Abstract
Tatton-Brown-Rahman (TBRS) syndrome is a recently described overgrowth syndrome caused by loss of function variants in the DNMT3A gene. This gene encodes for a DNA methyltransferase 3 alpha, which is involved in epigenetic regulation, especially during embryonic development. Somatic variants in DNMT3A have been widely studied in different types of tumors, including acute myeloid leukemia, hematopoietic, and lymphoid cancers. Germline gain-of-function variants in this gene have been recently implicated in microcephalic dwarfism. Common clinical features of patients with TBRS include tall stature, macrocephaly, intellectual disability (ID), and a distinctive facial appearance. Differential diagnosis of TBRS comprises Sotos, Weaver, and Malan Syndromes. The majority of these disorders present other clinical features with a high clinical overlap, making necessary a molecular confirmation of the clinical diagnosis. We here describe seven new patients with variants in DNMT3A, four of them with neuropsychiatric disorders, including schizophrenia and psychotic behavior. In addition, one of the patients has developed a brain tumor in adulthood. This patient has also cerebral atrophy, aggressive behavior, ID, and abnormal facial features. Clinical evaluation of this group of patients should include a complete neuropsychiatric assessment together with psychological support in order to detect and manage abnormal behaviors such as aggressiveness, impulsivity, and attention deficit-hyperactivity disorder. TBRS should be suspected in patients with overgrowth, ID, tall stature, and macrocephaly, who also have some neuropsychiatric disorders without any genetic defects in the commonest overgrowth disorders. Molecular confirmation in these patients is mandatory.
Subject terms: Genetics research, Medical genetics
Introduction
Overgrowth syndromes (OGS) comprise a heterogeneous group of disorders whose main characteristic is that either the weight, height, or head circumference, (often also occurring together) are above the 97th centile or 2–3 standard deviations (SD) above the mean for age and gender [1]. Most of the OGS are associated with other clinical features that sometimes overlap between them, making the clinical diagnosis a challenge for pediatricians and geneticists.
Tatton-Brown-Rahman syndrome (TBRS) (MIM 615879) is a relatively recent described OGS disorder [2]. Cardinal features of this condition include overgrowth (mainly tall stature and macrocephaly), intellectual disability (ID), and distinctive facial appearance consisting of an oval face, horizontal eyebrows, and narrow palpebral fissures (Supplementary Table 1) [3]. TBRS is caused by heterozygous loss of function variants in the methyltransferase 3 alpha (DNMT3A) gene located on chromosome band 2p23.3. DNMT3A encodes an enzyme involved in epigenetic programming by de novo methylation, maintenance, and remodeling of DNA methylation patterns, and a more recently uncovered function in transcriptional activation [4, 5]. The DNA methyltransferase family of proteins comprises a group of enzymes that catalyze the transfer of a methyl group to DNA. Three active DNA methyltransferases have been described in mammals (DNMT1, DNMT3A, and DNMT3B). DNMT1 is the gene responsible of ADCADN (Autosomal dominant cerebellar ataxia, deafness, and narcolepsy, MIM# 604121) [6]. Depression and psychosis have also been reported in patients with ADCADN [7], suggesting that variants in DNMT1 may result in an aberrant gene expression or silencing of particular neuronal cells. DNMT1 is also involved in sensory hereditary neuropathy type IE characterized by adult onset of progressive peripheral sensory loss associated with progressive hearing impairment and early onset dementia [8]. Immunodeficiency-centromeric instability-facial anomalies syndrome 1 (MIM# 242860) is caused by autosomal recessive variants in DNMT3B, being the most frequent clinical features the facial dysmorphism, ID, recurrent respiratory, skin, and gastrointestinal infections, and a variable immunodeficiency with constant decrease of IgA.
