancer borealis. J. Exp. Biol. 181, 1–26. [DOI] [PubMed] [Google Scholar]
- Weimann JM, Skiebe P, Heinzel HG, Soto C, Kopell N, Jorge-Rivera JC, Marder E, 1997. Modulation of oscillator interactions in the crab stomatogastric ganglion by crustacean cardioactive peptide. J. Neurosci. 17, 1748–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurch T, Lestienne F, Pauwels P, 1998. A modified overlap extension PCR method to create chimeric genes in the absence of restriction enzymes. Biotechnol. Tech. 12, 653–657. [Google Scholar]
- Yamanaka N, Hua YJ, Roller L, Spalovská-Valachová I, Mizoguchi A, Kataoka H, Tanaka Y, 2010. Bombyx prothoracicostatic peptides activate the sex peptide receptor to regulate ecdysteroid biosynthesis. Proc. Natl. Acad. Sci. USA 107, 2060–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka N, Yamamoto S, Zitnan D, Watanabe K, Kawada T, Satake H, Kaneko Y, Hiruma K, Tanaka Y, Shinoda T, Kataoka H, 2008. Neuropeptide receptor transcriptome reveals unidentified neuroendocrine pathways. PLoS One 3, e3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Wang J, Zhang Z, Jia C, Schmerberg C, Catherman AD, Thomas PM, Kelleher NL, Li L 2015. Defining the neuropeptidome of the spiny lobster Panulirus interruptus brain using a multidimensional mass spectrometry-based platform. J. Proteome Res. 14, 4776–4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.