Abstract
Quantitative systems pharmacology (QSP) models are often implemented using a wide variety of technical workflows and methodologies. To facilitate reproducibility, transparency, portability, and reuse for QSP models, we have developed gQSPSim, a graphical user interface–based MATLAB application that performs key steps in QSP model development and analyses. The capabilities of gQSPSim include (i) model calibration using global and local optimization methods, (ii) development of virtual subjects to explore variability and uncertainty in the represented biology, and (iii) simulations of virtual populations for different interventions. gQSPSim works with SimBiology‐built models using components such as species, doses, variants, and rules. All functionalities are equipped with an interactive visualization interface and the ability to generate presentation‐ready figures. In addition, standardized gQSPSim sessions can be shared and saved for future extension and reuse. In this work, we demonstrate gQSPSim’s capabilities with a standard target‐mediated drug disposition model and a published model of anti‐proprotein convertase subtilisin/kexin type 9 (PCSK9) treatment of hypercholesterolemia.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
☑ Quantitative systems pharmacology (QSP) models are a powerful tool for gaining insight into pharmacological effects in a disease setting. However, they are frequently generated using a mixture of custom methods in a variety of programming languages, hindering collaboration and reproducibility.
WHAT QUESTION DID THIS STUDY ADDRESS?
☑ gQSPSim is designed to provide the means for transparent, reproducible, and portable QSP modeling by extending the capabilities of SimBiology.
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
☑ gQSPSim is the first interactive graphical user interface that provides the capability for calibration of QSP models to aggregated standardized data as well as the generation, simulation, interactive visualization, and statistical calibration of virtual subjects. All generated results are stored in Excel files for easy reference and modular input to each of core functionalities within gQSPSim.
HOW MIGHT THIS CHANGE DRUG DISCOVERY, DEVELOPMENT, AND/OR THERAPEUTICS?
☑ gQSPSim will greatly enhance the ability to share and reproduce original QSP models and workflows, thereby accelerating model development, reuse, and distribution. This is expected to facilitate activities across all stages of drug research and development.
Pharmaceutical researchers are increasingly exploring modeling approaches such as quantitative systems pharmacology (QSP) to address current challenges in drug development.1 QSP models of varying complexity and biological focus have been successfully used in drug development applications2, 3, 4, 5, 6 in recent years. As QSP continues to gain traction, there is an increasing need for standards and tools that facilitate the efficient execution, review, and dissemination of the developed models and workflows. Although conceptual frameworks for QSP workflows have been proposed that address key aspects such as uncertainty in complex models, biological variability, and more, there still remains a need for standardized tools to execute them. For example, Cheng et al. 7 recently published a QSP toolbox for use with MATLAB‐based (MathWorks, Natick, MA) models, which provides many of the core functionalities of the typical QSP workflow including optimization, virtual population development, and the ability to perform and visualize simulations. This set of scripts provides a valuable resource for QSP modelers that can help to standardize the modeling workflow. However, the users should be well‐versed in Matlab scripting and have a good understanding of the built‐in toolbox functions in order to effectively use the capabilities for their QSP projects. Additionally, the toolbox does not provide a graphical interface for performing tasks or visualizing results.
To address this technology gap, we developed the Genentech QSP Simulator (gQSPSim; Genentech Inc., South San Francisco, CA) application, a tool designed to facilitate the development, exploration, and distribution of QSP models using an interactive graphical environment. gQSPSim is based on MATLAB, one of the most commonly used platforms for QSP and pharmacokinetic/pharmacodynamic (PK/PD) modelling,8 along with the SimBiology toolbox, which offers user‐friendly model development and simulation capability for biological models. gQSPSim extends SimBiology capabilities by, among others, enabling streamlined calibration of QSP models simultaneously to data collected across multiple experimental conditions or cohorts, exploration of parameter uncertainty and biological variability via generation of virtual populations composed of individual virtual subjects, and the generation of robust model‐based predictions with virtual populations aided by interactive visualizations. As such, gQSPSim has been designed to support the core QSP workflow, and especially stages 3–5 (“Representing the Biology,” “Capturing Behaviors & Building Confidence,” and “Exploring Knowledge Gaps & Variability”) in the six‐stage QSP workflow described by Gadkar et al. 9
gQSPSim uses elements of the SimBiology models, including Doses, Variants, Rules, and Reactions, as optional inputs to configure task‐specific simulation settings. This enables customizable, model‐based exploration for hypothesis testing, treatment comparisons, visual comparison to data, and model qualification tests. gQSPSim is designed for reproducibility, transparency (i.e., no additional knowledge or configuration is required to replicate and understand shared results), and robustness. To maximize project reuse and distribution, projects are encapsulated within cross‐platform compatible sessions files that can be easily shared between users. Beyond just model sharing, gQSPSim facilitates workflow sharing by including the functionalities inside the session, ensuring that the user of the model executes the simulations and functionalities in an identical way as the author. All input and result files are stored as Microsoft (Redmond, WA) Excel files for easy modification and interpretation. The entire QSP workflow from parameter optimization to novel predictions can be performed within gQSPSim using the contents of the session file and the SimBiology model. In the following sections, we describe the full capabilities of gQSPSim and provide two illustrative case studies demonstrating these capabilities.
Methods
Overview of software development
gQSPSim is a graphical user interface based on MATLAB. It uses functionalities from MathWorks MATLAB, the SimBiology and Statistics and Machine Learning Toolboxes, and the GUI Layout Toolbox version 2.3.3 and higher.10 Its use requires a MATLAB license with the aforementioned toolboxes. The Parallel Computing Toolbox can also be used to enable a parallel version of gQSPSim’s core functionalities for faster execution on local or remote clusters. Additional optimization algorithms are available if the Optimization Toolbox and/or Global Optimization Toolbox are installed.
