Abstract
Acute myeloid leukemia (AML) is an aggressive hematopoietic disease characterized by glutamine-dependent metabolism. A novel glutaminase (GLS) inhibitor, CB-839, is currently under evaluation for treatment of hematopoietic malignancies and solid tumors. Our purpose was to measure cellular changes in AML associated with CB-839 treatment and to test the ability of hyperpolarized pyruvate for interrogating these changes to OCI-AML3 cells. Our results show that treatment with CB-839 interfered with the citric acid cycle, reduced the NADH/NAD+ ratio and ATP levels, reduced cell proliferation and viability, and reduced the basal and maximal respiratory capacities (OCR). We observed a reduction of the conversion of hyperpolarized pyruvate to lactate in cell lines and in a mouse AML model after CB-839 treatment. Our in vitro and in vivo results support the hypothesis that, in AML, glutamine is utilized to generate reducing equivalents (NADH, FADH2) through the citric acid cycle and that reduction in redox state by glutaminase inhibition decreases the rate of pyruvate to lactate conversion catalyzed by lactate dehydrogenase. We propose hyperpolarized pyruvate/lactate measurement as a method for direct monitoring of metabolic changes occurring in AML patients receiving CB-839. With further optimization, this method may provide a non-invasive imaging tool to assess the early efficacy of therapeutic intervention with GLS inhibitors.
Keywords: Glutamine metabolism, acute myeloid leukemia, magnetic resonance, CB-839
INTRODUCTION
Acute myeloid leukemia (AML) is a hematopoietic malignancy that results in the accumulation and expansion of immature cells (blasts). AML patients usually have an initial response to standard-of-care chemotherapy, but a large subset of these patients will experience relapse. Often, relapsed disease is chemotherapy-resistant, with an ultimately fatal outcome. A significant number of studies focus on exploring cellular mechanism and pathways in AML with the overall goal of improving the outcome and increasing the overall survival of patients [1,2]. One of those mechanisms is metabolic reprogramming, which is a hallmark of cancer cells and is currently a major target for medicinal chemistry efforts [3].
Metabolic reprogramming is often linked with cellular changes such as accelerated tumor growth, tumor aggressiveness, and invasiveness or protection against environmental stresses such as chemotherapy (therapy resistance), nutrient deprivation (autophagy), oxygen limitation (tumor hypoxia), and others [3,4]. Otto Warburg’s paradigm-shifting work revealed that many cancer cells have greater glucose uptake than normal cells [5,6]. Recent studies have found that this increased glucose intake in many cancer cell types is not used for energy production but for biomass production (synthesis of amino acids, nucleotides, etc.); for energy, cancer cells can utilize other small molecules such as glutamine [7,8]. One of the critical steps in glutamine utilization in AML and other cancer types is its conversion to glutamate by glutaminase (GLS) enzymes [8–11]. In humans, the glutaminase family consists of two members which are encoded by separate genes on different chromosomes [12]. The GLS gene, located on chromosome 2, encodes two isoforms, known as kidney (k-type) glutaminase (KGA) and glutaminase C (GAC), both of which are expressed in the mitochondrial matrix (Figure 1). KGA and GAC are collectively referred to as GLS. The GLS2 gene, located on chromosome 12, codes for two liver (L-type) isozymes LGA, as well as the longer isoform GAB. These two isoforms are referred to as GLS2 and are expressed in the cytosol [12]. GLS has been shown to be overexpressed compared to GLS2 in several cancers, and specifically in several AML cell lines [11]. Analysis of The Cancer Genome Atlas database (TCGA) confirmed that GLS expression is significantly higher than GLS2 expression in AML cells. The mean expression of GLS is in the upper 4% of all genes expressed in the AML cohort, underlining the importance of GLS in leukemia metabolism [13]. This increased expression and utilization of GLS is correlated with the increased susceptibility of several AML cell lines to glutaminase inhibitors, such as the small molecule CB-839 [11,13]. In addition, glutaminolysis is important in T cell acute lymphoblastic leukemia (T-ALL) and is controlled by NOTCH1 signaling [14]. CB-839 is a potent inhibitor of both major isoforms of GLS, has good oral bioavailability, and is in phase I and phase II studies for solid tumors and hematological malignancies [15,16].
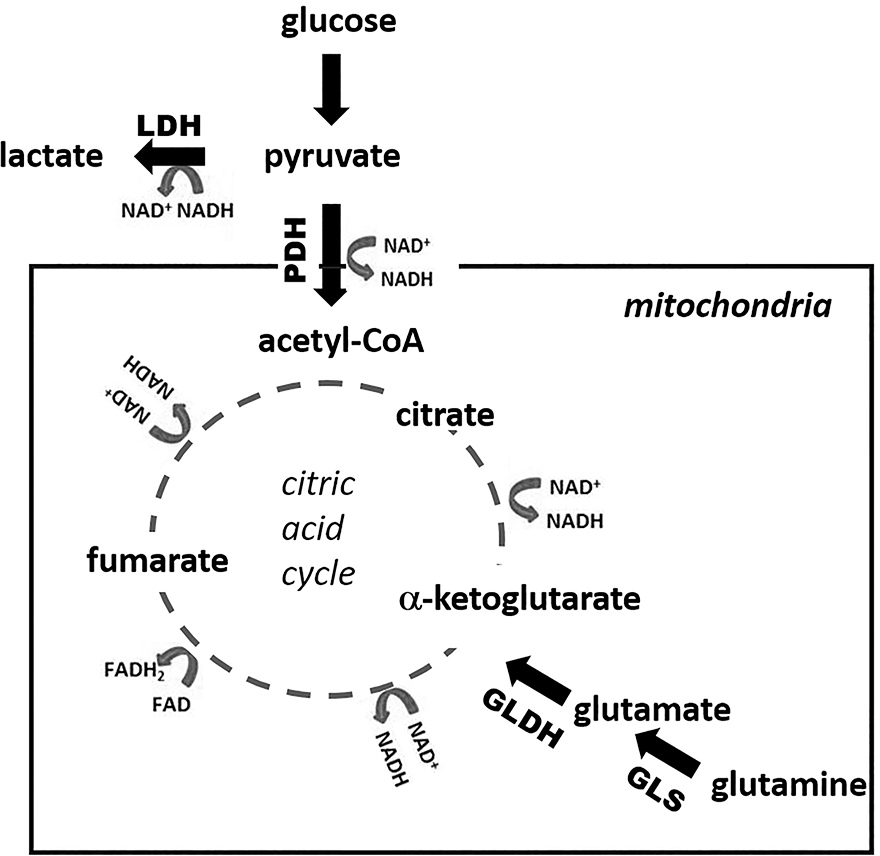
Figure 1. Metabolic pathways.
This schematic depicts some of the pathways of glycolysis, glutaminolysis, and the citric acid cycle with specific enzymes, conversion of cofactors such as NAD+, and specific metabolites labeled. Pyruvate can be utilized either by pyruvate dehydrogenase (PDH) in the mitochondria and then enter the citric acid cycle or by lactate dehydrogenase (LDH) in the cytosol to generate lactate. GLDH, glutamate dehydrogenase; GLS, glutaminase.
