Abstract
Drosophila Clueless (Clu) is a ribonucleoprotein that directly affects mitochondrial function. Loss of clu causes mitochondrial damage, and Clu associates with proteins on the mitochondrial outer membrane. Clu’s subcellular pattern is diffuse throughout the cytoplasm, but Clu also forms large mitochondria-associated particles. Clu particles are reminiscent of ribonucleoprotein particles such as stress granules and processing bodies. Ribonucleoprotein particles play critical roles in the cell by regulating mRNAs spatially and temporally. Here, we show that Clu particles are unique, highly dynamic and rapidly disperse in response to stress in contrast to processing bodies and autophagosomes. In addition, Clu particle formation is dependent on diet as ovaries from starved females no longer contain Clu particles, and insulin signaling is necessary and sufficient for Clu particle formation. Oxidative stress also disperses particles. Since Clu particles are only present under optimal conditions, we have termed them “bliss particles”. We also demonstrate that many aspects of Clu function are conserved in the yeast homolog Clu1p. These observations identify Clu particles as stress-sensitive cytoplasmic particles whose absence corresponds with altered cell stress and mitochondrial localization.
Keywords: mitochondria, Clueless, Drosophila, insulin, ribonucleoprotein particle
Introduction:
Clu encodes a large multi-domain protein that is directly involved in regulating mitochondrial function, although the molecular mechanisms are still not completely understood (Cox & Spradling, 2009; Sen & Cox, 2016; Sen, Kalvakuri, Bodmer, & Cox, 2015). Loss of clu in Drosophila is adult lethal, with flies surviving only 4-7 days post-eclosion (Sen & Cox, 2016; Sen, Damm, & Cox, 2013). clu mutants are male and female sterile, and mitochondria in female germ cells are clumped, mislocalized, and morphologically swollen which is a phenotypic hallmark of damaged, nonfunctional mitochondria (Cox & Spradling, 2009). Drosophila Clu also physically and genetically interacts with the PINK1/Parkin mitophagy complex and thus may play a role as a sensor linking mitochondrial function with mitophagy although what role Clu plays in mitophagy is not yet clear (Sen et al., 2015). Clu, CLUH and Clu1p, the Drosophila, human and yeast homologs, respectively, are ribonucleoproteins (Gao et al., 2014; Schatton et al., 2017; Sen & Cox, 2016). Clu and Clu1p bind to mRNA in Drosophila and yeast, respectively, and Drosophila Clu was shown to associate with the ribosome in Drosophila, potentially at the mitochondrial outer membrane (Sen & Cox, 2016). CLUH binds to and regulates mRNAs encoding proteins that will be imported into mitochondria and CLUH deficiency alters metabolism (Gao et al., 2014; Schatton et al., 2017; Wakim et al., 2017).
Ribonucleoprotein (RNP) granules are cytoplasmic, non-membranous aggregates which function in temporal and spatial post-transcriptional regulation of their mRNA cargos (Anderson & Kedersha, 2006, 2009). RNP granules are present in somatic and germ cells, and they serve to regulate gene expression by a variety of mechanisms (Anderson & Kedersha, 2009). Processing bodies (P-bodies) and stress granules, two major types of RNP granules in the cytoplasm, have well-characterized roles in cellular stress responses. During stress, both become more abundant in order to regulate the transport, translation, and stability of mature mRNAs (Decker & Parker, 2012; Namkoong, Ho, Woo, Kwak, & Lee, 2018; C. Wang et al., 2018). Clu forms large particles in Drosophila female germ cells that are closely juxtaposed with mitochondria (Cox & Spradling, 2009). Particles are also found in the surrounding somatic follicle cells, larval neuroblasts and other neuronal cell types and larval muscle (Sen et al., 2013; Z. H. Wang, Clark, & Geisbrecht, 2016). The Arabidopsis thaliana homolog, friendly mitochondria (FMT), forms particles, which are also found in close proximity to mitochondria (El Zawily et al., 2014). In addition, the vertebrate homolog CLUH has a granular cytoplasmic pattern in cultured COS7 cells (Gao et al., 2014).
Here, we examine the dynamic nature of Clu particle formation and disaggregation using live-imaging to show that Clu forms previously undescribed and highly stress-sensitive cytoplasmic particles. Clu particles are dynamic and require an intact microtubule cytoskeleton in order to move processively. The oocyte does not contain particles and has very low levels of Clu protein relative to the connected germ cells (called nurse cells). Clu particles do not colocalize with other well described cytoplasmic components that form under stress; thus, we believe these particles uniquely respond to stress by disaggregating. Additionally, we demonstrate a conserved role for Clu in Saccharomyces cerevisiae. Yeast Clu1p also forms particles, clu1 deletion causes decreased growth on a non-fermentable carbon source and increased petite colony formation, and Clu1p binds the ribosomal protein RpL3p. Clu particles in fly ovaries are highly sensitive to nutrition and insulin. Starved follicles no longer have particles in germ cells and surrounding somatic follicle cells even though Clu protein levels remain the same. This effect is at least partly regulated by insulin, as insulin is both necessary and sufficient for particle formation. Nutritional stress is not the only particle disruptor, as increased mitochondrial oxidation also causes particles to disperse. We have named Clu particles “bliss particles” because they are only present under stress-free, well-fed optimal conditions, unlike stress granules and processing bodies. Finally, we show mitochondrial localization in germ cells is completely dependent on well-fed, stress-free conditions, as any of the aforementioned stressors cause clumping and mislocalization. These observations shed light on how Clu’s subcellular localization is highly dependent on the cell’s nutritional status and this localization changes in response to insulin signaling.
Results:
Clu particles are abundant and highly dynamic
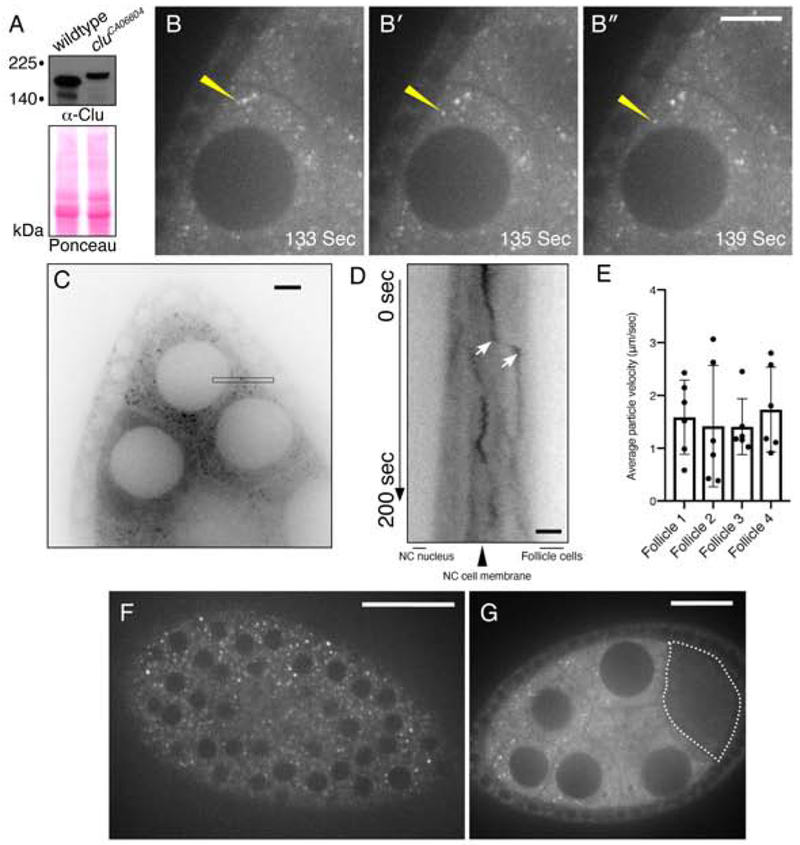
Clu protein forms particles in the cytoplasm of many cell types. Female germ cells have been an excellent tissue in which to study Clu particles as they are highly metabolically active, are very large and have abundant particles (Kato & Nakamura, 2012). To better understand Clu’s role in the cell, we used live-imaging to dissect Clu particle dynamics. To do this, we imaged Clu::GFP in the GFP trap line cluelessCA06604 and compared these results to our previous observations in fixed, wild-type ovaries labeled with anti-Clu antibodies (Cox & Spradling, 2009; Sen et al., 2013; Sen et al., 2015). cluCA06604 is homozygous viable with no apparent defects in oogenesis or lifespan (Buszczak et al., 2007; Cox & Spradling, 2009). Extract from cluCA06604 probed with anti-Clu antibody had only one higher migrating band indicating all Clu in the cells is GFP labeled (Fig. 1A). Clu particles appeared more abundant using live-imaging compared to the abundance of particles seen using anti-Clu antibody in fixed germ cells (Movie 1 vs. Fig. 4, (Cox & Spradling, 2009)). During live-imaging, Clu::GFP showed a mix of apparently random and directed movement. At any given time, approximately 12% of the particles appeared to move in a directed manner over the course of 200 seconds (Fig. 1B–B″, yellow arrows, Movie 1). For particles that move quickly, kymographic analysis supported that the average particle velocity is 1.5 μm/sec (Fig. 1C, D, E). The rest of the particles were either fairly stationary or appear to move randomly in the cytoplasm. Adding the microtubule destabilizer colcemide disrupted the microtubule cytoskeleton as expected and caused particles and mitochondria to remain stationary, indicating that particle movement is microtubule-based (Fig. S1, Movie 2, 3, 4). Clu particles were present in the surrounding somatic follicle cells, but did not appear to move as much, likely due to the restrictive size of the cells (Fig. 1F, Movie 5). In addition, Clu::GFP protein levels were very low in the oocyte relative to the nurse cells, and we never observed Clu::GFP particles in the oocyte (Fig. 1G, dotted line).
Figure 1.
Clu::GFP live-imaging shows robust, dynamic particles in germ cells and surrounding follicle cells. (A) Western blot analysis indicating that all Clu::GFP protein is GFP-tagged in follicles from cluCA06604 females. (B-B″) cluCA06604 female germ cells. Clu particles (white) are plentiful. Yellow arrow indicates example of processive movement. (C-E) A subset of particles move at speeds consistent with directed movement along microtubules. (C) Representative image showing a single still-frame. The black thin box shows the orientation and plane used to make the kymograph (D). (D) An example of directed movement is indicated between the white arrows of the kymograph. (E) Quantification of the average velocity of directed Clu::GFP particle movement. (F) En face optical section of the top of a cluCA06604 follicle showing the surrounding somatic follicle cells. The black circles are the nuclei. (G) Cross section of Clu::GFP follicle. Clu is decreased in the oocyte (dotted line) and does not contain particles. Details of n values and analysis are in the materials and methods. Scale bar: 20 μm in B″ for B-B″, 10 μm in C for C, 5 μm in D for D, 40 μm in F for F, 10 μm in G for G. For E, error bars = SD.