The implementation of the application is based on the model‐view controller or MVC architecture. The “model” contains the analysis or configuration. This can be executed from the MATLAB command‐line independently from the front‐end viewer. The model is represented by the back‐end objects within the “+QSP” MATLAB package folder. The “viewer” designates the graphical user interface (GUI), which is mostly contained within the “+QSPViewer” package folder. Implementation of the viewer is based on the GUI Layout Toolbox,10 a programmatic layout manager, from MATLAB Central. The “controller,” represented by the viewer's callback functions, responds to inputs by the user by updating the model and subsequently the viewer. The controller is also contained within the “+QSPViewer” package folder.
gQSPSim has been tested in MATLAB releases R2017b and R2018a. The software is not compatible with earlier MATLAB releases and currently has not been fully tested for releases after MATLAB R2018a. The application requires MATLAB and is not supported for distribution via MATLAB Compiler. Both Windows and Mac operating systems are supported. It is available to download from MATLAB Central and GitHub (https://www.mathworks.com/matlabcentral/fileexchange/73631-gqspsim).
GUI application overview
Figure 1 shows the framework for gQSPSim. Model construction is performed separately within SimBiology, and the functionalities that use the model are run in gQSPSim. Any subsequent changes to the SimBiology model, such as changes to species, tasks, doses, and so on, are automatically reflected in the gQSPSim session.
Figure 1.
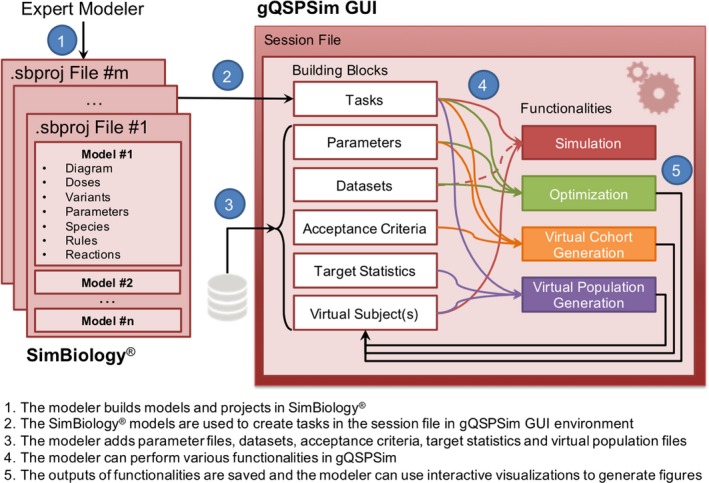
A schematic workflow of how modelers employ SimBiology and gQSPSim. An expert modeler builds the model diagram, variants, dosing objects, and so on in SimBiology. Simultaneously, the modeler can launch a new session in gQSPSim and use SimBiology models to create tasks with the settings specified in the Methods section. The modeler can also add parameter files, datasets, acceptance criteria, target statistics, and virtual subjects as building blocks to perform a variety of functionalities including simulation, Optimization, Virtual Cohort Generation, and Virtual Population Generation. Because the session file only stores the folder path to model files and datasets and does not store their content, the modeler can change models and files outside of gQSPSim, save the files, and seamlessly run the previously defined functionalities. All of the functionalities are supported by features such as interactive visualization and export of results as presentation‐ready figures and Excel files. The session file can be saved for future use. There is an option to autosave the session files and also to use parallel processing to speed up the execution of functionalities. GUI, graphical user interface.
gQSPSim organizes its building blocks and functionalities into “session files,” each containing the necessary elements for the development of a QSP project. The session file maintains the file names, relationships, and mappings for the project, but does not store the contents of the data files or SimBiology models. Instead, the most recently saved versions of the SimBiology model and input files will be loaded automatically at run time ensuring that the simulation dependencies are current when it is executed. In addition, gQSPSim can use multiple SimBiology project files and models simultaneously within a single session file. The building blocks and functionalities are described below. Table 1 shows a general description of gQSPSim components and features, and Figure 2 shows a screenshot of gQSPSim GUI.
Table 1.
gQSPSim components and features
| Items | Description |
|---|---|
| General | |
| SimBiology Project | A SimBiology .sbproj file containing the models of interest |
| gQSPSim GUI | Graphical user interface for gQSPSim, providing methods for development and utilization of SimBiology models |
| Session File | A MATLAB .qsp.mat file containing settings, building blocks, and functionalities created for one or more SimBiology projects |
| Building blocks | Tasks, parameters, datasets, acceptance criteria, target statistics, and virtual subjects |
| Functionalities | Simulation, Optimization, Virtual Cohort Generation, and Virtual Population Generation |
| Menus | |
| File |
|
| QSP |
|
| Features and functionalities | |
|
|
| Views | |
| Summary | Highlights the name, description, results path, and a high‐level overview of the dataset, tasks, parameters, species‐data mapping, initial conditions, and so forth, defined in each building block or functionality |
| Edit | Allows the user to specify/change the different inputs mentioned in the summary view specific to each building block or functionality |
| Visualization | Visualization of functionality outputs including simulated time profiles for selected species and diagnostic plots for Virtual Cohorts Generation |
GUI, graphical user interface; OS, operating system; QSP, quantitative systems pharmacology.
Figure 2.
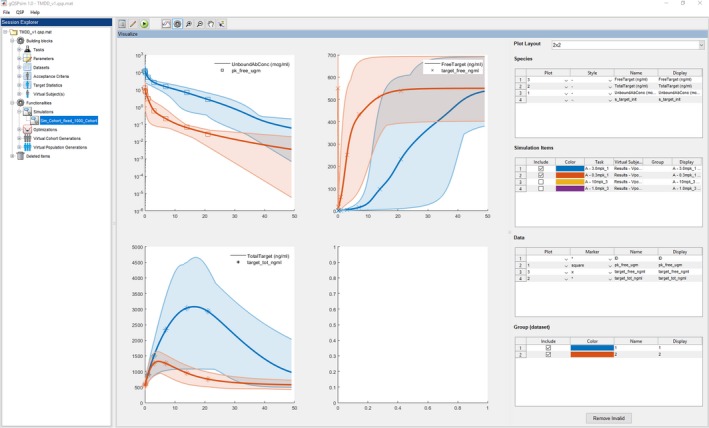
gQSPSim: Session explorer and visualization view for the simulation functionality. In gQSPSim there are two major sections: (i) Session Explorer, which is a tree structure on the left that shows the open session files as the root nodes and under each session it includes building blocks, functionalities; and deleted items and (ii) the views on the right, where by default all the nodes land on the summary view but the user can switch to the edit (Figures S3 –S6) or visualization view (above). The visualization view provides interactive plotting where the user can select what simulation and species to plot. Then the user selects data columns and group of data to be overlaid on the plots. The user can change colors, markers, and line styles and has access to extended plot settings (Figure S7) to modify different figure properties.
gQSPSim general settings
The root node in the gQSPSim Session Explorer tree view displays the session name and lets the user modify general settings including the following:
Defining the session's root directory and paths for custom objective functions and user‐defined functions. The root directory sets the absolute path for the top‐level folder containing all the building blocks and results produced by different functionalities. In the session, the location of every file is saved relative to the root path. When a session is opened with another computer and/or operating systems, the user only needs to change the root path to the correct location.