CB-839 has been studied in in vitro settings and in preclinical animal models in a vast number of tumors [11,13,15,17]. Bouscary at al. demonstrated significant differences in its inhibition of cell proliferation and reduction of viability, ranging from 0% to 50% after incubation with CB-839 for 72 hours, illustrating the differences in glutamine utilization and role of glutaminase among different AML cell lines [11]. In a mouse model of triple-negative breast cancer, CB-839 (200 mg/kg) reached plasma and tumor saturation >1.5 μmol/L 4 hours post oral administration, and, compared to vehicle, increased the glutamine/glutamate ratio in the tumor tissue [15]. Even with this rapid change in the concentration of glutamate and glutamine, metabolic changes as measured by oxygen consumption rate (OCR) were detected in both triple-negative breast cancer and AML cell lines after incubation with CB-839 for periods ranging from 80 minutes [15] to 6 hours [11]. This suggests that changes in metabolic flux measured by glutamine and glutamate might appear earlier than changes in oxidative phosphorylation reflected by OCR.
A novel spectroscopic imaging technique called hyperpolarization is an emerging method to measure metabolic flux. Hyperpolarization allows in vivo real-time assessment of metabolic flux by increasing the magnetic resonance (MR) signal intensity of a compound by as much as 10,000-fold [18–21]. This increase in signal is retained not only on the hyperpolarized compound but also on its downstream metabolites. The most utilized hyperpolarized imaging agent is 1-13C pyruvate, which was recently assessed in a phase 1 clinical trial for a safety and dose-escalation study of pyruvate ( NCT02526368) [19]. Hyperpolarized 1-13C pyruvate is quickly taken up by many cancer cells and converted to 1-13C lactate through lactate dehydrogenase (LDH) [18,19,22–24]. In a previous study, we measured the pyruvate to lactate flux in the bone marrow of AML-engrafted mice and normal controls using hyperpolarized pyruvate [25]. We found that the observed rate of conversion of hyperpolarized pyruvate in mice with AML1/ETO leukemia was double that in normal mice [25]. We continued this work in the current study, with the aim of determining whether hyperpolarized pyruvate conversion would be a useful imaging technique for real-time, in vivo monitoring of metabolic changes in AML-bearing mice after CB-839 treatment.
METHODS
Chemistry
The chemical structure of CB-839 is found in Supplementary Figure S1. CB-839 is used as provided by Calithera Bioscience, synthesized according to the method in reference [26]. The synthesis and chemical characterization can be found in [26], referenced as compound 670.
Cell Culture
HL-60 and OCI-AML3 cells lines were purchased from DSMZ (German Collection of Microorganisms and Cell Cultures). The cell lines were grown in suspension in RPMI medium supplemented with 10% fetal bovine serum, 10 U/mL penicillin, and 10 μg/mL streptomycin. All in vitro culture assays (except hyperpolarization) were performed in the same modified RPMI medium. Cells suspensions were split into new flasks when the concentration of cells reached 0.6–0.9 × 106 cells per milliliter. OCI-AML3 cells expressing luciferase and green fluorescent protein (GFP) were generated using standard transfection techniques. OCI-AML3 cells were stably transfected with pHUSH luciferase (control), pHUSH puro-KGA (KGA knockdown), pHUSH puro-GAC (GAC knockdown), or pHUSH puro-D3 (KGA and GAC knockdown). shRNA vectors were obtained from the Hatzivassiliou laboratory [27]. To induce GLS knockdown, cells in suspension (1 million per ml) were cultured in the presence or absence of 4 μg/ml doxycycline for 3 days. Knockdown was confirmed by western blot. All cell lines were tested and authenticated by The University of Texas MD Anderson Cancer Center Cell Line Validation Core Facility, through the short tandem repeat method.
Western Blot Analysis
The OCI-AML3 cells stably transfected with pHUSH luciferase control, pHUSH puro-KGA, pHUSH puro-GAC, or pHUSH puro-D3 shRNA were grown, collected, washed in PBS, and pelleted before lysing with RIPA buffer (PIERCE) containing protease and phosphatase inhibitors (Roche). After 30 min incubation on ice, cell lysates were subject to sonication (~2 min) and centrifugation (12,000 rpm x 10 min, 4° C). The supernatant was collected and the protein concentration quantified using the BCA Protein Assay (Thermo Scientific). Western blot analysis performed according to standard procedures. Antibodies used in western analyses: GLS (ab93434, Abcam), GAPDH (MAB374, MilliporeSigma), LDH (2012S, Cell Signal Technology), α-tubulin (ab7291, Abcam); and donkey anti-mouse antibody IRDye 800CW (C61116–02, LI-COR), donkey anti-rabbit antibody (C70601–05, LI-COR). GLS (ab93434) recognizes both the KGA and GAC isotypes but not GLS2 [28,29].
In Vitro Studies
Fresh glutamine was added to all in vitro assays prior to the experiment to a final concentration of 2 mM. CB-839 concentration was 1 μM for all experiments. To obtain this concentration, CB-839 was dissolved in dimethyl sulfoxide (DMSO) at a concentration of 10 mM. The 1 μM CB-839 solution was generated from separate aliquots of the 10 mM solution so that the CB-839 concentrated solution had to undergo only one freeze-thaw cycle. Vehicle control wells were treated with 0.1% DMSO.
For the ATP studies, HL-60 or OCI-AML3 cells were seeded (2000 cells per well) in a 96-well plate and were treated with CB-839 or DMSO for 72 hours. The cells were then directly treated with CellTiter-Glo Luminescent Cell Viability Assay (G7571; Promega). ATP levels were determined on a Synergy H4 microplate reader. For the cell proliferation assay, 20 million OCI-AML3 cells (1 million cells per ml) were treated with CB-839 or DMSO and the flasks incubated for 72 hours. Cells were counted using a Beckman Vicell. To examine the rescue effect of DMKG on the growth of cells, cells were simultaneously treated with 1 μM CB-839 and 1 mM dimethyl-α-ketoglutarate (DMKG). Cell growth in treated cells was determined after 72 hours by flow cytometry using an AnnexinV-DAPI assay.
To determine NADH and NAD+ concentrations, OCI-AML3 cells (2000 per well) were placed in a 96-well plate and incubated with 1 μM CB-839 for 12 hours and NADH and NAD+ levels were determined using the NAD/NADH Glo (G9071; Promega) luminescent assay kit. To determine the effect of rescue from glutaminase inhibition, OCI-AML3 cells were incubated with 1 μM CB-839 or 1 μM CB-839 + 4 mM DMKG for 72 hours prior to analysis with the NAD/NADH kit.
To determine GSH and GSSG concentrations, OCI-AML3 cells were grown with and without 1 μM CB-839 for 24 hours. After incubation, cells were pelleted and homogenized in 5% solution of 5-sulfo-salicylic acid dihydrate at a concentration of 5 million cells per ml. GSH and GSSG concentrations were determined using Invitrogen Glutathione Colorimetric Kit (EIAGSHC).
High-resolution MRS in cell culture
For all high-resolution MR spectroscopy (MRS), a 500 MHz Bruker Avance III HD NMR equipped with a Prodigy BBO cyroprobe (Bruker BioSpin MRI GmbH) was utilized. The cyroprobe increases the sensitivity of the measurement threefold to fourfold. All supplies (D2O, DSS-d6 [3-trimethylsilyl)-1-propanesulfonic acid-d6 sodium salt], phosphate buffer, DMKG) were purchased from Sigma-Aldrich and used without further purification.