Both DNMT3A and DNMT3B proteins have a key role in epigenetic regulation by de novo methylation in the human genome [9]. DNMT3A contains three well established functional domains consisting in a tetrapeptide PWWP (Pro-Trp-Trp-Pro motif) domain, a polybromo homology domain (PHD) and a methyltransferase domain (MTase).
TBRS is a rare, possibly underdiagnosed condition with an unknown prevalence. Up to date, there are 79 patients formally reported or mentioned in the literature (55 summarized in [10]), most of them diagnosed by means of NGS DNA sequencing technologies, which has made possible the diagnosis of individuals with ID and/or overgrowth. Reported pathogenic variants in TBRS include missense, nonsense, and frameshift changes in DNMT3A, and at least one case had a microdeletion of 2p23 including this gene, supporting haploinsufficiency as the pathogenetic mechanism of this disorder [11]. Isolated cases and vertical transmission in some pedigrees demonstrated autosomal dominant inheritance with variable expressivity [3].
Somatic variants in DNMT3A have been reported in acute myeloid leukemia (AML, MIM# 601626) [12, 13] and other hematologic malignancies by epigenetic changes due to alterations in the methyltransferase activity of the enzyme. Thus, abnormal methylation pattern of DNMT3A seems to be a major factor for cancer development. Functional analysis of the variant p.Arg882His, which is located in the MTase domain of DNMT3A, showed strong changes in the flanking sequence preference of DNTM3A in different CpG sites, which in turn alters the DNA interaction of DNMT3A [14]. There is only one reported case of TBRS with development of AML [15], while there are others TBRS patients with the same DNMT3A variants (commonly associated with cancer) that have not presented neoplasia up to date [16]. Thus, the relationship between TBRS and neoplasia due to constitutive variants in DNMT3A is not clear yet. Besides, germline gain of function variants in DNMT3A have been recently implicated in microcephalic dwarfism, a group of conditions of profound size reduction in humans [29].
Schizhophrenia (SZ) is a multifactorial neurological disorder with a heterogeneous complexity, making hard to establish the underlying genetic causative defect. Dysregulation in metabolic pathways represents an important and frequent mechanism in SZ. In fact, several reports have pointed out the importance of this mechanism in the physiopathogenesis of SZ [17–19]. Interestingly, DNMT3A is also involved in several metabolic pathways, including cysteine and methionine metabolism through the catalysis of the transfer of a methyl group from an S-adenosyl-L-methionine to S-adenosyl-L-homocysteine.
Herein, we report seven patients with novel disease-causing variants in DNMT3A, four of them with different degrees of neuropsychiatric disorders, encompassing psychotic behavior, schizophrenia, and hallucinations. We also briefly review all the TBRS cases reported so far.
Material and methods
Patients
Patients were selected from the Spanish Overgrowth Syndromes Registry (SOGRI), which includes about 2000 individuals and relatives with overgrowth disorders. This study has been approved by the ethical committee of Hospital Universitario La Paz and informed consent was obtained from all patients and/or parents. Neuropsychiatric findings were evaluated by an specialist for each patient.
In addition to the SOGRI patients, a revision of all previously reported individuals in the scientific literature has been made and the phenotypes of these cases have been compared with the SOGRI patients described in this report.
