Abstract
Animal embryogenesis is initiated by maternal factors, but zygotic genome activation (ZGA) shifts regulatory control to the embryo during blastula stages. ZGA is thought to be mediated by maternally provided transcription factors (TFs), but few such TFs have been identified in vertebrates. Here we report that NF-Y and TALE TFs bind zebrafish genomic elements associated with developmental control genes already at ZGA. In particular, co-regulation by NF-Y and TALE is associated with broadly acting genes involved in transcriptional control, while regulation by either NF-Y or TALE defines genes in specific developmental processes, such that NF-Y controls a cilia gene expression program while TALE controls expression of hox genes. We also demonstrate that NF-Y and TALE-occupied genomic elements function as enhancers during embryogenesis. We conclude that combinatorial use of NF-Y and TALE at developmental enhancers permits the establishment of distinct gene expression programs at zebrafish ZGA.
Keywords: transcription, embryogenesis, maternal, zygotic, enhancer, nucleosome
INTRODUCTION
In animal embryogenesis, initial development of the zygote is controlled by parental material deposited into sperm and oocytes during gametogenesis. The bulk of this material is provided by the oocyte and the duration of the maternally controlled period varies between different animal species. However, embryonic development in all animals eventually switches to zygotic control. This maternal-to-zygotic transition (MZT) is a complex process that involves changes in the cell cycle, chromatin state and gene expression (reviewed in [1]). Zygotic genome activation (ZGA) is a key component of MZT and establishes gene expression programs driving subsequent embryonic development during gastrulation and organogenesis stages. ZGA is thought to be initiated by the action of maternally provided transcription factors (TFs). Accordingly, the Zelda TF induces expression of many genes at ZGA in Drosophila [2–4], but there is no known Zelda ortholog in vertebrates. Instead, other TFs have been proposed to regulate ZGA in vertebrates. For instance, Nanog, SoxB1 and Oct4/Pou5f1 (a.k.a. Pou5f3 in zebrafish) are maternally provided in zebrafish and are required for gene expression at zebrafish ZGA [5, 6]. Since the half-life of these proteins is relatively short (1–2hrs; [7, 8]), ZGA is likely controlled by TFs translated from maternally deposited mRNA. Similarly, Dppa2 and Dppa4 are maternally transmitted and act via Dux TFs to activate large numbers of genes at ZGA in mouse and human [9–12]. However, subsequent genetic analyses indicated that Dux is not required for zygotic development in vivo [13] and that the requirement for zebrafish Nanog is likely indirect via an effect on extraembryonic tissues [14], suggesting that vertebrate ZGA is more complicated and that additional TFs are likely involved in this process.
De novo activation of the zygotic gene expression program requires that the maternally transmitted TFs are able to access their binding sites in compacted chromatin, a property associated with pioneer factors (reviewed in [15]). Accordingly, Zelda opens inaccessible genomic regions to permit binding by other TFs [16, 17] and both Oct4/Pou5f1 and Sox proteins also possess pioneer activity [18, 19]. Notably, pioneer factors are active at many stages of embryogenesis to establish tissue-specific gene expression programs. For instance, FoxA TFs control the initiation of hepatic gene expression [20, 21] and PU.1 controls myeloid and lymphoid development [22]. Since the initiation of tissue specific gene expression programs is conceptually similar to zygotic gene activation, it is possible that pioneer factors with later roles (e.g. in gastrulation or organogenesis) also act at the ZGA. Accordingly, we recently reported that two TFs of the TALE family (Prep1 and Pbx4) – that were originally defined as cooperating with Hox proteins in the activation of tissue-specific gene expression during organogenesis – actually occupy their genomic binding sites already during maternal stages of zebrafish embryogenesis [23, 24]. Many of these sites are also occupied by nucleosomes, consistent with previous reports that TALE TFs possess pioneer activity [25]. While we find maternally deposited TALE TFs bound at genomic elements near genes activated at ZGA, they are also bound near genes active later in development [23]. Notably, we further demonstrated that a subset of TALE-occupied sites is associated with nearby binding motifs for NF-Y. NF-Y was originally considered a ubiquitous basal transcription factor (reviewed in [26]), but has recently been shown to be maternally deposited in zebrafish, to form protein complexes with TALE TFs and to possess pioneer activity [23, 27, 28]. Previous work identified NF-Y binding sites near genes activated at ZGA in mouse embryos [29, 30], but NF-Y also binds near many genes acting at later stages of development [27]. Additionally, NF-Y disruption leads to embryonic lethality in mouse [31], but has only mild effects on gene expression at ZGA [30]. Hence, both NF-Y and TALE may act at ZGA, but their roles are not well defined and it remains unclear if they act together.
Here we report that NF-Y and TALE bind zebrafish genomic elements already at ZGA and have both shared and separate roles during embryogenesis. In particular, regulation by either NF-Y or TALE is associated with genes active during gastrulation and segmentation stages – such that NF-Y controls a cilia gene expression program while TALE TFs control expression of homeobox genes. In contrast, co-regulation by NF-Y/TALE appears selectively associated with early-expressed genes involved in transcriptional regulation. Accordingly, we find that disruption of NF-Y or TALE function produces phenotypes that share some features – particularly anterior deformations – but that also show unique defects, such as hindbrain abnormalities in TALE-deficient embryos. We further demonstrate that NF-Y and TALE-occupied genomic elements possess enhancer activity when tested in a transgenesis assay in vivo. We conclude that combinatorial use of NF-Y and TALE at ZGA defines distinct gene expression programs where co-occupied enhancers control early-acting transcriptional regulators, while enhancers individually occupied by NF-Y or TALE control later-acting cilia and hox genes, respectively.
MATERIALS AND METHODS
Animal Care
The Institutional Animal Care and Use Committee (IACUC) of the University of Massachusetts Medical School approved all procedures involving zebrafish. Adult zebrafish were maintained at 28°C in groups at a maximum density of 12 individuals per liter with constant flow. To collect embryos for timing-sensitive experiments, one adult male fish and one adult female fish were placed in separate chambers of a 500mL tank overnight then placed together the following morning for no more than 30 minutes. For experiments that were not timing-sensitive, both adults were placed in the same chamber overnight. Eggs were collected in 10cm dishes, immersed in egg water (60μg/mL Instant Ocean, 0.0002% methylene blue), and maintained in an incubator at 29°C. Dead and unfertilized eggs were manually removed after two hours.
Method Details
Generation of mRNA for injection
Capped messenger RNAs encoding the dominant negative NF-YA (NF-YA DN; [32]), dominant negative Pbx4 (PBCAB; [33]) proteins were generated from 2μg of Notl-digested linearized pCS2+ plasmids using the mMessage mMachine SP6 Transcription Kit (ThermoFisher Scientific) according to the manufacturer’s guidelines. The RNA was purified using the RNeasy column with DNase treatment (Qiagen) according to the manufacturer’s guidelines. RNA quality was assessed on a 1% agarose gel and its concentration was measured on a NanoDrop instrument. 300pg of RNA injection mix containing water and 0.1% phenol red was injected into zebrafish embryos at the 1-cell stage. Injected embryos were raised to the proper stage according to animal care guidelines.
Characterization of TALE and NF-Y phenotypes
For gross phenotype assessment, 24hpf zebrafish embryos were placed on glass slides in 80% glycerol. For alcian blue staining, all incubations and washes took place on a nutator. 5dpf zebrafish embryos were fixed overnight in 4% phosphate-buffered paraformaldehyde. Following fixation, the embryos were washed in 0.1% phosphate-buffered Tween-20 (PBST) and bleached in 30% hydrogen peroxide for 2 hours. Once bleached, the embryos were rinsed twice in PBST and then stained overnight in alcian blue solution (1% hydrochloric acid (HCl), 70% ethanol, 0.1% alcian blue). After staining, the embryos were washed five times in acidic ethanol (HCl-EtOH; 5% HCl, 70% ethanol) with the final wash lasting 20 minutes. The embryos were then rehydrated in a series of 10-minute incubations of 75% HCl-EtOH/25% water, 50% HCl-EtOH/50% water, 25% HCl-EtOH/75% water, and 100% water and imaged. For in situ hybridizations, all incubations and washes took place on a nutator. 24hpf zebrafish embryos were fixed overnight in 4% phosphate-buffered paraformaldehyde. Following fixation, the embryos were washed in a 1:1 methanol:PBST solution, then PBST, and then treated with 1 μg/mL Proteinase K in PBST for 2 minutes. The embryos were washed once with −20°C acetone and twice with PBST then incubated at 70°C for 1 hour in Hyb+tRNA Buffer (50% formamide, 5X saline sodium citrate (SSC), 9.2mM citric acid, 0.5% Tween-20, 50 μg/mL heparin, 500 μg/mL tRNA). Next, the embryos were transferred to pax2/krox20/hoxd4a probe solution and incubated at 70°C overnight. After probe incubation, the embryos were washed sequentially for 10 minutes each at 70°C in Hyb Wash Buffer (50% formamide, 5X saline sodium citrate (SSC), 9.2mM citric acid, 0.5% Tween-20, 50 μg/mL heparin), 2:1 Hyb:2xSSC, 1:2 Hyb:2xSSC, 2xSSC, 0.2xSSC, and 0.1xSSC, then blocked in Blocking Solution (2% lamb serum and 2 μg/μL bovine serum albumin in PBST) at 4°C for 1 hour. Th e embryos were then incubated with 0.01% anti-DIG antibody at 4°C overnight. Following antibody treatment, the embryos were washed four times with PBST and two times with Staining Buffer (0.1M Tris pH 9.5, 50mM MgCl2, 125mM NaCl, 0.5% Tween20) then stained with Staining Solution (100 mg/mL polyvinyl alcohol, 0.35% 5-Bromo-4-chloro-3-indolyl phosphate, 0.45% 4-Nitro blue tetrazolium) at 37°C until the color developed. The embryos were then washed four times in PBST and scored. Sample size for phenotypic analyses was based on previous published reports that these dominant negative constructs produce phenotypes in >85% of injected embryos [23, 33–35]. Embryos were randomly selected for inclusion in injected or control pools. No animals were excluded, and experiments were not blinded.
RNA extraction
Zebrafish embryos were injected with either PBCAB, NF-YA DN, GFP, or antisense NF-YA DN mRNA as described above. At the desired timepoint, embryos were collected into three biological replicates of 50–100 embryos per condition. Dead animals were counted, but excluded from RNA extraction procedures. No other animals were excluded, and selection was not blinded. Each sample was placed in 1mL of Trizol and frozen at −80°C to help break up embryos. Once thawed, the embryos were broken up by pipette and 250μL of chloroform was added to each sample followed by vigorous shaking and a 3-minute incubation at room temperature. The samples were then centrifuged at 12,000g for 15 minutes at 4°C and th e aqueous phase was transferred to a new tube with 500mL of isopropanol and 10μg of GlycoBlue (ThermoFisher Scientific). The samples were vortexed, incubated at room temperature for 10 minutes, and then centrifuged at 12,000g for 15 minutes at 4°C. The supernatant was removed, and the pellet washed in 75% ethanol then centrifuged at 7,500g for 5 minutes at room temperature. The supernatant was once again removed, and the pellet was air-dried at room temperature for 10 minutes before resuspension in 50μL of water. The samples were then further purified and treated with DNase using the RNeasy Column kit (Qiagen) and eluted in 30μL of water.
RNA-seq library preparation and deep sequencing
The concentration and quality of each sample was assessed on a Bioanalyzer (Agilent), with all samples having a minimum RNA Quality Number of 8.0 and 28S/18S ratio of 1.0. 4μg of each sample of RNA was shipped to BGI for library preparation and sequencing. Polyadenylated RNAs were selected using oligo dT beads and then fragmented. N6 random primers were then used to reverse transcribe the library into double-stranded cDNA. A minimum of 20 million single-end 50bp reads were then generated with the BGISEQ-500 platform.
RT-qPCR
The concentration of each sample was assessed on a NanoDrop instrument. 1μg of RNA was reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (ThermoFisher Scientific). To measure the quantity of select mRNAs, 25μL samples were prepared using 2μL of cDNA, 0.2mM of forward and reverse primer for each pair, and SYBR Green qPCR Master Mix (Bimake) according to the manufacturer’s guidelines. Measurements were made on a 7300 Real-Time PCR System (Applied Biosystems).
Generation of NF-YA Antiserum
Zebrafish NF-YA antiserum was prepared by ABClonal Technology. DNA encoding amino acids 1–328 of zebrafish NF-YA was cloned into the vector pET-28a-SUMO, containing a 12aa SUMO tag and a 6aa His tag. The vector was transformed into the E. coli Rosetta strain and the antigen peptide was induced with 0.8mM IPTG at 37°C for 4 hours. Small-scale antigen expression was confirmed by Western blot, showing a band at ~58kDa corresponding to the peptide. The peptide was purified, appearing in both the supernatant and inclusion bodies. The concentration in the supernatant was 2mg/mL, which was deemed appropriate for immunization. Two rabbits were used for immunization and serum was collected on Day 52. The antiserum was tested by ELISA and deemed of sufficient quality with an OD450 > 0.4 at a 1:64,000 dilution. The antibody was purified via antigen affinity purification, with the polyclonal antibody concentration from animal #E7260 at 4.25mg/mL and from animal #E7621 at 4.66mg/mL. The antibodies were tested via Western blot at a 1:1000 dilution with 10, 5, 1, and 0.5ng of antigen. Bands of ~60kDa were observed for antibodies from both animals at all four antigen concentrations.
