Abstract
We generated genome-wide ancient DNA data from the Balearic Islands, Sicily, and Sardinia, increasing the number of individuals with reported data from 5 to 66. The oldest individual from the Balearic Islands (~2400 BCE) carried ancestry from Steppe pastoralists that likely derived from west-to-east migration from Iberia, while two later Balearic individuals had less. In Sicily, Steppe pastoralist ancestry arrived by ~2200 BCE in part from Iberia; Iranian-related ancestry arrived by the mid-second millennium BCE contemporary to its previously documented spread to the Aegean; and there was large-scale population replacement following the Bronze Age. In Sardinia, nearly all ancestry derived from the island’s early farmers until the first millennium BCE, with an exception of a third millennium BCE outlier who had primarily North African ancestry and who along with an approximately contemporary Iberian documents widespread Africa-to-Europe gene flow in the Chalcolithic. Major immigration into Sardinia began in the first millennium BCE and today no more than 56–62% of Sardinian ancestry is from its first farmers, which is lower than previous estimates highlighting how Sardinia—like every other region in Europe—has been a stage for major movement and mixtures of people.
Introduction
After around 3000 BCE, people with ancestry similar to that in the Steppe north of the Black and Caspian Seas began moving westward, mixing with local central European farmers to contribute up to three quarters of the ancestry of peoples associated with the Corded Ware complex1–3, and spreading in western Europe in association with the Bell Beaker complex4,5. In Iberia, Steppe pastoralist-related ancestry (hereafter denoted “Steppe ancestry”) appeared in outlier individuals by ~2500 BCE5, and became fully mixed into the Iberian population by ~2000 BCE6. On Crete in the eastern Mediterranean, there was little if any Steppe ancestry in all published individuals from the Middle to Late Bronze Age “Minoan” culture (dating to 2400–1700 BCE), although these individuals derived about 15% of their ancestry from groups related to early Iranian herders (“Iranian-related ancestry”)7. However, Steppe ancestry arrived in Crete and nearby Greece by the time of the “Minoan” culture ~2400–1700 BCE and “Mycenaean” culture ~1600–1200 BCE (Fig. 1).
Figure 1: Geographical origins and temporal distribution of newly reported data.
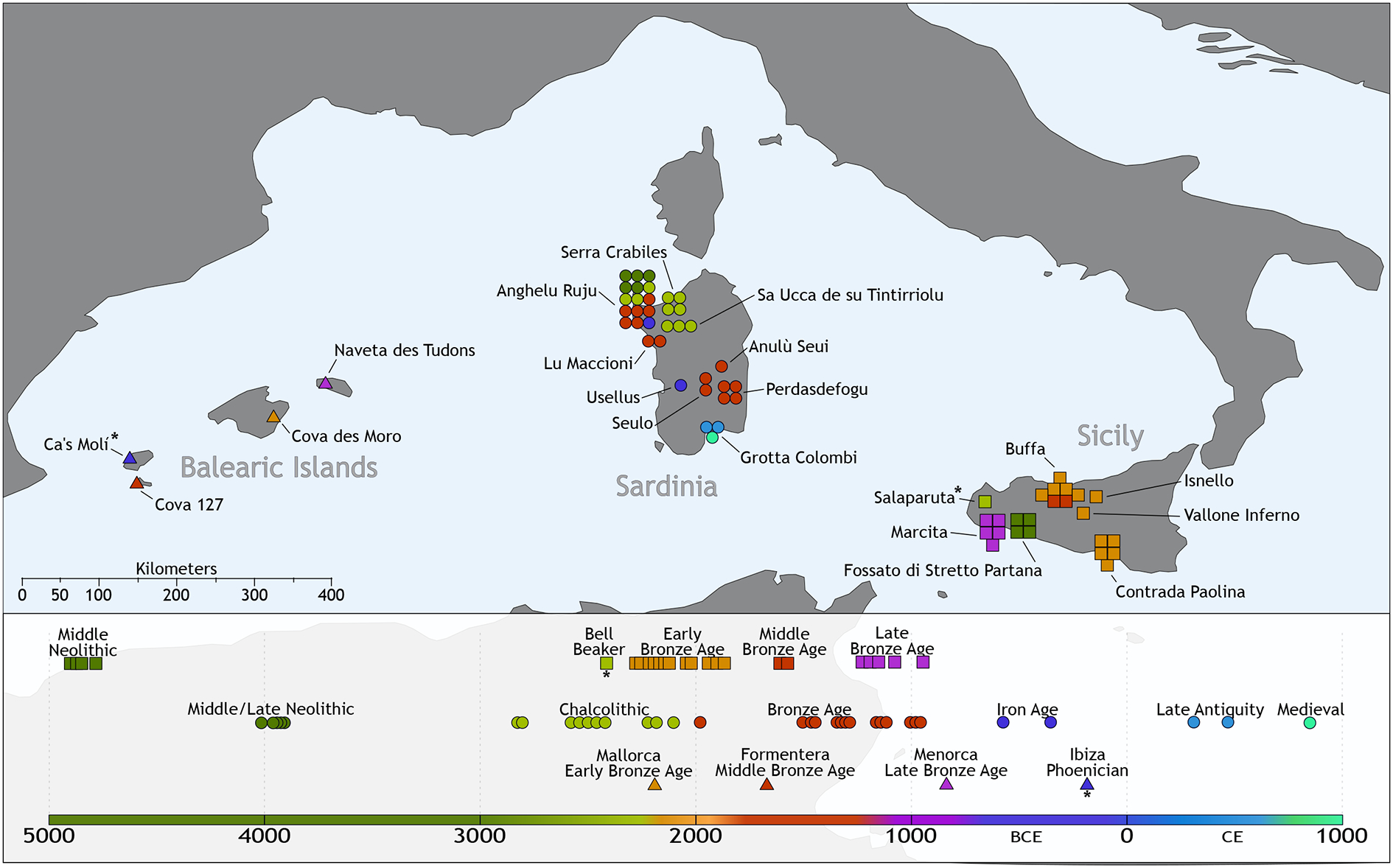
We present the 61 newly reported ancient individuals along with a previously reported Beaker-associated Sicilian individual for whom we increase data quality, and a reported Phoenician individual from Ibiza (both marked with an asterisk). Scale bar divided in 50 and 100 kilometer sections.
In the islands of the central and western Mediterranean, the Bronze Age transition has not been investigated with ancient DNA. The first permanent human presence in the Balearic Islands dates to ~2500–2300 BCE8,9. Around 1200 BCE, the Talaiotic culture was marked by intensified management of food resources and the appearance of monumental towers, the eponymous talaiots, some with suggestive similarities to the Sardinian nuraghi10,11,12. In turn, Bronze Age Nuragic Sardinian farmers also exchanged material goods with groups from the eastern Mediterranean.13 Sardinia and Sicily were affected by the spread of the Beaker complex after ~2500 BCE, while Sicily was affected by Aegean influences in the Mycenaean period14–16. An open question has been the extent to which these cultural exchanges were accompanied by movements of people, which we addressed by generating genome-wide ancient DNA data from 61 individuals.
Results
Data set
We prepared powder from petrous bones and teeth, extracted DNA17–20, and converted it into double-21 or single-stranded libraries21. We treated all libraries with Uracil-DNA Glycosylase (UDG)21 to greatly reduce the rate of cytosine-to-thymine errors characteristic of ancient DNA. We enriched for sequences overlapping approximately 1.24 million single nucleotide polymorphisms (SNPs)23,24, and after sequencing obtained genome-wide data from 61 individuals from the Balearic Islands, Sardinia, and Sicily while increasing data quality on a previously reported individual from Sicily (Fig. 1, Online Table 1, Supplementary Materials). We assembled direct radiocarbon dates on bone for 46 individuals (Online Table 2). We removed four individuals with evidence of contamination based on heterogeneity on the X chromosome (in males) or mitochondrial DNA, and one individual who was a first degree relative of another higher coverage individual25. For all but the by-sample analyses we removed individuals with data at <100,000 SNPs. For the 49 individuals that remained for analysis after this filtering, the median coverage on targeted SNPs on the autosomes was 2.91-fold (range 0.11–12.13), and the median rate of cytosine-to-thymine damage in the terminal nucleotides was 11.7% (range 3.7%−25.6%) in the range expected for authentic ancient DNA21 (Online Table 1). Genetic structure was similar when restricting to transversion SNPs not affected by characteristic ancient DNA errors (Supplementary Fig. 1).
