Abstract
Living cells regulate key cellular processes by spatial organisation of catalytically active proteins in higher-order signalling complexes. These act as organising centres to facilitate proximity-induced activation and inhibition of multiple intrinsically weakly associating signalling components, which makes elucidation of the underlying protein-protein interactions challenging. Here we show that DNA origami nanostructures provide a programmable molecular platform for the systematic analysis of signalling proteins by engineering a synthetic DNA origami-based version of the apoptosome, a multi-protein complex that regulates apoptosis by co-localizing multiple caspase-9 monomers. Tethering of both wildtype and inactive caspase-9 variants to a DNA origami platform demonstrates that enzymatic activity is induced by proximity-driven dimerization with half-of-sites reactivity, and additionally, reveals a multivalent activity enhancement in oligomers of three and four enzymes. Our results offer fundamental insights in caspase-9 activity regulation and demonstrate that DNA origami-based protein assembly platforms have the potential to inform the function of other multi-enzyme complexes involved in inflammation, innate immunity and cell death.
Nanoscale organisation of interacting proteins is a key regulatory principle in signalling pathways involved in all major cell events, including apoptosis, metabolism, inflammation, and immunity1,2. Inactive enzymes with a low intrinsic affinity and present at low intracellular concentrations, can be physically assembled into well-defined higher-order signalling complexes3 or open-ended assemblies4. Dedicated scaffold proteins serve as supramolecular organising centres (SMOCs), facilitating protein-protein interactions with precise control over the position and orientation of the individual components (Fig. 1a)5. Efforts to address and rewire SMOC-based signalling complexes have provided important structural and functional understanding into the underlying design principles6–8. In general, experimental and theoretical work illustrate that co-localization of signalling components promotes proximity-induced enzyme activation through weak non-covalent interactions, thereby overcoming signal thresholds, increasing pathway robustness, and shaping response dynamics9–11.
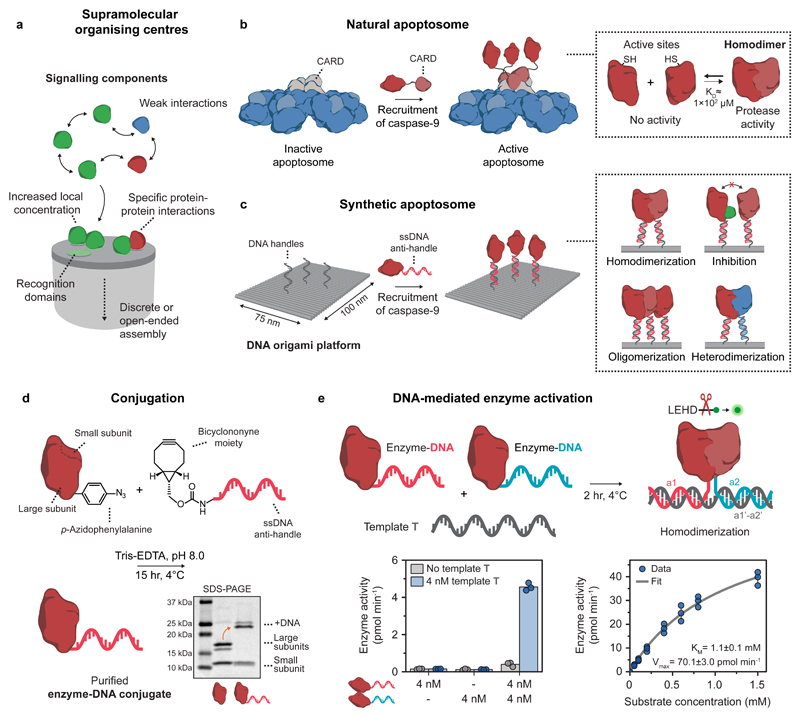
Figure 1. General concept and design elements for the construction of a DNA-based synthetic apoptosome.
a, Schematic concept of supramolecular organising centres (SMOCs). b, Schematic drawing of the natural apoptosome that functions by assembling inactive caspase-9 monomers through caspase recruitment domains (CARDs). The increase in local concentration induces caspase-9 dimerization, leading to proteolytic cleavage of downstream caspases and eventually apoptosis. c, Schematic drawing of the DNA origami-based synthetic apoptosome. Single-stranded DNA (ssDNA) handles on the DNA origami surface recruit caspase-9 monomers with full control over the number, position, and relative geometry, allowing for the characterisation of specific protein-protein interactions such as homodimerization, inhibition, oligomerization, and heterodimerization. d, Reaction scheme for the conjugation of caspase-9 to an oligonucleotide anti-handle. Caspase-9 was expressed in E. coli with unnatural amino acid p-azidophenylalanine at the N-terminus and reacted with a bicyclononyne-functionalized oligonucleotide (8 kDa). Purification with affinity chromatography afforded pure enzyme-DNA conjugates, as shown by SDS-PAGE analysis. Mature caspase-9 consists of a non-covalently bound N-terminal large (18.3 and 19.2 kDa) and C-terminal small subunit (12.8 kDa), which are separated during gel analysis (see also Supplementary Fig. 4). The identity of all protein fragments was confirmed using mass spectrometry analysis (Supplementary Fig. 2). e, Bivalent template T was used to induce dimerization of two identical caspase-9 monomers with different DNA sequences (a1 and a2, indicated in red and blue, respectively). Enzyme activity (left graph) was measured using stoichiometric amounts of all three components and 167 μM of the synthetic caspase substrate LEHD-AFC (Supplementary Fig. 8). Enzyme kinetics (right graph) were determined by measuring activity at 4 nM of each enzyme-DNA conjugate and varying concentrations of the substrate (0-1.5 mM). The data was fitted with the standard Michaelis-Menten expression (see Methods). Bars represent mean enzyme activity. All experiments were performed in independent triplicates.
Bottom-up approaches employing synthetic platforms enable systematic analysis and full control over the number, position, and orientation of interacting components, providing an excellent strategy to further unravel the molecular mechanisms behind spatial organisation in signalling pathways12,13. The programmability of DNA and its inherent biocompatibility enables rational design of defined synthetic architectures for the construction of protein-DNA hybrid systems14–17. DNA origami-based nanostructures, in particular, are well-suited as synthetic platforms as their unique addressability allows for precise assembly of multiple non-identical proteins with nanometre accuracy18–20. The DNA origami technique has found broad applicability as an experimental tool for spatial organisation of native multi-protein systems, such as amyloid fibrils21, membrane fusion proteins22, nucleosomes23,24, and intrinsically disordered proteins25. Additionally, these platforms have been used to engineer localized genetic circuits26, to study confinement-induced enzyme activity27,28, and to investigate scaffolded metabolic cascades29–33. Although these studies have elegantly applied the programmability of DNA nanotechnology to facilitate organisation of e.g. structural protein assemblies and metabolic enzymes with small-molecule substrates, a DNA nanostructure-based platform for directly probing protein-protein interactions between catalytically active intracellular signalling components is currently lacking.
Here, we present a DNA-based synthetic SMOC for studying proximity-induced protein-protein interactions involved in intracellular signal transduction. We construct DNA origami-based synthetic variants of the apoptosome, a 27-nm diameter, sevenfold-symmetric multi-protein complex involved in the intrinsic apoptotic pathway. In the cell, mitochondrial outer membrane permeabilization and subsequent release of cytochrome c induces the assembly of the apoptosome, which recruits the cysteine-dependent aspartic protease caspase-9 to initiate a cascade of proteolytic activity that eventually leads to programmed cell death34. Previous works suggest that the apoptosome recruits up to four caspase-9 monomers through caspase recruitment domains (CARDs), after which dimerization contributes to a dramatic increase in enzyme activity (Fig. 1b)35–37. By mimicking the scaffolding function of the apoptosome with a DNA origami platform18, we can assemble individual caspase-9 monomers with absolute control over their position, using the hybridization of DNA-functionalized enzymes to protruding single-stranded handles on the DNA origami surface (Fig. 1c). Using a bottom-up approach, our in vitro studies reveal that caspase-9 activity is induced by proximity-driven dimerization, driven by an increase in effective concentration as a result of tethering of the components to the platform. We then construct three- and four-enzyme systems to study the effect of higher-order clustering on caspase-9 activity. By combining experimental and theoretical results, we analyse kinetic data considering possible effects such as enzyme incorporation efficiency and statistical factors, and suggest a multivalent catalytic effect leading to enhanced activity in caspase-9 oligomers. Finally, we provide direct evidence that the enzymatic activity of a heterodimer consisting of a wildtype monomer and an active-site mutant, is similar to the activity of the wildtype homodimer, confirming the hypothesis that conformational changes in the active sites of a caspase-9 homodimer proceed via an asymmetric mechanism38. Our experimental platform demonstrates that systematic in vitro analysis of native protein-protein interactions using DNA-based synthetic SMOCs can facilitate the discovery of new molecular mechanisms in proximity-driven enzyme regulation.