Toggling parallel computation and the parallel cluster to be utilized (optional).
Toggling periodic autosave and/or autosave before each run (optional).
In the Session Explorer, right clicking on top‐level nodes allows users to add a new subnode of that type. Right‐clicking on subnodes also provides options to delete or duplicate the item. The duplicated item will have the same properties and settings, but there will be no results associated with it. Each session has a deleted items node that keeps all deleted items until they are permanently deleted. Deleted items can be restored to their original location in the tree.
gQSPSim building blocks and functionalities
gQSPSim fully supports and facilitates the “Six‐Stage Workflow” for QSP model development described by Gadkar et al.9 Following the “Information Gathering” and “Model Scoping” steps, a typical workflow would be to (i) use SimBiology to construct a model (“Representing the Biology”), (ii) use gQSPSim to optimize this model for all appropriate experimental conditions (“Capturing Behaviors & Building Confidence”), (iii) use gQSPSim to explore biological variability and/or model uncertainty through the construction of a virtual cohort (“Exploring Knowledge Gaps and Variability”), (iv) use gQSPSim to recalibrate the generated cohort to best match available statistical data through the use of prevalence weights using the Virtual Population Generation functionality, and (v) use the generated virtual cohort or virtual population and the defined tasks to perform simulation studies addressing questions or scenarios of interest (“Supporting Study Design”).
In this section, we introduce the basic building blocks defined in gQSPSim (see Table S1 and Figure S1 for a detailed description of building blocks and functionalities). Refer to Gadkar et al. 9 for a more detailed explanation of QSP‐related concepts.
Task: A simulated experiment with user‐defined selection of variants, doses, species, rules, reactions, and simulation settings. Each task can be thought of as an equivalent of a real‐world experimental condition/study with specific treatments (dosing level/regimen). Figure S2 shows the Edit view for this building block.
Parameters: A list of model parameters used for either Optimization or Virtual Cohort Generation, specifying upper and lower bounds for exploration, and initial estimates for each parameter.
DataSet: A preclinical or clinical dataset to be used for visual predictive checks and/or numerical optimization.
Acceptance Criteria: A set of upper and lower bound limits of observed measurements used to define acceptable virtual subjects for inclusion into a virtual cohort.
Target Statistics: A preclinical or clinical dataset containing statistics of observed measurements to be used for Virtual Population Generation.
Virtual Subject(s): A collection of one or more virtual subjects (also referred to as virtual patients). Each virtual subject is a vector of parameters and/or initial conditions of species. The collection of virtual subjects captures observed data variability. Virtual subjects can be either weighted or unweighted. Unweighted virtual subjects are referred to as virtual cohorts, whereas weighted virtual subjects are called virtual populations because they are typically prevalence weighted to resemble a real‐world population. The virtual subjects in a cohort or population can have an optional “Group” identifier to indicate a subset of subjects of a particular phenotype. The user can choose to simulate a selection of these groups from the virtual subjects file.
Gadkar et al. 9 provides more detailed definitions of the terms virtual subject, reference virtual subject, virtual cohort, virtual population, prevalence weighting, and acceptance criteria. Datasets, acceptance criteria, target statistics, parameters, and virtual subjects are all stored as Excel files with a defined column format (see Table S1 for details).
The building blocks are used to enable the different functionalities used in QSP model development workflows as described (see Table S2 for a detailed description):
Simulation: Runs simulations for a set of task‐virtual population pairs; each task is run for the specified virtual population. The choice of virtual population may also be the model default parameter values (see Figure S3 for the Edit view). The simulation results are saved as a .mat file in the specified results directory.
Optimization: Runs parameter estimation using either scatter search,11 particle swarm,12 or a local search method (depending on the available optimization toolboxes). A dataset and a parameter file are required for this functionality (Figure S4 ). Optimization produces a single virtual subject which is saved as an Excel file in the optimization results folder and automatically added to the available virtual subject(s).
Cohort Generation: Generates a virtual cohort using one of two methods: (i) dispersed random sampling (with user‐specified distribution) or (ii) likelihood‐free Markov Chain Monte Carlo method.13 The user needs to specify a parameter file and an acceptance criteria (Figure S5 ). The results are saved as an Excel file in the specified results folder and the outcome is automatically added to the virtual subject(s).
Virtual Population Generation: Estimates the optimal prevalence weights for each virtual subject within the specified virtual cohort using the provided target statistics (Figure S6 ). The results are saved under the virtual subject(s).
Results
Case study 1: A target‐mediated drug disposition (TMDD) model to evaluate antibody PK and target profiles
Antibody‐based drug development often involves estimating target neutralization as a function of dose level, regimen, and antibody affinity to determine the requirements for desired target engagement. In this case study, we consider a two‐compartment model with target‐mediated drug disposition in the central compartment.14 This model was selected because of its broad use and familiarity to the modeling community. A schematic of this model is shown in figure 1 of Hosseini et al. 15
In the following, we demonstrate how to use gQSPSim to (i) perform optimization to obtain reference parameter values to fit the mean of clinical data, (ii) develop a virtual cohort to capture the variability observed in the clinical patients, (iii) generate a virtual population that closely matches the clinical population statistics, and (iv) run model‐based predictions for novel scenarios using the virtual population. Detailed instructions for reproducing this case study are provided in Supplementary Instructions S1 .