Genetic analysis
NGS libraries were constructed from peripheral blood leukocyte DNA with the Roche SeqCap EZ Kit with a custom NGS panel which contains 212 genes and seven genomic loci related to OGS (overgrowth panel v.2.3). NGS sequencing has been performed using a NextSeq500 equipment (Illumina, USA). Data analysis consisted first, in transforming the bcl files from the NextSeq500 sequencer to fastq files by means of the Illumina des-multiplexing tool bcl2fastq. Then, the sequences were mapped to the UCSC human reference genome, hg19 (version February 2009) with Bowtie2 [20]. Duplicate reads were removed using Picard’s MarkDuplicates function (http://picard.sourceforge.net/). Indel realignment and base quality score recalibration was performed afterward (RealignerTargetCreator, IndelRealigner and BaseRecalibrator functions from the suite GATK) following the best practices proposed by GATK [21]. Variant calling was performed over the realigned and recalibrated BAM files. The variants characterized were the result of an in-house consensus criterion between the outputs of the GATK variant callers UnifiedGenotyper and HaplotypeCaller. The consensus vcf files were filtered and annotated with Annovar [22]. In addition, the vcf file was enriched with prediction tools of pathogenicity provided by the proxy dbNSFP [23] (release 3.0) together with population data (Exac Non-Finnish European data) [24] and the CIBERER Spanish Variant Server (http://csvs.babelomics.org/), clinical, and genomic information. Candidate variants were validated by Sanger sequencing according to the standard procedures and electropherograms were analyzed with Sequencher v4.1.4 (Genecodes, USA). For patient four, whole exome sequencing was performed with the Ion AmpliSeq exome (ThermoFisher, USA), which captures >97% of the consensus coding sequences, >90% base on-target, and >90% coverage uniformity. Sequencing was performed by an Ion Proton instrument (ThermoFisher, USA) and reads were then aligned against the human reference sequence built 37/hg19. Interpretation of variants was made according to the American College of Medical Genetics and Genomics (ACMG) guidelines [25]. These guidelines classify variants in five groups: benign, likely benign, variant of unknown significance, likely pathogenic, and pathogenic.
Results
Clinical description of patients
Patient 1
A 25 years old male son of a non-consanguineous couple. He was born at term by Cesarean section due to cephalo-pelvic disproportion. Preeclampsia and macrosomia during pregnancy were observed. At birth: weight: 4.58 kg (+3.11 SD), height: 56 cm (+2.23 SD), occipitofrontal circumference: 38 cm (+1.3 SD), Apgar 9–10. He had generalized hypotonia, left hip subluxation, and macroglossia. Sitting at 7 month, crawling at 13 months, walking at 23 months. First syllables at 6 month, first words at 13 months, first phrases of two words at 28 months and three or more at 33 months.
At 30 months old he was hospitalized due to febrile seizures. Generalized developmental delay was detected with abnormal behavior. Physical examination at 12 years old: weight: 79.2 kg (+3.24 SD), height: 162.5 cm (+1.77 SD), head circumference: 57.8 cm (+2.79 SD). He has macrocephaly and his facial features include a long oval face with large forehead, thick eyebrows, hypertelorism, downslanting and narrow palpebral fissures, broad nasal base, deep philtrum, and high palate. The hands had thin fingers and there was an enlarged circumference of the leg with marked hypertrophy and mild bilateral hallux valgus. His karyotype was 46,XY, without pathogenic CNVs in the microarray analysis. MS-MLPA for Prader-Willi/Angelman syndromes did not show any abnormality. He was enrolled at SOGRI as a patient with non-syndromic OGS with ID.
A neuropsychiatric evaluation revealed crisis of aggressiveness to objects in response to auditory hallucinations, visual and kinesthetic hallucinatory symptomatology that disappeared when readjusting antipsychotics. At 20 years he was referred to the hospital due to a heteroaggressive episode with severe behavior alterations and psychotic symptoms due to persistent auditory hallucinations.
Patient 2
A male aged 26 years, son of a non-consanguineous couple. His parents were 29 and 32 years old at the moment of the child’s birth, respectively. His maternal grandmother suffered from breast cancer. Pregnancy was controlled with maternal obesity. He was born at term; his mother suffered from hyperglycemia and eclampsia requiring Cesarean section. At birth: weight was 4.5 kg (+2.91 SD) and height 52 cm (+0.7 SD). Head circumference and Apgar information were not available. He was hospitalized at the neonatal period due to urinary infection with bilateral hydronephrosis demonstrated by dilatation of renal calyces. During his infancy he was operated of adenoid hypertrophy, strabismus, and ear tube drainage. He had mild ID. Physical examination at 14 years: weight: 73 kg (+1.6 SD), height: 184 cm (+2.59 SD), head circumference: 58 cm (+2.22 SD). Long oval face, large forehead, thick horizontal eyebrows, deep philtrum, scoliosis, enlarged breast button, and cutis laxa. A brain computed tomography evidenced discrete frontotemporal atrophy.