Results
Activity of DNA-functionalized caspase-9
To construct a DNA origami-based platform that allows for programmable caspase-9 organisation, enzyme-DNA conjugates were synthesized20,39. Full-length caspase-9 monomers consist of an N-terminal CARD connected to the catalytic domain via a long flexible linker (Fig. 1b)38. Crystal structures show that upon recruitment, CARDs on the apoptosome interact with caspase-9 CARDs to form well-defined three-dimensional complex, to which multiple caspase-9 catalytic domains are tethered flexibly36,37. Replacing the CARD domain of caspase-9 with an oligonucleotide enables incorporation of the catalytic domain onto DNA nanostructures, and mimics the recruiting function of the CARD using hybridization to complementary single-stranded handles on the DNA origami surface (Fig. 1b,c). The specific protein-protein interactions involved in caspase-9 activation warrant stoichiometric, site-specific oligonucleotide functionalization without the use of large protein helper domains. Therefore, we incorporated the non-canonical amino acid p-azidophenylalanine at the N-terminus of the catalytic domain of caspase-9 by using amber codon suppression in E. coli with an engineered orthogonal aminoacyl tRNA synthetase/tRNA pair from M. janaschii (see Methods and Supplementary Fig. 1a and 2a)40. The small bioorthogonal azide moiety was used for conjugation to a bicyclononyne-functionalized oligonucleotide (BCN-DNA) using strain-promoted azide-alkyne cycloaddition (Fig. 1d)41–43. The oligonucleotide consists of a 10-nucleotide (nt) single-stranded linker separating the enzyme and a 15-nt anti-handle used for hybridization to the handles (Supplementary Table 2 and Supplementary Fig. 3). Analysis using polyacrylamide gel electrophoresis confirmed successful conjugation of a single oligonucleotide to the N-terminus of caspase-9 and subsequent purification resulted in complete removal of all unreacted protein and excess BCN-DNA (Fig. 1d, Supplementary Fig. 4 and 6).
Since conjugation of oligonucleotides to enzymes can significantly influence catalytic behaviour44,45, we measured the activity of the caspase-9 enzyme-DNA conjugates. We employed a 30-nt single-stranded template T to bring two caspase-9 monomers into close proximity through DNA hybridization46, and followed proteolytic cleavage of synthetic caspase substrate LEHD-AFC47 over time (Fig. 1e and Methods). We observed a sharp increase in activity only when both enzyme-DNA conjugates and the template are present (Fig. 1e; left graph). This suggests that template T functions as a bivalent scaffold inducing dimerization of caspase-9 by increasing the effective concentration, in a similar manner as protein-based dimerizing scaffolds reported in literature48–50. We performed a quantitative kinetic analysis and determined the Michaelis constant KM of the ternary complex consisting of the caspase-9 conjugates and template T. The value of 1.1 ± 0.1 mM is in the same range as found for caspase-9 activation by the native apoptosome and by other, synthetic scaffold platforms (Fig. 1e; right graph)47. Collectively, these results confirm the successful synthesis of functionally active caspase-9 enzyme-DNA conjugates, and demonstrate that DNA can be used to facilitate proximity-induced activation.
DNA origami-mediated caspase-9 enzyme activity
The native apoptosome is an organising platform that induces the co-localization of caspase-9 enzymes through seven caspase recruitment domains (CARD). Flexible tethering of caspase-9 through CARD-CARD interactions results in dimerization of the catalytic domains and a subsequent 100-1000 fold increase in enzyme activity51,52. While crystal structures indicate that up to four caspase-9 monomers bind to the apoptosome simultaneously36,37, in vitro investigations have so far focused solely on dimerization by employing protein-based bivalent scaffolds or engineering chimeric proteins48–50. To assess the effects of multivalent enzyme clustering, we constructed a synthetic DNA-based version of the apoptosome using a twist-corrected rectangular DNA origami nanostructure18, on which the location of an arbitrary number of caspase-9 monomers can be tightly controlled (see Methods and Supplementary Methods (Self-assembly of DNA origami nanostructures)). In our design, we substitute the structural proteins of the apoptosome with a DNA nanostructure and replace the recruiting function of the CARDs by DNA-DNA hybridization. As such, several features of the native system that could modulate caspase-9 activation and regulation are not taken into account. For example, recent work has indicated that allosteric interactions of the catalytic domain of caspase-9 with its CARD, the flexible linkers, and the apoptosome itself can affect enzyme activity36,53,54. However, we argue that a modular DNA-based model system composed of a controlled number of catalytic domains with well-defined interactions can help isolate the key molecular determinants of caspase-9 regulation.
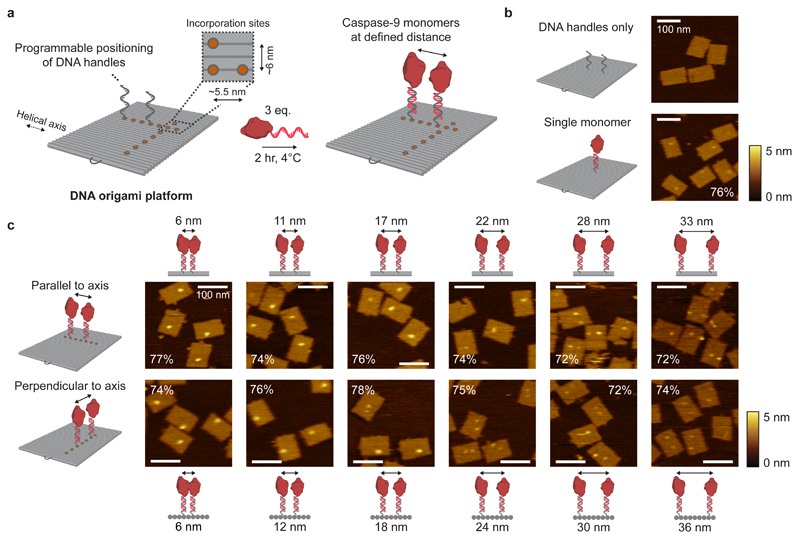
The unique addressability of DNA origami allows for programmable positioning of the handles with ~6 nm resolution across the entire platform (Fig. 2a). Atomic force microscopy (AFM) confirmed the self-assembly of 75×100-nm2 DNA origami nanostructures and revealed that individual 4.5-nm diameter, 32-kDa caspase-9 monomers can be observed on the DNA origami surface (Fig. 2b and Supplementary Fig. 13). Analysis of 260 well-formed one-enzyme DNA nanostructures revealed an average enzyme incorporation efficiency of 76% (Supplementary Fig. 16c), which is similar to values reported in literature55,56. Next, we designed two-enzyme DNA nanostructures with varying distance between two caspase-9 monomers arranged either parallel (Fig. 2c; top row) or perpendicular (bottom row) to the DNA helical axis, and used agarose gel electrophoresis (Supplementary Fig. 12) and AFM imaging (Fig. 2c, Supplementary Fig. 14 and 15) to analyse the integrity of the structures. When monomer separation is large (i.e. >20 nm), two individual spots can be distinguished in the AFM images, indicating faithful incorporation of caspase-9 at the pre-programmed positions, as well as physical separation of the monomers by the DNA nanostructure. In contrast, only a single spot is discerned at smaller separation distances (<20 nm), with the shape and intensity of these features suggesting the presence of two adjacent caspase-9 monomers (Supplementary Fig. 16a,b). We attribute this observation to a combination of the limited resolution of the imaging technique and intermolecular interactions (both specific and non-specific, see below) between the enzymes. Visual inspection of the AFM images allows straightforward identification of DNA nanostructures with either one or two enzymes (Fig. 2c; compare e.g. bottom row, 12 and 24 nm). The enzyme incorporation efficiency per handle was found to be approximately 75% per handle for all samples, irrespective of monomer separation (Fig. 2c, Supplementary Fig. 16c). Although this is in contrast with previous work that reported a diminished incorporation when two enzymes were brought into close proximity on a similar DNA origami scaffold30, we hypothesize that the attractive interaction between two caspase-9 monomers balances possible steric effects at small separation distances, leading to an overall constant incorporation efficiency. Taken together, the results of the AFM analysis confirm that DNA nanostructures containing two caspase-9 monomers can be constructed with tight control over the position of, and distance between, tethered monomers.
Figure 2. Characterisation of caspase-9 assembly onto DNA origami nanostructures.
a, Schematic of the general strategy for caspase-9 incorporation on DNA origami platforms. By including appropriate handle-extended staple strands during the self-assembly process the position of, and distance between, two ssDNA handles can be controlled. Specifically, the DNA origami technique enables a minimal distance between incorporation sites (orange circles) of 5.5 nm parallel, and 6 nm perpendicular to the DNA helical axis (Supplementary Methods (Self-assembly of DNA origami nanostructures)). Incubation of complementary enzyme-DNA conjugates leads to hybridization and incorporation of two caspase-9 monomers at defined distances. Typically, 4 nM DNA origami was incubated with 3 equivalents of enzyme-DNA conjugate per handle for 2 hr at 4°C. For AFM imaging, functionalized nanostructures were purified using 1.5% agarose gel extraction. b,c, Topographic AFM (tapping mode in solution) images of control samples (b) and DNA origami nanostructures functionalized with two 32-kDa caspase-9 monomers (c) at various distances parallel (top row) and perpendicular (bottom row) to the helical axis. The caspase-9 incorporation efficiency per handle is indicated in percentages, and was calculated based on at least 250 well-formed nanostructures using 4 different images per sample. Colour bars indicate height scale in AFM images. Scale bars, 100 nm.