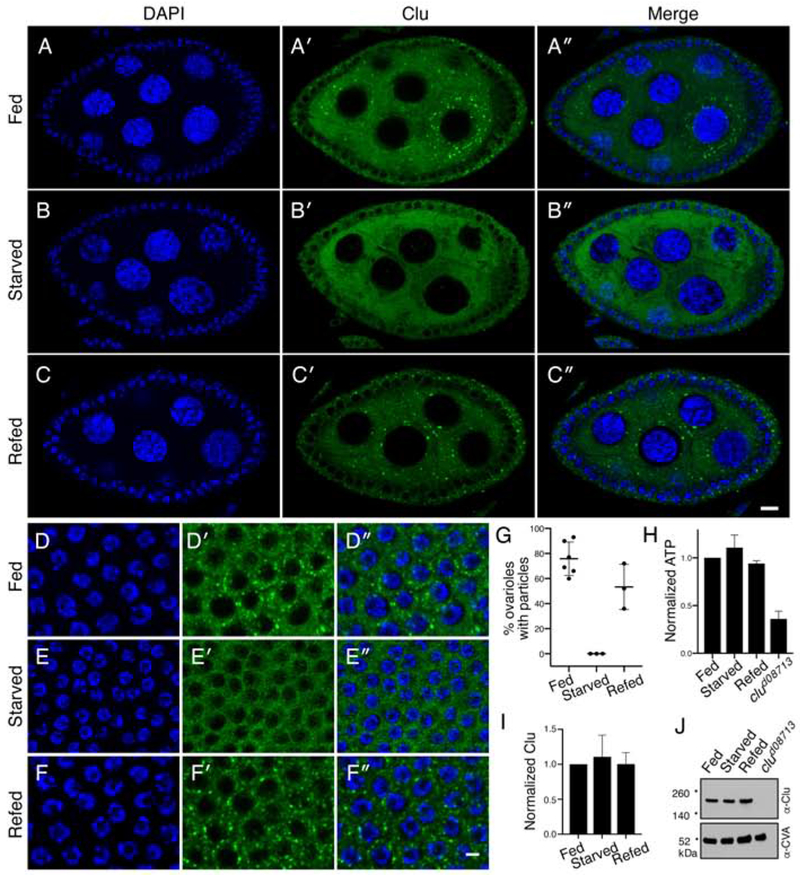
Figure 4.
Clu particles disaggregate in response to starvation. (A-C″). Follicles from w1118 females. Well-fed follicles contain many large particles in the germ cells (A′). Starvation for 5 hours on H20 causes the particles to disaggregate (B′). Re-feeding starved females causes Clu particles to reform (C′). Quantification is shown in (G). (D-F″) Surrounding somatic follicle cells show the same dynamic as the germ cells. ATP levels (H) and Clu levels (I, J) remain the same for all three conditions. clud08713 is a positive control. Details of n values are in the materials and methods. Error bars are S.E.M. Green = Clu (A-F″) and blue = DAPI (A-F″). Scale bar: 10 μm in C″ for A-C″ and in F″ for D-F″.
Clu particles do not colocalize with many known structures
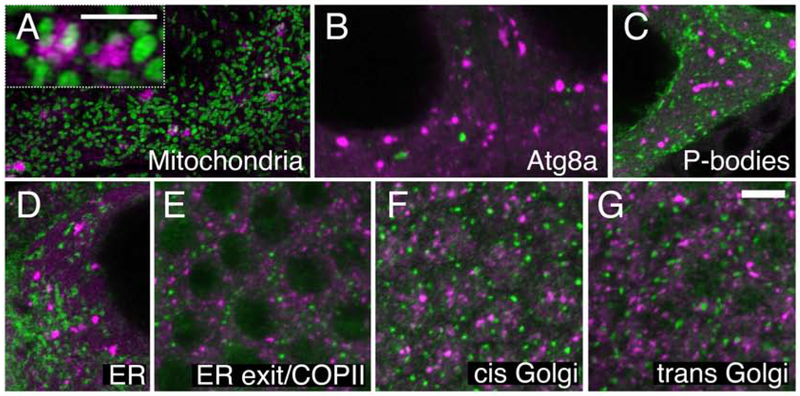
As we have previously shown, Clu particles tightly associate, but do not co-localize, with mitochondria (Fig. 2A) (Cox & Spradling, 2009). Every large particle associates with several mitochondria in fixed tissues, but many of the mitochondria present are not associated with particles. Initially, we thought this mitochondrial association may be due to Clu being involved in autophagosome formation and mitophagy (Kim, Rodriguez-Enriquez, & Lemasters, 2007). However, in germ cells labeled with anti-Clu antibody, Clu did not co-localize with the LC3 homolog Atg8a, a marker of autophagosomes whose number increases in response to stress. Clu particles also did not co-localize with a second stress-associated cytoplasmic body, processing bodies (Fig. 2B and 2C). Finally, it appeared that Clu associated with ER-exit sites in Drosophila larval muscle (Z. H. Wang, Rabouille, & Geisbrecht, 2015). However, we did not observe any co-localization with components of the secretory pathway. Clu particles did not show any particular association with endoplasmic reticulum in the germ cells (Fig. 2D).
Figure 2:
Clu particles are unique. (A) Structured illumination micrograph. Clu particles are granular and touch mitochondria in germ cells (inset), as previously shown. (B, C) Clu particles do not co-localize with autolysosomes (B) or Processing bodies (C) in germ cells. (D-G) Clu particles do not appear to associate with ER in germ cells (D), nor with ER-exit sites (E), cis-Golgi (F) or trans-Golgi (G) in surrounding somatic follicle cells. Green = ATP synthase (A), Atg8a (B), Tral::GFP (C), Sec61α::GFP (D), Sec23 (E), GMAP (F), and Golgin245 (G). Magenta = Clu (A-G). Scale bar: 5 μm in G for A-G, 2.5μm in A for inset.
Antibodies used to determine colocalization with components of ER exit sites/COPII vesicles, cis Golgi, and trans Golgi labeled surrounding follicle cells, but did not penetrate the female germ cells well (Fig. 2E–G). Thus, we examined the surrounding follicle cells for co-localization. In somatic follicle cells, Clu particles were distinct from ER exit sites/COPII vesicles (Fig. 2E), cis Golgi (Fig. 2F) and trans Golgi (Fig. 2G). Therefore, we believe Clu forms previously undescribed particles in the cytoplasm specific to mitochondrial function.
Clu particles are conserved
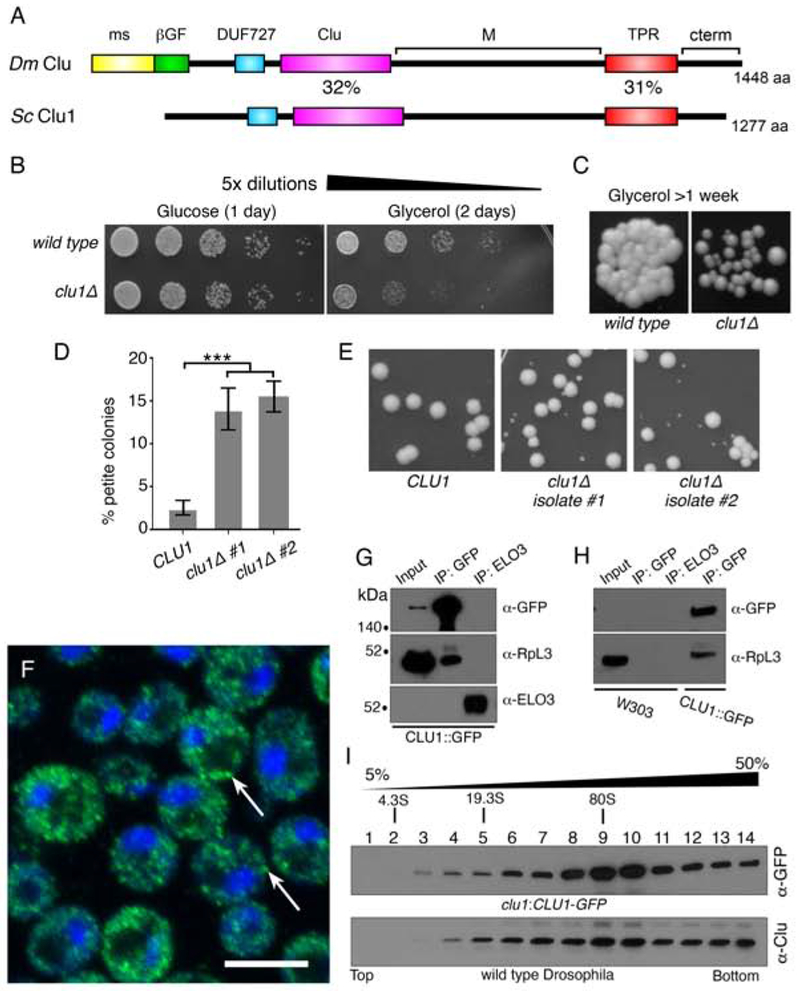
Clu has homologs in many species, including the poorly characterized Clu1p in Saccharomyces cerevisiae. Clu1p shares 39% amino acid identity overall with Drosophila Clu, 31% between the tetratricopeptide repeat (TPR) domains, and 32% between the Clu domains (Fig. 3A). Since Clu directly affects mitochondrial function in Drosophila, we tested whether a clu1Δ knockout yeast strain shows defects associated with mitochondrial dysfunction including loss of ability to grow on a non-fermentable carbon source and spontaneous petite colony formation. Wild type yeast normally use fermentation and grow well on glucose (Fig. 3B). clu1Δ grew equally as well as wild type yeast on media containing glucose (Fig. 3B). However, when grown on the non-fermentable carbon source glycerol which forces the cells to rely on oxidative phosphorylation, clu1Δ showed decreased growth compared to wild type (Fig. 3B and 3C). This was true in two different wild type backgrounds (BY4741 and W303) with three re-derived clu1::KANMX deletions (Fig. S2). The poor growth and small colonies could be readily seen after growing a small number of cells on glycerol for over a week (Fig. 3C). Petite colony formation occurs in mutants that are defective for oxidative phosphorylation even when they are grown on a fermentable carbon source. Newly derived clu1Δ in a BY4741 background had a significantly higher percent (~15%) petite formation vs wild type BY4741 (~2%) when grown on Yeast Extract-Peptone-Dextrose (YPDextrose) (Fig. 3D, E). The clu1:CLU1-GFP strain contains a GFP insertion at the endogenous CLU1 locus (Huh et al., 2003). Anti-GFP antibody labeling of this strain showed that Clu1p, similar to Clu, was punctate in the cytoplasm (Fig. 3F, arrows). As with Drosophila Clu, yeast Clu1p also associated with a ribosomal protein, RpL3p (Fig. 3G, H (Sen & Cox, 2016)). Drosophila Clu sediments in the heavier fractions on a sucrose gradient (Fig. 3I, (Sen & Cox, 2016)). Yeast Clu1p had a similar sedimentation pattern (Fig. 3I). Thus, many aspects of yeast Clu1p recapitulate those found in Drosophila, supporting a conserved function between the two species.
Figure 3.