He was initially enrolled at SOGRI with the diagnosis of Sotos syndrome (SoS) (MIM 117550) (Fig. 1). His karyotype was 46,XY. Previous MLPA and Sanger sequencing of the 23 exons of NSD1 gene did not find deletions/duplications or single nucleotide pathogenic variants.
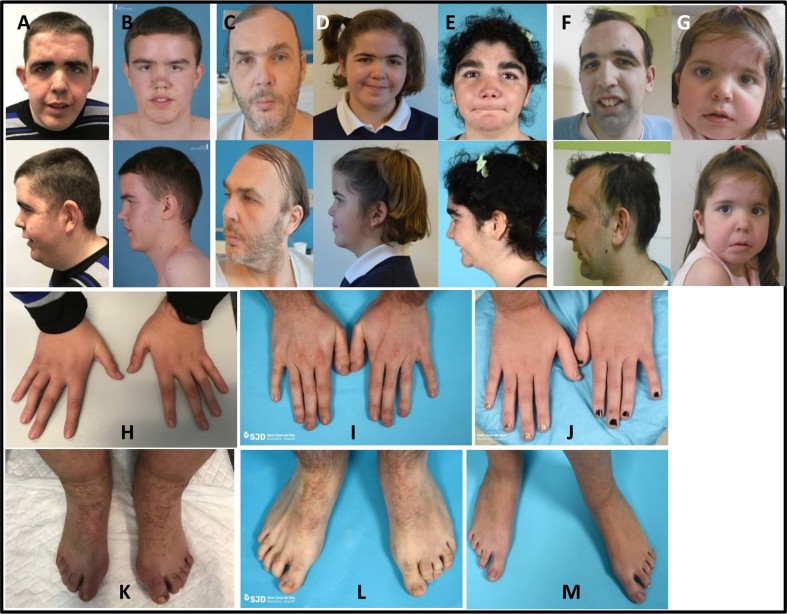
Fig. 1.
Phenotypic features of seven patients with variants in DNMT3A. a–g (Patients 1–7, respectively): Note facial dysmorphic features with macrocephaly, horizontal and thick eyebrows. Also, large hands with large fingers and abnormal feet. h and k: patient 1; i and l: patient 2; j and m: patient 5
Patient 3
A 48-year-old male, referred to Neurology Department because of paranoid schizophrenia, aggressiveness, and cerebral atrophy. He had been followed-up due to a diagnosis of paranoid behavior. He also has moderate ID with tall stature, joint hypermobility, multiple nevus, mitral regurgitation, abnormal facial shape with long oval face, thick eyebrows, and narrow palpebral fissures (Fig. 1c).
The patient was admitted at the hospital several times since 2010, because of lack of pharmacopsychiatric adherence. One year ago, a MRI screening showed abnormalities compatible with a cerebral tumor, which was later confirmed with a biopsy. The pathology report demonstrated that he had a benign glioma without histological signs of aggressiveness. He also presented impossibility to control the sphincters and difficult to sleep with prolonged drowsiness.
Patient 4
A 10-year-old female, third child of a non-consanguineous parents. Normal pregnancy, preterm birth at 36 weeks with 3300 g of weight (90th centile). Developmental delay was detected during first months of life. Autonomous ambulation was at 26 months of life. At 2 years she presented an episode of febrile seizure. At 6 years old, she suffered for speech delay without behavior abnormalities, needing a speech therapist. Anthropometric parameters were in the medium-high centile: 105.5 cm height (75–90 centile), 21.5 kg (97th centile). She has an abnormal facial shape including broad forehead, midface hypoplasia with short nasal bridge and short neck (Fig. 1d).