Overview of genetic structure
We carried out principal component analysis (PCA) of the newly reported ancient individuals merged with previously published data1,2,4–7,26–62, projected onto variation among 737 diverse present-day west Eurasians analyzed at ~600,000 SNPs7,40,63–65 (Fig. 2b, Online Table 3). We also performed unsupervised clustering with ADMIXTURE66 (Fig. 2a). We used qpWave2 to evaluate whether each individual in turn was consistent with being from the same group as others from the same time period and region (that is, we tested if they were consistent with forming a clade at p<0.01), allowing us to create analysis groupings (Fig. 3, Supplementary Fig. 4). Where qpWave was ambiguous, we carried out refined tests to determine groupings (Supplementary Materials).
Figure 2: Overview of genetic strucfture.

We show results for ancient Sardinians, Sicilians and Balearic islanders and other ancient and present-day populations according to a) unsupervised ADMIXTURE with K=10 clusters; and b) PCA with previously published data (non-filled symbols), projected onto variation from present-day populations shown in solid-color circles without outlines (maroon=Balearic, orange=Sicilians, blue=Sardinians, gray=all others).
Figure 3: Pairwise qpWave testing to group individuals.

Black lines represent groupings by location and period. Gray-colored models have a P-value below 0.01 and are rejected.
In the Balearic Islands, the three Bronze Age individuals fall between the European Neolithic and Bronze Age clusters in PCA; their evidence of Steppe ancestry is also evident in an ADMIXTURE component maximized in Yamnaya Steppe pastoralists. qpWave revealed significant ancestry differences between the Early Bronze Age individual Mallorca_EBA and the Middle and Late Bronze Age individuals, Formentera_MBA (p=0.001) and Menorca_LBA (p=0.001) (Supplementary Table 1). Taking into account the partial evidence of genetic heterogeneity and the very different archaeological contexts of the three individuals, we treated each separately for analysis.
In Sicily, the four Middle Neolithic individuals cluster with early European farmers and are genetically homogeneous so we group them as Sicily_MN (Fig. 2, Supplementary Figs. 1 & 3). Relative to Sicily_MN, all the Early Bronze Age Sicilians deviate in PCA and ADMIXTURE toward eastern groups with two Early Bronze Age outliers Sicily_EBA11443 and Sicily_EBA8561 clearly having Steppe pastoralist admixture, and the remaining individuals dividing into a main Sicily_EBA grouping of four individuals and two subtly differentiated individuals Sicily_EBA3123 (p=0.005) and Sicily_EBA3124 (p=0.001) (Supplementary Tables 2 and 3). The two Middle Bronze Age individuals were consistent with being a clade and differentiated from the Early Bronze Age individuals so we grouped them as Sicily_MBA (Supplementary Table 4). All five Late Bronze Age individuals were consistent with being a clade at p>0.01 (Sicily_LBA) (Supplementary Table 5).
In Sardinia we observe clustering with mainland European Early and Middle Neolithic farmers from the beginning of the Neolithic to the end of the Bronze Age; all but two individuals are indistinguishable in their ancestry components based on qpWave (Fig. 2). The three Neolithic individuals were homogeneous and so we grouped them (Sardinia_Neolithic) (Supplementary Table 6); all but one of the Chalcolithic individuals were homogeneous (Sardinia_Chalcolithic) (Supplementary Tables 7 and 8); and all but one of the Nuragic Bronze Age individuals were homogeneous (Supplementary Tables 9 and 10). The two outliers were Sardinia_Chalcolithic15940 (radiocarbon dated to 2345–2146 calBCE) who had significant affinity to Levantine and North African Neolithic individuals (p<10−12), and Sardinia_BA10365 (1643–1263 calBCE) who had subtle evidence of eastern Mediterranean ancestry and was significantly differentiated from others of this period (p=0.000024 (Supplementary Tables 9 and 10). The two Iron Age individuals did not form a clade (Supplementary Table 11): Sardinia_IA10366 (391–209 calBCE) has evidence of Iranian-related ancestry, whereas Sardinia_IAI16163 (762–434 calBCE) has evidence of Steppe ancestry (Fig. 3). The two Sardinia_LateAntiquity individuals were consistent with forming a clade with each other and Sardinia_EarlyMedieval (Fig. 3, Supplementary Tables 12 and 13), but Sardinia_EarlyMedieval separated from the earlier individuals in PCA (Fig. 2) and dated to significantly later, so we analyzed this individual separately.
Balearic Islands
We used qpAdm2,63 to decompose the ancestry of each analysis grouping into five “distal” sources dating to the Late Neolithic or earlier: Anatolia_Neolithic, Western Hunter-Gatherers (WHG), Iran_Ganj_Dareh_Neolithic, Yamnaya_Samara, and Morocco_LN. We represented each of these sources with published ancient DNA data. We caution that finding a fitting model in qpAdm should not be interpreted as implying that the true source population came from the same location. Instead, a fit implies that the modeled population is consistent with being a mixture of groups that are derived from the same ancestral populations as the groups we use as surrogates for them. We tested all possible 2, 3, 4 and 5-way mixture models, and when more than one model of the same rank produced valid fits we included the competing source among the set of outgroups. This “model competition” approach40,62 is designed to detect shared genetic drift between any test population and an outgroup that is not captured by one of the sources used in the model; this sometimes reveals that we are failing to correctly model a proximal ancestry source. We quote the most parsimonious model (as measured by the lowest number of ancestry sources) that fits at p>0.05, and when multiple models fit quote all of them (Fig. 4b and Supplementary Tables 14 and 16).
Figure 4: Distal modeling of ancestry proportions using qpAdm.

We report results a) by-individual, and b) analysis grouping (see Supplementary Tables 14 and 16 for actual numbers), with the P-values shown within the bars. We show all valid models (p>0.05) for the lowest fitting number of sources (some instances produced two valid models at p>0.05). In panel b) we add a published individual (Ibiza_Phoenician) and modern groups (Balearic, Sardinian, Sicilian). Asterisks denote models using Morocco_EN instead of Morocco_LN to improve fits (Supplementary Materials).
Mallorca_EBA dates to the earliest period of permanent occupation of the islands at ~2400 BCE10,68. We modeled 38.1 ± 4.4% of her ancestry as deriving from a source related to Yamnaya_Samara (Fig. 4, Supplementary Table 14). We next used qpAdm to identify “proximal” sources for Mallorca_EBA’s ancestry. Using our “model competition” approach, we tested a list of potential sources motivated by being close in space or time or showing cultural links, including all of them among the outgroups if they were not explicitly tested as sources. Mallorca_EBA can be modeled as a clade with the (small) subset of Iberian Beaker complex associated individuals who carried Steppe-derived ancestry5 (p=0.413). We reject models that do not involve an Iberian source, as we find no passing models when we use qpAdm to test non-Iberian sources for Mallorca_EBA with Iberians including among the outgroups. Thus, the movements of people that brought Steppe ancestry into Iberia overlapped those that settled the Balearic Islands. We caution that we only have data from a single individual from this period and her ancestry profile may not be representative of all the early settlers. Future ancient DNA sampling could reveal individuals with different proportions of Steppe ancestry and without a specific Iberian connection.
Our estimates of Steppe ancestry in the two later Balearic Islands individuals are lower at 20.4 ± 3.4% for Formentera_MBA and 20.8 ± 3.6% for Menorca_LBA (Supplementary Table 14) (Fig. 3, Supplementary Fig. 4, Supplementary Table 1). These two individuals thus represent mixtures of groups with a relatively large proportion of Steppe ancestry, plausibly the population of which Mallorca_EBA was a part, and other groups with more early European farmer-related ancestry. This could be the result of early immigrants to the islands harboring different proportions of Steppe ancestry and then mixing, or later waves of gene flow from groups with relatively more European first farmer-related ancestry. The proximal modeling of Formentera_MBA produced 2-way fits with one source always bearing Steppe ancestry (such as Mallorca_EBA, Iberia_Bell_Beaker_highSteppe, or France_Bell_Beaker), and the other always bearing large proportions of Anatolian Neolithic-related ancestry (fitting models include sources in mainland Italy, France, or Sardinia) (Supplementary Table 17). Menorca_LBA fit a 1-way model with Formentera_MBA (p=0.172), and thus may be directly continuous with that population (Supplementary Materials).