ChIP-seq
Groups of ~5,000 embryos (for Pbx4) and ~10,000 embryos (for NF-YA) were collected at 3.5hpf and dechorionated in 1X pronase. The embryos were then dissociated by pipette, fixed in 2% formaldehyde in PBS for 10 minutes at room temperature, quenched with 125mM glycine, and flash-frozen in liquid nitrogen. Processing of cell pellets followed the protocol previously described [36]. Nuclei were isolated in L1 Buffer (50mM Tris-HCl pH 8.0, 2mM EDTA, 0.1% NP-40, 10% glycerol, 1mM PMSF) then lysed in SDS Lysis Buffer (50mM Tris-HCl pH 8.0, 10mM EDTA, 1% SDS). Chromatin was sheared to an average length of 300bp using a Palmer immersion sonicator (Three 1-minute rounds of 10s on/2s off at 40% amplitude) and diluted 1:10 in ChIP Dilution Buffer (50 mM Tris-HCl pH8.0, 5 mM EDTA, 200 mM NaCl, 0.5% NP-40, 1 mM PMSF). The samples were pre-cleared with 50μL of Protein A Dynabeads (ThermoFisher Scientific) at 4°C for 3 hours, then Input samples were set aside and stored at −80°C. Next, 10μL of the appropriate antiserum was added (anti-Pbx4 or anti-NF-YA) and the samples were incubated rotating at 4°C overnight. The immune complexes were precipitated onto 50μL of Protein A Dynabeads, which were washed five times with Wash Buffer (20 mM Tris-HCl pH8.0, 2 mM EDTA, 500 mM NaCl, 1% NP-40, 0.1% SDS, 1 mM PMSF), three times with LiCl Buffer (20 mM Tris-HCl pH8.0, 2 mM EDTA, 500 mM LiCl, 1% NP-40, 0.1% SDS, 1 mM PMSF), and three times with TE Buffer (10 mM Tris-HCl pH8.0, 1 mM EDTA, 1 mM PMSF). To elute chromatin, the beads were incubated in 50μL of fresh Elution Buffer with shaking at 1,500 RPM for 15 minutes at 25°C then 15 minutes at 65°C. To reverse crosslinks, 2μL of 5M sodium chloride was added to the samples, which were then incubated at 65°C overnight. Purification of the DNA was accomplished using the MicroChIP Dia Pure Column kit (Diagenode) according to the manufacturer’s guidelines with an 11μL elution. To quantify the concentration of DNA, 1μL of each sample was passed through the dsDNA HS Assay (ThermoFisher Scientific) according to the manufacturer’s guidelines and quantified on a Qubit device.
ChIP-seq library preparation and deep sequencing
ChIP-seq libraries were prepared using the MicroPlex Library Preparation Kit v2 (Diagenode) according to the manufacturer’s guidelines. The entirety of each ChIP sample was used and Input samples were either diluted to the same concentration as their corresponding ChIP sample or, if the concentration of the corresponding ChIP sample was below the Qubit’s range, diluted to 0.2 ng/μL. Following library synthesis, an Illumina HiSeq4000 Sequencer was used to sequence the libraries.
E1b-GFP-Tol2 cloning
Putative enhancers of ~500bp centered on Prep1 peaks near DECA sites and CCAAT boxes were amplified via PCR from 24hpf wild-type zebrafish genomic DNA using specific primers with XhoI sites (tcf3a, tle3a, dachb, fgf8a, pax5, her6, prdm14) or BglII sites (yap1) flanking either end (Table 1). The fragments were ligated into the E1b-GFP-tol2 [37, 38] empty backbone digested with XhoI or BglII and transformed into competent DH5alpha E. coli cells (New England Biolabs). The amplified vector was validated by Sanger sequencing and purified using the Plasmid Maxi Kit (Qiagen).
TABLE 1:
Primer sequences used to amplify putative enhancers from zebrafish genomic DNA.
| Primer | Sequencea |
|---|---|
| Tcf3a-enh1F1 | ATGCCTCGAGTACTGCGTTAATCGCGCGTT |
| Tcf3a-enh1R1 | ATGCCTCGAGGTTAGTGTGATATAATCTGT |
| Tle3a-enh1F1 | ATGCCTCGAGGAAAAAAATAGATGACATTAC |
| Tle3a-enh1R1 | ATGCCTCGAGGCTAGCGCTGGGAATACGA |
| Dachb-enh1F1 | ATGCCTCGAGCGGTTTCTTTGCCATTCTTT |
| Dachb-enh1R1 | ATGCCTCGAGAACTAAGAACAATGTACG |
| Fgf8a-XhoIenhF1 | ATGCCTCGAGGGAGGTCGTTTGCGTATTTG |
| Fgf8a-XhoIenhR1 | ATGCCTCGAGCTTGTCAATCCACCCTGCTT |
| Yap1-enhF1 | |
| Yap1-enhR1 | |
| Pax5-enh1F1 | ATGCCTCGAGGCAAACGGATATTTTAAAAT |
| Pax5-enh1R1 | ATGCCTCGAGGTGCGTAAAAATCCAAGTAA |
| Her6-enh1F1 | ATGCCTCGAGTTCTTTTATAATTGTACTG |
| Her6-enh1R1 | ATGCCTCGAGTGATGTAAATAGAAATACTG |
| Prdm14-XhoIenhF1 | ATGCCTCGAGCCCCTCTTCTTTGTCCCTTG |
| Prdm14-XhoIenhR1 | ATGCCTCGAGGGTAGGCTATCTGGACGGATAAT |
Each primer contains an XhoI site (CTCGAG) to allow it to be cloned into the E1b-GFP-Tol2 vector with the exception of yap1, which contained BglII sites (AGATCT).
Generation of pTransgenesis donor vectors
Mutant enhancers were generated by changing DECA sites contained within each enhancer to the sequence CGGTTGGTGC, which has been shown to prevent TALE binding [39], and CCAAT boxes to the sequence ATGCG. Both mutant and wild-type versions of each enhancer were generated using gBlock technology (Integrated DNA Technologies; Table 2). Due to limitations in gBlock synthesis, a 34bp AT-rich region at the 3’ end of the tcf3a enhancer could not be included compared to the E1b-GFP-tol2 version. A-tails were added to each end of the gBlock fragments using OneTaq Hot Start DNA Polymerase (NEB) (50ng of gBlock DNA, 1 unit of OneTaq Hot Start DNA Polymerase, 1X OneTaq Standard Reaction Buffer, 0.05mM dATP, 1.5mM MgCl2) and incubating the samples at 70°C for 30 minutes. 1μL of A-tailed gBlock fragment solution was then cloned into the pCR8 vector using the pCR8/GW/TOPO TA Cloning Kit (ThermoFisher Scientific) according to the manufacturer’s guidelines. The product was transformed into TOP10 chemically competent cells, validated by Sanger sequencing, and then purified using the Plasmid Midi Kit (Qiagen).
Table 2:
Sequences of gBlocks used to generate pTransgenesis vectorsa.
| tcf3a-WT enhancer TACTGCGTTAATCGCGCGTTTACTTTGATATTTAATCCACAACCAACACAATTAAAACGCCAAACATCAGC GACGACAGTATATGTAACTTTATCCTGATATTTCCCGATTGTGCTTTAAATCACGCAGTACTAGACTCGCG CGCGGAATGACACGACGCACTGTTGAAGAGCGATGGACTGAGAAAAAGTGCGAGATGGCACGATAGA CCCACTGAGCGGACCAATAGCGATCGGGGAAAGTTTGATTGACGTATTCGGTGGCCAATCGAAGATCGT GTTAACACGAAAGCCAAGCCTCTCTTCCATGCACACCCTAGCCAGGTTTTAAAAGAATGGCAACAGGAA GCCATGGAATACTGTTGTGTTTTGTTGTTTGGTAAATGCTAATGTTTACCGCTAACCGCTCAAACTAACTT CAAATGAATTCGACTCGAAACATAACATTGTTATTATTACATTTAGAC |
| tcf3a-mut enhancer TACTGCGTTAATCGCGCGTTTACTTTGATATTTAATCCACAACCAACACAATTAAAACGCCAAACATCAGC GACGACAGTATATGTAACTTTATCCTGATATTTCCCGATTGTGCTTTAAATCACGCAGTACTAGACTCGCG CGCGGAATGACACGACGCACTGTTGAAGAGCGATGGACTGAGAAAAAGTGCGAGATGGCACGATAGA CCCACTGAGCGGAATGCGAGCGATCGGGGAAAGTTCGGTTGGTGCATTCGGTGGATGCGCGAAGATCG TGTTAACACGAAAGCCAAGCCTCTCTTCCATGCACACCCTAGCCAGGTTTTAAAAGAATGGCAACAGGA AGCCATGGAATACTGTTGTGTTTTGTTGTTTGGTAAATGCTAATGTTTACCGCTAACCGCTCAAACTAACT TCAAATGAATTCGACTCGAAACATAACATTGTTATTATTACATTTAGAC |
| tle3a-WT enhancer ATAGATGACATTACCAGGACTGTATTGTTATATGGGTAACATGCGATTATGAGTGAGGGCTTTTTTTAAT GTTATTAAGTGTTTGCATGCTCCTTTGCTCCTTTGTTTTATGTAAGGCTCTCATTACCACGTGGTAGTAAC AGATTGTTTGAACTGGAAAGAAAAGCCATTCGAAGCTAATTAAGCAGCCATTCCAGGCACTATTCACGG GCAGAAGAGCGAGAAGCACAGGCATTTGTCAGCGCTTGACCCCGCGTGGTATTGATTGACAACAAACCT TCTTGAATGACAGCCTTAACCTTTCCCGTCCAATTGCAGTCGAGAGAATATAGATGCTGCTCTGCGATTG GCTGAGAAGCTGTAAAGCCGCAAAGGGATCCCACGTGGGTGCAGCAGAAGAAACGGCACAGGATTGG CCGCTTCTTCTGAGTTCAGACATGGCCGTTGTTCACGGAGATCAAACCTGAACAATCATCGTATTCCCAG CGCTAGC |
| tle3a-mut enhancer ATAGATGACATTACCAGGACTGTATTGTTATATGGGTAACATGCGATTATGAGTGAGGGCTTTTTTTAAT GTTATTAAGTGTTTGCATGCTCCTTTGCTCCTTTGTTTTATGTAAGGCTCTCATTACCACGTGGTAGTAAC AGATTGTTTGAACTGGAAAGAAAAGCCATTCGAAGCTAATTAAGCAGCCATTCCAGGCACTATTCACGG GCAGAAGAGCGAGAAGCACAGGCATTTGTCAGCGCTTGACCCCGCGTGGTATTGATTGACAACAAACCT TCTCGGTTGGTGCCCTTAACCTTTCCCGTATGCGTGCAGTCGAGAGAATATAGATGCTGCTCTGCGCGCA TCTGAGAAGCTGTAAAGCCGCAAAGGGATCCCACGTGGGTGCAGCAGAAGAAACGGCACAGCGCATCC CGCTTCTTCTGAGTTCAGACATGGCCGTTGTTCACGGAGATCAAACCTGAACAATCATCGTATTCCCAGC GCTAGC |
| sv40 minimal promoter aaagatctGCGATCTGCATCTCAATTAGTCAGCAACCATAGTCCCGCCCCTAACTCCGCCCATCCCGCCCCT AACTCCGCCCAGTTCCGCCCATTCTCCGCCCCATCGCTGACTAATTTTTTTTATTTATGCAGAGGCCGAGG CCGCCTCGGCCTCTGAGCTATTCCAGAAGTAGTGAGGAGGCTTTTTTGGAGGCCTAGGCTTTTGCAAAA AGCTTGGCATTCCGGTACTGTTGGTAAAggatccaa |
Sequences of gBlocks encoding wildtype and mutant tcf3a enhancers, wildtype and mutant tle3a enhancers, and the minimal SV40 promoter. Green highlights indicate wildtype DECA sites, blue indicate wildtype CCAAT boxes, purple indicate mutant DECA sites and red indicate mutant CCAAT boxes.
