Abstract
The use of circulating microRNAs as biomarkers opens up new opportunities for the diagnosis of cardiovascular diseases because of their specific expression profiles. The aim of the present study was to identify circulating microRNAs in human plasma as potential biomarkers of heart failure and related diseases. We used real-time quantitative PCR to screen microRNA in plasma samples from 62 normal controls and 62 heart failure samples. We found that circulating miR-21-5p, miR-30a-3p, miR-30a-5p, miR-155-5p, miR-216a and miR-217 expressed differently between healthy controls and heart failure patients. Plasma levels of miR-21-5p, miR-30a-3p, miR-30a-5p, miR-155-5p, miR-216a and miR-217 were unaffected by hemolysis. Correlation analysis showed any two of these miRNAs possess a strong correlation, indicating a possibility of combined analysis. MiR-21-5p, miR-30a-3p, miR-30a-5p, miR-155-5p, miR-216a and miR-217 could be combined in two or three or more combinations. The results suggest that miR-21-5p, miR-30a-3p, miR-30a-5p, miR-155-5p, miR-216a and miR-217 may be a new diagnostic biomarker for heart failure and related diseases.
Keywords: Combined detection, diagnostic biomarker, early heart failure diseases, microRNAs
Introduction
In recent years, cardiovascular diseases still rank the first killer of human death and heart failure accounts one of them. Heart failure is not an independent disease but the end stage of the development of heart diseases. Although the trend of younger heart failure group rises substantial in recent years, no enough research attention has been paid to the younger generation [1–5]. Therefore, effective diagnosis and prevention of heart failure is an urgent problem in medical and biological researches [6–9].
MicroRNAs (miRNAs) are a class of non-protein encoded small RNAs that widely exist in eukaryotes and have a length of 21–25 nucleotides. They are highly stable in the blood circulation and can regulate gene expression in a sequence-specific manner. They play an important role in development, apoptosis, metabolism and human diseases. The physiological and pathological regulation mechanism of miRNA is a new discipline which has been highly valued in recent years [10–12].
Recent studies have shown that cardiovascular diseases can cause significant changes in the expression level of specific miRNAs in the body. Therefore, detection of specific miRNAs in body fluids can play an essential role in the diagnosis and prevention of cardiovascular diseases [13–16]. However, there is still insufficient research on specific miRNAs in heart failure [17–20].
Several studies showed that the dynamics of many miRNA expression are closely related to the occurrence of diseases; for example, many heart failure-related miRNA expression changes in vivo related to the occurrence of heart failure. To this end, we selected six miRNAs from previous studies, namely, miR-21-5p, miR-30a-3p, miR-30a-5p, miR-155-5p, miR-216a and miR-217, which showed an upward trend with the occurrence of heart failure. In combination with other researchers ‘statistics on sample size, we selected more than 60 healthy samples and more than 60 cases of heart failure. Exhaustion samples were analyzed [21–38].
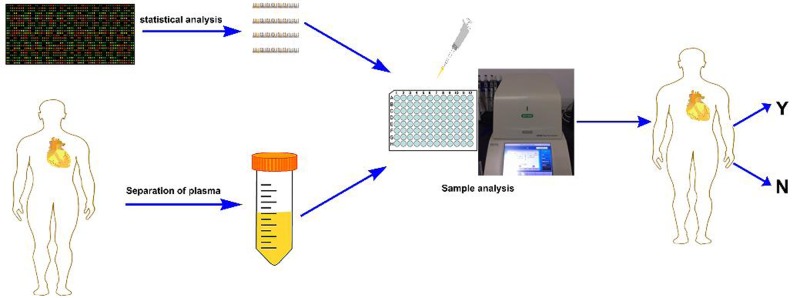
As shown in Figure 1, the aim of the present study was to identify circulating miRNAs in human plasma as biomarkers for the diagnosis of heart failure and its related diseases, to assess the appropriate biomarkers for identifying key characteristics of miRNAs, and to analyze their performance.
Figure 1. Schematic maps of screening and identifying microRNAs as potential biological targets for heart failure detection.
Among them, Y means that the test result is positive, and N means that the test result is negative.
Materials and methods
Study population
Patients with heart failure were recruited at the Affiliated Hospital of Qingdao University, Qingdao, China. The healthy control group was from the physical examination center of Qingdao University affiliated hospital. Through similar experiments by other researchers, we finally identified more than 60 healthy control samples and more than 60 heart failure disease samples. Detailed characteristics of heart failure group and control group were listed in Table 1.