The clinical dataset available for this case study includes time‐course measurements for free antibody PK concentrations, free target concentrations, and total target concentrations available for two dose groups at 0.3 and 3.0 mg/kg (Table S3 ). These data are linked to gQSPSim by creating a dataset building block and specifying the input file. To perform an optimization of the model parameters, the user defines a subset of parameters to be explored along with a specified range and initial estimate for each model parameter stored in an Excel file. Within the Optimization functionality, the user configures the settings and selects the objective function calculated using the user‐specified mappings between individual tasks and experimental groups within the selected dataset. Figure 3 a–c shows the optimization results for the free drug (PK), free target, and total target for the reference virtual subject matching the mean values of the measured data. Optimization using either particle swarm or scatter search gave identical performances. The final estimate of each parameter and the initial values are shown in Table S4 . A comparison of simulations using the optimized parameters and the initial parameter values highlights the significant improvement in the model fits (Figure S8 ).
Figure 3.
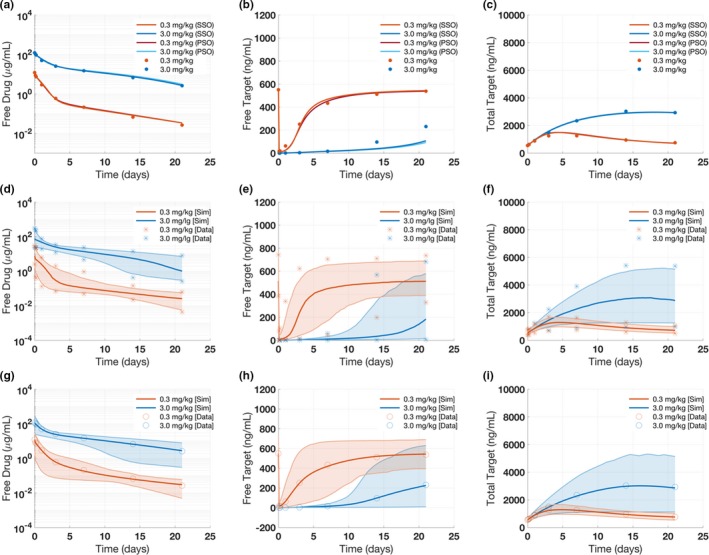
Case study 1: Results for target‐mediated drug disposition model Optimization, Virtual Cohort, and Virtual Population Generation. The results show (a–c) the output of Optimization, (d–f) Virtual Cohort, and (g–i) Virtual Population Generation for three species: free drug, free target, and total target concentrations. (a–c) Two Optimization algorithms were used: particle swarm optimization and scatter search methods. Note that the results for the two Optimization methods gave identical performances. (d–f) For the Virtual Cohort Generation, we used the random sampling method. The solid line represents the weighted median and the shaded region represents the 2.5th and 97.5th percentiles of the virtual cohort. The symbols represent the lower and upper bounds of the observed data, suggesting that the virtual cohort captures the range of observed data fairly well. (g–i) The virtual population was generated by prevalence weighting of the virtual cohort shown previously. The solid line represents the weighted mean, and the symbols represent the observed mean data. PSO, particle swarm optimization; SSO, scatter search.
The Virtual Cohort Generation functionality in gQSPSim produces alternate parameterizations of the model so that the model outputs collectively span the inter‐patient range of the clinical measurements. The range from the clinical data is included in an Excel file and linked to an acceptance criteria object in gQSPSim. Figure 3 d–f shows the corresponding plots for the virtual cohort subjects at the dose levels specified previously. The solid line represents the median, and the shaded region represents 95% of the virtual cohort (the 2.5th and 97.5th percentiles of the virtual cohort). The ‘*’ symbols represent the acceptance criteria, that is, the lower and upper bounds of the clinical data. Here, we observe that all the virtual cohort subjects are within the acceptance criteria and together capture the variability of the clinical data. The Virtual Cohort Generation is run until either the desired minimum number of virtual subjects are generated or a user‐defined maximum number of simulations are performed.
The diagnostic plots generated within Cohort Generation provide insight into the robustness of the cohort generated. In particular, one can see whether the virtual subjects span the range of the observed data; if not, one may wish to adjust the range of parameter values used for Virtual Cohort Generation. Figure S9 displays the simulated time profiles for each valid virtual subject in the cohort compared with the acceptance criteria. Figure S10 shows the distribution of the parameters for the valid virtual subjects in the cohort along with the prespecified ranges for each parameter. From this view, it is evident whether valid virtual subjects tend to be biased toward particular parameter values and whether the ranges should be extended to improve the chance of generating new valid virtual subjects. One may also visualize the simulated outputs for invalid virtual subjects (i.e., where the acceptance criteria were not satisfied) to inform subsequent iterations of Virtual Cohort Generation if required (Figure S11 ; invalid patients shown in gray).
The Virtual Cohort Generation functionality also includes the flexibility to generate virtual cohorts for groups that have different initial conditions for some measurements. In the example shown in Figure S12 , the target levels are different between the two dose groups (560 and 1120 ng/mL for groups 1 and 2, respectively). This information can be included in the Virtual Cohort Generation step as described in Supplementary Instructions S1 .
Although the virtual cohort spans the range of the clinical data, it does not necessarily capture the statistics of the dataset. For this, virtual subjects within the virtual cohort must be assigned prevalence weights to create a virtual population (see Stage 5 “Exploring Knowledge Gaps & Variability” of Gadkar et al.9). gQSPSim provides the Virtual Population Generation functionality for this purpose. The target statistics file contains the mean or mean and standard deviations for each time point, species, and group combination. The prevalence weighting is performed such that the difference between the model output statistics and the target statistics is minimized (see Klinke16 for a description of prevalence weighting). This optimization is performed by constructing and solving a quadratic objective function with linear constraints.