Generation of pTransgenesis vectors
pTransgenesis vectors were assembled using the LR Clonase II Plus enzyme mix (ThermoFisher Scientific). Four cassettes were assembled in one reaction, with gamma-crystallin:venusGFP as the p1 cassette (European Xenopus Resource Center (EXRC)), gBlock enhancers in pCR8 as the p2 cassette and Tol2/I-SceI-CH4-SAR/I-SceI/Tol2/P-element (EXRC) as the pDest-4 cassette. The p3.13 cassette was generated by ligating a BamHI/Bglll-digested gBlock (containing the SV40 minimal promoter; Table 2) into BamHI-digested p3.13 Katushka-RFP plasmid (EXRC). 10fmol of each of the p1, p2, and p3 cassettes were combined with 20fmol of p4 cassette and 2μL of LR Clonase II Plus enzyme mix for the LR reaction in 10μL. The reaction was incubated at 25°C for 16 hours then treated with Proteinase K at 37°C for 10 minutes. 2μL of LR reaction product was transformed into Top10 chemically competent cells, validated by Sanger sequencing, and then purified using the Plasmid Maxi Kit (Qiagen).
Generation and observation of transgenic animals
Injection mixes containing 100ng/μL of E1b-GFP-Tol2 or pTransgenesis vector, 100ng/μL of Tol2 mRNA, and 0.1% phenol red were injected into wild-type zebrafish embryos at the 1-cell stage. The animals were observed for transient fluorescence for the first week, then raised to adulthood. Mature fish were crossed with wild-type fish and the offspring were observed for fluorescence. For E1b-GFP-Tol2 fish, GFP was observed as early as 18hpf. For pTransgenesis fish, RFP expression and GFP expression overlap was best observed at 32hpf, with RFP being apparent sooner and disappearing by ~48hpf while GFP persisted after that time. Thus, any fish that appeared to be RFP+/GFP- were separated and observed for GFP expression at a later timepoint.
Quantification and Statistical Analysis
RNA-seq analysis
RNA-seq analysis was performed using the University of Massachusetts Medical School Dolphin web interface. Ribosomal RNA reads were filtered out and FastQC was used to assess the quality of the remaining reads. RSEM_v1.2.28 with parameters -p4 –bowtie-e 70 --bowtie-chunkmbs 100 [40] was used to align the reads to the DanRer10 zebrafish transcriptome and normalize gene expression to transcripts per million (TPM). This revealed that PBCAB replicate 2 underperformed relative to the other samples and was excluded from further analysis. DeSeq2 [41] was used to identify differentially-expressed genes between three independent biological replicates of 12hpf embryos injected with GFP and three independent biological replicates of 12hpf embryos injected with NF-YA DN or between three independent biological replicates of 1 hpf embryos injected with GFP and two independent biological replicates of 12hpf embryos injected with PBCAB. DEBrowser was used to identify outliers among the replicates. To compensate for the exclusion of one replicate in GFP versus PBCAB analysis, only differentially expressed genes with a p-adj ≤0.01 (Benjamini and Hochberg FDR) were considered for analysis.
ChIP-seq Data Processing
All eight ChIP-seq fastq files (two independent 3.5hpf Pbx4 biological replicates, two independent 3.5hpf NF-YA biological replicates, and matched input DNA controls for each) contained 76bp paired-end sequences. The raw sequence quality was assessed with FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and Fastq-screen (https://www.bioinformatics.babraham.ac.uk/projects/fastq_screen/). Next, remaining adapter reads were filtered out and poor-quality 3’ end sequences were trimmed with Trimmomatic version 0.36 [42] using default parameters for ILLUMINACLIP and SLIDINGWINDOW and MINLENGTH set to 50bp. Using Bowtie2 version 2.2.3 [43], the processed reads were then mapped to UCSC browser zebrafish genome release GRCz10 (danRer10/September 2014) [44], and the mapped reads were further filtered with SAMtools view version 0.1.19 [45] (with flags used -f 2 -q30) to remove reads with poor mapping quality and discordant mapped read pairs. To call peaks, the data, excluding reads that mapped to the mitochondrial genome and unassembled contigs in the assembly, was next passed through MACS2 version 2.1.0.20140616 [46] with the q-value threshold set to 0.05 and default parameters except that the effective genome size was set to 1.03e9 (this equates to 75% of the total genome sequence, excluding ‘N’ bases).
ChIP-seq Analysis
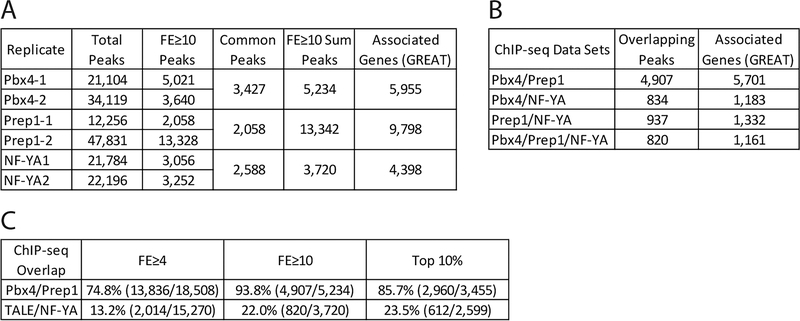
Since the biological replicates for each factor demonstrated robust overlap, the sum of the two replicates was used for all subsequent analyses, by including all peaks meeting the selected cutoff in at least one of the biological replicates. Three different cutoffs were considered: all peaks with a fold enrichment (FE) ≥ 10, all peaks with a FE ≥ 4, and the top 10% of all peaks in each data set. The FE ≥ 10 cutoff showed the highest overlap between Pbx4 and Prep1 peaks as a percentage of the total Pbx4 peaks and was selected as the best cutoff for ChIP-seq analysis (Figure 5C). For a larger set of peaks, FE ≥ 4 peaks were considered for comparison to RNA-seq data (Figure 6).
Figure 5: Identification of genomic binding sites for NF-Y and Pbx4 in 3.5hpf zebrafish.
(A) Table showing data for Pbx4 and NF-YA ChIP-seq biological replicates with our previous Prep1 ChIP-seq data REF included as reference. (B) Table showing number of peaks that overlap (at least 1bp shared between 200bp fragments centered on peaks) between Prep1, Pbx4 and NF-YA ChIP-seq data sets. Only peaks with a 10-fold or greater enrichment over input were considered. (C) Table showing extent of overlap of Pbx4 peaks with Prep1 peaks and TALE peaks with NF-YA peaks at three different cutoffs (FE≥4, FE≥10 and top 10% of peaks).
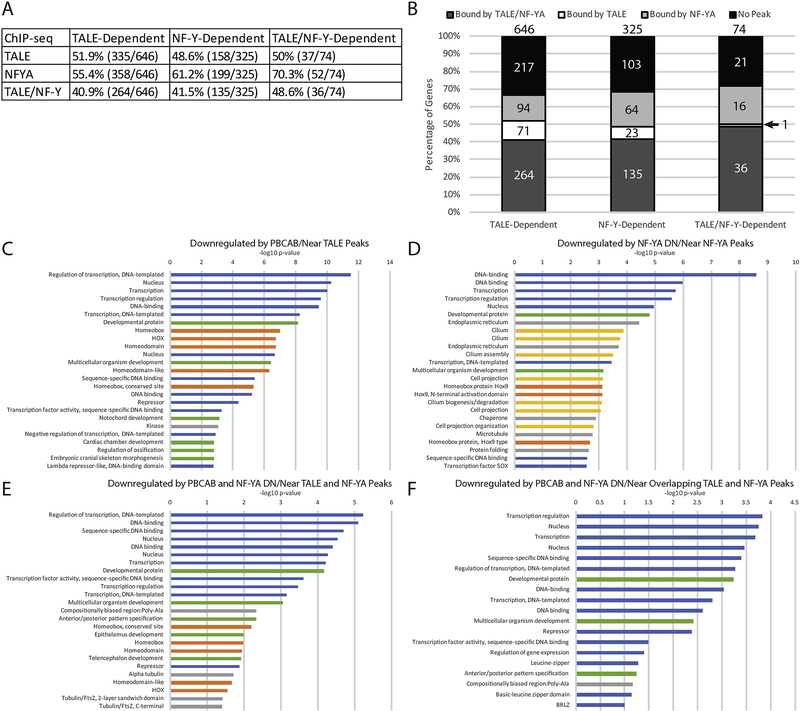
Figure 6: Combinatorial function of NF-Y and TALE defines distinct gene expression programs.
(A) Table showing correlation between NF-Y and/or TALE-dependent genes and binding by the corresponding TF at a nearby site (ChIP peaks enriched by 4-fold or greater over input were considered). (B) Graphical breakdown of NF-Y and/or TALE occupancy near NF-Y and/or TALE-dependent genes. (C-F) Top 25 GO terms returned by DAVID for TALE-dependent genes associated with TALE peaks (C), NF-Y dependent genes associated with NF-YA peaks (D), TALE/NF-Y dependent genes associated with both NF-YA and TALE peaks (E), and TALE/NF-Y dependent genes associated with overlapping TALE and NF-YA peaks (F). Blue bars correspond to transcription-related, green to embryogenesis-related, orange to homeodomain-related, yellow to cilia-related, and gray bars to other ontologies.
ChIP peak overlap analysis
In the text, we use the term ‘overlap’ to indicate peaks identified as follows: ChIP peaks shared between different data sets were identified with the Intersect tool and exclusive peaks were identified using the Subtract tool in Galaxy [47]. All coordinates used were 200bp in length centered on peak summits and considered overlapping if they shared one or more base pairs.
qPCR Analysis
ddCt values were calculated from raw Ct values according to the formula 0.5Ct. Average ddCt values were then calculated by taking the mean of all three biological replicates. The ddCt of each GFP replicate was then normalized to the average gapdh ddCt according to the formula ddCtGFP/average ddCtgapdh and then the mean of the normalized values was determined. Error bars were calculated based on the standard deviation of the three normalized GFP replicates in Excel. To determine whether the dominant negative conditions were significantly different from the control condition, an unpaired t-test was used in Excel, with p-values < 0.05 considered significant.
Determination of nearest genes to ChIP peaks
The number of Ensembl zebrafish transcription start sites within 5kb or 30kb of the summit of ChIP peaks was determined using the bedtools suite [48] in the Galaxy toolshed [47]. ChIP peak coordinates in danrer10 were converted to danrer7 (Zv9) using the LiftOver tool in the UCSC browser. The identities of genes near ChIP peaks were determined by the GREAT software version 3.0.0 [49, 50] using the he default settings of basal plus extension with proximal set to 5kb upstream and 1kb downstream and distal set to 1,000kb.
GO term analysis
Gene ontology (GO) terms enriched within different sets of genes were determined using DAVID version 6.8 [51, 52]. GO terms were ranked according to the EASE score, which was calculated based on a modified Fisher’s exact p-value, and graphed as the −log10 of that value.
DNA binding motif analysis
Significantly enriched binding motifs were identified using MEME and DREME within the MEME-Suite version 4.11.1 [53, 54]. Both MEME and DREME were run according to their default settings. CENTRIMO was also run with default settings to determine the frequency of discovered motifs within ChIP peaks.
Chromatin feature analysis
Version 2.0 of the Deeptools [55] toolset in the Galaxy toolshed was used to create mean score profiles and heatmaps. Using the computeMatrix tool with region inputs of BED files containing ChIP coordinates and sample inputs of wiggle files from previously published data sets downloaded from GEO (Key Resources Table), signal matrices were generated in reference-point mode with the center set as the reference point. The distance upstream of the start sites and downstream of the end sites were set to 1000bp with a bin size of 25bp and ranked by mean signal when necessary. Heatmaps and profiles were generated from the matrices using the plotHeatmap and plotProfile tools respectively. The previously-published H3K27ac, H3K4me1, H3K4me3, and MNase data sets were all performed at 4.3hpf, which is somewhat later than the Pbx4, NF-YA, and Prep1 ChIP-seq experiments performed at 3.5hpf; however, asynchronous development in zebrafish embryos and large sample sizes make considerable overlap likely.