To identify differences in intracellular metabolite levels after glutaminase inhibition, cells (approximately 1 million cells per mL in a total volume of 22.5 mL) were incubated with either 0.1% DMSO (control) or 1 μM CB-839 for 4 hours. Four replicates were performed for each treatment. 1-13C pyruvate was then added to the cell suspensions to a final concentration of 2 mM. Aliquots of the suspensions were taken at time zero (addition of 1-13C pyruvate) and at 2 and 4 hours after this addition. At the 4-hour time point (after a cumulative 8 hours incubation with CB-839), cells were pelleted and washed with phosphate-buffered saline solution, and metabolites were extracted using 3 mL of a 2:1 solution of ice-cooled methanol to water and ~500 μL of lysing matrix D beads (MP Biomedicals, Santa Ana, CA) per 107 cells. The homogenates were then subjected to centrifugation for 10 minutes at 4000g and the supernatant removed; the samples were lyophilized overnight, and the remaining metabolites were dissolved in D2O with 0.5 mM DSS-d6 and 50 mM K2HPO4 (pH 7.4). The 1D 1H-NMR spectra used 256 scans, a relaxation delay of 6 seconds, and a spectral width of 10245 Hz and were referenced to DSS-d6 at 0.00 ppm. Water suppression was performed with presaturation. Data were processed/analyzed with Chenomx software (Chenomx Inc., Edmonton, Alberta) or MestReNova software (Mestrelab Research, Santiago, Spain). Integrated values of intracellular metabolites were determined by taking the ratio of the resonance for each metabolite over the DSS-d6 peak. For extracellular lactate measurements similar 1D 1H-NMR spectroscopy of the cell media was performed using 152 scans, a relaxation delay of 6 seconds, and a spectral width of 10245 Hz and referenced to DSS-d6 at 0.00 pm. All samples were normalized to cell count. Proton decoupled 1-D 13C spectroscopy was performed (4096 averages, relaxation delay 6 seconds, spectral width 29760 Hz, 30° flip angle) on each sample in 1-13C pyruvate feeding study. Metabolite resonances were identified through reference to either of two online metabolomics databases, Human Metabolome Database (http://www.hmdb.ca)[30] or Biological Magnetic Resonance Bank (http://www.bmrb.wisc.edu/metabolomic) and, when necessary, confirmed by spiking the sample with a known amount of the metabolite in question.
Seahorse oxygen consumption rate
OCI-AML3 cells were treated with 0.1% DMSO; 1 μM CB-839; 0.5, 1, 2, or 4 mM DMKG; or 1 μM CB-839 + 0.5, 1, 2, or 4 mM of DKMG for 6, 12, or 24 hours in RPMI medium containing 10% fetal bovine serum. Cells were washed with phosphate-buffered saline solution; resuspended in XF Assay medium supplemented with fresh glucose 10 mM, glutamine 2 mM, and pyruvate 1 mM; plated in XF96e Seahorse Biosciences plates pre-coated with Cell-Tak (BD Biosciences, San Jose, CA) at a density of 3 × 105 cells per well; and spun down on the plate to allow cells to adhere equally. Six replicate wells were analyzed for each treatment.
Mouse AML Models
All experiments that involved mice were performed, and mice were maintained, in accordance with the guidelines, regulations, and animal use protocols approved by the MD Anderson Cancer Center Institutional Animal Care and Use Committee. Male NSG mice were injected with ~5 × 106 OCI-AML3 cells via the tail vein. To ensure engraftment, the animals were imaged approximately 2 to 3 weeks later on an IVIS Spectrum bioluminescence imager 10 minutes after intraperitoneal injection of 10 μL/g of a 15 mg/mL solution of luciferin-D. Animals with a high bioluminescence signal were then utilized in experiments with hyperpolarized pyruvate. Prior to hyperpolarization experiments, animals were anesthetized with 3% isoflurane mixed with oxygen; anesthesia was maintained at 0.5–1% isoflurane and a tail vein catheter was placed. Animals were placed on a heated pad and their respiration and heart rate monitored during imaging procedures.
Imaging Procedures and Hyperpolarized Pyruvate Animal Studies
Ox063 trityl radical (GE Healthcare) was mixed with neat 1-13C pyruvic acid (Sigma-Aldrich) to a concentration of 15 mM. This solution (20 μL) with 0.4 μL of 50 mM Gd3+ (Magnevist, Bayer Healthcare) was loaded into a DNP commercial HyperSense polarizer (Oxford Instruments) and irradiated at a microwave frequency of 94.100 GHz for 30–40 minutes (until polarization plateau) and then dissolved in 4 mL buffer solution containing 40 mM Tris (7.6 pH preset), 80 mM NaOH, 0.1 g/L EDTA, and 50 mM NaCl. After dissolution, the neutral (pH 7–8) 80 mM hyperpolarized 1-13C pyruvate solution was injected into each mouse via a tail vein catheter.
All imaging and spectroscopy were performed with a dual tuned 1H/13C volume coil (OD: 75mm, ID:40mm, Bruker BioSpin MRI GmbH) in a 7T Bruker Biospec horizontal bore MR scanner (Bruker BioSpin MRI GmbH) equipped with a single channel for carbon excitation/reception. Proton anatomic images were taken using a multi-slice T2-weighted RARE sequence. A small 8 M 13C-urea phantom doped with gadolinium-DPTA was placed between the legs to allow for chemical shift referencing. For 13C spectroscopy, a series of slice-selective 13C spectra (field-of-view 40 × 40 mm, slice-thickness 12–15 mm) were collected right after injection of hyperpolarized 1-13C pyruvate. The single slice was placed over the legs using the multi-slice proton imaging sequence for placement. A total of 90 transients were acquired at 2-second intervals (total time 3 minutes). Each transient utilized a 15–20° degree flip angle excitation pulse (gauss pulse) and 2048 data points. Data were processed by MATLAB (MathWorks Inc.), TopSpin (Bruker BioSpin GmbH), and MestReNova (Mestrelab Research) software. The dynamic spectra were manually phased, and line-broadening was applied (10 to 15 Hz). The areas under the spectral peaks for pyruvate and lactate were integrated over the whole array. Normalized lactate (nLac) ratio was calculated as lactate signal divided by the sum of pyruvate and lactate signals [31].
Hyperpolarized Pyruvate Cell Studies
Pyruvate was hyperpolarized by a method similar to that used in animal experiments. All experiments were performed on a vertical bore 7T Bruker BioSpin NMR with 10 mm broadband probe and running TopSpin3.5 software. OCI-AML3 cells (2.5 mL of a suspension in RPMI [dry powder mixed with 90% water and 10% D2O]) were added to 10-mm NMR tubes. The probe was heated to 37°C and was maintained at this temperature throughout the experiment. After auto-locking, tuning, and shimming were performed, 500 μL of the hyperpolarized compound was injected into the suspended cells using an injection port. The final concentration of hyperpolarized pyruvate in the NMR reactor was approximately 8.7 mM. Single 13C transients were taken using Waltz decoupling (zgdc pulse) every 6 seconds with a 15° flip angle. The nLac ratio was calculated as the total lactate signal divided by the sum of pyruvate and lactate signals over the first 2 minutes of the experiment. Cell viability was analyzed using trypan blue staining and counted on a BioRad TC20 automated cell counter. The percentages of viable cells were similar before and after the experiments (88–96% viable). Table 2 presents the values per million cells times 1000.
Table 2.
