Molecular responses of fishes to a heatwave are species specific and do not concur with measures of oxygen uptake in captivity.
Abstract
The marine heatwave of 2016 was one of the longest and hottest thermal anomalies recorded on the Great Barrier Reef, influencing multiple species of marine ectotherms, including coral reef fishes. There is a gap in our understanding of what the physiological consequences of heatwaves in wild fish populations are. Thus, in this study, we used liver transcriptomes to understand the molecular response of five species to the 2016 heatwave conditions. Gene expression was species specific, yet we detected overlap in functional responses associated with thermal stress previously reported in experimental setups. The molecular response was also influenced by the duration of exposure to elevated temperatures. This study highlights the importance of considering the effects of extreme warming events when evaluating the consequences of climate change on fish communities.
INTRODUCTION
Human-induced climate change is one of the main threats to the persistence of marine organisms. For instance, ocean warming can lead to physiological challenges for marine ectotherms (1), which, in turn, can lead to large-scale alterations at the population, community, and ecosystem levels (2). Studies on the long-term effects of ocean warming suggest that increasing temperatures have already caused shifts in species’ distribution, changes in behavior, and population declines in many marine taxa (3, 4). However, global warming following the Industrial Revolution has also led to an increase in the frequency and intensity of marine heatwaves, particularly during El Niño Southern Oscillations (ENSOs), over the past 40 years (5–7). A notable example was the ENSO of the austral summer of 2015–2016, which resulted in high temperatures that lasted longer than any other recorded marine heatwave to date in the Great Barrier Reef (GBR) Australia (6), leading to widespread coral bleaching and mortality (8). Studies suggest that heatwaves, like the one in 2016, can lead to population declines in fish species that directly depend on reef-building corals for food, shelter, and/or reproduction (9). Moreover, long-term shifts in the composition of fish communities (10) and changes in fish behavior can arise after acute heat stress (11). However, a complete picture of the response of fishes to marine heatwaves remains elusive. Much less attention has been given to the direct physiological response of wild fish populations to warming events, although laboratory experiments show that high temperatures can reduce individual performance (1), with potential detrimental consequences for population dynamics and sustainability (12).
Tropical ectotherms are thought to be more sensitive to higher temperatures when compared to temperate species, because the former evolved under relatively stable thermal conditions (13). Laboratory studies show that acute warming leads to an increase in resting oxygen uptake rates in fishes, likely triggered by the increase in resting metabolism (1). In tropical fishes, this increase in resting oxygen uptake is not matched by an increase in maximum oxygen uptake at higher temperature, leading to a collapse of aerobic metabolic scope. It has been suggested that the collapse of aerobic scope at extreme temperatures is at the level of the cardiorespiratory system, which is collectively termed the oxygen- and capacity-limited thermal tolerance hypothesis (14). When temperatures are too high, the cardiorespiratory system is no longer able to fulfill additional oxygen demands for aerobic activities, leading to a reduction in aerobic performance (15, 16), alteration of growth rates (17), and reduction in swimming speeds (18). Previous studies focusing on gene expression of captive fishes have shown that increased oxygen demands at higher temperatures can lead to activation of genes related to mitochondrial activity, the electron-transport chain, fatty acid metabolism, and cellular stress response (19, 20). Furthermore, reproductive hormones can be affected by warmer conditions, leading to changes in reproductive behavior and declines in reproductive output (21). However, it remains to be determined whether these molecular responses and their accompanying physiological challenges are also observed in wild fish populations during marine heatwaves. Considering that extreme temperatures could have greater impacts on the performance of ectotherms, when compared with slight thermal increments over years or decades (2), a molecular approach would complement our understanding of how acute warming events directly affect reef fish communities in nature.
In this study, we evaluate the molecular responses to the 2016 marine heatwave in five species of coral reef fishes from the Lizard Island, Australia: the damselfishes Acanthochromis polyacanthus and Pomacentrus moluccensis (Pomacentridae) and the cardinalfishes Ostorhinchus cyanosoma, Ostorhinchus doederleini, and Cheilodipterus quinquelineatus (Apogonidae; Fig. 1). The selected species are highly abundant on reefs of Lizard Island and represent families of high diversity and ecological relevance in coral reef ecosystems around the world (22). Furthermore, these species have previously been studied in laboratory settings for their response to acute warming, showing differences in their thermal sensitivity based on measures of oxygen uptake (15, 16). Here, we focus on changes of the transcriptional program of the liver, as previous studies have successfully used this organ to understand the effects of elevated temperatures on metabolic conditioning (19). For the five species, patterns of gene expression of ~16,000 orthologous genes were analyzed from samples collected over four time points across the heatwave. Specifically, we sought to (i) determine whether the transcriptional responses in the liver are influenced by collection time and temperature, (ii) identify common molecular mechanisms associated with the response to acute warming, and (iii) assess differences between initial and prolonged exposure to heatwave conditions.
Fig. 1. Summary of the sampling design.
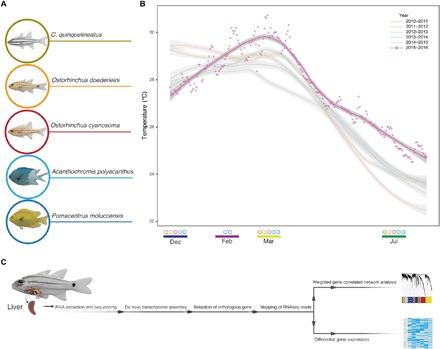
(A) Five fish species analyzed in this study. (B) Temperature records for the sampling location from 2010 to 2016. Dots are daily averages in 2015–2016. The trendlines representing the 5 years before the heatwave, as well as 2015–2016, are based on the t-based approximation of the daily temperature averages. The circles at the bottom represent the species collected in that month (i.e., only A. polyacanthus and P. moluccensis were collected in February). Average temperatures for each month were as follows: December, 27.81°C (SD ±0.45°C); February, 29.55°C (SD ±0.69°C); March, 29.70°C (SD ±0.61°C); July, 24.96°C (SD ±0.28°C). (C) Schematic representation of the sample processing from liver extraction to gene expression analysis. RNASeq, RNA sequencing.
RESULTS
Liver transcriptional programs of five fish species
To understand the molecular response of coral reef fishes to heatwave conditions, we sequenced liver transcriptomes of fishes from wild populations throughout the heatwave. De novo assembled transcriptomes for the five reef fish species contained an average of 166,000 transcripts per species (±51,000; data file S1 and fig. S1). These transcripts were then clustered into orthologous groups, which included one or more contigs from each species. To compare the same genes across the five species, these orthologous groups were filtered on the basis of sequence similarity, resulting in a total of 16,494 curated proteins (data file S1). This reference set of orthologous genes allowed patterns of gene expression to be compared across samples collected at different time points through the heatwave. These assemblies also represent the first published transcriptome assemblies for the three cardinalfishes: O. cyanosoma, O. doederleini, and C. quinquelineatus.
Gene expression divergence is driven by interspecific differences and thermal conditions
To explore the molecular basis of the reaction to a heatwave in natural fish populations, we investigated the gene expression patterns of the five species. For this, we examined the relative contributions of interspecific differences and collection times and temperatures to the patterns of gene expression. Clustering of biological samples, as well as differential gene expression, revealed that the largest differences were between the five species, suggesting that the dominant factor influencing expression was the specific molecular patterns exhibited by each species (Fig. 2). To understand how gene expression varies based on phylogenetic relatedness of the studied species, we performed an analysis of Expression Variance and Evolution (EVE) (23). The EVE analysis resulted in 145 genes with significantly diverged expression profiles (i.e., higher differences between species than within samples of one species; data file S2), including some associated to energy metabolism and stress response (e.g., HSP7C, FA10, CO9, CP2U1, ADT2, and CRBB; fig. S2). Many of these genes were also differentially expressed in comparisons with samples collected in different months and temperatures. These divergent genes showed significant enrichment for 39 Gene Ontology (GO) categories including catalytic activity, gene expression, response to stress, enzyme regulator activity, translation regulator activity, guanosine triphosphatase (GTPase) activity, and lipid metabolic process (data file S3). Meanwhile eight genes showed diverse expression profiles (i.e., higher variance of expression within species), most notably DDX46, GADD45, DIP2A, and EMC8 (data file S4).
Fig. 2. Molecular response to the 2015–2016 marine heatwave in five coral reef fishes.
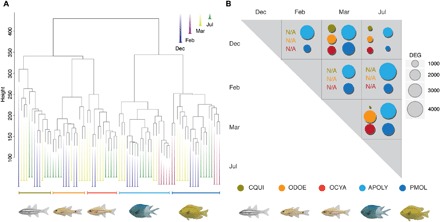
(A) Clustering analysis across all biological replicates revealing samples grouped tightly by species and collection months represented by different colors. For all analyses, comparisons with samples from February were only possible for damselfishes A. polyacanthus and P. moluccensis. (B) Number of DEGs between samples collected at different time points of the heatwave. (CQUI, C. quinquelineatus; ODOE, O. doederleini; OCYA, O. cyanosoma; APOLY, A. polyacanthus; PMOL, P. moluccensis); N/A, not available.