Key Resources Table.
| Reagent or Resource | Source/Reference | Identifier |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-zebrafish Pbx | [77] | N/A |
| Rabbit polyclonal anti-zebrafish NF-YA | This paper | N/A |
| Bacterial and Virus Strains | ||
| Subcloning Efficiency DH5α competent cells | ThermoFisher Scientific | 18265–017 |
| OneShot Top10 chemically competent cells | ThermoFisher Scientific | C404003 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Protein-A Dynabeads | ThermoFisher Scientific | 10001D |
| NotI | New England Biolabs | R0189S |
| XhoI | New England Biolabs | R0146S |
| BglII | New England Biolabs | R0144S |
| Critical Commercial Assays | ||
| mMESSAGE mMACHINE SP6 Transcription Kit | ThermoFisher Scientific | AM1340 |
| RNeasy Mini Kit | Qiagen | 74104 |
| DIG DNA Labeling Mix | Millipore Sigma | 11277065910 |
| Trizol | ThermoFisher Scientific | 15596026 |
| GlycoBlue | ThermoFisher Scientific | AM9515 |
| MicroPure DiaChIP Columns | Diagenode | C03040001 |
| dsDNA HS Assay | ThermoFisher Scientific | Q32851 |
| MicroPlex Library Preparation Kit v2 | Diagenode | C05010012 |
| OneTaq Hot Start DNA Polymerase | New England Biolabs | M0481L |
| pCR8/GW/TOPO | ThermoFisher Scientific | 45–0642 |
| Plasmid Midi/Maxi Kit | Qiagen | 12143/12163 |
| LR Clonase II Plus Enzyme Mix | ThermoFisher Scientific | 12538–120 |
| High Capacity cDNA Reverse Transcription Kit | ThermoFisher Scientific | 4368814 |
| SYBR Green qPCR Master Mix | Bimake | B21203 |
| Deposited Data | ||
| Pbx4 ChIP-seq and Inputs in 3.5 hpf zebrafish embryos | This paper | E-MTAB-8137 |
| NF-YA ChIP-seq and Inputs in 3.5 hpf zebrafish embryos | This paper | E-MTAB-8137 |
| PBCAB and GFP RNA-seq in 12 hpf zebrafish embryos | This paper | GSE133459 |
| NF-YA DN and GFP RNA-seq in 12 hpf zebrafish embryos | This paper | GSE133459 |
| Prep1 ChIP-seq and Inputs in 3.5 hpf zebrafish embryos | [23] | E-MTAB-5967 |
| H3K27ac ChIP-seq in dome zebrafish embryos, WIG files | [79] | GSM915197 |
| H3K4me1 ChIP-seq in dome zebrafish embryos, WIG files | [79] | GSM915193 |
| H3K4me3 ChIP-seq in dome zebrafish embryos, WIG files | [79] | GSM915189 |
| H3K27ac ChIP-seq in 80% epiboly zebrafish embryos, WIG files | [79] | GSM915198 |
| H3K4me1 ChIP-seq in 80% epiboly zebrafish embryos, WIG files | [79] | GSM915194 |
| H3K4me3 ChIP-seq in 80% epiboly zebrafish embryos, WIG files | [79] | GSM915190 |
| MNase-seq in 4.5hpf zebrafish embryos, WIG files | [80] | GSM1081554 |
| Experimental Models: Organisms/Strains | ||
| Strain EKW | Ekkwill breeders | http://www.ekkwill.com/ |
| Oligonucleotides | ||
| Gapdh forward primer TGCTGGTATTGCTCTCAACG | N/A | |
| Gapdh reverse primer AACAGCAAAGGGGTCACATC | N/A | |
| GFP forward primer ATGGTGAGCAAGGGCGAGGAG | N/A | |
| GFP reverse primer TTACTTGTACAGCTCGTCCATG | N/A | |
| Recombinant DNA | ||
| NF-YA DN in pCS2+ | [23] | N/A |
| PBCAB in pCS2+MT | [33] | N/A |
| tcf3a element in E1b-GFP-Tol2 | This paper | N/A |
| tle3a element in E1b-GFP-Tol2 | This paper | N/A |
| dachb element in E1b-GFP-Tol2 | This paper | N/A |
| fgf8a element in E1b-GFP-Tol2 | This paper | N/A |
| yap1 element in E1b-GFP-Tol2 | This paper | N/A |
| pax5 element in E1b-GFP-Tol2 | This paper | N/A |
| her6 element in E1b-GFP-Tol2 | This paper | N/A |
| prdm14 element in E1b-GFP-Tol2 | This paper | N/A |
| tcf3a element in pCR8 | This paper | N/A |
| tle3a element in pCR8 | This paper | N/A |
| Mutant tcf3a element in pCR8 | This paper | N/A |
| Mutant tle3a element in pCR8 | This paper | N/A |
| pTransgenesis p1 gamma-crystallin::venus GFP | [81] | N/A |
| pTransgenesis p3 sv40 minimal promoter:: Katushka RFP | This paper | N/A |
| pTransgenesis pDest4 Tol2/I-SceI-CH4-SAR/I-SceI/Tol2/P-element | [81] | N/A |
| Software and Algorithms | ||
| FastQC | Babraham Institute | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ RRID:SCR_014583 |
| FastQ Screen | Babraham Institute | https://www.bioinformatics.babraham.ac.uk/projects/fastq_screen/ RRID:SCR_000141 |
| Trimmomatic v0.32 | [42] | http://github.com/timflutre/trimmomatic RRID:SCR_011848 |
| Bowtie v2.2.3 | [43] | http://github.com/BenLangmead/bowtie2 RRID:SCR_005476 |
| SAMtools v0.1.19 | [45] | http://github.com/samtools/samtools RRID:SCR_002105 |
| MACS v2.1.0.20140616 | [46] | http://github.com/taoliu/MACS |
| RSEM v1.2.28, Dolphin, Biocore, University of Massachusetts Medical School | [40] | http://www.umassmed.edu/biocore/introducing-dolphin/ RRID:SCR_013027 |
| DESeq2, Dolphin, Biocore, University of Massachusetts Medical School | [41] | http://www.umassmed.edu/biocore/introducing-dolphin/ RRID:SCR_015687 |
| DEBrowser v v1.12.2 | [82] | http://github.com/UMMS-Biocore/debrowser |
| Galaxy web interface | [47] | http://usegalaxy.org RRID:SCR_006281 |
| BedTools, Galaxy | [48] | http://usegalaxy.org RRID:SCR_006646 |
| DeepTools, Galaxy | [55] | http://usegalaxy.org |
| MEME-ChIP | [53, 54] | http://meme-suite.org/tools/meme-chip RRID:SCR_00178 |
| DAVID v6.8 | [51, 52] | http://david.ncifcrf.gov/ RRID:SCR_001881 |
| GREAT v3.0.0 | [49, 50] | http://bejerano.stanford.edu/great/public/html RRID:SCR_005807 |
Data and Code Availability
RNA-seq data is available in GEO under accession number GSE133459 and ChIP-seq data is available in ArrayExpress under accession number E-MTAB-8137.
RESULTS
NF-Y and TALE TFs are required for formation of anterior embryonic structures
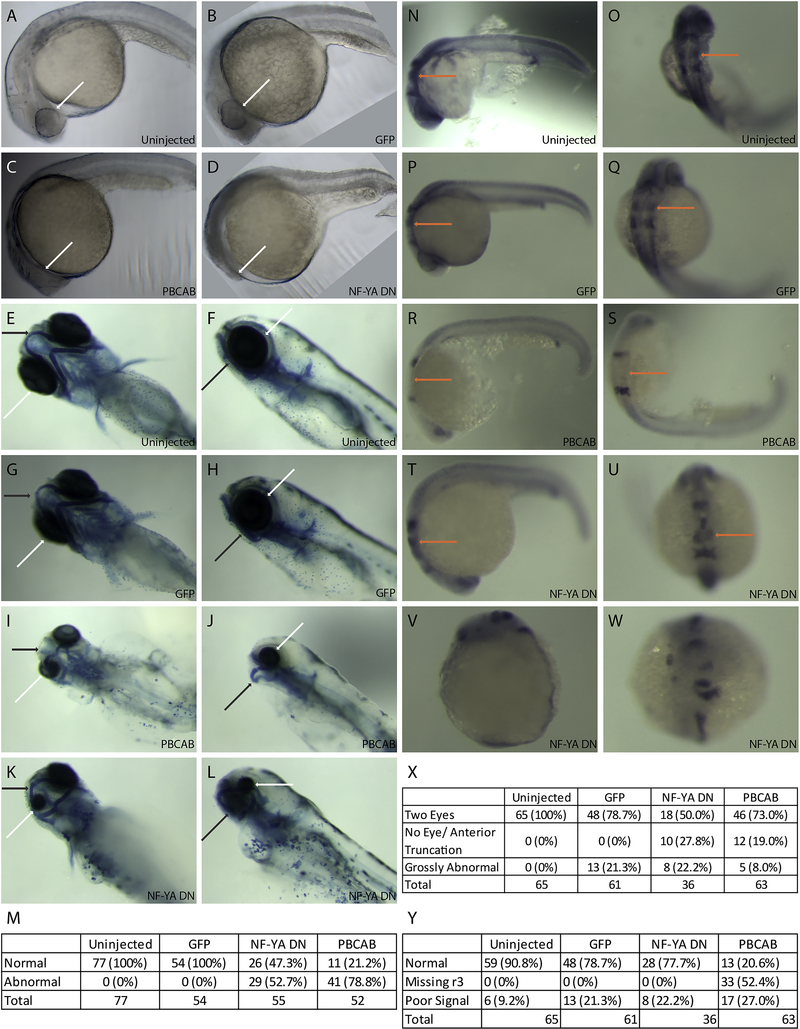
In order to assess the roles of NF-Y and TALE during embryogenesis, we first set out to disrupt their function. Previous work demonstrated that TALE TFs are required for formation of anterior embryonic structures, such that loss of various combinations of TALE factors (using germline mutants, antisense approaches and dominant negative constructs) results in animals with small heads, small (or absent) eyes, cardiac edema and hindbrain defects [23, 33–35, 56–58]. This similarity in phenotypes is likely due to the fact that multiple TALE factors act together in larger protein complexes, which are rendered ineffective when one or more TALE factors are disrupted (reviewed in [59, 60]). In preliminary experiments we recently observed abnormal anterior development also upon disruption of NF-Y function [23], consistent with a previous report using antisense morpholino oligos to target NF-Y in zebrafish [61]. Here we extend this analysis to directly compare disruption of TALE factors (using the dominant negative PBCAB construct reported previously; [33]) to disruption of NF-Y function (using a previously reported NF-YA dominant negative construct; [32]) and find smaller heads in both cases (Figure 1A–D). A more detailed examination revealed abnormal head cartilage formation (53% of animals with disrupted NF-Y and 79% of animals with disrupted TALE function; Figure 1E–M) as well as loss of eyes (28% of animals with disrupted NF-Y and 19% of animals with disrupted TALE function; Figure 1X). Using in situ hybridization to detect expression of pax2 (at the midbrain-hindbrain boundary), krox20 (in rhombomeres 3 and 5 of the hindbrain) and hoxd4 (in the spinal cord) in 24hpf embryos, we observed loss of r3 krox20 expression upon TALE disruption (52% of embryos; as reported previously; [33–35, 56, 57]), but did not detect any effects of NF-Y disruption (Figure N-U, Y). We conclude that both NF-Y and TALE function in formation of the anterior embryo and that TALE TFs have a distinct role in patterning of the hindbrain.
Figure 1: Disruption of NF-Y or TALE function affects anterior embryonic development.
Zebrafish embryos were left uninjected (A, E, F, N, O) or injected with either control mRNA (GFP; B, G, H, P, Q), mRNA encoding a TALE dominant negative construct (PBCAB; C, I, J, R, S) or mRNA encoding an NF-Y dominant negative construct (NF-YA DN; D, K, L, T-W) at the 1–2 cell stage and raised to 24hpf (N-W), 28hpf (A-D) or 5dpf (E-L). Embryos were either left untreated (A-D), stained with alcian blue (E-L) or processed for detection of pax2 (at the mid/hindbrain boundary), krox20 (in rhombomeres 3 and 5) and hoxd4 (in the spinal cord) transcripts by in situ hybridization (N-W). White arrows highlight differences in eye morphology (A-L), black arrows highlight differences in head cartilage formation (E-L) and orange arrows indicate differences in rhombomere 3 krox20 expression (N-U). Tables summarize effects of TALE or NF-Y disruption on head cartilage formation (M), eye formation (X) and gene expression (Y). Panels V and W show representative images of embryos scored as having gross abnormalities in panel X. Embryos are shown in lateral (A-D, F, H, J, L, N, P, R, T, V) or dorsal (E, G, I, K, O, Q, S, U, W) views.