To test for evidence for genetic links between peoples of the Talaiotic culture of the Balearic Islands and the Nuragic culture of Sardinia, we examined the fitting 2-way admixture models for the Talaiotic culture-associated individual Menorca_LBA. While some of the models involved a Sardinian population as a fitting source, we also found fits for models that included as sources France_Bell_Beaker_lowSteppe, Italy_Bell_Beaker, Italy_Remedello.SG, or Italy_MN_Iceman.SG with Nuragic Sardinians included among the outgroups (Supplementary Table 18). Thus, our analysis does not produce any specific evidence of a genetic link between people of these two cultures. A caveat is that we only have access to data from a single Talaiotic-associated individual; it is also important to remember that there can be cultural contact without movement of people.
Phoenician colonies were established in the Balearic Islands in the Iron Age. The Ibiza individual published in59 from a collective burial in a Punic hypogeum and dated to 361–178 calBCE is not consistent with forming a clade with any of the Bronze Age Balearic individuals and has a qualitatively different ancestry profile; for example, a North African source of ancestry is required to obtain a fit (our model is 10.8 ± 2.7% Iran_Ganj_Dareh_Neolithic and 89.2 ± 2.7% Morocco_LN ancestry (Morocco Late Neolithic)) (Fig. 4, Supplementary Table 14). In proximal modeling, Ibiza_Phoenician also always requires Morocco_LN as one of the sources. While some of these models include a Balearic Island Bronze Age source, it is possible that the Ibiza Phoenician individual has no ancestry at all from earlier Balearic peoples, as we fit her with models that have all the Balearic Bronze Age individuals among the outgroups (e.g. 17.0 ± 3.1% France_Bell_Beaker and 83.0 ± 3.1% Morocco_LN) (Supplementary Table 19).
Modern Balearic individuals can only be fit with 5 distal sources, as a mixture of Steppe, Iranian-related, and North African-related ancestry in addition to Anatolian farmer-related and WHG. Thus, the Balearic Islands today reflect a mixture of disparate ancestry sources some of which are likely to reflect movements in the past from the southern and eastern Mediterranean (Fig. 4, Supplementary Table 14). Among the uniparental haplogroups found in present day Balearic populations are all the ones found in the ancient individuals (mitochondrial DNA J2, H, U5, and Y chromosome R1b)69,70, although none of these are unique to the ancient Balearic populations and so cannot be taken as a proof of local population continuity (Online Table 1).
Sicily
In the Middle Neolithic, Sicilians harbored ancestry typical of early European farmers, which we can fit as a mixture of Anatolia_Neolithic and WHG (Fig. 2, Fig. 4, Supplementary Table 14). We also approximately tripled coverage on a previously reported Beaker complex-associated individual, and our reanalysis of these data confirmed the previous finding of no evidence of Steppe ancestry.5
In the Early Bronze Age, we find evidence of Steppe ancestry by ~2200 BCE. In distal qpAdm, the two outliers with the strongest evidence have 22.1 ± 3.6% Steppe ancestry (Sicily_EBA8561) and 39.0 ± 3.5% Steppe ancestry (Sicily_EBA11443); the latter individual is consistent with forming a clade with Mallorca_EBA (p=0.245) which suggests that they may harbor ancestry from a similar source (most plausibly Iberian, see below) (Fig. 4a, Supplementary Table 14). For the main Early Bronze Age cluster of four individuals and two other outliers, we also fit Steppe ancestry albeit at lower proportions of 14.1 ± 3.4% in Sicily_EBA3123, 13.5 ± 3.4% in Sicily_EBA3124, and 9.9 ± 2.2% in the main cluster of Sicily_EBA (Fig. 4, Supplementary Table 14, Supplementary Materials). For Sicily_EBA and Sicily_EBA3123 we could not rule out an alternative model of Iranian-related ancestry rather than Steppe as a third ancestry source, although we favor Steppe models because of the results from the proximal modeling and the definitive presence of this ancestry in the two extreme outlier individuals. The presence of Steppe ancestry in Early Bronze Age Sicily is also evident in Y chromosome analysis, which reveals that 3 of the 5 Early Bronze Age males carried haplogroup R1b1a1a2a1a2 (R1b-M269) associated with the first western Europeans who carried significant proportions of Steppe ancestry (Online Table 1). Two of these four individuals carried the Y haplogroup subtype R1b1a1a2a1a2a1 (Z195), which today is largely restricted to Iberia and has been hypothesized to have originated there 2500–2000 BCE71. A parsimonious scenario is that west-to-east gene flow from Iberia introduced these haplogroups into Sicily as well as to the Balearic Islands where Y haplogroup R1b-M269 is also found in Menorca_LBA.
We detect Iranian-related ancestry in Sicily by the Middle Bronze Age 1800–1500 BCE, consistent with the directional shift of these individuals toward Minoans and Mycenaeans in PCA (Fig. 2b); in distal modeling, Sicily_MBA requires 15.7 ± 2.6% of Iran_Ganj_Dareh_Neolithic-related ancestry (p=0.060) (Fig. 4, Supplementary Table 14). Sources closer in time always require Minoan_Lassithi or Anatolia_EBA as a source (Supplementary Table 21). Modern southern Italians harbor Iranian-related ancestry,74 and our results show this ancestry must have reached Sicily before the period of Greek political control when Sicily and southern Italy were part of Magna Graecia.
We can model Sicily_LBA as 81.5 ± 1.6% Anatolia_Neolithic, 5.9 ± 1.6% WHG, and 12.7 ± 2.1% Yamnaya_Samara (Fig. 4b, Supplementary Table 14). Although this distal modeling provides no hint of Iranian-related ancestry, modeling with sources closer in time supports Sicily_LBA having such ancestry via groups like Anatolia_EBA or Minoan_Lassithi (Supplementary Table 22).
Our distal modeling of modern Sicilians requires not only the two eastern ancestry sources that we have shown were present by the Bronze Age—10.0 ± 2.6% Yamnaya_Samara and 19.9 ± 1.4% Iran_Ganj_Dareh_Neolithic— but also a predominant component of North African ancestry (46.9 ± 5.6% Morocco_LN) (Fig. 4, Supplementary Table 14). These results are consistent with most of the North African-related ancestry having come into Sicily in the Iron Age and afterward, a scenario that is further supported by our observation that modern Sicilians form a clade with Ibiza_Phoenician (p=0.060) and the three most recent Sardinian individuals in our time series (Supplementary Materials). Although these results are consistent in principle with a nearly complete ancestry turnover on the island since the Bronze Age, we cannot rule out the possibility that Bronze Age Sicilians made a more modest ancestry ancestry contribution to modern Sicilians. Uniparental markers in modern Sicilians overlap those from the Bronze Age with Y haplogroup R1b-M269 occurring at high frequency (~25%72) and mitochondrial haplogroups H, T, U and K also being present in the Bronze Age as well as in present-day groups.73
Sardinia
In qpAdm, almost all Neolithic, Chalcolithic, and Bronze Age individuals fit as descending from the same two deep ancestral sources (Anatolia_Neolithic and WHG) with similar and indistinguishable mixture proportion estimates of 82.9% - 86.0% (Fig. 4, Supplementary Table 14). Sardinia_Neolithic and Sardinia_Chalcolithic and consistent with being a clade (p=0.285), as are Sardinia_Chalcolithic and Sardinia_BA (p=0.173), further supporting a high degree of continuity (Supplementary Materials). When we consider sources closer in time and space, Neolithic Sardinians can be modeled as a mixture of 74–77% France_MN, with the rest 26% from Hungary_EN (p=0.051) or 23% from Croatia_Cardial_Impressa_EN (p=0.051) (Supplementary Materials). The true source for early Sardinian farmers is likely an unsampled group, potentially from an unsampled region such as northern Italy or Corsica where there is little or no available ancient DNA data. We detect no evidence of Steppe ancestry detected even in individuals buried in a Beaker context, which is similar to the pattern observed in Beaker-associated burials in Sicily and most in Iberia, a pattern that contrasts sharply with that in central and northern Europe where Beaker-associated individuals carried significant Steppe ancestry5 (Supplementary Table 16, Online Table 1).
Despite evidence of extraordinary ancestry continuity in Sardinia from the Neolithic through the Bronze Age, there are two notable outliers.
The most surprising is Sicily_Chalcolithic15940 from the site of Anghelu Ruju, for whom we obtained a radiocarbon of 2345–2146 calBCE on the same bone we analyzed for DNA. We model this individual as 22.7 ± 2.4% Anatolia_Neolithic and 77.3 ± 2.4% Morocco_EN (p=0.321). This individual is similar in ancestry composition to the approximately contemporary Iberian individual I4246 from the site of Camino de las Yeseras radiocarbon dated to 2473–2030 calBCE, who also had North African-related ancestry as well as the same mitochondrial DNA haplogroup M1a1b1 and Y haplogroup E1b1b1 which are both typical of North Africans25 (Supplementary Table 14). The finding of African-to-European gene flow in both individuals shows that such movement was widespread across the Mediterranean long before the classical period when such gene flows became intensive and the ancestries made a larger demographic impact.