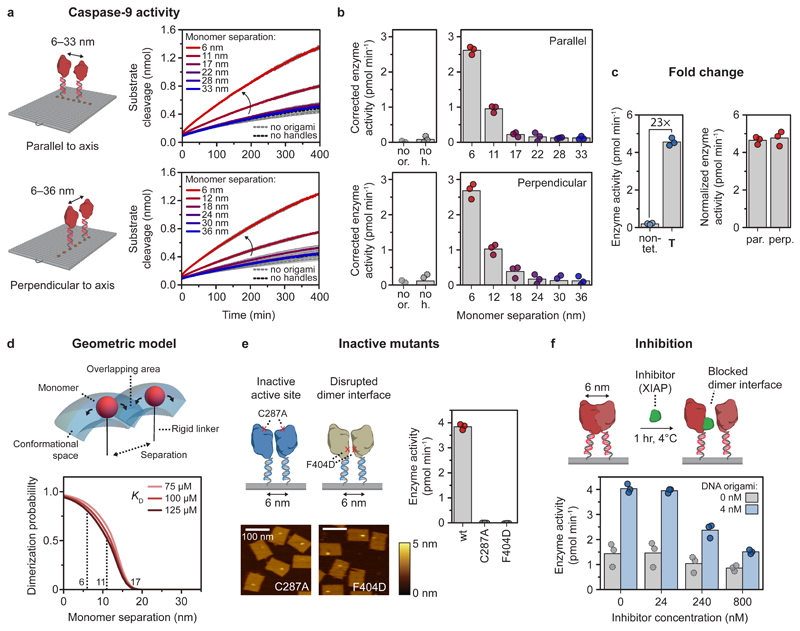
We then assessed if caspase-9 displays functional enzymatic activity when assembled on DNA origami nanostructures. Since proximity-induced activation of caspase-9 involves dimerization, we envisioned that the activity can be tuned by varying the distance between the monomers. To this end, we assembled two-enzyme DNA nanostructures with monomer separation varying between 6 and 36 nm, and followed proteolytic activity over time (Fig. 3a,b and Methods). Highest enzyme activity was observed when the monomers were in closest proximity, after which the activity dropped sharply and approached background activity levels for separations >20 nm (Fig. 3b). Although we use a twist-corrected version of the DNA origami rectangle18 that adopts the designed shape when deposited on a surface for AFM imaging (Fig. 2), theoretical and experimental studies have determined that the single-layer DNA nanostructure can still exhibit moderate flexibility in solution (Supplementary Fig. 9)57,58. To exclude that this structural flexibility has an effect on the distance-dependent performance of our system, we compared enzyme activity in two different arrangements of the caspase-9 monomers (Fig. 3b). Enzyme monomers arranged either parallel to the rigid DNA helical axis (with an expected persistence length >40 nm59) or perpendicular to the axis displayed similar distance-dependent behaviour (Fig. 3b; compare top and bottom graphs), indicating that local fluctuations in protein positions do not have an influence on enzymatic activity. Increasing the single-stranded DNA linker between enzyme and handle from 10 to 15 nt slightly decreased the maximum activity while retaining distance-dependent behaviour, illustrating that enzyme activation requires tight co-localization of the monomers and that the DNA-based assembly method does not introduce adverse steric effects (Supplementary Fig. 18).
Figure 3. Activation of caspase-9 occurs by distance-dependent dimerization of tethered monomers.
Reactions were carried out with 4 nM DNA origami (unless indicated otherwise) and 3 equivalents of enzyme-DNA conjugate per handle, and incubated for 2 hr at 4°C. Activity was determined by monitoring cleavage of 167 μM LEHD-AFC at 18°C. Bars represent mean activity. All experiments were performed in independent triplicates. a,b, Distance-dependent enzyme activity of two caspase-9 monomers on DNA origami nanostructures parallel (top) and perpendicular (bottom) to the DNA helical axis. Data in a is represented as mean ± s.d. of three independent experiments. Enzyme activity (b) was calculated from the initial slope of the time traces in a and corrected by subtracting the mean background activity (no or.). Labels: no or., no DNA origami present; no h., DNA origami without handles. c, Fold change in enzyme activity for template T, and 6-nm samples parallel (par.) and perpendicular (perp.) to DNA origami helical axis was calculated by comparing with 4 nM non-tethered caspase-9 in buffer (non-tet.). The activity of DNA origami samples was normalized based on an incorporation efficiency per handle of 75%. d, Three-dimensional geometric model assuming free movement for each tethered monomer in the conformational space, determined by molecular dynamics simulations. The tethered dimerization probability was plotted as a function of the KD of caspase-9 dimerization in solution and the separation between the tethered monomers. e, AFM images show correct incorporation of caspase-9 point mutants C287A and F404D on DNA origami nanostructures at 6 nm monomer separation, but both mutants exhibit no enzymatic activity. Colour bars indicate height scale in AFM images. Scale bars, 100 nm. f, Inhibition of caspase-9 by various concentrations of X-linked inhibitor of apoptosis protein (XIAP), in the absence (gray) or presence (blue) of 4 nM 6-nm two-enzyme DNA nanostructures.
After correcting for background activity and incorporation efficiency as determined by AFM, DNA origami-mediated caspase-9 activation at 6-nm monomer separation resulted in a 23-fold increase in enzyme activation, equivalent to dimerization enforced by the bivalent template T (Fig. 3c; see Methods for data correction). This activity increase is in the same range as reported in literature for other engineered systems for caspase-9 activation, which obtained fold change values in the range of 3-9036,47,49,50, indicating that DNA-based induction of caspase-9 activity represents a viable strategy for investigating its catalytic function. Although in the cell apoptosome-mediated activation of caspase-9 results in a two to three orders of magnitude upregulation of activity51, our results are comparable with reported activity profiles of in vitro reconstituted native apoptosomes, when using the same synthetic substrate47. Taken together, even though the synthetic DNA-based design does not capture all molecular determinants contributing to caspase-9 activation by the apoptosome, our results demonstrate that proximity-induced dimer formation and subsequent induction of catalytic activity plays a major role in the native system.
To validate DNA origami nanostructures as inert assembly platforms we performed several essential control experiments. First, a control in which DNA origami without handles was used exhibited only background activity, confirming that the DNA nanostructures do not influence caspase-9 function (Fig. 3b; compare dotted gray and black). Next, we assessed the influence of an altered pH near the surface of the negatively-charged DNA origami structure60, by measuring enzyme activity both in solution and mediated by the 6-nm two-enzyme DNA nanostructure at varying pH levels. Both experiments revealed a bell-shaped pH dependence with an optimum at pH 7.061, suggesting that the behaviour of tethered caspase-9 near the surface of the DNA origami platform is not affected by local changes in pH (Supplementary Fig. 19). Finally, we determined the kinetic parameters of the 6-nm two-enzyme DNA nanostructure and found a KM of 1.8 ± 0.1 mM and a Vmax of 51.5±2.1 pmol min-1 (Supplementary Fig. 20). Although the KM is slightly higher and the Vmax is slightly lower than the values reported previously for the bivalent template T (Fig. 1e), it is known that immobilization of enzymes on a surface can affect the kinetic parameters, which can be attributed to a lower affinity of the substrate for the enzyme due to diffusional limitations or conformational changes of the enzyme near the surface62. Collectively, these results establish DNA origami nanostructures as a platform for the assembly of physically interacting enzymes, such as caspase-9, and the systematic analysis of the effects of relative geometry on enzyme activity.
To further rationalize the experimental results, we constructed a geometric model for two tethered interacting enzymes based on the concept of effective concentration63 and calculated the fraction of tethered dimer as a function of the distance between the anchor points (Fig. 3d and Methods). Coarse-grained molecular dynamics simulations were employed to define a conformational volume in which the tethered enzymes can move freely (Supplementary Fig. 21 and Supplementary Methods (Molecular dynamics simulations)). The model allowed us to calculate the dimerization probability as a function of monomer separation and the KD of non-tethered caspase-9 (see Methods)64. The results show that at monomer separation >20 nm, the calculated fraction of dimer approaches zero as the tethered monomers cannot physically interact (Fig. 3d). In accordance with the experimental data, a sharp drop in the dimerization probability is observed in the regime between 5 and 15 nm, decreasing from 90% at 5 nm to less than 10% at 15 nm. Although the model does not consider steric effects or the specific mutual orientation of the monomers, it describes the experimental data well, suggesting that caspase-9 dimerization on the DNA origami platform originates from an increase in effective concentration. Taken together, these results confirm that DNA origami-mediated activation of caspase-9 is consistent with proximity-induced homodimerization, and that the extent of activity can be tuned by varying the separation between interacting monomers.
To illustrate the functionality of the DNA origami platforms for studying protein-protein interactions, we investigated the behaviour of caspase-9 mutants and the effect of inhibition on enzyme activity using a biologically relevant inhibitor. First, we expressed and conjugated two caspase-9 point mutants to DNA (Supplementary Fig. 1b,c, 2b,c and 5), resulting in enzyme-DNA conjugates with either a disabled active site (C287A mutant) or a disrupted dimer interface (F404D mutant). While AFM imaging revealed correct assembly of 6-nm-spaced two-enzyme DNA nanostructures, both mutants did not exhibit enzymatic activity, reaffirming that DNA origami-mediated caspase-9 activation proceeds via a homodimerization mechanism (Fig. 3e and Supplementary Fig. 17). Second, we investigated the response of DNA origami-mediated caspase-9 activity to the BIR3 domain of X-linked inhibitor of apoptosis protein (XIAP), an important human regulatory protein that strongly binds to the C-terminal small subunit of caspase-9 (Ki < 20 nM), forming a heterodimeric complex preventing caspase-9 dimerization65,66. After assembly of 6-nm two-enzyme wildtype caspase-9 DNA nanostructures, increasing concentrations of inhibitor were added and protease activity was measured (Fig. 3f). Although the binding affinity of the inhibitor to caspase-9 is in the low nanomolar range, the experiments reveal that very high concentrations (60 equivalents and higher) are needed to effectively inhibit enzyme activity, illustrating the increased effective concentration of caspase-9 on the DNA origami platform. Combined, these results demonstrate that our DNA origami platform can serve as a versatile tool for biochemical analysis of intracellular signalling components and their regulation by other proteins, such as inhibitors.