NF-Y and TALE TFs have both shared and independent transcriptional targets
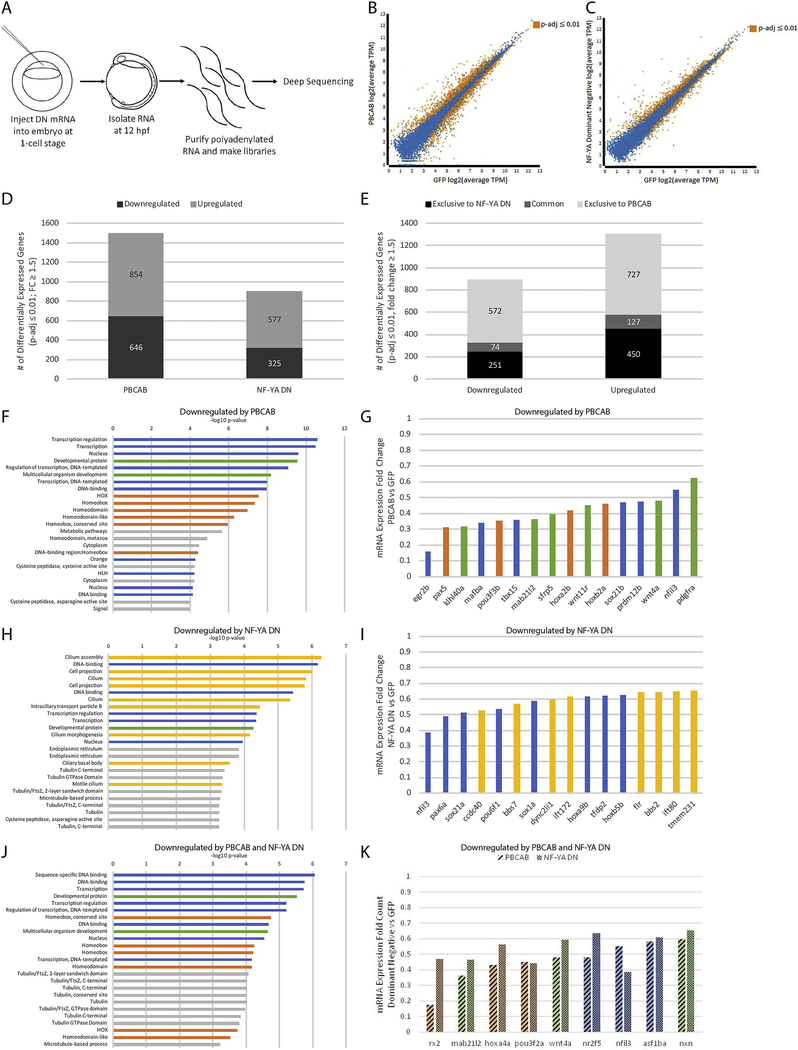
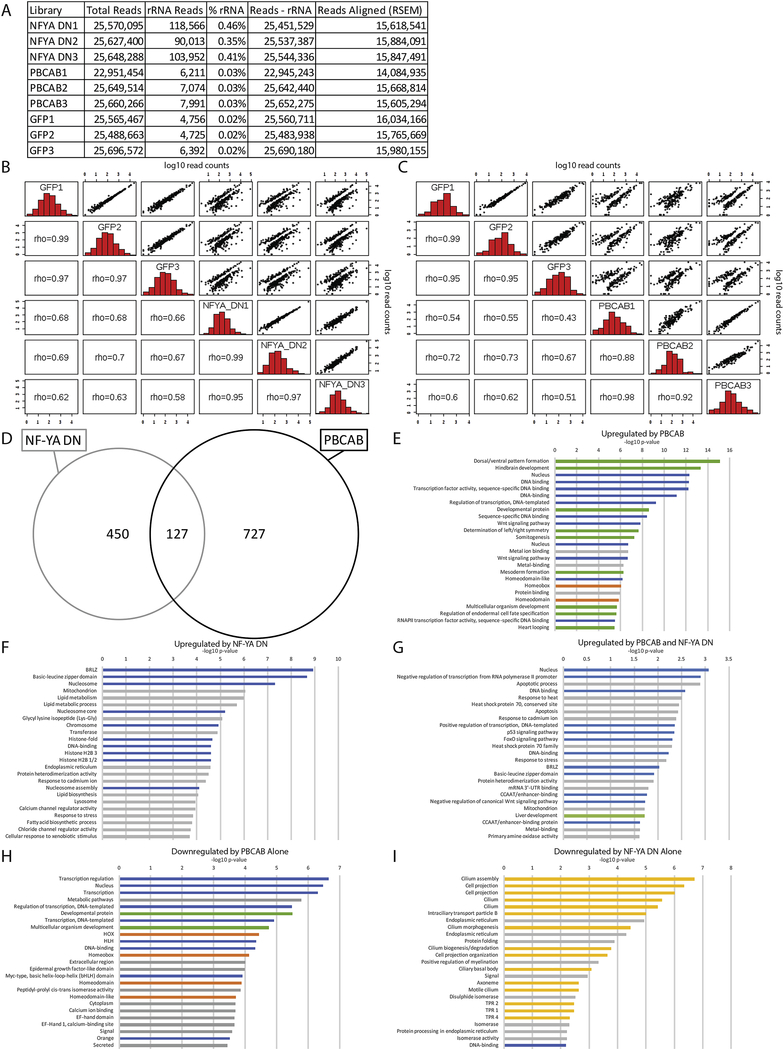
To identify transcriptional targets of NF-Y and TALE, we next carried out RNA-seq at 12hpf of zebrafish development (Figure 2A–C; Figure 3A–C). This time-point was selected in order to ensure broad capture of gene expression changes resulting from disruption of NF-Y and TALE function. We find that disruption of TALE function affects the expression of 1,500 genes (646 downregulated, 854 up-regulated; FC≥1.5, padj≤0.01; Figure 2B, D) at 12hpf. Since TALE factors are thought to act primarily as activators of transcription, we focused on genes downregulated upon disruption of TALE function. Applying the DAVID functional annotation tool, we find that TALE-dependent genes are enriched for functions related to transcription (particularly hox genes) as well as for factors controlling embryogenesis (Figure 2F; GO-terms for genes up-regulated upon disruption of TALE function are shown in Figure 3E). Accordingly, an examination of individual TALE-dependent genes identified members of several classes of TFs and developmental control genes (Figure 2G), consistent with the phenotype observed in figure 1. We next examined the effect of disrupting NF-Y function and find that 902 genes are affected (325 downregulated, 577 upregulated; Figure 2C, D) at 12hpf. An analysis of the GO terms associated with NF-Y-dependent genes revealed high enrichment in functions related to cilia and, to a lesser extent, in genes broadly controlling transcription and development (Figure 2H; GO-terms for up-regulated genes are shown in Figure 3F). In particular, different classes of TFs, as well as both structural and motor proteins found in cilia are downregulated upon disruption of NF-Y function (Figure 2I).
Figure 2: NF-Y and TALE TFs have both shared and independent transcriptional targets.
(A) Schematic of RNA-seq experiments. (B-C) Scatterplots of gene expression in PBCAB vs GFP-injected (B) and NF-YA DN vs GFP-injected (C) zebrafish embryos (expression presented as log2 of average TPM for multiple replicates; see methods). Expression of genes highlighted in orange is significantly different at 12hpf (padj≤0.01; Wald test in DESeq2). (D) Number of genes differentially expressed in PBCAB (left) or NF-YA DN (right) relative to GFP-injected embryos (p-adj ≤ 0.01; fold-change ≥ 1.5). (E) Breakdown of downregulated (left) and upregulated (right) genes exclusive or common to each experimental condition. (F, H, J) DAVID analyses showing the 25 most significant GO terms (EASE Score) associated with genes downregulated by PBCAB (F), NF-YA DN (H), and common to both (J). Blue bars correspond to transcription-related, green to embryogenesis-related, orange to homeodomain-related, yellow to cilia-related, and gray to other ontologies. (G, I, K) Selected genes downregulated by PBCAB (G), NF-YA DN (I), or both (K). Color coding is the same as in (F, H, J).
Figure 3: Identification of NF-Y and/or TALE-dependent genes in zebrafish.
(A) Read counts for the RNA-seq analysis. (B, C) Histograms, scatter plots, and Spearman’s rank correlation coefficient comparing each biological replicate of NF-YA DN with GFP (B) or PBCAB with GFP (C). (D) Venn diagram showing upregulated genes (p-adj ≤ 0.01; FC ≥ 1.5) in embryos injected with PBCAB or NF-YA DN. (E-I) GO terms associated with genes upregulated (p-adj ≤ 0.01, FC ≥ 1.5) by PBCAB (E), upregulated by NF-YA DN (F), upregulated by both PBCAB and NF-YA DN (G), downregulated exclusively by PBCAB (H) or downregulated exclusively by NF-YA DN (I). In E-I, blue bars correspond to transcription-related, green to embryogenesis-related, orange to homeodomain-related, yellow to cilia-related, and gray bars to other ontologies.
Since disruption of either NF-Y or TALE factors produces embryos with shared phenotypes, we next identified genes whose expression is dependent on both NF-Y and TALE function. We find that there are 201 such genes (74 downregulated, 127 upregulated; Figure 2E). Strikingly, the annotation of genes downregulated both upon disruption of TALE function and upon disruption of NF-Y function identifies transcriptional and developmental roles, but not roles associated with cilia (though several terms associated with tubulin function are retained; Figure 2J, K; GO-terms for up-regulated genes are shown in Figure 3G). Lastly, we analyzed genes regulated exclusively by each TF. We find that genes regulated exclusively by NF-Y revealed strong enrichment for cilia terms (Figure 3I), while genes exclusively dependent on TALE function return GO terms enriched for transcriptional regulation such as hox genes (Figure 3H). These results indicate that NF-Y and TALE co-regulate a set of transcriptional and developmental control genes that is distinct from genes regulated by either NF-Y or TALE alone.
NF-Y and TALE TFs broadly occupy genomic elements associated with developmental and transcriptional control genes at zebrafish ZGA
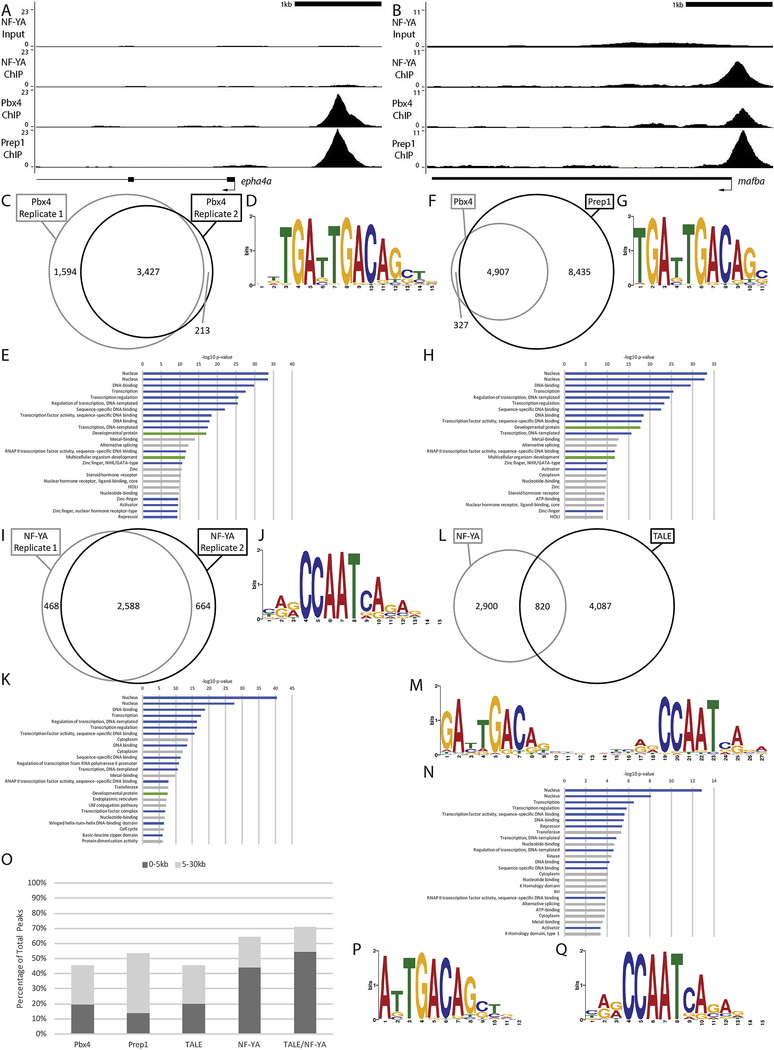
Given that NF-Y and TALE appear to have both shared and independent functions, we next examined binding of these TFs across the zebrafish genome. In order to determine if they have a role at ZGA, we focused our analysis at 3.5hpf – when zygotic genes are becoming active in zebrafish embryos. We previously used chromatin immunoprecipitation (ChIP) to characterize Prep1 occupancy and found that this TF is bound at many genomic elements at ZGA and earlier [23, 24], consistent with reports that TALE factors are maternally transmitted in zebrafish [33, 34, 56]. Specifically, our analysis identified a 10bp motif (TGATTGACAG; termed the ‘DECA motif’; [62, 63]) as the predominant element occupied by Prep1 at ZGA in zebrafish embryos. The DECA motif contains two half-sites – one for Pbx proteins (TGAT) and one for Prep factors (TGACAG) – and Pbx factors are known to form dimers with Prep proteins (reviewed in [59]). Accordingly, using ChIP-qPCR we demonstrated that zebrafish Pbx4 occupied 11 of 12 tested DECA sites in 3.5hpf zebrafish embryos [23]. We have now extended this analysis to the entire zebrafish genome by performing ChIP-seq for Pbx4 at 3.5hpf (Figure 4A–C; Figure 5A). We find that the majority of Pbx4 peaks overlap with a Prep1 peak (94% overlap at FE≥10; Figure 4A, B, F; Figure 5B, C) and that the predominant sequence motif at Pbx4 binding sites is indistinguishable from the DECA motif observed at Pbx4/Prep1 co-occupied sites (Figure 4D, G). We also find that the distribution of Pbx4 peaks relative to TSSs is similar to that for Prep1 (Figure 4O), with ~50% of all binding sites located within 30kb of an annotated promoter element [64] in both cases, and that sites co-occupied by Pbx4/Prep1 show a similar distribution (Figure 4O). GO-term analyses (Figure 4E) revealed that genes associated with Pbx4 binding sites are enriched for the same transcriptional regulation and embryogenesis functions that we previously identified for genes associated with Prep1 bound sites at 3.5hpf [23]. As expected, genes associated with Prep1/Pbx4 co-occupied sites return essentially the same GO terms (Figure 4H). Notably, a large number of Prep1 binding sites do not overlap with Pbx4 peaks (Figure 4F). While this could indicate that Prep1 has functions independent of Pbx4, it may also be a reflection of different affinities of the two antisera. Nevertheless, our observations indicate that Pbx4 binding takes place primarily at DECA sites in the context of Pbx4/Prep1 heterodimers at this stage of embryogenesis. Here we focus on these Prep1/Pbx4 co-occupied sites and refer to them as ‘TALE sites’.