Table 1. Partial indexes of heart failure group and healthy control group.
| Variable | Heart failure | Control | P-value |
|---|---|---|---|
| N = 62 | N = 62 | ||
| Age (years) | 62 ± 8.89 | 60 ± 11.80 | 0.55 |
| Men | 40 | 42 | |
| BMI (kg/m2) | 26.07 ± 3.36 | 24.76 ± 3.83 | 0.13 |
| Total cholesterol (mg/dl) | 4.57 ± 1.56 | 4.08 ± 1.51 | 0.23 |
| Triglycerides (g/l) | 1.62 ± 0.81 | 1.14 ± 0.67 | 0.0047** |
| LDL cholesterol (mg/dl) | 2.86 ± 1.41 | 2.52 ± 1.12 | 0.29 |
| HDL cholesterol (mg/dl) | 1.15 ± 0.29 | 1.05 ± 0.53 | 0.49 |
| Creatinine | 62.7 ± 16.76 | 76.0 ± 18.46 | 0.0018** |
| NT-proBNP | 1809.6 ± 1465 | 100.1 ± 82.36 | 0.0384* |
| CKMB | 12.7 ± 27.46 | 11.8 ± 8.93 | 0.87 |
| MYO | 245 ± 441.16 | 40.4 ± 13.35 | 0.36 |
| HsTnT | 0.467 ± 1.16 | 0.005 ± 0.0055 | 0.43 |
BMI: body mass index; CKMB: creatine kinase-MB; HDL: high-density lipoprotein; HsTnT: hight-sensitivity troponin T; LDL: low-density lipoprotein; MYO: myoglobin; NT-proBNP: N-terminal pro-B-type natriuretic peptide. Data are shown as mean±SE; *P<0.05 and **P<0.01.
Blood collection
Peripheral blood from patients with heart failure and healthy people was collected in 10 ml EDTA anticoagulation tube. After 30 min of blood collection, the samples were centrifuged at 3000 g and 4°С for 10 min. After centrifugation, the plasma was separated and frozen immediately until RNA was separated.
miRNA extraction
RNA isolation was advanced by using the traditional Triazole method. The specific method was: 250 μl plasma was mixed with 750 μl Triazole, then 10 μl and 50 pM of coccidian RNA was added. In the experiment, the RNA of nematode was used as the standard external reference, and 200 μl chloroform was added to the plasma and mixed with severe vibration for 15 s. Centrifuge for 10 min at the condition of 4°С and 12,000 g, removed 550 μl from the lower layer and added isopropanol of the same volume, added 5 μl glycogen, mixed upside down, rest overnight at the condition of −20°С, centrifuge for 10 min at the condition of 4°С and 12,000 g, discard the supernatant, added 75% ethanol without RNase enzyme of 1 ml into the tube, and separated at the condition of 4°С and 12,000 g. After 10 min, the supernatant was discarded and 75% ethanol without RNase enzyme was added into the tube. The supernatant was discarded and then centrifuged for 5 min at 4°С and 12,000 g for 10 min at room temperature. Water with 20 μl of Ranse without enzyme was added into the tube and dissolved at 4°С. The RNA was quantified and quantified by RNA electrophoresis gel and measuring concentration.
The extracted RNA was inverted by TaKaRa, the Mir-X miRNA First-Strand Synthesis Kit (Clontech). The specific method of inversion was the standard method of TaKaRa reverse transcription.
Quantitative PCR experiment
We selected TaKaRa quantitative PCR kit and Bio-Rad real-time quantitative PCR instrument to process and analyze the samples we collected. According to the instructions of TaKaRa products, the instrument was programmed to react in three steps, including the first step of 95°С reaction for 15 min, the second step of reaction including 94°С reaction for 20 s, 63°С reaction for 30 s, 72°С reaction for 34 s, the whole process was repeated five times, the third step of reaction including 94°С reaction for 20 s, 63°С reaction for 34 s and collection. The fluorescence signal was repeated 45 times, and then the reaction was completed after 5 s of 65°С reaction and 5 s of 95°С reaction. All data were repeated three times in the experiment.