The second largest driver of divergence between groups was the patterns of expression between time points, that is, between contrasting thermal conditions (Fig. 2). This is illustrated by the clustering of samples in the cladogram (Fig. 2A), as well as the number of differentially expressed genes (DEGs) between samples collected at particular time points (Fig. 2B). When examining the pairwise comparisons of the collection time points, there were no DEGs shared across all species (fig. S3). Furthermore, clustering algorithms revealed large gene networks that were highly correlated by species (fig. S4). These results suggest that there is a molecular response to the heatwave conditions, but there are differences in the composition and magnitude of the response between species across collection times throughout the heatwave.
Despite the lack of shared DEGs across all five species, there was a phylogenetic signal underlying the observed gene expression response across the heatwave (Fig. 2A). We found gene clusters that significantly correlated with the warmest months (February, only damselfishes; March, both families) when the two different fish families were separated in the analysis (figs. S5 and S6). This analysis demonstrates that damselfishes and cardinalfishes reacted differently, because the former showed a higher number of DEGs between the different collection times (Fig. 2B). Furthermore, there was considerable overlap in the response of the two species of damselfishes (both gene and functional categories; fig. S5). Overall, there was a gradient in the patterns of gene expression, with A. polyacanthus showing the largest number of DEGs across comparisons followed by P. moluccensis, O. cyanosoma, and O. doederleini, which had a more intermediate reaction, and C. quinquelineatus presented the smallest numbers of DEGs between all time points (Fig. 2B and data file S5).
Initial response to heatwave conditions
Between December 2015 and February 2016, water temperatures at Lizard Island went from an average of 27.81°C (SD ±0.35 ° C) to 29.50°C (SD ±0.51 ° C). Animals typically experience an increase in temperature with the onset of summer, but warming in 2016 was faster and more intense than previous years (Fig. 1B). Because of sampling constraints (see Materials and Methods), we only have February samples for the two damselfishes, but both species show large differences in gene expression during the onset of the heatwave (fig. S7). Comparing A. polyacanthus samples from December and February resulted in 3024 DEGs, whereas P. moluccensis exhibited 993 DEGs (Fig. 2B). Of these DEGs, 158 had a common pattern of expression in both damselfishes, which were also exclusive to the onset of the heatwave (i.e., not differentiated in other comparisons). This set of genes showed GO enrichment for cholesterol metabolic processes. Because of the relatively small number of overlapping genes, six gene network clusters (i.e., modules) correlated significantly for the February collection point when both species were included (fig. S5). There were also differences in the functional responses to initial elevated temperatures between the two damselfishes: A. polyacanthus showed enrichment for mitochondrial activity, chaperone-mediated protein folding, immunity, DNA replication, and activity of endoplasmic reticulum, whereas P. moluccensis showed enrichment for apoptosis, heme activity, and oxidoreductase activity (data files 6 and 7).
The response to the peak temperatures in March was evaluated for all five species. For the comparisons between December (before the heatwave) and March (peak warming), A. polyacanthus showed 3251 DEGs, and P. moluccensis exhibited a similar magnitude in response (2236 DEGs) with an overlap of 860 DEGs between the two species. There were shared functional responses between the two damselfishes in this comparison, including enrichment of gene categories associated with mitochondrial activity, adenosine triphosphate synthesis, negative regulation of glucocorticoid signaling, protein translation, and oxidoreductase activity (data file S8). These same functional enrichments were found in various modules of the correlated gene network analysis. In addition, multiple genes associated with histone modifications and chromatin remodeling were up-regulated in the two damselfishes during the hottest month of the heatwave (data files S9 and S10). Both of these molecular processes are potentially related to the change in epigenetic regulation, because they can influence DNA transcription, repair, and replication when organisms are exposed to environmental stressors (24).
In general, cardinalfishes displayed smaller numbers of DEGs than damselfishes (Fig. 2B), which also resulted in the enrichment of fewer functional categories. The response between December and March resulted in 1118 DEGs for O. cyanosoma, showing enrichment for the synthesis of glycerol and phosphoenolpyruvate carboxykinase activity (data file S11). For O. doederleini, there were 1332 DEGs, revealing enrichment for mitochondrial activity, detoxification, cholesterol, and fatty acid metabolism. An interesting gene that was up-regulated in December for both Ostorhinchus species was the cold shock domain-containing protein E1 (CSDE1), an RNA binding protein aiding in cell survival at suboptimal temperatures. Lastly, C. quinquelineatus exhibited only 992 DEGs, with enrichment for activity of the endoplasmic reticulum and mediated unfolded protein response. Twenty of these genes overlapped for all three cardinalfishes: 17 change their expression in the same direction (either up- or down-regulated), while the other 3 genes show opposing but significant changes (data file S12). Thus, we observed up-regulation of exostosin 1B (EXT1B) and syntenin (SDCB1) for O. cyanosoma and reduced expression of these genes for O. doederleini and C. quinquelineatus between December and March. The opposite occurs for core 1 synthase glycoprotein-N-acetylgalactosamine 3-β-galactosyltransferase 1 (C1GALT1), which is up-regulated at high temperatures for O. doederleini and C. quinquelineatus but down-regulated for O. cyanosoma. One of the gene network clusters that correlated with the March samples of cardinalfishes (cyan module; fig. S6) showed significant enrichment for inflammatory and immune response (data file S13). Further evidence of immune response activation during warming is that genes of the complement system (CO3, CO7, and CO9) showed significant differences in the gene network and gene expression analyses in the March samples relative to December. DEP domain-containing protein 5 (DEPDC5), a negative regulator of the mammalian target of rapamycin (mTOR) complex, was one of the most highly differentiated genes for all three cardinalfishes in March, potentially playing an important role during the warmest period. Although damselfishes and cardinalfishes did not show much overlap of DEGs in the different comparisons, similar molecular pathways were activated in samples collected during the warmest period. For example, one module (red module; fig. S8) showed enrichment of molecular processes that were up-regulated in damselfishes, including mitochondrial electron transport and histone modification (data file S14).
To get a more holistic understanding of the effects of acute warming on these fish species, we calculated the temperature quotient (Q10) of resting oxygen uptake rates (MO2Rest) using previously calculated values for these species (15, 16). Here, we focused on the change of oxygen uptake rates from 29° to 31°C, because this is representative of the heatwave conditions of 2016. The results indicate that O. cyanosoma (Q10 = 10.46) and A. polyacanthus (Q10 = 8.66) showed the most temperature sensitivity in MO2Rest across this temperature range. Meanwhile, P. moluccensis (Q10 = 2.05) and O. doederleini (Q10 = 1.97; data file S15) exhibited some levels of aerobic compensation. The species with the least temperature dependence in terms of changes in resting oxygen uptake rates was C. quinquelineatus (Q10 = 0.23).
Consequences of exposure to a prolonged heatwave
Our sampling strategy allowed us to compare patterns of gene expression between samples collected at the beginning of the heatwave in February and after 4 weeks of exposure to elevated temperatures until mid-March. The DEGs revealed through this comparison can be interpreted as the molecular response to prolonged warming, as between the February and March collections, average water temperatures were 29.77°C (SD ±0.54). We grouped the DEGs from the different pairwise comparisons between sampling times by the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and plotted these comparisons using a principal components analysis. Clustering by KEGG pathways showed that “prolonged exposure” was the most distinct category among all other comparisons (Fig. 3A). As mentioned previously, this contrast was only possible for damselfishes (see Materials and Methods for details), where the number of DEGs was 2706 for A. polyacanthus and 1720 for P. moluccensis (1003 in common). The slightly lower number in DEGs for P. moluccensis is most likely a result of the intraspecific variance observed among biological replicates of this species (fig. S7). Despite the differences in the number of DEGs, there was considerable overlap in the functional response of damselfishes, as they both showed enrichment for mitochondrial activity, respiratory chain complex, synthesis of fatty acids, and oxidoreductase activity (data file S16). Another common response to prolonged warming was the differential regulation of genes encoding for heat shock proteins (HSPs). HSPs were activated at the beginning of the heatwave in February, but they were down-regulated in the March samples for both A. polyacanthus (HSP13, HSP70, HSP74, and HSPB1) and P. moluccensis (HSP7E, HSP13, and HSP74), although average water temperature was higher in March (fig. S9). Only HSPB7 was up-regulated in A. polyacanthus for March in comparison to February (data file S17). A gene network cluster of genes for prolonged exposure between February and March (green module; fig. S10) showed differences for RNA processing and ribonucleoprotein complex biogenesis between the beginning of the heatwave and after prolonged exposure (data file 18). Finally, 360 overlapping DEGs were exclusively found in the comparisons of prolonged exposure in damselfishes (i.e., not differentially expressed with the onset of the heatwave, from December to February). These were genes associated with biotin metabolic process, cellular oxidant detoxification, aerobic respiration, fatty acid biosynthesis, and cellular ketone metabolic processes (data file S19).
