Abstract
The ability to define cell types and how they change during organogenesis is central to our understanding of animal development and human disease. Despite the crucial nature of this knowledge, we have yet to fully characterize all distinct cell types and the gene expression differences that generate cell types during development. To address this knowledge gap, we produced an atlas using single-cell RNA-sequencing methods to investigate gene expression from the pharyngula to early larval stages in developing zebrafish. Our single-cell transcriptome atlas encompasses transcriptional profiles from 44,102 cells across four days of development using duplicate experiments that confirmed high reproducibility. We annotated 220 identified clusters and highlighted several strategies for interrogating changes in gene expression associated with the development of zebrafish embryos at single-cell resolution. Furthermore, we highlight the power of this analysis to assign new cell-type or developmental stage-specific expression information to many genes, including those that are currently known only by sequence and/or that lack expression information altogether. The resulting atlas is a resource for biologists to generate hypotheses for functional analysis, which we hope integrates with existing efforts to define the diversity of cell-types during zebrafish organogenesis, and to examine the transcriptional profiles that produce each cell type over developmental time.
INTRODUCTION
During animal development, stem cells generate a vast diversity of differentiated cells to form functioning tissues and organs. The specification of cell fates during development and the ability to function properly during adult life are determined by the RNA messages these cells express and the functions and quantities of proteins produced from these messages. A central question in biology is how gene expression unfolds over time to precisely coordinate the development of adult organisms. Such coordinated gene expression programs are critical given that disruptions to the expression of cell-type specific RNAs can lead to abnormal function and the progression of degeneration and disease. A major obstacle for regenerative therapies is not knowing the full system of genes normally expressed in a given cell type, thus preventing the development of cell-type specific genetic and pharmaceutical interventions as well as inhibiting the efficient reprogramming of stem cells to regenerate damaged tissue. Furthermore, genes associated with genetic disorders are often poorly described in terms of cell and developmental stage. To understand development, and to provide a foundation for inventing therapeutics, we must comprehend the systems-level phenomenon of how genes and proteins are used across cell type and over time.
Characterizing the in toto systems of a developing animal is a challenge. However, recent advances in single-cell RNA-seq (scRNAseq) provide a platform to track transcriptional changes across thousands of cells simultaneously. Capturing all transcriptional changes across all of the cells of a developing animal is a powerful step towards identifying the molecular and genetic basis of cell-type specification, organogenesis, and adult homeostasis, which is evident in a recent expansion of efforts to undertake this work (Briggs et al., 2018; Cao et al., 2019; Farrell et al., 2018; Fincher et al., 2018; Pandey et al., 2018; Regev et al., 2017; Tang et al., 2017; Wagner et al., 2018). Here we set out to fill a critical gap in the understanding of zebrafish and generate a single-cell RNA-seq atlas of organogenesis. Our overarching goal is to encourage collective efforts to compile these data for zebrafish, to transcriptionally identify cell types, and to afford researchers the opportunity to efficiently identify de novo candidates for genetic analysis in tissues, cell types, and developmental gene expression programs of interest. We have drafted an atlas from whole zebrafish embryos and larvae where single-cell transcriptomic data are compiled from samples that span four days of development (1–5 days post fertilization).
Zebrafish is an excellent model system for producing an atlas of vertebrate gene expression over organogenesis. Zebrafish development is rapid, with free-swimming animals emerging within the first three days after fertilization, accompanied by a system-wide expansion and differentiation of organ specific cell types including: neurogenesis (and associated complex behaviors), maturation of blood, muscle, gastro-intestinal tissues, germ cells, and the specification of pigmented skin epithelium, vasculature, cartilage, and bone . Here, we present an atlas and demonstrate how to use it for insight into several of these developmental processes. We identify hundreds of transcriptionally-defined cell types and their corresponding gene expression developmental trajectories, and we anchor them to zebrafish anatomy by comparison with RNA in situ expression patterns. This atlas provides a rich resource of transcriptionally-defined cell types across zebrafish organogenesis. Researchers can mine this resource for transcripts that were previously not attributed to specific cell types of interest (de novo gene expression analysis) as well as probe for temporal changes in gene expression that underly cell-type specification during development.
RESULTS
The zebrafish scRNA-seq atlas across organogenesis
We designed the atlas to be a tool for investigating changes in gene expression associated with cell-fate specification during animal development with the goal of providing new insight into vertebrate organogenesis. To generate high-quality single-cell transcriptional profiles, we examined the reproducibility of our cell dissociation and scRNAseq methods by performing replicate dissociations derived from independent matings, cDNA library preparations, and sequencing for each stage of development profiled in this study (1, 2, and 5 days post fertilization (dpf)). These data combined to total 44,012 cells, 74,914 mean reads per cell, 1807 median genes per cell, and a median of 8345 unique transcript molecules per cell (Figure S1A,B). We used Seurat (Butler et al., 2018) to cluster transcriptionally similar cells together and compared the results of replicate experiments to each other in terms of cluster diversity and the representation of cells from each duplicate within all clusters. We found that each independent experiment contributed to the computationally identified clusters at each age, showing that our dissociation methods were reproducibly sampling across all accessible cell types (Figure S1C–H). Thus, our methodology supports a robust and reproducible single-cell transcriptomic analysis and clustering, enabling investigation of cell type, state, and developmental transitions through organogenesis
Cell-type annotation and de novo gene identification for clusters within the atlas
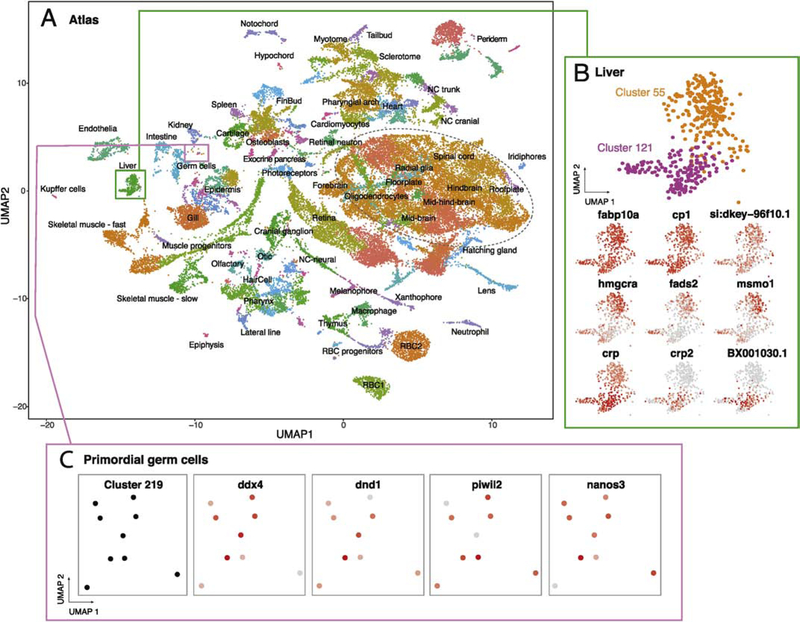
The Zebrafish scRNAseq atlas should offer the ability to produce accurate gene expression profiles for cell types of interest at multiple developmental stages. We therefore aggregated the results of all six experiments from three distinct developmental stages (1, 2, and 5 dpf) to examine the diversity of transcriptional cell types across developmental time (Figure 1A). Aggregation resulted in 220 computationally identified clusters (Figure S2) and for each cluster we sought to assign it to the most likely cell type.
Figure 1.
scRNA-seq atlas of developing zebrafish embryos during organogenesis. (A) Clustering of cell types enables gene expression analysis across transcriptional cells types over developmental time. Colors correspond to labels which indicate a grouping of clusters and annotations. Green box describes clusters enlarged in B; pink box describes cluster enlarged in C. Dashed oval approximates the region described in Figure 2. (B) Heterogeneity within hepatocytes is revealed by the identification of two clusters (55 and 121) with different but related gene expression profiles. Common and differential gene expression between these clusters are plotted using red to indicate high levels of expression and grey for low expression (normalized for total expression of the gene, see methods). (C) Rare cells types, including primordial germ cells (PGCs), can be efficiently profiled and are restricted to a single cluster (219). Four markers of PGCs show high levels of expression within this cluster.