The second outlier is Sardinia_BA10365 (1643–1263 calBCE) (Fig. 2b), which we fit as a mixture of a local Sardinian source and a second source carrying eastern Mediterranean-related (e.g Mycenaean or Jordan_EBA) or Steppe ancestry (Italy_Bell_Beaker or France_Bell_Beaker) (Supplementary Table 23). Uniparental marker analyses provide additional hints that the material culture exchange between Sardinians and eastern Mediterranean was accompanied by some movement of people67. Sardinia_Chalcolithic15943 from Anghelu Ruju carried a rare U1a mitochondrial haplogroup, known from ancient individuals from the Early Bronze Age Balkans and Western Asia51,54,62 (Online Table 1). Sardinia_BA10553 carried Y haplogroup J2b2a (Online Table 1), which today occurs at highest frequencies in the Balkans and the Middle East75, and which was nearly unique to these regions in the Bronze Age and earlier40,76,44,54.
The earliest definitive evidence in Sardinia of Steppe and Iranian-related ancestries comes from two Iron Age individuals, Sardinia_IA16163 (762–434 calBCE) with 22.5 ± 3.6% Yamnaya-related ancestry, and Sardinia_IA10366 (391–209 calBCE) with 12.7 ± 3.5.% Iran_Ganj_Dareh_Neolithic related ancestry (Fig. 4, Supplementary Table 14, Supplementary Table 24, Supplementary Materials). The qpAdm models fit even when including Bronze Age Sardinians in the outgroups, consistent with the hypothesis that these individuals, like the Phoenician from Ibiza in the Balearic Islands, had little ancestry from preceding local peoples. Iranian-related ancestry was even higher in at least some individuals in Late Antiquity, with the Sardinia_LateAntiquity cluster harboring 29.3 ± 4.1.% Iran_Ganj_Dareh_Neolithic-related ancestry (p=0.009 for rejection of the alternative model that attempts to model ancestry as derived from the Yamnaya, Supplementary Table 14). Sardinia_LateAntiquity is consistent with being a clade with Ibiza_Phoenician (p=0.238) as would be expected if this ancestry began to be introduced with the Phoenicians77 (Supplementary Table 25). Although we do not model Sardinia_EarlyMedieval due to his limited SNP coverage, his Y haplogroup, E1b1b1b2, belongs to the same branch (E1b1b) as Sardinia_Chalcolithic15940 and Hellenistic period Egyptians48, consistent with a source in the eastern Mediterranean. Taken together, these results show that the five most recent Sardinians in our time series—from the Iron Age (n=2), Late Antiquity (n=2), and Early Medieval period (n=1)—harbored minimal ancestry from Bronze Age, Chalcolithic, or Neolithic Sardinians. All five individuals were from coastal sites, suggesting immigration from groups outside of Sardinia. Unsampled regions of Sardinia (possibly on the coast and almost certainly in the interior) likely retained high proportions of ancestry from pre-Iron Age Sardinians in the Iron Age and later, as pre-Iron Age Sardinians remain the single largest contributor to modern Sardinians (see below).
We are only able to model modern Sardinians by invoking 4- and 5-way distal admixture models, which include Iranian-related and North African-related sources and thus are substantially more complex than the 3-way model for Sardinia proposed in the original paper that introduced the qpAdm method2 (that study was not able to reject a 3-way admixture model because it did not have access to our reference dataset of large numbers of ancient West Eurasians). With our more refined modeling, the proportion of ancestry from the first farmers of Sardinia (comprised of sources from Anatolian farmers and western hunter-gatherers)2 is around 72.2% (= 62.5% Anatolian Neolithic + 9.7% WHG) (Fig. 4, Supplementary Table 14). This is much less than the first qpAdm study of ancestry proportions in Sardinians estimated that 87% of the island’s ancestry could derive from Neolithic farmers. It is also much less than the ~100% that was inferred in a study that found modern Sardinians to be a clade with ancient DNA from the Late Neolithic Tyrolean Iceman78. It is finally much less than the proportions of 90–100% inferred were estimated in a study of more than 3500 modern Sardinians (with the estimates varying across geographic regions)79.
In fact, the true degree of population turnover was even larger. To test how much ancestry in modern Sardinians could be derived from local Sardinians from the Bronze Age and earlier, we examined the 27 modern Sardinians that have been the source of many of these previous estimates63, who are from the Gennargentu region and have among the least Steppe ancestry among modern Sardinians who vary in their ancestry proportions reflecting the geographic substructuring of modern Sardinians among different valleys and coastal and inland sites79. These modern Sardinians fit a 4-way distal model with a local Sardinian ancestry source and three further sources, Iran_Ganj_Dareh_Neolithic, Yamnaya_Samara, and Morocco_LN (Supplementary Table 26). We estimate that modern Sardinians retained between 56.3 ± 8.1% (when using Sardinia_Neolithic) to 62.2 ± 6.6% (with Sardinia_BA) of ancestry from local populations, along with a substantial proportion of North African Morocco_LN-related ancestry that ranges between 22.7 ± 9.9% (when using Sardinia_Neolithic) and 17.1 ± 8.0% (when using Sardinia_BA). This North African-related mixture is a plausible source for the sub-Saharan African admixture that has been detected in modern people from the island in multiple previous studies79–82. Even this range of estimates of ~56–62% is an upper bound, as we are forcing in pre-Iron Age Sardinians as the only source of European farmer ancestry on the island in all of these models.
Taken together, these results confirm that the proportion of ancestry derived from first farmers is higher in Sardinia than elsewhere in Europe, while revealing a previously unknown major contribution from groups that arrived later. The ancestry from multiple sources is also evident in uniparental markers; for example, modern Sardinians carry haplogroups common on the island from the Neolithic through to the Bronze Age period (Y haplogroup R1b1a[xR1b1a1a] and mtDNA haplogroups HV, JT, and U83), as well as a high-frequency of Y haplogroup R1b-M269 that was absent in the Bronze Age and earlier79,84. An ancient DNA study of Sardinia that appears alongside this one concurs that modern Sardinians harbor large proportions of ancestry from early Sardinian farmers as well as significant contributions from later periods that are larger than previously estimated. That second study also agrees with our finding that many coastal Sardinian sites harbored people with little ancestry from earlier local Sardinians in the Iron Age and antiquity85.
Discussion
We conclude by highlighting five observations.
First, we identify Iberia as a key ancestry source for Bronze Age peoples of both the Balearic islands and Sicily. Some early residents of the Balearic Islands likely derived at least part of their ancestry from Iberia as proximal models favor ancient Iberian sources. We also observe the characteristically Iberian Y chromosome haplogroup R1b1a1a2a1a2a1 (Z195) in two Early Bronze Age Sicilians71. Thus, Iberia was not just a destination of east-to-west human movement, but also an important source for west-to-east reflux86. However the demographic history of Iberia in this period was also distinctive from the Mediterranean islands25; for example, no nearly complete replacement of paternal lineages occurred in Sicily, where both Neolithic and Steppe-associated haplogroups persisted.
Second, our analysis shows that Iranian-related ancestry, which was widespread in the Aegean by the Middle Bronze Age in association with the Minoan and Mycenaean cultures, had also spread as west as Sicily in substantial proportion at least by the time of the Mycenaeans. One possibility is this ancestry spread west along with the Mycenaean cultural expansion87–90. However, the presence of artifacts associated with the Sicilian Castellucian culture in Malta, Greece, and Anatolia before the peak of the Mycenaean culture leaves open the possibility of earlier gene flows, a hypothesis that could be investigated once pre-Minoan data from the Aegean become available87,91. We find no evidence of substantial proportions of Iranian-related ancestry in the Balearic islands or Sardinia until the Phoenician period, but this does not mean that these islands were isolated from the eastern Mediterranean, and indeed the opposite is true. Archaeological evidence shows that this was a period of unprecedented material culture exchange, with substantial westward flows of goods as reflected for example in the importation of Cypriot copper in Late Bronze Age Sardinia92.