Fig. 3. Comparison of species functional responses across the heatwave.
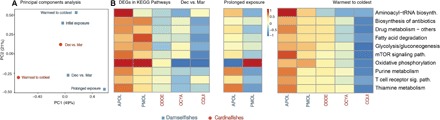
(A) Principal Component Analysis of relative expression of KEGG functional pathways underlying the weighted gene correlation network analysis. (B) Heatmap representing the number of DEGs associated with specific KEGG pathways on 10 categories associated with heat stress, across different collection times for each species. Red colors represent higher numbers of DEGs in a KEGG pathway, while blue colors represent few DEGs. For (A) and (B), “Initial exposure” represents the comparison between fish collected in December and February; “Dec vs. Mar” is the comparison of samples from December and March; “Prolonged exposure” represents the comparison between February and March; and “Warmest to coldest” is the comparison between March and July.
Readjustment from warmest to coldest temperatures
Between sampling points in March and July, the average water temperature decreased from 29.70°C (SD ±0.53 ° C) to 24.96°C (SD ±0.24 ° C; Fig. 1B). However, the GBR was considerably warmer in July 2016 when compared with previous winters (Fig. 1B). Contrasting the fishes’ responses across this ~5°C decrease resulted in a high number of DEGs for damselfishes: 3855 for A. polyacanthus and 2227 for P. moluccensis. The molecular response in the liver showed enrichment for functions associated to detoxification, immunity, mitochondrial activity, lipid synthesis, regulation of apoptosis, and oxidation-reduction process (red module; fig. S11 and data file S20). In addition, P. moluccensis showed increased activity for functions such as aerobic respiration, energy storage, and antioxidant activity, none of which were observed in A. polyacanthus for that comparison.
For cardinalfishes sampled between March and July, the gene network analysis showed enrichment for mitochondrial electron transport, Wnt signaling, and T cell receptor signaling pathways (black module; fig. S6 and data file S21). The comparison between the hottest and coldest months for O. cyanosoma and O. doederleini resulted in more than 1500 DEGs, many of which were associated with mitochondrial activity and lipid metabolism (data file S22). In addition, O. cyanosoma showed increased expression of genes related to detoxification and gametogenesis, while O. doederleini had activation of genes related to oxidoreductase activity and response to leptin. Across all comparisons in our study, the lowest number of DEGs was for C. quinquelineatus (466 DEGs; Fig. 2B). These genes are mainly involved in metabolism of peptides and synthesis of organonitrogen compounds (data file S23). This indicates that C. quinquelineatus exhibits less variation in gene expression than any other species in this study, even when facing the substantial change in conditions from the hottest to the coldest months (Fig. 3B).
The change from heatwave conditions in March to winter conditions in July was the only comparison with overlapping DEGs for all five species. Fifteen genes overlapped, of which five are expressed at higher levels in July including functions such as oxidative stress–induced survival signaling and transport of glucose (data file S24). The 11 common genes with decreased expression in July are mainly involved in functions such as translation (ribosomal proteins), alternative splicing, and mitochondrial respiration.
DISCUSSION
Our study shows how marine heatwaves, such as the one experienced in the austral summer of 2015–2016 in the northern GBR, can have direct consequences on the molecular response and physiology of coral reef fishes. Gene expression is a powerful tool to survey physiological processes and limitations in the face of an environmental stressor, and a plastic transcriptional response can be indicative of thermal tolerance (25). Hence, we used liver transcriptomes to evaluate the reaction of fishes across multiple sampling times during the heatwave, sampling five fish species of two different families to gather a broad multispecies perspective. The largest difference in gene expression was observed between different fish species, followed by divergence between individuals collected in different months and temperatures. This pattern is concurrent with our analysis of gene expression variance, which showed significant divergence in expression for genes involved in the stress response and enzymatic activity, between the species in this study. However, our analyses also indicate that interspecific differences in liver gene expression are maintained when fishes are subject to acute thermal stress. As such, we saw no overlap of DEGs across all species in the comparisons associated with the initial stages of warming (i.e., increase in temperature from December to February or March; ~1.5°C difference). Overlap of all species was only observed in the comparison between the hottest and the coldest months (March and July; ~5°C difference; 15 genes). To the best of our knowledge, differences in gene expression among closely related species in the context of a natural stressor have rarely been studied in wild populations, and on the basis of our results, it is clear that impacts of a temperature increase will depend on species-specific physiological tolerance and behavior. Although gene expression was largely species specific, we did see a phylogenetic signal where damselfishes exhibited a pattern of gene expression defined by the different collection points, which was less apparent for cardinalfishes.
We found a gradient of molecular reactions among the five studied species, from highly reactive with large changes in gene expression throughout the heatwave (A. polyacanthus) to an intermediate response (P. moluccensis, O. cyanosoma, and O. doederleini) to less reactive (C. quinquelineatus). The measures of change in resting oxygen uptake with increasing temperature are also species specific, but both lines of evidence only agree on the extreme responses. Thus, the most responsive species molecularly, A. polyacanthus, also shows significant changes in aerobic scope under similar thermal conditions (2, 22) and showed one of the highest Q10 for MO2Rest in our analysis. Meanwhile, the least reactive species in gene expression analysis, C. quinquelineatus, did not show a significant decrease in aerobic scope (16) and showed the lowest Q10 for MO2Rest across this temperature range in laboratory experiments (15). The relationship between oxygen uptake and molecular response is less clear for the species with intermediate molecular response. The damselfish P. moluccensis and the cardinalfish O. doederleini both showed comparable changes in oxygen uptake at warming. However, the former showed activation of a great number of genes associated with the electron-transport chain and mitochondrial activity as a result of temperature increase; yet, this molecular activation was not observed in the latter. The clearest discrepancy between measurements was for O. cyanosoma, because this species showed the largest Q10 for MO2Rest between 29° and 31°C but showed an intermediate molecular response with no activation of functions related to aerobic metabolism. Overall, the combined phenotypic and molecular evidence shows that different tropical fishes respond to warming in different ways, as indicated by both oxygen uptake and gene expression. Moreover, there is little concordance between these two measurements for species with intermediate responses, suggesting that measures of oxygen uptake (i.e., MO2Rest and aerobic scope), by themselves, may be inadequate indicators of the overall physiological responses of wild populations of reef fish exposed to elevated temperatures (1, 26).
In general, the molecular response of both species of damselfishes showed clear activation of molecular pathways associated to aerobic metabolism and cellular stress response, a pattern not observed in cardinalfishes. There is no clear indicator of what could be driving this pattern; yet, this observation could be associated to the particular life history traits of the analyzed species. For example, damselfishes are diurnal and feed over coral and rubble habitats by day (27), whereas cardinalfishes are nocturnal and shelter in the reef matrix by day, dispersing over a variety of habitats to feed at night (28). Furthermore, studies have suggested that there is a correlation between thermal tolerance and geographic range, as species with broader physiological niches can occupy larger areas (29). Our study partly supports this observation, as the least responsive species, C. quinquelineatus, has a much broader distribution [from Red Sea and Eastern Coast of Africa to the Pitcairn islands in the Central Pacific; (30)] than the species with the highest molecular response, A. polyacanthus [Indonesia, Philippines, northeast Australia, and Melanesia; (30)]. This relationship, however, is less clear for the species with intermediate responses, which also have relatively broad distributions throughout the Indo-West Pacific. Furthermore, multiple studies indicate that geographic range is not always a reliable predictor of thermal tolerance (31). Another factor that could be playing a role is the developmental differences between groups, as the most responsive species is one of the few coral reef fishes with direct development, where conditions experienced by the parents can directly influence the tolerance of the next generation (19).
The unique responses of the evaluated species were particularly apparent during the onset of the heatwave, where the reaction in damselfishes involved a change in the transcriptional program of genes associated with cholesterol metabolism. Previous studies have shown changes in cholesterol metabolism during heat stress in P. moluccensis (32) and relate change in cholesterol concentration at different temperatures to liver metabolism (33). Thus, activation of cholesterol metabolism (here only found in damselfishes) is important in the maintenance of cellular metabolism and not exclusively due to dietary changes. Meanwhile, specific responses in cardinalfishes included the up-regulation of CSDE1 in December (O. cyanosoma and O. doederleini) and July (O. doederleini). This highly conserved RNA binding protein has been associated to cellular growth in zebrafishes exposed to cold conditions [17°C; (34)], suggesting that conditions in July (austral winter) had an effect on the cellular metabolism of cardinalfishes. Furthermore, all cardinalfishes showed significant up-regulation of DEPDC5 in March, a negative regulator of the signaling pathway mTOR complex that is differentially expressed in zebrafish exposed to heat stress (35). Another response only found in cardinalfishes was the activation of the complement system during the heatwave, an important component of the innate immune response that promotes inflammation during cellular stress (36).