Clustering of multiple caspase-9 enhances enzymatic activity
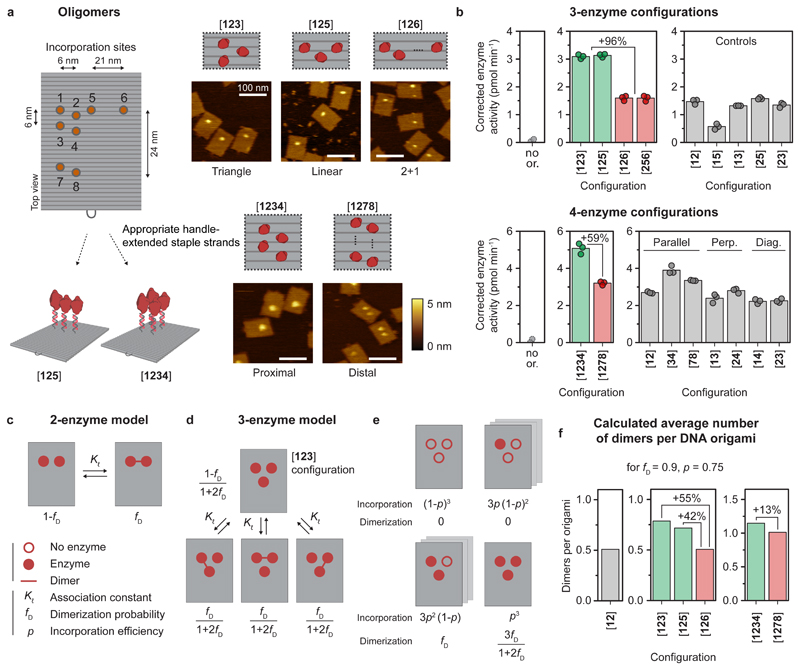
We have demonstrated that caspase-9 dimerization is sufficient to induce activity, however, the apoptosome is hypothesized to bind up to four enzymes simultaneously35,37,53. We therefore wondered how clustering of enzymes influences their catalytic activity. While multivalent effects on enzyme catalysis have not been described in biochemical literature, previous work by Prins et al. on zinc-based catalysts has revealed that clustering of dimerizing subunits can lead to activity enhancement67. The authors developed a theoretical model to show that the activity increase is correlated with a statistical increase in the number of active catalytic units upon clustering of subunits in a multivalent system. To investigate the effect of oligomerization, we constructed several multivalent caspase-9 DNA nanostructures, including linear ([125]) and triangular ([123]) three-enzyme configurations, and a four-enzyme variant ([1234]), and confirmed successful assembly using agarose gel electrophoresis (Supplementary Fig. 22) and AFM imaging (Fig. 4a and Supplementary Fig. 23). For both three-enzyme systems, enzymatic activity increased 96% compared to control configurations [126] and [256], in which one monomer is positioned such that it cannot interact with the other two monomers (Fig. 4b; top). As expected, the enzyme activity of these control 2+1-configurations was similar to the 6-nm two-enzyme controls. Activity of the four-enzyme [1234] proximal configuration increased by 59% compared to the [1278] distal control (Fig. 4b; bottom). Since the latter can be viewed as a non-interacting pair of two-enzyme systems on the same DNA origami, it exhibited similar activity compared to the two-enzyme controls, as expected (Fig. 4b; bottom). Collectively, these experimental results demonstrate an increase in proteolytic activity when more than two caspase-9 monomers are brought into close proximity.
Figure 4. Co-localization of more than two caspase-9 monomers leads to enhanced enzymatic activity.
Reactions were performed as described in Fig. 3. The DNA origami concentration was adjusted to keep the total concentration of enzyme at 24 nM (see Methods and Supplementary Table 3). All experiments were performed in triplicate. a, Schematic overview of possible incorporation sites for handle-extended staple strands (orange circles) for constructing three- and four-enzyme DNA nanostructures. Topographic AFM images show successful incorporation of caspase-9 according to the pre-programmed positions. The bracket notation indicates enzyme configuration as determined by the number and location of the indicated incorporation sites. Colour bars indicate height scale. Scale bars, 100 nm. b Enzymatic activity measurements for three-enzyme (top) and four-enzyme (bottom) DNA nanostructures (green and red) and two-enzyme controls (gray). Activity was corrected by subtracting the mean background activity (no or.) in all samples. Labels: no or., no DNA origami present; perp., perpendicular arrangement; diag., diagonal arrangement. c, Schematic depiction of the mass-balance model for a tethered two-enzyme system, with an equilibrium between a monomeric (left) and dimeric (right) state defined by effective association constant Kℓ, leading to the dimerization probability fD. d, Model for the tethered three-enzyme system in triangular configuration ([123]) (see Supplementary Note 1 for models of other configurations). One monomeric and three symmetric dimeric states can be defined, with corresponding state fractions expressed as a function of fD. e, Schematic depiction of the possible configurations in the triangular three-enzyme system with incomplete enzyme incorporation. f, When taking into account all possible states and incomplete incorporation, the models predict the average number of enzyme dimers per DNA origami for all two-, three-, and four-enzyme systems.
To dissect the potential factors contributing to the observed activity increase, we developed a thermodynamic model describing dimerization of tethered enzymes in two-, three-, and four-enzyme configurations. The underlying principle of the model is that two tethered enzymes exist in an equilibrium between two concentration-independent states, either as inactive monomers or as an active dimer, while higher-order interactions are not possible (Fig. 4c and Supplementary Note 1). This allows us to calculate, for each enzyme configuration, the average number of dimers per DNA origami, which we assume is proportional to the experimentally measured caspase-9 activity. The average number of dimers for the 6-nm two-enzyme DNA nanostructure is simply given by the dimerization probability fD (with a value of approximately 0.9 at 6 nm monomer separation, see Fig. 3c,d), allowing us to express the number of dimers in the other enzyme configurations as a function of system parameter fD. Applying this approach to, for example, the triangular three-enzyme system (which reflects the experimental [123] configuration) we can define four distinct states, one fully monomeric and three symmetric dimeric states, for each of which we derive an expression for the dimer fraction as a function of fD (Fig. 4d). However, in practice enzyme incorporation onto DNA nanostructures is not 100%, resulting in a distribution of species with varying enzyme occupancy, i.e. eight different species with 0, 1, 2, or 3 enzymes (Fig. 4e). We systematically derived expressions for the contribution of each species as a function of fD and incorporation efficiency p (as determined in Fig. 2), with the sum of all contributions representing the average number of dimers per DNA nanostructure (see Supplementary Note 1 for the derivation of all models). Although the model only considers dimerization and does not include any higher-order allosteric effects, the three- and four-enzyme configurations exhibit a moderate increase in the average number of dimers per DNA origami (Fig. 4f; compare green bars to red control bars). This effect has a statistical origin and is correlated to an increase in the number of dimerization possibilities compared to a two-enzyme situation. Interestingly, the models predict that the number of dimers in the [123] three-enzyme system is only 55% higher compared to the two-enzyme system, while experimentally an approximately two-fold increase in activity was observed (Fig. 4f). Similarly, the computed number of dimers of the proximal four-enzyme system is only 13% higher than the distal configuration, while experiments indicated a 59% increase (Fig. 4f; right graph). In addition to statistical effects related to an increased number of dimerization possibilities, we speculate that the discrepancy between theoretical and experimental results points to an allosteric effect in oligomers of three or more caspase-9 enzymes, leading to an additional enhancement in activity.
Taken together, this combined experimental and theoretical approach allowed us to determine the factors contributing to caspase-9 activation in multi-enzyme assemblies, demonstrating an increase in enzyme activity in oligomers of more than two monomers. Importantly, we found that the presence of multiple co-localized binding sites leads to a statistical enhancement in activity by increasing the probability of interaction between tethered enzymes when enzyme occupancy is incomplete. We envision that this principle, facilitated by spatial organisation of enzymes on SMOCs, could represent a general regulatory mechanism for inducing proximity-driven protein-protein interactions4,5.
Enzymatic activity originates from a single catalytic site
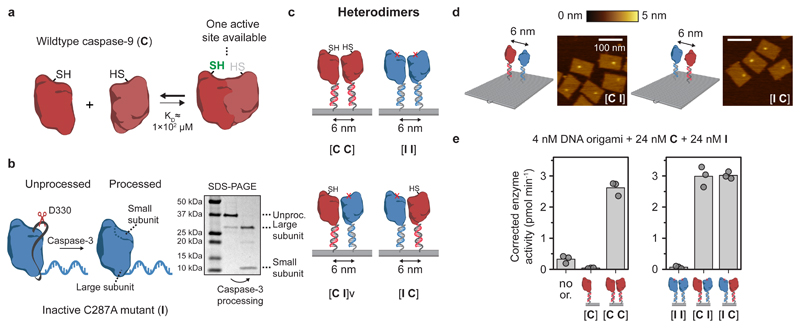
Finally, we used the modularity of the DNA origami method to investigate assembly and activity of caspase-9 heterodimers. Because the crystal structure of the caspase-9 homodimer reveals that only one of the two active sites is in an accessible, open conformation38, we hypothesized that a heterodimer consisting of a wildtype monomer and a mutant with a disabled active site would still display enzymatic activity (Fig. 5a). Recent work suggests that autoproteolytic processing of the intersubunit linker in wildtype caspase-9 is essential for the correct formation of an active dimeric state51. Catalytically inactive point mutant C287A does not undergo autoproteolytic processing, and therefore we induced processing using caspase-3, which is able to cleave caspase-9 in the intersubunit linker generating the large and small enzyme subunit (Fig. 5b and Supplementary Fig. 5)54. We then used wildtype caspase-9 monomer C and inactive monomer I to assemble homo- and heterodimeric two-enzyme DNA nanostructures, and confirmed that the heterodimeric variants assembled correctly using agarose gel electrophoresis (Supplementary Fig. 24) and AFM imaging (Fig. 5c,d and Supplementary Fig. 25). Remarkably, the level of protease activity of both heterodimeric systems [C I] and [I C] is equivalent to the activity of the homodimeric wildtype system [C C] (Fig. 5e). Control experiments, in which similar heterodimeric configurations were tested with unprocessed I and non-dimerizing point mutant F404D, exhibited background activity levels (Supplementary Fig. 26). These results suggest that (1) the formation of an active caspase-9 dimer proceeds through an asymmetric mechanism in which only a single active site is brought into an active conformation, and (2) that cleavage of the intersubunit linker of both monomers is strictly necessary for enzymatic activity. One of the possible mechanisms behind this half-of-sites reactivity is an absolute form of negative cooperativity between the enzyme monomers, where substrate binding in one the active sites abrogates the catalytic activity of the other site through induced conformational changes68–70. These experiments illustrate that the modularity of DNA origami-based platforms can be harnessed to investigate relevant biological questions concerning the molecular mechanisms behind interacting signalling proteins.