To annotate each of the clusters with cell-type information, we used the expression of unambiguous, previously identified marker genes and accessed this information using the Zebrafish Information Network (ZFIN)(Howe et al., 2013). For example, genes associated with hepatocytes, fabp10a and cp (Her et al., 2003; Korzh et al., 2001), were among the most differentially expressed in our entire data set, as demonstrated by their enrichment in the first principal component (PC) of the PC analysis (PCA) used to cluster cells (Figure S3). We plotted expression of these genes within the atlas and identified two clusters in close proximity that also expressed additional transcripts expressed in and associated with liver development and function (Figure 1B, Table S1, see clusters 55 and 121). Examining the top 30 most differentially expressed genes in these clusters, we found that 15 that have been described to be expressed in the hepatocytes (ZFIN), strongly supporting the annotation of these clusters (Figure 1B, Table S2, see clusters 55 and 121). Additionally, we find 7 genes that are poorly characterized, including si:dkey-96f10.1 (Fig. 1B), that are expressed in both hepatocyte clusters, highlighting the power of the atlas to provide information for genes known only by sequence. Furthermore, our analysis revealed several genes that were differentially expressed between these two clusters of putative hepatocytes, including hmgcra, fads2, msmo1, in cluster 55 and crp2 and BX001030.1, a poorly characterized gene in cluster 121 (Figure 1B), demonstrating a molecular basis for cellular diversity among zebrafish hepatocytes (see Discussion). It is unlikely that differences in gene expression between these two hepatocyte clusters are developmental stage-specific, as all cells from these clusters come from 5 dpf embryos (Table S2).
Next, we wondered whether these experiments captured rare cell types. Zebrafish embryos each have only about 30 primordial germ cells (PGCs) from 1 to 5 dpf (Tzung et al., 2015), thus providing a useful test case for detecting rare cells. We found that one computationally defined cluster (cluster 219) expressed the PGC markers ddx4, nanos3, dnd1, and piwi2 (Houwing et al., 2008, 2007; Köprunner et al., 2001; Pandey et al., 2018; Weidinger et al., 2003; Yoon et al., 1997) indicating that PGCs were profiled and clustered in the atlas (Figure 1C). The recovery of PGCs suggests that we have likely profiled most cell types present at the developmental stages analyzed in this study and provides an estimate of the lower threshold for detection of rare cell types. This finding does not rule out the possibility that some cell types may fail to be detected due to lack of dissociation, cell death, or inadequate sample size.
Finally, we sought to annotate all 220 clusters within the atlas. We first identified the top 16 most differentially expressed genes for every cluster as having the highest ratio of expression among cells within a cluster relative to all other cells in the atlas. Using RNA in situ expression patterns found in public databases we annotated the most likely cell type and corresponding tissue for each cluster (Fig 1A). These annotations occupy a nested hierarchy that contains information about germ layer, tissue type, and ultimately cell type (Table S2). Both unambiguous marker genes, with clear gene expression patterns referenced in ZFIN, as well as genes with no information on gene expression patterns, appeared within the top 16 most differentially expressed genes for each cluster (Table S2). For most clusters (169/220), genes that have little or no previous expression information (e.g. genes beginning with “si:” or “zgc:” on ZFIN) appeared among the top sixteen most differentially expressed genes with an average of two (mean = 2.2, range = 0:9, SD = 2.1, for all clusters, (Table S2). For example, in the liver, notochord, and retina, the genes si-dkey-96f10.1, si-ch1073–45j12.1, and si-dkey-16p21.8 were differentially expressed (respectively) and represent putative marker genes for these organs (Table S2). Thus, marker gene expression, coupled with analysis of RNA expression patterns in vivo, is an efficient means to annotate discrete clusters within the atlas and provides a resource for helping to assign cell type information to poorly characterized genes.
Combinatorial codes of gene expression inform anatomical position and identity.
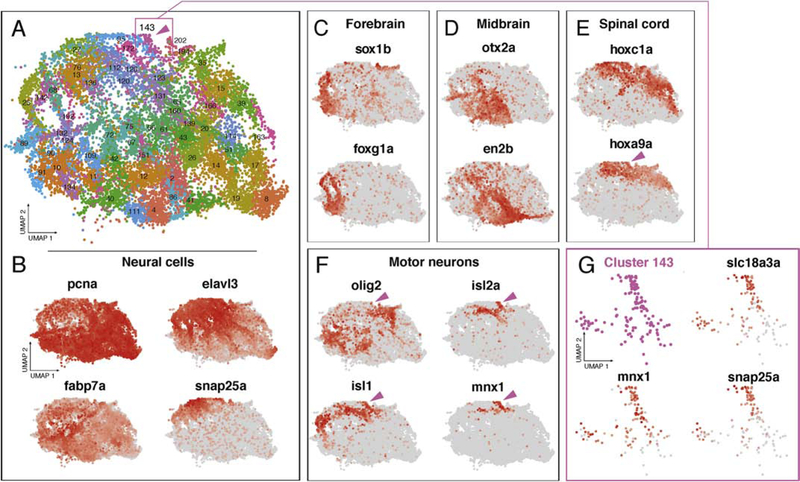
A cell-type atlas that uses transcriptomic data is more useful if it can provide spatial information. This is a major challenge in scRNA-seq data derived from whole zebrafish because the anatomical position of any profiled cell is lost upon dissociation. For some cells, such as hepatocytes (see above), transcriptional profiles are distinct, and their anatomical location in the animal is restricted, making their identification relatively simple. However, many tissues, including the nervous system which serves as an extreme example due to its staggering functional and cellular diversity, present unique challenges because many of the cell types are transcriptionally similar, yet occupy anatomically disparate locations in the animal. For example, excitatory and inhibitory neurons reside throughout the nervous system, and the neurotransmitters and vesicular transporters that distinguish these two cell-types demonstrated broad expression across many discrete neural clusters in the atlas (Figure S4), similar to what has been found in mouse (Cembrowski and Menon, 2018). We asked if spatially restricted expression domains would provide information about the subtypes of neural cells within our atlas.
Using clusters that corresponded to the nervous system (Fig. 2A), we found gene expression profiles that were associated with neural stem cells, differentiating, post-mitotic neurons, and mature neurons (Fig. 2B, Table S2) (Good, 1995; Ito et al., 2010; Wei et al., 2013). Within this subset of clusters from the atlas, we examined genes known to have spatially restricted expression profiles along the A/P axis of the central nervous system and found that the clusters were organized in UMAP space in a manner that mimicked the in vivo A/P relationship of these genes (Fig. 2C–E) (Amores et al., 1998; Duggan et al., 2008; McClintock et al., 2001; Mori et al., 1994; Pandey et al., 2018; Scholpp et al., 2007; Scholpp and Brand, 2001; Woolfe and Elgar, 2007). The nervous system is well known for integrating both A/P and D/V information to generate particular cell fates. One of the best studied examples is spinal cord motorneuron specification where the so-called pMN ventral progenitor cells within the neural tube receive high levels of Sonic Hedgehog signaling turning on transcription factors such as hoxa9a, olig2, isl1, isl2a, and mnx1 (Hutchinson and Eisen, 2005; Ravanelli and Appel, 2015). To identify spinal cord motorneurons we first used hoxa9a (Figure 2E), along with hoxb9a, hoxc8a, hoxb8a, hoxc6b (e.g. Cluster 15 Table S2), which are restricted to the posterior spinal cord (Prince et al., 1998) and thus distinguishes spinal cord neural cells. Next, we examined pMN markers that result from positional information from the D/V axis and found that they were expressed in increasingly restricted regions of the nervous system (Figure 2F). Strikingly, only a single cluster of neural cells mapped to the intersection of the A/P and D/V markers (Fig. 2A,F – magenta arrow head, and 2G), and this cluster expresses genes such as slc18a3a, the acetylcholine neurotransmitter transporter, mnx1, a motor neuron marker gene, and snap25a, which marks differentiated neurons (Bayés et al., 2017; Nakano et al., 2005; Wei et al., 2013). These results indicate that a combinatorial code of spatially confined transcription factors with motorneuron fate determining molecules is sufficient to identify anatomically and functionally distinct populations of neurons in the atlas. This approach was applied to all central nervous system clusters of the atlas (Table S2) and facilitates analysis of the regional transcriptional differences amongst anatomically and functionally diverse neurons. We conclude that spatially restricted gene expression can be mapped to the atlas even given complicated and similar transcriptional profiles, enhancing our knowledge of cell-type-specific gene expression within these domains.