Figure 3 g–i shows the weighted average free antibody, free target, and total target concentrations for the virtual population along with the means from the data. Through visual comparison, it is evident that this procedure achieves good agreement with the provided target statistics. Following the prevalence weighting, the virtual population can then be used to make prospective predictions using the Simulation functionality. As a demonstration of this capability, four different dosing scenarios are simulated using the virtual population: 0.3 mg/kg single dose, 3 mg/kg single dose, 1 mg/kg every 2 weeks, and 10 mg/kg every 2 weeks. These scenarios are created using the task building block. As seen in Figure 4 a–c, when compared with the lower doses, the 10 mg/kg dose binds the target for a relatively long time and achieves nearly complete neutralization for the first ~30 days. The shaded regions contain the 95% of the virtual population (taking into account the prevalence weights), and the line represents the weighted median. Note that the simulation functionality allows the flexibility to simulate tasks with a variety of choices for virtual subjects including the model default parameter values as a single virtual subject, user‐defined virtual subjects with or without prevalence weights, and virtual subjects produced by Optimization, Virtual Cohort Generation, or Virtual Population Generation. In addition, the virtual cohorts/populations may contain group information, which can be used to classify them based on observed measurements, such as when groups in the dataset have different baseline levels for a specific measurement. For example, Figure S13 shows a simulation for a virtual cohort containing two groups with different baseline free target levels.
Figure 4.
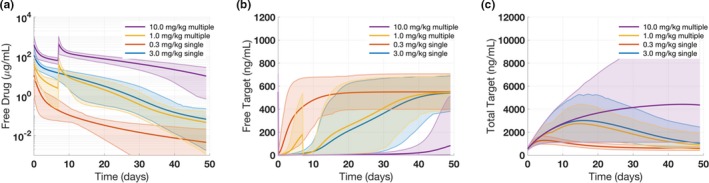
Case study 1: Target‐mediated drug disposition model simulation results for alternative dosing regimens. The results show (a) free drug, (b) free target, and (c) total target concentrations of four different dosing regimens using the generated virtual population. In each plot, the solid line represents the weighted mean and the shaded region represents the 2.5th and 97.5th percentiles.
The case study exemplifies how scientists can use gQSPSim to perform the technical workflow associated with QSP modeling approaches, such as generation of a cohort or population that agrees with measured PK/pharmacodynamics, evaluation of different phenotypes within the population, and predictions for novel contexts. The results produced by the various functionalities can be explored through the interactive plots and easily exported for additional analyses and dissemination.
Case study 2: A mechanistic systems model of anti‐proprotein convertase subtilisin/kexin type 9 (PCSK9) to evaluate clinical dosing regimens
The calibration and validation of QSP models often require clinical data from multiple studies as well as published literature. It is often challenging, especially in more complex models, to report the specific details of the calibration and validation process. With the gQSPSim app, all steps of QSP model development, including calibration, virtual population development, and simulation, are made transparent and reproducible, greatly facilitating model distribution. The goal of this case study is to reproduce the Virtual Population Generation and validation steps contained in a published QSP model, showcasing how the app could be used in the development of a clinically impactful QSP model. We decided to use the anti‐PCSK9 model17 because this model has been shown to be fully reproducible on the basis of its published details and supplementary materials.18
The model describes the mechanism of action of anti‐PCSK9 in lowering low density lipoprotein (LDL) cholesterol. LDL receptors on hepatocytes are responsible for the removal of LDL particles from circulation. However, circulating PCSK9 proteins can bind to these LDL receptors, targeting them for degradation and decreasing the number of receptors available for LDL uptake. Treatment with anti‐PCSK9 monoclonal antibodies reduces binding of PCSK9 to LDL receptor and consequently improves LDL particle uptake by hepatocytes. The QSP model includes these mechanisms to evaluate anti‐PCSK9 therapy. Details on the model structure can be found in Gadkar et al.,17 and instructions for reproducing the case study are given in Supplementary Instructions S2 .
In gQSPSim, we constructed a Simulation that outputs the concentrations of total anti‐PCSK9, LDL, and total PCSK9 using the .sbproj file from Gadkar et al.17 Simulating 100 virtual subjects within physiological ranges of parameter values showed that LDL and PCSK9 concentrations typically reached steady state by day 300 (Figure S14 ). Therefore, in all subsequent simulations, the model was run to steady state for 300 days before any subsequent perturbations using the steady‐state feature in the task building block.
The model was calibrated using phase I clinical data where a single dose of anti‐PCSK9 was administered at six dose levels ranging from 10 to 800 mg (Table S5 ). The calibration included three measurements: LDL, total anti‐PCSK9 (free drug + drug‐target complex), and total PCSK9 (target + drug‐target complex). The following three steps were performed in this case study: optimization to obtain a reference virtual subject, generation of a virtual cohort, and generation of a virtual population. To generate the reference virtual subject in gQSPSim, we used the Optimization functionality with the scatter search algorithm. Simulations performed with the optimized parameter values captured the dynamics of LDL, total anti‐PCSK9, and PCSK9 at all dose levels (Figure 5). The same clinical data (Table S5 ) were then used to define the acceptance criteria for the three measurements at all six dose levels for Virtual Cohort Generation. The resulting virtual cohort successfully spans the range of the observed data as shown in Figure S15 . Finally, the virtual population was generated from the virtual cohort by fitting the baseline LDL and PCSK9 means and standard deviations to the clinical data (Figure 6). The virtual population generated using the aforementioned workflow is comparable to that of the original publication (see figure 2 in Gadkar et al.17).
Figure 5.