Reduced hyperpolarized pyruvate to lactate conversion in culture after CB-839 treatment
| OCI-AML3 Cells | Hyperpolarized nLac/million cells* |
|---|---|
| CB-839 Treated (n =6) | 0.12 + 0.02 |
| Control (n = 4) | 0.21 + 0.02 |
To normalize for cell count, the nLac value (lactate signal/pyruvate + lactate signal) was divided by the number of cells in each experiment and then multiplied by 1000.
Statistical Analysis
All graphs, Student t-tests, and ANOVA analyses were performed in GraphPad Prism software (GraphPad Software). All experimental data are shown as mean ± standard error of the mean.
RESULTS
CB-839 treatment reduced cell viability, which was rescued with DMKG
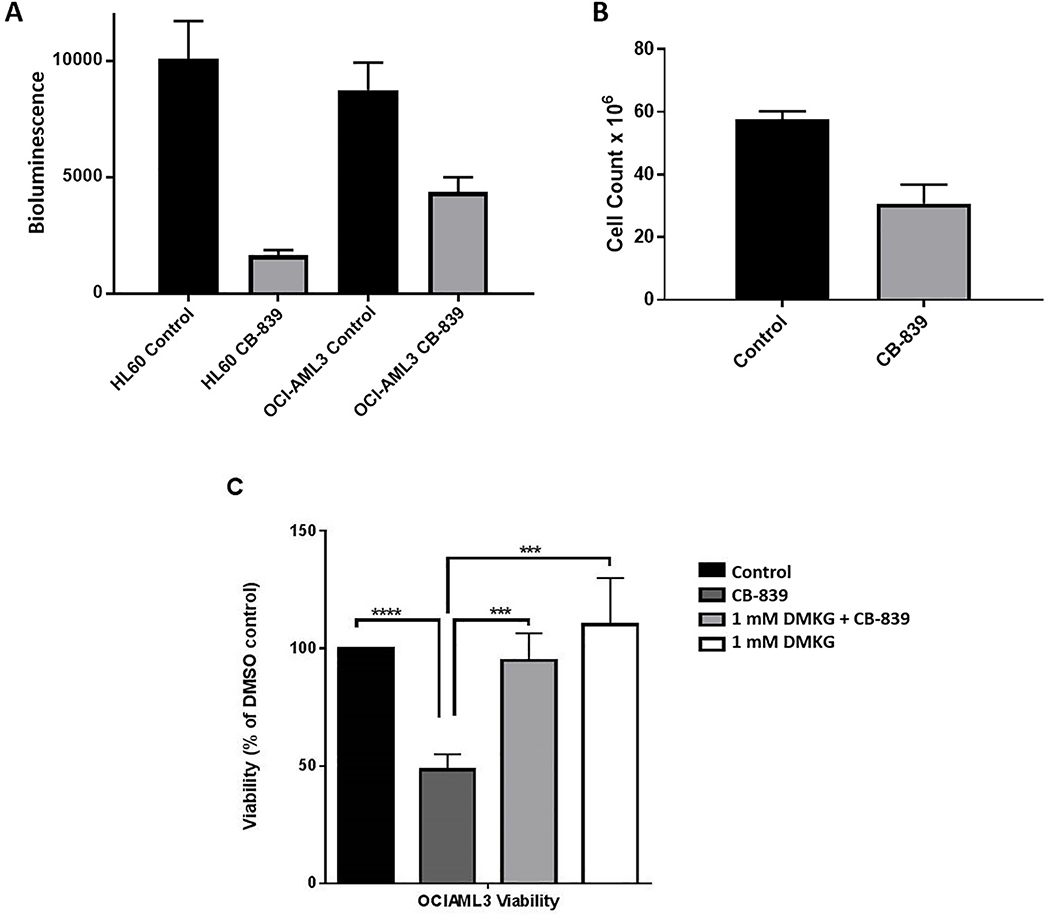
The first aim of this study was to investigate the impact of CB-839 on AML cell proliferation and ATP production. In an ATP luminescence assay, ATP levels were significantly reduced in AML cell lines HL-60 and OCI-AML3 incubated with 1 μM CB-839 for 72 hours (Figure 2A). Proliferation of OCI-AML3 cells also was reduced after incubation with 1 μM CB-839 for 72 hours (Figure 2B). To test the hypothesis that reduction of ATP and reduction of cell proliferation in AML was due to decreased flux of the citric acid cycle and oxidative phosphorylation (Figure 1), rescue experiments with dimethyl-α-ketoglutarate (DMKG) were performed. DMKG, a cell-permeable analog of α-ketoglutarate, increases the citric acid cycle flux and enables rescue of GLS inhibition by directly restoring the pool of α-ketoglutarate [11,15]. Figure 2C depicts the viability of cells after 72 hours incubation with 0.1% dimethyl sulfoxide (DMSO; control), 1 μM CB-839, or a combination of CB-839 + 1 mM DMKG. Compared to vehicle, CB-839 significantly reduced the viability of cells, but this reduced viability was completely rescued with the addition of 1 mM DMKG (Figure 2C).
Figure 2. Viability and ATP levels lowered in AML cells by CB-839 treatment.
(A) ATP levels were quantified by using a CellTiter-Glo assay and are expressed as bioluminescence values. Cells were treated with either 0.1% DMSO or 1 μM CB-839 for 72 hours. Six to seven replicates of each condition were run. The differences in ATP levels between treated and untreated cells were analyzed for statistical significance by an unpaired, two-tailed t-test (P <0.002 for both cell types). (B) OCI-AML3 cells (20 million cells/flask) were treated with either 1 μM CB-839 or 0.1% DMSO for 72 hours (n = 3) and then counted. The difference in cell count between the treated and control groups was analyzed for statistical significance by unpaired, two-tailed t-test (P <0.002). (C) OCI-AML3 cells were treated with 0.1% DMSO, 1 μM CB-839, or a combination of 1 μM CB-839 + 1 mM DKMG for 72 hours. Cell viability was evaluated by flow cytometry with AnnexinV/ROS/DAPI staining. The cell numbers per 300 beads were recorded. Viability was reduced by CB-839 treatment alone compared to controls but was rescued with the addition of DMKG. ***P <0.005, ****P <0.0001, as analyzed by one-way ANOVA.
Metabolites reduced in OCI-AML3 cells after CB-839 treatment
OCI-AML3 were treated with CB-839 or vehicle (0.1% DMSO) for 4 hours prior to addition of 1-13C pyruvate. Extracellular 1-13C lactate values were determined 2 hours after addition of labeled pyruvate. Reduced lactate production was observed in CB-839 treated cells (Table 1, n = 4 of each condition). Similar reduction in lactate is observed when GLS is knocked down after 72 hours of doxycycline induction (Supplementary Table 1, Supplementary Figure S3). A similar reduction in lactate is observed when the GAC isotype is knocked down in HT29 and HCT116 colorectal cells [32].
Table 1.
Reduced metabolites observed in OCI-AML3 cells after CB-839 treatment
| In vitro treatment with 1 μM CB-839 or DMSO | ||||
|---|---|---|---|---|
| OCI-AML3 Cells | Extracellular 1-13C Lactate | Lactate (intracellular) | Alanine (intracellular) | Glutamate (intracellular) |
| CB-839 Treated (n = 4) | 0.43 ± 0.09 | 0.88 ± 0.04 | 0.03 ± 0.01 | 0.14 ± 0.05 |
| Control (n = 4) | 0.75 ± 0.21 | 1.92 ± 0.39 | 0.13 ± 0.02 | 0.36 ± 0.02 |
Intracellular lactate, alanine, glutamate values were determined after 8 hours of treatment with CB-839 or vehicle. Metabolic values from CB-839–treated cells differ significantly (Table 1) from those of controls by two-way ANOVA (P < 0.0001). This result corresponds to our previously reported liquid chromatography-mass spectroscopy measurements where levels of glutamate and citric acid cycle intermediates such as fumarate, malate, and citrate and glutathione were reduced after CB-839 treatment in OCI-AML3 cells [13].