Yeast Clu1p accumulates as cytoplasmic particles and associate with RpL3p. (A) A cartoon diagram of the shared domain structure between Drosophila Clu (Dm) and yeast Clu1p (Sc). (B) Serially diluted clu1Δ cultures grow normally on glucose, but not on glycerol. (C) clu1Δ forms small colonies after one week of growth on glycerol compared to its parental wild type strain. (D) Two independent isolates of clu1Δ. (BY4741 genetic background) form significantly higher percentages of petite colonies. (E) Representative micrographs showing petite colony formation. (F) Fixed and anti-GFP labeled log-phase yeast cells of strain clu1::CLU1-GFP. Clu1p puncta are indicated by arrows. (G) Co-immunoprecipitation shows that Clu1p associates with the ribosomal protein RpL3. Elongation of fatty acids protein 3 (Elo3p) serves as a negative control. Pulldowns were from clu1::CLU1-GFP extract. (H) The anti-GFP antibody is specific for clu1::CLU1-GFP. GFP pulldown from a wild type strain (W303) indicates there is no non-specific cross-reactivity. (I) Sucrose gradient using extract from yeast strain clu1::CLU1-GFP (top) and Drosophila wild type ovaries (bottom). Green = Clu1p::GFP, blue = DAPI. Scale bar: 5 μm in F for F. For D, p < 0.001 using a two-way ANOVA test.
Clu particles are sensitive to nutritional stress
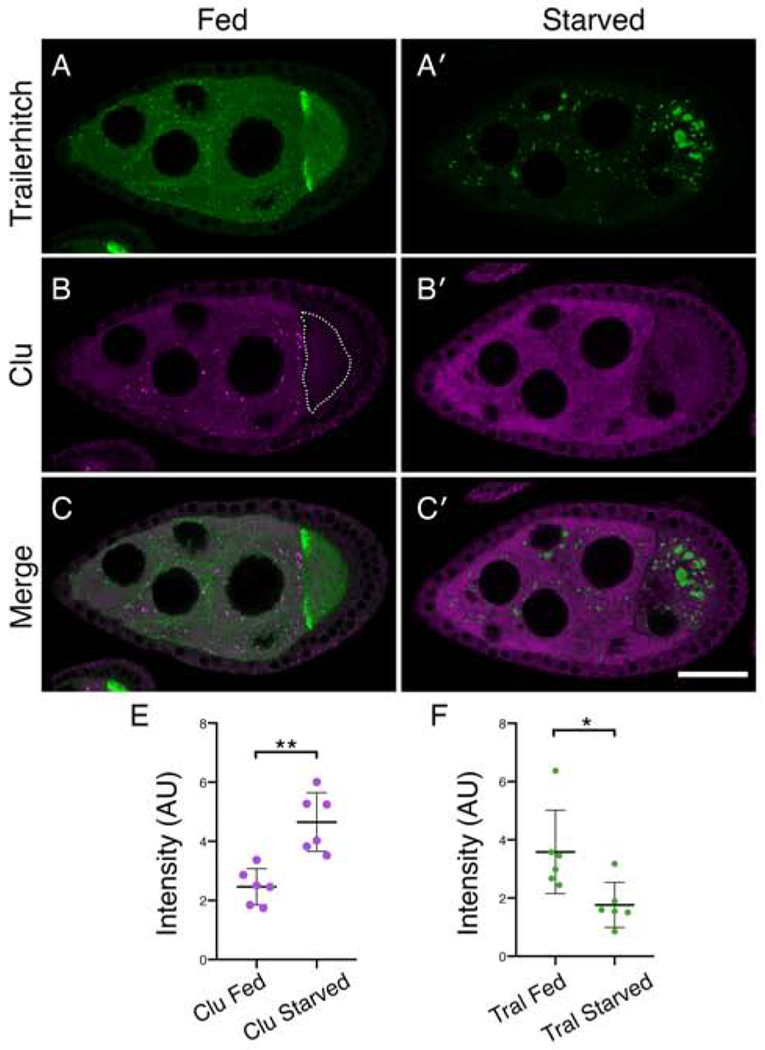
In Drosophila, we have observed that only well-fed females reproducibly have robust Clu particles. This suggests that ample access to yeast is important and that the distribution of Clu is sensitive to nutritional stress. To analyze the effect of nutritional stress on Clu particle dynamics, we starved and refed wild type flies then labeled the ovaries with anti-Clu antibodies. Wild type females fed wet yeast paste always had robust Clu particles (Fig. 4A′–A″). However, starving the same well-fattened flies for five hours on water completely abolished particles, resulting in dispersed Clu (Fig. 4B′–B″). Re-feeding yeast paste for as little as two hours completely reversed this effect (Fig. 4C′–C″, G). These results were the same for the surrounding somatic follicle cells (Fig. 4D–F″). The short five-hour starvation (typical starvation protocols last >24 hours (Barth, Szabad, Hafen, & Kohler, 2011; Nezis et al., 2009)) did not result in any behavioral defects and levels of ATP remained the same (Fig. 4H). Particle disaggregation was not due to protein degradation as Clu protein levels remained the same for all three conditions (Fig. 4I, J). These results support nutrition as an important regulator of Clu particle formation, and that their formation and disaggregation are highly dynamic and reversible.
After starvation, Clu protein levels remain the same (Fig. 4I, J) but Clu particles disperse. To confirm that dispersed cytoplasmic Clu levels increase when particles disperse, we measured the amount of diffuse Clu in the nurse cell cytoplasm and compared this to a protein found in processing bodies. Trailer hitch (Tral) is a ribonucleoprotein that is a component of processing bodies (Barbee et al., 2006; Wilhelm, Buszczak, & Sayles, 2005). Under well-fed conditions, tralCA06517 follicles expressing Tral::GFP and subsequently labeled with anti-GFP antibody formed characteristic small Tral-containing particles in the cytoplasm (Fig. 5A). Tral labeling was also concentrated at the anterior of the oocyte and was diffusely cytoplasmic (Fig. 5A) (Wilhelm et al., 2005). In the same follicle, anti-Clu antibody labeling showed Clu particles (Fig. 5B). Similar to our result using live-imaging, Clu protein levels were very low in the oocyte relative to the nurse cells, and we never observed Clu particles in the oocyte (Fig. 5B, dotted line). After five-hour starvation, Tral::GFP formed very large processing bodies in the nurse cells and oocyte with a significant decrease in the diffuse cytoplasmic staining (Fig. 5A′, C′, F) (Burn et al., 2015). In contrast, in the same follicle Clu particles dispersed completely with a significant rise in diffuse cytoplasmic staining (Fig. 5B′,C′, E).
Figure 5.
Clu particles and the Processing body component Trailer hitch respond oppositely to starvation. (A-C′) Follicles from Tral::GFP females. (A, C) Tral::GFP forms small aggregates in the nurse cells, homogeneous dispersed staining, and is concentrated at the anterior of the oocyte (A). Upon starvation, Tral::GFP forms very large processing bodies, the diffuse Tral signal decreases (A′, C′), whereas Clu particles disaggregate with Clu becoming diffuse (B′, C′). The dotted line indicates the oocyte (B). (E) Quantification of homogeneously dispersed Clu intensity in nurse cells under fed and starved conditions. (F) Quantification of Tral::GFP homogeneously dispersed intensity in nurse cells under fed and starved conditions. Details of n values are in the materials and methods. Green = Tral::GFP (A-A′, C-C′ ), magenta = Clu (B-B′, C-C′). Scale bar: 40μm in C′ for A-C′. Error bars are S.E.M. * p = 0.015, ** p = 0.0016 using a Student’s t-test with Welch’s correlation.
Insulin is necessary and sufficient to induce Clu particle formation
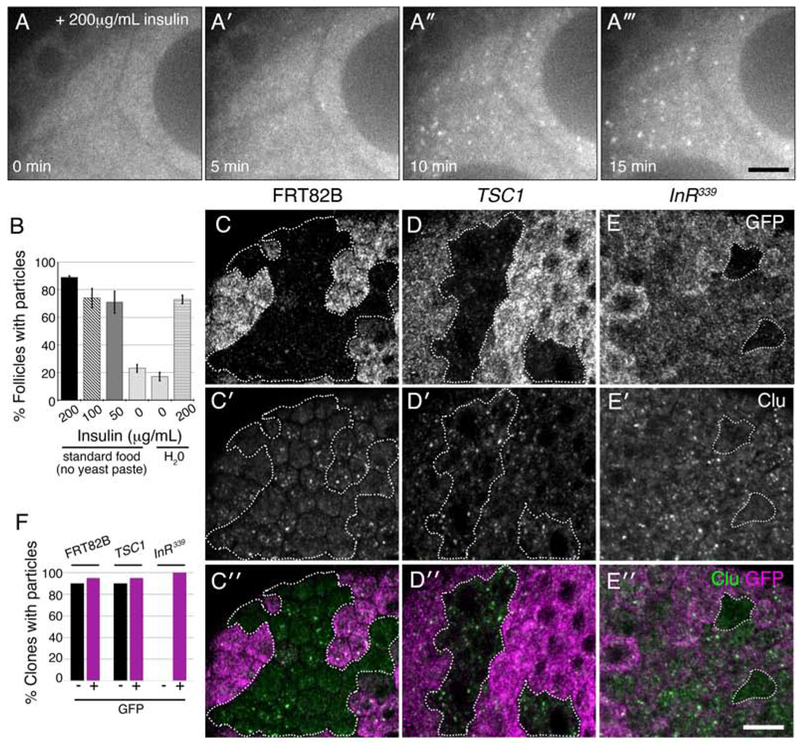
Since nutrition availability affects Clu particle formation, we examined the role of insulin signaling on this process. To determine if insulin is sufficient to induce particle formation, we used live-imaging. Well-fed Clu::GFP expressing cluCA06604 females were transferred to either standard food (no yeast paste) or H20 for five hours, followed by dissection in insulin-free media. Insulin addition caused robust Clu particle formation within ten minutes, with 80% of the follicles containing particles after 15 minutes (Fig. 6A–A‴, B, Movie 6). The percentage of individual follicles that formed particles in response to insulin was similar whether the females were starved on water or fed only standard food (no yeast paste) (Fig. 6B) and was also similar to fixed samples from well-fed females (Fig. 6B vs 4G). Using this technique, both starvation conditions resulted in a baseline of 20% individual follicles with particles (Fig. 6B, 0μg/mL insulin bars). These results support that insulin is sufficient to induce particle formation.
Figure 6.
Insulin is necessary and sufficient for Clu particle formation. (A-A‴) Stills from live-imaging of starved cluCA06604 germ cells expressing Clu::GFP. Females were raised on standard fly food (no yeast paste) and dissected in Complete Schneider’s. After adding insulin at time zero (A), particles start forming within five minutes (A′). (B) Quantification of the percent single follicles containing Clu particles for females starved on standard food (no yeast paste) or H2O. (C-E″) Clonal analysis in follicle cells. Wild type control clones (FRT82B, GFP−, dotted white line) have Clu particles as expected (C-C″). Clones mutant for TSC1Q87X also have Clu particles (D-D″, dotted white lines). Clones mutant for InR339 lack particles (dotted white lines) (E-E″). (F) Quantification of the number of GFP+ and GFP− clones containing particles for control FRT82B (n = 20), TSC1 (n = 20), and InR339 (n = 11) mutant clones. White = Clu::GFP (A-A‴), GFP (C, D, E), and Clu, (C′, D′, E′), green = Clu, magenta = GFP. For B, error bars = S.D. Scale bars: 20 μm in A‴ for A-A‴, 10 μm in E″ for C-E′.