Data analysis and statistical methods
Quantitative miRNA expression data ABI SDS software (Life Technologies) was used to estimate the cyclic threshold (CT) with an average threshold of 0.2 (30). The CT value > 35 is considered below the detection limit [39] for calculation, the CT value > 35 is labeled 35 [40]. The level of miRNA is 2− Ct [41].
Then, we used SPSS software, MedCalc software, Prism software to further analyze the data of fluorescence quantitative PCR.
Results
Expression profile analyses of candidate miRNAs
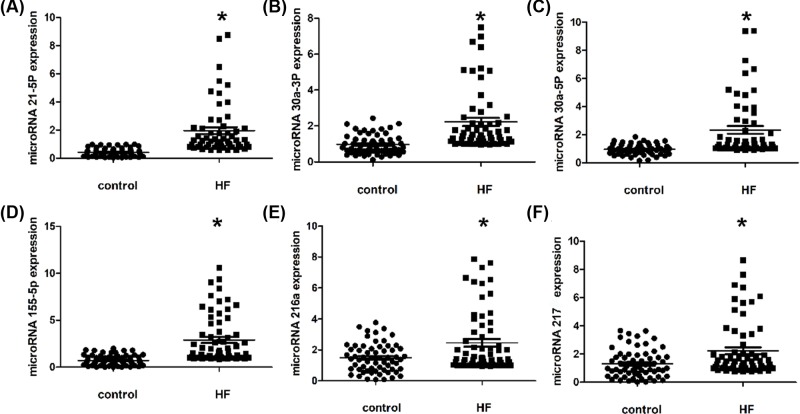
In order to identify the most stable and reliable candidate miRNAs as the biological target molecule. Based on the literature search, we selected 30 miRNAs as candidate target molecules. Through the preliminary experiments and analysis of the relationship between miRNA expression and heart failure disease, we finally selected six miRNAs, namely, microRNA-21-5p, microRNA-30a-3p, microRNA-30a-5p, microRNA-155-5p, microRNA-216a and microRNA-217. We used internal reference evaluation software RefFinder (http://www.leonxie.com/referencegene.php) to carry out online evaluation of internal reference genes [42], and finally selected the sequence of miR-39 of the nematode as a reference. Related content has been added to the article. RefFinder is a method of assigning weights based on the geometric mean of four commonly used algorithms (geNorm, BestKeeper, NormFinder, and comparativedelta-CT) to rank gene stability [43–46]. We used the prism data analysis software to analyze the data of fluorescence quantitative PCR, and the results were shown in Figure 2. From the analysis results, we can see that the six kinds of miRNA statistics show that in the heart failure samples, the expression of microRNA - 21-5p, microRNA 30a-3p, microRNA 30a-5p, microRNA 155-5p, microRNA 216a and microRNA217 have a significant upward trend. This is also consistent with the conclusions drawn by other research groups, which ensures the accuracy of the experimental results.
Figure 2. Plasma miRNA levels in the validation population.
The scatter plots show the expression levels of (A) miR-21-5p, (B) miR-30a-3p, (C) miR-30a-5p, (D) miR-155-5p, (E) miR-216a and (F) miR-217 measured by quantitative real-time polymerase chain reaction (qRT-PCR) in patients with HF and control subjects (n = 62). From the analysis results, we can see that the six kinds of miRNA statistics show that in the heart failure samples have a significant upward trend. The relative miRNA expression levels were normalized to cel-miR-39 and calculated by -ΔΔCt. Differences between each groups were compared by Kruskal–Wallis ANOVA test; ∗P < 0.05.
ROC analysis of selected miRNAs
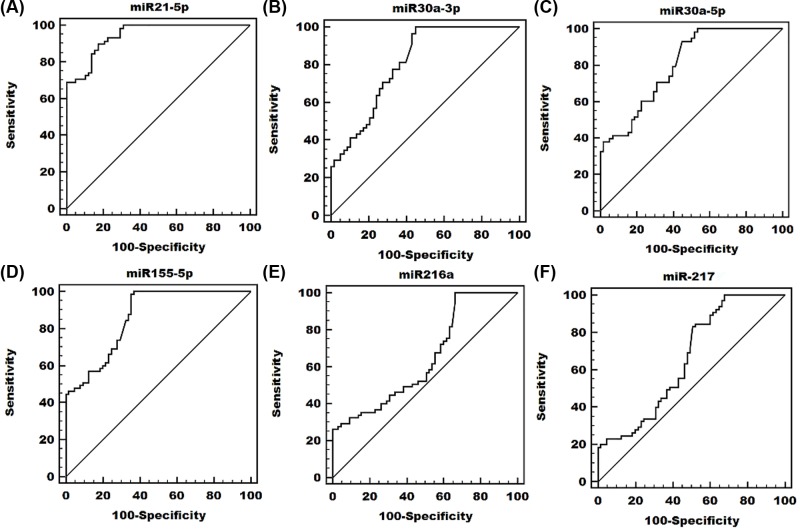
We used MedCalc data analysis software to perform ROC analysis of quantitative fluorescent PCR data. The results were shown in Figure 3 and Table 2.