Patient 5
A 17-year-old female, the daughter of a non-consanguineous couple with neither history of cancer nor ID. The couple has had three previous spontaneous abortions. Delivery was at term. Birth weight, height, and head circumference were 2.9 kg (50th centile), 51 cm (75th centile), and 35 cm (75th centile), respectively.
Since birth, overgrowth was evident with tall stature and macrocephaly. She also presented facial dysmorphic features such as anteverted nares, downslanting palpebral fissures, ocular hypertelorism, long philtrum, low anterior hairline, thick eyebrow, thin upper lip vermilion, large earlobes, and hypertrichosis. Abnormalities of the skeletal system included genu valgum and recurvatum, broad hallux, joint hypermobility with laxity and pes planus (Fig. 1i, l). She presented moderate ID with cognitive impairment, motor delay, delayed gross motor development with delayed speech and language development, seizures, and stereotypic behavior (Fig. 1e).
Patient 6
A 30-year-old male patient who was referred to the Department of Clinical Genetics due to Marfanoid-like features (tall stature and aortic root dilatation), ID, and schizophrenic behavior. Pregnancy was normal and delivery at term with a weight of 4000 g (90th centile), height of 55 cm (>97th centile); head circumference was not reported. Family history revealed that the father suffered for aortic root dilatation and lung cancer and the mother presented lupus erythematosus without any cardiac defect. During the early childhood, mild to moderate global developmental delay was detected with speech and language delay. Therefore, he attended to a special school. He was operated of both umbilical hernia and micrognathia. Tall stature and macrocephaly were evident from infancy and facial dysmorphic features included high forehead with high anterior hairline, deeply set eyes, downslanted palpebral fissures, ocular hypertelorism, low-set ears, micrognathia, and central incisor macrodontia (Fig. 1). He also suffered for myopia. He manifested psychotic episodes with schizophrenia symptoms since 18 years old. MRI did not show any abnormality, and echocardiogram confirmed the artery root dilatation of 57 mm, requiring replacement surgery. Initial molecular tests discarded Marfan syndrome (FBN1 variants negative), TGFRB1 and TGFRB2 negative (both Sanger sequencing and MLPA were normal) (data not shown), SKY negative (only exon 1). CGH microarrays did not show any rearrangement. Due to the suspicious of an OGS, whole exome sequencing was requested.
Patient 7
This girl is the second child of a non-consanguineous Caucasian couple. Her father and her older sister presented macrocephaly without ID. Pregnancy without complications. She was born at term by Cesarean section because of macrosomia. An abdominal mass was detected at 2 months of age; final diagnosis was Neuroblastoma. She undertook surgery at 6 months of age. Developmental assessment at 8 month-old showed remarkable hypotonia and global developmental delay. She also has marked joint laxity and pes planus. She walked independently at 24 months of age. Language delay was also noted with dysarthria and hypophonia. At 5-year-old she attends ordinary school with support.
The patient was referred to the clinical genetics department when she was 2 years and 6 months of age. At that moment anthropometric parameters showed weight 13.9 kg (0 SD), height 94.7 cm (+2 SD), and OFC 52.5 cm (>+2 SD). She presented macrocephaly with turricephaly and broad forehead, downslanted palpebral fissures, and midface hypoplasia.
Comprehensive clinical examination of the patient suggested the possibility of an OGS. To uncover the underlying molecular cause of the phenotype displayed by the patient, whole exome sequencing was performed. A variant in DNMT3A (NM_175629.2:c.919 C>T):p.(Pro307Ser) was detected. The variant has not been previously reported in public databases (dbSNP, 1000 Genomes Project, Exome Variant Server, Genome Aggregation Database). Segregation analysis showed that this variant was a de novo event in the proband.