Figure 2.
Spatially restricted gene expression patterns mapped to the atlas. (A) Clusters from the atlas with annotated neural cell identifies. The cells represented correspond to the dashed oval in Figure 1A. Cluster 143 is marked by magenta arrowhead. Magenta box describes location of cluster 143 enlarged in G. (B) Neural maker genes show regions of progenitor (pcna, fabp7a) and differentiated neural identities (elavl3, and snap25a). (C-E) Spatially restricted transcription factors show discrete domains of expression across neural clusters in the atlas. Cells with expression of genes related to the forebrain (sox1b, foxg1a)(C), midbrain (otx2a, en2b)(D), and spinal cord (hoxc1a, hoxa9a)(E), cluster together in the UMAP projection. (F) Combinatorial code of gene expression within hoxa9a expressing domain reveals cluster of putative spinal cord motor neurons expressing markers of the progenitor domain (olig2) and differentiation (isl1, isl2a, mnx1). Cluster 143 marked by magenta arrowhead. (G) Magnified view of cluster 143 shows high levels of motor neuron marker genes slc18a3a and mnx1 and the differentiated neuronal marker snap25a.
Cell-type specific gene expression changes over time are mapped in the atlas
A primary goal of this atlas is to provide temporal information about gene expression changes across four days of zebrafish development. This approach is critical for understanding the genetic basis of how stem cells and progenitor cells produce diverse progeny, and the pathways along which differentiated cells mature during organogenesis to form functioning tissues.
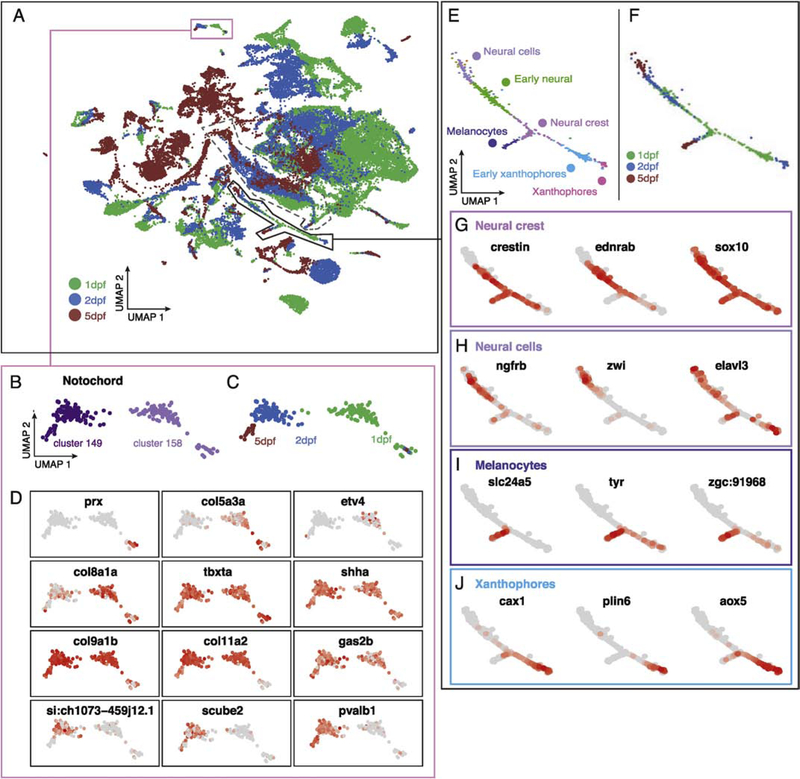
By aggregating our replicate experiments from three developmental stages, we sought to determine which changes in gene expression were associated with cell-type specification and organogenesis during this period of development. Within the transcriptional mapping space of the atlas, cell clusters often separated according to developmental age (Fig. 3A). Although a major discretizing factor across the atlas is time of origin, within any particular transcriptionally identified cell type there are examples of apparent transcriptional continuity that connect developmental timepoints.
Figure 3.
Temporal gene expression analysis in the atlas using cell age. (A) Aggregating cells from 1, 2, 5 dpf embryos reveals age-related changes in gene expression for discrete clusters. Pink box describes notochord clusters enlarged in B-D; solid black line shows approximate region of neural crest lineages enlarged in E-J. Dashed line shows approximate region of retinal cells described in Figure 4. (B-C) Notochord clusters contain cells of distinct ages. (D) Differential gene expression analysis in developing the notochord. (E) Discrete clusters corresponding to cell type specification in neural crest lineages are annotated with cell-type and color coded by cluster. (F) Age of cells associated with neural crest lineage. (G-J) Differential gene expression analysis over time in neural crest progenitors (G), neural cells (H), melanocytes (I), and xanthophores (J) across developmental time.
For example, the notochord formed several discrete clusters across developmental time (Figure 3B,C). We identified notochord cells based on the exclusive expression of tbxta (Odenthal et al., 1996), and in addition, expression of col2a1a, col9a2 and matn4 (Figure S5) (Garcia et al., 2017; Halpern et al., 1997; KO et al., 2005). Thus we found that the UMAP transcriptional space effectively clustered cells of the notochord together and also preserved transcriptional differences associated with developmental time.
Next, we asked which genes were differentially expressed between notochord clusters of progressing developmental age. We found col5a3a, prx, and etv4 to have enriched expression in 1 dpf notochord, a poorly characterized gene, si-ch1073–45j12.1 expressed in 2 dpf notochord, but repressed at 5 dpf, while scube2, pvalvb1, gas2b were specifically expressed in older, 2–5 dpf notochord (Figure 3D). Importantly, we identified genes that are expressed in notochord throughout the developmental stages analyzed in this study (common to both notochord clusters), including col9a1b, col11a2, and shha (Figure 3D). Mining these data suggests functional experiments to understand the mechanisms that drive notochord differentiation. We conclude that temporal changes in gene expression are efficiently captured in the atlas by comparing transcriptional profiles from cells of the same tissue at progressing developmental ages.
Next, we analyzed transcriptional trajectories of cell type specification across developmental time in related cell types. A striking example appeared in the neural crest lineage (Fig. 3E,F). Clusters corresponding to neural crest progenitors were identified using crestin, ednrab, and sox10 (Figure 3G) (Bonano et al., 2008; Dutton et al., 2001; Luo et al., 2001). Neural progeny of the neural crest were characterized by expression of ngfrb, zwi, and elavl3 (Schaefer and Brösamle, 2009), melanocytes by slc24a5 and tyr expression (Braasch et al., 2009, 2007; Kelsh et al., 2000), and xanthophores by cax1, plin6, and aox5 expression (Granneman et al., 2017; Manohar et al., 2010) (Figure 3G–J)(putative xanthophores from 5 dpf embryos were separated from this region in the UMAP projection). Cells with these distinct neural crest derivative transcriptional profiles were proximal to the neural crest progenitors in UMAP coordinates and connected by a continuum that was arranged according to developmental stage (Figure 3E–F).
Interestingly, a poorly described gene, zgc:91968, demonstrated high levels of expression in the melanocyte cluster (Figure 3I and Table S2) confirming recent expression analysis of this gene (Heffer et al., 2017), and si:dkey-25li10.2 and si:dkey-129g1.9 in xanthophore clusters (Table S2), expanding our knowledge of cell-type specific expression patterns for these transcripts. Thus, the atlas is a useful tool for investigating gene expression changes associated with the differentiation of multiple cell types from a lineage of common progenitors.