To further assess the role of insulin in Clu particle regulation, we used clonal analysis to increase and decrease insulin signaling in the surrounding somatic follicle cells. In a FRT82B control wild type background, GFP− and GFP+ clones contained particles indicating the presence or absence of GFP has no effect on Clu particle formation (Fig. 6C–C″, dotted outline, F). To determine whether an increase in insulin signaling affects Clu particle formation, we made follicle cell clones mutant for TSC1Q87X, a negative effector of insulin signaling. TSC1 mutant clones contained particles at the same frequency as the sibling TSC1/+ control clones (Fig. 6D–D″, dotted outlines, F). To test if insulin signaling is necessary for Clu particle formation, we made clones for two alleles of the Insulin-like Receptor (InR). InR mutant cells divide slowly and our heat shock protocol was short from clone induction to dissection, thus follicle cell clone size and frequency were small (Chen, Jack, & Garofalo, 1996; Oldham, Montagne, Radimerski, Thomas, & Hafen, 2000; Oldham et al., 2002). Loss of InR resulted in a loss of Clu particles in the mutant clones compared to InR/+ sibling clones (Fig. 6E–E″, dotted outlines, F, Fig. S3). These results support that upregulating insulin signaling does not affect Clu particle formation and that loss of insulin signaling causes loss of particles, indicating insulin is required for particle formation.
Clu particle formation is sensitive to mitochondrial oxidative stress
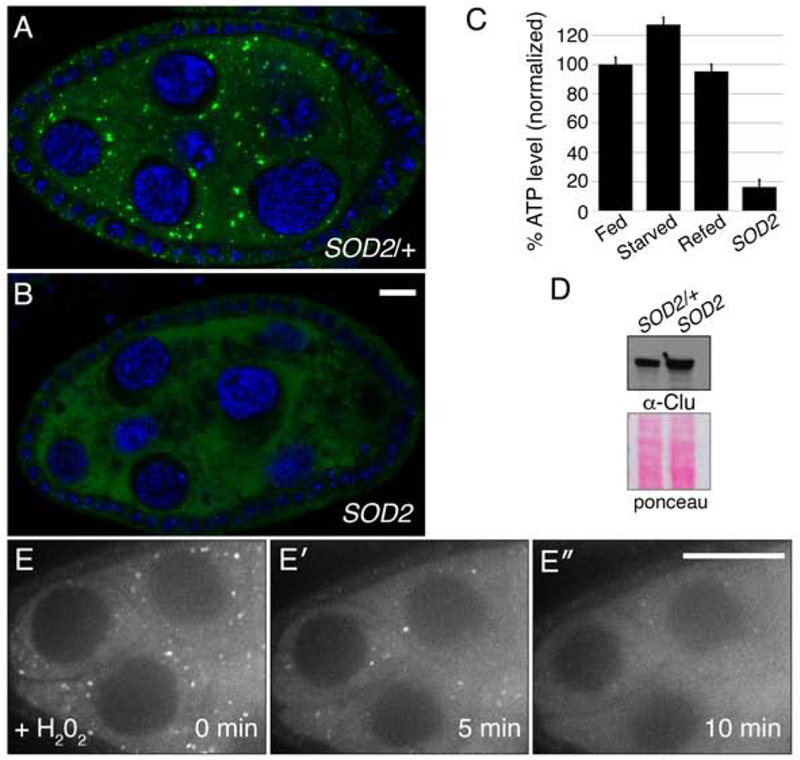
Nutritional stress and lack of insulin caused particles to disaggregate. To determine whether more general oxidative stress has the same effect, we examined Superoxide Dismutase 2 (SOD2) mutants. SOD2 scavenges the free-radical superoxide in the mitochondrial matrix (Fridovich, 1995). Loss of SOD2 causes an increase in mitochondrial oxidation (Paul et al., 2007; Sen et al., 2013). SOD2 mutant flies eclose at normal numbers and appear healthy upon eclosion, but die within 24 hours (Paul et al., 2007; Sen et al., 2013). Their ovaries appeared to develop relatively normally; however, they completely lacked Clu particles (Fig. 7A, B). We have shown previously that SOD2 mutant adults have low levels of ATP (Fig. 7C) (Sen et al., 2013). However, Clu levels were not reduced (Fig. 7D). SOD2 mutants test the effect of systemic loss of an important free-radical scavenging enzyme throughout development. However, superoxide is known to function in multiple cell signaling pathways and loss of SOD2 in Drosophila has been shown to cause defects in behavior, axonal targeting and neurodegeneration (Celotto, Liu, Vandemark, & Palladino, 2012; Y. Wang, Branicky, Noe, & Hekimi, 2018). To determine if the effect of SOD2 loss on Clu particles is due to increased oxidative damage, we added H202 to insulin-containing culture media on follicles dissected from cluCA06604 well-fed females. Clu::GFP particles started to disaggregate in as quickly as five minutes after addition of H202 indicating acute oxidative stress can quickly disperse Clu particles (Fig. 7E–E″, Movie 7).
Figure 7.
Oxidative stress disperses Clu particles. (A, B) Clu particles are always present in wild type siblings (n = 44) (A) but always missing in SOD2 mutant follicles (n = 51). (C, D) SOD2 mutants have decreased levels of ATP (C), but not decreased levels of Clu protein (D). (E-E″) Live-image stills of a well-fed cluCA06604 follicle. Addition of H202 causes particles to disperse (n = 11). Green = Clu (A, B), blue = DAPI (A, B), white = Clu::GFP (E-E″). Error bars are S.E.M. Scale bar: 10μm in B for A, B, 40μm in E′ for E-E′.
Mitochondrial mislocalization in Drosophila female germ cells is downstream of stress
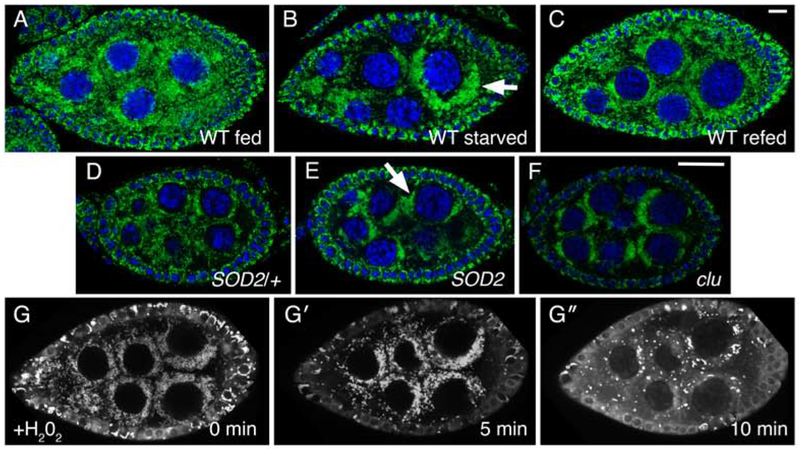
We have shown that mitochondria mislocalize in female germ cells due to a variety of mitochondrial dysfunction, including loss of clu, PINK1 and parkin (Cox & Spradling, 2009; Sen et al., 2015). Under well-fed conditions, mitochondria are evenly dispersed in developing follicles in the ovary as previously described (100%: 68/68 individual follicles) (Fig. 8A) (Cox & Spradling, 2003). However, after five-hour starvation, mitochondria clumped in the nurse cell cytoplasm in a manner reminiscent of clu mutants (82%: 75/92 individual follicles) (Fig. 8B, arrow, F). After two hours re-feeding, mitochondria evenly dispersed back to the pattern seen in well-fed flies (70%: 67/97 individual follicles) (Fig. 8C).
Figure 8.
Stress causes mitochondrial mislocalization in Drosophila germ cells. (A-C) Follicles from w1118 females. Well-fed females have evenly dispersed mitochondria in germ cells (A). Starvation causes mitochondria to clump (arrow) (B). Refeeding yeast paste for two hours post-starvation causes mitochondria to disperse (C). (D, E) Mitochondria in SOD2 mutant germ cells also form clumps (E, arrow) compared to wild type SOD2/+ sibling follicles (D). This clumping is reminiscent of loss of Clu, as previously published (F). (G- G″) Stills from live-imaging well-fed cluCA06604. Adding H202 during imaging to initiate oxidative stress also causes mitochondria to clump. TMRE labeling of mitochondria indicates that mitochondria are initially dispersed at time zero (G), and that mitochondria start to clump after H202 addition (G′) (n = 6). At a later time-point, the TMRE labeling becomes spotty due to mitochondria losing their membrane potential and therefore their ability to sequester the dye (G″). Green = ATP synthase (A-F), blue = DAPI (A-F), white = TMRE (G-G″). Scale bars: 10 μm in C for A-C, G-G″, 10 μm in F for D-F.
SOD2 mutants also showed a similar mitochondrial clumping phenotype (100%: n = 44/44) compared to wild type SOD2/+ siblings (0% clumped: n = 51/51) (Fig. 8D, E, arrow). To examine the effect of oxidative damage on mitochondrial localization using live-imaging, we utilized TMRE, a cell-permeant cationic dye that preferentially sequesters in mitochondria with high membrane potential. At time zero, TMRE-labeled mitochondria were evenly distributed and thin and rod-shaped (Fig. 8G, Movie 8). After incubating follicles in H202 for five minutes, mitochondria started to clump in the cytoplasm and became more swollen and rounded (Fig. 8G′). After ten minutes, oxidative damage had accumulated to a level where most mitochondria are quite dim with only a small subset still fluorescing (Fig. 8G″). These results indicate that once the cells undergo stress including oxidative damage, nutritional stress, or the presence of mutations causing mitochondrial dysfunction, mitochondria no longer retain their proper dispersion and localization. This effect occurs quite quickly, and with respect to nutrition, can be readily reversed.
Discussion
Here, we show that Clu bliss particles are dynamic cytoplasmic bodies whose formation and dispersal is highly dependent on nutritional and oxidative stress. Dynamic cytoplasmic movement is a common feature of RNP particles (reviewed in (Schisa, 2012)). Clu particle processive movement occurs at a speed consistent with mitochondrial movement in neurons (Allen, Metuzals, Tasaki, Brady, & Gilbert, 1982; Hollenbeck, 1996; Ligon & Steward, 2000). The 12% directed Clu particle movement we observed is likely an under-estimate since we only examined one focal plane during live-imaging. Mitochondria are highly dynamic in Drosophila nurse cells; however, neither the changes in mitochondrial movement during development and cell stress nor their cytoplasmic localization has been systematically investigated using live-imaging. A potential function of Clu particles is to sequester mRNAs and proteins that are required for mitochondrial oxidative phosphorylation. For processively moving Clu particles, cotransport of Clu particles with mitochondria could be directed to parts of the cytoplasm with high ATP requirements. As smaller and larger Clu particles appeared to move in a directed fashion, size does not seem to dictate particle movement (Movie 1). The directed movement appears to rely on the microtubule cytoskeleton, as disruption from colcemide treatment causes particles to become stationary (Fig. S1, Movie 4). Clu particles also appear to undergo random movement similar to what has been observed for other proteins at this developmental stage (Shimada, Burn, Niwa, & Cooley, 2011). Furthermore, Clu protein labeling is low in the oocyte, and Clu particles are never observed there (Fig. 1G, 5A). This finding may be due to the metabolic differences between the oocyte and nurse cells (Sieber, Thomsen, & Spradling, 2016). It also suggests that either Clu is not moving into the oocyte from the nurse cells during follicle development or that it may be actively degraded in the oocyte during these stages.