Despite the lack of shared differential expression for specific genes, multiple functional pathways did overlap across the different species. At warm temperatures, damselfishes showed activation of molecular functions such as immune response, mitochondrial activity, and toxin metabolism for most comparisons. Cardinalfishes collected in March also showed activation of these pathways when compared with samples from colder months (December for O. doederleini and July for both O. doederleini and O. cyanosoma), albeit less prominently. The only species that did not show activation of similar functional responses was C. quinquelineatus. These results are consistent with previous laboratory studies in tropical fishes, where elevated water temperatures led to higher energetic demands, causing an increase in the resting oxygen uptake rates and reduction in the aerobic scope (15, 19). In turn, increased oxygen uptake leads to up-regulation of genes associated with mitochondrial activity, immune response, and toxin metabolism in the liver (19, 20), a link that is well established in both terrestrial and aquatic organisms (1). The side effects of increased oxygen uptake rates include the production of reactive oxygen species and metabolic waste, which leads to up-regulation of pathways associated with cellular stress response (37). This relationship of stress response with warming conditions has been observed multiple times in both marine and freshwater fishes (38). Hence, there were some similarities in the functional response to warming for both damselfishes and cardinalfishes (with the exception of C. quinquelineatus), and this general response coincides with previous studies of marine fishes in captivity, including A. polyacanthus (19, 20), even though the relative magnitude of changes in gene expression and MO2Rest did not exhibit close concordance among the five species.
For the two damselfishes, it was possible to contrast patterns of gene expression during the initial stages of the heatwave (February) and the extended exposure to warm waters (March). This comparison revealed large differences in the transcriptional program between two time points (separated by approximately 4 weeks) with comparable temperatures, emphasizing the importance of considering the duration of warming when evaluating molecular responses. For example, February showed higher levels of expression for HSPs when compared with March samples, for both species of damselfishes. These genes are associated with proper folding of proteins when encountering heat stress and are typically up-regulated during the initial stages of warming, but their activation is metabolically costly and not maintained for extended periods of times (39). Similarly, laboratory experiments in A. polyacanthus showed differential expression for four HSPs at elevated temperatures (40), yet their activation was not maintained over long-term exposure to warm conditions (20). These results highlight the importance of temporal scales in the response to warming.
The comparison between fish collected in March and July is where we found the only overlap in differential gene expression between all five species (15 genes), suggesting a common mechanism of readjustment from the warmest to the coldest conditions. This common response included the deactivation of mitochondrial genes associated with the electron transport chain. In March, all species also showed up-regulation of the gene encoding the transcriptional regulator chromodomain helicase DNA binding protein 7 (COMD7), an inactivator of the nuclear factor κB complex, a protein influencing cell cycle regulation during stress (41). Previous studies on warm- and cold-water acclimation in zebrafish suggest that the most drastic response to stress will be found for the temperature that is close to the maximum tolerance of each species (42). Thus, it is possible that this common response was triggered by the readjustment to temperatures closer to their optimal range, leading to decreased oxygen uptake rates at lower temperatures (i.e., reduced mitochondrial activity) and recovery after cellular damage produced during the heatwave (i.e., deactivation of the cellular stress response). Nevertheless, these findings should be interpreted with caution, because this large change in temperature between the two points (~5°C) not only is a readjustment after the heatwave but also represents a change from summer to winter conditions.
The major caveat of this study is that we lack a reference for the measures of liver gene expression throughout nonheatwave years. This study was initiated late in 2015 in response to the unprecedented heatwave conditions that oceanographic models projected would occur on the northern GBR during the austral summer of 2015–2016. Consequently, we did not have a preestablished baseline of summer gene expression patterns for the study species. Ideally, we would have a complete understanding of how liver gene expression fluctuates throughout the year for the five analyzed species, as liver gene expression can be influenced by multiple other environmental and biological factors [e.g., reproduction, diet (43)]. Furthermore, heatwave conditions continued into the summer of 2016–2017, rendering this unsuitable for summer baseline sampling. One additional challenge is that seasonal food availability and trophic changes associated with the heatwave (i.e., increase in algal cover, J. L. Rummer, personal observation) could have influenced the patterns of gene expression we observed. We detected differences in fatty acid metabolism, glucose transport, and cholesterol, which could be associated with dietary change, but it remains unclear whether these changes also occur between seasons during regular years. Unfortunately, obtaining a “control” year was logistically impossible because we could not predict the heatwave conditions sufficiently in advance, and due to an already very ambitious sampling scheme for species with no previous genomic resources (with the exception of A. polyacanthus). Despite these limitations, the functions of genes that showed expression changes over the duration of the heatwave concur with previous laboratory experiments on A. polyacanthus, one of the most studied tropical marine fish species in the context of global change (19, 20).
One of the central questions in the study of ecological genomics is the relative importance of phenotypic plasticity with respect to divergent selection. The analyses of differential expression show that within-species plasticity plays an important role in the response to acute warming events. Yet, this multispecies dataset allowed us to confirm that differences between species also play a role in the response to acute warming, as the analysis of expression variance revealed a considerable number of genes associated to cellular stress response with elevated divergence between species. Thus, our results confirm that both mechanisms are important in tropical marine fishes, depending on the time scale: Plasticity aids in the acute thermal response of heatwaves, but as these events become more frequent, a heritable signal of expression could lead species toward new fitness peaks (44).
This is the first time that gene expression patterns have been directly evaluated in wild fish populations during a marine heatwave. Using liver transcriptomes, we demonstrated that there is a species-specific molecular response associated with temperature increases in the wild, which appears to be influenced by both the intensity as well as the duration of warming. We detected a clear signal of activation of molecular pathways associated with increased oxygen uptake, activation of the electron-transport chain, and corresponding cellular stress responses, all of which have been previously linked to temperature increase in multiple species. However, comparisons with oxygen uptake rates indicate that these laboratory measures, by themselves, do not provide a complete picture of how marine fishes respond to heatwave conditions, as there was little concordance between these estimates and patterns of gene expression of wild fish samples. Questions remain as to how repeated episodes of acute warming could influence the condition and fitness of fishes and how adaptation will influence long-term response. Considering that marine heatwaves are increasing in frequency, duration, and intensity (5, 7), our results highlight the importance of recognizing the impacts of extreme temperature events on reef fishes in conjunction with the average warming currently being caused by climate change.
MATERIALS AND METHODS
Sampling and sample preparation
Fish were collected in front of Palfrey Island, near Lizard Island in the northern GBR, Australia (14°41′39.1″S, 145°27′05.3″E). We collected samples at four different time points: before the heatwave (8 to 10 December 2015), at the beginning of the heatwave (20 to 22 February 2016), during the extended period of warming (18 to 20 March 2016), and during the Austral winter (18 to 20 July 2016; Fig. 1B). Water temperature records from 2010 to 2016 were obtained from a sensor at 0.6 m at Lizard Island, operated by the Australian Institute of Marine Science (http://weather.aims.gov.au/#/station/1166; Fig. 1B). Average temperatures for all days in the calendar month were as follows: December, 27.81°C (SD±0.45°C; summer); February, 29.55°C (SD ±0.69°C; summer); March 2016, 29.70°C (SD ±0.61°C; fall); July 2016, 24.96°C (SD ±0.28°C; winter). Average temperatures 7 days before sampling were as follows: December 2015, 27.50°C (SD ±0.09°C); February 2016, 29.60°C (SD ±0.61°C); March 2016, 29.50°C (SD ±0.24°C); July 2016, 24.72°C (SD ±0.13°C).
We collected two species of damselfishes [family Pomacentridae; the spiny chromis (A. polyacanthus) and the lemon damselfish (P. moluccensis)] and three species of cardinalfishes [family Apogonidae; the five-lined cardinalfish (C. quinquelineatus), the yellow-striped cardinalfish (O. cyanosoma), and Doederlein’s cardinalfish (O. doederleini; Fig. 1A]. Cardinalfish samples collected in February did not yield good-quality RNA, making it impossible to develop RNA sequencing libraries. Thus, for the three cardinalfishes, collection points were December, March, and July. For each collection point, three to five fish were collected and sequenced for each of the analyzed species (data file S25).
Fish were collected using clove oil anesthetic and hand nets on scuba, and they were euthanized by severing the spinal cord once transferred to the boat (James Cook University Animal Ethics approval A2408). Liver tissues were dissected out immediately on the boat, snap frozen in liquid nitrogen, and stored in −80°C until processing. Total RNA was extracted by homogenizing the whole liver for 1 min in a Fisherbrand Bead Beater with single-use silicon beads and using up to 30 mg in RNeasy Mini Kits (Qiagen). DNA contamination was removed with DNAse I (Qiagen) treatment for 15 min in a column, following the manufacturer’s instructions. RNA quality was evaluated with an Agilent Bioanalyzer. Paired-end fragments of 150 base pairs were sequenced at Macrogen, South Korea, with an Illumina HiSeq 4000. Ten individuals were sequenced per Illumina lane, and samples from species and seasons were randomly assigned to different lanes to avoid any potential biases during sequencing.