Pseudotemporal analysis using Monocle bolsters temporal gene expression analysis during development
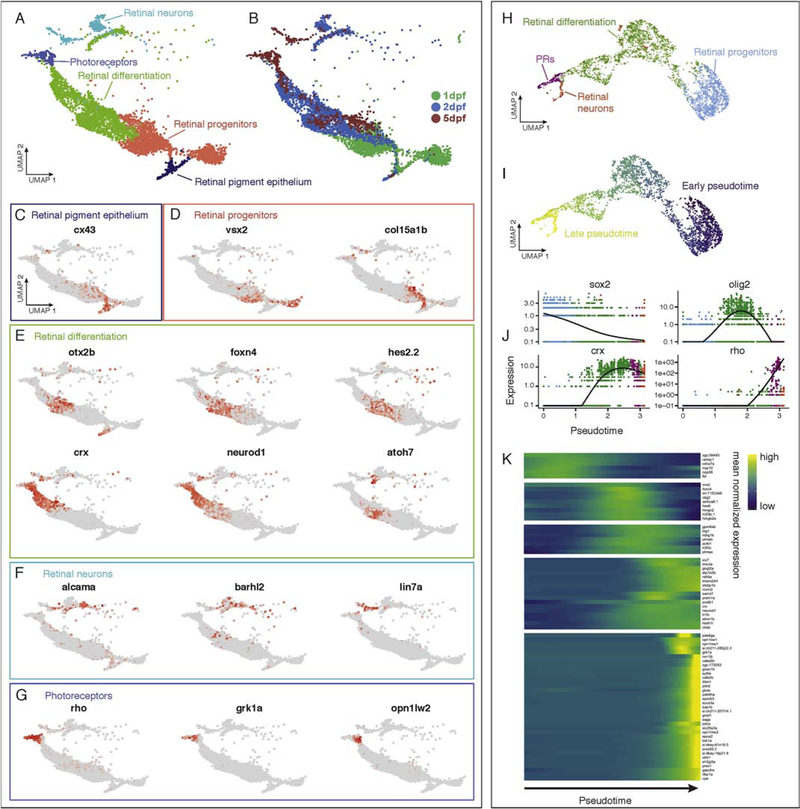
To validate changes in gene expression identified by comparing cells of similar types but from different developmental stages, we focused on developing retinal lineages as they provide a striking example (Figure 4A,B). We identified clusters of retinal pigment epithelium based on expression of cx43 (Figure 4C) (Dermietzel et al., 2000); retinal progenitors using vsx2 and col15a1b (Figure 4D) (Pujic et al., 2006; Vitorino et al., 2009); differentiating retinal cells using crx and neurod1 (Hitchcock and Kakuk-Atkins, 2004; Ochocinska and Hitchcock, 2009, 2007; Shen and Raymond, 2004) and also found differential expression of otx2b, foxn4, hes2.2 and ntoh7 (Figure 4E); retinal neurons using lin7a (Wei et al., 2006) and also found differential expression of alcama and barhl2 (Figure 4F); photoreceptors using rho (Vihtelic et al., 1999) and also found differential expression of grk1a and opn1lw2 (Figure 4G). Interestingly, the proximity in UMAP coordinates of these cell types suggested that their transcriptomes were related in a continuous fashion (Figure 4A), similar to the notochord and neural crest lineages (see above). Furthermore, the youngest cells (1 dpf) were strongly enriched within the retinal progenitor cluster, whereas photoreceptors and retinal neuron clusters mostly contained older cells (5 dpf) (Figure 4A–B). Between these progenitors and differentiated cells, we found cells primarily from 2 dpf embryos (Figure 4B). While there was a general progression from younger to older cells across the UMAP space, we also found cells derived from 2 and 5 dpf larvae within the clusters defined as retinal progenitors, and 5 dpf cells within the retinal differentiation clusters (Fig. 4A–B). This was expected as retinal development continues throughout life (Malicki et al., 2016) and there is a large amount of proliferation and specification occurring over the time period assayed in our experiments.
Figure 4.
Temporal and pseudotemporal gene expression analysis of the retina. (A) Retinal cells form continuous transitions in UMAP space. Colors correspond to groupings of clusters and annotations with the cells represented corresponding to the dashed oval in Figure 3A. (B) Age of cells associated with retinal cell clusters. (C-H) Differential expression analysis over time during retina development in clusters of putative (C) retinal pigment epithelium, (D) retinal progenitors, (E) differentiating retinal cells, (F) retinal neurons, and (G) photoreceptors. (H-K) Pseudotemporal analysis of retina neuron development in Monocle. (H) Monocle reconstruction of retinal neural lineages agrees with UMAP analysis and annotation (compare to A). (I) Monocle reconstruction of retinal lineage colored by pseudotemporal age. (J) Monocle analysis reveals progressive repression of sox2, activation of crx and rho, and transient activation of olig2 across pseudotime. (K) Heatmap of gene expression analysis associated with retinal cell type specification in Monocle.
We sought to compare relationships based on known ages to an unsupervised, pseudotemporal analysis of changes in gene expression across the retinal lineage. We omitted the clusters associated with retinal pigment epithelium from Monocle analysis, as these cells differentiate from retinal progenitors independently of retinal neurons (Fuhrmann et al., 2014). This analysis using Monocle (Cao et al., 2019) revealed many de novo predicted genes associated with retinal development, advancing our investigation of changes in gene expression associated with distinct developmental stages. Strikingly, the unsupervised pseudotemporal analysis strongly correlated with the developmental stage of each cell type within the retinal lineages (Figure 4H). Specifically, Monocle described a pseudotemporal order of retinal cells with striking fidelity to our annotations, and in concert with their developmental age (compare Figure 4H with Figure 4A–B). This result suggests that temporal changes in gene expression occur in a continuous fashion from progenitor to differentiated photoreceptors and retinal neurons. This pseudotemporal analysis helped identify genes that are downregulated during retinal differentiation, such as sox2 and also genes that are upregulated during retinal differentiation, such as crx, olig2 and rho (Figure 4J). Additional suites of genes with temporally restricted expression occurring throughout retinal differentiation were identified by Monocle providing a rich resource for understanding genes associated with retinal progenitors and photoreceptor specification during development (Figure 4K). These results highlight the usefulness of the atlas for investigating temporal changes in gene expression during zebrafish development.
DISCUSSION
The atlas of zebrafish organogenesis presented here is a tool for researchers to investigate gene expression, developmental trajectories, and identify cell types in developing zebrafish. Our profiling of 44,102 cells spanning one to five days post fertilization provide transcriptional insight into how all of the major organs form in this vertebrate system. We annotated all 220 bioinformatically identified clusters and highlighted several strategies for interrogating changes in gene expression associated with the development of zebrafish embryos at single cell resolution. Specifically, we demonstrate that: 1) algorithmically derived clusters can be annotated using marker gene expression, 2) expression analysis can advance knowledge for genes previously known only by sequence, and 3) temporal gene expression changes track continuous changes in gene expression patterns during differentiation of specific cell-types.
The clusters presented in this study require additional work to improve the accuracy and utility of this organogenesis atlas. Specifically, researchers with deep expertise regarding cell-type specific gene expression can help modify our tentative cluster annotations, which will improve this initial version, thus making it more useful for hypothesis testing. We envision this as an ongoing process with the community and make the current raw data, analysis methods, and our current draft of cluster identification available to the community (http://www.adammillerlab.com), with ongoing work and feedback continually improving the utility of the atlas going forward. Fundamentally, we view gene expression analysis in the atlas as a hypothesis-generating process that will require subsequent functional experiments to test the accuracy of the in silico predictions presented here. For example, the diversity of zebrafish liver cells suggested in this work is reminiscent of recent work in mammalian systems (Jenne and Kubes, 2013; MacParland et al., 2018; Tripathi et al., 2018). Furthermore, the differential expression of metabolic enzymes (hmgcra, fads2, msmo1) in one cluster of liver cells (cluster 55) and immunity-related, pro-inflammatory gene expression (crp, crp2) in another cluster of liver cells, suggests that developing zebrafish subfunctionalize distinct cell-types to engage the immune and metabolic tasks required of this tissue. Such predictions require RNA in situ hybridization and also loss of function analysis to examine their function in development and homeostasis. The results of such experiments will be forthcoming from our and collaborating research groups. To facilitate access to the processed data presented in this study, they are hosted at the UCSC Cell Browser (http://zebrafish-dev.cells.ucsc.edu).