GLS inhibition reduced oxidative phosphorylation in OCI-AML3 cell line
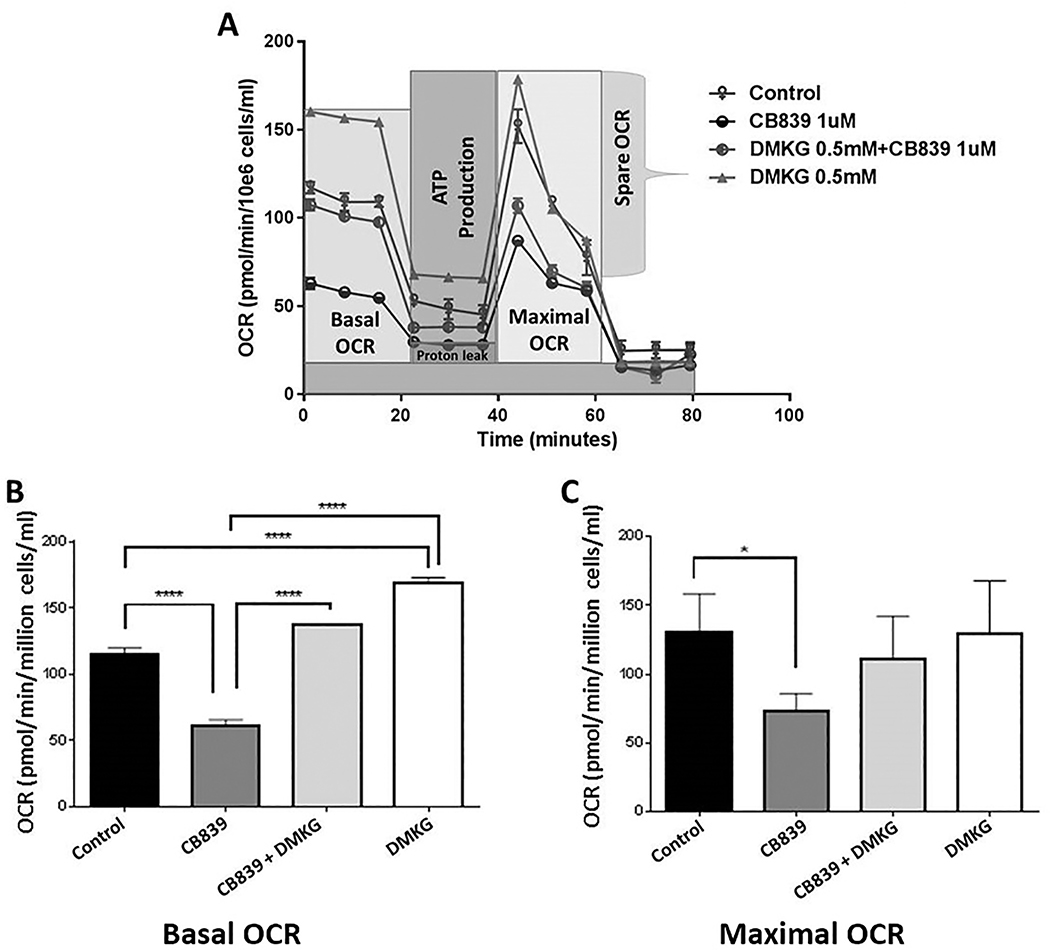
To better understand the impact of GLS inhibition on oxidative phosphorylation, OCR was measured in OCI-AML3 cells with and without CB-839 treatment using a Seahorse XFe Bioanalyzer (Figure 3A). The basal and maximal OCR values were recorded after 6 hours (Figure 3B, 3C), 12 hours, and 24 hours (Supplementary Figure S2A, S2B) of incubation with 1 μM CB-839. Both basal and maximal OCRs were significantly reduced after 6 hours incubation with CB-839 compared to controls, and these reductions persisted in cells incubated with CB-839 for as long as 24 hours.
Figure 3. Oxygen consumption rate in AML cells reduced after incubation with CB-839.
The effects of GLS inhibitor CB-839 and DKMG, a citric acid cycle fuel, on oxygen consumption rate (OCR) in OCI-AML3 cells as measured by a Seahorse Bioscience XFe96 Extracellular Flux Analyzer. (A) Representative example of five independent experiments is shown. (B, C) Basal (B) and maximal (C) OCRs are shown. OCI-AML3 cells were treated with 1 μM CB-839, 0.1% DMSO (control), 1 mM DMKG, or the combination of 1 μM CB-839 + 1 mM DMKG for 6 hours. Six replicate wells per treatment condition were analyzed, and the OCR values were normalized to cell number. Differences between treatment groups were analyzed for statistical significance by one-way ANOVA. Significant differences were observed in basal (B) and maximal (C) OCRs between CB-839– and control-treated cells. DMKG induced statistically significant rescue of basal OCR but not maximal OCR compared to cells treated with CB-839 alone. **** P <0.0001 for basal OCRs; * P <0.03 for maximal OCR, as analyzed by one-way ANOVA.
In the time interval between 6 and 24 hours, OCR reduction by CB-839 could be successfully overcome by the addition of 0.5 mM to 4 mM DMKG (Figure 3B, 3C and Supplementary Figure S2C). The OCRs in cells treated simultaneously with CB-839 and DKMG were close to or higher than those in the controls. DMKG, as a fuel specific to the citric acid cycle, increases both the baseline and stressed respiratory capacities of cells. This increase in respiratory activity was observed with both increasing concentration of DMKG and increasing duration of incubation.
