SUMMARY
• Plant immune responses need to be tightly controlled for growth-defense balance. The mechanism underlying this tight control is not fully understood. Here we identify epigenetic regulation of nucleotide-binding leucine rich repeat or Nod-Like Receptor (NLR) genes as an important mechanism for immune responses.
• Through a sensitized genetic screen and molecular studies, we identified and characterized HOS15 and its associated protein HDA9 as negative regulators of immunity and NLR gene expression.
• The loss-function of HOS15 or HDA9 confers enhanced resistance to pathogen infection accompanied with increased expression of one-third of the 207 NLR genes in Arabidopsis thaliana. HOS15 and HDA9 are physically associated with some of these NLR genes and repress their expression likely through reducing the acetylation of H3K9 at these loci. In addition, these NLR genes are repressed by HOS15 under both pathogenic and non-pathogenic conditions but by HDA9 only under infection condition.
• Together, this study uncovers a previously uncharacterized histone deacetylase complex in plant immunity and highlights the importance of epigenetic regulation of NLR genes in modulating growth-defense balance.
Keywords: Arabidopsis thaliana, histone deacetylation, HOS15, HDA9, NLR genes, plant immunity
INTRODUCTION
Plants are exposed to various abiotic and biotic stresses and have evolved elaborate mechanisms to adapt to unfavorable environment. In response to microbial pathogens, plants utilize at least two layers of innate immune systems (Jones, et al., 2006). The first layer is triggered by the recognition of conserved microbe features called pathogen-associated molecular patterns (PAMP) through plasma membrane-localized pattern recognition receptors (PRR). This first layer is therefore named pattern-triggered immunity (PTI). Adapted pathogens could suppress PTI by secreting effector proteins to help pathogens invade plants. To fight back invaded pathogens, plants have evolved the second layer of plant innate immunity called effector-triggered immunity (ETI). During ETI, the effectors are recognized directly or indirectly by intracellular disease resistance gene-encoded receptors, most of which are nucleotide-binding leucine rich repeat/ NOD-like receptor (NB-LRR or NLR) proteins (Meyers et al., 2003; Urbach et al., 2017). NB-LRRs mainly fall into two classes by the difference in their N-terminus: CC-NB-LRRs contain a coiled-coil domain, whereas TIR-NB-LRRs share homology with the cytoplasmic domains of the Drosophila Toll and mammalian interleukin-1 transmembrane receptors (Meyers et al., 2003). NLRs, upon recognizing effectors, change their protein conformation and activate downstream events to defend against various pathogens (DeYoung et al., 2006). PTI and ETI induce a diverse array of immune responses such as Ca2+ burst, reactive oxygen species burst, mitogen-activated protein kinase cascade activation, hormone signaling and transcriptional reprogramming (Tsuda et al., 2010; Bigeard et al., 2015; Tsuda et al., 2015). One key feature of immune responses is the massive gene expression changes or transcriptional reprogramming that is critical for plant defense against pathogens (Tao et al., 2003; Moore et al., 2011; Tsuda et al., 2015).
NLR genes needs to be tightly regulated for balancing plant growth and immunity response. On the one hand, they often have a very low expression under non-pathogenic conditions as constitutive expression of NLR genes leads to autoimmunity and retarded plant growth (Palma et al., 2010; Gou et al., 2012). For example, the bon1 mutant in Arabidopsis is an autoimmune mutant resulted from the upregulation of a NLR gene SNC1 (SUPPRESSOR OF npr1-1, CONSTITUTIVE 1) (Yang et al., 2004). Introduction of the wild-type SNC1 gene from the Col-0 accession into the wild-type Ws accession leads to autoimmune due to an higher expression of SNC1 in Ws than in Col-0, indicating a strong repression at the endogenous locus of SNC1 (Li et al., 2007). On the other hand, NLR genes need to be expressed at a proper level for pathogen recognition and defense signaling (Bieri et al., 2004; Holt et al., 2005; Mohr et al., 2010). The mechanisms for transcriptional control of NLR genes include alternative splicing, alternative polyadenylation, small RNA induced post-transcriptional regulation, and transcription factors controlled gene regulation (Lai et al., 2018; Zhang et al., 2018). Recent evidences indicate that chromatin modifications and remodeling are also important for the expression of NLR genes including SNC1 (Li et al., 2010; Palma et al., 2010; Xia et al., 2013; Zou et al., 2014; Zou et al., 2017; Zhang et al., 2018). A number of positive regulators of SNC1 expression at the chromatin level have been reported. For example, in the autoimmune mutants such as snc1-1 (a gain of function mutant) and bon1, both ARABIDOPSIS TRITHORAX-RELATED 7 (ATXR7) and HISTONE MONO-UBIQUITINATION 1 (HUB1) are required for the SNC1 induction through depositing H3K4me3 and H2Bub marks at SNC1 locus, respectively (Xia et al., 2013; Zou et al., 2014). CHROMATIN REMODELING 5 (CHR5), a member of Chd subfamily of chromatin remodeler, is also shown to activate SNC1 gene expression by changing the nucleosome occupancy in its promoter region (Zou et al., 2017). In addition, SNC1 is regulated by MODIFIER OF snc1 (MOS1) potentially occurring at the chromatin level, and the mos1 mutation could suppress the dwarf phenotypes of snc1-1 and bon1 (Li et al., 2010; Bao et al., 2014). This regulation may come from the physical interaction between MOS1 and TCP transcription factors that directly bind to the promoter of SNC1 to promote its expression (Zhang et al., 2018). Despite the increasing evidence supporting the positive role of chromatin modifications and remodeling in SNC1 regulation, the precise mechanism of SNC1 and NLR repression in general is largely uncharacterized.
Histone acetylation removes positive charges by adding an acetyl group to the lysine residues of histone proteins, thereby reducing the histone-DNA affinity and resulting in chromatin decondensation and active transcription (Bannister et al., 2011). Histone acetylation level is dynamically regulated by the combined actions of various histone acetyltransferases (HATs) and histone deacetylases (HDACs). HDACs are conserved enzymes in eukaryotes and play critical roles in diverse biological processes, including transcription, genome stability, development, abiotic and biotic stress responses (Haberland et al., 2009; Bosch-Presegué et al., 2014; Seto et al., 2014). In Arabidopsis thaliana, a total of 18 HDAC genes are divided into three groups: 12 REDUCED POTASSIUM DEPENDENCE 3 (RPD3)-like genes, 2 SILENT INFORMATION REGULATOR 2 (SRT) homologs, and 4 plant-specific HISTONE DEACETYLASE 2 (HD2) genes (Hollender et al., 2008). Among these 18 HDACs, four of them have been implicated in plant immunity. HDA6 in the RPD3 group binds to and represses the expression of defense responsive genes such as PR1, PR2, and WRKY38 accompanied with decreased H3 acetylation at these loci (Wang et al., 2017). HDA19, in the same group as HDA6, is found to inhibit salicylic acid mediated defense responses (Choi et al., 2012), although another study shows that it promotes the basal resistance against the virulent pathogen Pseudomonas syringae pv.tomato (Pst) DC3000 (Kim et al., 2008). The loss of function mutant of SRT2 in the second group is more resistant to Pst DC3000 than the wild type, which is associated with increased expression of defense responsive genes including SID2, EDS5, and PAD4 (Wang et al., 2010). HD2B is a plant-specific HDAC reported to be important for flg22-induced basal defense response via its deacetylation activity (Latrasse et al., 2017). Despite the correlative studies between the mutant phenotype and the expression of defense response genes, the direct targets of these HDACs in plant immunity are largely unknown.
HISTONE DEACETYLASE 9 (HDA9) is a member of RPD3-like group and plays important roles in flowering, leaf senescence, seed germination, and stress response ( Kim et al., 2013; van Zanten et al., 2014; Kang et al., 2015; Chen et al., 2016; Zheng et al., 2016; Park et al., 2019). HDA9 interacts with POWERDRESS (PWR) and WRKY53 to regulate leaf aging, and the hda9 loss-of-function mutant exhibited early senescence (Chen et al., 2016). The hda9 mutant also exhibited enhanced resistance to salt and osmotic stresses compared to the wild type (Zheng et al., 2016). Recent studies showed that HDA9 forms a complex with a WD40-repeat containing protein HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES 15 (HOS15) to regulate leaf development and flowering (Mayer et al., 2019; Park et al., 2019). The loss-of-function hos15 mutant was hypersensitive to freezing temperatures (Zhu et al., 2008). Recently, HOS15 was shown to induce the expression of COLD RESPONSIVE (COR) genes by hyperacetylation of COR chromatin through degradation of HD2C in response to cold stress (Park et al., 2018b). In addition, the wheat homolog of HOS15, TaHOS15, was found to interact with HDA6 to fine-tune defense responses to powdery mildew (Liu et al., 2018).
Here, we uncovered HOS15 and HDA9 as two new players in plant immunity. A sensitized genetic screen led to the isolation of HOS15 as a negative regulator of expression of a NLR gene SNC1 and plant immunity in Arabidopsis. Strikingly, one third of the 207 NLR genes in Arabidopsis showed increased expression without HOS15 under non-pathogenic conditions. We found that HOS15 acts via HDA9 to directly repress the expression of SNC1 through decreasing H3K9ac abundance at the SNC1 locus. Similar regulation was observed in other NLR genes. Furthermore, HOS15 constitutively represses SNC1 expression while HDA9 represses SNC1 expression under pathogen attack. Together, our study established a critical role for epigenetic regulation of NLR genes in immunity and growth-defense balance.
MATERIALS AND METHODS
Plant materials
Arabidopsis thaliana accession Col-0 was used as control for this study. Seeds of hos15-3 (GK_785B10), hos15-2 (SALKseq_064435) and hda9-1 (SALK_007123) were obtained from the Arabidopsis Biological Resource Center (ABRC). All plants were grown at 22°C under constant light and 50% humidity conditions unless specified. Two-week-old plants were used for experiments unless otherwise specified. HDA9-FLAG transgenic plants were reported previously (Chen et al., 2016).