Our data presented here add to the extensive work ongoing in the zebrafish field that has used scRNAseq to analyze changes during earlier stages in zebrafish embryogenesis (up to 24 hpf) (Farrell et al., 2018; Wagner et al., 2018). These data, and the ever-increasing body of cell-type and transcriptomic data with accompanying tools for analysis, represent some of the first system-level molecular and genetic insight to be leveraged toward understanding animal development (Karaiskos et al., 2017; Macaulay et al., 2016; McKenna et al., 2016; Raj et al., 2018; Tang et al., 2017). Ongoing work will expand the dataset presented here, and integrate data with those from other researchers to span zebrafish development and to include adult tissues, which should continue to be made available via public repositories such as the UCSC cell browser and ZFIN. It is our hope that data generated across the community will contribute to a comprehensive cell atlas of zebrafish, providing a framework for experimentation that will greatly enhance efforts toward unifying and translational applications of biology.
METHODS
Fish were maintained by the University of Oregon Zebrafish facility using standard husbandry techniques (Westerfield, 2007). Embryos were collected from natural matings, staged and pooled (n=15 per replicate). Animals used in this study were: Tg(olig2:GFP)vu12 for 1 dpf, n=2; 2 dpf, n=1; 5 dpf, n=2 samples and Tg(elavl3:GCaMP6s) for 2 dpf, n=1 sample (Figure S1).
Embryo dissociation
Collagenase P was prepared to a 100mg/mL stock solution in HBSS. Chemical dissociation was performed using 0.25% Trypsin, Collagenase P (2mg/mL), 1 mM EDTA (pH 8.0), and PBS for 15min at 28C with gently pipetting every 5 min. Dissociation was quenched using 5% calf serum, 1mM CaCl2, and PBS. Cells were washed and resuspended in chilled (4C), 1% calf serum, 0.8 mM CaCl2, 50 U/mL penicillin, 0.05 mg/mL streptomycin, and DMEM and passed through a 40uM cell strainer (Falcon) and diluted into PBS + 0.04% BSA to reduce clumping. A final sample cell concentration of 2000 cells per microliter, as determined on a Biorad TC20 cell counter, was prepared in PBS + 0.04% BSA for cDNA library preparation.
Single-cell cDNA library preparation
Sample preparation was performed by the University of Oregon Genomics and Cell Characterization core facility (https://gc3f.uoregon.edu/). Dissociated cells were run on a 10X Chromium platform using 10x v.2 chemistry targeting 10,000 cells. Dissociated samples for each time point (1, 2 and 5 dpf) were submitted in duplicate to determine technical reproducibility. Replicates for 1 and 5 dpf samples were from different clutches, but prepared in tandem, on the same day. Replicates for 2 dpf samples were prepared from different transgenic backgrounds on different days. The resulting cDNA libraries were amplified with 15 cycles of PCR and sequenced on either an Illumina Hi-seq (5/6 samples) or an Illumina Next-seq (n=1, 48h dpf sample) (Figure S1).
Computational analysis
The resulting sequencing data were analyzed using the 10X Cellranger pipeline, version 2.2.0 (Zheng et al., 2017) and the Seurat (Satija et al., 2015) software package for R, v3.4.4 (R Core Team, 2018) using standard quality control, normalization, and analysis steps. Briefly, we aligned reads to the zebrafish genome, GRCz11_93, and counted expression of protein coding reads. The resulting matrices were read into Seurat where we performed PCA using 178 PCs based on a Jack Straw determined significance of P < 0.01. UMAP analysis was performed on the resulting dataset with 178 dimensions and a resolution of 18.0, which produced 220 clusters and one singleton. For each gene, expression levels are normalized by the total expression, multiplied by a scale factor (10,000) and log-transformed. Differential gene expression analysis was performed using the FindAllMarkers function in Seurat using Wilcoxon rank sum test. Cells were subsetted based on gene expression and cluster for Monocle analysis (see below). We compared the correspondence between our replicates by counting the number of cells from each sample, in every cluster, and plotting these values in a scatterplot (x,y = fraction of cells from sample in a single cluster from replicate one, fraction of cells in a single cluster from replicate two) and performed a regression analysis. Additional code is provided on our website (https://www.adammillerlab.com/).
Pseudotemporal Trajectory analysis
We subsetted the retinal progenitors, retinal neurons and photoreceptor cells and applied UMAP dimensionality reduction to project cells in three dimensions using the preprocessCDS and reduceDimension functions in Monocle (v.2.99.1) (Cao et al., 2019) using default parameters (except for preprocessCDS: num_dim = 20; reduceDimension: reduction_method=UMAP, metric=cosine, n_neighbors=20, mid_dist=0.2). To learn the pseudotemporal trajectory, we then used the partitionCells, learnGraph, and orderCells functions using default parameters (except for partitionCells: k = 15; learnGraph: close_loop = TRUE, ncenter=400). To determine differentially expressed genes over pseudotime, we filtered the data set for genes expressed in at least 5 cells and performed differential expression analysis using a full model of sm.ns(Pseudotime, df=3). The top 80 DEGs were selected by q-value and plotted using the plot_pseudotime_heatmap function in Monocle.
Cluster annotation
We identified the top 16 most differentially expressed genes for every cluster as having the highest ratio of expression among cells within a cluster relative to all other cells in the atlas. Using RNA in situ expression patterns found in public databases, particularly ZFIN, we annotated the most likely cell type and corresponding tissue for each cluster. These annotations occupy a nested hierarchy that contains information about germ layer, tissue type, and ultimately cell type (Table S2).
Supplementary Material
Figure S1. Summary of statistics related to 10x Chromium cDNA library preparation and sequencing. (A) The sample name, genotype, age, reads per cell and sequencing platform used for individual experiments. (B) Summary of read, transcript, gene and cellular diversity and abundance for aggregated data used to produce the atlas. Replicate experiments for 1 dpf (C), 2 dpf (D) and 5 dpf (E) show mutual representation across clusters and indicate high reproducibility. We analyzed the correspondence between our replicates by counting the proportion of cells in each independent replicate, in every cluster and plotting these values in a scatterplot (x,y = proportion of all cells in a single cluster from replicate one, proportion of all cells in a single cluster from replicate two) and found a high level of correlation with R2 values of 0.8 (1 dpf), 0.4 (2 dpf) and 0.8 (5 dpf). Several representative clusters that do not show high correlation between replicates are labeled with their cluster number. (F-H) Replicate experiments for each developmental stage presented in this study are plotted together to show their relative contribution in the UMAP projection.
Figure S2. UMAP plot of all 220 clusters in the atlas. Replicate experiments from 1, 2 and 5 dpf embryos are aggregated into a single plot. Cluster numbers are applied to centroid of each cluster and correspond to Table S1 and Table S2 and cluster names referenced throughout this work.
Figure S3. Heatmap representation of genes represented in the first principal component (PC1) following PCA analysis. The hepatocyte markers cp and fabp10a are among the most differentially expressed genes. Yellow = high expression, magenta = low expression. 500 cells are plotted.
Figure S4. Genes encoding neurotransmitter vesicular transporters and biosynthesis pathways are markers of neuronal subtypes including, slc17a6a / slc17a6b for glutamatergic neurons, and gad1a / gad1b for gamma-aminobutyric acid (GABA), GABAergic neurons. Red = high expression, gray = low expression.
Figure S5. Marker genes tbxta, matn4, col9a2, and col2a1a, enable annotation of discrete clusters of notochord cells. Red = high expression, gray = low expression.
Table S1. List of differentially expressed genes for each cluster. The table was generated using the FindAllMarkers command of Seurat, using the Wilcoxon rank sum test. Pct.1 and Pct.2 reflect the fraction of cells expressing each marker gene (Boolean) within each cluster and for all other cells, respectively.