To compare the molecular response of the analyzed fishes with their phenotypic reaction, values of resting oxygen uptake (MO2Rest) at 29°C (T1) and 31°C (T2) were used to calculated the temperature quotient (Q10) (15, 16). The Q10 provides a standardized way to compare the change in MO2Rest with increasing temperature across different species. By definition, biochemical rate functions should double for every 10°C increase in temperature (i.e., Q10 = 2), and deviations from this indicate thermal independence (i.e., values lower than 2) or dependence (i.e., values higher than 2). The values of Q10 were calculated with the equation Q10 (MO2Rest2/MO2Rest1)[10/T2 − T1] (16). Only measures between 29° and 31°C are presented in this comparison, as these resemble the temperatures reported for the 2016 heatwave. With the exception of samples of C. quinquelineatus (Nago Island, Papua New Guinea), all values represent samples from Lizard Island, Australia.
Transcriptome assembly, annotation, and ortholog selection
The RNA sequencing reads were quality trimmed with Trimmomatic v0.33 (45) by using the parameters “ILLUMINACLIP:TruSeq3-PE-2.fa:2:30:10, LEADING:4, TRAILING:3, SLIDINGWINDOW:4:15, MINLEN:40,” retaining 3.1 billion high-quality reads. Putative contamination was removed with Kraken 0.10.6 (46), which resulted in the removal of 3.3% of all reads. The complete table of read numbers per individual before and after trimming is available in data file S26. The remaining 484 million reads for P. moluccensis, 457 million reads for A. polyacanthus, 437 million reads for O. cyanosoma, 313 million reads for O. doederleini, and 421 million reads for O. quinquelineatus were used to assemble species-specific transcriptomes with Trinity v2.4.0 (47) using default parameters. The de novo assemblies of transcriptomes with high-quality reads resulted in 569,734 contigs for P. moluccensis, 521,812 contigs for A. polyacanthus, 401,607 contigs for O. doederleini, 2,729,489 contigs for O. cyanosoma, and 493,866 contigs for C. quinquelineatus (fig. S1). This large number of transcripts was reduced using TransDecoder v3.0.1 (48) with default parameters to retain only those contigs that contained predicted open reading frames (fig. S1). The percentage of assembled contigs that passed open reading frame filtering were 24% for P. moluccensis, 32% for A. polyacanthus, 33% for O. doederleini, 9% for O. cyanosoma, and 28% for C. quinquelineatus. The average number of transcripts for orthologous gene predictions was 166,143 contigs (±51,099). The completeness of the de novo transcriptome assemblies was assessed by determining the number of missing benchmarking universal single-copy orthologous (BUSCO) genes with BUSCO 2.0 (49) using the Actinopterygii database (data file S27 and fig. S12) and ranged from 13.5% (A. polyacanthus) to 16.3% (O. doederleini), while the fraction of complete genes ranged from 80.1% (A. polyacanthus) to 75.2% (C. quinquelineatus) in the primary Trinity assemblies.
To be able to compare the molecular response between different species, we first grouped the protein sequences predicted by TransDecoder into clusters of orthologous genes across the five species using OrthoFinderv1.1.2 (50) with default parameters. This initial assessment resulted in 57,396 groups, which, on average, contained 10.5 transcripts. These groups were further filtered to keep only groups that contained at least one transcript from each of the five species, resulting in 19,333 groups. This inclusive set of orthologs was annotated with the UniProt database (release 2017_06) using BLASTP (Blast+ v2.6.0). Only UniProt accessions that were one of the top 10 hits for each of the five species were retained, and the final UniProt ID was selected on the basis of the number of species that had it as a best match. These filtering steps resulted in 18,416 annotated orthologous groups. However, some groups showed large variation in the quality of matches to the selected UniProt ID across species. Thus, we removed ortholog groups where the average e value of the species was larger than e−30, resulting in the selection of 16,494 annotated orthologous groups across five species for further analysis. For each orthologous group, a representative protein sequence from each species was selected as the species-specific reference. This sequence was chosen on the basis of the best match to the selected UniProt ID, as determined first by the e value and then by the bit score. The final set of representative transcripts was then annotated with GO terms, KEGG, and InterPro pathways in Blast2GO (51).
To assess quality of the assembled contigs, an analysis with Transrate was performed for both the Trinity-assembled and TransDecoder-filtered transcripts, as well as the contigs retained after filtering of orthologs with OrthoFinder. The results showed that the contigs representing the orthologous groups (i.e., those used as reference for mapping) ranged between 83 and 93% good-quality contigs (data file S28).
Gene expression analyses
Gene expression levels were obtained by mapping the high-quality reads against the representative transcript sequence (data file S29) obtained in the previous orthologous gene filtering step with Bowtie 2 v2.2.9 (52) by using the “--highly-sensitive” parameter and analyzing the alignment files with RSEM v1.2.26 (default settings). Mapping success of high-quality reads back to the representative ortholog sequences was, on average, 29.8% (±10%). Gene expression was then analyzed via two different methods: gene network analysis and differential gene expression analysis. For the weighted gene correlation network analysis, the package WGCNA (53) was used, and differential expression was analyzed using the program DESeq2 version 1.18 (54), both in R.
WGCNAs were performed using all biological replicates of all species, as well as subsets that included only the 2 damselfish species (with 4 time points each), and only the cardinalfish species (with 3 time points each). WGCNA requires large numbers of biological replicates to be powerful; therefore, individual species analysis (ranging from 13 to 20 biological samples) were inconclusive and not included. Hierarchical clustering was performed with all datasets for outlier detection and clustering of samples into dendrograms. Threshold power tests for the WGCNAs were performed, and modules were computed (parameters = blockwiseModules; power = 12; mergeCutHeight = 0.3; min ModuleSize = 30; TOMType = signed). Resulting modules were exported (data file S30) and annotated. Correlations were calculated with provided trait data. Traits of each biological replicate were (i) the species or the family (Pomacentridae or Apagonidae) to correlate with a species-specific or phylogenetic signal and (ii) 0 or 1 values for collections in December, February, March, or July, as well as February and March combined to identify correlations with length of exposure to warm conditions. Heatmaps with module-trait relationships were elaborated, modules with significant trait correlations were further investigated, and module gene lists were used to perform functional enrichment tests (see below).
Differential gene expression analyses were performed independently for each of the five species, contrasting samples of damselfishes collected in four different months and cardinalfishes collected on three different months. We performed a likelihood ratio test with DESeq2 for each species to identify DEGs between samples collected across all time points. This analysis allows discerning genes that are differentially expressed between all months (i.e., differences in temperature) from those differentiated within a particular month (i.e., individual variance within samples of a month). Only genes with a multiple test corrected significance cutoff of P < 0.01 (i.e., adjusted P value) for at least one of the thermal treatments were considered to be differentially expressed. For all individuals of each species, the variance-stabilized data of the raw read counts of the DEG was extracted with the package Vegan (55) to make a principal coordinate analysis. To determine the DEGs between samples of two specific months, pairwise analyses were performed with the “Contrast” function of DESeq2. At least four samples were required to replace an outlier of expression for a particular gene, and if multiple outliers were detected on the basis of Cook’s distances, that particular gene would not be considered for downstream analyses.
Functional enrichment and pathway analyses were performed for all gene sets of interest (either WGCNA modules or DEGs). Enrichment of GO categories for each pairwise comparison was done with a Fisher’s exact test using Blast2GO (51). For this, a list of genes of interest was contrasted to the total list of genes in the analysis (16,494 for all species). The false discovery rate of the analysis was set to 5%, and the resulting GO terms were summarized with the option of “Reduce to most specific terms” of Blast2GO. The resulting GO terms were divided by their corresponding domains: biological process, cellular component, and metabolic function, according to their function. To understand the relevance of functional pathways across time points, we annotated the genes in the WGCNA clusters and the DEGs to the KEGG Pathway Database (56). We estimated the proportion of sequences that correspond to a particular KEGG Pathway, and we visualized their similarity across collection time points with a principal components analysis. In addition, the numbers of DEGs that belong to a particular pathway are presented in a heatmap using the R package pheatmap 1.0.10.
To understand the variation in the response to warming from a phylogenetic perspective, we performed, an analysis of EVE (23), based on a maximum likelihood (ML) tree of the five species. For this, the assembled contigs were translated into protein coding sequences using TransDecoder, and OrthoFinder was used to define orthogroups of transcripts based on the protein sequences between all species, generating a concatenated multiple sequence alignment of 1142 orthogroups (268,959 base pairs, 31.01% gaps, and 84.53% invariant sites). The program RAxML-NG (57) was used to infer the best ML tree based on 50 random and 50 parsimony-based starting tree topologies and the LG + G4m model (LG substitution matrix with discrete GAMMA N categories, ML estimate of alpha). The 1% threshold of nonparametric bootstrapping was reached at 1700 iterations. Only one unique topology was detected among all the ML trees (fig. S13). Furthermore, the topological Robinson-Foulds distance of the obtained candidate trees showed that average absolute and relative distances were zero, suggesting that an optimal tree topology was reached.