EMS mutagenesis
Approximately 7,000 seeds were treated with 0.25% Ethyl methanesulfonate (EMS) for 12h on a shaker. Seeds were washed with water in a 15 ml tube for at least 5 times and suspended in 0.1% agar. Suspended seeds were sown directly on soil under light without stratification and about 3000 M1 plants were survived. 10 M1 plants were pooled and M2 seeds were collected together. Approximately 100 seeds from each pool were used to screen bon1-like mutants.
Plasmid construction and generation of transgenic plants
For complementation analysis, genomic DNA of HOS15 was first cloned into pCR8/GW/TOPO vector (Invitrogen, K250020), and recombined into binary vector pMDC99 by LR reactions (Invitrogen, 11791-020). For HOS15-HA, a genomic fragment of HOS15 from approximately 900bp upstream of ATG start codon to stop codon (without stop codon) was cloned into pDONR222 vector by BP reactions (Invitrogen, 11789020) and then into binary vectors pGWB413 (Nakagawa et al., 2007) to create pHOS15::HOS15:HA. All constructs were transformed into GV3101 strains for agrobacterium-mediated infection into bon1-1 mos1-6 hos15-4 and hos15-4 mutant background. Transgenic plants were selected on ½ MS plates with respective antibiotic markers. Details of the primers used for plasmid construction can be found in Supplemental Table 1.
Mapping-by-Sequencing
The mapping-by-sequencing was mainly carried out as described by Hua et al., 2017. To be short, F2 progenies from a cross of bon1 mos1 smo1 and bon1 mos1 segregated bon1 mos1 -like (non-mutant) and bon1 mos1 smo1 -like (mutant) dwarf plants. Equal amounts of leaf tissues were collected from each plant and were pooled together from at least 50 plants with similar phenotype. Mutant and non-mutant pools were used for genomic DNA extraction using Total DNA Purification Kit (Invitrogen, 45-7004). DNA libraries and Illumina Nextseq500 sequencing were done by Institute of Biotechnology, Cornell University. Data analysis was performed as described by Hua et al., 2017. The sequencing data analysis showed each pool had around 35x coverage of Arabidopsis genome. All the sequence reads from the two pools were aligned to Col-0 reference genome. The mapping was based on the frequency of the non-reference allele of a SNP from the two pools.
RNA extraction, quantitative RT-PCR and RNA sequencing
Two-week-old plants were used for total RNA extraction by Trizol Reagent (Invitrogen, 15596026). RNA was treated with DNase before used for cDNA synthesis by AffinityScript QPCR cDNA synthesis kit (Agilent Technologies, 600559). Quantitative RT-PCR was performed on CFX96™Real-Time System (BIO-RAD) using iQ SYBR Green supermix (BIO-RAD, 1708880). At least two biological experiments were performed for each gene expression analysis.
For RNA sequencing, total RNA was extracted from two-week-old plants using Trizol Reagent and then purified by RNA Clean and Concentrator-5 (ZYMO RESEARCH, R1014). cDNA libraries were constructed using NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (NEB, E7760). All samples were run on Illumina NextSeq500 platform. Three biological replicates were performed for both Col-0 and hos15-4 mutant. The raw reads were aligned to TAIR 10 transcriptome using STAR and differentially expressed genes were identified in R with edgeR package.
3,3’-Diaminobenzidine (DAB) staining
DAB staining was performed exactly according to Daudi et al., 2012. At least six plants were stained at the same time for each genotype and photographed the most representative one.
Chromatin Immunoprecipitation (ChIP)
ChIP of HOS15-HA was performed as previously described (Chen et al., 2016) with modifications. The same amount of sheared human chromatin isolated from HEK293 cells expressing H3-FLAG-HA were added into each sample as spike-in before taking input sample and adding antibody. After sequential washes with ChIP Dilution Buffer, High Salt Dilution Buffer (ChIP Dilution buffer with 350 mM NaCl), LiCl Buffer (0.25M LiCl, 1% NP-40, 1% sodium deoxycholate, 1mM EDTA, 10mM Tris-HCl pH 8) and TE Buffer (10mM Tris-HCl pH 8, 1mM EDTA), the DNA-protein complex was eluted with ChIP Elution Buffer (1% SDS, 0.1M NaHCO3) and reverse cross-linked at 65 °C for over 6 hours. After proteinase K and RNase treatment, DNA was purified by standard phenol–chloroform method for qPCR.
ChIP-H3K9ac
ChIP experiments were conducted as described in Desvoyes et al., 2018 with minor modifications. At least 0.5g 2-week-old plants were cross-linked in ice-cold 1x PBS buffer with 1% formaldehyde by vacuum for 18min in total. 2.5ng H3K9ac antibodies (Anti-H3K9ac from Millipore, catalog number: 07-352) were added to 1mL dilution chromatin and 25μL dynabeads protein G (Invitrogen, 10004D) were used for pulling down the antibody-associated chromatin. Primers used for PCR can be found in Table S1.
Pst DC3000 growth assay
Pseudomonas syringae pv. tomato (Pst) DC3000 was grown for 2 days at 28 °C on King’s B medium plates with rifamycin. Then bacteria were regrown overnight on a new King’s B medium plate. In the next morning, bacteria were suspended in 10 mM MgCl2 and 0.02% Silwet L-77 to OD600 of 0.05. Two-week-old plants grown under 12h:12h, light: dark were dipped in the bacterial suspension for 20 seconds. Afterwards, plants were put back to growth chamber and covered with a dome for 1h. Bacteria growth was analyzed at 1 hour post inoculation (0 DPI) and 3 days post inoculation (3 DPI). Each time, 3 plants were pooled together as one biological replicate and 3 biological replicates were measured at the same time. Plants were homogenized in 10 mM MgCl2 using pestles and then diluted to 10, 102, 103, 104, 105 and 106 times with 10 mM MgCl2. Each diluted bacterial suspension was spotted on plates and grown for 2 days to measure pathogen growth. At least three independent experiments were done for each figure.
Pst DC3000 inoculation for gene induction, western blotting and ChIP
Two-week-old plants growing under constant light were dipped with Pst DC3000 (OD of 0.05). Plants dip-inoculated with 10 mM MgCl2 were collected right after inoculation as samples before inoculation (0h) while plants dip-inoculated with Pst DC3000 were collected at 4 hours post-inoculation as samples after inoculation (4h).
Co-immunoprecipitation (co-IP) in Nicotiana benthamiana
Agrobacteria strains containing HOS15-HA and HDA9-FLAG were co-infiltrated into two-week-old Nicotiana benthamiana plants for 4 days. At the same time, infiltrate HDA9-FLAG into plants as a control. At day 4, some of the plants co-expressing HOS15-HA and HDA9-FLAG were infiltrated with Pst DC3000 (OD 0.02). Samples were collected at 4 hours post-inoculation. At least 1 gram of tissues were ground to fine power and resuspended in 3ml IP buffer (50mM Tris-HCl pH 7.5, 150 mM NaCl, 5 mM MgCl2, 5% glycerol, 0.1% NP-40, 1mM PMSF and cocktail complete). The homogenates were transferred to a Dounce tissue grinder, 15-20 times with “tight”, then subjected to centrifuge at 10,000g for 15 min at 4°C. The supernatant was incubated with dynabeads protein G (Invitrogen, 10004D) coated with anti-HA antibody (Enzo Life Sciences, ENZ-ABS118-0200) for 3 hours at 4 °C. Wash the beads with IP buffer at least 4 times. Boil beads with 30 μl SDS loading buffer at 95°C for 10mins. The eluted proteins were subjected to immunoblotting.
Cytoplasm-Nucleus fractionation
The Cytoplasm-Nucleus fractionation was performed as previous described (Mayer et al., 2019). Immunoblots were performed with anti-FLAG (Sigma, A8592), anti-H3 (Abclonal, A2348) and anti-Tubulin (Abclonal, AC021) antibodies.
Data Availability
RNA-seq data is available at the Gene Expression Omnibus and can be found with the GEO accession number GSE131227.
Sequence data from this article can be found through TAIR (https://www.arabidopsis.org) under the following accession numbers:
HOS15: AT5G67320, HDA9: AT3G44680; SNC1: AT4G16890; PR1: AT2G14610; MOS1: AT4G24680; BON1: AT5G61900; PAD4: AT3G52430.
RESULTS
The smo1 mutation activates SNC1-mediated defense responses in the bon1 mos1 mutant
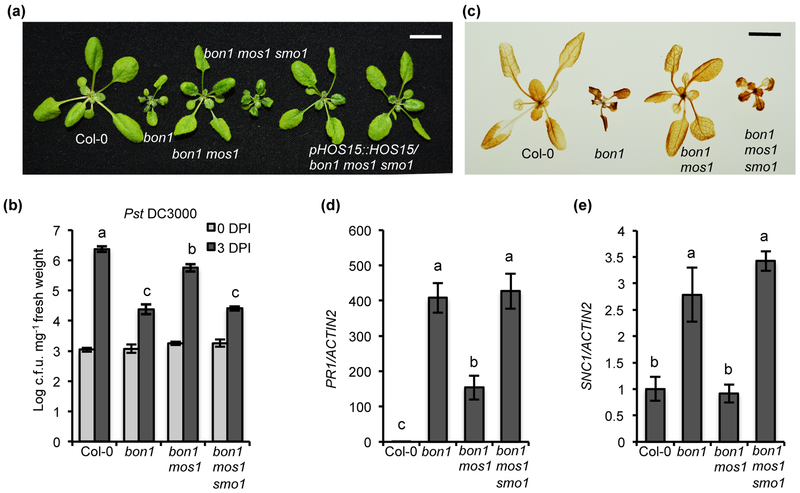
To identify additional repressors of SNC1 expression and regulators of plant immunity, we performed a suppressor screen of the bon1 mos1 mutant, in which autoimmunity due to SNC1 upregulation in bon1 is inhibited by the mos1 mutation. We screened plants exhibiting bon1-like autoimmunity from the M2 plants and named these putative suppressor mutations as smo (suppressor of mos1 bon1) (Fig. 1a). One such smo mutant bon1 mos1 smo1 was further characterized for its immunity phenotype. The bon1 mos1 smo1 plants, along with Col-0, bon1 and bon1 mos1, were infected by the virulent pathogen Pst DC3000. Consistent with previous study (Bao et al., 2014), pathogen grew significantly less in bon1 compared to Col-0 and bon1 mos1 had more pathogen growth compared to bon1 (Fig. 1b). Most importantly, bon1 mos1 smo1 supported significantly less pathogen growth than bon1 mos1 (Fig. 1b), indicating a restoration of upregulation of immunity by smo1. In addition, bon1 mos1 smo1 had a higher expression of defense response marker gene PR1 and a higher accumulation of defense activation associated H2O2 compared to bon1 mos1 (Fig. 1c, d). These data suggested that the smo1 mutation restored autoimmunity of bon1. Furthermore, the analysis of gene expression by RT-qPCR revealed that bon1 mos1 smo1 has an increased SNC1 RNA transcript compared to bon1 mos1 (Fig. 1e). These analyses indicated that the smo1 mutation restored NLR-mediated defense responses in bon1 mos1.