Figure 5. Enzymatic activity of the caspase-9 dimer originates from a single catalytic site.
a, Schematic overview of wildtype caspase-9 (C) homodimerization. Crystal structures of dimeric caspase-9 indicate that only one of the two active sites is in an active, accessible conformation. b, Wildtype caspase-9 undergoes autoproteolytic processing at three cleavage sites in the intersubunit linker region (black ribbon), but mutant C287A (I) is inactive and therefore does not undergo autoprocessing. To mimic processing, enzyme-DNA conjugates were incubated with caspase-3, a constitutively active protease that also cleaves caspase-9 in the linker region. Purification was performed as described in Fig. 1, and confirmed by SDS-PAGE under reducing conditions. Label: unproc., unprocessed enzyme-DNA conjugate. c, The modularity of DNA origami was exploited to construct both homo- and heterodimeric 6-nm two-enzyme DNA nanostructures consisting of combinations of wildtype and inactive monomers. The specific configuration of wildtype (C) and inactive (I) mutants is denoted using bracket notation. d, Topographic AFM images of [I C] and [C I] heterodimers. Colour bar indicates height scale. Scale bars, 100 nm. e, Enzymatic activity measurements were performed as described in Fig. 3. In all samples, both 24 nM C and 24 nM processed I were added (for data with unprocessed I, see Supplementary Fig. 26). Activity was corrected by subtracting the mean background activity in all samples. All experiments were performed in triplicate. Label: no or., no DNA origami present.
Conclusions
Many intracellular signalling proteins assemble into multi-molecular complexes composed of unique combinations of pathway components. Co-localization of proteases, kinases, and phosphatases via their association to dedicated scaffold proteins results in the assembly of higher-order signalling machines such as the myddosome, the apoptosome, and the necrosome, that are able to efficiently control signal transmission via proximity-driven enzyme activation4,5,71. Here, we show that DNA origami can be used as a unique in vitro platform for constructing synthetic higher-order signalling machines. As a proof-of-principle, we engineered synthetic DNA origami-based variants of the apoptosome and revealed how the distance and number of caspase-9 monomers influence enzymatic activity. Our results reveal a multivalent catalytic effect as evidenced by an increase in catalytic activity in three- and four-enzyme systems compared to a two-enzyme configuration. A thermodynamic model based on tethered dimerization revealed that the observed activity enhancement partially originates from a statistical increase in the number of active catalytic units in higher-order enzyme configurations. We envision that clustering of catalytic enzymatic subunits into higher-order complexes, either through SMOC-based assembly4,5, functional homotypic interactions72,73, or via liquid phase separation74,75, could represent a general mechanism for enzymatic activity enhancement or regulation in various intracellular processes.
In contrast to other available platforms for engineering higher-order signalling machines, such as synthetic protein scaffolds7 or leucine zipper-induced assemblies47, DNA origami allows oligomerization of non-identical signalling proteins and user-defined control over their number, position, and relative geometry. The construction of higher-order signalling complexes using DNA origami-based SMOCs allows a detailed analysis of their function and can be used to probe unresolved molecular mechanisms in intracellular signalling, such as for example the multivalent enhancement of catalytic activity as reported in this work. Recent work has shown the possibility of genetically encoded DNA and RNA nanostructures and revealed successful intracellular assembly of a simple DNA cross-over nanostructure on which proteins could be organised using orthogonal zinc fingers76–78. As such, in vivo production and assembly of DNA-based SMOCs, analogous to those developed by us, is a realistic possibility and could find application as modular synthetic control elements for diversifying signalling dynamics of existing pathways. We anticipate that DNA origami platforms will find broad use to inform the function of many other important SMOCs for which oligomerization-driven allosteric regulation of non-identical enzymes, such as for example kinases, is a common regulatory principle.
Methods
Expression and purification of caspase-9
The catalytic domain of human caspase-9 (140-416) with an N-terminal amber stop codon was encoded on a pET28a plasmid and synthesized by GenScript. The construct contains an N-terminal His-SUMO tag (the SUMO tag was included to improve stability and solubility during expression) and a C-terminal Strep-tag (see Supplementary Methods (DNA and protein sequence caspase-9)). C287A and F404D mutants were generated using the QuikChange Lightning Multi Site-Directed Mutagenesis kit (Agilent), according to the manufacturer’s instructions and using the primers in Supplementary Table 1. The pEVOL-pAzF vector, encoding for the orthogonal aminoacyl tRNA synthetase/tRNA pair, was a kind gift from Peter Schultz (Addgene plasmid #31186). Both plasmids were transformed into E. coli BL21(DE3) competent bacteria (Novagen) and cultured at 37°C in 500 mL 1xTB (terrific broth, VWR) supplemented with 0.4% (v/v) glycerol, 25 μg/mL kanamycin and 25 μg/mL chloramphenicol. Protein expression was induced at OD600=0.6 by addition of 1 mM β-D-1-thiogalactopyranoside and 0.02% (w/v) arabinose. Simultaneously, the non-canonical amino acid p-azidophenylalanine was added directly to the culture medium at a final concentration of 1 mM. Expression was carried out for ~16 h at 18°C. Cells were harvested by centrifugation at 10,000 g for 10 min at 4°C, the pellet resuspended in lysis buffer (10 mL per gram pellet, 1x PBS, 370 mM NaCl, 10% (v/v) glycerol, 20 mM imidazole, Benzonase nuclease (25 U per 10 mL buffer, Novagen), pH 7.4) and the cells were lysed using an EmulsiFlexC3 High Pressure homogenizer (Avestin) at 15,000 psi for two rounds. The soluble fraction (cleared lysate) containing caspase-9 was collected by centrifugation at 40,000 g for 30 min at 4°C.
Typically, non-canonical amino acids are not fully incorporated leading to the presence of truncated protein fragments. Therefore, purification was performed by both Ni2+-affinity chromatography and Strep-tactin affinity chromatography, using the N- and C-terminal affinity tags on caspase-9, respectively. The cleared lysate was loaded on a Ni-charged column (His-Bind® Resin, Novagen), washed with wash buffer (1x PBS, 370 mM NaCl, 10% (v/v) glycerol, 20 mM imidazole, pH 7.4), and eluted with elution buffer (1x PBS, 370 mM NaCl, 10% (v/v) glycerol, 250 mM imidazole, pH 7.4). Cleavage of the N-terminal His-SUMO tag was performed by adding SUMO protease dtUD1 (1:500, purified according to standard procedure79) and 2 mM EDTA to the elution fractions, while dialyzing (MWCO 3.5 kDa, Thermo Fisher) against 4 L of dialysis buffer (50 mM Tris, 150 mM NaCl, pH 8.0) for ~16 hr at 4°C. Finally, the concentrate was applied to a Strep-tactin column (Superflow® resin, IBA Life Sciences). The column was washed with wash buffer (100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH 8.0) and the protein eluted with wash buffer supplemented with 2.5 mM desthiobiotin. Elution fractions were combined and concentrated using Amicon 10 kDa MWCO centrifugal filters (Merck Millipore) to a final concentration of ~1.5 mg/mL (~47 μM), and then snap frozen in liquid nitrogen and stored in 100 μL aliquots at -80°C. The concentration of caspase-9 was determined by measuring the absorption at 280 nm (NanoDrop 1000, Thermo Scientific) assuming an extinction coefficient of 3.1×104 M-1 cm-1 80. Total yield after purification typically was ~4 mg/L culture medium. Purity of caspase-9 was assessed using SDS-PAGE under reducing conditions (Supplementary Fig. 1). Molecular weight was confirmed using liquid chromatography quadrupole time-of-flight mass spectrometry (Waters ACQUITY UPLC I-Class System coupled to a Xevo G2 Q-ToF) by injecting a 0.1 μL sample into an Agilent Polaris C18A RP column with a flow of 0.3 mL/min and a 15-60% acetonitrile gradient containing 0.1% formic acid (Supplementary Fig. 2).
Enzyme-DNA conjugation
Functionalization of amino-modified ODNs with bicyclononyne (BCN) is described in the Supplementary Methods (Functionalization of oligonucleotides). Before conjugation, 100 μL caspase-9 aliquots were buffer exchanged to wash buffer (100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH 8.0) using Zeba desalting columns (7 kDa MWCO, 0.5 mL, Thermo Scientific) according to the manufacturer’s instructions, to remove all remaining desthiobiotin. Typically, conjugation reactions were carried out on a 500 μL scale using 10 μM protein and 30 μM BCN-DNA in reaction buffer (100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 0.1% CHAPS (w/v), pH 8.0) for ~16 hr at 4°C. Competing thiol-yne reactions of BCN-DNA with the 11 cysteines in caspase-9 were suppressed by pre-incubating the protein with 1 mM β-mercaptoethanol for 30 min at 4°C81. For proteolytic processing of C287A and F404D mutants, 0.5 μM active caspase-3 (expressed and purified as described82) was added directly after the conjugation reaction and incubated for 2 hr at 18°C.