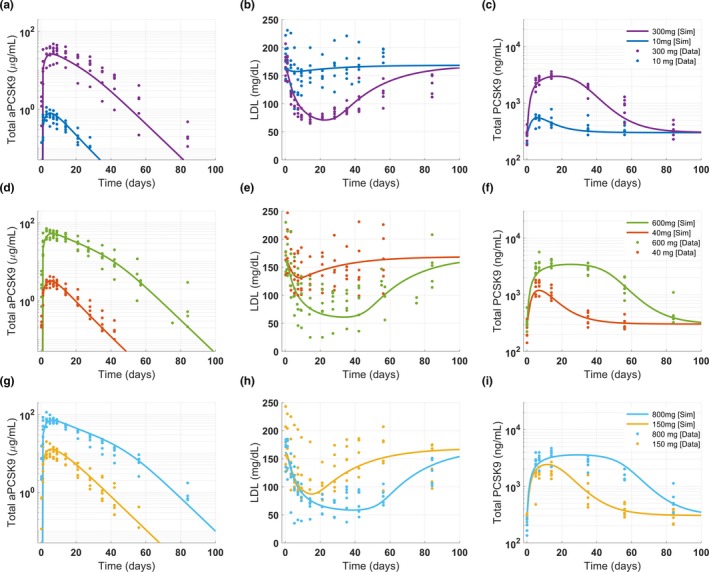
Case study 2: Reference virtual subject for anti‐PCSK9 model optimized using scatter search. The results show model calibration to single dose clinical data at six dose levels (10, 40, 150, 300, 600, 800 mg) and three measurements: total anti‐PCSK9 (a, d, g), LDL (b, e, h), and total PCSK9 concentrations (c, f, i). Each plot contains data and reference virtual subject simulation for two dose levels as shown in the legends. PCSK9, proprotein convertase subtilisin/kexin type 9.
Figure 6.
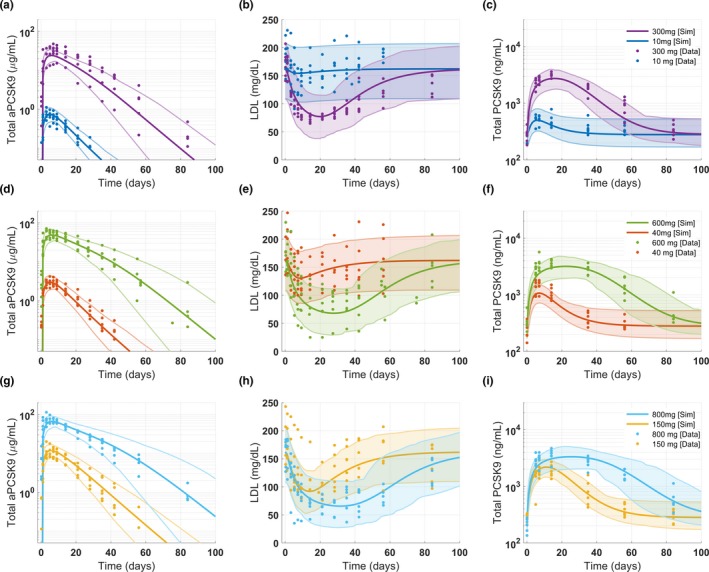
Case study 2: Virtual population generated for anti‐PCSK9 model. The range of observed clinical data for (a, d, g) total anti‐PCSK9, (b, e, h) LDL concentration and (c, f, i) total PCSK9 is well captured by the virtual population generated using the workflow in gQSPSim. The shaded region represents 2.5–97.5% percentiles of the virtual subjects, and the thicker line represents the median. PCSK9, proprotein convertase subtilisin/kexin type 9.
A separate clinical trial data with weekly dose of anti‐PCSK9 with and without statin treatment (see Table S6 for dosing information) were used to validate the virtual population. Simulation of the four treatment groups within the clinical trial using the virtual population captured the observed clinical data on LDL reduction (Figure S16 A–D, corresponding to figure 3 in Gadkar et al.17).
Finally, the validated virtual population was used to simulate potential different dosing levels and schedules to determine the scenario with the least amount of LDL fluctuation for a future phase II study. The following four scenarios were simulated: 400 mg every 4 weeks, 800 mg every 8 weeks, 400 mg every 8 weeks, and 200 mg every 8 weeks. Based on the simulation results (Figure S16 E–F), 400 mg every 4 weeks and 800 mg every 8 weeks are the two scenarios that show the least fluctuation, as was shown in figure 4 of Gadkar et al.17 Note that we used the generated virtual population in this case study for the phase II predictions, whereas in the original article,17 the virtual population used for the phase II prediction was further modified to capture a change in the inclusion criteria of the study.
Using the published anti‐PCSK9 model, we illustrated how the app can be a valuable tool for QSP model development, utilization, and novel prediction to support clinical development of a molecule. Moreover, the entire QSP workflow from model calibration to prediction was stored in a single session file in gQSPSim, rendering the reproduction of model predictions an efficient process for other scientists in academia and industry.
Discussion
QSP models are seeing increased utilization and impact in pharmaceutical research and development across therapeutic areas. Although proposals and recommendations for standardized workflows exist for model development, qualification criteria, and reporting,19 there is a lack of accessible and user‐friendly tools and algorithms to execute the QSP workflow. Furthermore, there are several issues with QSP models, including (i) lack of quantitative information in the model and/or the publication, (ii) failing to provide the associated model files or programming codes along with the article in which the model is published, (iii) lack of proper documentation of the workflow, and (iv) inconsistencies between model behavior and what is presented in the article. We have attempted to address these issues by developing gQSPSim to be used in QSP model development and applications. Besides adding new functionality to SimBiology, gQSPSim also provides a standardized platform for the creation of extensible, customizable QSP workflows for adoption in the QSP community. We envision that the combination of gQSPSim and SimBiology will provide the means necessary to facilitate scientists to develop robust QSP models impacting all stages of drug development.
A common theme of QSP model development is the need to integrate data from disparate sources that are then employed for calibration, testing, and the exploration of biological variability. However, these data may be obtained from a wide array of in vitro, in vivo, ex vivo, and clinical sources and typically lack a cohesive structure. gQSPSim remedies this by providing a standard format for including these datasets for integration into the QSP workflow. One may use gQSPSim to fully leverage available data. For example, in the calibration process, in vitro data are often used to determine the initial guesses for model parameters, whereas the preclinical and clinical data are used for subsequent parameter fitting. Data obtained from different literature sources can have a high degree of variability. This information can be used within gQSPSim by creating a user‐defined objective function that weights each dataset appropriately. Furthermore, the observed variability in the data is used to determine the acceptance criteria in gQSPSim for Virtual Cohort Generation ensuring the simulations of the virtual subjects span a similar range as the clinical data, providing a robust framework for the efficient exploration of model parameters consistent with the data. In addition, the statistical information obtained from clinical measurements can be formatted as target statistics to be used in Virtual Population Generation functionality to calculate prevalence weights for virtual subjects and ensure model output statistics match the observed statistics.