Figure 3. Receiver operating characteristic (ROC) curves analysis of (A) miR-21-5p, (B) miR-30a-3p, (C) miR-30a-5p, (D) miR-155-5p, (E) miR-216a and (F) miR-217.
ROC curve evaluation statistics show that all AUC values are greater than 0.5, indicating that the detection method is effective.
Table 2. The areas under the curves (AUC), 95% CI, sensitivity, criterion and specificity of miR-21-5p, miR-30a-3p, miR-30a-5p, miR-155-5p, miR-216a and miR-217.
| AUC | 95% confidence interval | Sensitivity | Criterion | Specificity | |
|---|---|---|---|---|---|
| miR−21-5p | 0.944 | 0.886–0.978 | 89.7 | >0.7201 | 82.8 |
| miR-30a-3p | 0.809 | 0.726–0.876 | 100.00 | >0.9544 | 55.2 |
| miR-30a-5p | 0.801 | 0.716–0.869 | 93.1 | >0.9988 | 55.2 |
| miR-155-5p | 0.861 | 0.790–0.916 | 98.5 | >0.8591 | 64.6 |
| miR-216a | 0.648 | 0.559–0.729 | 100.00 | >0.9178 | 33.8 |
| miR-217 | 0.660 | 0.571–0.740 | 83.1 | >0.9969 | 49.2 |
The area under the ROC curve is between 1.0 and 0.5. miR-21-5p, miR-30a-3p, miR-30a-5p, miR-155-5p, miR-216a and miR-217 detection results are accurate.
ROC curve evaluation statistics show that all AUC values are greater than 0.5, indicating that the detection method is effective, and most results greater than 0.7 are more accurate, and the MIC-21-5p AUC value is higher than 0.9 has higher accuracy.
In addition, we carried out a combination test of several miRNAs, and the results of the combination test were shown in the Supplementary Data. From the results of each combination, we can see that the detection results of multiple small RNAs, especially more than three small RNAs, will be more accurate and more reliable.
Similarly, we performed ROC analysis on some of the clinical data collected. The relevant data were listed in the Supplementary Data.
Correlation analysis
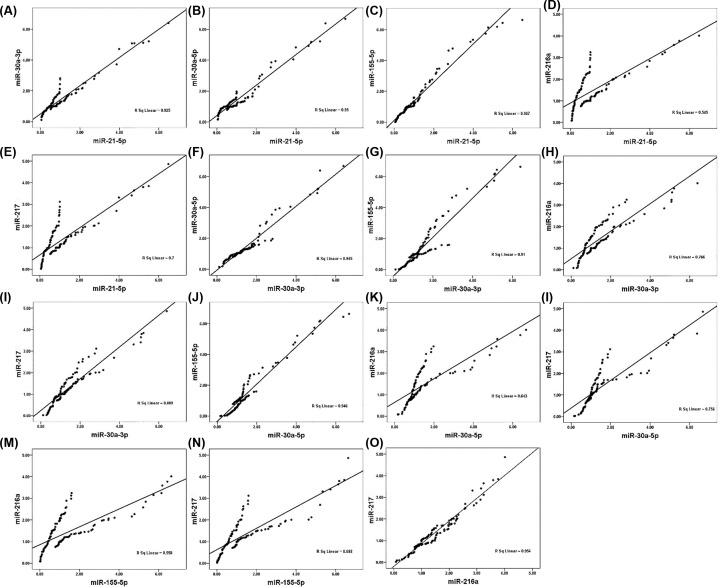
We used SPSS analysis software to analyze the correlation of the quantitative data. The results were shown in Figure 4 and Table 3.
Figure 4. Spearman correlations between the circulating miRNAs in patients with HF.