Figure 4: NF-Y and TALE occupy genomic elements associated with developmental and transcriptional control genes at ZGA.
(A-B) Representative UCSC Genome Browser tracks for NF-YA, Pbx4 and Prep1 ChIP-seq analyses at 3.5hpf. (C, F, I, L) Venn diagrams showing the overlap (at least 1bp shared between 200bp fragments centered on peaks) of two Pbx4 ChIP-seq replicates (C), the overlap of Pbx4 and Prep1 ChIP-seq peaks (F), the overlap of two NF-YA ChIP-seq replicates (I) and the overlap of TALE and NF-Y ChIP-seq peaks (L). (D, G, J, M) The top sequence motif returned by MEME for Pbx4-occupied sites (D), Pbx4/Prep1 co-occupied sites (G), NF-YA occupied sites (J) and TALE/NF-YA co-occupied sites (M). (E, H, K, N) The top 25 gene ontology (GO) terms returned by the GREAT analysis tool for genes associated with Pbx4-occupied sites (E), Pbx4/Prep1 co-occupied sites (H), NF-YA occupied sites (K) and TALE/NF-YA co-occupied sites (N). (O) Chart showing percent of ChIP-seq peaks found within 5kB or 30kB of a promoter. (P, Q) Top sequence motif returned by MEME for peaks bound by TALE, but not NF-YA (P) and peaks bound by NF-YA, but not TALE (Q). Only peaks with a 10-fold or greater enrichment over input (FE≥10) were considered for the analyses in C-Q.
We previously reported that approximately 30% of TALE-occupied DECA sites observed at 3.5hpf have a CCAAT sequence motif nearby – usually at a distance of ~20bp [23]. In other systems, such CCAAT boxes serve as binding sites for the heterotrimeric NF-Y transcription factor. Since NF-Y is maternally deposited in zebrafish [61], we previously used a commercial NF-Y antiserum for ChIP-qPCR to test 15 CCAAT boxes located near DECA sites and found that nine were occupied by NF-Y [23]. To examine NF-Y binding genome-wide, we have now raised antiserum to zebrafish NF-YA (the sequence-specific DNA binding component of the NF-Y heterotrimer; see Methods section) and carried out ChIP-seq on 3.5hpf zebrafish embryos (Figure 4A, B, I; Figure 5A). As expected, NF-Y-occupied genomic sites are highly enriched for the CCAAT box sequence motif (Figure 4J; 86% of NF-Y peaks contain a CCAAT box), but we find that the distribution of NF-Y peaks in the genome is somewhat different than the distribution of TALE peaks, such that NF-Y appears to be preferentially bound closer to promoters (Figure 4O). Further, we find that NF-Y-bound genomic elements are associated with genes enriched for functions related to transcriptional regulation and embryogenesis (Figure 4K), similar to the TALE-associated genes.
To further address the potential cooperation between NF-Y and TALE, we next examined if NF-Y and TALE peaks co-localize in the zebrafish genome by determining if 200bp sequences centered on each peak overlapped by at least 1bp (see Methods section). Using this criterium, we find that approximately 22% of the NF-Y occupied sites overlap with a TALE-occupied site (corresponding to 17% of the TALE bound sites; Figure 4A, B, L; Figure 5B, C). Strikingly, motif analyses identified a ~27bp sequence motif encompassing both a DECA motif and a CCAAT box associated with NF-Y/TALE co-occupied sites (Figure 4M; 97% of overlapping peaks contain this extended motif). In contrast, sites occupied by TALE alone display a DECA motif (Figure 4P; 88% of peaks contain the DECA motif) and those occupied by NF-Y alone contain a CCAAT box (Figure 4Q; 79% of peaks contain a CCAAT box). GO terms for genes associated with co-occupied sites are again enriched for functions related to transcriptional control, but less so for functions controlling embryogenesis (Figure 4N). We conclude that, at zebrafish ZGA, NF-Y and TALE individually occupy genomic regions associated with both developmental and transcriptional regulators, but also co-occupy an extended binding motif that appears more selectively associated with transcriptional control genes.
Combinatorial function of NF-Y and TALE defines distinct gene expression programs
Our RNA-seq analysis identified shared and independent targets of NF-Y and TALE, but it is not clear how direct this regulation might be. To address this question, we first examined whether TALE-dependent genes are associated with genomic elements bound by TALE TFs. We find that, of the 646 genes we identified as being TALE-dependent, 52% (335/646) are found near (as defined using default parameters in GREAT; see Methods section) a TALE-occupied element (Figure 6A, B) and these genes are enriched for functions related to embryonic development and transcriptional regulation with a specific emphasis on hox genes (Figure 6C). Similarly, of the 325 genes our RNA-seq analysis showed to be downregulated upon disruption of NF-Y function, we find that 61% (199/325) are near a NF-Y occupied element (Figure 6A, B). The GO terms for these genes are enriched for functions related to transcriptional regulation, as well as for cilia structure/function (Figure 6D). Hence, 50–60% of NF-Y and TALE-dependent genes are associated with a binding site for the corresponding TF and the functional annotations of these genes show specific enrichment for the same terms as we observed in our RNA-seq analysis – cilia structure/function for NF-Y dependent genes and hox TFs for TALE-dependent genes.
To assess co-regulation, we focused on genes dependent on both NF-Y and TALE (as defined in figure 2E, J, K). We find that of the 74 genes in this category, 70% are associated with an NF-Y (52/74) occupied site and 50% (37/74) with a TALE-occupied site (Figure 6A, B). In fact, 49% of co-regulated genes are found near both NF-Y and TALE occupied sites (36/74). Strikingly, the top 25 GO terms of co-regulated genes associated with both NF-Y and TALE-occupied elements are enriched for transcriptional regulation and embryonic development, while hox terms are less represented and cilia terms are absent (Figure 6E). Further, if we specifically focus on genes that are near regulatory elements with overlapping NF-Y/TALE peaks (as defined in Figure 4L–N), we find that their functions converge on transcription and regulation of development, but the categories related to either cilia or hox functions are no longer detected (Figure 6F). Hence, our results indicate that genes co-regulated by NF-Y and TALE act primarily in transcriptional control, while NF-Y and TALE independently control cilia and hox gene expression programs, respectively.
Genomic elements occupied by NF-Y and TALE TFs act as enhancers in vivo
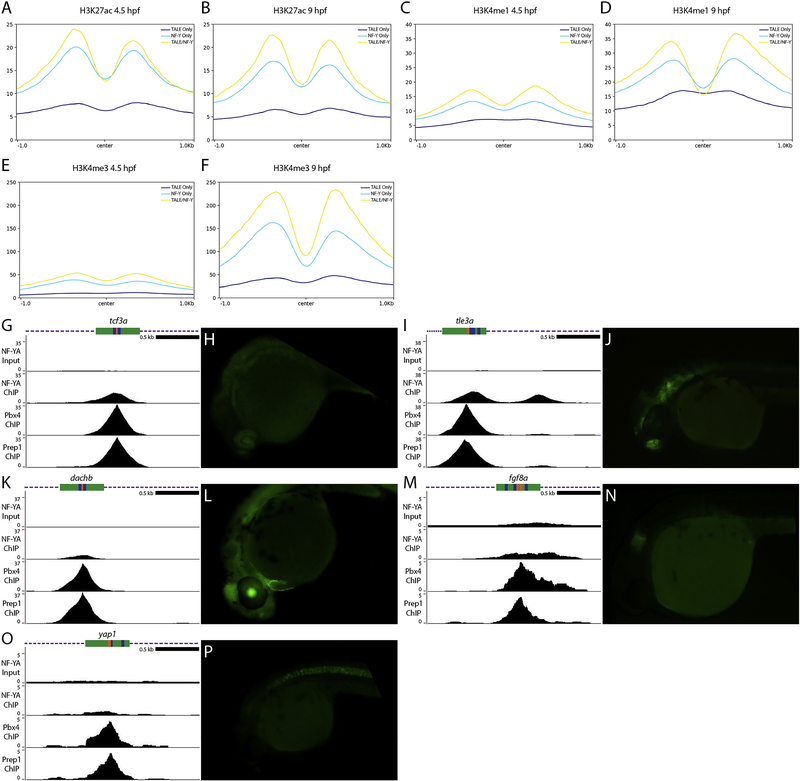
Our data show that many genomic elements occupied by TALE and NF-Y are found near promoters, but TALE TFs are known to act at enhancers [58, 65–70]. Further, while NF-Y was originally identified as acting at promoters (reviewed in [71]), more recent work has revealed an important role for NF-Y at tissue-specific enhancers [27]. To explore these relationships in greater detail, we examined the chromatin state at NF-Y and TALE-occupied elements. We find that both H3K4me1 and H3K27ac (that mark enhancers and promoters) are highly enriched already at 4.3hpf and persist at 9hpf at co-occupied elements (Figure 7A–D). We also note that elements bound by NF-Y alone and, to a lesser extent, TALE alone have the same characteristics. In agreement with NF-Y and TALE occupied elements driving gene expression at this stage of development, we also find a dramatic increase in H3K4me3 modifications (a mark of active promoters) between 4.3hpf and 9hpf (Figure 7E, F).
Figure 7: Genomic elements occupied by NF-Y and TALE act as enhancers in vivo.
(A-F) Average histone mark signals at genomic regions containing only TALE peaks (dark blue), only NF-YA peaks (light blue), or NF-YA/TALE peaks (yellow) for H3K27ac at 4.3hpf (A) and 9hpf (B), H3K4me1 at 4.3hpf (C) and 9hpf (D), H3K4me3 at 4.3hpf (E) and 9hpf (F). (G, I, K, M, O) UCSC Genome Browser tracks showing NF-YA, Pbx4, and Prep1 ChIP-seq data for the tcf3a (G), tle3a (I), dachb (K), fgf8a (M) and yap1 (O) loci. The diagrams above the tracks show the putative enhancer region in green, DECA motifs in orange and CCAAT boxes in blue. (H, J, L, N, P) GFP expression in 24hpf F1 tcf3a:E1b-GFP (H), tle3a:E1b-GFP (J), dachb:E1b-GFP (L), fgf8a:E1b-GFP (N) and yap1:E1b-GFP (P) transgenic embryos resulting from crosses between male founders and wild type females.