To remove excess BCN-DNA, Strep-tactin affinity chromatography was performed as described above. Then, ion-exchange chromatography was performed to remove unreacted protein. After equilibration of the ion-exchange column (0.5 mL strong anion-exchange spin columns, Thermo Scientific) with purification buffer (100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.1% CHAPS (w/v), pH 8.0), the protein mixture was directly loaded onto the column in 400 μL fractions, according to the manufacturer’s instructions. Elution was performed by stepwise increase of the NaCl concentration in the buffer (200, 300, 400, 500, 600 mM, respectively). Typically, the protein eluted at <300 mM NaCl, while enzyme-DNA conjugates eluted at 500-600 mM NaCl (Supplementary Fig. 4 and 5). Elution fractions containing pure enzyme-DNA conjugates were pooled, supplemented with glycerol (5% (v/v) final concentration), snap frozen in liquid nitrogen, and stored at -80°C in 5 μL aliquots. The concentration of purified enzyme-DNA conjugates was determined with gel densitometry on SDS-PAGE. To this end, conjugate gel band intensity was determined using the ImageJ (v1.52n) gel analysis plugin and then compared to a calibration curve of known concentrations of protein (Supplementary Fig. 7).
Assembly of caspase-9 on template T
The linear 30-nt single-stranded template T was designed to act as a bivalent scaffold for caspase-9 (5’ to 3’: TCATACGACTCACTCCTGACTGACTGACTG), simultaneously hybridizing to enzyme-DNA conjugates with anti-handle sequences a1 and a2 (Supplementary Table 2), similar to designs used in literature46,83. Activation of caspase-9 with template T was performed by adding the two caspase-9 enzyme-DNA conjugates and template T in equimolar amounts (4 nM) for 2 hr at 4°C in activity buffer (10 mM Tris, 1 mM EDTA, 10 mM MgCl2, 100 mM NaCl, 1 mM DTT, 0.1% (w/v) CHAPS, pH 7.5). Enzyme activity was measured as described below.
Assembly of caspase-9 on DNA origami
The DNA origami rectangle used in this study was based on the tall rectangle design by Rothemund18. Staple strands and handle-extended staple strands were obtained in desalted form from Integrated DNA Technologies and dissolved at a stock concentration of 500 μM in DNase/RNase-free water. M13mp18 scaffold was purchased from Eurofins. Folding was performed on a 25-nM scaffold scale and purification was performed according to standard procedure using 100 kDa MWCO 0.5 mL Amicon centrifugal filters (Merck Millipore). For detailed notes on design and assembly of DNA origami nanostructures, and characterisation with gel electrophoresis and AFM imaging, see Supplementary Methods.
Incorporation of enzyme-DNA conjugates onto purified DNA origami nanostructures was performed by incubating DNA origami with 3 molar equivalents of caspase-9 DNA conjugate per handle for 2 hr at 4°C in activity buffer (10 mM Tris, 1 mM EDTA, 10 mM MgCl2, 100 mM NaCl, 1 mM DTT, 0.1% (w/v) CHAPS, pH 7.6). The total concentration of caspase-9 DNA conjugate was kept constant at 24 nM for all experiments, reflecting typical concentrations in the cytosol34. As a result, 4 nM, 2.67 nM, and 2 nM DNA origami was used for two-, three-, and four-enzyme DNA nanostructures, respectively. To allow comparison of the activity of three-enzyme DNA nanostructures with two-enzyme controls, all experiments in the top row of Fig. 4b including the two-enzyme configurations, were performed with 2.67 nM DNA origami (for an overview of component concentrations, see Supplementary Table 3). For heterodimer experiments, the concentration of each enzyme-DNA conjugate was kept at 24 nM, which results in the same background activity originating from wildtype caspase-9. For inhibition experiments, varying concentrations of XIAP (human recombinant BIR3-XIAP, R&D Systems) were added after caspase-9 incorporation, and the reaction was incubated additionally for 1 hr at 4°C before measuring enzyme activity at 18°C, as described below.
Activity assays
Enzyme activity was measured using the synthetic tetrapeptide caspase-9 substrate LEHD (dissolved in dry DMSO at 10 mM)47, which is cleaved by caspase-9 after the aspartic acid residue releasing and unquenching the fluorescent dye 7-amino-4-(trifluoromethyl)coumarin (AFC). After assembly of caspase-9 on DNA origami, substrate was added to a final concentration of 167 μM and proteolytic cleavage was monitored over time in 384-well plates (60 μL reaction volume) at 18°C by measuring fluorescence (ex.: 400 nm, em.: 505 nm) in a Tecan Spark 10M platereader. Fluorescence units were converted to concentration using a calibration curve (Supplementary Fig. 8). To this end, varying concentrations of free AFC (dissolved in dry DMSO at 10 mM) were added to activity buffer, and fluorescence was measured as described. Raw data of all activity assays were extracted, converted, and formatted using in-house MATLAB scripts (R2015a). Enzyme activity (in pmol min-1) was determined by fitting the initial slope (20-60 min) of the kinetic trace to a linear curve. Kinetic parameters were determined by measuring enzyme activity as a function of varying LEHD concentration between 0-1.5 mM, and fitting the results with the standard Michaelis-Menten equation:
| (1) |
with Vmax the maximum rate (in pmol min-1) and KM the Michaelis constant (in mM). Because high concentrations of the substrate (>0.5 mM) strongly influence the pH, kinetic analyses were performed at a higher buffer concentration (50 mM HEPES, 5 mM Tris, 1 mM EDTA, 10 mM MgCl2, 100 mM NaCl, 1 mM DTT, 0.1% (w/v) CHAPS, pH 7.6).
Data processing and correction
Typically, an excess of caspase-9 enzyme-DNA conjugate was used to incorporate caspase-9 onto DNA nanostructures, leading to background activity originating from untethered enzymes remaining in solution. In some cases background correction was performed by measuring the activity of untethered caspase-9 without DNA origami, at the same concentration as in measurements with DNA origami, in triplicate and in parallel for each experiment (indicated by no origami). Subsequently, the mean activity was subtracted from the enzyme activity of other samples, leading to the corrected enzyme activity, as reported in Fig. 3b, 4b, and 5e. The corrected enzyme activity for one-enzyme DNA nanostructures (see Fig. 5e) and two-enzyme DNA nanostructures at large monomer separation (>20 nm, see Fig. 3b) is similar to the negative controls (no origami and no handles; see Fig. 3b), suggesting that untethered caspase-9 in solution does not interact with or influence the behaviour of caspase-9 on DNA nanostructures.
To compare activation of caspase-9 on DNA nanostructures with bivalent template T as reported in Fig. 3c, activity normalization was performed. Due to incomplete enzyme incorporation (75% per handle, as determined in Fig. 2), only ~56% (or ~2.3 nM) of the two-enzyme DNA nanostructures contain two enzymes and are therefore in an active state. Because the enzyme activity of caspase-9 on DNA nanostructures is concentration independent, the corrected enzyme activity was normalized to 100% enzyme incorporation (or 4 nM DNA origami), leading to the normalized enzyme activity as reported in Fig. 3c. Assuming quantitative assembly of caspase-9 on template T (melting temperature >40°C for 15-nt handle-anti-handle duplexes at 4 nM84), this allows comparison of the activity of caspase-9 assembled on either DNA origami or 4 nM template T. The fold change in Fig. 3c was calculated based on the untethered enzyme activity of 4 nM caspase-9 in solution (indicated by non-tet.).
Geometric model
Molecular dynamics simulations were performed to determine tethered handle-anti-handle movement on the DNA origami surface using oxDNA (v2.2.2), a robust coarse-grained molecular dynamics model specifically optimized for the accurate modeling of DNA nanostructures (see Supplementary Methods (Molecular dynamics simulations)58. Based on the results of these simulations, a three-dimensional geometric model was constructed using Mathematica (v10, Wolfram). The model includes two tethered particles separated by a distance s, reflecting the experimental setup detailed in Fig. 2 and Fig. 3a,b. The diameter of a caspase-9 monomer was estimated to be 4.5 nm based on the crystal structure (PDB: 1JXQ). The tether connecting the enzyme to the DNA origami scaffold consists of the 15-bp handle-anti-handle DNA duplex, the BCN-azide moiety, single-stranded DNA linkers, and a short peptide linker, and was estimated to have a total length of approximately 8 nm (Supplementary Fig. 21). By following the approach of Van Valen et al.64, the model allows us to calculate the probability that the tethered particles form a dimer (expressed as the dimerization probability fD) as a function of monomer separation s and the KD of dimerization in solution. The approach relies on treating the particles as point objects and assuming that the particles are free to dimerize when they are within each other’s vicinity. The probability that the particles are close depends on the exact conformational movement of both particles. A particle’s conformational space with volume vs is defined based on a hemi-shell bounded by an angle of 35° with the scaffold, as determined by the overall movement of the handle-anti-handle duplex in the coarse-grained simulations (Supplementary Fig. 21). The intersection with volume vi(s) between two conformational spaces can then be calculated as a function of the separation between the two particles. Assuming the particles can move freely and homogeneously in their conformational spaces and independently of each other, we can calculate the probability p1(s) that one particle is at the intersection as
| (2) |
and probability that two particles are at the intersection. This allows us to define a probability density function
| (3) |
as the concentration of tethered particles in each other’s vicinity. Note that J is expressed in units of concentration, and can therefore be viewed as an effective concentration. With this, we can write an expression for the dimerization probability fD based on statistical mechanical treatment of the system64, as
| (4) |
This expression was used for the graph shown in Fig. 3d. The KD of non-tethered caspase-9 dimerization in solution is not known precisely but reported in the high micromolar range (> 50 μM)34, and therefore results are shown for several values of KD in this range.