Third, our study highlights widespread human mobility from North Africa to Europe in the Chalcolithic and Bronze Age. Specifically, we identify an outlier individual in Sardinia with a large proportion of North African-derived ancestry who dates to 2345–2146 calBCE and has an ancestry profile similar to an approximately contemporary central Iberian individual dated to 2473–2030 calBCE25 (and a Bronze Age individual from Iberia dated to 1932–1697 calBCE who carried North African-related ancestry in admixed form25). Altogether, 1.6% of the 191 individuals from Mediterranean Europe in our analysis dataset from between 5000–3000 years ago have evidence of ancestry from North African migrants in the few generations before they lived61.
Fourth, our analysis documents the impact of the Phoenician and Greek colonial periods as well as subsequent immigration on the islands of the western Mediterranean.93 For example, all six individuals in our analysis dataset from the Balearic islands and post-Bronze Age period have no evidence of ancestry from earlier local groups. In conjunction with previous findings, the emerging picture is that from the Iron Age onward, the coastal regions of the western Mediterranean were characterized by ethnically segregated populations of immigrants and local groups that co-existed in geographic proximity; indeed, there is direct documentation of this from the Greek colony of Empúries in northeast Iberia, where two clusters of genetically distinct individuals co-existed consistent with the historical descriptions by Strabo.25 In some regions such as the Balearic islands and Sicily our data are consistent a nearly complete replacement of the pre-Iron Age populations (although we cannot rule out a degree of local continuity in either set of islands).
Fifth and finally, our co-analysis of modern and ancient Sardinians questions the common view that Sardinians are well described as an isolated descendants of Europe’s first farmers78. Our ancient DNA time transect does reveals that Neolithic, Chalcolithic, and Bronze Age Sardinians had a typical early European farmer ancestry profile that persisted longer on the island than anywhere else in Europe studied to date, and in this sense our study supports previous findings that Sardinia is special in Europe in retaining more of its first farmer ancestry. However, immigrants from the eastern Mediterranean and North Africa made a substantial demographic impact on Sardinia from the Iron Age onward just as they did on other parts of the coastal western Mediterranean. Thus, all five of the post-Bronze Age individuals from Sardinia in our time series (all from coastal sites) have no evidence of pre-Bronze Age Sardinian ancestry, and they clearly mixed with the previously established populations to contribute at least ~38–44% of the ancestry of modern Sardinians (with North African-derived ancestry estimated at ~17–23%). Thus, rather than being fully sheltered from admixture and migration since the Neolithic, Sardinia, like almost all other regions in Europe, has been a major stage for movement and mixture of peoples.
Materials and Methods
Laboratory work and bioinformatic analysis
We extracted powder from skeletal samples in dedicated ancient DNA facilities at the University of Vienna in Austria, University College Dublin in Ireland, the University of Florence in Italy, the University of Palermo in Italy, and Harvard Medical School in Boston USA (Online Table 1)17,19,20,94,95. We treated all but one DNA extract (S4420.E1.L1) with Uracil-DNA Glycosylase (UDG) to remove characteristic ancient DNA damage, thereby greatly reducing the rate of damage-induced errors96. For all but one sample, we performed DNA extraction at Harvard Medical School, sometimes using silica coated magnetic beads to support robotic cleanups (instead of silica column cleanups that were used for manual DNA extraction)17,19,20. We converted these DNA extracts to individually double-barcoded (or double-indexed) libraries either using double-strand ligation21 or single-strand ligation22, in most cases assisted by a robotic liquid handler (see Online Table 1 for details). For one of the samples, we performed DNA extraction17,18 and double-indexed library preparation using double-stranded ligation in Florence21.
We initially screened some libraries by enriching them for the human mitochondrial genome97 and about 3000 nuclear SNPs (Online Table 1, mtDNA+3000SNPs) using synthesized baits (CustomArray Inc.). We sequenced on an Illumina NextSeq500 instrument with 2×76 cycles and read the indices with 2×7 cycles; for some libraries, we sequenced on a HiSeq X10 with 2×101 cycles and read the indices with 2×8 cycles. We assigned sequences based on the library-specific barcodes/indices. We used one of two bioinformatics pipelines to process the data, as specified in Online Table 1. In “Pipeline 1,” we merged read pairs that overlapped by at least 15 base pairs allowing up to one mismatch using SeqPrep (https://github.com/jstjohn/SeqPrep) (representing each overlapping base by the higher quality base), whereas for “Pipeline 2” we allowed one mismatch when the forward and reverse base had quality ≥20, or 3 mismatches when quality was <20 (always retaining the base with higher quality, but when mismatches were found base quality was defined as the difference of base qualities). We used either SeqPrep (Pipeline 1) or a custom script (https://github.com/DReichLab/ADNA-Tools) (Pipeline 2) to computationally trim adapters and barcodes. We mapped the merged sequences to the reconstructed human mitochondrial DNA consensus sequence98 using bwa (Pipeline 1: v.0.6.1, Pipeline 2: v.0.7.15-r1140)99 with the parameters –n 0.01 and –l 16500 (Pipeline 1) or -n 0.01, -o 2 and -l 16500 (Pipeline 2). We removed duplicate sequences that had the same orientation and same start and stop positions using a custom script (Pipeline 1) or the Broad Institute’s Picard MarkDuplicates tool (http://broadinstitute.github.io/picard/). We restricted to sequences of at least 30 base pairs with a mapping quality≥30 and to bases with base quality≥30. We assessed the data for authenticity by computing the damage rate at the terminal cytosines21, and by estimating the rate of mismatches to the consensus mitochondrial sequence using contamMix23 (v.1.0–10 for Pipeline 1 and v.1.0–12 for Pipeline 2). We determined mitochondrial haplogroups using HaploGrep2100 (Online Table 4).
To evaluate whether differences between the two bioinformatics pipelines are introducing artifactual into our analysis—for example by causing sequences processed using Pipeline 1 to match each other at a higher rate than those sequenced by Pipeline 2—we carried out two sets of tests. For the first set of tests, we separated the Sardinia_BA individuals by processing pipeline (3 from Pipeline 1, and 10 from Pipeline 2). We computed symmetry f4-statistics of the form f4(Mbuti.DG, Test, Sardinia_BA_Pipeline1, Sardinia_BA_Pipeline2) using as Test 12 groups including modern Sardinian, Mycenaean, Iberia_BA.SG, Menorca_LBA (Online Table 5). None of the tests produced a significant deviation from zero (the largest had a |Z|=1.126 deviation from zero, showing that at least for Sardinia_BA, the processing of samples with different pipelines does not affect results.
For our second set of tests, we analyzed data for 20 previously published individuals25,62 whose sequencing data we processed with both pipelines. To test for artifactual attraction of samples processed by Pipeline 1 and samples processed by Pipeline 2, we computed “ABBA” and “BABA” counts and associated P-values for differences in the rates of these counts for the set of samples (individual1_pipeline1, individual1_pipeline2; individual2_pipeline1, individual2_pipeline2). For example, a BABA count corresponds to a case where the sequences match for the two individuals processed using Pipeline 1, and mismatch the sequences at the same position for the two individuals processed using Pipeline 2 (a BABA count is the direction expected for a bias, and hence we expect to observe an excess of BABA over ABBA counts if there is a bias). We computed ABBA and BABA counts for all 190 possible comparisons, and assessed statistical significance based on a one-sided binomial test for an excess of BABA over ABBA counts (assuming conservatively that the counts are independent). Only one test produced a p-value below 0.05 (p=0.022), which is not significant after Bonferroni correction for the 89 pairwise tests where we had sufficient coverage for the pair of samples to have power in principle detect a signal at the p<0.05 level (that is, ABBA+BABA was at least 5). The results are shown in the newly added Online Table 6.
We enriched the screened libraries with promising quality for 1,233,013 targeted SNPs (“1240K” target capture)2,24 and newer libraries for the mitochondrial genome together with the 1240k targets (“1240k+” target capture). We sequenced and processed the data as for the mitochondrial DNA with the difference that we mapped to the human reference genome hg19. We estimated contamination based on the ratio of Y to X chromosome sequences (confirming that all individuals in the dataset had a ratio consistent with a male or a female) as well as the rate of heterozygosity at X chromosome positions (only valid as an estimate of contamination in males who should have no X chromosome variation101). For Pipeline 1 we determined each SNP in each individual by a randomly sampled sequence covering the nucleotide with mapping quality ≥30 and base quality ≥30, whereas for Pipeline 2 we used mapping quality ≥10 and base quality ≥20. We restricted analysis to individuals with coverage of at least 20,000 SNPs (when analyzing populations represented by a single person we required a coverage of at least 100,000 SNPs).