Table S2. The top 16 most differentially expressed genes for each cluster that were used for annotation. Our annotation includes the germ layer, tissue and cell type that correspond to each cluster. Also noted is the presence of cells from each of the developmental time points profiled in this study (1, 2, and 5 days post fertilization). These parameters are displayed as column headings.
A cell-type atlas of organogenesis from 1–5 dpf zebrafish embryos.
We annotated 220 distinct cell-type clusters.
Transcriptional changes are mapped across space and time.
These data are available for public access.
Acknowledgements
We thank Charles Kimmel, Monte Westerfield, Kryn Stankunas, Judith Eisen, John Postlethwait, Travis Wiles, and Chris Q. Doe for many helpful conversations during the conceptualization of the project and preparation of the manuscript. We thank Maximilian Haeussler and Mathew Speir for hosting data on the UCSC Cell Browser. We thank the University of Oregon Fish Facility for superb animal care. Funding was provided by the NIH Resource-Related Research Project Grant R24OD026591, NIH NINDS R01NS105758, and the University of Oregon to A.C.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession of data:
Sequences used in this study were deposited to the NCBI SRA and can be found using: PRJNA564810. The UCSC Cell Browser provides access to the data presented here at: http://zebrafish-dev.cells.ucsc.edu. Additional data and code relevant to this paper can be accessed at: https://www.adammillerlab.com/.
REFERENCES
- Amores A, Force A, Yan YL, Joly L, Amemiya C, Fritz A, Ho RK, Langeland J, Prince V, Wang YL, Westerfield M, Ekker M, Postlethwait JH, 1998. Zebrafish hox clusters and vertebrate genome evolution. Science (80-. ). 10.1126/science.282.5394.1711 [DOI] [PubMed]
- Bayés À, Collins MO, Reig-Viader R, Gou G, Goulding D, Izquierdo A, Choudhary JS, Emes RD, Grant SGN, 2017. Evolution of complexity in the zebrafish synapse proteome. Nat. Commun 10.1038/ncomms14613 [DOI] [PMC free article] [PubMed]
- Bonano M, Tríbulo C, De Calisto J, Marchant L, Sánchez SS, Mayor R, Aybar MJ, 2008. A new role for the Endothelin-1/Endothelin-A receptor signaling during early neural crest specification. Dev. Biol 10.1016/j.ydbio.2008.08.007 [DOI] [PubMed]
- Braasch I, Brunet F, Volff J-N, Schartl M, 2009. Pigmentation Pathway Evolution after Whole-Genome Duplication in Fish. Genome Biol. Evol 10.1093/gbe/evp050 [DOI] [PMC free article] [PubMed]
- Braasch I, Schartl M, Volff JN, 2007. Evolution of pigment synthesis pathways by gene and genome duplication in fish. BMC Evol. Biol 10.1186/1471-2148-7-74 [DOI] [PMC free article] [PubMed]
- Briggs JA, Weinreb C, Wagner DE, Megason S, Peshkin L, Kirschner MW, Klein AM, 2018. The dynamics of gene expression in vertebrate embryogenesis at single-cell resolution. Science (80-. ). 10.1126/science.aar5780 [DOI] [PMC free article] [PubMed]
- Butler A, Hoffman P, Smibert P, Papalexi E, Satija R, 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed]
- Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, Zhang F, Mundlos S, Christiansen L, Steemers FJ, Trapnell C, Shendure J, 2019. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 10.1038/s41586-019-0969-x [DOI] [PMC free article] [PubMed]
- Cembrowski MS, Menon V, 2018. Continuous Variation within Cell Types of the Nervous System. Trends Neurosci. 10.1016/j.tins.2018.02.010 [DOI] [PubMed]
- core Team R., 2018. R: A Language and Environment for Statistical Computing R Found. Stat. Comput Vienna, Austria. [Google Scholar]
- Dermietzel R, Kremer M, Paputsoglu G, Stang A, Skerrett IM, Gomès D, Srinivas M, Janssen-Bienhold U, Weiler R, Nicholson BJ, Bruzzone R, Spray DC, 2000. Molecular and Functional Diversity of Neural Connexins in the Retina. J. Neurosci 10.1523/jneurosci.20-22-08331.2000 [DOI] [PMC free article] [PubMed]
- Duggan CD, DeMaria S, Baudhuin A, Stafford D, Ngai J, 2008. Foxg1 is required for development of the vertebrate olfactory system. J. Neurosci 10.1523/JNEUROSCI.1134-08.2008 [DOI] [PMC free article] [PubMed]
- Dutton KA, Pauliny A, Lopes SS, Elworthy S, Carney TJ, Rauch J, Geisler R, Haffter P, Kelsh RN, 2001. Zebrafish colourless encodes sox10 and specifies non-ectomesenchymal neural crest fates. Development. [DOI] [PubMed]
- Farrell JA, Wang Y, Riesenfeld SJ, Shekhar K, Regev A, Schier AF, 2018. Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science (80-. ). 10.1126/science.aar3131 [DOI] [PMC free article] [PubMed]
- Fincher CT, Wurtzel O, de Hoog T, Kravarik KM, Reddien PW, 2018. Cell type transcriptome atlas for the planarian Schmidtea mediterranea. Science (80-. ). 360 10.1126/science.aaq1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrmann S, Zou CJ, Levine EM, 2014. Retinal pigment epithelium development, plasticity, and tissue homeostasis. Exp. Eye Res 10.1016/j.exer.2013.09.003 [DOI] [PMC free article] [PubMed]
- Garcia J, Bagwell J, Njaine B, Norman J, Levic DS, Wopat S, Miller SE, Liu X, Locasale JW, Stainier DYR, Bagnat M, 2017. Sheath Cell Invasion and Trans-differentiation Repair Mechanical Damage Caused by Loss of Caveolae in the Zebrafish Notochord. Curr. Biol 10.1016/j.cub.2017.05.035 [DOI] [PMC free article] [PubMed]
- Good PJ, 1995. A conserved family of elav-like genes in vertebrates. Proc. Natl. Acad. Sci. U. S. A 10.1073/pnas.92.10.4557 [DOI] [PMC free article] [PubMed]
- Granneman JG, Kimler VA, Zhang H, Ye X, Luo X, Postlethwait JH, Thummel R, 2017. Lipid droplet biology and evolution illuminated by the characterization of a novel perilipin in teleost fish. Elife. 10.7554/eLife.21771 [DOI] [PMC free article] [PubMed]
- Halpern ME, Hatta K, Amacher SL, Talbot WS, Yan YL, Thisse B, Thisse C, Postlethwait JH, Kimmel CB, 1997. Genetic interactions in zebrafish midline development. Dev. Biol 10.1006/dbio.1997.8605 [DOI] [PubMed]
- Heffer A, Marquart GD, Aquilina-Beck A, Saleem N, Burgess HA, Dawid IB, 2017. Generation and characterization of Kctd15 mutations in zebrafish. PLoS One. 10.1371/journal.pone.0189162 [DOI] [PMC free article] [PubMed]
- Her GM, Yeh YH, Wu JL, 2003. 435-bp liver regulatory sequence in the liver fatty acid binding protein (L-FABP) gene is sufficient to modulate liver regional expression in transgenic zebrafish. Dev. Dyn 10.1002/dvdy.10324 [DOI] [PubMed]
- Hitchcock P, Kakuk-Atkins L, 2004. The basic helix-loop-helix transcription factor neuroD is expressed in the rod lineage of the teleost retina. J. Comp. Neurol 10.1002/cne.20244 [DOI] [PubMed]
- Houwing S, Berezikov E, Ketting RF, 2008. Zili is required for germ cell differentiation and meiosis in zebrafish. EMBO J. 10.1038/emboj.2008.204 [DOI] [PMC free article] [PubMed]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, Plasterk RHA, Hannon GJ, Draper BW, Ketting RF, 2007. A Role for Piwi and piRNAs in Germ Cell Maintenance and Transposon Silencing in Zebrafish. Cell. 10.1016/j.cell.2007.03.026 [DOI] [PubMed]