Fig. 1. Identification of HOS15 as a new negative regulator of immunity in Arabidopsis thaliana.
(a) Morphology of wild-type Col-0, bon1, bon1 mos1, bon1 mos1 smo1 and two independent lines of pHOS15::HOS15 in bon1 mos1 smo1. (b) Growth of bacterial pathogen Pst DC3000 in Col-0, bon1, bon1 mos1 and bon1 mos1 smo1. Statistical analysis was performed with infected plants at 3 DPI (Days Post-Inoculation). (c) DAB staining of Col-0, bon1, bon1 mos1 and bon1 mos1 smo1. (d, e) Analysis of PR1 (d) and SNC1 (e) gene expression in Col-0, bon1, bon1 mos1, bon1 mos1 smo1 by qRT-PCR. Error bars represent S.D. from three biological replicates for (b) or two biological replicates for (d) and (e). Different letters indicate significant difference tested by One-way ANOVA/ Duncan (P < 0.05). Scale bar for (a) and (c), 1 cm.
The SMO1 gene is HOS15
To identify the SMO1 gene, we utilized the Mapping-by-Sequencing method to identify casual mutations (Zhu et al., 2012; Hua et al., 2017). The most promising SNP for the smo mutation resides in the 7th exon of HOS15 (AT5G67320), causing a premature stop codon (Fig. S1a). The HOS15 gene encodes a protein with LisH domain at the N terminal and eight WD40 repeats at the C terminal (Zhu et al., 2008) (Fig. S1b). The mutation was within the second WD40 repeat which may cause a truncated protein depleting seven WD40 repeats (Fig. S1b).
We verified this mutation in HOS15 to be SMO1 by two methods. First, a complementation analysis was performed by transforming the wild-type genomic fragment of HOS15 (pHOS15::HOS15) into bon1 mos1 smo1. 32 out of the 39 T1 plants exhibited bon1 mos1 phenotype (Fig. 1a) and a co-segregation of the wild-type looking (bon1mos1-like) phenotype with the pHOS15::HOS15 transgene in six independent T2 populations was observed. Second, a T-DNA loss-of-function mutant allele hos15-3 (Park et al., 2018b) (Fig. S1a) was introduced into bon1 mos1 by crossing and the bon1 mos1 hos15 mutant showed a similar growth phenotype to bon1 mos1 smo1 (Fig. S1c). In addition, the bon1 mos1 hos15 mutant exhibited autoimmunity similarly to bon1 mos1 smo1, including an enhanced resistance to Pst DC3000, an accumulation of H2O2, and a higher expression of PR1 and SNC1 compared to bon1 mos1 (Fig. S1d-g). These analyses confirm that HOS15 is the SMO1 gene.
HOS15 negatively regulates immune responses partially through SNC1
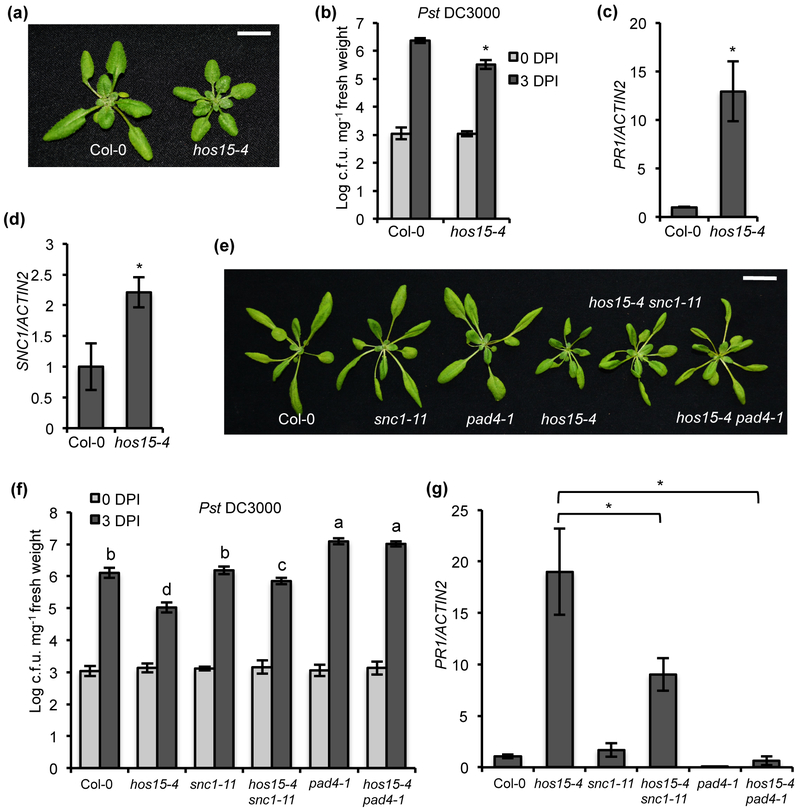
To investigate how HOS15 influences immunity, we isolated hos15 single mutant (hereafter hos15-4) from the F2 progenies of a cross between bon1 mos1 smo1 and Col-0. The hos15-4 single mutant had more compact and smaller rosette leaves than Col-0 (Fig. 2a; Fig. S2a). When infected with Pst DC3000, the hos15-4 single mutant supported significantly less pathogen growth compared to Col-0 (Fig. 2b). It also had more H2O2 accumulation and increased PR1 gene expression compared to Col-0 (Fig. 2c; Fig. S2b). Furthermore, the SNC1 gene expression was significantly increased in hos15-4 compared to Col-0 (Fig. 2d). To further confirm that HOS15 is a negative regulator of immunity, we tested the immune phenotypes of other known knockout hos15 alleles. Both the hos15-2 and hos15-3 have the T-DNA inserted in introns and they have been shown to be knockout mutants (Chen et al., 2016; Park et al., 2018b) (Fig. S1a). The hos15-2 and hos15-3 displayed the similar growth phenotype as hos15-4 (Fig. S3a). In addition, they have increased SNC1 and PR1 gene expression, and reduced pathogen growth compared to Col-0, to a similar level as hos15-4 (Fig. S3b-3d). These results indicate that HOS15 is a negative regulator of plant immunity and hos15 mutant is an autoimmune mutant that has upregulated defense responses in the absence of pathogen infection.
Fig. 2. HOS15 acts partially through SNC1 to regulate immunity in Arabidopsis.
(a) Morphology of wild-type Col-0 and hos15-4 at two weeks old grown under constant light. Scale bar, 1 cm. (b) Growth of bacterial pathogen Pst DC3000 in Col-0 and hos15-4 mutant. Statistical analysis was performed with infected plants at 3 DPI (Days Post-Inoculation). (c, d) Analysis of PR1 (c) and SNC1 (d) gene expression in Col-0 and hos15-4 by qRT-PCR. (e) Morphology of Col-0, snc1-11, pad4-1, hos15-4, hos15-4 snc-11 and hos15-4 pad4-1 grown for three weeks under constant light. Scale bar, 2 cm. (f) Growth of bacterial pathogen Pst DC3000 in Col-0, hos15-4, snc1-11, hos15-4 snc1-11, pad4-1 and hos15-4 pad4-1. Different letters indicate significant difference tested by One-way ANOVA/ Duncan (P < 0.05). Statistical analysis was performed with infected plants at 3 DPI. (g) Analysis of PR1 gene expression in Col-0, hos15-4, snc1-11, hos15-4 snc1-11, pad4-1 and hos15-4 pad4-1 by qRT-PCR. The expression of Col-0 was set to 1 for (c), (d) and (g). Different letters indicate significant difference tested by One-way ANOVA/ Duncan (P < 0.05). Error bars represent S.D. from three biological replicates for (b), (d), (f) and (g) or two biological replicates for (c). * indicates significant difference tested by Student’s t-test (P < 0.05).
Because hos15-4 mutant was isolated for restoring autoimmunity associated with SNC1 in bon1 mos1, we asked whether SNC1 contributes to the hos15-4 mutant phenotype. Double mutant of hos15-4 snc1-11 was generated and it exhibited a milder growth defect compared to hos15-4 (Fig. 2e; Fig. S2c), suggesting that SNC1 partially contributed to the growth defect of hos15-4. Mutation of PAD4, a mediator of most NLR genes (Cui et al., 2017), was subsequently introduced into hos15-4, and the hos15-4 pad4-1 exhibited a wild-type growth phenotype (Fig. 2e; Fig. S2c). Defense responses to Pst DC3000 were subsequently analyzed in these two double mutants. Both hos15-4 snc1-11 and hos15-4 pad4-1 double mutants had significant more pathogen growth compared to the hos15-4 single mutant, with snc1-11 mutation partially and the pad4-1 mutation fully suppressing the enhanced resistance in hos15-4 compared to Col-0 (Fig. 2f). Consistent with the pathogen growth phenotype, expression of the defense response marker gene PR1 in hos15-4 mutant was significantly decreased by snc1-11 and pad4-1 (Fig. 2g), indicating that SNC1 and PAD4 are essential for immune repsonses in hos15-4.