Radiocarbon dating and quality control
We obtained 43 accelerator mass spectrometry (AMS) radiocarbon dates (14C) at the Pennsylvania State University (PSU) Radiocarbon Laboratory, as well as an additional 3 direct dates from other laboratories (the dates were from 48 distinct samples as we dated two samples twice). Here we provide a description of sample processing at PSU, as it is the source of most of our dates (the sample description is adapted from the standard description of this processing; for the other samples, we refer readers to the published protocols). As a precaution at PSU, we removed possible contaminants (conservants/adhesives) by sonicating all bone samples in successive washes of ACS grade methanol, acetone, and dichloromethane for 30 minutes each at room temperature, followed by three washes in Nanopure water to rinse. We extracted bone collagen and purified using a modified Longin method with ultrafiltration (>30kDa gelatin102). If collagen yields were low and amino acids poorly preserved we used a modified XAD process (XAD Amino Acids103). For quality assurance, we measured carbon and nitrogen concentrations and C/N ratios of all extracted and purified collagen/amino acid samples with a Costech elemental analyzer (ECS 4010). We evaluated sample quality by % crude gelatin yield, %C, %N and C/N ratios before AMS 14C dating. C/N ratios for all directly radiocarbon samples fell between 3.1 and 3.3, indicating excellent preservation104. We combusted collagen/amino acid samples (~2.1 mg) for 3 h at 900°C in vacuum-sealed quartz tubes with CuO and Ag wires. Sample CO2 was reduced to graphite at 550°C using H2 and a Fe catalyst, and we drew off reaction water with Mg(ClO4)2105. We pressed graphite samples into targets in Al boats and loaded them onto a target wheel and made all 14C measurements on a modified National Electronics Corporation compact spectrometer with a 0.5 MV accelerator (NEC 1.5SDH-1). We corrected the 14C ages for mass-dependent fractionation with measured δ13C values106 and compared with samples of Pleistocene whale bone (backgrounds, >48,000 14C BP), late Holocene bison bone (~1,850 14C BP), late 1800s CE cow bone, and OX-2 oxalic acid standards. We calibrated 14C ages with OxCal version 4.3107 and the IntCal13 northern hemisphere curve108. The stable carbon and nitrogen isotope measurements we obtained do not indicate a large marine dietary component in these individuals despite their coming from island populations and hence we did not perform a correction of the dates for marine reservoir effect.
Uniparental haplogroup determination
We determined mitochondrial haplogroups using HaploGrep2100 and phylotree109 (build 17) on the data from the mitochondrial enrichment experiment105 (Online Table 4). We restricted sequences and base qualities to values of ≥30, and built a consensus mitochondrial genome sequence with samtools and bcftools110 (v.0.1.19–96b5f2294a), using a majority rule and minimum coverage of 1, trimming 2 basepairs from the end of each sequence. We further restricted the data for each sample to the damaged reads as determined by pmdtools (using a minimum pmdscore of 3) and repeated the calling. In almost every case where there was sufficient post-damage restricted coverage to give a confident haplogroup call, the calls for the damage-restricted data matched the calls obtained when we did not perform damage-restricted. For Y chromosome haplogroup assessment we only used base qualities ≥30, identifying the most derived mutations using the nomenclature of the International Society of Genetic Genealogy (http://www.isogg.org) v. 11.110.
Dataset assembly
Our basic dataset included 3359 individuals, of which 2182 were modern7,40,63–65 and 1177 were ancient individuals from previous publications1,2,4–7,26–62, which we combined with the newly reported 57 samples that passed screening (Online Table 3). We performed all subsequent analysis on autosomal data.
Principal component analysis
We used a subset of 591 modern and 1266 ancient West Eurasians for principal component analysis (PCA) using smartpca from the EIGENSOFT package111(v.7.2.1), using default parameters with two exceptions. We used the option lsqproject: YES to project all ancient individuals onto the eigenvectors computed from modern vectors. The approach of projecting each ancient sample onto patterns of variation learned from modern samples is useful for ancient DNA analysis, as it means that we can use data from a large fraction of SNPs covered in each individual, thereby maximizing the information about ancestry that would be lost in approaches that require restriction to a potentially smaller number of SNPs for which there is intersecting data across low coverage ancient individuals112. We used the option shrinkmode: YES to remap the points for the samples used to generate the PCA onto the positions where they would be expected to fall if they had been projected (thereby allowing the projected and non-projected samples to be appropriately co-visualized). We used a dataset containing only transversions to assess the robustness of our qualitative inferences to bias due to ancient DNA damage-induced errors (Supplementary Fig. 1).
Population structure analysis
We ran ADMIXTURE66 (v.1.2.3) after pruning to remove one SNP each in pairs of SNPs in linkage disequilibrium, using PLINK1.9113 and the option --indep-pairwise 200 25 0.4, leaving 315,235 SNPs. We ran ADMIXTURE from K=5 to K=15, with 10 random-seeded replicates for each value of K. We used cross validation by adding the option --cv to find the runs with the lowest errors (Supplementary Fig. 2). For each K, we kept the replicate with lowest error. We present results for K=10, as we empirically found that this is the value of K with lowest cross-validation error that also showed clear distinctions between ancient Western, Eastern, and Caucasus Hunter-Gatherer backgrounds, while maximizing the Neolithic Anatolian-associated component. We also performed ADMIXTURE restricting to transversion SNPs and obtained qualitatively similar results suggesting that ancient DNA damage is unlikely to be strongly biasing our findings (Supplementary Fig. 1).
f4-statistics
We used ADMIXTOOLS63 (v.4.1) to compute f4 symmetry statistics (qpDstat). We used Mbuti.DG as the outgroup, and computed statistics of the form f4(Mbuti.DG, X; Y, Z), where X is our test population (or individual) and Y/Z are pairs of populations (or individuals) that we tested for sharing alleles at an equal rate with X. We used the options f4mode: YES and printsd: YES.
qpWave/qpAdm
We used qpWave/qpAdm from ADMIXTOOLS63 (v.4.1) to estimate admixture coefficients and to model our individuals/populations as mixtures of groups that we propose as clades with the true source population. We used a base outgroup set including the following individuals/populations: Mbuti.DG, Ust_Ishim, CHG, EHG, ElMiron, Vestonice16, MA1, Israel_Natufian, Jordan_PPNB. Extra populations were included in each test to improve accuracy when using populations with similar ancestries (see Supplementary Materials for a detailed description). When visualizing the results we present the ones with the fewest sources that also has the highest probability (although we also report alternative models with the same number of sources that fit at P>0.05). We used the option allsnps: YES to specify that the f4-statistics that are the basis of qpWave/qpAdm should be computed using the intersection of SNPs with coverage in all four groups that contribute to each f4-statistic. We set a minimum threshold of 100,000 SNPs when modeling single individuals.
Data Availability
All raw data are available at the European Nucleotide Archive under the accession number PRJEB35980 and at https://reich.hms.harvard.edu/datasets.
Code Availability
All custom code can be found at https://github.com/DReichLab/ADNA-Tools.
Supplementary Material
Acknowledgements
This manuscript is dedicated to the memory of Sebastiano Tusa of the Soprintendenza del Mare in Palermo, who would have been an author of this study had he not tragically died in the crash of Ethiopia Airlines flight 302 on March 10 2019. We thank Zhao Zhang for database support. We thank the Soprintendenza BBCCAA Palermo and Rosario Schicchi (director of Museum of Castelbuono) for facilitating access to important skeletal materials. D.F. was supported by an Irish Research Council grant GOIPG/2013/36. Radiocarbon work was supported in part by the NSF Archaeometry program BCS-1460369 to D.J.K. and B.J.C. C.L.-F. was supported by Obra Social La Caixa and by FEDER-MINECO (BFU2015- 64699-P and PGC2018-095931-B-100). D.C. was supported by grant 20177PJ9XF MIUR PRIN 2017. D.Re. is an Investigator of the Howard Hughes Medical Institute and his ancient DNA laboratory work was supported by National Science Foundation HOMINID grant BCS-1032255, by National Institutes of Health grant GM100233, by an Allen Discovery Center grant, and by grant 61220 from the John Templeton Foundation.
Footnotes
Competing Interests
The authors declare no competing financial interests.