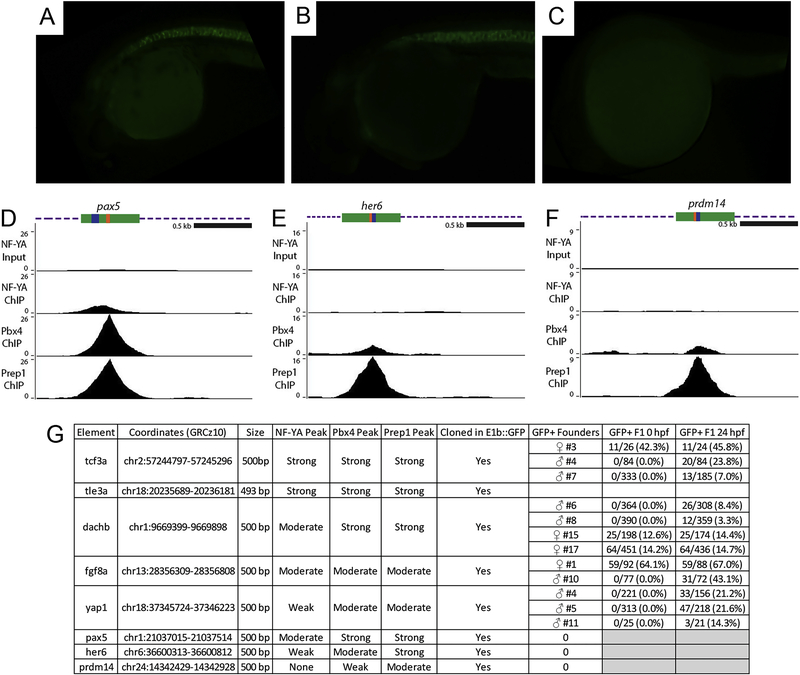
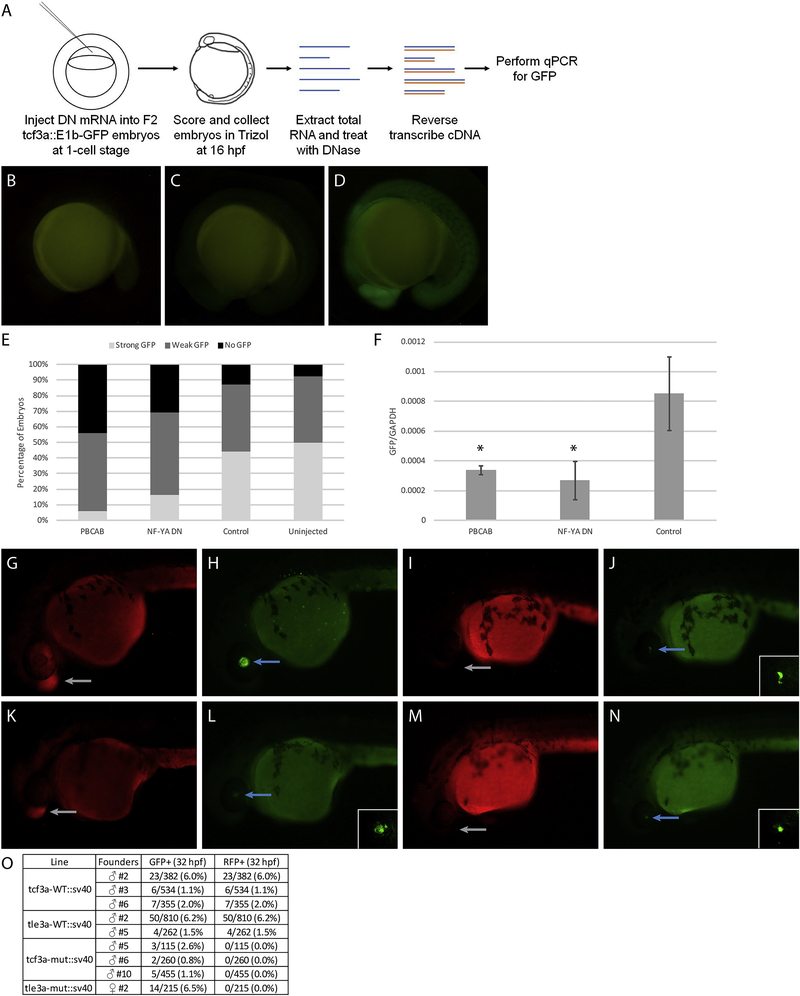
To directly test if NF-Y and TALE-occupied elements act as enhancers in vivo, we used a previously published enhancer assay [37] and inserted individual genomic elements upstream of the E1b minimal promoter and the GFP reporter gene. We selected eight genomic elements that contain adjacent NF-Y/TALE motifs (as in Figure 4L–N) and that are associated with genes expressed in the anterior embryo (Figure 7G, I, K, M, O; Figure 8D–G) and used these to generate transgenic zebrafish. Of the eight constructs (named after the identity of the nearest gene), five showed expression in the F0 generation and GFP-positive embryos were raised to generate stable lines (summarized in Figure 8G). The remaining three constructs did not show F0 expression and were not considered further. In stable lines for each of the five constructs we detected tissue restricted GFP expression with each construct producing a distinct pattern (Figure 7H, J, L, N, P). We screened at least two independent founders for each stable line and find that GFP expression is indistinguishable between founders carrying the same construct (Figure 8A, B, G), indicating that each element imparts a unique tissue specificity to the basal E1b-GFP reporter that is independent of its integration site. In some instances, the observed expression pattern is comparable to that of the nearest gene (e.g. fgf8a; Figure 7N), suggesting that it represents an enhancer element controlling expression of the nearby gene. In other instances, the enhancer drives expression in a novel pattern (e.g. yap1; Figure 7P), suggesting that it may control a gene further away, or that the enhancer element tested (which is ~500bp in length) lacks some inputs required for proper expression of the nearby gene. These results indicate that NF-Y and TALE-occupied elements act as enhancers in vivo.
Figure 8: Characterization of NF-Y/TALE-regulated enhancers in zebrafish.
(A-C) GFP expression in F1 embryos from yap1:E1b-GFP founder #11 (A), founder #5 (B) and representative image of a 24hpf GFP-negative embryo (C). (D-F) UCSC Genome Browser tracks showing NF-YA, Pbx4, and Prep1 ChIP-seq data for the pax5 (D), her6 (E) and prdm14 (F) loci. The diagrams above the tracks show the putative enhancer region in green, DECA motifs in orange and CCAAT boxes in blue. (G) Table summarizing information about each enhancer element. Note that embryos that inherited the transgene from a female founder were GFP positive already at fertilization, indicating that these enhancer elements are active in the female germline. For this reason, all images in figures 7 and 8 are of embryos that inherited the transgene from a male founder.
We next took two approaches to confirm that the observed expression patterns are dependent on NF-Y and TALE function. First, we expressed the dominant negative NF-Y and TALE constructs in embryos from a cross of the tcf3a:E1b-GFP transgenic line (Figure 9A). We find that GFP expression is dramatically reduced in embryos expressing either dominant negative construct (Figure 9B–E), indicating that expression from the tcf3a genomic element requires both TALE and NF-Y function. This observation was further confirmed by qRT-PCR analysis (Figure 9F). Second, we made use of a distinct transgenesis strategy that allows us to test the effect of mutating the TALE and NF-Y binding sites in a given enhancer element. Specifically, our transgenic construct includes the gamma-crystallin promoter driving GFP along with the candidate enhancer element (Table 2) driving RFP. We find that the wildtype tcf3a and tle3a enhancers drive tissue-specific RFP expression (Figure 9G, K, O), as expected based on our results in Figure 7. However, when we test mutated versions of these elements (where the TALE and NF-Y binding sites have been disrupted), we find that transgenic animals (defined by GFP expression in the eye; Figure 9J, N) lack RFP expression (Figure 9I, M, O). We conclude that NF-Y and TALE-occupied elements possess enhancer activity and that this activity requires NF-Y and TALE function.
Figure 9: Disruption of TALE and NF-Y function reduces enhancer activity.
(A) Schematic showing workflow for dominant negative disruption of tcf3a:E1b-GFP. (B-D) Representative images showing no GFP (B), weak GFP (C), and strong GFP (D) of dominant negative-injected embryos. (E) Distribution of GFP expression in uninjected embryos and embryos injected with PBCAB, NF-YA DN or control RNA. (F) RT-qPCR-based detection of GFP expression in embryos injected with PBCAB, NF-YA DN or control RNA. Data are shown as mean +/− SEM. Statistical test: unpaired t-test. (G-N) Representative examples of RFP (G, K, I, M) and GFP (H, L, J, N) signal in tcf3a-WT:sv40 (G, H), tcf3a-mut:sv40 (I, J), tle3a-WT:sv40 (K, L) and tle3a-mut:sv40 (M, N) embryos at 32hpf. Insets in panels L, J, N show higher magnification of GFP expression in lens. Note that embryo in panels G, H is at a later stage than embryos in panels I-N. (O) Table quantifying results from experiment in panels G-N.
DISCUSSION
Our results indicate that combinatorial use of NF-Y and TALE TFs permits regulation of distinct gene expression programs during zebrafish embryogenesis. In particular, co-operation between NF-Y/TALE regulates expression of transcriptional control genes. These co-regulated genes are associated with an extended binding motif containing both TALE binding sites (DECA sites) and NF-Y binding sites (CCAAT boxes). While this extended motif has been observed before [72], it had not been assigned a function previously. The structure of this motif is consistent with our previous finding that TALE and NF-Y proteins interact to form a complex [23]. We also find that NF-Y and TALE act separately to regulate genes that have more specific functions in embryogenesis and that are associated with separate CCAAT and DECA motifs, respectively. This is particularly clear for NF-Y, which regulates a cilia-related gene expression program, but also for TALE that appears preferentially associated with hox gene expression.
We show that NF-Y and TALE occupy their genomic binding sites already at ZGA (3.5hpf), but since NF-Y and TALE are maternally deposited, they may be bound at these sites even earlier during embryogenesis. Indeed, we have previously shown that TALE binding can be detected as early as 2hpf [24]. Importantly, transcriptional control genes become expressed at ZGA [5, 73] – consistent with NF-Y/TALE occupancy at this stage – but initial cilia formation and hox expression occurs at later gastrula and segmentation stages in zebrafish [74–76]. Hence, genes separately regulated by each TF appear to be expressed several hours after TF occupancy is first detected at nearby regulatory elements. These elements may be continuously occupied by NF-Y and TALE TFs from 3.5hpf until the stage when the corresponding genes are expressed. Indeed, our previous work showed that DECA sites remain occupied by Prep from 3.5hpf at least until 12hpf [23]. Strikingly, we also observed a large increase in total TALE binding sites from 3.5hpf to 12hpf and noted that the sites newly established by 12hpf are enriched near genes involved in hox-dependent functions, suggesting that specific gene expression programs become reinforced following ZGA.
Given that NF-Y/TALE co-regulate early transcriptional control genes, it is unclear why disruption of NF-Y or TALE function does not produce a more severe phenotype. For instance, disruption of Nanog, SoxB1 and Oct4/Pou5f3, which are reported to act at ZGA in zebrafish, causes embryogenesis to stall at blastula stages [5]. However, complete developmental blockade is achieved only when all three of these factors are disrupted, indicating that they act in combination to drive gene expression at ZGA. Additionally, recent work suggests that disruption of Nanog, which produces the most severe phenotype of the three TFs, may not affect ZGA directly, but instead block formation of essential extraembryonic tissues [14]. Hence, it may be the case that vertebrate ZGA requires the action of multiple TFs and that disruption of any one TF is not sufficient to block ZGA. In agreement with such a model, recent work revealed that disruption of Dux, a TF implicated in murine ZGA, does not block embryonic development [13].
Our results indicate that the genomic regions occupied by NF-Y and TALE possess enhancer activity. As early as 4.3hpf (the earliest stage for which such data are available), these regions are enriched for histone modifications (H3K4me1 and H3K27ac) indicative of enhancer elements. H3K4me3 modifications (indicative of active transcription) are low at 4.3hpf, but increase by 9hpf, consistent with these enhancer elements being located near genes that are activated shortly after ZGA. We also demonstrate that both NF-Y and TALE activity is required for the enhancer elements to drive gene expression in vivo, but it is not yet clear what specific functions are contributed by each TF. Previous work has suggested that both NF-Y and TALE may represent pioneer factors [25, 28]. Accordingly, we recently showed that many 3.5hpf TALE-bound sites are also occupied by nucleosomes [23], suggesting that these TFs may be able to access their binding sites in nucleosomal DNA. Previous work has also demonstrated that TALE TFs can recruit histone modifying enzymes [24, 77, 78] and may therefore promote the deposition of histone marks. Notably, we previously tested several of the NF-Y and TALE-occupied elements for enhancer activity in HEK293 cells and failed to detect activity [23]. Furthermore, both NF-Y and TALE are ubiquitously expressed during embryogenesis and therefore unlikely to mediate the tissue-specific expression we observe in the transgenic lines. Hence, it is possible that NF-Y and TALE are generally required for enhancer activity (possibly by rearranging nucleosomes and promoting histone modifications), but that additional tissue-specific TFs (that are present in embryos, but not in HEK293 cells) act at these enhancers to drive expression in specific patterns.
We conclude that combinatorial use of NF-Y and TALE at ZGA defines distinct gene expression programs where co-occupied enhancers control early-acting transcriptional regulators, while enhancers individually occupied by NF-Y or TALE control later-acting cilia and hox genes, respectively.
Supplementary Material
Supplemental File 1: Excel file containing a full list of all GO terms identified for each data set. The file is arranged such that each tab contains the GO terms from the analysis in one figure panel. Each tab is labeled with the name of the corresponding figure panel. GO term analysis was carried out as describe in the Materials and Methods section.
HIGHLIGHTS.