HOS15 preferentially represses the expression of NLR genes
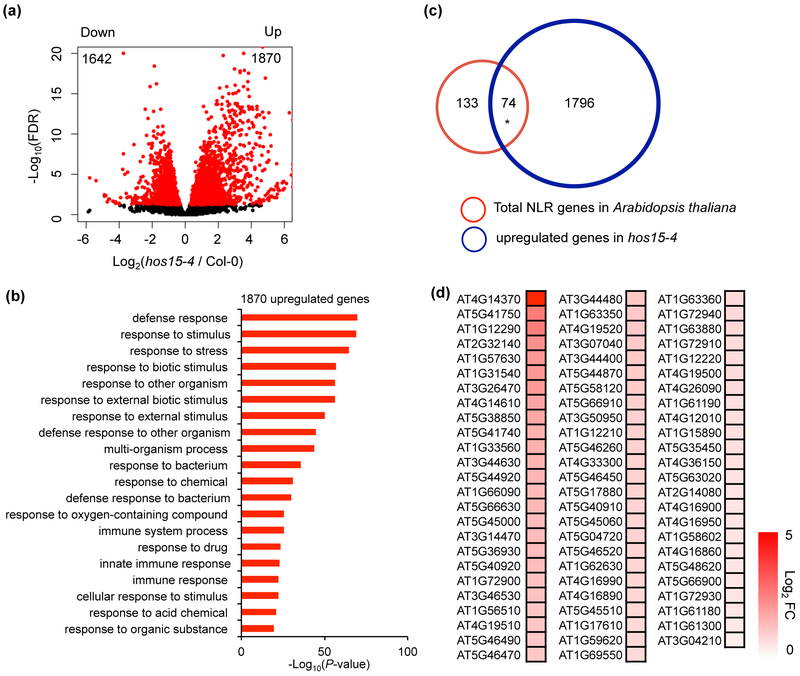
To further investigate the role of HOS15 in immune response, we performed whole genome transcriptome (RNA-seq) analysis of the hos15-4 mutant. A total of 3,512 differentially expressed genes (DEGs), with 1,870 upregulated and 1,642 downregulated, were found in hos15-4 mutant compared to wild type with three biological repeats (Fig. 3a; Dataset S1). Gene Ontology (GO) enrichment analysis revealed that upregulated DEGs were significantly enriched in processes such as defense response, response to biotic stimulus, and response to other organism and immune system process (Fig. 3b). Known genes in salicylic acid pathway were found in the upregulated DEG, including SID2, PAD4, PR1, NPR1, and SNC1. On the other hand, genes involved in photosynthesis, response to abiotic stimulus, nucleic acid metabolic process, and response to cold are over represented among downregulated DEGs (Fig. S4). Our RNA-seq data indicated an important role of HOS15 in suppressing the expression of many defense response genes.
Fig. 3. HOS15 represses the expression of a large number of defense response genes including many NLR genes in Arabidopsis.
(a) Volcano plot showing significantly upregulated and downregulated genes (red dots, FDR < 0.05) in hos15-4 mutant compared to wild type. (b) Gene Ontology analysis of significantly upregulated genes (Fold change > 2, FDR < 0.05) using Panther (http://www.pantherdb.org). Shown is the Top 20 most significant groups. (c) Overlap of total NLR genes in Arabidopsis thaliana and upregulated genes in hos15-4. * indicates significant difference. P < 2.923e-22 was calculated by Fisher’s exact test. (d) Heatmap of 74 upregulated NLR genes in hos15-4. FC, Fold Change.
Next, we asked whether HOS15 specifically regulates the expression of SNC1 or generally acts on other NLR genes. We examined the expression of all NLR genes in Arabidopsis in hos15-4. There are a total of 207 annotated NLR genes in Arabidopsis thaliana (Meyers et al., 2003) (Dataset S2). Although some of them are atypical NLRs that do not have all the three domains of typical NLRs, they might still have the ability to function as immune regulators (Zhao et al., 2015; Nishimura et al., 2017). Interestingly, 74 out of 207 NLR genes were upregulated and none was downregulated in hos15-4 mutant, and therefore NLR genes are significantly enriched in hos15-4 DEGs (Fig. 3c,d; Dataset S3). These analyses suggested that HOS15 represses a large number of NLR genes in Arabidopsis.
HOS15 and HDA9 act in the same pathway to regulate plant immunity
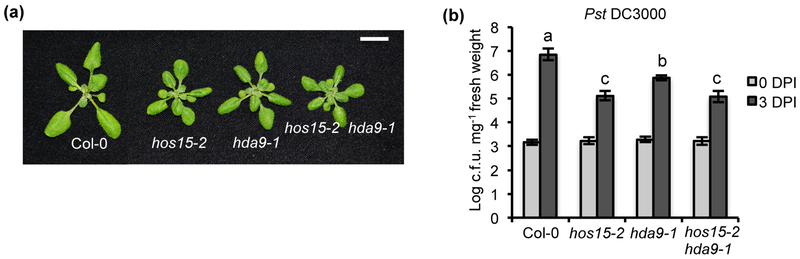
It was reported that HOS15 interacts with HDA9 to regulate transcription and developmental processes (Suzuki et al., 2018; Mayer et al., 2019; Park et al., 2019). We therefore determined whether HDA9 also plays a role in immune response together with HOS15. We examined the hda9-1 mutant (SALK_007123) which was shown previously to be a knockout mutant (van Zanten et al., 2014; Chen et al., 2016). This hda9-1 mutant had a more compact rosette than the wild type, similarly to the hos15-2 mutant (Fig. 4a). Importantly, when inoculated with Pst DC3000, the hda9-1 mutant showed significantly enhanced disease resistance to Pst DC3000 compared to the wild-type Col-0 (Fig. 4b), indicating that HDA9 is a negative regulator of plant immunity. We subsequently analyzed the hos15-2 hda9-1 (hereafter hos15 hda9) double mutant. The hos15 hda9 mutant exhibited a growth phenotype very similar to those of hos15 and hda9 single mutants (Fig. 4a). In addition, the double mutant had an enhanced disease resistance to a similar extent as the hos15-2 single mutant to Pst DC3000 (Fig. 4b), suggesting that HOS15 and HDA9 do not have additive functions but rather function in the same pathway in plant immunity regulation.
Fig. 4. HDA9 in Arabidopsis is a negative regulator of defense response, similar to HOS15.
(a) Morphology of Col-0, hos15-2, hda9-1 and hos15-2 hda9-1. Scale bar, 1 cm. (b) Growth of bacterial pathogen Pst DC3000 in Col-0, hos15-2, hda9-1 and hos15-2 hda9-1. Error bars represent S.D. from three biological replicates. Statistical test was performed with infected plants at 3 DPI (Days Post Inoculation). Different letters indicate significant difference tested by One-way ANOVA/ Duncan (P < 0.05).
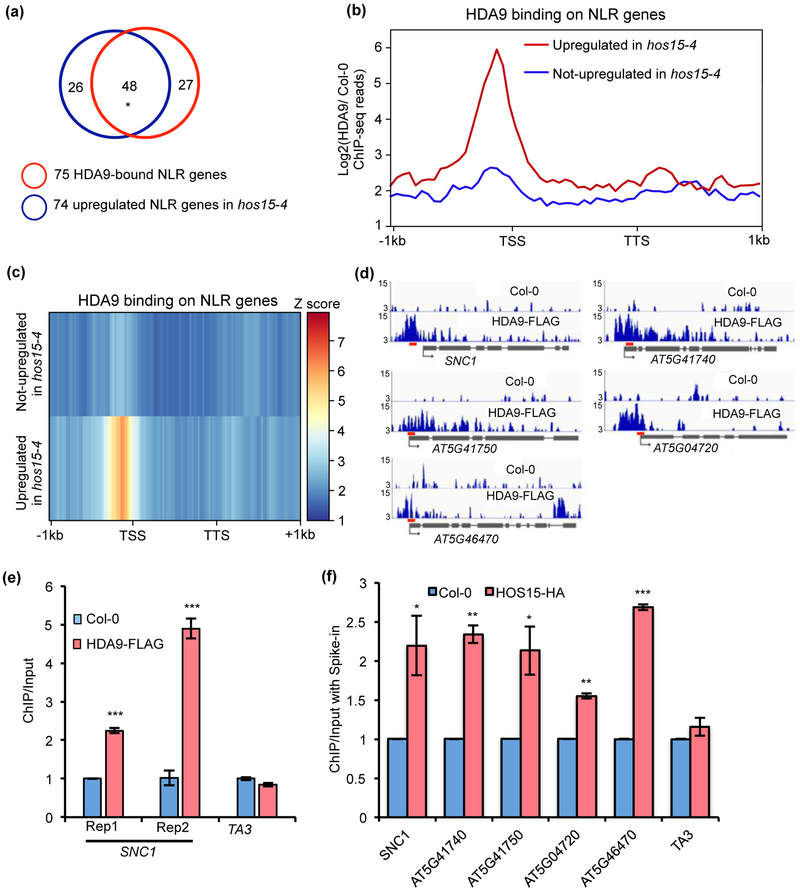
HDA9 and HOS15 directly bind to the same subset of NLR genes
Because a large number of NLR genes were upregulated in the hos15-4 mutant (Fig. 3d), we examined whether HDA9 plays a role in regulating the expression of these HOS15-repressed NLR genes. Analysis on the overlapping genes revealed that 48 of 74 NLR genes upregulated in hos15-4 were directly bound by HDA9 as reported in our ChIP (Chromatin Immunoprecipitation)-seq analysis (Chen et al., 2016) (Fig. 5a; Dataset S4). In addition, we noted a significant enrichment of HDA9 protein over promoters of NLR genes that were upregulated in hos15-4 compared to those unaltered in hos15-4 (Fig. 5b,c). Among them, we found a strong enrichment of HDA9 in promoter region of SNC1 gene (Fig. 5d). To further confirm this association, we performed two independent ChIP experiments using FLAG-tagged HDA9 transgenic plants driven by its native promoter (pHDA9::HDA9:FLAG in hda9, HDA9-FLAG) (Chen et al., 2016). The ChIP-qPCR results showed that HDA9 indeed binds to SNC1 (Fig. 5e).