There are several well-described RNP particles in the cytoplasm, including stress granules and processing bodies, which are known to regulate mRNA biology (Buchan & Parker, 2009; Guzikowski, Chen, & Zid, 2019). These RNP particles generally respond to stress by becoming larger and more plentiful (Fig. 5B’, (Burn et al., 2015)). Here, we demonstrate Clu particles have the opposite response. The dispersion of Clu particles is complete upon nutritional stress and reversible. Re-feeding flies yeast paste or adding insulin to the culture media quickly induces particle reformation. Our live-imaging and clonal analysis indicates insulin is both necessary and sufficient for Clu particle formation (Fig. 6). Upregulated insulin signaling did not appear to affect Clu particle formation however follicle cell clonal analysis of InR mutant clones indicated loss of Clu particles (Fig. 6). The standard method used by the field to induce germline stem cell clones (3x heat shock, 7-10 days on yeast paste before dissection) produced mutant germline clones but did not show consistent, reproducible Clu particles in the germ cells of controls. Thus, the results were not interpretable and we did not use this protocol (see Materials and Methods). We assume the inconsistent Clu particle loss in the controls using the standard longer protocol was due to stress from the addition heat shock combined with aging. Instead, we reduced the number of heat shocks and the time after heat shock for dissection, which resulted in small InR follicle cell clones. In addition, visualizing Clu particles live without insulin resulted in a higher percentage of follicles with particles compared to dissecting and fixing wild type starved flies (20% vs 0%) (Fig. 6B vs 4G). This difference is likely due to the fact that we counted individual follicles at high magnification using live-imaging whereas with fixed imaging we used a high-throughput method and counted ovarioles (strings of developing follicles). Oxidative stress also causes particles to disperse in addition to nutritional stress. Producing oxidative stress systemically using SOD2 mutants or acutely with the addition of H202 causes either a complete lack of Clu particles (Fig. 7B) or real-time particle dispersion (Fig. 7F-F”, Movie 7). In all the conditions we tested, the change in cytoplasmic localization with altered nutrition and increased oxidative stress is independent of Clu protein levels and is thus not due to protein degradation (Fig. 4I, J). This is the first time to our knowledge that this dynamic for a cytoplasmic particle has been described.
Clu directly effects mitochondrial function. Clu particles are closely associated with mitochondria in the cytoplasm (Fig. 2A, (Cox & Spradling, 2009)), Clu associates with mitochondrial outer membrane proteins (Sen & Cox, 2016; Sen et al., 2015), and it associates with at least one ribosomal protein preferentially in mitochondrial fractions (Sen & Cox, 2016). Clu particles do not co-localize with processing bodies or autophagosomes (Fig. 2). This was surprising to us because Clu genetically and physically interacts with the PINK1/Parkin mitophagy complex (Sen et al., 2015; Z. H. Wang et al., 2016). Since only a subset of Clu particles associates with mitochondria in fixed antibody-labeled tissues, we expected particles would increase in response to stress, and perhaps be involved in autophagosome formation and culling damaged mitochondria (Kim et al., 2007). However, to our surprise, the opposite occurs. Thus, not only do Clu particles have a previously undescribed dynamic, they are also a novel cytoplasmic particle consisting of a protein with a direct effect on, and an association with, mitochondria. Given that their presence is strictly dependent on stress-free conditions, we have named them “bliss particles” to differentiate their cytoplasmic dynamic from the many other RNP particles that form in response to stress.
Clu and Clu1p bind mRNA, and CLUH preferentially binds transcripts encoding proteins destined for mitochondrial import (Gao et al., 2014; Sen & Cox, 2016). There is evidence that in the absence of CLUH, target mRNAs undergo increased degradation, suggesting that CLUH is required for mRNA stability (Schatton et al., 2017). Clu and Clu1p both bind ribosomal proteins and eukaryotic initiation factors (Fig. 3, (Sen & Cox, 2016; Vornlocher, Hanachi, Ribeiro, & Hershey, 1999)). Because insulin and nutrition regulate Clu particle formation, this supports the notion that the presence or absence of Clu particles could also affect mitochondrial function and perhaps translation and import of mitochondrial proteins.
Clu contains multiple putative domains based on sequence alignment between species and literature searches (Fig. 3A) (Cox & Spradling, 2009; Sen et al., 2015). The protein contains tetratricopeptide repeats (TPR) and Clu domains which show 55% and 85% amino acid identity, respectively, between Clu and CLUH (Sen & Cox, 2016). Based on several available online algorithms (Iupred2a and Globplot), the M domain is not predicted to have structure and does not contain any sequences associated with intrinsically disordered domains (IDDs). IDD-containing proteins have been shown to be involved in liquid-liquid phase separation as well as mRNA association, two traits that control membraneless organelle formation, including some RNP particles (Uversky, 2017). For Clu, the TPR domain, not the M domain, was determined to be functionally important for mRNA binding (Sen et al., 2015). We have previously shown that ectopically expressing deletion constructs for each of Clu’s domains in a clu null mutant background does not rescue pupation delays, adult lethality, mitochondrial localization defects, or reduced ATP levels (Sen & Cox, 2016). The resulting adults that hatch die within approximately four days and are as weak and sick as clu null adult flies. Thus, we were unable to assess which Clu domain, including the M domain, is required for Clu particle formation in vivo as the flies are systemically stressed. clu mutant phenotypes were only rescued by expressing full-length Drosophila Clu, melanogaster specific (ms) domain deletion, or CLUH. Furthermore, Clu associates with itself, further complicating deletion analysis to determine which domain is necessary for Clu particle formation (Sen & Cox, 2016).
Mitochondria are plentiful in the cytoplasm of developing germ cells, and they undergo characteristic localization, shape, and number changes during oogenesis (Cox & Spradling, 2003, 2006). In well-fed wild type females, mitochondria in the germ cells are evenly dispersed throughout the cytoplasm. Through mutation analysis, we previously described that mitochondria mislocalize into clumps in clu, PINK1 and parkin mutant germ cells (Cox & Spradling, 2009; Sen et al., 2015). Fields et al. also showed evidence that Clu1p influences mitochondrial distribution and morphology, and in a large screen examining mitochondrial localization Dimmer et al. showed that clu1Δ knockout yeast have mislocalized and aggregated mitochondria (Dimmer et al., 2002; Fields, Conrad, & Clarke, 1998). In this work, we demonstrate this clumping is present in SOD2 mutants and starvation causes the same mislocalization. From these data, it is clear that mitochondrial mislocalization in germ cells is likely an indirect, downstream effect of general cellular stress. Thus, experiments examining mitochondrial localization in germ cells should take into account the culture and fixation conditions in order to ensure that low-stress conditions and normal mitochondrial distribution are maintained.
Drosophila Clu and vertebrate CLUH are ribonucleoproteins, and CLUH was shown to preferentially bind mRNAs encoding nucleus-encoded mitochondrial proteins. Loss of Clu/CLUH disrupts mitochondrial function by reducing ATP levels, causing improper mitochondria localization and mitochondrial ultrastructural changes, and by altering metabolism (Cox & Spradling, 2009; Gao et al., 2014; Schatton et al., 2017; Sen & Cox, 2016; Wakim et al., 2017). Drosophila Clu associates with the ribosome and the mitochondrial outer membrane proteins TOM20 and Porin, suggesting it may have a role in site-specific or co-translational import of nucleus-encoded proteins (Sen & Cox, 2016). Since the particles respond quickly to nutritional cues, they may also represent an additional acute control of mRNA translation, with differing translation rates in the presence and absence of particles. We do not yet know if Clu particles are the sites of translation for Clu-bound mRNAs, or whether they are enriched with mRNAs and/or ribosomes. This hypothesis remains to be tested. Clu associates with TOM20 and Porin and also self-associates; however, it is unclear how Clu particles associate with mitochondria, whether they directly associate with microtubules, and how this association changes with stress (Sen et al., 2015). Attempts to simultaneously visualize mitochondria and Clu particles using live-imaging have been stymied by sensitivity to TMRE addition (the Clu particles disperse); however, there is evidence that Clu particles do move with mitochondria in Arabidopsis (El Zawily et al., 2014).
There have not yet been patients identified with mitochondrial disorders who harbor mutations in CLUH. This may be because any perturbations in CLUH are either not compatible with life or cause very early lethality. This is supported by evidence that in mice knockout of the vertebrate homolog CLUH causes post-natal lethality between P0-1, with no apparent respiratory failure and a concomitant 10-15% decrease in weight (Schatton et al., 2017). Schatton et al found post-natal death is due to metabolic disruption in hepatocytes which have mislocalized mitochondria and are depleted of key enzymes used in catabolic energy pathways (Schatton et al., 2017). Additionally, in human development, comparison of preterm to term newborn umbilical cord mesenchymal stem cells shows that changes in CLUH expression correspond with changes in the mitochondrial network and cellular metabolic changes in response to an oxygen-rich environment (Ravera et al., 2018). Nonetheless, given that Clu forms dynamic cytoplasmic particles highly sensitive to stress and that it is an important RNP critical for directly affecting mitochondrial function and biogenesis, understanding Clu’s molecular role and how its mRNA binding is related to cytoplasmic localization will be important in the future.
Materials and Methods:
Fly stocks:
The following stocks were used for experiments: w1118 , cluelessd08713/CyO P(ActGFP)JMR1 (Cox & Spradling, 2009), trailer hitchCA06517, Sec61αCC00735, cluelessCA06604 (Buszczak et al., 2007), SOD2Δ2/ CyO P(ActGF)}JMR1, P(ry[+t7.2]=neoFR)82B ry[506] , w *; P{ry[+t7.2]=neoFRT}82B P(w[+mC]=Ubi-GFP.D)83 (Bloomington Drosophila Stock Center), FRT82B TSC1Q87X/ TM3 (Tapon, Ito, Dickson, Treisman, & Hariharan, 2001), FRT82B InR339/TM3, FRT82B InRE19/ TM3 (LaFever & Drummond-Barbosa, 2005), Ubi-GFP::tubulin/ CyO; Pri/TM6, Tb (Rebollo, Llamazares, Reina, & Gonzalez, 2004). Flies were reared on either standard cornmeal fly media or standard cornmeal fly media supplemented with yeast paste at 22° or 25° C.
Clone generation:
0-24hr females of the appropriate genotype were fed yeast paste for 24 hr, then heat shocked 2x 1 hour in a 37°C water bath, morning and evening. Except for during heat shock, the flies were kept undisturbed on a quiet shelf at room temperature. After heat shock, the flies were fed for an additional 24 hr, then 3x ten flies were dissected in parallel with controls and fixed and labeled as described below. Clones were distinguished by absence of GFP antibody labeling.