The scatter plots show the marked correlation in the expression values between (A) miR-21-5p and miR-30a-3p, (B) miR-21-5p and miR-30a-5p, (C) miR-21-5p and miR-155-5p, (D) miR-21-5p and miR-216a, (E) miR-21-5p and miR-217, (F) miR-30a-3p and miR-30a-5p, (G) miR-30a-3p and miR-155-5p, (H) miR-30a-3p and miR-216a, (I) miR-30a-3p and miR-217, (J) miR-30a-5p and miR-155-5p, (K) miR-30a-5p and miR-216a, (L) miR-30a-5p and miR-217, (M) miR-155-5p and miR-216a, (N) miR-155-5p and miR-217, (O) miR-216a and miR-217, in the population (n = 62) with AMI.
Table 3. Spearman correlations between the circulating miRNAs in patients with HF, list of P and r result.
| miR−21-5p | miR-30a-3p | miR-30a-5p | miR-155-5p | miR-216a-5p | miR-217-5p | ||
|---|---|---|---|---|---|---|---|
| miR−21-5p | r | 0.976 | 0.987 | 0.983 | 0.738 | 0.836 | |
| P | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | ||
| miR-30a-3p | P | 0.976 | 0.981 | 0.954 | 0.854 | 0.943 | |
| sig | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | ||
| miR-30a-5p | P | 0.987 | 0.981 | 0.973 | 0.802 | 0.870 | |
| sig | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | ||
| miR-155-5p | P | 0.983 | 0.954 | 0.973 | 0.747 | 0.832 | |
| sig | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | ||
| miR-216a | P | 0.738 | 0.875 | 0.802 | 0.747 | 0.977 | |
| sig | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | ||
| miR-217 | P | 0.836 | 0.943 | 0.870 | 0.832 | 0.977 | |
| sig | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | ||
From the experimental results, we can see that there is a significant correlation between the six small RNAs, which proves that the results are related. The consistency of test results is ensured.
Similarly, we analyzed the correlation between the miRNA test results and the clinical gold standard test results. The results were shown in Supplementary Data.
Summary
Up to now, many studies have shown that miRNAs as biomarkers have important clinical applications, but it is still necessary to study whether miRNAs can establish reliable diagnostic and potential biomarkers for heart failure to verify their accuracy and repeatability. We found that plasma levels of miR-21-5p, miR-30a-3p, miR-30a-5p, miR-155-5p, miR-216a and miR-217 were not affected by hemolysis, age, and gender when used to diagnose heart failure. miR-21-5p, miR-30a-3p, miR-30a-5p, miR-155-5p, miR-216a and miR-217 may be a new biomarker for the diagnosis of heart failure and related diseases. These microRNAs have potential as diagnostic and prognostic biomarkers and will be used in clinical diagnosis and treatment of heart failure diseases in conjunction with existing gold standards.
Supplementary Material
Abbreviations
- CT
cyclic threshold
- miRNA
microRNA
- qRT-PCR
quantitative real-time polymerase chain reaction
Contributor Information
Han Ding, Email: dinghan2011@163.com.
Yi An, Email: any2018@qq.com.
Peifeng Li, Email: peifli@hotmail.com.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by the Shandong university scientific research project [grant number J18KA127]; Qingdao University Research Fund and National Natural Science Foundation of China [grant numbers 81741173, 31430041 and 91849209].
Author Contribution
Han Ding, Yin Wang and Sheng Xue for isolation and extraction of RNA. Yu Wang, Lei Zhang, Yuan Zhang and Hongzhao Qi for Reverse transcription and quantitative PCR. Longgang Hu and Hua Yu for sample collection. Yi An for clinical diagnosis. Han Ding, Lynn Htet Htet Aung and Peifeng Li for concept and manuscript writing.
Ethics Approval
All participants in the study provided informed consent. Rules and approvals for patient privacy were obtained from the ethics committee of the medical department of Qingdao University.