NF-Y and TALE factors occupy genomic elements during zygotic genome activation
Genomic elements defined by NF-Y and TALE occupancy act as enhancers in vivo
NF-Y and TALE have both shared and independent transcriptional targets
Combinatorial use of TALE and NF-Y factors controls distinct genetic programs
Acknowledgments
FUNDING
This work was supported by National Institutes of Health grant NS038183 to CGS, grant DP3DK111898 to RM and BBSRC grant BB/N00907X/1 to NB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Vastenhouw NL, Cao WX, and Lipshitz HD, The maternal-to-zygotic transition revisited. Development, 2019. 146(11). [DOI] [PubMed] [Google Scholar]
- 2.Nien CY, et al. , Temporal coordination of gene networks by Zelda in the early Drosophila embryo. PLoS Genet, 2011. 7(10): p. e1002339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrison MM, et al. , Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLoS Genet, 2011. 7(10): p. e1002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liang HL, et al. , The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature, 2008. 456(7220): p. 400–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee MT, et al. , Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature, 2013. 503(7476): p. 360–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leichsenring M, et al. , Pou5f1 transcription factor controls zygotic gene activation in vertebrates. Science, 2013. 341(6149): p. 1005–9. [DOI] [PubMed] [Google Scholar]
- 7.Saxe JP, et al. , Post-translational regulation of Oct4 transcriptional activity. PLoS One, 2009. 4(2): p. e4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramakrishna S, et al. , PEST motif sequence regulating human NANOG for proteasomal degradation. Stem Cells Dev, 2011. 20(9): p. 1511–9. [DOI] [PubMed] [Google Scholar]
- 9.Eckersley-Maslin M, et al. , Dppa2 and Dppa4 directly regulate the Dux-driven zygotic transcriptional program. Genes Dev, 2019. 33(3–4): p. 194–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Iaco A, et al. , DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat Genet, 2017. 49(6): p. 941–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Iaco A, et al. , DPPA2 and DPPA4 are necessary to establish a 2C-like state in mouse embryonic stem cells. EMBO Rep, 2019. 20(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hendrickson PG, et al. , Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat Genet, 2017. 49(6): p. 925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Z and Zhang Y, Loss of DUX causes minor defects in zygotic genome activation and is compatible with mouse development. Nat Genet, 2019. 51(6): p. 947–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gagnon JA, Obbad K, and Schier AF, The primary role of zebrafish nanog is in extra-embryonic tissue. Development, 2018. 145(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iwafuchi-Doi M and Zaret KS, Cell fate control by pioneer transcription factors. Development, 2016. 143(11): p. 1833–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun Y, et al. , Zelda overcomes the high intrinsic nucleosome barrier at enhancers during Drosophila zygotic genome activation. Genome Res, 2015. 25(11): p. 1703–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schulz KN, et al. , Zelda is differentially required for chromatin accessibility, transcription factor binding, and gene expression in the early Drosophila embryo. Genome Res, 2015. 25(11): p. 1715–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soufi A, et al. , Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell, 2015. 161(3): p. 555–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soufi A, Donahue G, and Zaret KS, Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell, 2012. 151(5): p. 994–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gualdi R, et al. , Hepatic specification of the gut endoderm in vitro: cell signaling and transcriptional control. Genes Dev, 1996. 10(13): p. 1670–82. [DOI] [PubMed] [Google Scholar]
- 21.Cirillo LA, et al. , Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell, 2002. 9(2): p. 279–89. [DOI] [PubMed] [Google Scholar]
- 22.Heinz S, et al. , Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell, 2010. 38(4): p. 576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ladam F, et al. , TALE factors use two distinct functional modes to control an essential zebrafish gene expression program. Elife, 2018. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choe SK, Ladam F, and Sagerstrom CG, TALE factors poise promoters for activation by Hox proteins. Dev Cell, 2014. 28(2): p. 203–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magnani L, et al. , PBX1 genomic pioneer function drives ERalpha signaling underlying progression in breast cancer. PLoS Genet, 2011. 7(11): p. e1002368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dolfini D, Gatta R, and Mantovani R, NF-Y and the transcriptional activation of CCAAT promoters. Crit Rev Biochem Mol Biol, 2012. 47(1): p. 29–49. [DOI] [PubMed] [Google Scholar]
- 27.Oldfield AJ, et al. , Histone-fold domain protein NF-Y promotes chromatin accessibility for cell type-specific master transcription factors. Mol Cell, 2014. 55(5): p. 708–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nardini M, et al. , Sequence-specific transcription factor NF-Y displays histone-like DNA binding and H2B-like ubiquitination. Cell, 2013. 152(1–2): p. 132–43. [DOI] [PubMed] [Google Scholar]
- 29.Gao L, et al. , Chromatin Accessibility Landscape in Human Early Embryos and Its Association with Evolution. Cell, 2018. 173(1): p. 248–259 e15. [DOI] [PubMed] [Google Scholar]
- 30.Lu F, et al. , Establishing Chromatin Regulatory Landscape during Mouse Preimplantation Development. Cell, 2016. 165(6): p. 1375–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhattacharya A, et al. , The B subunit of the CCAAT box binding transcription factor complex (CBF/NF-Y) is essential for early mouse development and cell proliferation. Cancer Res, 2003. 63(23): p. 8167–72. [PubMed] [Google Scholar]
- 32.Mantovani R, et al. , Dominant negative analogs of NF-YA. J Biol Chem, 1994. 269(32): p. 20340–6. [PubMed] [Google Scholar]
- 33.Choe SK, Vlachakis N, and Sagerstrom CG, Meis family proteins are required for hindbrain development in the zebrafish. Development, 2002. 129(3): p. 585–95. [DOI] [PubMed] [Google Scholar]
- 34.Deflorian G, et al. , Prep1.1 has essential genetic functions in hindbrain development and cranial neural crest cell differentiation. Development, 2004. 131(3): p. 613–27. [DOI] [PubMed] [Google Scholar]
- 35.Waskiewicz AJ, et al. , Zebrafish Meis functions to stabilize Pbx proteins and regulate hindbrain patterning. Development, 2001. 128(21): p. 4139–51. [DOI] [PubMed] [Google Scholar]
- 36.Amin S, et al. , Hoxa2 selectively enhances Meis binding to change a branchial arch ground state. Dev Cell, 2015. 32(3): p. 265–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Q, et al. , A systematic approach to identify functional motifs within vertebrate developmental enhancers. Dev Biol, 2010. 337(2): p. 484–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Birnbaum RY, et al. , Coding exons function as tissue-specific enhancers of nearby genes. Genome Res, 2012. 22(6): p. 1059–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vlachakis N, Ellstrom DR, and Sagerstrom CG, A novel pbx family member expressed during early zebrafish embryogenesis forms trimeric complexes with Meis3 and Hoxb1b. Dev Dyn, 2000. 217(1): p. 109–19. [DOI] [PubMed] [Google Scholar]
- 40.Li B and Dewey CN, RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics, 2011. 12: p. 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anders S and Huber W, Differential expression analysis for sequence count data. Genome Biol, 2010. 11(10): p. R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bolger AM, Lohse M, and Usadel B, Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, 2014. 30(15): p. 2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Langmead B and Salzberg SL, Fast gapped-read alignment with Bowtie 2. Nat Methods, 2012. 9(4): p. 357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tyner C, et al. , The UCSC Genome Browser database: 2017 update. Nucleic Acids Res, 2017. 45(D1): p. D626–D634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H, et al. , The Sequence Alignment/Map format and SAMtools. Bioinformatics, 2009. 25(16): p. 2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, et al. , Model-based analysis of ChIP-Seq (MACS). Genome Biol, 2008. 9(9): p. R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goecks J, et al. , Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol, 2010. 11(8): p. R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quinlan AR and Hall IM, BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics, 2010. 26(6): p. 841–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McLean CY, et al. , GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol, 2010. 28(5): p. 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hiller M, et al. , Computational methods to detect conserved non-genic elements in phylogenetically isolated genomes: application to zebrafish. Nucleic Acids Res, 2013. 41(15): p. e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang da W, Sherman BT, and Lempicki RA, Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc, 2009. 4(1): p. 44–57. [DOI] [PubMed] [Google Scholar]
- 52.Huang da W, Sherman BT, and Lempicki RA, Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res, 2009. 37(1): p. 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Machanick P and Bailey TL, MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics, 2011. 27(12): p. 1696–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bailey TL, et al. , MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res, 2009. 37(Web Server issue): p. W202–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ramirez F, et al. , deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res, 2014. 42(Web Server issue): p. W187–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waskiewicz AJ, Rikhof HA, and Moens CB, Eliminating zebrafish pbx proteins reveals a hindbrain ground state. Dev Cell, 2002. 3(5): p. 723–33. [DOI] [PubMed] [Google Scholar]
- 57.Popperl H, et al. , lazarus is a novel pbx gene that globally mediates hox gene function in zebrafish. Mol Cell, 2000. 6(2): p. 255–67. [DOI] [PubMed] [Google Scholar]
- 58.Popperl H, et al. , Segmental expression of Hoxb-1 is controlled by a highly conserved autoregulatory loop dependent upon exd/pbx. Cell, 1995. 81(7): p. 1031–42. [DOI] [PubMed] [Google Scholar]
- 59.Ladam F and Sagerstrom CG, Hox regulation of transcription: more complex(es). Dev Dyn, 2014. 243(1): p. 4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Merabet S and Mann RS, To Be Specific or Not: The Critical Relationship Between Hox And TALE Proteins. Trends Genet, 2016. 32(6): p. 334–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen YH, Lin YT, and Lee GH, Novel and unexpected functions of zebrafish CCAAT box binding transcription factor (NF-Y) B subunit during cartilages development. Bone, 2009. 44(5): p. 777–84. [DOI] [PubMed] [Google Scholar]
- 62.Chang CP, et al. , Meis proteins are major in vivo DNA binding partners for wild type but not chimeric Pbx proteins. Mol Cell Biol, 1997. 17(10): p. 5679–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Knoepfler PS and Kamps MP, The highest affinity DNA element bound by Pbx complexes in t(1;19) leukemic cells fails to mediate cooperative DNA-binding or cooperative transactivation by E2a-Pbx1 and class I Hox proteins - evidence for selective targetting of E2a-Pbx1 to a subset of Pbx-recognition elements. Oncogene, 1997. 14(21): p. 2521–31. [DOI] [PubMed] [Google Scholar]
- 64.Nepal C, et al. , Dynamic regulation of the transcription initiation landscape at single nucleotide resolution during vertebrate embryogenesis. Genome Res, 2013. 23(11): p. 1938–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ferretti E, et al. , Segmental expression of Hoxb2 in r4 requires two separate sites that integrate cooperative interactions between Prep1, Pbx and Hox proteins. Development, 2000. 127(1): p. 155–66. [DOI] [PubMed] [Google Scholar]
- 66.Ferretti E, et al. , Hoxb1 enhancer and control of rhombomere 4 expression: complex interplay between PREP1-PBX1-HOXB1 binding sites. Mol Cell Biol, 2005. 25(19): p. 8541–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jacobs Y, Schnabel CA, and Cleary ML, Trimeric association of Hox and TALE homeodomain proteins mediates Hoxb2 hindbrain enhancer activity. Mol Cell Biol, 1999. 19(7): p. 5134–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grieder NC, et al. , Synergistic activation of a Drosophila enhancer by HOM/EXD and DPP signaling. EMBO J, 1997. 16(24): p. 7402–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ryoo HD and Mann RS, The control of trunk Hox specificity and activity by Extradenticle. Genes Dev, 1999. 13(13): p. 1704–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tumpel S, et al. , Expression of Hoxa2 in rhombomere 4 is regulated by a conserved cross-regulatory mechanism dependent upon Hoxb1. Dev Biol, 2007. 302(2): p. 646–60. [DOI] [PubMed] [Google Scholar]
- 71.Maity SN and de Crombrugghe B, Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends Biochem Sci, 1998. 23(5): p. 174–8. [DOI] [PubMed] [Google Scholar]
- 72.Penkov D, et al. , Analysis of the DNA-binding profile and function of TALE homeoproteins reveals their specialization and specific interactions with Hox genes/proteins. Cell Rep, 2013. 3(4): p. 1321–33. [DOI] [PubMed] [Google Scholar]
- 73.Aanes H, et al. , Zebrafish mRNA sequencing deciphers novelties in transcriptome dynamics during maternal to zygotic transition. Genome Res, 2011. 21(8): p. 1328–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Prince VE, et al. , Zebrafish hox genes: expression in the hindbrain region of wild-type and mutants of the segmentation gene, valentino. Development, 1998. 125(3): p. 393–406. [DOI] [PubMed] [Google Scholar]
- 75.Prince VE, et al. , Zebrafish hox genes: genomic organization and modified colinear expression patterns in the trunk. Development, 1998. 125(3): p. 407–20. [DOI] [PubMed] [Google Scholar]
- 76.Essner JJ, et al. , Conserved function for embryonic nodal cilia. Nature, 2002. 418(6893): p. 37–8. [DOI] [PubMed] [Google Scholar]
- 77.Choe SK, et al. , Meis cofactors control HDAC and CBP accessibility at Hox-regulated promoters during zebrafish embryogenesis. Dev Cell, 2009. 17(4): p. 561–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Saleh M, et al. , Cell signaling switches HOX-PBX complexes from repressors to activators of transcription mediated by histone deacetylases and histone acetyltransferases. Mol Cell Biol, 2000. 20(22): p. 8623–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bogdanovic O, et al. , Dynamics of enhancer chromatin signatures mark the transition from pluripotency to cell specification during embryogenesis. Genome Res, 2012. 22(10): p. 2043–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang Y, et al. , Canonical nucleosome organization at promoters forms during genome activation. Genome Res, 2014. 24(2): p. 260–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Love NR, et al. , pTransgenesis: a cross-species, modular transgenesis resource. Development, 2011. 138(24): p. 5451–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kucukural A, et al. , DEBrowser: interactive differential expression analysis and visualization tool for count data. BMC Genomics, 2019. 20(1): p. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental File 1: Excel file containing a full list of all GO terms identified for each data set. The file is arranged such that each tab contains the GO terms from the analysis in one figure panel. Each tab is labeled with the name of the corresponding figure panel. GO term analysis was carried out as describe in the Materials and Methods section.
Data Availability Statement
RNA-seq data is available in GEO under accession number GSE133459 and ChIP-seq data is available in ArrayExpress under accession number E-MTAB-8137.