Fixed Image Immunofluorescence:
0-24 hr flies were fattened with yeast paste for 3-7 days. 60 female flies were then transferred to an empty vial containing a Kimwipe soaked in water for five hours to induce starvation. 30 starved flies were then transferred back to a vial with fresh fly food and yeast paste for two hours for re-feeding. 30 female flies remained feeding on yeast paste throughout as controls. From each condition, 3x 10 flies were then dissected at room temperature (RT) in Grace’s Insect Medium (modified) (BioWhittaker, Lonza, Cologne, Germany). Ovaries were fixed for 20 minutes in 4% paraformaldehyde and 20mM formic acid solution (Sigma, St. Louis, MO) made in Grace’s. Tissues were washed two times (40 minutes each) with Antibody wash buffer (Ab) (1X PBS:0.1% Triton X-100:1% BSA), then incubated in primary antibody over night at 4°C. They were then washed two times (40 minutes each) and incubated overnight at 4°C in secondary antibody. After washing, DAPI (1:1000) was added for five minutes then removed, and Vectashield (Vector Laboratories, Inc., Burlingame, CA) was added. For yeast staining, log phase Clu1p::GFP yeast grown in Yeast-Peptone-Dextrose (YPD). 20% paraformaldehyde was added to the culture to a final concentration of 4% for 20 min. The cells were spun, washed with 1.2M sorbitol, 50mM KPOH, spun twice, then resuspended in 500 uL of the same buffer. The cells were treated with Zymolase 100T (MP Biomedicals, LLC, Irving CA), for 35 min at 37°C, then very gently washed 2x with Ab wash, followed by a 2 hour incubation with α-GFP sitting at room temperature with occasional gentle inversion. The cells were then washed 2x with Ab wash, incubated with α -rabbit Alexa-488 in Ab wash for 1 hour, then washed gently 2x with DAPI addition in the second wash. The cells were then mounted in Vectashield and imaged using a Leica AF6000 Time-lapse Imaging System. The following primary antibodies were used: guinea pig anti-Clu N-terminus (1:2000) (Cox & Spradling, 2009), mouse anti-Complex V (ATP synthase (CVA)) (1:1000, AbCam, Cambridge, MA, cat# ab14748), rabbit anti-GFP (1:2000, AbCam, Cambridge, MA, cat# ab290), chicken anti-GFP (1:1000, AbCam, Cambridge, MA, cat# ab13970), GABARAP (Atg8a) (1:200, AbCam, Cambridge, MA, cat# ab109364), Sec23 (1:500), GMAP (1:1000, Developmental Studies Hybridoma Bank, Iowa City, Iowa), Golgin245 (1:1000, Developmental Studies Hybridoma Bank, Iowa City, Iowa), . The following secondary antibodies were used: goat anti-mouse IgG2b Alexa 568 (1:500), goat anti-guinea pig Alexa 488 (1:1000), and goat anti-chicken Alexa 568 (1:1000) (Invitrogen, Waltham, MA), donkey anti-guinea pig Cy3 (1:1000) and donkey anti-rabbit Alexa 488 (1:500) (Jackson ImmunoResearch Laboratories, Inc, West Grove, PA). Samples were imaged using a Zeiss 700 confocal microscope and 63x Plan Apo NA 1.4 lens.
Image Quantification:
For Clu particle quantification, slides were scanned at 40x using a Zeiss Axio Scan.Z1 slide scanner. The number of ovarioles per slide was counted (87-170 ovarioles per slide, average = 132 ± 16 ovarioles) using the Cell Counter tool in ImageJ. Each ovariole on a slide was subsequently scored using the Zeiss Zen blue software; an ovariole was scored as having particles if at least one follicle in the ovariole contained particles and as having no particles if no follicles contained particles. The mean percentage of ovarioles with particles in each experiment (n = 6 groups of 10 flies for well-fed group, n = 3 groups of 10 flies for starved group, n = 3 groups of 10 flies for refed group) is represented in the graph with error bars representing mean ± SD. The amount of Clu::GFP and Trailer hitch::GFP was quantified by measuring the fluorescence intensity (integrated density) using ImageJ (n = 6 images, (same images for both)). Taking care to avoid Clu and Tral particles three equally sized ROIs were randomly placed within the nurse cells. Significance was calculated using an unpaired t-test with Welch’s correlation. Kymographs were generated using ImageJ from videos of cluelessCA06604 well-fed follicles taken at 60x, with one frame captured every second for 200 seconds. To measure average particle velocity, six randomly chosen particles which showed processive movement were analyzed from four follicles (n = 24 particles). A representative image was used in the figure (1D). For quantification of the percentage of clones with particles in control FRT82B clones, TSC1 mutant clones and InR339 mutant clones, samples were imaged using a Zeiss 700 confocal microscope and 63x Plan Apo NA 1.4 lens. Twenty clones (FRT82B, TSC1) and 11 clones (InR339) from individual images were scored for the presence or absence of particles in the GFP− and GFP+ regions to generate the graph.
Live Imaging Microscopy for Drosophila Tissue Samples:
cluelessCA06604 females that had been either fattened with yeast paste, fed only standard cornmeal fly media (no yeast paste), or starved on water for 5 hours were dissected at room temperature (RT) in either Complete Schneider’s Media (Schneider’s Drosophila media (ThermoFisher Scientific, Waltham, MA) with 15% Fetal Bovine Serum and 0.6X Pen-Strep solution) or Complete Schneider’s Media supplemented with 200 μg/mL of insulin (insulin which did not readily dissolve in media was allowed to settle overnight at 4°C prior to use). Ovaries were removed and further teased apart into single, isolated ovarioles. Ovarioles were then loaded onto a glass bottom 35mm dish (MatTek Corporation, Ashland, MA) and live videos of a single focal plane were collected using a Nikon spinning disk/TIRF/3D-STORM microscope at 60x at room temperature. For experiments with insulin addition, ovarioles were prepared as above in Complete Schneider’s Media. Complete Schneider’s Media containing 400 μg/mL insulin would be added to the dish with a 3mL syringe to dilute the sample 1:1, resulting in an insulin concentration of 200 μg/mL. For the Insulin response graph, flies were dissected in Complete Schneider’s Media containing the amount of insulin indicated and viewed on the Nikon spinning disk microscope as described above.
Individual follicles were counted (16-62 follicles per experiment, n values below) to either have particles (if at least one nurse cell in follicle had particles) or no particles after focusing throughout entire follicle. The mean percentage of total number of follicles with particles in each culture condition is represented in the graph with error bars representing mean ± S.E. The following follicles counted per experiment:
Non-fattened flies in plus 200ug/mL insulin – 16, 39 and 62;
Non-fattened flies plus 100ug/mL insulin – 27, 39, and 39;
Non-fattened flies plus 50ug/mL insulin – 29, 27, and 26;
Non-fattened flies in 0ug/mL insulin – 34 and 16;
Starved flies in 0ug/mL insulin – 19 and 23;
Starved flies plus 200ug/mL insulin – 29 and 26.
For colcemide treatment, follicles were dissected in Complete Schneider’s Media plus 200μg/mL insulin and 100 μg/mL colcemide (Sigma-Aldrich, St. Louis, MO) then incubated for 3 hours at room temperature before imaging. Tetramethylrhodamine, ethyl ester, perchlorate (TMRE, AnaSpec cat# AS88061, ThermoFisher, Waltham, MA, 100 mM stock in DMSO) was used at a concentration of 1:10,000. Follicles were dissected in Complete Schneider’s Media plus 200μg/mL insulin and TMRE, incubated for 20 min then directly imaged with no washes. For H2O2 treatments, follicles were dissected in Complete Schneider’s Media plus 200μg/mL insulin and transferred to 100 μL of the same media in a MatTek dish (MatTek Corporation, Ashland, MA). 100 μL of additional media + 2mM H2O2 was added during imaging to make the final concentration 1mM. Treatment lasted for 15 minutes while filming. To determine the percent Clu particles that undergo directed vs apparently random movement, Clu-particle containing follicles were imaged in the presence of insulin for 200 seconds and particles were counted by hand in each frame for three movies. Each movie contained 4 complete or partial nurse cells. Upon overlaying the image with a 1.5 μm by 1.5 μm grid, a particle was scored as moving directionally if it moved in one direction across two grid boxes over a minimal of 6 frames. These parameters were chosen based on the maximum, minimum and average particle speed (Fig. 1E). This analysis gave an average of 12% moving particles from an average of 174 total particles per follicle (moving particles/total particles per follicle: 24/152, 12/170, & 30/200). To analyze the effect of nutrition on mitochondrial mislocalization, control (well-fed) and experimental (starved on water and refed) ovaries were labeled for mitochondria (anti-ATP synthase) and anti-Clu. All ovarioles on the slide were imaged through three Z-sections. Using just the mitochondria channel, each follicle in the ovariole was scored for completely dispersed or clumped mitochondria.
Yeast Growth Assays:
Saccharomyces cerevisiae strain BY4741 (his3 leu2 met15 ura3) and two clu1Δ derivatives (clu1::kanMX; strain numbers FPS674 and FPS675) were used in this study. clu1 deletions were generated using the S288C-derived BY4741 strain background (MATa his3Δ leu2Δ met15Δ ura3Δ clu1::kanMX) (Giaever et al., 2002) obtained from Horizon Discovery, Cambridge, UK, (formally Open Biosystems). Yeast chromosomal DNA was purified from strain BY4741 using the YeaStar Genomic DNA Kit (Zymo Research, Irvine, CA), according to the manufacturer’s protocol. The clu1::kanMX cassette was then PCR-amplified using the following primers, which are respectively approximately 250 base pairs up- and down-stream of the cassette: GTGTAACGGCTATCACATC (CLU1-250F) and CTTTAGAGGGAACTCTTGCG (CLU1-250R). Amplified DNA was purified using the GeneJET PCR Purification Kit (ThermoFisher, Waltham, MA), according to the manufacturer’s protocol. The purified DNA was used to freshly delete CLU1 in strain backgrounds BY4741 (MATa his3Δ leu2Δ met15Δ ura3Δ) and W303 (leu2 ade2-1 ura3 can1 trp1 his3) using a previously described method (Baudin, Ozier-Kalogeropoulos, Denouel, Lacroute, & Cullin, 1993). For percent petite colony formation, cells from each yeast strain were dispersed on Yeast-Peptone-Dextrose (YPD) plates (~500 cells per plate; performed in quadruplicate) and grown at 30° C for ~1 week. Total normal (rho+) and petite (rho−) colonies were counted. For the glycerol growth assay, log-phase BY4741 wild type and clu1Δ strains were grown overnight in YPGlucose, then diluted the next morning and grown until achieving log-phase. The cells were then serially diluted on both YPGlucose agar and YPGlycerol agar and grown overnight. To image individual colonies, BY4741 wild type and clu1Δ strains were grown overnight in YPGlucose, then diluted and grown the next morning until log-phase. Approximately the same number of cells was spotted on YPGlycerol and allowed to grow for a week before imaging.
ATP Assays:
0-24 hr flies were fattened with yeast paste for 3-7 days. 30 female flies were then transferred for five hours to an empty vial containing a Kimwipe soaked in water. 15 starved flies were then transferred back to a vial with fresh fly food and yeast paste for two hours for re-feeding. 15 female flies continued feeding on yeast paste throughout as a control. Flies were then homogenized in extraction buffer (100 mM Tris-Cl, pH 8.0, 4 mM EDTA, pH 8.0; 6 M guanidine hydrochloride) (3 groups of 5 flies/50 μL extraction buffer), boiled for 4 minutes, then centrifuged at 8000 g for 5 minutes at RT. The protein concentration of the samples was determined using a Bradford assay. The ATP concentration was determined using an ATP Determination Kit (Molecular Probes, Invitrogen, Waltham, MA) according to the manufacturer’s directions. 100 μl assays were performed in a 96 well plate and the luminescence was measured using a BioTek Synergy H1 luminometer (BioTek Instruments, Winooski, VT). Each sample was read in triplicate. The amount of ATP was normalized against protein concentration.