References
- 1.Allentoft ME et al. Population genomics of Bronze Age Eurasia. Nature 522, 167–172 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Haak W et al. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 522, 207–211 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kristiansen K Re-theorising Mobility and the Formation of Culture and Language Among the Corded Ware Culture in Europe. Antiquity 91, 334–347 (2017). [Google Scholar]
- 4.Cassidy LM et al. Neolithic and Bronze Age migration to Ireland and establishment of the insular Atlantic genome. Proc. Natl. Acad. Sci. U. S. A 113, 368–373 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olalde I et al. The Beaker phenomenon and the genomic transformation of northwest Europe. Nature 555, 190–196 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martiniano R et al. The population genomics of archaeological transition in west Iberia: Investigation of ancient substructure using imputation and haplotype-based methods. PLoS Genet. 13, e1006852 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lazaridis I et al. Genetic origins of the Minoans and Mycenaeans. Nature 548, 214–218 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alcover JA The First Mallorcans: Prehistoric Colonization in the Western Mediterranean. Journal of World Prehistory 21, 19–84 (2008). [Google Scholar]
- 9.Ramis D Early Island Exploitations: Productive and Subsistence Strategies on the Prehistoric Balearic Islands in The Cambridge Prehistory of the Bronze and Iron Age Mediterranean (eds. Knapp AB & van Dommelen P) 40–56 (Cambridge University Press, 2014). [Google Scholar]
- 10.Ramis D Animal Exploitation in the Early Prehistory of the Balearic Islands. The Journal of Island and Coastal Archaeology 13, 269–282 (2018). [Google Scholar]
- 11.Plantalamor L & Van Strydonck M La cronologia de la prehistòria de Menorca. (Treballs del Museu de Menorca 20, 1997).
- 12.Lull V, Mico R, Rihuete CI & Risch R Los cambios sociales en las islas Baleares a lo largo del II milenio. Cypsela 15, 123–148 (2004). [Google Scholar]
- 13.Holt E Mobility and meaning in the Nuragic culture of Bronze Age Sardinia in Forging Identities. The Mobility of Culture in Bronze Age Europe (eds. Suchowska-Ducke P & Reiter SS & Vandkilde) 1, 193–202 (British Archaeological Reports, 2015). [Google Scholar]
- 14.Ugas G L’alba dei nuraghi (Fabula, 2005; ). [Google Scholar]
- 15.Sestieri AMB The Bronze Age in Sicily in The Oxford Handbook of European Bronze Age (eds. Harding A & Fokkens H) 653–667 (Oxford University Press, 2013). [Google Scholar]
- 16.Sarno S et al. Ancient and recent admixture layers in Sicily and Southern Italy trace multiple migration routes along the Mediterranean. Sci. Rep 7, 1984 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dabney J et al. Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc. Natl. Acad. Sci. U. S. A 110, 15758–15763 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Damgaard PB et al. Improving access to endogenous DNA in ancient bones and teeth. Sci. Rep 5, 11184 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korlević P et al. Reducing microbial and human contamination in DNA extractions from ancient bones and teeth. Biotechniques 59, 87–93 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Rohland N, Glocke I, Aximu-Petri A & Meyer M Extraction of highly degraded DNA from ancient bones, teeth and sediments for high-throughput sequencing. Nat. Protoc 13, 2447–2461 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Rohland N, Harney E, Mallick S, Nordenfelt S & Reich D Partial uracil-DNA-glycosylase treatment for screening of ancient DNA. Philos. Trans. R. Soc. Lond. B Biol. Sci 370, 20130624 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gansauge M-T et al. Single-stranded DNA library preparation from highly degraded DNA using T4 DNA ligase. Nucleic Acids Res. 45, e79 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu Q et al. A revised timescale for human evolution based on ancient mitochondrial genomes. Curr. Biol 23, 553–559 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu Q et al. An early modern human from Romania with a recent Neanderthal ancestor. Nature 524, 216–219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olalde I et al. The genomic history of the Iberian Peninsula over the past 8000 years. Science 363, 1230–1234 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keller A et al. New insights into the Tyrolean Iceman’s origin and phenotype as inferred by whole-genome sequencing. Nat. Commun 3, 698 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Lazaridis I et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 513, 409–413 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gamba C et al. Genome flux and stasis in a five millennium transect of European prehistory. Nature Communications 5, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olalde I et al. Derived immune and ancestral pigmentation alleles in a 7,000-year-old Mesolithic European. Nature 507, 225–228 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raghavan M et al. Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans. Nature 505, 87–91 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skoglund P et al. Genomic diversity and admixture differs for Stone-Age Scandinavian foragers and farmers. Science 344, 747–750 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Günther T et al. Ancient genomes link early farmers from Atapuerca in Spain to modern-day Basques. Proc. Natl. Acad. Sci. U. S. A 112, 11917–11922 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones ER et al. Upper Palaeolithic genomes reveal deep roots of modern Eurasians. Nat. Commun 6, 8912 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mathieson I et al. Genome-wide patterns of selection in 230 ancient Eurasians. Nature 528, 499–503 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olalde I et al. A Common Genetic Origin for Early Farmers from Mediterranean Cardial and Central European LBK Cultures. Mol. Biol. Evol 32, 3132–3142 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Broushaki F et al. Early Neolithic genomes from the eastern Fertile Crescent. Science 353, 499–503 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fu Q et al. The genetic history of Ice Age Europe. Nature 534, 200–205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hofmanová Z et al. Early farmers from across Europe directly descended from Neolithic Aegeans. Proc. Natl. Acad. Sci. U. S. A 113, 6886–6891 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kılınç GM et al. The Demographic Development of the First Farmers in Anatolia. Curr. Biol 26, 2659–2666 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lazaridis I et al. Genomic insights into the origin of farming in the ancient Near East. Nature 536, 419–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martiniano R et al. Genomic signals of migration and continuity in Britain before the Anglo-Saxons. Nat. Commun 7, 10326 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schiffels S et al. Iron Age and Anglo-Saxon genomes from East England reveal British migration history. Nat. Commun 7, 10408 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.González-Fortes G et al. Paleogenomic Evidence for Multi-generational Mixing between Neolithic Farmers and Mesolithic Hunter-Gatherers in the Lower Danube Basin. Curr. Biol 27, 1801–1810.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haber M et al. Continuity and Admixture in the Last Five Millennia of Levantine History from Ancient Canaanite and Present-Day Lebanese Genome Sequences. Am. J. Hum. Genet 101, 274–282 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones ER et al. The Neolithic Transition in the Baltic Was Not Driven by Admixture with Early European Farmers. Current Biology 27, 576–582 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lipson M et al. Parallel palaeogenomic transects reveal complex genetic history of early European farmers. Nature 551, 368–372 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saag L et al. Extensive Farming in Estonia Started through a Sex-Biased Migration from the Steppe. Curr. Biol 27, 2185–2193.e6 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Schuenemann VJ et al. Ancient Egyptian mummy genomes suggest an increase of Sub-Saharan African ancestry in post-Roman periods. Nat. Commun 8, 15694 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Unterländer M et al. Ancestry and demography and descendants of Iron Age nomads of the Eurasian Steppe. Nat. Commun 8, 14615 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Amorim CEG et al. Understanding 6th-century barbarian social organization and migration through paleogenomics. Nat. Commun 9, 3547 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Damgaard P et al. The first horse herders and the impact of early Bronze Age steppe expansions into Asia. Science 360, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fernandes DM et al. A genomic Neolithic time transect of hunter-farmer admixture in central Poland. Sci. Rep 8, 14879 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fregel R et al. Ancient genomes from North Africa evidence prehistoric migrations to the Maghreb from both the Levant and Europe. Proc. Natl. Acad. Sci. U. S. A 115, 6774–6779 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mathieson I et al. The genomic history of southeastern Europe. Nature 555, 197–203 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mittnik A et al. The genetic prehistory of the Baltic Sea region. Nat. Commun 9, 442 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Valdiosera C et al. Four millennia of Iberian biomolecular prehistory illustrate the impact of prehistoric migrations at the far end of Eurasia. Proc. Natl. Acad. Sci. U. S. A 115, 3428–3433 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van de Loosdrecht M et al. Pleistocene North African genomes link Near Eastern and sub-Saharan African human populations. Science 360, 548–552 (2018). [DOI] [PubMed] [Google Scholar]