- Howe DG, Bradford YM, Conlin T, Eagle AE, Fashena D, Frazer K, Knight J, Mani P, Martin R, Moxon SAT, Paddock H, Pich C, Ramachandran S, Ruef BJ, Ruzicka L, Schaper K, Shao X, Singer A, Sprunger B, Van Slyke CE, Westerfield M, 2013. ZFIN, the Zebrafish Model Organism Database: Increased support for mutants and transgenics. Nucleic Acids Res. 10.1093/nar/gks938 [DOI] [PMC free article] [PubMed]
- Hutchinson SA, Eisen JS, 2005. Zebrafish {\em{Isl1}} promotes motoneuron formation and inhibits interneuron formation. Dev. Biol
- Ito Y, Tanaka H, Okamoto H, Ohshima T, 2010. Characterization of neural stem cells and their progeny in the adult zebrafish optic tectum. Dev. Biol 10.1016/j.ydbio.2010.03.008 [DOI] [PubMed]
- Jenne CN, Kubes P, 2013. Immune surveillance by the liver. Nat. Immunol 10.1038/ni.2691 [DOI] [PubMed]
- Karaiskos N, Wahle P, Alles J, Boltengagen A, Ayoub S, Kipar C, Kocks C, Rajewsky N, Zinzen RP, 2017. The Drosophila embryo at single-cell transcriptome resolution. Science (80-. ). 10.1126/science.aan3235 [DOI] [PubMed]
- Kelsh RN, Schmid B, Eisen JS, 2000. Genetic analysis melanophore development in zebrafish embryos. Dev. Biol 10.1006/dbio.2000.9840 [DOI] [PubMed]
- KO Y-P, KOBBE B, PAULSSON M, WAGENER R, 2005. Zebrafish ( Danio rerio ) matrilins: shared and divergent characteristics with their mammalian counterparts . Biochem. J 10.1042/bj20041486 [DOI] [PMC free article] [PubMed]
- Köprunner M, Thisse C, Thisse B, Raz E, 2001. A zebrafish nanos-related gene is essential for the development of primordial germ cells. Genes Dev. [DOI] [PMC free article] [PubMed]
- Korzh S, Emelyanov A, Korzh V, 2001. Developmental analysis of ceruloplasmin gene and liver formation in zebrafish. Mech. Dev 10.1016/S0925-4773(01)00330-6 [DOI] [PubMed]
- Luo R, An M, Arduini BL, Henion PD, 2001. Specific pan-neural crest expression of zebrafish crestin throughout embryonic development. Dev. Dyn [DOI] [PubMed]
- Macaulay IC, Svensson V, Labalette C, Ferreira L, Hamey F, Voet T, Teichmann SA, Cvejic A, 2016. Single-Cell RNA-Sequencing Reveals a Continuous Spectrum of Differentiation in Hematopoietic Cells. Cell Rep. 10.1016/j.celrep.2015.12.082 [DOI] [PMC free article] [PubMed]
- MacParland SA, Liu JC, Ma XZ, Innes BT, Bartczak AM, Gage BK, Manuel J, Khuu N, Echeverri J, Linares I, Gupta R, Cheng ML, Liu LY, Camat D, Chung SW, Seliga RK, Shao Z, Lee E, Ogawa S, Ogawa M, Wilson MD, Fish JE, Selzner M, Ghanekar A, Grant D, Greig P, Sapisochin G, Selzner N, Winegarden N, Adeyi O, Keller G, Bader GD, McGilvray ID, 2018. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat. Commun 10.1038/s41467-018-06318-7 [DOI] [PMC free article] [PubMed]
- Malicki J, Pooranachandran N, Nikolaev A, Fang X, Avanesov A, 2016. Analysis of the retina in the zebrafish model. Methods Cell Biol. 10.1016/bs.mcb.2016.04.017 [DOI] [PubMed]
- Manohar M, Mei H, Franklin AJ, Sweet EM, Shigaki T, Riley BB, MacDiarmid CW, Hirschi K, 2010. Zebrafish (Danio rerio) endomembrane antiporter similar to a yeast Cation/H+ transporter is required for neural crest development. Biochemistry. 10.1021/bi100362k [DOI] [PubMed]
- McClintock JM, Carlson R, Mann DM, Prince VE, 2001. Consequences of Hox gene duplication in the vertebrates: An investigation of the zebrafish Hox paralogue group 1 genes. Development. [DOI] [PubMed]
- McKenna A, Findlay GM, Gagnon JA, Horwitz MS, Schier AF, Shendure J, 2016. Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science (80-. ). 10.1126/science.aaf7907 [DOI] [PMC free article] [PubMed]
- Mori H, Miyazaki Y, Morita T, Nitta H, Mishina M, 1994. Different spatio-temporal expressions of three otx homeoprotein transcripts during zebrafish embryogenesis. Mol. Brain Res 10.1016/0169-328X(94)90004-3 [DOI] [PubMed]
- Nakano T, Windrem M, Zappavigna V, Goldman SA, 2005. Identification of a conserved 125 base-pair Hb9 enhancer that specifies gene expression to spinal motor neurons. Dev. Biol 10.1016/j.ydbio.2005.04.017 [DOI] [PubMed]
- Ochocinska MJ, Hitchcock PF, 2009. NeuroD regulates proliferation of photoreceptor progenitors in the retina of the zebrafish. Mech. Dev 10.1016/j.mod.2008.11.009 [DOI] [PMC free article] [PubMed]
- Ochocinska MJ, Hitchcock PF, 2007. Dynamic expression of the basic helix-loop-helix transcription factor neuroD in the rod and cone photoreceptor lineages in the retina of the embryonic and larval zebrafish. J. Comp. Neurol 10.1002/cne.21150 [DOI] [PubMed]
- Odenthal J, Haffter P, Vogelsang E, Brand M, Van Eeden FJM, Furutani-Seiki M, Granato M, Hammerschmidt M, Heisenberg C-P, Jiang Y-J, Kane DA, Kelsh RN, Mullins MC, Warga RM, Allende ML, Weinberg ES, Nüsslein-Volhard C, 1996. Mutations affecting the formation of the notochord in the zebrafish, Danio rerio. Development. [DOI] [PubMed]
- Pandey S, Shekhar K, Regev A, Schier AF, 2018. Comprehensive Identification and Spatial Mapping of Habenular Neuronal Types Using Single-Cell RNA-Seq. Curr. Biol 10.1016/j.cub.2018.02.040 [DOI] [PMC free article] [PubMed]
- Prince VE, Joly L, Ekker M, Ho RK, 1998. Zebrafish hox genes: genomic organization and modified colinear expression patterns in the trunk. Development. [DOI] [PubMed]
- Pujic Z, Omori Y, Tsujikawa M, Thisse B, Thisse C, Malicki J, 2006. Reverse genetic analysis of neurogenesis in the zebrafish retina. Dev. Biol 10.1016/j.ydbio.2005.12.056 [DOI] [PubMed]
- Raj B, Wagner DE, McKenna A, Pandey S, Klein AM, Shendure J, Gagnon JA, Schier AF, 2018. Simultaneous single-cell profiling of lineages and cell types in the vertebrate brain. Nat. Biotechnol 10.1038/nbt.4103 [DOI] [PMC free article] [PubMed]
- Ravanelli AM, Appel B, 2015. Motor neurons and oligodendrocytes arise from distinct cell lineages by progenitor recruitment. Genes Dev. 10.1101/gad.271312.115 [DOI] [PMC free article] [PubMed]
- Regev A, Teichmann SA, Lander ES, Amit I, Benoist C, Birney E, Bodenmiller B, Campbell P, Carninci P, Clatworthy M, Clevers H, Deplancke B, Dunham I, Eberwine J, Eils R, Enard W, Farmer A, Fugger L, Göttgens B, Hacohen N, Haniffa M, Hemberg M, Kim S, Klenerman P, Kriegstein A, Lein E, Linnarsson S, Lundberg E, Lundeberg J, Majumder P, Marioni JC, Merad M, Mhlanga M, Nawijn M, Netea M, Nolan G, Pe’er D, Phillipakis A, Ponting CP, Quake S, Reik W, Rozenblatt-Rosen O, Sanes J, Satija R, Schumacher TN, Shalek A, Shapiro E, Sharma P, Shin JW, Stegle O, Stratton M, Stubbington MJT, Theis FJ, Uhlen M, Van Oudenaarden A, Wagner A, Watt F, Weissman J, Wold B, Xavier R, Yosef N, 2017. The human cell atlas. Elife. 10.7554/eLife.27041 [DOI] [PMC free article] [PubMed]
- Satija R, Farrell JA, Gennert D, Schier AF, Regev A, 2015. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol 10.1038/nbt.3192 [DOI] [PMC free article] [PubMed]