Western blotting and Immunoprecipitation:
0-24 hr flies were fattened with yeast paste for 3-7 days. 30 female flies were then transferred for five hours to an empty vial containing a Kimwipe soaked in water. 15 starved flies were then transferred back to a vial with fresh fly food and yeast paste for two hours for re-feeding. 15 female flies continued feeding on yeast paste throughout as a control. Ovaries were then removed at room temperature (RT) in Grace’s Insect Medium (modified) (BioWhittaker, Lonza, Cologne, Germany). Ovaries were homogenized in 150 μL of sample buffer and the extract run on a 4-20% polyacrylamide gel using a standard SDS-PAGE protocol for Western blotting. After electrophoresis, proteins were transferred onto a Hybond-ECL nitrocellulose membrane (GE Healthsciences, Inc., Chicago, IL) then soaked with Ponceau S for 10 min and briefly rinsed. Blots were exposed to the following antibodies: anti-Clu (1:20,000) (Cox & Spradling, 2009) and anti-α-tubulin (1:5000, AA4.3, Developmental Studies Hybridoma Bank, University of Iowa, Iowa City). Quantification of Western blots was performed with ImageJ, and the amount of Clu protein was normalized against α-tubulin (n = 3). Immunoprecipitation (IP) from adult flies was performed as previously described (Sen et al., 2015). For IP from yeast, an equal volume of silica beads and 1X PBS was added to the yeast pellet. The tube was vortexed frequently for up to 5 minutes. The slurry was resuspended in IP buffer (20 mM HEPES, pH 7.4; 50 mM KCl, 0.02% Triton X-100, 1% NP-40 (sub), 1 mM EDTA, 0.5 mM EGTA, 5% glycerol) supplemented with 1 mM DTT and Protease inhibitor cocktail (Roche, Basel, Switzerland). The following antibodies were used for IP and western blots from yeast samples; anti-ScRPL3 (1:1000, Developmental Studies Hybridoma Bank, University of Iowa, Iowa city), anti-ELO3 (1:2500, a gift from Dr. Teresa Dunn, Uniformed Services University). For the anti-ELO3 blot, the IP was done using GFP-Trap magnetic beads (ChromoTek GmbH, Planegg-Martinsried, Germany) not anti-GFP antibody since ELO3 migrates at a similar position to IgG heavy chain.
Sucrose gradients:
Sucrose gradients for fly extract were performed as previously described (Sen & Cox, 2016). The modifications for yeast were the following: yeast from a 5 ml YPD overnight culture was pelleted and transferred to an Eppendorf tube. An equal volume of silica beads was added to the pellet plus 50μl of 1X mDXB (25 mM HEPES pH 6.8, 50 mM KCl, 1 mM MgCl2, 125 mM Sucrose, 1 mM DTT and 1x protease inhibitor cocktail). The tube was vortexed frequently for up to 5 minutes. 300μl 1X mDXB was added to the mixture and spun at 2000g, twice for 5 minutes to collect the supernatant. Half the supernatant was loaded on the sucrose column.
Supplementary Material
Figure S1. Colcemide treatment disrupts microtubules. Ubi-GFP::tubulin expressing follicles. Follicles cultured in Complete Schneider’s Media supplemented with 200 μg/mL of insulin display an intact microtubule cytoskeleton (A). Follicles after colcimide treatment show the microtubule cytoskeleton has been disrupted (B). Scale bar: 40μm in B for A, B.
Movie 5. Clu particles in surrounding follicle cells are more stationary than in germ cells. Follicle from cluCA06604 (Clu-GFP) female. Imaged one frame per second for 120 seconds. Video is 24 frames per second.
Movie 6. Clu particle formation is induced with insulin. Follicle from cluCA06604 (Clu-GFP) female. Imaged one frame per minute for 45 minutes. Video is 10 frames per second.
Movie 7. Oxidative stress disperses Clu particles. Follicle from cluCA06604 (Clu-GFP) female. Imaged one frame per 15 seconds for 15 minutes. Video is 10 frames per second.
Movie 8. Oxidative stress causes mitochondrial mislocalization in Drosophila germ cells. Follicle from cluCA06604 (Clu-GFP) female. Imaged one frame per 15 seconds for 20 minutes. Video is 10 frames per second.
Figure S2. Multiple Δclu1 strains derived in two different wild type genetic backgrounds have reduced growth on glycerol. Each plate shows three newly generated deletions of CLU1 (clu1::KanMX) in two wild type strain backgrounds (W303, left plate and BY4741, right plate). After one week on glycerol, the wild type strain grows at a normal rate whereas colonies of three newly derived Δclu1 strains are smaller and less robust.
Figure S3. InRE19 mutant follicle cell clones do not contain Clu particles. (A-C) The surrounding somatic follicle cells of a follicle. The mutant cells are indicated by lack of GFP antibody labeling (dotted outline, A). Clu particles are not present in cells mutant for InRE19 (dotted outline, B, C). White = GFP (A) and Clu (B). Magenta = GFP (C). Green = Clu (C). Scale bar = 10 μm in C for A-C. n=7.
Movie 1. Clu live-imaging shows robust, dynamic particles in germ cells. Follicle from cluCA06604 (Clu-GFP) female. Imaged one frame per second for 200 seconds. Video is 36 frames per second.
Movie 2. Mitochondria movement is normal without colcemide treatment. Follicle from cluCA06604 (Clu-GFP) female treated with TMRE. Imaged one frame per second for 120 seconds. Video is 10 frames per second.
Movie 3. Mitochondrial movement ceases with treatment of the microtubule destabilizer colcemide. Follicle from cluCA06604 (Clu-GFP) female treated with TMRE. Imaged one frame per second for 120 seconds. Video is 10 frames per second.
Movie 4. Clu particle movement ceases with treatment of the microtubule destabilizer colcemide. Follicle from cluCA06604 (Clu-GFP) female. Imaged one frame per second for 120 seconds. Video is 24 frames per second.
Highlights:
Drosophila Clu is a conserved ribonucleoprotein required for mitochondrial function
Clu protein forms new dynamic, mitochondria-associated particles in the cytoplasm
Clu particle formation is controlled by nutrition and insulin signaling
Nutritional and oxidative stress cause particles to disperse
Acknowledgements:
We would like to thank the USUHS Biomedical Instrumentation Core and Dr. Dennis McDaniel for imaging support and The Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology. We thank Ms. Hala Wyne, Department of Biochemistry, USU, for assistance with the yeast petite colony assay. This work was supported by the National Institutes of Health (1R21NS085730 and 1R01GM127938 to R.T.C. and 1R35GM119790-01 to F.S.) and Uniformed Services University (USUHS BIO-71-3019) to R.T.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allen RD, Metuzals J, Tasaki I, Brady ST, & Gilbert SP (1982). Fast axonal transport in squid giant axon. Science, 218(4577), 1127–1129. doi: 10.1126/science.6183744 [DOI] [PubMed] [Google Scholar]
- Anderson P, & Kedersha N (2006). RNA granules. J Cell Biol, 172(6), 803–808. doi:jcb.200512082 [pii] 10.1083/jcb.200512082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, & Kedersha N (2009). RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol, 10(6), 430–436. doi: 10.1038/nrm2694 [DOI] [PubMed] [Google Scholar]
- Barbee SA, Estes PS, Cziko AM, Hillebrand J, Luedeman RA, Coller JM, … Ramaswami M (2006). Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron, 52(6), 997–1009. doi:S0896-6273(06)00827-0 [pii] 10.1016/j.neuron.2006.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth JM, Szabad J, Hafen E, & Kohler K (2011). Autophagy in Drosophila ovaries is induced by starvation and is required for oogenesis. Cell Death Differ, 18(6), 915–924. doi:cdd2010157 [pii] 10.1038/cdd.2010.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, & Cullin C (1993). A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res, 21(14), 3329–3330. doi: 10.1093/nar/21.14.3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, & Parker R (2009). Eukaryotic stress granules: the ins and outs of translation. Mol Cell, 36(6), 932–941. doi:S1097-2765(09)00861-2 [pii] 10.1016/j.molcel.2009.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burn KM, Shimada Y, Ayers K, Vemuganti S, Lu F, Hudson AM, & Cooley L (2015). Somatic insulin signaling regulates a germline starvation response in Drosophila egg chambers. Dev Biol, 398(2), 206–217. doi: 10.1016/j.ydbio.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buszczak M, Paterno S, Lighthouse D, Bachman J, Planck J, Owen S, … Spradling AC (2007). The carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics, 175(3), 1505–1531. doi:genetics.106.065961 [pii] 10.1534/genetics.106.065961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celotto AM, Liu Z, Vandemark AP, & Palladino MJ (2012). A novel Drosophila SOD2 mutant demonstrates a role for mitochondrial ROS in neurodevelopment and disease. Brain Behav, 2(4), 424–434. doi: 10.1002/brb3.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Jack J, & Garofalo RS (1996). The Drosophila insulin receptor is required for normal growth. Endocrinology, 137(3), 846–856. doi: 10.1210/endo.137.3.8603594 [DOI] [PubMed] [Google Scholar]
- Cox RT, & Spradling AC (2003). A Balbiani body and the fusome mediate mitochondrial inheritance during Drosophila oogenesis. Development, 130(8), 1579–1590. Retrieved from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12620983 [DOI] [PubMed] [Google Scholar]
- Cox RT, & Spradling AC (2006). Milton controls the early acquisition of mitochondria by Drosophila oocytes. Development, 133(17), 3371–3377. doi:dev.02514 [pii] 10.1242/dev.02514 [DOI] [PubMed] [Google Scholar]
- Cox RT, & Spradling AC (2009). Clueless, a conserved Drosophila gene required for mitochondrial subcellular localization, interacts genetically with parkin. Dis Model Mech, 2(9-10), 490–499. doi:dmm.002378 [pii] 10.1242/dmm.002378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, & Parker R (2012). P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb Perspect Biol, 4(9), a012286. doi: 10.1101/cshperspect.a012286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimmer KS, Fritz S, Fuchs F, Messerschmitt M, Weinbach N, Neupert W, & Westermann B (2002). Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol Biol Cell, 13(3), 847–853. doi: 10.1091/mbc.01-12-0588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Zawily AM, Schwarzlander M, Finkemeier I, Johnston IG, Benamar A, Cao Y, … Logan DC (2014). FRIENDLY regulates mitochondrial distribution, fusion, and quality control in Arabidopsis. Plant Physiol, 166(2), 808–828. doi: 10.1104/pp.114.243824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields SD, Conrad MN, & Clarke M (1998). The S. cerevisiae CLU1 and D. discoideum cluA genes are functional homologues that influence mitochondrial morphology and distribution. J Cell Sci, 111 (Pt 12), 1717–1727. Retrieved from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9601101 [DOI] [PubMed] [Google Scholar]
- Fridovich I (1995). Superoxide radical and superoxide dismutases. Annu Rev Biochem, 64, 97–112. doi: 10.1146/annurev.bi.64.070195.000525 [DOI] [PubMed] [Google Scholar]