Supplementary Material
Acknowledgements
We thank Joost van Dongen for help with the mass spectrometry analyses, Nick van der Zon for initial protein expression experiments, and Glenn Cremers for helpful discussions. The ICMS Animation Studio contributed the cartoons of DNA strands and the DNA origami structure. This work was supported by the European Research Council, ERC (project n. 677313 BioCircuit), an NWO-VIDI grant from the Netherlands Organisation for Scientific Research (NWO, 723.016.003) and funding from the Ministry of Education, Culture and Science (Gravity programs, 024.001.035 & 024.003.013).
Footnotes
Data availability
The data that support the plots within this paper and other findings of this study are available from the corresponding author (t.f.a.d.greef@tue.nl) upon reasonable request.
Code availability
Custom-written code for the computer models and simulations that support the experimental findings in this study is available from the corresponding author (t.f.a.d.greef@tue.nl) upon reasonable request.
Author contributions
B.R. designed the study, performed experiments, developed the geometric model, analysed the data, and wrote the manuscript. A.M. developed and derived the thermodynamic model and analysed the data. B.G.A. performed and analysed all AFM measurements. J.R. performed molecular dynamics simulations. A.d.H. performed initial protein expression and provided critical input for the experiments. L.B. supervised the study and provided critical feedback on the manuscript. T.d.G. conceived, designed, and supervised the study, analysed the data, and wrote the manuscript. All authors discussed the results and commented on the manuscript.
Competing interests
The authors declare no competing interests.
References
- 1.Bhattacharyya RP, Reményi A, Yeh BJ, Lim WA. Domains, motifs, and scaffolds: the role of modular interactions in the evolution and wiring of cell signaling circuits. Annu Rev Biochem. 2006;75:655–680. doi: 10.1146/annurev.biochem.75.103004.142710. [DOI] [PubMed] [Google Scholar]
- 2.Park HH, et al. The death domain superfamily in intracellular signaling of apoptosis and inflammation. Annu Rev Immunol. 2007;25:561–586. doi: 10.1146/annurev.immunol.25.022106.141656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Good MC, Zalatan JG, Lim WA. Scaffold proteins: hubs for controlling the flow of cellular information. Science. 2011;332:680–686. doi: 10.1126/science.1198701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu H. Higher-order assemblies in a new paradigm of signal transduction. Cell. 2013;153:287–292. doi: 10.1016/j.cell.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kagan JC, Magupalli VG, Wu H. SMOCs: Supramolecular organizing centres that control innate immunity. Nat Rev Immunol. 2014;14:821–826. doi: 10.1038/nri3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin S-C, Lo Y-C, Wu H. Helical assembly in the MyD88–IRAK4–IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465:885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park S-H, Zarrinpar A, Lim WA. Rewiring MAP kinase pathways using alternative scaffold assembly mechanisms. Science. 2003;299:1061–1064. doi: 10.1126/science.1076979. [DOI] [PubMed] [Google Scholar]
- 8.Tan Y, Kagan JC. Innate immune signaling organelles display natural and programmable signaling flexibility. Cell. 2019;177:384–398.e11. doi: 10.1016/j.cell.2019.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kholodenko BN, Hancock JF, Kolch W. Signalling ballet in space and time. Nat Rev Mol Cell Biol. 2010;11:414–426. doi: 10.1038/nrm2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castellana M, et al. Enzyme clustering accelerates processing of intermediates through metabolic channeling. Nat Biotechnol. 2014;32:1011–1018. doi: 10.1038/nbt.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim WA. Designing customized cell signalling circuits. Nat Rev Mol Cell Biol. 2010;11:393–403. doi: 10.1038/nrm2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conrado RJ, Varner JD, DeLisa MP. Engineering the spatial organization of metabolic enzymes: mimicking nature’s synergy. Curr Opin Biotechnol. 2008;19:492–499. doi: 10.1016/j.copbio.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 13.Idan O, Hess H. Engineering enzymatic cascades on nanoscale scaffolds. Curr Opin Biotechnol. 2013;24:606–611. doi: 10.1016/j.copbio.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 14.Chen Y-J, Groves B, Muscat RA, Seelig G. DNA nanotechnology from the test tube to the cell. Nat Nanotechnol. 2015;10:748–760. doi: 10.1038/nnano.2015.195. [DOI] [PubMed] [Google Scholar]
- 15.Seeman NC, Sleiman HF. DNA nanotechnology. Nat Rev Mater. 2017;3 17068. [Google Scholar]
- 16.Engelen W, Janssen BMG, Merkx M. DNA-based control of protein activity. Chem Commun. 2016;52:3598–3610. doi: 10.1039/c5cc09853j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu J, Liu M, Liu Y, Yan H. Spatially-interactive biomolecular networks organized by nucleic acid nanostructures. Acc Chem Res. 2012;45:1215–1226. doi: 10.1021/ar200295q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rothemund PWK. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 19.Voigt NV, et al. Single-molecule chemical reactions on DNA origami. Nat Nanotechnol. 2010;5:200–203. doi: 10.1038/nnano.2010.5. [DOI] [PubMed] [Google Scholar]
- 20.Saccà B, Niemeyer CM. Functionalization of DNA nanostructures with proteins. Chem Soc Rev. 2011;40:5910–5921. doi: 10.1039/c1cs15212b. [DOI] [PubMed] [Google Scholar]
- 21.Udomprasert A, et al. Amyloid fibrils nucleated and organized by DNA origami constructions. Nat Nanotechnol. 2014;9:537–541. doi: 10.1038/nnano.2014.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu W, et al. A programmable DNA origami platform to organize SNAREs for membrane fusion. J Am Chem Soc. 2016;138:4439–4447. doi: 10.1021/jacs.5b13107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Funke JJ, et al. Uncovering the forces between nucleosomes using DNA origami. Sci Adv. 2016;2:e1600974. doi: 10.1126/sciadv.1600974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le JV, et al. Probing nucleosome stability with a DNA origami nanocaliper. ACS Nano. 2016;10:7073–7084. doi: 10.1021/acsnano.6b03218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ketterer P, et al. DNA origami scaffold for studying intrinsically disordered proteins of the nuclear pore complex. Nat Commun. 2018;9 doi: 10.1038/s41467-018-03313-w. 902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masubuchi T, et al. Construction of integrated gene logic-chip. Nat Nanotechnol. 2018;13:933–940. doi: 10.1038/s41565-018-0202-3. [DOI] [PubMed] [Google Scholar]
- 27.Zhao Z, et al. Nanocaged enzymes with enhanced catalytic activity and increased stability against protease digestion. Nat Commun. 2016;7 doi: 10.1038/ncomms10619. 10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Linko V, Eerikäinen M, Kostiainen MA. A modular DNA origami-based enzyme cascade nanoreactor. Chem Commun. 2015;51:5351–5354. doi: 10.1039/c4cc08472a. [DOI] [PubMed] [Google Scholar]
- 29.Wilner OI, et al. Enzyme cascades activated on topologically programmed DNA scaffolds. Nat Nanotechnol. 2009;4:249–254. doi: 10.1038/nnano.2009.50. [DOI] [PubMed] [Google Scholar]
- 30.Fu J, Liu M, Liu Y, Woodbury NW, Yan H. Interenzyme substrate diffusion for an enzyme cascade organized on spatially addressable DNA nanostructures. J Am Chem Soc. 2012;134:5516–5519. doi: 10.1021/ja300897h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Timm C, Niemeyer CM. Assembly and purification of enzyme-functionalized DNA origami structures. Angew Chem Int Ed. 2015;54:6745–6750. doi: 10.1002/anie.201500175. [DOI] [PubMed] [Google Scholar]
- 32.Ngo TA, Nakata E, Saimura M, Morii T. Spatially organized enzymes drive cofactor-coupled cascade reactions. J Am Chem Soc. 2016;138:3012–3021. doi: 10.1021/jacs.5b10198. [DOI] [PubMed] [Google Scholar]
- 33.Fu J, et al. Multi-enzyme complexes on DNA scaffolds capable of substrate channelling with an artificial swinging arm. Nat Nanotechnol. 2014;9:531–536. doi: 10.1038/nnano.2014.100. [DOI] [PubMed] [Google Scholar]
- 34.Riedl SJ, Salvesen GS. The apoptosome: signalling platform of cell death. Nat Rev Mol Cell Biol. 2007;8:405–413. doi: 10.1038/nrm2153. [DOI] [PubMed] [Google Scholar]
- 35.Hu Q, et al. Molecular determinants of caspase-9 activation by the Apaf-1 apoptosome. Proc Natl Acad Sci USA. 2014;111:16254–16261. doi: 10.1073/pnas.1418000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y, et al. Mechanistic insights into caspase-9 activation by the structure of the apoptosome holoenzyme. Proc Natl Acad Sci USA. 2017;114:1542–1547. doi: 10.1073/pnas.1620626114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng TC, Hong C, Akey IV, Yuan S, Akey CW. A near atomic structure of the active human apoptosome. eLife. 2016;5:e17755. doi: 10.7554/eLife.17755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Renatus M, Stennicke HR, Scott FL, Liddington RC, Salvesen GS. Dimer formation drives the activation of the cell death protease caspase 9. Proc Natl Acad Sci USA. 2001;98:14250–14255. doi: 10.1073/pnas.231465798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niemeyer CM. Semisynthetic DNA-protein conjugates for biosensing and nanofabrication. Angew Chem Int Ed. 2010;49:1200–1216. doi: 10.1002/anie.200904930. [DOI] [PubMed] [Google Scholar]