Fig. 5. HOS15 and HDA9 are associated with some NLR genes in Arabidopsis.
(a) Overlap of 75 HDA9-bound NLR genes and 74 upregulated NLR genes in hos15-4 mutant. * indicates significant difference. P < 2.457e-103 was calculated by Fisher’s exact test. (b, c) Metaplots (b) and heatmaps (c) showing HDA9 enrichment on NLR genes upregulated and not upregulated in hos15-4 mutant. Log2 value of HDA9 ChIP-seq signal normalized to wild type control was calculated for each gene, and then averaged within gene group respectively. The averaged value was used for drawing metaplots and heatmaps. TSS and TTS represent transcription start site and transcription termination site, respectively. −1kb and +1kb represent 1kb upstream of TSS and 1kb downstream of TTS, respectively. The color bar on the right of heatmaps indicates the Z-score. Y-axis of metaplots represents HDA9 ChIP-seq read density. (d) Snapshots of IGV views of HDA9 binding on NLR genes. Red bars represent the positions of primers used in Fig. 5f. (e) ChIP-qPCR showing HDA9 binding on SNC1 promoter. Rep1 and Rep2 represent two biological replicates. ChIP was normalized to Input and then normalized to Col-0. TA3 served as negative control. Error bars represent S.D. from three technical replicates. (f) ChIP-qPCR shows that HOS15-HA protein is enriched at NLR genes upregulated in hos15-4 mutant. After normalized to respective spike-in, ChIP was normalized to Input. Error bars represent S.D. from two biological replicates. * P < 0.05, **P < 0.01, ***P < 0.001.
Next, we investigated whether HOS15 co-occupied the same set of NLR genes as HDA9. We generated transgenic plants expressing HA-tagged HOS15 driven by its native promoter in hos15-4 mutant background (pHOS15::HOS15:HA in hos15-4, HOS15-HA). The HOS15-HA construct rescued both the hos15 single and the bon1 mos1 hos15 triple mutant, indicating that this fusion protein is functional (Fig. S5). Besides SNC1, we tested four additional NLR genes (AT5G41740, AT5G41750, AT5G04720, and AT5G46470) that were randomly chosen from those bound by HDA9 (Fig. 5d) in the previous ChIP-seq experiment (Chen et al., 2016). These four NLR genes were upregulated in hos15-4 mutant and evenly distributed in the fold change range among the top 50% upregulated NLR genes in hos15-4 mutant (Fig. 3d). ChIP-seq data revealed that HDA9 was enriched near transcription start site (TSS) of these NLR genes (Fig. 5d). ChIP-qPCR using the HOS15-HA line revealed that HOS15 protein is enriched at the same region at these NLR genes as HDA9 (Fig. 5f; Fig. S6). These data suggest that HOS15 and HDA9 bind and regulate the same subset of NLR genes.
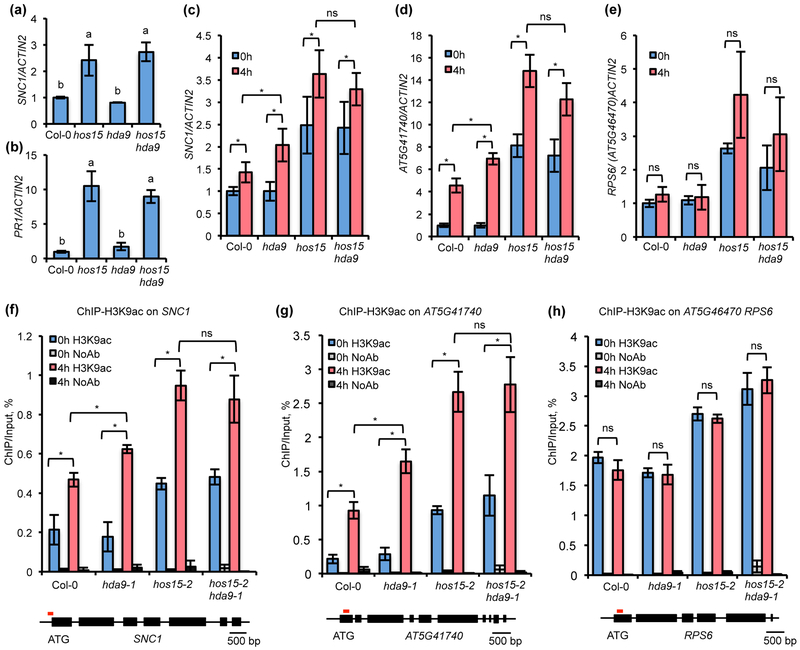
HOS15 and HDA9 regulate the expression and acetylation of SNC1
Previous RNA-seq analysis did not reveal a significant enrichment of defense responses among DEGs of hda9-1 grown under normal conditions (Chen et al., 2016), unlike the hos15-4 mutant. Consistently, the analysis of the SNC1 and PR1 transcript levels by RT-qPCR in hda9-1 mutant showed no significant difference compared to WT under normal conditions (Fig. 6a,b). These data suggest that hda9 does not have constitutive defense response without pathogen infection. However, it might have a heightened defense response upon pathogen infection. We therefore analyzed the expression of SNC1 in hda9 mutant after pathogen infection. As expected, SNC1 transcript level was significantly increased upon four-hour infection in Col-0 as reported previously (Zou et al., 2014) (Fig. 6c). This induction was also observed in hda9, hos15, and hos15 hda9 (Fig. 6c). SNC1 gene expression was induced to a greater extent in the hda9 mutant compared to wild-type Col-0 (Fig. 6c). The SNC1 gene expression was higher in hos15 and hos15 hda9 compared to the wild type before infection, and was further induced upon pathogen infection (Fig. 6c). In addition, there is no statistically significant difference of the SNC1 induction in hos15 and hos15 hda9 (Fig. 6c). We subsequently analyzed another NLR gene which is bound by both HOS15 and HDA9 and is also induced by PstDC3000 infection (Mohr et al., 2010) (Fig. 5d-f). This AT5G41740 showed an upregulation in hos15 but not hda9 under non-pathogenic condition and a higher expression after infection in both hda9 and hos15 mutants (Fig. 6d). In contrast, another NLR gene AT5G46470 (RPS6) that is bound by HOS15 and HDA9 (Fig. 5d-f) was not significantly induced by Pst DC3000 (Mohr et al., 2010) (Fig. 6e). Its expression is similar in hda9 and the wild type with or without pathogen infection (Fig. 6e). This analysis suggested an association of pathogen responsiveness with its regulation by HDA9 for NLR genes that are bound by HOS15 and HDA9.
Fig. 6. HOS15 and HDA9 repress some NLR loci by influencing histone acetylation in Arabidopsis.
(a, b) Analysis of SNC1 (a) and PR1 (b) gene expression in Col-0, hos15-2, hda9-1 and hos15-2 hda9-1 by qRT-PCR. (c-e) Analysis of SNC1 (c), AT5G41740 (d) and AT5G46470 (e) gene expression in Col-0, hda9-1, hos15-2 and hos15-2 hda9-1 before (0h) and after (4h) pathogen infection by qRT-PCR. (f-h) ChIP-qPCR analysis of H3K9ac enrichment at SNC1 (f), AT5G41740 (g) and RPS6 (h) in Col-0, hda9-1, hos15-2 and hos15-2 hda9-1. Samples were collected before and after 4h post inoculation of Pst DC3000. All samples were normalized to the input. The red line above SNC1 gene is the region for detecting H3K9ac enrichment. Error bars represent S.D. from three biological replicates for (a), two biological replicates for (b, f-h), or four biological replicates for (c-e). Different letters indicate significant difference tested by One-way ANOVA/ Duncan (P < 0.05). * indicates significant difference while n.s. indicates no statistic difference tested by Student’s t-test (P < 0.05). “H3K9ac” means samples with antibodies while “NoAb” means samples without antibodies.
Because HDA9 has been reported to influence gene expression by regulating histone H3 acetylation, including H3K9ac (Chen et al., 2016), we determined whether H3K9ac abundance at these NLR genes is affected by HOS15 and HDA9 by ChIP using anti-H3K9ac antibody. Because the histone deacetylation site of HDA9 close to the 3’ end of TSS (Chen et al., 2016), we detected the acetylation status of these NLR genes in regions immediately 3’ to TSS. Under normal conditions, the H3K9ac levels at both SNC1 and AT5G41740 were similar between Col-0 and hda9 but were higher in hos15 and hos15 hda9 (Fig. 6f,g). Under infection by Pst DC3000, H3K9 acetylation at both SNC1 and AT5G41740 loci was increased compared to non-infection conditions in the Col-0, and the increase of H3K9ac by infection was even higher in hda9 than that in Col-0 (Fig. 6f,g). The hos15 hda9 double mutant had a similar increase of H3K9ac by Pst DC3000 treatment as the single mutant hos15 at these two loci (Fig. 6f,g). The AT5G46470 (RPS6) gene, unlike SNC1 and AT5G41740, does not have an increased expression after pathogen infection (Mohr et al., 2010). No increase of H3K9ac was observed for RPS6 after pathogen induction, but H3K9ac level was higher in hos15 and hos15 hda9 compared to wild type or hda9 (Fig. 6h). Therefore, there was a close correlation of expression and H3K9ac levels of these three NLR genes, suggesting that HDA9 and HOS15 regulating NLR expression through histone deacetylation.