References
- 1.Khan S.S., Shah S.J., Colangelo L.A., Panjwani A., Liu K., Lewis C.E. et al. (2018) Association of Patterns of Change in Adiposity with Diastolic Function and Systolic Myocardial Mechanics from Early Adulthood to Middle Age: The Coronary Artery Risk Development in Young Adults Study. J. Am. Soc. Echocardiog. 31, 1261–1269 10.1016/j.echo.2018.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pareek N., Cevallos J., Moliner P., Shah M., Tan L.L., Chambers V. et al. (2018) Activity and outcomes of a cardio-oncology service in the United Kingdom-a five-year experience. Eur. J. Heart. 20, 1721–1731 10.1002/ejhf.1292 [DOI] [PubMed] [Google Scholar]
- 3.Ikegami R., Shimizu I., Yoshida Y. and Minamino T. (2017) Metabolomic Analysis in Heart Failure. Circ. J. 82, 10–16 10.1253/circj.CJ-17-1184 [DOI] [PubMed] [Google Scholar]
- 4.Nayar P., Yu F., Chandak A., Kan G.L., Lowes B. and Apenteng B.A. (2018) Risk Factors for In-Hospital Mortality in Heart Failure Patients: Does Rurality, Payer or Admission Source Matter? J. Rural. Hwalth. 34, 103–108 10.1111/jrh.12186 [DOI] [PubMed] [Google Scholar]
- 5.Braunwald E. (2013) Heart Failure. JACC-Hwart. Fail. 1, 1–20 10.1016/j.jchf.2012.10.002 [DOI] [PubMed] [Google Scholar]
- 6.Almeida A.G. (2017) NT-proBNP and Myocardial Fibrosis the Invisible Link Between Health and Disease. J. Am. Coll. Cardiol. 70, 3110–3112 10.1016/j.jacc.2017.11.010 [DOI] [PubMed] [Google Scholar]
- 7.Kato M., Komamura K., Kitakaze M. and Hirayama A. (2017) The Impact of Bronchodilator Therapy on Systolic Heart Failure with Concomitant Mild to Moderate COPD. Diseases 6, 1–10 10.3390/diseases6010004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu X.L., Zeng W.J., Li M.P., Yang Y.L., Kuang D.B., Li H. et al. (2017) AGXT2 rs37369 polymorphism predicts the renal function in patients with chronic heart failure. Gene 637, 145–151 10.1016/j.gene.2017.09.038 [DOI] [PubMed] [Google Scholar]
- 9.Naseroleslami M., Aboutaleb N. and Parivar K. (2018) the effects of superparamagnetic iron oxide nanoparticles-labeled mesenchymal stem cells in the presence of a magnetic field on attenuation of injury after heart failure. Drug. Deliv. Transl. Re. 8, 1214–1225 10.1007/s13346-018-0567-8 [DOI] [PubMed] [Google Scholar]
- 10.Montazerian M., Yasari F. and Aghaalikhani N. (2018) Ovarian extracellular MicroRNAs as the potential non-invasive biomarkers: An update. Biomed. Pharmacother. 106, 1633–1640 10.1016/j.biopha.2018.07.073 [DOI] [PubMed] [Google Scholar]
- 11. Puneet, Kazmi H.R., Kumari S., Tiwari S., Khanna A. and Narayan G. (2018) Epigenetic Mechanisms and Events in Gastric Cancer-Emerging Novel Biomarkers. Pathol. Oncol. Res. 24, 757–770 10.1007/s12253-018-0410-z [DOI] [PubMed] [Google Scholar]
- 12.Nadeem A., Ashraf M.R., Javed M., Hussain T., Tariq M.S. and Babar M.E. (2018) Review - MicroRNAs: A new paradigm towards mechanistic insight of diseases. Pak. J. pharm. Sci. 31, 2017–2026 [PubMed] [Google Scholar]
- 13.Lee Y., Ahn C., Han J.J., Choi H., Kim J., Yim J. et al. (2003) The nuclear RNase III Drosha initiates microRNA processing. Nature 425, 415–419 10.1038/nature01957 [DOI] [PubMed] [Google Scholar]
- 14.Basson M. (2007) MicroRNAs loom large in the heart. Nat. Med. 13, 541 10.1038/nm0507-541 [DOI] [PubMed] [Google Scholar]
- 15.Callis T.E. and Wang D.Z. (2008) Taking microRNAs to heart. Trends. Mol. Med. 14, 254–260 10.1016/j.molmed.2008.03.006 [DOI] [PubMed] [Google Scholar]
- 16.Care A., Catalucci D., Felicetti F., Bonci D., Addario A., Gallo P. et al. (2007) MicroRNA-133 controls cardiac hypertrophy. Nat. Med. 13, 613–618 10.1038/nm1582 [DOI] [PubMed] [Google Scholar]