We used the phylogenetic tree and normalized read counts of all samples from DEseq2 (16,494 genes) to run the EVE model. Postprocessing of the obtained file including all theta values was performed following (58) to obtain P values for each expressed transcript. The false discovery rate was used to correct P values for multiple testing. Significant genes (false discovery rate < 0.05) were those that showed similar patterns of expression within a lineage but were highly differentiated between lineages (i.e., diverged expression profiles), as well as genes with higher differences in expression within than between lineages (i.e., diverse expression profiles). An analysis of enrichment of GO categories was done for genes with significantly diverged expression profiles using a Fisher’s exact test in Blast2GO (51).
Supplementary Material
Acknowledgments
We thank J. Johansen, T. Nay, and M. McCormick for help with the sampling, as well as the Lizard Island Research Station for logistical support. We also thank A. Budd for help in the molecular laboratory as well as the Integrative Systems Biology Laboratory (KAUST). Figures 1 and 2 were created by H. Hwang, scientific illustrator at the KAUST. Funding: This study was supported by the Office of Competitive Research Funds OSR-2015-CRG4-2541 from the King Abdullah University of Science and Technology (KAUST) (to T.R., P.L.M., C.S., and J.L.R.), the Australian Research Council (ARC), and the ARC Centre of Excellence for Coral Reef Studies (to P.L.M. and J.L.R.). Author contributions: P.L.M., J.L.R., H.D.V., and B.J.M.A. initiated and managed the fish sample collection. C.S. extracted the RNA and with M.A.B. prepared the samples for sequencing. R.L. assembled the transcriptomes and mapped the reads. D.J.L. assigned the ortholog groups. M.A.B. and C.S. analyzed and interpreted the transcriptome expression data. M.A.B., C.S., P.L.M., and T.R. wrote the paper. All authors read, revised, and approved the final manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the manuscript are present in the paper and/or the Supplementary Materials. Sequencing data are archived in NCBI BioProject PRJNA489934 and the sequences read archive (SRA) SRP160415. The assembled sequences used for the ML tree are available at https://github.com/roblehmann/marineHeatwave. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/12/eaay3423/DC1
Fig. S1. Number of transcripts obtained after de novo transcriptome assembly with Trinity (gray) and retained transcripts after predicting open reading frames with TransDecoder (black).
Fig. S2. Normalized read counts for six of the genes that showed divergent expression (i.e., larger differences between species) in the analysis of EVE.
Fig. S3. Venn diagram showing the overlap of differential expressed genes in the pairwise comparisons between December and March and December versus July for the five species of interest.
Fig. S4. Module-trait correlation matrix for all species showing correlation value with P value in brackets for each module and trait.
Fig. S5. Module-trait correlation matrix for A. polyacanthus and P. moluccensis (damselfish species).
Fig. S6. Module-trait correlation matrix for specimen of all three cardinalfish species (C. quinquelineatus, O. doederleini, and O. cyanosoma).
Fig. S7. Principal Coordinate Analysis for damselfishes, based on liver gene expression.
Fig. S8. Eigenvalue of expression data for each biological replicate in the red module for cardinalfishes.
Fig. S9. Heatmaps for the expression of heat-shock proteins in damselfishes.
Fig. S10. Eigenvalue of expression data for each biological replicate in the green module for damselfishes.
Fig. S11. Eigenvalue of expression data for each biological replicate in the red module for damselfishes.
Fig. S12. Transcriptome completeness estimates for all five species with BUSCO.
Fig. S13. ML tree showing the relationship between the five species analyzed in this study, generated with RAXML.
Data file S1. Final list of orthologous genes shared across the five sampled species and their corresponding annotation.
Data file S2. Significantly diverged genes (i.e. higher difference between species) in the Expression Variance and Evolution (EVE) analysis for all samples collected in our study, and their corresponding annotation.
Data file S3. Enriched Gene Ontology Categories for the significantly diverged genes (i.e. higher difference between species) in the Expression Variance and Evolution (EVE) analysis.
Data file S4. Genes with significant plasticity (i.e. higher difference within individuals of a species) in the Expression Variance and Evolution (EVE) analysis, for all samples collected in our study.
Data file S5. Differentially expressed genes (DEGs) between fish collected at different months for the five focal species of the study. The dashes represent comparisons that were not possible, as samples from February for three of the species (C. quinquelineatus, O. cyanosoma, O. doederleini) were not available.
Data file S6. Functional enrichment for Acanthochromis polyacanthus, based on the DEGs between samples from December and February.
Data file S7. Functional enrichment of GO categories for Pomacentrus moluccensis, based on the DEGs between samples from December and February.
Data file S8. Overlap in the enriched Gene Onthology categories based on the differentially expressed genes of A. polycanthus and P. moluccensis between samples from December and March.
Data file S9. Functional enrichment of GO terms for the two species of damselfishes, based on genes belonging to the correlated network “BLUE”.
Data file S10. Functional enrichment of GO terms for the two species of damselfishes, based on genes belonging to the correlated network “YELLOW”.
Data file S11. Significantly enriched Gene Ontology terms between December and March for O. cyanosoma, O. doederleini and C. quinquelineatus, based on the analysis of differential expression.
Data file S12. Overlapping differentially expressed genes between the three species of cardinalfishes when comparing samples collected in December and March.
Data file S13. Significantly enriched GO terms for the “CYAN” gene module, for March samples of three species of cardinalfishes.
Data file S14. Significantly enriched Gene Ontology processes for the “RED” gene module for March samples of three species of cardinalfishes.
Data file S15. Average Resting Oxygen consumption (MO2Rest), Standard Deviation (SD in parenthesis), number of samples and temperature quotients (Q10), from studies by Nilsson et al. (2009) and Rummer et al. (2014; for C. quinquelineatus). Estimates for four of the species correspond to Lizard Island populations (i.e. same location as genetic analyses in the main text), while the samples of C. quinquelineatus correspond to Nago Island, Papua New Guinea. Only measures between 29°C and 31°C are presented in this comparison, as these resemble the temperatures during the 2016 heatwave. The values of Q10 were calculated with the equation Q10 = (MO2Rest2/MO2Rest1)[10/T2-T1].
Data file S16. Overlap in the enriched Gene Onthology categories based on the differentially expressed genes of A. polyacanthus and P. moluccensis between samples collected in February and March.
Data file S17. Heat shock proteins that were differentially expressed in the comparison of samples from February and March for A. polyacanthus and P. moluccensis.
Data file S18. Significantly enriched GO terms for the “GREEN” gene module, corresponding to samples of damselfishes collected in March.
Data file S19. Overlapping differentially expressed genes that were exclusively found in the comparisons of prolonged exposure (February vs. March) in A. polyacanthus and P. moluccensis.
Data file S20. Significantly enriched GO terms for the “RED” gene module, resulting from the contrast of damselfishes collected in March and July.
Data file S21. Significantly enriched GO terms for the “BLACK” gene module, corresponding to samples of cardinalfishes collected in March.
Data file S22. Differentially expressed genes between samples of March and July for O. cyanosoma (OCYA) and O. doederleini (ODOE).
Data file S23. Differentially expressed genes between samples of March and July of C. quinquelineatus.
Data file S24. Common differentially expressed genes across all five species, when comparing March and July samples.
Data file S25. Table of collected individuals, dates and fish sizes.
Data file S26. Number of reads per individual before and after trimming and decontamination.
Data file S27. Statistics of de novo assemblies of transcriptomes determined with BUSCO.
Data file S28. Results from the quality assessment using the program Transrate, for contigs assembled with Trinity and summarized with Transdecoder (A) and the orthologous sequences (B).
Data file S29. Representative transcript for each species in our study, for all orthologous genes.
Data file S30. Number of transcripts in WGCNA modules.