CB-839 treatment reduced hyperpolarized pyruvate-to-lactate conversion in leukemia-bearing mice and AML cells
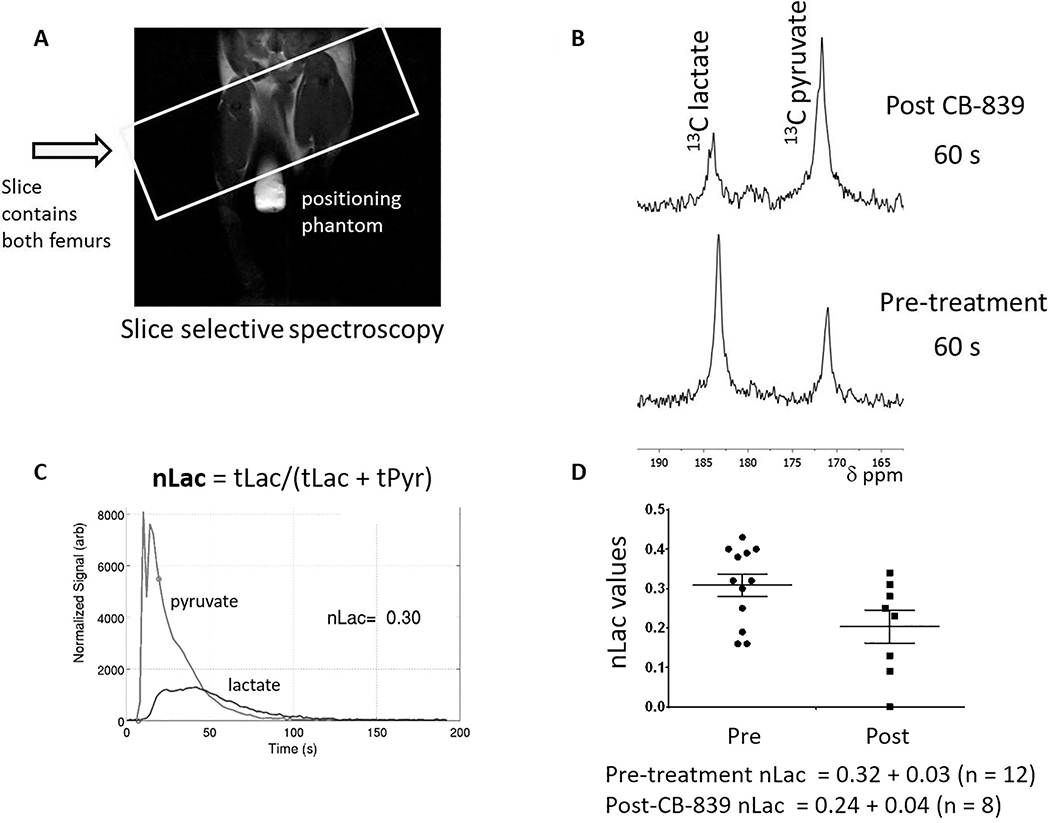
NSG mice were injected via the tail vein with OCI-AML3 cells stably transfected with luciferase and green fluorescent protein. After 2 to 3 weeks, the animals were imaged with luciferin-D and those with the highest bioluminescence in the legs, pelvis, and lower torso consistent with leukemia engraftment, were utilized for hyperpolarization studies (n=8–12/group). Animals were injected with 200–250 μL of 80 mM (16 μmol) hyperpolarized 1-13C pyruvate. A series of slice-selective 13C spectra were collected right after injection of hyperpolarized 1-13C pyruvate to observe the arrival of the compound to the femurs and knee cap and its conversion to lactate (Figure 4A). Animals were allowed to recuperate from isoflurane anesthesia (30 minutes to several hours) and then administered CB-839 (200 mg/kg) by oral gavage. Animals were re-imaged with hyperpolarized pyruvate 4 hours later. The conversion was quantified by integrating the resonance for pyruvate at 173 ppm and lactate at 185 ppm for each spectrum and plotting these values over time (Figure 4B). The hyperpolarized lactate pool produced in each experiment (normalized Lac, or nLac) was determined by calculating the area under the time course curve of 13C signal for lactate divided by the combined 13C signal for lactate and pyruvate (Figure 4C) [33]. Treatment with CB-839 reduced hyperpolarized pyruvate-to-lactate conversion (nLac Pre-treatment 0.32 ± 0.03 (n = 12), nLac Post-treatment 0.24 ± 0.04 (n = 8), P < 0.05, Mann-Whitney two-tailed Student t-test).
Figure 4. Pyruvate-to-lactate conversion reduced in vivo by CB-839 treatment.
(A) A characteristic proton image of an OCI-AML3-bearing mouse with an 8 mM 1-13C urea phantom placed between the legs. The box represents the single slice placement for 13C spectroscopy. (B) The pyruvate-to-lactate resonances found 60 s after injection of hyperpolarized 1-13C pyruvate prior to treatment (lower spectrum) and 4 hours after CB-839 treatment (top spectrum). (C) Representative normalized lactate (nLac) measurement from a pre-treatment animal. nLac was determined as the area under the lactate resonance cure (blue) divided by the total area under the lactate and pyruvate curves. (D) Pre-treatment and post-treatment pyruvate-to-lactate conversion rates prior to treatment and 4 hours after treatment with CB-839. To establish whether the change in pyruvate-to-lactate conversion after CB-839 treatment was significantly different from the pre-treatment value, a Mann-Whitney two-tailed Student t-test was used (P <0.05).
To confirm the in vivo mouse data, we measured the conversion of hyperpolarized pyruvate to lactate in vitro in OCI-AML3 cells treated for 24 hours with vehicle (buffer control) or 1 μM CB-839 in cell culture. The data confirmed that pyruvate-to-lactate conversion (nLac) was reduced by CB-839 (Table 2). Statistical significance in nLac between treated and untreated cells was determined by unpaired t-test with Welch correction (P <0.03). The viability of cells was determined before and after the hyperpolarized experiment by using trypan blue staining. The differences in percent viability before and after the polarization experiment were quite low, (0.4% ± 0.8 for CB-839 and 3% ± 3 for control experiments), indicating that the assay reflected true metabolic conversion changes rather than simply a drop in viability.
CB-839 reduced lactate levels and NADH/NAD+ ratio in OCI-AML3 cells
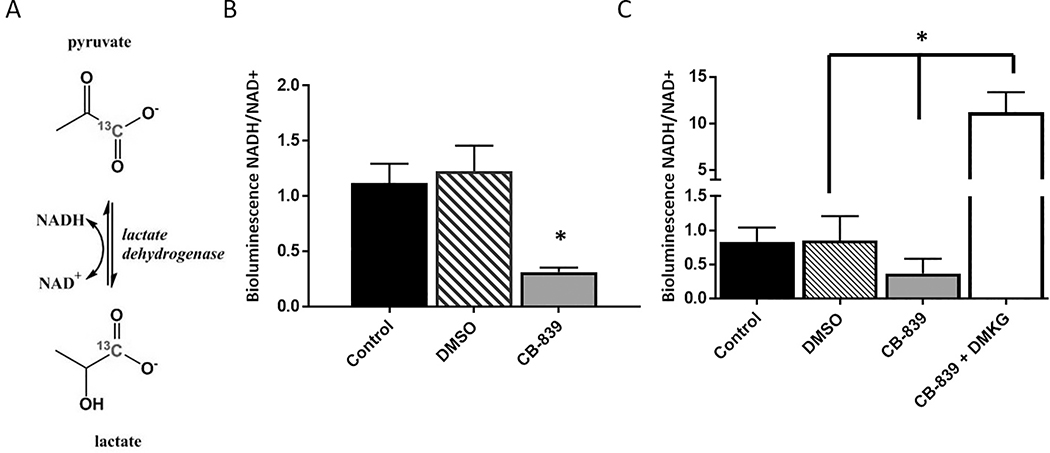
The reduction of pyruvate-to-lactate conversion by LDH is dependent on the co-factor NADH, which is oxidized to NAD+ during the reaction (Figure 5A). Therefore, the LDH activity rate is highly dependent on the intracellular concentrations of NADH and NAD+ [34,35]. To determine whether a reduction in the ratio of NADH/NAD+ might be the underlying reason for the reduction of pyruvate-to-lactate conversion after CB-839 treatment, the relative concentrations of NAD+ and NADH were measured using bioluminescence assay 12 hours after CB-839 treatment (Figure 5B). CB-839 treatment significantly reduced the NADH/NAD+ ratio in OCI-AML3 cells, and this ratio was increased when DMKG was added to CB-839 (Figure 5C). These results suggest that there is a change in the redox state of the cell after CB-839 treatment and that the change in pyruvate conversion we observed after treatment might be reflective of this effect.
Figure 5. NADH/NAD+ ratio reduced by incubation with CB-839 was rescued by DMKG.