- 17.Mann D.L. (2007) MicroRNAs and the failing heart. New. Engl. J. Med. 356, 2644–2645 10.1056/NEJMcibr072068 [DOI] [PubMed] [Google Scholar]
- 18.Thum T., Galuppo P., Wolf C., Fiedler J., Kneitz S., van Laake L.W. et al. (2007) MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation 116, 258–267 10.1161/CIRCULATIONAHA.107.687947 [DOI] [PubMed] [Google Scholar]
- 19.Chien K.R. (2007) Molecular medicine-microRNAs and the tell-tale heart. Nature 447, 389–390 10.1038/447389a [DOI] [PubMed] [Google Scholar]
- 20.Wang H.J. and Cai J. (2017) The role of microRNAs in heart failure. BBA-Mol. Basis. Dis. 1863, 2019–2030 10.1016/j.bbadis.2016.11.034 [DOI] [PubMed] [Google Scholar]
- 21.De Rosa S., Eposito F., Carella C., Strangio A., Ammirati G., Sabatino J. et al. (2018) Transcoronary concentration gradients of circulating microRNAs in heart failure. Eur. J. Heart. Fail. 20, 1000–1010 10.1002/ejhf.1119 [DOI] [PubMed] [Google Scholar]
- 22.Marketou M.E., Kontaraki J.E., Maragkoudakis S., Patrianakos A., Konstantinou J., Nakou H. et al. (2018) MicroRNAs in Peripheral Mononuclear Cells as Potential Biomarkers in Hypertensive Patients with Heart Failure with Preserved Ejection Fraction. Am. J. Hypertens. 31, 651–657 10.1093/ajh/hpy035 [DOI] [PubMed] [Google Scholar]
- 23.Schneider S.I.D., Silvello D., Martinelli N.C., Garbin A., Biolo A., Clausell N. et al. (2018) Plasma levels of microRNA-21, -126 and-423-5p alter during clinical improvement and are associated with the prognosis of acute heart failure. Mol. Med. Rep. 17, 4736–4746 [DOI] [PubMed] [Google Scholar]
- 24.Zhao Y., Li Y.F., Tong L., Liang X.Y., Zhang H., Li L. et al. (2018) Analysis of microRNA Expression Profiles Induced by Yiqifumai Injection in Rats with Chronic Heart Failure. Front. Physiol. 8, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomaniak M., Sygitowicz G., Blaszczyk O., Koltowski L., Puchta D., Malesa K. et al. (2018) miR-1, miR-21, and galectin-3 in hypertensive patients with symptomatic heart failure and left ventricular hypertrophy. Kardiol. Pol. 76, 1009–1011 10.5603/KP.2018.0117 [DOI] [PubMed] [Google Scholar]
- 26.Dubois-Deruy E., Cuvelliez M., Fiedler J., Charrier H., Mulder P., Hebbar E. et al. (2017) MicroRNAs regulating superoxide dismutase 2 are new circulating biomarkers of heart failure. Sci. Rep-U.K. 7, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang J.H., Xing Q., Zhou X.H., Li J.X., Li Y.D., Zhang L. et al. (2017) circulating miRNA-21 is a promising biomarker for heart failure. Mol. Med. Rep. 16, 7766–7774 10.3892/mmr.2017.7575 [DOI] [PubMed] [Google Scholar]
- 28.Jing J.N., Li F.L., Wang X., Wan X., Zhao Q.Q. and Cui X.L. (2017) SR 59230A on the expression of MicroRNAs in myocardium of heart failure rats. Chinese J. Applied Physiol. 33, 456–460 [DOI] [PubMed] [Google Scholar]
- 29.Marques F.Z., Vizi D., Khammy O., Mariani J.A. and Kaye D.M. (2016) The transcardiac gradient of cardio-microRNAs in the failing heart. Eur. J. Heart. Fail. 18, 1000–1008 10.1002/ejhf.517 [DOI] [PubMed] [Google Scholar]
- 30.Xia X. (2014) The relationship of serum miR-30a and heart failure in children with congenital heart disease. J. Chinical Pediatrics 32, 607–609 [Google Scholar]
- 31.Zhao D.S., Chen Y., Jiang H., Lu J.P., Zhang G. and Geng J. (2013) Serum miR-210 and miR-30a expressions tend to revert to fetal levels in Chinese adult patients with chronic heart failure. Cardiovasc. Pathol. 22, 444–450 10.1016/j.carpath.2013.04.001 [DOI] [PubMed] [Google Scholar]
- 32.Chen Y., Dawson K., Wu C.T., Wakili R., Luo X.B. and Kaab S. (2012) Differential Chamber and Cell-specific Expression of MicroRNAs in Experimental Heart Failure. Circulation 126, 18802 [Google Scholar]