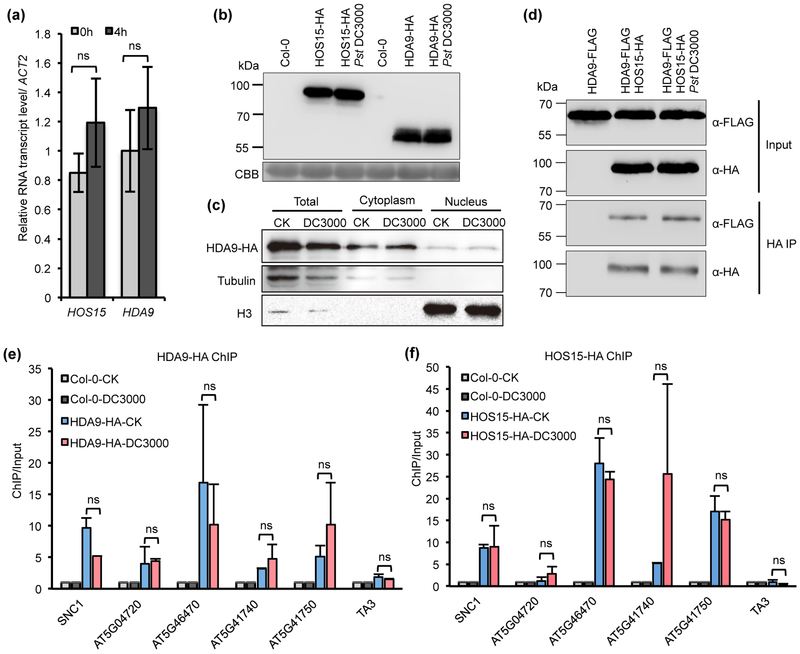
The protein accumulation, protein interaction, and chromatin association of HOS15 and HDA9 were not affected by Pst DC3000 invasion
We further investigate whether or not the regulation of NLR genes by HDA9 and HOS15 is altered during pathogen infection. Wild type Col-0 plants were inoculated with Pst DC3000 and expression of HOS15 and HDA9 were analyzed by RT-qPCR. No significant change of transcript levels of these two genes were observed 4 hours after pathogen infection (Fig. 7a). The protein levels of HOS15 and HDA9 were analyzed by using HOS15-HA and HDA9-HA (pHDA9::HDA9:HA in hda9, HDA9-HA, Mayer et al., 2019) transgenic plants, and no obvious changes were found for HOS15 or HDA9 before and after pathogen infection (Fig. 7b). We further determined the cellular dynamics of the HDA9 protein in response to pathogen inoculation by fractionation. Consistent with previous study (Chen et al., 2016), HDA9 is localized to both cytosol and nucleus (Fig. 7c). No significant changes in the distribution of HDA9 protein between nuclear and cytosol was observed after Pst DC3000 treatment (Fig. 7c). These data indicate that pathogen infection does not significantly alter the amount of HOS15 and HDA9 or the subcellular localization of HDA9. Next, we determined whether or not the interaction between HOS15 and HDA9 was altered by Pst DC3000 inoculation. The HDA9-FLAG and HOS15-HA proteins were transiently co-expressed in Nicotiana benthamiana via Agro-infiltration for 4 days before plants were inoculated by Pst DC3000. The HDA9-FLAG protein was co-IPed with HOS15-HA as expected (Mayer et al., 2019) (Fig. 7d) and the interaction was similar before and after pathogen infection (Fig. 7d). We further analyzed the chromatin association of HOS15 and HDA9 with the NLR loci upon pathogen infection by using transgenic plants carrying both HOS15-HA and HDA9-HA. ChIP analysis was carried out to assess the association of these proteins with NLR genes before and 4 hours after pathogen infection. Both HOS15 and HDA9 were enriched at these NLR genes but not the control TA3 loci before infection (Fig. 5d-f; Fig. 7e,f), and no significant change was observed for the associations after pathogen infection (Fig. 7e,f). The only potential exception is SNC1 where HDA9 but not HOS15 had a reduced association (Fig. 7e,f). All together, these data indicate that pathogen infection does not significantly alter the regulation of NLR genes by HOS15 or HDA9.
Fig. 7. The protein accumulation, protein interaction and chromatin association of HOS15 and HDA9 in Arabidopsis were not affected by Pst DC3000 invasion.
(a) Analysis of HOS15 and HDA9 gene expression in Col-0 treated with or without Pst DC3000 by RT-qPCR. The expression of HDA9 before infection was set to “1”. Error bars represent S.D. from three biological replicates. ‘ns’ indicates no statistical difference tested by Student’s t-test (P < 0.05). (b) Immunoblotting of HOS15 and HDA9 protein level after Pst DC3000 treatment. Samples were collected 4h post-inoculation of Pst DC3000. CBB, coomassie brilliant blue. Experiments were repeated four times with similar results. (c) Immunoblotting of HDA9-HA protein in total cell extract (Total), cytoplasmic (Cytoplasm), and nuclear (Nucleus) fractions. Immunoblotting of tubulin and histone H3 serve as controls for cytoplasmic and nuclear fractions, respectively. Two biological replicates were performed with similar results. (d) co-IP of the interaction between HOS15 and HDA9 after Pst DC3000 treatment. HDA9-FLAG was served as a control. This experiment was repeated once with similar results. (e,f) ChIP-qPCR analysis of HDA9 (e) and HOS15 (f) enrichment at target loci in normal condition (CK) and pathogen treatment (DC3000). ChIP was normalized to Input. Negative control (Col-0) was set as 1 for both CK and DC3000 conditions. Data are represented as mean ± S.D. with two biological replicates. ‘ns’ indicates no statistical difference tested by Student’s t-test (P < 0.05).
HOS15 regulates flowering time and silique development independent of immunity
In addition to its role in immunity, HOS15 regulates other processes including flowering time and silique development (Mayer et al., 2019; Park et al., 2019). We asked whether HOS15 regulates these developmental processes independently of immune response by using snc1-11 and pad4-1 to inhibit immune responses in hos15-4. The hos15-4 mutant had an early flowering phenotype compared to the wild-type Col-0 (Fig. S7a,c). The double mutant hos15-4 snc1-11 and hos15-4 pad4-1 exhibited the same early flowering phenotype as hos15-4 (Fig. S7c), indicating that HOS15 regulates flowering time independently of its regulation of immunity. The hos15-4 mutant had enlarged silique tips compared to the wild type (Mayer et al., 2019) (Fig. S7b), and the hos15-4 snc1-11 and hos15-4 pad4-1 mutants had a similarly enlarged silique tips in hos15-4 (Fig. S7d), indicating that silique development function of HOS15 is separate from the immunity function of HOS15.
DISCUSSION
This study has identified HOS15 and HDA9 as two new negative regulators of plant immunity. The loss of HOS15 function leads to autoimmunity, mounting of defense responses even in the absence of pathogen invasion, and the consequent reduced plant growth. This indicates that HOS15 plays a critical role in repressing immune responses and thus enabling growth under non-pathogenic conditions. The loss of HDA9 function does not induce autoimmunity but leads to heightened immune responses during pathogen infection. This indicates that HDA9 is critical in damping defense responses after pathogen invasion likely to prevent over-stimulating defense responses and/or confine defense responses to keep a balance of immunity and survival/growth. HOS15 has a similar role to HDA9 during pathogen infection as its loss of function leads to a further heightened defense response on top of the already upregulated defense response. Together, this study shows that immune responses are repressed by HOS15 and HDA9 under both pathogenic and non-pathogenic conditions.
HOS15 and HDA9 are found to regulate a large number of intracellular immune receptor NLR genes that are key components in plant immunity. Analysis of ChIP-seq data reveals that HDA9 is physically associated with a large amount of NLR genes (75 out of a total of 207) including AT5G41740, AT5G41750, AT5G04720, AT5G46470, and SNC1 in Arabidopsis (Fig. 5a,d,e; Dataset S5). These genes overlap with those that are regulated by HOS15, indicating that HOS15 interacts with HDA9 at these NLR loci to regulate their expression. This is supported by the association of HOS15 with all randomly selected NLR genes that are bound by HDA9 (Fig. 5f). In addition, HOS15 and HDA9 are found to interact with each other under both pathogenic and non-pathogenic conditions (Fig. 7d). HOS15 and HDA9 have been shown previously to work collaboratively in developmental processes (Park et al., 2019; Mayer et al., 2019). Therefore, this study identified NLR genes as new targets of HOS15 and HDA9. Importantly, we demonstrate that SNC1 is a direct target of HOS15 (Fig. 5f) and it contributes significantly to the immunity function of HOS15. The loss-of-function mutation of SNC1 partially suppressed the autoimmune phenotype of the hos15-4 mutant (Fig. 2f,g), indicating a substantial contribution from SNC1 expression regulation in the immunity function of HOS15. This also implies that SNC1 is not the only target of HOS15 in immunity regulation. Additional NLRs among those that are repressed by HOS15 could also be direct targets of HOS15 and contribute to its immunity function (Fig. 3d).
This study indicates that HOS15 and HDA9 regulate NLR gene expression through histone deacetylation (Fig. S8). HDA9 is a key HDAC for NLR gene repression under pathogen infection, which is shown by its physical association with the NLR loci and the increased histone acetylation at those loci in hda9 mutant. Under non-pathogenic conditions, the loss of HDA9 function does not significantly alter NLR gene expression or histone acetylation at those loci although the HDA9 protein is associated with the NLR loci. This suggests that the HDA9 has no significant role in NLR regulation without pathogen invasion or it has an overlapping function with other HDACs under this condition. Another possibility is that HDA9 is pre-deposited on these loci but does not repress their expression until pathogen invasion. Previous studies did find that HDAC proteins bind to many loci but do not regulate their expression under normal conditions (Yang et al., Plant Cell, 2016; Chen et al., eLife, 2016). HOS15 appears to have a larger role than HDA9 in plant immunity regulation. The loss of HOS15 leads to increased histone acetylation at the NLR loci under both pathogenic and non-pathogenic conditions, indicating its function in histone deacetylation. HOS15 does not have demonstrated enzymatic activities, but it has eight WD40 repeats which serve as the scaffold for multi-protein assemblies (Zhu et al., 2008). HOS15 was found to potentially interact with multiple HDACs such as class I type HDACs (HDA6, HDA17, HDA19), class II type HDAC (HDA18), and plant-specific HD2 type HDACs (HD2A, HD2B, HD2C) in addition to HDA9 (Yu et al., 2017; Park et al., 2018a, b; Mayer et al., 2019). Additionally, some HDACs are implicated in plant immunity regulation (Kim et al., 2008; Latrasse et al., 2017; Wang et al., 2017). Thus, HOS15 is likely required for particular HDACs to function at the NLR loci and the loss of HOS15 function compromises the activity of HDACs at the NLR loci. In addition, HOS15 is required for proper HDA9 accumulation in nuclei and enrichment on chromatin (Meyers, et al., 2019). It will be interesting to determine whether or not additional HDACs might function together with HOS15 in repressing NLR gene expression under non-pathogenic conditions. These findings illustrate that histone deacetylation conferred by HDACs including HDA9 and its interacting HOS15 is an important epigenetic regulatory mechanism for proper NLR expression under both normal and pathogenic conditions.