- Gao J, Schatton D, Martinelli P, Hansen H, Pla-Martin D, Barth E, … Rugarli EI (2014). CLUH regulates mitochondrial biogenesis by binding mRNAs of nuclear-encoded mitochondrial proteins. J Cell Biol, 207(2), 213–223. doi:jcb.201403129 [pii] 10.1083/jcb.201403129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, … Johnston M (2002). Functional profiling of the Saccharomyces cerevisiae genome. Nature, 418(6896), 387–391. doi: 10.1038/nature00935 [DOI] [PubMed] [Google Scholar]
- Guzikowski AR, Chen YS, & Zid BM (2019). Stress-induced mRNP granules: Form and function of processing bodies and stress granules. Wiley Interdiscip Rev RNA, 10(3), e1524. doi: 10.1002/wrna.1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenbeck PJ (1996). The pattern and mechanism of mitochondrial transport in axons. Front Biosci, 1, d91–102. doi: 10.2741/a118 [DOI] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, & O’Shea EK (2003). Global analysis of protein localization in budding yeast. Nature, 425(6959), 686–691. doi: 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- Kato Y, & Nakamura A (2012). Roles of cytoplasmic RNP granules in intracellular RNA localization and translational control in the Drosophila oocyte. Dev Growth Differ, 54(1), 19–31. doi: 10.1111/j.1440-169X.2011.01314.x [DOI] [PubMed] [Google Scholar]
- Kim I, Rodriguez-Enriquez S, & Lemasters JJ (2007). Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys, 462(2), 245–253. doi: 10.1016/j.abb.2007.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFever L, & Drummond-Barbosa D (2005). Direct control of germline stem cell division and cyst growth by neural insulin in Drosophila. Science, 309(5737), 1071–1073. doi: 10.1126/science.1111410 [DOI] [PubMed] [Google Scholar]
- Ligon LA, & Steward O (2000). Movement of mitochondria in the axons and dendrites of cultured hippocampal neurons. J Comp Neurol, 427(3), 340–350. doi: [DOI] [PubMed] [Google Scholar]
- Namkoong S, Ho A, Woo YM, Kwak H, & Lee JH (2018). Systematic Characterization of Stress-Induced RNA Granulation. Mol Cell, 70(1), 175–187 e178. doi: 10.1016/j.molcel.2018.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezis IP, Lamark T, Velentzas AD, Rusten TE, Bjorkoy G, Johansen T, … Brech A (2009). Cell death during Drosophila melanogaster early oogenesis is mediated through autophagy. Autophagy, 5(3), 298–302. doi:7454 [pii] [DOI] [PubMed] [Google Scholar]
- Oldham S, Montagne J, Radimerski T, Thomas G, & Hafen E (2000). Genetic and biochemical characterization of dTOR, the Drosophila homolog of the target of rapamycin. Genes Dev, 14(21), 2689–2694. doi: 10.1101/gad.845700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham S, Stocker H, Laffargue M, Wittwer F, Wymann M, & Hafen E (2002). The Drosophila insulin/IGF receptor controls growth and size by modulating PtdInsP(3) levels. Development, 129(17), 4103–4109. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/12163412 [DOI] [PubMed] [Google Scholar]
- Paul A, Belton A, Nag S, Martin I, Grotewiel MS, & Duttaroy A (2007). Reduced mitochondrial SOD displays mortality characteristics reminiscent of natural aging. Mech Ageing Dev, 128(11-12), 706–716. doi:S0047-6374(07)00164-9 [pii] 10.1016/j.mad.2007.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravera S, Podesta M, Sabatini F, Fresia C, Columbaro M, Bruno S, … Frassoni F (2018). Mesenchymal stem cells from preterm to term newborns undergo a significant switch from anaerobic glycolysis to the oxidative phosphorylation. Cell Mol Life Sci, 75(5), 889–903. doi: 10.1007/s00018-017-2665-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo E, Llamazares S, Reina J, & Gonzalez C (2004). Contribution of noncentrosomal microtubules to spindle assembly in Drosophila spermatocytes. PLoS Biol, 2(1), E8. doi: 10.1371/journal.pbio.0020008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatton D, Pla-Martin D, Marx MC, Hansen H, Mourier A, Nemazanyy I, … Rugarli EI (2017). CLUH regulates mitochondrial metabolism by controlling translation and decay of target mRNAs. J Cell Biol, 216(3), 675–693. doi: 10.1083/jcb.201607019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schisa JA (2012). New insights into the regulation of RNP granule assembly in oocytes. Int Rev Cell Mol Biol, 295, 233–289. doi: 10.1016/B978-0-12-394306-4.00013-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen A, & Cox RT (2016). Clueless is a conserved ribonucleoprotein that binds the ribosome at the mitochondrial outer membrane. Biol Open, 5(2), 195–203. doi: 10.1242/bio.015313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen A, Damm VT, & Cox RT (2013). Drosophila clueless is highly expressed in larval neuroblasts, affects mitochondrial localization and suppresses mitochondrial oxidative damage. PLoS One, 8(1), e54283. doi: 10.1371/journal.pone.0054283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen A, Kalvakuri S, Bodmer R, & Cox RT (2015). Clueless, a protein required for mitochondrial function, interacts with the PINK1-Parkin complex in Drosophila. Dis Model Mech, 8(6), 577–589. doi:dmm.019208 [pii] 10.1242/dmm.019208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada Y, Burn KM, Niwa R, & Cooley L (2011). Reversible response of protein localization and microtubule organization to nutrient stress during Drosophila early oogenesis. Dev Biol, 355(2), 250–262. doi: 10.1016/j.ydbio.2011.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber MH, Thomsen MB, & Spradling AC (2016). Electron Transport Chain Remodeling by GSK3 during Oogenesis Connects Nutrient State to Reproduction. Cell, 164(3), 420–432. doi: 10.1016/j.cell.2015.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapon N, Ito N, Dickson BJ, Treisman JE, & Hariharan IK (2001). The Drosophila tuberous sclerosis complex gene homologs restrict cell growth and cell proliferation. Cell, 105(3), 345–355. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/11348591 [DOI] [PubMed] [Google Scholar]
- Uversky VN (2017). Intrinsically disordered proteins in overcrowded milieu: Membrane-less organelles, phase separation, and intrinsic disorder. Curr Opin Struct Biol, 44, 18–30. doi: 10.1016/j.sbi.2016.10.015 [DOI] [PubMed] [Google Scholar]
- Vornlocher HP, Hanachi P, Ribeiro S, & Hershey JW (1999). A 110-kilodalton subunit of translation initiation factor eIF3 and an associated 135-kilodalton protein are encoded by the Saccharomyces cerevisiae TIF32 and TIF31 genes. J Biol Chem, 274(24), 16802–16812. Retrieved from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10358023 [DOI] [PubMed] [Google Scholar]
- Wakim J, Goudenege D, Perrot R, Gueguen N, Desquiret-Dumas V, Chao de la Barca JM, … Khiati S (2017). CLUH couples mitochondrial distribution to the energetic and metabolic status. J Cell Sci, 130(11), 1940–1951. doi: 10.1242/jcs.201616 [DOI] [PubMed] [Google Scholar]
- Wang C, Schmich F, Srivatsa S, Weidner J, Beerenwinkel N, & Spang A (2018). Context-dependent deposition and regulation of mRNAs in P-bodies. Elife, 7. doi: 10.7554/eLife.29815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Branicky R, Noe A, & Hekimi S (2018). Superoxide dismutases: Dual roles in controlling ROS damage and regulating ROS signaling. J Cell Biol, 217(6), 1915–1928. doi: 10.1083/jcb.201708007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZH, Clark C, & Geisbrecht ER (2016). Drosophila clueless is involved in Parkin-dependent mitophagy by promoting VCP-mediated Marf degradation. Hum Mol Genet, 25(10), 1946–1964. doi: 10.1093/hmg/ddw067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZH, Rabouille C, & Geisbrecht ER (2015). Loss of a Clueless-dGRASP complex results in ER stress and blocks Integrin exit from the perinuclear endoplasmic reticulum in Drosophila larval muscle. Biol Open, 4(5), 636–648. doi: 10.1242/bio.201511551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm JE, Buszczak M, & Sayles S (2005). Efficient protein trafficking requires trailer hitch, a component of a ribonucleoprotein complex localized to the ER in Drosophila. Dev Cell, 9(5), 675–685. doi:S1534-5807(05)00377-1 [pii] 10.1016/j.devcel.2005.09.015 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Colcemide treatment disrupts microtubules. Ubi-GFP::tubulin expressing follicles. Follicles cultured in Complete Schneider’s Media supplemented with 200 μg/mL of insulin display an intact microtubule cytoskeleton (A). Follicles after colcimide treatment show the microtubule cytoskeleton has been disrupted (B). Scale bar: 40μm in B for A, B.
Movie 5. Clu particles in surrounding follicle cells are more stationary than in germ cells. Follicle from cluCA06604 (Clu-GFP) female. Imaged one frame per second for 120 seconds. Video is 24 frames per second.
Movie 6. Clu particle formation is induced with insulin. Follicle from cluCA06604 (Clu-GFP) female. Imaged one frame per minute for 45 minutes. Video is 10 frames per second.
Movie 7. Oxidative stress disperses Clu particles. Follicle from cluCA06604 (Clu-GFP) female. Imaged one frame per 15 seconds for 15 minutes. Video is 10 frames per second.
Movie 8. Oxidative stress causes mitochondrial mislocalization in Drosophila germ cells. Follicle from cluCA06604 (Clu-GFP) female. Imaged one frame per 15 seconds for 20 minutes. Video is 10 frames per second.
Figure S2. Multiple Δclu1 strains derived in two different wild type genetic backgrounds have reduced growth on glycerol. Each plate shows three newly generated deletions of CLU1 (clu1::KanMX) in two wild type strain backgrounds (W303, left plate and BY4741, right plate). After one week on glycerol, the wild type strain grows at a normal rate whereas colonies of three newly derived Δclu1 strains are smaller and less robust.
Figure S3. InRE19 mutant follicle cell clones do not contain Clu particles. (A-C) The surrounding somatic follicle cells of a follicle. The mutant cells are indicated by lack of GFP antibody labeling (dotted outline, A). Clu particles are not present in cells mutant for InRE19 (dotted outline, B, C). White = GFP (A) and Clu (B). Magenta = GFP (C). Green = Clu (C). Scale bar = 10 μm in C for A-C. n=7.
Movie 1. Clu live-imaging shows robust, dynamic particles in germ cells. Follicle from cluCA06604 (Clu-GFP) female. Imaged one frame per second for 200 seconds. Video is 36 frames per second.
Movie 2. Mitochondria movement is normal without colcemide treatment. Follicle from cluCA06604 (Clu-GFP) female treated with TMRE. Imaged one frame per second for 120 seconds. Video is 10 frames per second.
Movie 3. Mitochondrial movement ceases with treatment of the microtubule destabilizer colcemide. Follicle from cluCA06604 (Clu-GFP) female treated with TMRE. Imaged one frame per second for 120 seconds. Video is 10 frames per second.
Movie 4. Clu particle movement ceases with treatment of the microtubule destabilizer colcemide. Follicle from cluCA06604 (Clu-GFP) female. Imaged one frame per second for 120 seconds. Video is 24 frames per second.