- 40.Chin JW, et al. Addition of p-azido-l-phenylalanine to the genetic code of Escherichia coli. J Am Chem Soc. 2002;124:9026–9027. doi: 10.1021/ja027007w. [DOI] [PubMed] [Google Scholar]
- 41.Chin JW. Expanding and reprogramming the genetic code of cells and animals. Annu Rev Biochem. 2014;83:379–408. doi: 10.1146/annurev-biochem-060713-035737. [DOI] [PubMed] [Google Scholar]
- 42.Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu Rev Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 43.Marth G, et al. Precision templated bottom-up multiprotein nanoassembly through defined click chemistry linkage to DNA. ACS Nano. 2017;11:5003–5010. doi: 10.1021/acsnano.7b01711. [DOI] [PubMed] [Google Scholar]
- 44.Glettenberg M, Niemeyer CM. Tuning of peroxidase activity by covalently tethered DNA oligonucleotides. Bioconjug Chem. 2009;20:969–975. doi: 10.1021/bc800558g. [DOI] [PubMed] [Google Scholar]
- 45.Trads JB, Tørring T, Gothelf KV. Site-selective conjugation of native proteins with DNA. Acc Chem Res. 2017;50:1367–1374. doi: 10.1021/acs.accounts.6b00618. [DOI] [PubMed] [Google Scholar]
- 46.Sancho Oltra N, Bos J, Roelfes G. Control over enzymatic activity by DNA-directed split enzyme reassembly. ChemBioChem. 2010;11:2255–2258. doi: 10.1002/cbic.201000517. [DOI] [PubMed] [Google Scholar]
- 47.Yin Q, et al. Caspase-9 holoenzyme is a specific and optimal procaspase-3 processing machine. Mol Cell. 2006;22:259–268. doi: 10.1016/j.molcel.2006.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pop C, Timmer J, Sperandio S, Salvesen GS. The apoptosome activates caspase-9 by dimerization. Mol Cell. 2006;22:269–275. doi: 10.1016/j.molcel.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 49.Chao Y, et al. Engineering a dimeric caspase-9: a re-evaluation of the induced proximity model for caspase activation. PLoS Biol. 2005;3:e183. doi: 10.1371/journal.pbio.0030183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Den Hamer A, et al. Small-molecule-induced and cooperative enzyme assembly on a 14-3-3 scaffold. ChemBioChem. 2017;18:331–335. doi: 10.1002/cbic.201600631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu Q, Wu D, Chen W, Yan Z, Shi Y. Proteolytic processing of the caspase-9 zymogen is required for apoptosome-mediated activation of caspase-9. J Biol Chem. 2013;288:15142–15147. doi: 10.1074/jbc.M112.441568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shiozaki EN, Chai J, Shi Y. Oligomerization and activation of caspase-9, induced by Apaf-1 CARD. Proc Natl Acad Sci USA. 2002;99:4197–4202. doi: 10.1073/pnas.072544399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu C-C, et al. The Apaf-1 apoptosome induces formation of caspase-9 homo- and heterodimers with distinct activities. Nat Commun. 2016;7 doi: 10.1038/ncomms13565. 13565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huber KL, Serrano BP, Hardy JA. Caspase-9 CARD : core domain interactions require a properly formed active site. Biochem J. 2018;475:1177–1196. doi: 10.1042/BCJ20170913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saccà B, et al. Orthogonal protein decoration of DNA origami. Angew Chem Int Ed. 2010;49:9378–9383. doi: 10.1002/anie.201005931. [DOI] [PubMed] [Google Scholar]
- 56.Rosier BJHM, et al. Incorporation of native antibodies and Fc-fusion proteins on DNA nanostructures via a modular conjugation strategy. Chem Commun. 2017;53:7393–7396. doi: 10.1039/c7cc04178k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baker MAB, et al. Dimensions and global twist of single-layer DNA origami measured by small-angle X-ray scattering. ACS Nano. 2018;12:5791–5799. doi: 10.1021/acsnano.8b01669. [DOI] [PubMed] [Google Scholar]
- 58.Snodin BEK, Schreck JS, Romano F, Louis AA, Doye JPK. Coarse-grained modelling of the structural properties of DNA origami. Nucleic Acids Res. 2019;47:1585–1597. doi: 10.1093/nar/gky1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sa-Ardyen P, Vologodskii AV, Seeman NC. The flexibility of DNA double crossover molecules. Biophys J. 2003;84:3829–3837. doi: 10.1016/S0006-3495(03)75110-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Y, Tsitkov S, Hess H. Proximity does not contribute to activity enhancement in the glucose oxidase–horseradish peroxidase cascade. Nat Commun. 2016;7 doi: 10.1038/ncomms13982. 13982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stennicke HR, Salvesen GS. Biochemical characteristics of caspases-3, -6, -7, and -8. J Biol Chem. 1997;272:25719–25723. doi: 10.1074/jbc.272.41.25719. [DOI] [PubMed] [Google Scholar]
- 62.Wingard LB, Jr, Katchalski-Katzir E, Goldstein L. Applied Biochemistry and Bioengineering. Volume 1: Immobilized Enzyme Principles. Academic Press; New York: 1976. [Google Scholar]
- 63.Page MI, Jencks WP. Entropic contributions to rate accelerations in enzymic and intramolecular reactions and the chelate effect. Proc Natl Acad Sci USA. 1971;68:1678–1683. doi: 10.1073/pnas.68.8.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Van Valen D, Haataja M, Phillips R. Biochemistry on a leash: the roles of tether length and geometry in signal integration proteins. Biophys J. 2009;96:1275–1292. doi: 10.1016/j.bpj.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Salvesen GS, Duckett CS. IAP proteins: blocking the road to death’s door. Nat Rev Mol Cell Biol. 2002;3:401–410. doi: 10.1038/nrm830. [DOI] [PubMed] [Google Scholar]
- 66.Shiozaki EN, et al. Mechanism of XIAP-mediated inhibition of caspase-9. Mol Cell. 2003;11:519–527. doi: 10.1016/s1097-2765(03)00054-6. [DOI] [PubMed] [Google Scholar]
- 67.Zaupa G, Scrimin P, Prins LJ. Origin of the dendritic effect in multivalent enzyme-like catalysts. J Am Chem Soc. 2008;130:5699–5709. doi: 10.1021/ja7113213. [DOI] [PubMed] [Google Scholar]
- 68.Hill TL, Levitzki A. Subunit neighbor interactions in enzyme kinetics: half-of-the-sites reactivity in a dimer. Proc Natl Acad Sci USA. 1980;77:5741–5745. doi: 10.1073/pnas.77.10.5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Biemann HP, Koshland DE. Aspartate receptors of Escherichia coli and Salmonella typhimurium bind ligand with negative and half-of-the-sites cooperativity. Biochemistry. 1994;33:629–634. doi: 10.1021/bi00169a002. [DOI] [PubMed] [Google Scholar]
- 70.Vivoli M, Pang J, Harmer NJ. A half-site multimeric enzyme achieves its cooperativity without conformational changes. Sci Rep. 2017;7 doi: 10.1038/s41598-017-16421-2. 16529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Csizmok V, Follis AV, Kriwacki RW, Forman-Kay JD. Dynamic protein interaction networks and new structural paradigms in signaling. Chem Rev. 2016;116:6424–6462. doi: 10.1021/acs.chemrev.5b00548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Levy ED, Erba EB, Robinson CV, Teichmann SA. Assembly reflects evolution of protein complexes. Nature. 2008;453:1262–1265. doi: 10.1038/nature06942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bergendahl LT, Marsh JA. Functional determinants of protein assembly into homomeric complexes. Sci Rep. 2017;7 doi: 10.1038/s41598-017-05084-8. 4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shin Y, Brangwynne CP. Liquid phase condensation in cell physiology and disease. Science. 2017;357 doi: 10.1126/science.aaf4382. eaaf4382. [DOI] [PubMed] [Google Scholar]
- 76.Elbaz J, Yin P, Voigt CA. Genetic encoding of DNA nanostructures and their self-assembly in living bacteria. Nat Commun. 2016;7 doi: 10.1038/ncomms11179. 11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Praetorius F, Dietz H. Self-assembly of genetically encoded DNA-protein hybrid nanoscale shapes. Science. 2017;355 doi: 10.1126/science.aam5488. eaam5488. [DOI] [PubMed] [Google Scholar]
- 78.Delebecque CJ, Lindner AB, Silver PA, Aldaye FA. Organization of intracellular reactions with rationally designed RNA assemblies. Science. 2011;333:470–474. doi: 10.1126/science.1206938. [DOI] [PubMed] [Google Scholar]
- 79.Weeks SD, Drinker M, Loll PJ. Ligation independent cloning vectors for expression of SUMO fusions. Protein Expr Purif. 2007;53:40–50. doi: 10.1016/j.pep.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gasteiger E, et al. The Proteomics Protocols Handbook. Humana Press; Totowa: 2005. Protein identification and analysis tools on the ExPASy server; pp. 571–607. [Google Scholar]
- 81.Tian H, Sakmar TP, Huber T. A simple method for enhancing the bioorthogonality of cyclooctyne reagent. Chem Commun. 2016;52:5451–5454. doi: 10.1039/c6cc01321j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Den Hamer A, et al. Bright bioluminescent BRET sensor proteins for measuring intracellular caspase activity. ACS Sensors. 2017;2:729–734. doi: 10.1021/acssensors.7b00239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Janssen BMG, Engelen W, Merkx M. DNA-directed control of enzyme–inhibitor complex formation: a modular approach to reversibly switch enzyme activity. ACS Synth Biol. 2015;4:547–553. doi: 10.1021/sb500278z. [DOI] [PubMed] [Google Scholar]
- 84.Zadeh JN, et al. NUPACK: Analysis and design of nucleic acid systems. J Comput Chem. 2011;32:170–173. doi: 10.1002/jcc.21596. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.