REFERENCES AND NOTES
- 1.Schulte P. M., The effects of temperature on aerobic metabolism: Towards a mechanistic understanding of the responses of ectotherms to a changing environment. J. Exp. Biol. 218, 1856–1866 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Smale D. A., Wernberg T., Oliver E. C. J., Thomsen M., Harvey B. P., Straub S. C., Burrows M. T., Alexander L. V., Benthuysen J. A., Donat M. G., Feng M., Hobday A. J., Holbrook N. J., Perkins-Kirkpatrick S. E., Scannell H. A., Gupta A. S., Payne B. L., Moore P. J., Marine heatwaves threaten global biodiversity and the provision of ecosystem services. Nat. Clim. Chang. 9, 306–312 (2019). [Google Scholar]
- 3.Poloczanska E. S., Brown C. J., Sydeman W. J., Kiessling W., Schoeman D. S., Moore P. J., Brander K., Bruno J. F., Buckley L. B., Burrows M. T., Duarte C. M., Halpern B. S., Holding J., Kappel C. V., O’Connor M. I., Pandolfi J. M., Parmesan C., Schwing F., Thompson S. A., Richardson A. J., Global imprint of climate change on marine life. Nat. Clim. Chang. 3, 919–925 (2013). [Google Scholar]
- 4.Sunday J. M., Pecl G. T., Frusher S., Hobday A. J., Hill N., Holbrook N. J., Edgar G. J., Stuart-Smith R., Barrett N., Wernberg T., Watson R. A., Smale D. A., Fulton E. A., Slawinski D., Feng M., Radford B. T., Thompson P. A., Bates A. E., Species traits and climate velocity explain geographic range shifts in an ocean-warming hotspot. Ecol. Lett. 18, 944–953 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Oliver E. C. J., Donat M. G., Burrows M. T., Moore P. J., Smale D. A., Alexander L. V., Benthuysen J. A., Feng M., Gupta A. S., Hobday A. J., Holbrook N. J., Perkins-Kirkpatrick S. E., Scannell H. A., Straub S. C., Wernberg T., Longer and more frequent marine heatwaves over the past century. Nat. Commun. 9, 1324 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.L’Heureux M. L., Takahashi K., Watkins A. B., Barnston A. G., Becker E. J., Di Liberto T. E., Gamble F., Gottschalck J., Halpert M. S., Huang B., Mosquera-Vásquez K., Wittenberg A. T., Observing and predicting the 2015/16 El Niño. Bull. Am. Meteorol. Soc. 98, 1363–1382 (2017). [Google Scholar]
- 7.H. O. Pörtner et al., in IPCC, 2019: Summary for Policymakers. In: IPCC Special Report on the Ocean and Cryosphere in a Changing Climate.
- 8.Hughes T. P., Anderson K. D., Connolly S. R., Heron S. F., Kerry J. T., Lough J. M., Baird A. H., Baum J. K., Berumen M. L., Bridge T. C., Claar D. C., Eakin C. M., Gilmour J. P., Graham N. A. J., Harrison H., Hobbs J.-P. A., Hoey A. S., Hoogenboom M., Lowe R. J., McCulloch M. T., Pandolfi J. M., Pratchett M., Schoepf V., Torda G., Wilson S. K., Spatial and temporal patterns of mass bleaching of corals in the Anthropocene. Science 359, 80–83 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Munday P. L., Jones G. P., Pratchett M. S., Williams A. J., Climate change and the future for coral reef fishes. Fish Fish. 9, 261–285 (2008). [Google Scholar]
- 10.Pratchett M. S., Hoey A. S., Wilson S. K., Messmer V., Graham N. A. J., Changes in biodiversity and functioning of reef fish assemblages following coral bleaching and coral loss. Diversity 3, 424–452 (2011). [Google Scholar]
- 11.Keith S. A., Baird A. H., Hobbs J.-P. A., Woolsey E. S., Hoey A. S., Fadli N., Sanders N. J., Synchronous behavioural shifts in reef fishes linked to mass coral bleaching. Nat. Clim. Chang. 8, 986–991 (2018). [Google Scholar]
- 12.Pörtner H. O., Knust R., Climate Change affects marine fishes through the oxygen limitation of thermal tolerance. Science 315, 95–97 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Deutsch C. A., Tewksbury J. J., Huey R. B., Sheldon K. S., Ghalambor C. K., Haak D. C., Martin P. R., Impacts of climate warming on terrestrial ectotherms across latitude. Proc. Natl. Acad. Sci. U.S.A. 105, 6668–6672 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pörtner H.-O., Bock C., Mark F. C., Oxygen- and capacity-limited thermal tolerance: Bridging ecology and physiology. J. Exp. Biol. 220, 2685–2696 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Nilsson G. E., Crawley N., Lunde I. G., Munday P. L., Elevated temperature reduces the respiratory scope of coral reef fishes. Glob. Chang. Biol. 15, 1405–1412 (2009). [Google Scholar]
- 16.Rummer J. L., Couturier C. S., Stecyk J. A. W., Gardiner N. M., Kinch J. P., Nilsson G. E., Munday P. L., Life on the edge: Thermal optima for aerobic scope of equatorial reef fishes are close to current day temperatures. Glob. Chang. Biol. 20, 1055–1066 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Munday P. L., Kingsford M. J., O’Callaghan M., Donelson J. M., Elevated temperature restricts growth potential of the coral reef fish Acanthochromis polyacanthus. Coral Reefs 27, 927–931 (2008). [Google Scholar]
- 18.Johansen J. L., Jones G. P., Increasing ocean temperature reduces the metabolic performance and swimming ability of coral reef damselfishes. Glob. Chang. Biol. 17, 2971–2979 (2011). [Google Scholar]
- 19.Bernal M. A., Donelson J. M., Veilleux H. D., Ryu T., Munday P. L., Ravasi T., Phenotypic and molecular consequences of stepwise temperature increase across generations in a coral reef fish. Mol. Ecol. 27, 4516–4528 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Veilleux H. D., Ryu T., Donelson J. M., van Herwerden L., Seridi L., Ghosheh Y., Berumen M. L., Leggat W., Ravasi T., Munday P. L., Molecular processes of transgenerational acclimation to a warming ocean. Nat. Clim. Chang. 5, 1074–1078 (2015). [Google Scholar]
- 21.Pankhurst N. W., Munday P. L., Effects of climate change on fish reproduction and early life history stages. Mar. Freshw. Res. 62, 1015–1026 (2011). [Google Scholar]
- 22.Frédérich B., Sorenson L., Santini F., Slater G. J., Alfaro M. E., Iterative ecological radiation and convergence during the evolutionary history of damselfishes (Pomacentridae). Am. Nat. 181, 94–113 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Rohlfs R. V., Nielsen R., Phylogenetic ANOVA: The expression variance and evolution model for quantitative trait evolution. Syst. Biol. 64, 695–708 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eirin-Lopez J. M., Putnam H. M., Marine environmental epigenetics. Annu. Rev. Mar. Sci. 11, 335–368 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Evans T. G., Hofmann G. E., Defining the limits of physiological plasticity: How gene expression can assess and predict the consequences of ocean change. Philos. Trans. R. Soc. B Biol. Sci. 367, 1733–1745 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Norin T., Malte H., Clark T. D., Aerobic scope does not predict the performance of a tropical eurythermal fish at elevated temperatures. J. Exp. Biol. 217, 244–251 (2014). [DOI] [PubMed] [Google Scholar]
- 27.B. Frédérich, E. Parmentier, Biology of Damselfishes (CRC Press, 2015). [Google Scholar]
- 28.Marnane M. R., Bellwood D. R., Diet and nocturnal foraging in cardinalfishes (Apogonidae) at one tree reef, great barrier reef, Australia. Mar. Ecol. Prog. Ser. 231, 261–268 (2002). [Google Scholar]
- 29.Calosi P., Bilton D. T., Spicer J. I., Votier S. C., Atfield A., What determines a species’ geographical range? Thermal biology and latitudinal range size relationships in European diving beetles (Coleoptera: Dytiscidae). J. Anim. Ecol. 79, 194–204 (2010). [DOI] [PubMed] [Google Scholar]
- 30.R. Froese, D. Pauly, Search FishBase. FishBase (2019); www.fishbase.org.