Current study did not reveal a regulation on HOS15 or HDA9 activities by pathogen infection. The expression and localization of these two proteins do not appear to change after infection. The interaction between HOS15 and HDA9 or their association with NLR genes do not appear to alter in response to pathogen invasion either. It is therefore likely that HOS15 and HDAC exert a constitutive repression on NLR genes irrespective of the pathogenic conditions. Some NLR genes have higher expression at the loci upon pathogen invasion and the increased expression could be conferred by a higher histone acetylase activity in response to pathogen. However, the current data do not exclude the possibility that HOS15 and/or HDA9 may undergo post-translational modifications which might occur upon pathogen invasion and activate HDA9 for histone deacetylation.
Because HOS15 and HDA9 do not have DNA binding domains, it is unknown how they are recruited to target genes. The only known cis-element that is enriched in the promoter regions of NLR genes is W-box, the consensus binding site for WRKY transcription factors (Mohr et al., 2010). WRKY transcription factors are shown to be important to plant immunity (Birkenbihl et al., 2018), but it is not known whether or not NLR genes are their regulatory target genes. Several W-boxes are present in the promoter regions of SNC1 and the other four NLRs bound by HOS15 and HDA9, and these W-boxes are in the same or close to the association sites of HOS15 and HDA9 at NLR genes (Fig. S6). Therefore, WRKY factors are candidate transcription factors that bring HOS15 and HDA9 to the NLR genes. In addition, one of the WRKY factors WRKY53 was found to be associated with HDA9 (Chen et al., 2016). It would be of interest to test whether or not WRKYs are transcription factors recruiting HOS15 and HDA9 to NLR genes.
In summary, HOS15 and HDA9 are physically associated with a significant fraction of NLR genes and repress their expression which is positively correlated with histone acetylation. This study highlights the direct involvement of histone modifying enzymes in fine-tuning defense responses. Knowledge gained from this study will shed light on how immune receptor genes are precisely regulated to balance plant growth and defense. In addition, the underling mechanism of HOS15 and HDA9 on NLR genes in Arabidopsis may also apply to their homologs in crop plants, providing new insights on the regulation of disease resistance in crops.
Supplementary Material
Fig. S1 The hos15-3 mutant suppressed bon1 mos1.
Fig. S2 The hos15-4 is dwarf and has H2O2 accumulation.
Fig. S3 The hos15-2, hos15-3 and hos15-4 showed similar growth and immune phenotypes.
Fig. S4 GO term enrichment analysis of downregulated genes in hos15-4.
Fig. S5 pHOS15::HOS15:HA complements hos15-4 and bon1 mos1 hos15.
Fig. S6 Predicted WRKY53 binding sites on NLR genes.
Fig. S7 HOS15 regulates flowering time and silique development independent of its regulation on immunity.
Fig. S8 Working model for HOS15 and HDA9 in plant immunity.
Table S1 Primers used in this study.
Dataset S1. Differentially expressed genes in hos15-4 mutant
Dataset S2. Total NLRs in Arabidopsis thaliana
Dataset S3. NLRs upregulated in hos15-4 mutant
Dataset S4. HDA9 bound NLRs with increase gene expression in hos15-4 mutant
Dataset S5. NLRs bound by HDA9
ACKNOWLEDGMENTS
We thank the Arabidopsis Biological Resource Center for providing T-DNA lines. We would like to acknowledge technical advice and assistance by Dr. Dean Sanders, Dr. Jiapei Yan, Dr. Lilan Hong, Dr. Adele Zhou, Dr. Adrienne Roeder, Dr. Wojtek Pawlowski, Dr. Eric Richards, Dr. Olena Vatamaniuk and PhD candidate Mingyuan Zhu and Yingyu Liu. Work in JH’s laboratory was supported by NSF IOS-1353738. Work in XZ’s laboratory was supported by NSF CAREER award (MCB-1552455), USDA (Hatch 1012915), and NIH-MIRA (R35GM124806).
REFERENCES
- Bannister AJ, and Kouzarides T. 2011. Regulation of chromatin by histone modifications. Cell research 21(3): 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Z, Zhang N, and Hua J. 2014. Endopolyploidization and flowering time are antagonistically regulated by checkpoint component MAD1 and immunity modulator MOS1. Nature communications 5: 5628. [DOI] [PubMed] [Google Scholar]
- Bieri S, Mauch S, Shen QH, Peart J, Devoto A, Casais C, Ceron F, Schulze S, Steinbiss HH, … and Schulze-Lefert P 2004. RAR1 positively controls steady state levels of barley MLA resistance proteins and enables sufficient MLA6 accumulation for effective resistance. The Plant Cell 16(12): 3480–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigeard J, Colcombet J, and Hirt H. 2015. Signaling mechanisms in pattern-triggered immunity (PTI). Molecular plant 8(4): 521–539. [DOI] [PubMed] [Google Scholar]
- Birkenbihl RP, Kracher B, Ross A, Kramer K, Finkemeier I, and Somssich IE. 2018. Principles and characteristics of the Arabidopsis WRKY regulatory network during early MAMP-triggered immunity. The Plant Journal 96(3): 487–502. [DOI] [PubMed] [Google Scholar]
- Bosch-Presegué L, and Vaquero A. 2015. Sirtuin-dependent epigenetic regulation in the maintenance of genome integrity. The FEBS journal 282(9): 1745–1767. [DOI] [PubMed] [Google Scholar]
- Chen X, Lu L, Mayer KS, Scalf M, Qian S, Lomax A, Smith LM, and Zhong X. 2016. POWERDRESS interacts with HISTONE DEACETYLASE 9 to promote aging in Arabidopsis. Elife 5: e17214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SM, Song HR, Han SK, Han M, Kim CY, Park J, Lee YH, Jeon JS, Noh YS, and Noh B. 2012. HDA19 is required for the repression of salicylic acid biosynthesis and salicylic acid-mediated defense responses in Arabidopsis. The Plant Journal 71(1): 135–146. [DOI] [PubMed] [Google Scholar]
- Cui H, Gobbato E, Kracher B, Qiu J, Bautor J, and Parker JE. 2017. A core function of EDS1 with PAD4 is to protect the salicylic acid defense sector in Arabidopsis immunity. New Phytologist 213(4): 1802–1817. [DOI] [PubMed] [Google Scholar]
- Daudi A, and O’Brien JA. 2012. Detection of Hydrogen Peroxide by DAB Staining in Arabidopsis Leaves. Bio-protocol 2(18): e263. [PMC free article] [PubMed] [Google Scholar]
- Desvoyes B, Vergara Z, Sequeira-Mendes J, Madeira S, and Gutierrez C. 2018. A rapid and efficient ChIP protocol to profile chromatin binding proteins and epigenetic modifications in Arabidopsis In Marian Bemer and Célia Baroux (Eds), Plant Chromatin Dynamics. New York, NY: Humana Press, 71–82. [DOI] [PubMed] [Google Scholar]
- DeYoung BJ, and Innes RW. 2006. Plant NBS-LRR proteins in pathogen sensing and host defense. Nature immunology 7(12): 1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou M, and Hua J. 2012. Complex regulation of an R gene SNC1 revealed by autoimmune mutants. Plant signaling and behavior 7(2): 213–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland M, Montgomery RL, and Olson EN. 2009. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nature Reviews Genetics 10(1): 32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollender C, and Liu Z. 2008. Histone deacetylase genes in Arabidopsis development. Journal of integrative plant biology 50(7): 875–885. [DOI] [PubMed] [Google Scholar]
- Holt BF, Belkhadir Y, and Dangl JL. 2005. Antagonistic control of disease resistance protein stability in the plant immune system. Science 309(5736): 929–932. [DOI] [PubMed] [Google Scholar]
- Hua J, Wang S, and Sun Q. 2017. Mapping and Cloning of Chemical Induced Mutations by Whole-Genome Sequencing of Bulked Segregants In Libo Shan and Ping He (Eds), Plant Pattern Recognition Receptors. New York, NY: Humana Press, 285–289. [DOI] [PubMed] [Google Scholar]
- Jones JD, and Dangl JL. 2006. The plant immune system. Nature 444(7117): 323–329. [DOI] [PubMed] [Google Scholar]
- Kang MJ, Jin HS, Noh YS, and Noh B. 2015. Repression of flowering under a noninductive photoperiod by the HDA9-AGL19-FT module in Arabidopsis. New Phytologist 206(1): 281–294. [DOI] [PubMed] [Google Scholar]
- Kim KC, Lai Z, Fan B, and Chen Z. 2008. Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. The Plant Cell 20(9): 2357–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Latrasse D, Servet C, and Zhou DX. 2013. Arabidopsis histone deacetylase HDA9 regulates flowering time through repression of AGL19. Biochemical and Biophysical Research Communications 432(2): 394–398. [DOI] [PubMed] [Google Scholar]
- Lai Y, and Eulgem T. 2018. Transcript-level expression control of plant NLR genes. Molecular plant pathology 19(5): 1267–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latrasse D, Jégu T, Li H, de Zelicourt A, Raynaud C, Legras S, Gust A, Samajova O, Veluchamy A, … and Hirt H 2017. MAPK-triggered chromatin reprogramming by histone deacetylase in plant innate immunity. Genome biology 18(1): 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Tessaro MJ, Li X, and Zhang Y. 2010. Regulation of the expression of plant resistance gene SNC1 by a protein with a conserved BAT2 domain. Plant physiology 153(3): 1425–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Yang S, Yang H, and Hua J. 2007. The TIR-NB-LRR gene SNC1 is regulated at the transcript level by multiple factors. Molecular plant-microbe interactions 20(11): 1449–1456. [DOI] [PubMed] [Google Scholar]