- 33.Yan H.L., Li Y.F., Wang C., Zhang Y., Liu C., Zhou K.Y. et al. (2017) Contrary microRNA Expression Pattern Between Fetal and Adult Cardiac Remodeling: Therapeutic Value for Heart Failure. Cardiovasc. Toxicol. 17, 267–276 10.1007/s12012-016-9381-z [DOI] [PubMed] [Google Scholar]
- 34.Lok S.I., de Jonge N., van Kuik J., van Geffen A.J.P., Huibers M.M.H., van der Weide P. et al. (2015) MicroRNA Expression in Myocardial Tissue and Plasma of Patients with End-Stage Heart Failure during LVAD Support: Comparison of Continuous and Pulsatile Devices. Plos. One. 10, 1–13 10.1371/journal.pone.0136404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ikitimur B., Cakmak H.A., Coskunpinar E., Barman H.A. and Vural V.A. (2015) the relationship between circulating microRNAs and left ventricular mass in symptomatic heart failure patients with systolic dysfunction. Kardiol. Pol. 73, 740–746 10.5603/KP.a2015.0082 [DOI] [PubMed] [Google Scholar]
- 36.Fan K.L., Zhang H.F., Shen J., Zhang Q. and Li X.L. (2013) Circulating microRNAs levels in Chinese heart failure patients caused by dilated cardiomyopathy. Indian Heart J. 65, 6–12 10.1016/j.ihj.2012.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blanco R.R., Austin H., Vest R.N., Valadri R., Li W., Lassegue B. et al. (2012) Angiotensin Receptor Type 1 Single Nucleotide Polymorphism 1166A/C is Associated with Malignant Arrhythmias and Altered Circulating miR-155 Levels in Patients with Chronic Heart Failure. J. Card. Fail. 18, 717–723 10.1016/j.cardfail.2012.06.531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greco S., Fasanaro P., Castelvecchio S., D'Alessandra Y., Arcelli D., Di Donato M. et al. (2012) MicroRNA Dysregulation in Diabetic Ischemic Heart Failure Patients. Diabetes 61, 1633–1641 10.2337/db11-0952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guthrie J.L., Seah C., Brown S., Tang P., Jamieson F. and Drews S.J. (2008) Use of Bordetella pertussis BP3385 to establish a cutoff value for an IS481-targeted real-time PCR assay. J. Clin. Microbiol. 46, 3798–3799 10.1128/JCM.01551-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ning B., Dial S., Sun Y., Wang J., Yang J. and Guo L. (2008) Systematic and simultaneous gene profiling of 84 drug-metabolizing genes in primary human hepatocytes. J. Biomol. Screen. 13, 194–201 10.1177/1087057108315513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Livak K.J. and Schmittgen T.D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2 (T) (-Delta Delta C) method. Methods 25, 402–408 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 42.Thomas K.C., Zheng X.F., Suarez F.G., Raftery J.M., Quinlan K.G.R., Yang N. et al. (2014) Evidence Based Selection of Commonly Used RT-qPCR Reference Genes for the Analysis of Mouse Skeletal Muscle. PLoS. One. 9, e88653 10.1371/journal.pone.0088653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pfaffl M.W., Tichopad A., Prgomet C. and Neuvians T.P. (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper - Excel-based tool using pair-wise correlations. Biotechnol. Lett. 26, 509–515 10.1023/B:BILE.0000019559.84305.47 [DOI] [PubMed] [Google Scholar]
- 44.Andersen C.L., Jensen J.L. and Ørntoft T.F. (2004) Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer. Res. 64, 5245–5250 10.1158/0008-5472.CAN-04-0496 [DOI] [PubMed] [Google Scholar]
- 45.Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A. et al. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome. Biol. 3, research0034 10.1186/gb-2002-3-7-research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Silver N., Best S., Jiang J. and Thein S.L. (2006) Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC. Mol. Biol. 7, 33 10.1186/1471-2199-7-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.