- 31.Addo-Bediako A., Chown S. L., Gaston K. J., Thermal tolerance, climatic variability and latitude. Proc. Biol. Sci. 267, 739–745 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kassahn K. S., Crozier R. H., Ward A. C., Stone G., Caley M. J., From transcriptome to biological function: Environmental stress in an ectothermic vertebrate, the coral reef fish Pomacentrus moluccensis. BMC Genomics 8, 358 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wedemeyer G., Some physiological aspects of sublethal heat stress in the juvenile steelhead trout (Salmo gairdneri) and coho salmon (Oncorhynchus kisutch). J. Fish. Res. Board Can. 30, 831–834 (1973). [Google Scholar]
- 34.Kulkeaw K., Ishitani T., Kanemaru T., Fucharoen S., Sugiyama D., Cold exposure down-regulates zebrafish hematopoiesis. Biochem. Biophys. Res. Commun. 394, 859–864 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Long Y., Li L., Li Q., He X., Cui Z., Transcriptomic characterization of temperature stress responses in larval zebrafish. PLOS ONE 7, e37209 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holland M. C. H., Lambris J. D., The complement system in teleosts. Fish Shellfish Immunol. 12, 399–420 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Madeira D., Costa P. M., Vinagre C., Diniz M. S., When warming hits harder: Survival, cellular stress and thermal limits of Sparus aurata larvae under global change. Mar. Biol. 163, 91 (2016). [Google Scholar]
- 38.Makrinos D. L., Bowden T. J., Natural environmental impacts on teleost immune function. Fish Shellfish Immunol. 53, 50–57 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Feder M. E., Hofmann G. E., Heat-shock proteins, molecular chaperones, and the stress response: Evolutionary and ecological physiology. Annu. Rev. Physiol. 61, 243–282 (1999). [DOI] [PubMed] [Google Scholar]
- 40.Veilleux H. D., Ryu T., Donelson J. M., Ravasi T., Munday P. L., Molecular response to extreme summer temperatures differs between two Genetically differentiated populations of a coral reef fish. Front. Mar. Sci. 5, 349 (2018). [Google Scholar]
- 41.Burstein E., Hoberg J. E., Wilkinson A. S., Rumble J. M., Csomos R. A., Komarck C. M., Maine G. N., Wilkinson J. C., Mayo M. W., Duckett C. S., COMMD proteins, a novel family of structural and functional homologs of MURR1. J. Biol. Chem. 280, 22222–22232 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Vergauwen L., Benoot D., Blust R., Knapen D., Long-term warm or cold acclimation elicits a specific transcriptional response and affects energy metabolism in zebrafish. Comp. Biochem. Physiol. 157, 149–157 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Panserat S., Hortopan G. A., Plagnes-Juan E., Kolditz C., Lansard M., Skiba-Cassy S., Esquerré D., Geurden I., Médale F., Kaushik S., Corraze G., Differential gene expression after total replacement of dietary fish meal and fish oil by plant products in rainbow trout (Oncorhynchus mykiss) liver. Aquaculture 294, 123–131 (2009). [Google Scholar]
- 44.Munday P. L., Donelson J. M., Domingos J. A., Potential for adaptation to climate change in a coral reef fish. Glob. Chang. Biol. 23, 307–317 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wood D. E., Salzberg S. L., Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 15, R46 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., Chen Z., Mauceli E., Hacohen N., Gnirke A., Rhind N., di Palma F., Birren B. W., Nusbaum C., Lindblad-Toh K., Friedman N., Regev A., Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.A. Haas, B. Papanicolaou, Transdecoder (Find Coding Regions within Transcripts; http//transdecoder.github.io.
- 49.Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., Zdobnov E. M., BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Emms D. M., Kelly S., OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16, 157 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Conesa A., Götz S., García-Gómez J. M., Terol J., Talón M., Robles M., Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Langmead B., Trapnell C., Pop M., Salzberg S. L., Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Langfelder P., Horvath S., WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.J. Oksanen, F. G. Blanchet, M. Friendly, R. Kindt, P. Legendre, D. M. Glinn, P. R. Minchin, R. B. O’Hara, G. L. Simpson, P. Solymos, M. Henry H. Stevens, E. Szoecs, H. Wagner, Package “vegan” Title Community Ecology Package (2018); https://cran.ism.ac.jp/web/packages/vegan/vegan.pdf.
- 56.Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M., KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kozlov A. M., Darriba D., Flouri T., Morel B., Stamatakis A., RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35, 4453–4455 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brauer C. J., Unmack P. J., Beheregaray L. B., Comparative ecological transcriptomics and the contribution of gene expression to the evolutionary potential of a threatened fish. Mol. Ecol. 26, 6841–6856 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/12/eaay3423/DC1
Fig. S1. Number of transcripts obtained after de novo transcriptome assembly with Trinity (gray) and retained transcripts after predicting open reading frames with TransDecoder (black).
Fig. S2. Normalized read counts for six of the genes that showed divergent expression (i.e., larger differences between species) in the analysis of EVE.
Fig. S3. Venn diagram showing the overlap of differential expressed genes in the pairwise comparisons between December and March and December versus July for the five species of interest.
Fig. S4. Module-trait correlation matrix for all species showing correlation value with P value in brackets for each module and trait.
Fig. S5. Module-trait correlation matrix for A. polyacanthus and P. moluccensis (damselfish species).
Fig. S6. Module-trait correlation matrix for specimen of all three cardinalfish species (C. quinquelineatus, O. doederleini, and O. cyanosoma).
Fig. S7. Principal Coordinate Analysis for damselfishes, based on liver gene expression.
Fig. S8. Eigenvalue of expression data for each biological replicate in the red module for cardinalfishes.
Fig. S9. Heatmaps for the expression of heat-shock proteins in damselfishes.
Fig. S10. Eigenvalue of expression data for each biological replicate in the green module for damselfishes.
Fig. S11. Eigenvalue of expression data for each biological replicate in the red module for damselfishes.
Fig. S12. Transcriptome completeness estimates for all five species with BUSCO.
Fig. S13. ML tree showing the relationship between the five species analyzed in this study, generated with RAXML.
Data file S1. Final list of orthologous genes shared across the five sampled species and their corresponding annotation.
Data file S2. Significantly diverged genes (i.e. higher difference between species) in the Expression Variance and Evolution (EVE) analysis for all samples collected in our study, and their corresponding annotation.
Data file S3. Enriched Gene Ontology Categories for the significantly diverged genes (i.e. higher difference between species) in the Expression Variance and Evolution (EVE) analysis.
Data file S4. Genes with significant plasticity (i.e. higher difference within individuals of a species) in the Expression Variance and Evolution (EVE) analysis, for all samples collected in our study.
Data file S5. Differentially expressed genes (DEGs) between fish collected at different months for the five focal species of the study. The dashes represent comparisons that were not possible, as samples from February for three of the species (C. quinquelineatus, O. cyanosoma, O. doederleini) were not available.
Data file S6. Functional enrichment for Acanthochromis polyacanthus, based on the DEGs between samples from December and February.
Data file S7. Functional enrichment of GO categories for Pomacentrus moluccensis, based on the DEGs between samples from December and February.
Data file S8. Overlap in the enriched Gene Onthology categories based on the differentially expressed genes of A. polycanthus and P. moluccensis between samples from December and March.
Data file S9. Functional enrichment of GO terms for the two species of damselfishes, based on genes belonging to the correlated network “BLUE”.
Data file S10. Functional enrichment of GO terms for the two species of damselfishes, based on genes belonging to the correlated network “YELLOW”.
Data file S11. Significantly enriched Gene Ontology terms between December and March for O. cyanosoma, O. doederleini and C. quinquelineatus, based on the analysis of differential expression.
Data file S12. Overlapping differentially expressed genes between the three species of cardinalfishes when comparing samples collected in December and March.
Data file S13. Significantly enriched GO terms for the “CYAN” gene module, for March samples of three species of cardinalfishes.
Data file S14. Significantly enriched Gene Ontology processes for the “RED” gene module for March samples of three species of cardinalfishes.
Data file S15. Average Resting Oxygen consumption (MO2Rest), Standard Deviation (SD in parenthesis), number of samples and temperature quotients (Q10), from studies by Nilsson et al. (2009) and Rummer et al. (2014; for C. quinquelineatus). Estimates for four of the species correspond to Lizard Island populations (i.e. same location as genetic analyses in the main text), while the samples of C. quinquelineatus correspond to Nago Island, Papua New Guinea. Only measures between 29°C and 31°C are presented in this comparison, as these resemble the temperatures during the 2016 heatwave. The values of Q10 were calculated with the equation Q10 = (MO2Rest2/MO2Rest1)[10/T2-T1].
Data file S16. Overlap in the enriched Gene Onthology categories based on the differentially expressed genes of A. polyacanthus and P. moluccensis between samples collected in February and March.
Data file S17. Heat shock proteins that were differentially expressed in the comparison of samples from February and March for A. polyacanthus and P. moluccensis.
Data file S18. Significantly enriched GO terms for the “GREEN” gene module, corresponding to samples of damselfishes collected in March.
Data file S19. Overlapping differentially expressed genes that were exclusively found in the comparisons of prolonged exposure (February vs. March) in A. polyacanthus and P. moluccensis.
Data file S20. Significantly enriched GO terms for the “RED” gene module, resulting from the contrast of damselfishes collected in March and July.
Data file S21. Significantly enriched GO terms for the “BLACK” gene module, corresponding to samples of cardinalfishes collected in March.
Data file S22. Differentially expressed genes between samples of March and July for O. cyanosoma (OCYA) and O. doederleini (ODOE).
Data file S23. Differentially expressed genes between samples of March and July of C. quinquelineatus.
Data file S24. Common differentially expressed genes across all five species, when comparing March and July samples.
Data file S25. Table of collected individuals, dates and fish sizes.
Data file S26. Number of reads per individual before and after trimming and decontamination.
Data file S27. Statistics of de novo assemblies of transcriptomes determined with BUSCO.
Data file S28. Results from the quality assessment using the program Transrate, for contigs assembled with Trinity and summarized with Transdecoder (A) and the orthologous sequences (B).
Data file S29. Representative transcript for each species in our study, for all orthologous genes.
Data file S30. Number of transcripts in WGCNA modules.