- Liu J, Zhi P, Wang X, Fan Q, and Chang C. 2018. Wheat WD40-repeat protein TaHOS15 functions in a histone deacetylase complex to fine-tune defense responses to Blumeria graminis f. sp. tritici. Journal of Experimental Botany 70(1): 255–268. [DOI] [PubMed] [Google Scholar]
- Mayer KS, Chen X, Sanders D, Chen J, Jiang J, Nguyen P, Scalf M, Smith LM, and Zhong X. 2019. HDA9-PWR-HOS15 is a core histone deacetylase complex regulating transcription and development. Plant physiology 180(5): 342–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Kozik A, Grieg A, Kuang H, and Michelmore RW. 2003. Genome-wide analysis of NBS-LRR–encoding genes in Arabidopsis. The Plant Cell 15(4): 809–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr TJ, Mammarella ND, Hoff T, Woffenden BJ, Jelesko JG, and McDowell JM. 2010. The Arabidopsis downy mildew resistance gene RPP8 is induced by pathogens and salicylic acid and is regulated by W box cis elements. Molecular plant-microbe interactions 23(10): 1303–1315. [DOI] [PubMed] [Google Scholar]
- Moore JW, Loake GJ, and Spoel SH. 2011. Transcription dynamics in plant immunity. The Plant Cell 23(8): 2809–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, Tabata R, Kawai T, Tanaka K, … and Ishiguro S. 2007. Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Bioscience, biotechnology, and biochemistry 71(8): 2095–2100. [DOI] [PubMed] [Google Scholar]
- Nishimura MT, Anderson RG, Cherkis KA, Law TF, Liu QL, Machius M, Nimchuk ZL, Yang L, Chung EH, … and Dangl JL. 2017. TIR-only protein RBA1 recognizes a pathogen effector to regulate cell death in Arabidopsis. Proceedings of the National Academy of Sciences 114(10): E2053–E2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma K, Thorgrimsen S, Malinovsky FG, Fiil BK, Nielsen HB, Brodersen P, Hofius D, Petersen M, and Mundy J. 2010. Autoimmunity in Arabidopsis acd11 is mediated by epigenetic regulation of an immune receptor. PLoS pathogens 6(10): e1001137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HJ, Baek D, Cha JY, Liao X, Kang SH, McClung CR, Lee SY, Yun DJ, and Kim WY. 2019. HOS15 Interacts with the Histone Deacetylase HDA9 and the Evening Complex to Epigenetically Regulate the Floral Activator GIGANTEA. The Plant Cell 31(1): 37–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Lim CJ, Khan IU, Jan M, Khan HA, Park HJ, Guo Y, and Yun DJ. 2018a. Identification and Molecular Characterization of HOS15-interacting Proteins in Arabidopsis thaliana. Journal of Plant Biology 61(5): 336–345. [Google Scholar]
- Park J, Lim CJ, Shen M, Park HJ, Cha JY, Iniesto E, Rubio V, Mengiste T, Zhu JK, … and Yun DJ. 2018b. Epigenetic switch from repressive to permissive chromatin in response to cold stress. Proceedings of the National Academy of Sciences 115(23): E5400–E5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto E, and Yoshida M. 2014. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harbor perspectives in biology, 6(4): a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Shinozuka N, Hirakata T, Nakata MT, Demura T, Tsukaya H, and Horiguchi G. 2018. OLIGOCELLULA1/HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES15 promotes cell proliferation with HISTONE DEACETYLASE9 and POWERDRESS during leaf development in Arabidopsis thaliana. Frontiers in plant science, 9: 580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Xie Z, Chen W, Glazebrook J, Chang HS, Han B, Zhu T, Zou G, and Katagiri F. 2003. Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. The Plant Cell 15(2): 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, and Katagiri F. 2010. Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Current opinion in plant biology 13(4): 459–465. [DOI] [PubMed] [Google Scholar]
- Tsuda K, and Somssich IE. 2015. Transcriptional networks in plant immunity. New Phytologist 206(3): 932–947. [DOI] [PubMed] [Google Scholar]
- Urbach JM, and Ausubel FM. 2017. The NBS-LRR architectures of plant R-proteins and metazoan NLRs evolved in independent events. Proceedings of the National Academy of Sciences 114(5): 1063–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zanten M, Zöll C, Wang Z, Philipp C, Carles A, Li Y, Kornet NG, Liu Y, and Soppe WJ. 2014. HISTONE DEACETYLASE 9 represses seedling traits in Arabidopsis thaliana dry seeds. The Plant Journal 80(3): 475–488. [DOI] [PubMed] [Google Scholar]
- Wang C, Gao F, Wu J, Dai J, Wei C, and Li Y. 2010. Arabidopsis putative deacetylase AtSRT2 regulates basal defense by suppressing PAD4, EDS5 and SID2 expression. Plant and cell physiology 51(8): 1291–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Hu Q, Wu Z, Wang H, Han S, Jin Y, Zhou J, Zhang Z, Jiang J, and Yang W. 2017. HISTONE DEACETYLASE 6 represses pathogen defence responses in Arabidopsis thaliana. Plant, cell and environment 40(12): 2972–2986. [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, and Provart NJ. 2007. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PloS one 2(8): e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S, Cheng YT, Huang S, Win J, Soards A, Jinn TL, Jones JD, Kamoun S, Chen S, and Li X. 2013. Regulation of transcription of nucleotide-binding leucine-rich repeat-encoding genes SNC1 and RPP4 via H3K4 trimethylation. Plant physiology 162(3): 1694–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, and Hua J. 2004. A haplotype-specific Resistance gene regulated by BONZAI1 mediates temperature-dependent growth control in Arabidopsis. The Plant Cell 16(4): 1060–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CW, Tai R, Wang SC, Yang P, Luo M, Yang S, Cheng K, Wang WC, Cheng YS and Wu K. 2017. HISTONE DEACETYLASE6 acts in concert with histone methyltransferases SUVH4, SUVH5, and SUVH6 to regulate transposon silencing. The Plant Cell 29(8): 1970–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Wang Z, Bao Z, Yang L, Wu D, Shu X, and Hua J. 2018. MOS1 functions closely with TCP transcription factors to modulate immunity and cell cycle in Arabidopsis. The Plant Journal 93(1): 66–78. [DOI] [PubMed] [Google Scholar]
- Zhao T, Rui L, Li J, Nishimura MT, Vogel JP, Liu N, Liu S, Zhao Y, Dangl JL, and Tang D. (2015). A truncated NLR protein, TIR-NBS2, is required for activated defense responses in the exo70B1 mutant. PLoS genetics 11(1): e1004945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Ding Y, Sun X, Xie S, Wang D, Liu X, Su L, Wei W, Pan L, and Zhou DX. 2016. Histone deacetylase HDA9 negatively regulates salt and drought stress responsiveness in Arabidopsis. Journal of experimental botany 67(6): 1703–1713. [DOI] [PubMed] [Google Scholar]
- Zhu J, Jeong JC, Zhu Y, Sokolchik I, Miyazaki S, Zhu JK, Hasegawa PM, Bohnert HJ, Shi H, … and Bressan RA. 2008. Involvement of Arabidopsis HOS15 in histone deacetylation and cold tolerance. Proceedings of the National Academy of Sciences 105(12): 4945–4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Mang HG, Sun Q, Qian J, Hipps A, and Hua J. 2012. Gene discovery using mutagen-induced polymorphisms and deep sequencing: application to plant disease resistance. Genetics 192(1): 139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou B, Sun Q, Zhang W, Ding Y, Yang DL, Shi Z, and Hua J. 2017. The Arabidopsis chromatin-remodeling factor CHR5 regulates plant immune responses and nucleosome occupancy. Plant and Cell Physiology 58(12): 2202–2216. [DOI] [PubMed] [Google Scholar]
- Zou B, Yang DL, Shi Z, Dong H, and Hua J. 2014. Monoubiquitination of histone 2B at the disease resistance gene locus regulates its expression and impacts immune responses in Arabidopsis. Plant physiology 165(1): 309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 The hos15-3 mutant suppressed bon1 mos1.
Fig. S2 The hos15-4 is dwarf and has H2O2 accumulation.
Fig. S3 The hos15-2, hos15-3 and hos15-4 showed similar growth and immune phenotypes.
Fig. S4 GO term enrichment analysis of downregulated genes in hos15-4.
Fig. S5 pHOS15::HOS15:HA complements hos15-4 and bon1 mos1 hos15.
Fig. S6 Predicted WRKY53 binding sites on NLR genes.
Fig. S7 HOS15 regulates flowering time and silique development independent of its regulation on immunity.
Fig. S8 Working model for HOS15 and HDA9 in plant immunity.
Table S1 Primers used in this study.
Dataset S1. Differentially expressed genes in hos15-4 mutant
Dataset S2. Total NLRs in Arabidopsis thaliana
Dataset S3. NLRs upregulated in hos15-4 mutant
Dataset S4. HDA9 bound NLRs with increase gene expression in hos15-4 mutant
Dataset S5. NLRs bound by HDA9
Data Availability Statement
RNA-seq data is available at the Gene Expression Omnibus and can be found with the GEO accession number GSE131227.
Sequence data from this article can be found through TAIR (https://www.arabidopsis.org) under the following accession numbers:
HOS15: AT5G67320, HDA9: AT3G44680; SNC1: AT4G16890; PR1: AT2G14610; MOS1: AT4G24680; BON1: AT5G61900; PAD4: AT3G52430.