Calibration and generation of reference virtual subjects is a key step in QSP modeling (see Stage 4 “Capturing Behaviors & Building Confidence” of Gadkar et al.9). However, this is often a challenge as models are commonly overparameterized. Overcoming this challenge requires sophisticated methods such as global stochastic optimization. Alternatively, for QSP models that may have a fairly rich dataset for calibration and limited number of unknown parameters, gradient‐based optimization methods may be more efficient in terms of computation time and accuracy. In gQSPSim, we have implemented two of the most commonly used global optimization methods—scatter search and particle swarm optimization—in addition, we have a gradient‐based method that uses nlinfit from the Statistics and Machine Learning Toolbox in MATLAB. In principle, gQSPSim may be used with any optimization algorithm included in the Optimization Toolbox or Global Optimization Toolbox, and future versions will expand the available selection.
gQSPSim addresses a significant need for QSP model development and application, which is to explore underlying biological uncertainty and to capture the impact of data variability in making model‐based decisions. Through the generation of virtual cohorts and weighted virtual populations that systematically explore uncertainty, the app adds confidence to subsequent QSP model‐based predictions, especially for dose‐response predictions in clinical trials, identifying diagnostic markers for patient subtypes, deciding clinical study design, and optimizing patient inclusion criteria.
Although gQSPSim is sufficient for many QSP workflows, it is presently limited in some ways. It is based on MATLAB and SimBiology and will require any QSP modelers using other environments to adopt this workflow. It currently only supports a small range of interactive plotting functions such as line plots and diagnostic distribution plots. However, the simulation data can be easily loaded into MATLAB for manipulations in a scripting environment, and future versions of gQSPSim will provide interfaces for incorporating user‐defined functions to be performed on simulation outputs. Furthermore, gQSPSim is only compatible with ordinary differential equation‐based models built in SimBiology and does not currently support stochastic models. Future versions will enable more advanced features such as code plugins, sensitivity analyses, advanced plotting capabilities, activity logging, and more.
The software source code is freely available to the QSP community to use. Users who would like to contribute to the development of gQSPSim are invited to contact the corresponding author. Open sharing of the tools and software enables easy adoption for both academic and industry purposes. With these features, we hope gQSPSim and its future versions will become the standard platform for QSP model development and sharing within the broader QSP modeling community.
Funding
No funding was received for this work.
Conflict of Interest
I.H., J.F., M.S., V.R., S.R., and K.G. are full‐time employees at Genentech Inc. A.G. is a full‐time employee at MathWorks.
Author Contributions
All authors wrote the manuscript. I.H., J.F., A.G., and K.G. designed the research. I.H., J.F., A.G., and K.G. performed the research. All authors analyzed the data.
Supporting information
File S1. Supplement instructions, figures and tables for case studies.
Instructions S1. TMDD model.
Instructions S2. anti‐PCSK9 QSP model.
Figure S1. Quantitative systems pharmacology model development workflow.
Figure S2. gQSPSim: Edit view of the task building block.
Figure S3. gQSPSim: Edit view of the Simulation functionality.
Figure S4. gQSPSim: Edit view of the Optimization functionality.
Figure S5. gQSPSim: Edit view of Virtual Cohort Generation functionality.
Figure S6. gQSPSim: Edit view of Virtual Population Generation functionality.
Figure S7. gQSPSim: Extended plot settings window.
Figure S8. Case study 1: Simulation results with model default parameter values.
Figure S9. Case study 1: Virtual Cohort Generation diagnostic view.
Figure S10. Case study 1: Virtual Cohort Generation parameter distribution view.
Figure S11. Case study 1: Virtual Cohort Generation results including invalid virtual subjects.
Figure S12. Case study 1: Virtual Cohort Generation results with group‐specific initial target concentrations.
Figure S13. Case study 1: Simulation results with group‐specific virtual population.
Figure S14. Case study 2: Simulation results to find the time to steady state.
Figure S15. Case study 2: Virtual cohort generated for anti‐proprotein convertase subtilisin/kexin type 9 (PCSK9) model.
Figure S16. Case study 2: The validation and prediction results for the anti‐PCSK9 model.
Table S1. A detailed summary of gQSPSim building blocks.
Table S2. A detailed summary of gQSPSim functionalities.
Table S3. Case study 1: List of dosing levels and regimens used in the target‐mediated drug disposition model.
Table S4. Case study 1: List of parameters estimated in the target‐mediated drug disposition model.
Table S5. Case study 2: Clinical study design for the single dose intravenous study used to calibrate the anti‐PCSK9 model.
Table S6. Case study 2: Clinical study design for the multiple intravenous dosing study used to validate the anti‐PCSK9 model.
Table S7. Case study 2: Description of model parameters.
Table S8. Case study 2: Parameter file used in optimization.
Table S9. Case study 2: Parameter file used for cohort generation.
Table S10. Case study 2: Target statistics used for Virtual Population Generation.
File S2. Template_ExcelFiles.zip.
File S3. Zip file for case study 1: Target‐mediated drug disposition model.
File S4. Zip file for case study 2: Anti‐PCSK9 model.
Acknowledgments
The authors would like to thank Ricardo Paxson, Robyn Jackey, Henry Mattingly, Dan Lu, Jennifer Wilson, Greg Ferl, Vincent Lemaire, James Lu, and MathWorks Consulting for their input to this work.