(A) A schematic depiction of the reduction of 1-13C pyruvate to 1-13C lactate through lactate dehydrogenase. (B) The NADH/NAD+ ratio was quantified by using bioluminescence assay after no treatment (control) or treatment with 0.1% DMSO or 1 μM CB-839 for 12 hours. The experiment included six replicates of the CB-839 treatment and four to six replicates for no treatment or 0.1% DMSO controls. Differences between treatment groups were analyzed for statistical significance by one-way ANOVA. (C) The NADH/NAD+ value was quantified after no treatment or treatment with 0.1% DMSO, 1 μM CB-839, or 1 μM CB-839 + 4 mM DMKG for 72 hours. The experiment included seven replicates per treatment group. Statistical significance of differences between treatment groups were determined by one-way ANOVA, *P < 0.0001.
CB-839 reduces the GSH/GSSG ratio in OCI-AML3 cells
The ratio of free glutathione (GSH) versus its oxidized form (GSSG) is a marker of oxidative stress [36]. The relative concentrations of intracellular GSH and GSSG were measured using a colorimetric assay in OCI-AML3 cells incubated with and without 1 μM CB-839 for 24 hours. The ratio is approximately halved after CB-839 treatment (Supplementary Figure S4). This result is similar to the observed reduction in the GSH/GSSG ratio after CB-839 treatment in renal cell carcinoma [37] and pancreatic cancer cell lines [38].
DISCUSSION
Our results depict the cellular changes that occur in glutamine dependent AML cell lines when treated with glutaminase inhibitor CB-839. Lactate production is observed to be reduced both by genetic and drug targeting of GLS. Lactate production is reduced in both in vitro and hyperpolarized in vivo assays after CB-839 treatment. In addition, we observe a reduction in ATP, OCR, metabolites, cell viability, cell proliferation, GSH/GSSG ratio, and NADH/NAD+ ratio when cells are incubated with 1 μM CB-839. In addition, many of these reductions can be rescued by the addition of DMKG, which can directly fuel the citric acid cycle by restoring the pool of intracellular α-ketoglutartate. Using hyperpolarized-MR, we were able to observe metabolic changes due to CB-839 treatment in real time both in cell culture and in vivo animal models.
AML is typically diagnosed by bone marrow aspiration and biopsy. By the time the patient has symptoms and their disease is diagnosed by biopsy, the bone marrow throughout the body contains large numbers of leukemia cells. Thus, an in vivo imaging method would have the highest utility not in diagnosis but determining the efficacy of treatment. With the new metabolic inhibitors under clinical development, a robust, direct, real-time flux measurement of metabolic activity could determine quickly if the drug treatment is inducing desired therapeutic in vivo metabolic changes. In this study, we observed a reduction in hyperpolarized pyruvate-to-lactate conversion just 4 hours after treating OCI-AML3–bearing animals with CB-839. Our in vitro results reveal reduction of the NADH/NAD+ ratio in OCI-AML3 cells after treatment with CB-839. Our data suggest that CB-839 treatment reduces NADH levels, which in turn might lower the conversion of pyruvate to lactate by LDH both in vitro (Table 2) and in vivo (Figure 4). Our results differ slightly from those of previous studies of glutaminase inhibition using hyperpolarization imaging. In previous studies, conversion of pyruvate to lactate did not change in lymphoma animal models after treatment with BPTES (bis-2-(5-phenylacetamido-1,2,4-diathiazol-2-yl) ethyl sulfide), a micromolar GLS inhibitor [39]. The differences between our results with CB-839 and the BPTES results might be due to CB-839’s low nanomolar potency, lack of dependency on glutamine concentration, and slower reversible kinetics compared to BPTES [15]. In our studies, furthermore, post-treatment experiments were always performed 4 hours after oral gavage of CB-839 to ensure that the concentration of CB-839 in the blood stream was at its maximum [15]. When we carried out pilot experiments ~18 hours post treatment, changes in the conversion of pyruvate to lactate were not observed (nLac = 0.45 ± 0.03 [n = 3]; post nLac = 0.40 ± 0.07 [n =2]). For reversible metabolic inhibitors, inhibition occurs only when the drug is present.
With the advent of the clinical DNP polarizer (GE SPINlab™), the ability to utilize real-time metabolic imaging to monitor metabolic inhibition therapy has become a reality. Hyperpolarized pyruvate is an endogenous compound has been shown to be quite safe in patient use [19,40]. Our data reveal that, with further optimization, hyperpolarized pyruvate might be an effective method for direct measurement of the metabolic changes that occur with CB-839 in AML patients or in patients with other malignancies; we further conclude that this method could be used in the future to gauge the metabolic efficacy of novel agents affecting metabolism in leukemia.
Supplementary Material
ACKNOWLEDGMENTS
We want to thank the Department of Scientific Publications and Dr. Anna Romanowska-Pawliczek at MD Anderson Cancer Center for reading and editing the manuscript. In addition, we thank Calithera Biosciences for CB-839.
Financial Support: CPRIT grant RP150701, MDACC Institutional Research Grants, MDACC Institutional Startup, NCI U54 CA151668, P50 CA083639, NCI R01 CA206210, NCI R21 CA185536, John S. Dunn Foundation Collaborative Research Award administered by the Gulf Coast Consortia, NCI Cancer Center Support Grant CA016672.
Conflict of Interest: Part of this study was funded by Calithera Biosciences. Calithera Biosciences provided drug and vehicle control.
ABBREVIATIONS
- AML
acute myeloid leukemia
- ATP
adenosine triphosphate
- DMKG
dimethyl α-ketoglutarate
- DMSO
dimethyl sulfoxide
- GLS
glutaminase
- LDH
lactate dehydrogenase
- MR
magnetic resonance
- NAD+
oxidized form of nicotinamide adenine dinucleotide
- NADH
reduced form of nicotinamide adenine dinucleotide
- OCR
oxygen consumption rate
- GSH
glutathione
- GSSG
glutathione disulfide
REFERENCES
- 1.Reikvam H, Hauge M, Brenner AK, Hatfield KJ & Bruserud O Emerging therapeutic targets for the treatment of human acute myeloid leukemia (part 1) - gene transcription, cell cycle regulation, metabolism and intercellular communication. Expert review of hematology 2015; 8: 299–313. [DOI] [PubMed] [Google Scholar]
- 2.Fathi AT, Wander SA, Faramand R & Emadi A Biochemical, Epigenetic, and Metabolic Approaches to Target IDH Mutations in Acute Myeloid Leukemia. Seminars in hematology 2015; 52: 165–171. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D & Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674. [DOI] [PubMed] [Google Scholar]
- 4.DeBerardinis RJ & Thompson CB. Cellular metabolism and disease: what do metabolic outliers teach us? Cell 2012; 148: 1132–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Warburg O On the origin of cancer cells. Science 1956; 123: 309–314. [DOI] [PubMed] [Google Scholar]
- 6.Warburg O On respiratory impairment in cancer cells. Science 1956; 124: 269–270. [PubMed] [Google Scholar]
- 7.Vander Heiden MG, Cantley LC & Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 2009; 324: 1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wise DR & Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci 2010; 35: 427–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A 2007; 104: 19345–19350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheong H, Lu C, Lindsten T & Thompson CB. Therapeutic targets in cancer cell metabolism and autophagy. Nat Biotechnol 2012; 30: 671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacque N, Ronchetti AM, Larrue C, Meunier G, Birsen R, Willems L et al. Targeting glutaminolysis has antileukemic activity in acute myeloid leukemia and synergizes with BCL-2 inhibition. Blood 2015; 126: 1346–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mates JM, Campos-Sandoval JA & Marquez J Glutaminase isoenzymes in the metabolic therapy of cancer. Biochim Biophys Acta Rev Cancer 2018; 1870: 158–164. [DOI] [PubMed] [Google Scholar]
- 13.Matre P, Velez J, Jacamo R, Qi Y, Su X, Cai T et al. Inhibiting glutaminase in acute myeloid leukemia: metabolic dependency of selected AML subtypes. Oncotarget 2016; 7: 79722–79735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herranz D, Ambesi-Impiombato A, Sudderth J, Sanchez-Martin M, Belver L, Tosello V et al. Metabolic reprogramming induces resistance to anti-NOTCH1 therapies in T cell acute lymphoblastic leukemia. Nat Med 2015; 21: 1182–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gross MI, Demo SD, Dennison JB, Chen L, Chernov-Rogan T, Goyal B et al. Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol Cancer Ther 2014; 13: 890–901. [DOI] [PubMed] [Google Scholar]
- 16.Clinical Trials: NCT02071862, N, NCT03047993, NCT02861300, NCT03528642, NCT02771626, NCT02071888, NCT03163667, NCT03263429, NCT03428217, NCT03057600.