- Schaefer K, Brösamle C, 2009. Zwilling-A and -B, two related myelin proteins of teleosts, which originate from a single bicistronic transcript. Mol. Biol. Evol 10.1093/molbev/msn298 [DOI] [PubMed]
- Scholpp S, Brand M, 2001. Morpholino-induced knockdown of zebrafish engrailed genes eng2 and eng3 reveals redundant and unique functions in midbrain-hindbrain boundary development. Genesis. 10.1002/gene.1047 [DOI] [PubMed]
- Scholpp S, Foucher I, Staudt N, Peukert D, Lumsden A, Houart C, 2007. Otx1l, Otx2 and Irx1b establish and position the ZLI in the diencephalon. Development. 10.1242/dev.001461 [DOI] [PMC free article] [PubMed]
- Shen YC, Raymond PA, 2004. Zebrafish cone-rod (crx) homeobox gene promotes retinogenesis. Dev. Biol 10.1016/j.ydbio.2004.01.037 [DOI] [PubMed]
- Tang Q, Iyer S, Lobbardi R, Moore JC, Chen H, Lareau C, Hebert C, Shaw ML, Neftel C, Suva ML, Ceol CJ, Bernards A, Aryee M, Pinello L, Drummond IA, Langenau DM, 2017. Dissecting hematopoietic and renal cell heterogeneity in adult zebrafish at single-cell resolution using RNA sequencing. J. Exp. Med 10.1084/jem.20170976 [DOI] [PMC free article] [PubMed]
- Tripathi A, Debelius J, Brenner DA, Karin M, Loomba R, Schnabl B, Knight R, 2018. The gut-liver axis and the intersection with the microbiome. Nat. Rev. Gastroenterol. Hepatol 10.1038/s41575-018-0011-z [DOI] [PMC free article] [PubMed]
- Tzung KW, Goto R, Saju JM, Sreenivasan R, Saito T, Arai K, Yamaha E, Hossain MS, Calvert MEK, Orbán L, 2015. Early depletion of primordial germ cells in zebrafish promotes testis formation. Stem Cell Reports. 10.1016/j.stemcr.2014.10.011 [DOI] [PMC free article] [PubMed]
- Vihtelic TS, Doro CJ, Hyde DR, 1999. Cloning and characterization of six zebrafish photoreceptor opsin cDNAs and immunolocalization of their corresponding proteins. Vis. Neurosci 10.1017/S0952523899163168 [DOI] [PubMed]
- Vitorino M, Jusuf PR, Maurus D, Kimura Y, Higashijima SI, Harris WA, 2009. Vsx2 in the zebrafish retina: Restricted lineages through derepression. Neural Dev. 10.1186/1749-8104-4-14 [DOI] [PMC free article] [PubMed]
- Wagner DE, Weinreb C, Collins ZM, Briggs JA, Megason SG, Klein AM, 2018. Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science (80-. ). 10.1126/science.aar4362 [DOI] [PMC free article] [PubMed]
- Wei C, Thatcher EJ, Olena AF, Cha DJ, Perdigoto AL, Marshall AF, Carter BD, Broadie K, Patton JG, 2013. miR-153 Regulates SNAP-25, Synaptic Transmission, and Neuronal Development. PLoS One. 10.1371/journal.pone.0057080 [DOI] [PMC free article] [PubMed]
- Wei X, Luo Y, Hyde DR, 2006. Molecular cloning of three zebrafish lin7 genes and their expression patterns in the retina. Exp. Eye Res 10.1016/j.exer.2005.05.009 [DOI] [PubMed]
- Weidinger G, Stebler J, Slanchev K, Dumstrei K, Wise C, Lovell-Badge R, Thisse C, Thisse B, Raz E, 2003. dead end, a novel vertebrate germ plasm component, is required for zebrafish primordial germ cell migration and survival. Curr. Biol 10.1016/S0960-9822(03)00537-2 [DOI] [PubMed]
- Westerfield M, 2007. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio), 5th Edition. Univ. Oregon Press; Eugene. [Google Scholar]
- Woolfe A, Elgar G, 2007. Comparative genomics using Fugu reveals insights into regulatory subfunctionalization. Genome Biol. 10.1186/gb-2007-8-4-r53 [DOI] [PMC free article] [PubMed]
- Yoon C, Kawakami K, Hopkins N, 1997. Zebrafish vasa homologue RNA is localized to the cleavage planes of 2- and 4-cell-stage embryos and is expressed in the primordial germ cells. Development. [DOI] [PubMed]
- Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, Gregory MT, Shuga J, Montesclaros L, Underwood JG, Masquelier DA, Nishimura SY, Schnall-Levin M, Wyatt PW, Hindson CM, Bharadwaj R, Wong A, Ness KD, Beppu LW, Deeg HJ, McFarland C, Loeb KR, Valente WJ, Ericson NG, Stevens EA, Radich JP, Mikkelsen TS, Hindson BJ, Bielas JH, 2017. Massively parallel digital transcriptional profiling of single cells. Nat. Commun 10.1038/ncomms14049 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Summary of statistics related to 10x Chromium cDNA library preparation and sequencing. (A) The sample name, genotype, age, reads per cell and sequencing platform used for individual experiments. (B) Summary of read, transcript, gene and cellular diversity and abundance for aggregated data used to produce the atlas. Replicate experiments for 1 dpf (C), 2 dpf (D) and 5 dpf (E) show mutual representation across clusters and indicate high reproducibility. We analyzed the correspondence between our replicates by counting the proportion of cells in each independent replicate, in every cluster and plotting these values in a scatterplot (x,y = proportion of all cells in a single cluster from replicate one, proportion of all cells in a single cluster from replicate two) and found a high level of correlation with R2 values of 0.8 (1 dpf), 0.4 (2 dpf) and 0.8 (5 dpf). Several representative clusters that do not show high correlation between replicates are labeled with their cluster number. (F-H) Replicate experiments for each developmental stage presented in this study are plotted together to show their relative contribution in the UMAP projection.
Figure S2. UMAP plot of all 220 clusters in the atlas. Replicate experiments from 1, 2 and 5 dpf embryos are aggregated into a single plot. Cluster numbers are applied to centroid of each cluster and correspond to Table S1 and Table S2 and cluster names referenced throughout this work.
Figure S3. Heatmap representation of genes represented in the first principal component (PC1) following PCA analysis. The hepatocyte markers cp and fabp10a are among the most differentially expressed genes. Yellow = high expression, magenta = low expression. 500 cells are plotted.
Figure S4. Genes encoding neurotransmitter vesicular transporters and biosynthesis pathways are markers of neuronal subtypes including, slc17a6a / slc17a6b for glutamatergic neurons, and gad1a / gad1b for gamma-aminobutyric acid (GABA), GABAergic neurons. Red = high expression, gray = low expression.
Figure S5. Marker genes tbxta, matn4, col9a2, and col2a1a, enable annotation of discrete clusters of notochord cells. Red = high expression, gray = low expression.
Table S1. List of differentially expressed genes for each cluster. The table was generated using the FindAllMarkers command of Seurat, using the Wilcoxon rank sum test. Pct.1 and Pct.2 reflect the fraction of cells expressing each marker gene (Boolean) within each cluster and for all other cells, respectively.
Table S2. The top 16 most differentially expressed genes for each cluster that were used for annotation. Our annotation includes the germ layer, tissue and cell type that correspond to each cluster. Also noted is the presence of cells from each of the developmental time points profiled in this study (1, 2, and 5 days post fertilization). These parameters are displayed as column headings.