- 58.Veeramah KR et al. Population genomic analysis of elongated skulls reveals extensive female-biased immigration in Early Medieval Bavaria. Proc. Natl. Acad. Sci. U. S. A 115, 3494–3499 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zalloua P et al. Ancient DNA of Phoenician remains indicates discontinuity in the settlement history of Ibiza. Sci. Rep 8, 17567 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Feldman M et al. Late Pleistocene human genome suggests a local origin for the first farmers of central Anatolia. Nat. Commun 10, 1218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.González-Fortes G et al. A western route of prehistoric human migration from Africa into the Iberian Peninsula. Proc. Biol. Sci 286, 20182288 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Narasimhan VM et al. The formation of human populations in South and Central Asia. Science 365, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Patterson N et al. Ancient Admixture in Human History. Genetics 192, 1065–1093 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pickrell JK et al. The genetic prehistory of southern Africa. Nat. Commun 3, 1143 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qin P & Stoneking M Denisovan Ancestry in East Eurasian and Native American Populations. Mol. Biol. Evol 32, 2665–2674 (2015). [DOI] [PubMed] [Google Scholar]
- 66.Alexander DH, Novembre J & Lange K Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Holt EM Economy and Environment in Complex Societies: A Case Study from Bronze Age Sardinia. (University of Michigan, 2013). [Google Scholar]
- 68.Ramis D, Alcover JA, Coll J & Trias M The Chronology of the First Settlement of the Balearic Islands. Journal of Mediterranean Archaeology 15, (2002). [Google Scholar]
- 69.Picornell A, Gómez-Barbeito L, Tomàs C, Castro JA & Ramon MM Mitochondrial DNA HVRI variation in Balearic populations. Am. J. Phys. Anthropol 128, 119–130 (2005). [DOI] [PubMed] [Google Scholar]
- 70.Adams SM et al. The genetic legacy of religious diversity and intolerance: paternal lineages of Christians, Jews, and Muslims in the Iberian Peninsula. Am. J. Hum. Genet 83, 725–736 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Solé-Morata N et al. Analysis of the R1b-DF27 haplogroup shows that a large fraction of Iberian Y-chromosome lineages originated recently in situ. Sci. Rep 7, 7341 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Di Gaetano C et al. Differential Greek and northern African migrations to Sicily are supported by genetic evidence from the Y chromosome. Eur. J. Hum. Genet 17, 91–99 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sarno S et al. An ancient Mediterranean melting pot: investigating the uniparental genetic structure and population history of sicily and southern Italy. PLoS One 9, e96074 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Raveane A et al. Population structure of modern-day Italians reveals patterns of ancient and archaic ancestries in Southern Europe. bioRxiv (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Magoon GR et al. Generation of high-resolution a priori Y-chromosome phylogenies using ‘next-generation’ sequencing data. bioRxiv (2013). doi: 10.1101/000802 [DOI] [Google Scholar]
- 76.Wang C-C et al. Ancient human genome-wide data from a 3000-year interval in the Caucasus corresponds with eco-geographic regions. Nat. Commun 10, 590 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Matisoo-Smith E et al. Ancient mitogenomes of Phoenicians from Sardinia and Lebanon: A story of settlement, integration, and female mobility. PLoS One 13, e0190169 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sikora M et al. Population genomic analysis of ancient and modern genomes yields new insights into the genetic ancestry of the Tyrolean Iceman and the genetic structure of Europe. PLoS Genet. 10, e1004353 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chiang CWK et al. Genomic history of the Sardinian population. Nat. Genet 50, 1426–1434 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Moorjani P et al. The history of African gene flow into Southern Europeans, Levantines, and Jews. PLoS Genet. 7, e1001373 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Loh P-R et al. Inferring admixture histories of human populations using linkage disequilibrium. Genetics 193, 1233–1254 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hellenthal G et al. A genetic atlas of human admixture history. Science 343, 747–751 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Olivieri A et al. Mitogenome Diversity in Sardinians: A Genetic Window onto an Island’s Past. Mol. Biol. Evol 34, 1230–1239 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Morelli L et al. A Comparison of Y-Chromosome Variation in Sardinia and Anatolia Is More Consistent with Cultural Rather than Demic Diffusion of Agriculture. PLoS ONE 5, e10419 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Marcus JH et al. Population history from the Neolithic to present on the Mediterranean island of Sardinia: An ancient DNA perspective. bioRxiv (2019). doi: 10.1101/583104 [DOI] [Google Scholar]
- 86.Sangmeister E Die Datierung des Rickstroms der Glockenbecker und ihre Auswirkung auf die Chronologie der Kupferzeit in Portugal. Palaeohistoria 12, 395–407 (1966). [Google Scholar]
- 87.Holloway R The Archaeology of Ancient Sicily. (Routledge, 2000). [Google Scholar]
- 88.D’Agata AL Interactions between Aegean groups and local communities in Sicily in the Bronze Age: The evidence from pottery. Studi micenei ed egeo-anatolici 42, 61–83 (2000). [Google Scholar]
- 89.Shelton K Mainland Greece. in The Oxford Handbook of the Bronze Age Aegean (ed. Kline E) 139–148 (Oxford University Press, 2012). [Google Scholar]
- 90.Alberti G Issues in the absolute chronology of the Early-Middle Bronze Age transition in Sicily and southern Italy: a Bayesian radiocarbon view. J. Quat. Sci 28, 630–640 (2013). [Google Scholar]
- 91.Heyd V Europe 2500 to 2200 BC: Between Expiring Ideologies and Emerging Complexity in The Oxford Handbook of the European Bronze Age (ed. Harding A) 47–67 (Oxford University Press, 2013). [Google Scholar]
- 92.Sabatini S Late Bronze Age Oxhide and oxhide-like ingots from areas other than the Mediterranean: problems and challenges. Oxford Journal of Archaeology 35, 29–45 (2016). [Google Scholar]
- 93.Aubet ME & Turton M The Phoenicians and the West: Politics, Colonies and Trade. Journal of the American Oriental Society 117, 212 (1997). [Google Scholar]
- 94.Pinhasi R et al. Optimal Ancient DNA Yields from the Inner Ear Part of the Human Petrous Bone. PLoS One 10, e0129102 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pinhasi R, Fernandes DM, Sirak K & Cheronet O Isolating the human cochlea to generate bone powder for ancient DNA analysis. Nat. Protoc 14, 1194–1205 (2019). [DOI] [PubMed] [Google Scholar]
- 96.Briggs AW et al. Removal of deaminated cytosines and detection of in vivo methylation in ancient DNA. Nucleic Acids Res. 38, e87 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Maricic T, Whitten M & Pääbo S Multiplexed DNA sequence capture of mitochondrial genomes using PCR products. PLoS One 5, e14004 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Behar DM et al. A ‘Copernican’ reassessment of the human mitochondrial DNA tree from its root. Am. J. Hum. Genet 90, 675–684 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li H & Durbin R Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Weissensteiner H et al. HaploGrep 2: mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic Acids Res. 44, W58–63 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Korneliussen TS, Albrechtsen A & Nielsen R ANGSD: Analysis of Next Generation Sequencing Data. BMC Bioinformatics 15, 356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kennett DJ et al. Archaeogenomic evidence reveals prehistoric matrilineal dynasty. Nat. Commun 8, 14115 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lohse JC, Culleton BJ, Black SL & Kennett DJ, A. A Precise Chronology of Middle to Late Holocene Bison Exploitation in the Far Southern Great Plains. Journal of Texas Archeology and History 1, 94–126 (2014). [Google Scholar]
- 104.van Klinken GJ Bone Collagen Quality Indicators for Palaeodietary and Radiocarbon Measurements. Journal of Archaeological Science 26, 687–695 (1999). [Google Scholar]
- 105.Santos GM, Southon JR, Druffel-Rodriguez KC, Griffin S & Mazon M Magnesium Perchlorate as an Alternative Water Trap in AMS Graphite Sample Preparation: A Report On Sample Preparation at Kccams at the University of California, Irvine. Radiocarbon 46, 165–173 (2004). [Google Scholar]
- 106.Stuiver M & Polach HA Discussion Reporting of 14C Data. Radiocarbon 19, 355–363 (1977). [Google Scholar]
- 107.Ramsey CB & Lee S Recent and Planned Developments of the Program OxCal. Radiocarbon 55, 720–730 (2013). [Google Scholar]
- 108.Reimer PJ et al. IntCal13 and Marine13 Radiocarbon Age Calibration Curves 0–50,000 Years cal BP. Radiocarbon 55, 1869–1887 (2013). [Google Scholar]
- 109.van Oven M & Kayser M Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum. Mutat 30, E386–94 (2009). [DOI] [PubMed] [Google Scholar]
- 110.Li H et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Patterson N, Price AL & Reich D Population structure and eigenanalysis. PLoS Genet. 2, e190 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Skoglund P et al. Origins and genetic legacy of Neolithic farmers and hunter-gatherers in Europe. Science 336, 466–469 (2012). [DOI] [PubMed] [Google Scholar]
- 113.Chang CC et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data are available at the European Nucleotide Archive under the accession number PRJEB35980 and at https://reich.hms.harvard.edu/datasets.