References
- 1. Leil, T.A. & Bertz, R. Quantitative systems pharmacology can reduce attrition and improve productivity in pharmaceutical research and development. Front. Pharmacol. 5, 247 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ramanujan, S. , Gadkar, K. & Kadambi, A. Quantitative systems pharmacology: applications and adoption in drug development In: Systems Pharmacology and Pharmacodynamics. (eds. Mager D.E. & Kimko H.H.C.) 27–52 (Springer, Basel, 2016). [Google Scholar]
- 3. Peterson, M.C. & Riggs, M.M. A physiologically based mathematical model of integrated calcium homeostasis and bone remodeling. Bone 46, 49–63 (2010). [DOI] [PubMed] [Google Scholar]
- 4. Demin, O. et al Systems pharmacology models can be used to understand complex pharmacokinetic‐pharmacodynamic behavior: an example using 5‐lipoxygenase inhibitors. CPT Pharmacometrics Syst Pharmacol 2, 1–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schoeberl, B. et al Therapeutically targeting ErbB3: A key node in ligand‐induced activation of the ErbB receptor–PI3K axis. Sci Signal 2, ra31 (2009). [DOI] [PubMed] [Google Scholar]
- 6. Chen, X. , Hickling, T. & Vicini, P. A mechanistic, multiscale mathematical model of immunogenicity for therapeutic proteins: part 2—model applications. CPT Pharmacometrics Syst Pharmacol 3, 1–10 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cheng, Y. et al QSP Toolbox: computational implementation of integrated workflow components for deploying multi‐scale mechanistic models. AAPS J 19, 1002–1016 (2017). [DOI] [PubMed] [Google Scholar]
- 8. Ermakov, S. , Schmidt, B.J. , Musante, C.J. & Thalhauser, C.J. A survey of software tool utilization and capabilities for quantitative systems pharmacology: what we have and what we need. CPT Pharmacometrics Syst Pharmacol 8, 62–76 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gadkar, K. , Kirouac, D. , Mager, D. , van der Graaf, P.H. & Ramanujan, S. A six‐stage workflow for robust application of systems pharmacology. CPT Pharmacometrics Syst Pharmacol 5, 235–249 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sampson, D. GUI layout toolbox <https://www.mathworks.com/matlabcentral/fileexchange/47982-gui-layout-toolbox> (2019).
- 11. Martí, R. , Laguna, M. & Glover, F. Principles of scatter search. Eur. J. Oper. Res. 169, 359–372 (2006). [Google Scholar]
- 12. Eberhart, R. & Kennedy, J. Particle swarm optimization. IEEE International Conference on Neural Networks: proceedings, The University of Western Australia, Perth, Western Australia, November 27–December 1, 1995. Volume 2. 1995.
- 13. Marjoram, P. , Molitor, J. , Plagnol, V. & Tavaré, S. Markov chain Monte Carlo without likelihoods. Proc. Natl Acad. Sci. 100, 15324–15328 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mager, D.E. & Jusko, W.J. General pharmacokinetic model for drugs exhibiting target‐mediated drug disposition. J. Pharmacokinet. Pharmacodyn. 28, 507–532 (2001). [DOI] [PubMed] [Google Scholar]
- 15. Hosseini, I. et al gPKPDSim: a SimBiology®‐based GUI application for PKPD modeling in drug development. J. Pharmacokinet. Pharmacodyn. 45, 259–275 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Klinke, D.J. Integrating epidemiological data into a mechanistic model of type 2 diabetes: validating the prevalence of virtual patients. Ann. Biomed. Eng. 36, 321–334 (2008). [DOI] [PubMed] [Google Scholar]
- 17. Gadkar, K. , Budha, N. , Baruch, A. , Davis, J. , Fielder, P. & Ramanujan, S. A mechanistic systems pharmacology model for prediction of LDL cholesterol lowering by PCSK9 antagonism in human dyslipidemic populations. CPT: Pharmacometrics Syst Pharmacol 3, 1–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kirouac, D.C. , Cicali, B. & Schmidt, S. Reproducibility of quantitative systems pharmacology models: current challenges and future opportunities. CPT Pharmacometrics Syst Pharmacol 8, 205–210 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cucurull‐Sanchez, L. et al Practices to maximize the use and reuse of quantitative and systems pharmacology models: recommendations from the United Kingdom quantitative and systems pharmacology network. CPT Pharmacometrics Syst. Pharmacol. 8, 259–272 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
File S1. Supplement instructions, figures and tables for case studies.
Instructions S1. TMDD model.
Instructions S2. anti‐PCSK9 QSP model.
Figure S1. Quantitative systems pharmacology model development workflow.
Figure S2. gQSPSim: Edit view of the task building block.
Figure S3. gQSPSim: Edit view of the Simulation functionality.
Figure S4. gQSPSim: Edit view of the Optimization functionality.
Figure S5. gQSPSim: Edit view of Virtual Cohort Generation functionality.
Figure S6. gQSPSim: Edit view of Virtual Population Generation functionality.
Figure S7. gQSPSim: Extended plot settings window.
Figure S8. Case study 1: Simulation results with model default parameter values.
Figure S9. Case study 1: Virtual Cohort Generation diagnostic view.
Figure S10. Case study 1: Virtual Cohort Generation parameter distribution view.
Figure S11. Case study 1: Virtual Cohort Generation results including invalid virtual subjects.
Figure S12. Case study 1: Virtual Cohort Generation results with group‐specific initial target concentrations.
Figure S13. Case study 1: Simulation results with group‐specific virtual population.
Figure S14. Case study 2: Simulation results to find the time to steady state.
Figure S15. Case study 2: Virtual cohort generated for anti‐proprotein convertase subtilisin/kexin type 9 (PCSK9) model.
Figure S16. Case study 2: The validation and prediction results for the anti‐PCSK9 model.
Table S1. A detailed summary of gQSPSim building blocks.
Table S2. A detailed summary of gQSPSim functionalities.
Table S3. Case study 1: List of dosing levels and regimens used in the target‐mediated drug disposition model.
Table S4. Case study 1: List of parameters estimated in the target‐mediated drug disposition model.
Table S5. Case study 2: Clinical study design for the single dose intravenous study used to calibrate the anti‐PCSK9 model.
Table S6. Case study 2: Clinical study design for the multiple intravenous dosing study used to validate the anti‐PCSK9 model.
Table S7. Case study 2: Description of model parameters.
Table S8. Case study 2: Parameter file used in optimization.
Table S9. Case study 2: Parameter file used for cohort generation.
Table S10. Case study 2: Target statistics used for Virtual Population Generation.
File S2. Template_ExcelFiles.zip.
File S3. Zip file for case study 1: Target‐mediated drug disposition model.
File S4. Zip file for case study 2: Anti‐PCSK9 model.