- 17.Seltzer MJ, Bennett BD, Joshi AD, Gao P, Thomas AG, Ferraris DV et al. Inhibition of glutaminase preferentially slows growth of glioma cells with mutant IDH1. Cancer Res 2010; 70: 8981–8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Golman K, in ‘t Zandt R & Thaning M Real-time metabolic imaging. Proc Natl Acad Sci U S A 2006; 103: 11270–11275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nelson SJ, Kurhanewicz J, Vigneron DB, Larson PE, Harzstark AL, Ferrone M et al. Metabolic imaging of patients with prostate cancer using hyperpolarized [1-(1)(3)C]pyruvate. Sci Transl Med 2013; 5: 198ra108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee Y, Zacharias NM, Piwnica-Worms D & Bhattacharya PK. Chemical reaction-induced multi-molecular polarization (CRIMP). Chem Commun (Camb) 2014; 50: 13030–13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zacharias NM, Chan HR, Sailasuta N, Ross BD & Bhattacharya P Real-time molecular imaging of tricarboxylic acid cycle metabolism in vivo by hyperpolarized 1-(13)C diethyl succinate. J Am Chem Soc 2012; 134: 934–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen AP, Albers MJ, Cunningham CH, Kohler SJ, Yen YF, Hurd RE et al. Hyperpolarized C-13 spectroscopic imaging of the TRAMP mouse at 3T-initial experience. Magn Reson Med 2007; 58: 1099–1106. [DOI] [PubMed] [Google Scholar]
- 23.Albers MJ, Bok R, Chen AP, Cunningham CH, Zierhut ML, Zhang VY et al. Hyperpolarized 13C lactate, pyruvate, and alanine: noninvasive biomarkers for prostate cancer detection and grading. Cancer Res 2008; 68: 8607–8615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hurd RE, Yen YF, Chen A & Ardenkjaer-Larsen JH. Hyperpolarized 13C metabolic imaging using dissolution dynamic nuclear polarization. J Magn Reson Imaging 2012; 36: 1314–1328. [DOI] [PubMed] [Google Scholar]
- 25.Benito J, Ramirez MS, Millward NZ, Velez J, Harutyunyan KG, Lu H et al. Hypoxia-Activated Prodrug TH-302 Targets Hypoxic Bone Marrow Niches in Preclinical Leukemia Models. Clin Cancer Res 2016; 22: 1687–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J, Chen L, Goyal B, Laidig G, Stanton T & Sjogren E Calithera Biosciences I. Heterocyclic inhibitors of glutaminase. United States Patent 2013 Dec 10, US8604016.
- 27.Daemen A, Liu B, Song K, Kwong M, Gao M, Hong R et al. Pan-Cancer Metabolic Signature Predicts Co-Dependency on Glutaminase and De Novo Glutathione Synthesis Linked to a High-Mesenchymal Cell State. Cell Metab 2018; 28: 383–399 e389. [DOI] [PubMed] [Google Scholar]
- 28.Jiang S, Yan W, Wang SE & Baltimore D Let-7 Suppresses B Cell Activation through Restricting the Availability of Necessary Nutrients. Cell Metab 2018; 27: 393–403 e394. [DOI] [PubMed] [Google Scholar]
- 29.Kim S, Kim DH, Jung WH & Koo JS. Expression of glutamine metabolism-related proteins according to molecular subtype of breast cancer. Endocr Relat Cancer 2013; 20: 339–348. [DOI] [PubMed] [Google Scholar]
- 30.Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y et al. HMDB 3.0--The Human Metabolome Database in 2013. Nucleic Acids Res 2013; 41: D801–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J, Ramirez MS, Walker CM, Chen Y, Yi S, Sandulache VC et al. High-throughput hyperpolarized (13)C metabolic investigations using a multi-channel acquisition system. J Magn Reson 2015; 260: 20–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song Z, Wei B, Lu C, Li P & Chen L Glutaminase sustains cell survival via the regulation of glycolysis and glutaminolysis in colorectal cancer. Oncol Lett 2017; 14: 3117–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walker CM, Lee J, Ramirez MS, Schellingerhout D, Millward S & Bankson JA. A catalyzing phantom for reproducible dynamic conversion of hyperpolarized [1-(1)(3)C]-pyruvate. PLoS One 2013; 8: e71274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masters C The turnover characteristics of lactate dehydrogenase. Curr Top Cell Regul 1982; 21: 205–259. [DOI] [PubMed] [Google Scholar]
- 35.Simpson RJ, Brindle KM, Brown FF, Campbell ID & Foxall DL. Studies of lactate dehydrogenase in the purified state and in intact erythrocytes. Biochem J 1982; 202: 581–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones DP. Redox potential of GSH/GSSG couple: assay and biological significance. Methods Enzymol 2002; 348: 93–112. [DOI] [PubMed] [Google Scholar]
- 37.Abu Aboud O, Habib SL, Trott J, Stewart B, Liang S, Chaudhari AJ et al. Glutamine Addiction in Kidney Cancer Suppresses Oxidative Stress and Can Be Exploited for Real-Time Imaging. Cancer Res 2017; 77: 6746–6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Biancur DE, Paulo JA, Malachowska B, Quiles Del Rey M, Sousa CM, Wang X et al. Compensatory metabolic networks in pancreatic cancers upon perturbation of glutamine metabolism. Nat Commun 2017; 8: 15965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dutta P, Le A, Vander Jagt DL, Tsukamoto T, Martinez GV, Dang CV et al. Evaluation of LDH-A and glutaminase inhibition in vivo by hyperpolarized 13C-pyruvate magnetic resonance spectroscopy of tumors. Cancer Res 2013; 73: 4190–4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miloushev VZ, Granlund KL, Boltyanskiy R, Lyashchenko SK, DeAngelis LM, Mellinghoff IK et al. Metabolic Imaging of the Human Brain with Hyperpolarized (13)C Pyruvate Demonstrates (13)C Lactate Production in Brain Tumor Patients. Cancer Res 2018; 78: 3755–3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.