Molecular results
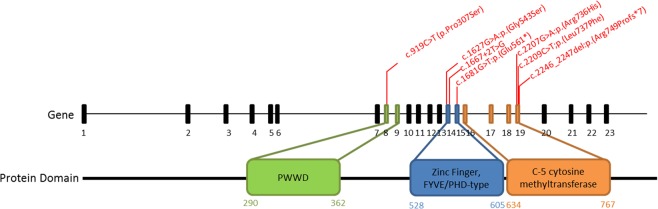
Seven variants in seven unrelated patients were detected by NGS analysis (Table 1 and Fig. 2) (one frameshift, one splice site, one nonsense, and four missense). Three of them are localized within the S-adenosyl-L-methionine (SAM)-dependent MTase c5-type protein domain, two are located within the DNMT1 and DNMT3b interaction motif (Zinc Finger, FYVE/PHD-type) (Table 1) and the last one is located within the PWWD domain. Segregation analysis in available families showed that all of the variants were de novo. None of the variants except the p.(Arg736His) were previously described in the control population databases (gnomAD exomes, gnomAD genomes, Kaviar, 1000 G, ESP). Therefore, this variant was classified as VUS (“variant of unknown significance”). The rest of variants were classified as pathogenic or likely pathogenic. For variant p.(Leu737Phe), the same change has been previously detected in somatic cells in an individual with ovarian serous carcinoma [26].
Table 1.
List of variants detected in DNMT3A in patients
| Variant ID | Pathogenic prediction | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Family | Genomic coordinate (hg19) | cDNA and protein location | Exon/ Intron | mutation type | Protein domain | Population frequencya | COSMIC | Pathogenic predictorsb | ACMG prediction | Inheritance | LOVD ID |
| Family I | chr2:g.25463246_25463246del | NC_000002.12 (NM_022552.4):c.2246_2247del:p.(Arg749Profs*7) | 19 | Frameshift | SAM dependent MTase c5-type | 0 | N/A | (2/2)+35 | P | de novo | #00231357 |
| Family II | chr2:g.25467407T>G | NC_000002.12 (NM_022552.4):c.1667+2T>G | IVS10 | Splicing | Zinc Finger, FYVE/PHD-type | 0 | N/A | (2/2)+29.6 | LP | de novo | #00231367 |
| Family III | chr2:g.25463284C>T | NC_000002.12 (NM_022552.4):c.2209C>T:p.(Leu737Phe). | 19 | Missense | SAM dependent MTase c5-type | 0 | COSM74415c | (8/9)+27.9 | LP | ND | #00231370 |
| Family IV | chr2:g.25463286C>T | NC_000002.12 (NM_022552.4):c.2207G>A:p.(Arg736His) | 19 | Missense | SAM dependent MTase c5-type | 4,81E–02 | N/A | (6/9)+24.7 | VUS | de novo | #00231373 |
| Family V | chr2:g.25467449C>T | NC_000002.12 (NM_022552.4):c.1627G>A:p.(Gly543Ser) | 14 | Missense | Zinc Finger, FYVE/PHD-type | 0 | N/A | (8/10)+25.1 | LP | de novo | #00231374 |
| Family VI | chr2:g.25467194C>A | NC_000002.12 (NM_022552.4):c.1681G>T:p.(Glu561*) | 15 | Nonsense | Zinc Finger, FYVE/PHD-type | 0 | N/A | (4/4)+41 | P | de novo | #00231375 |
| Family VII | chr2:g.25470555G>A | NC_000002.12 (NM_022552.4):c.919C>T p.(Pro307Ser) | 8 | Missense | PWWD | 0 | N/A | (9/9)+27.3 | LP | de novo | #00245882 |
Details of SNVs detected in six patients with TBRS. Variants were classified according to the ACGM guidelines. Reference sequence for variants nomenclature is NC_000002.12 (NM_022552.4). Database for DNMT3A uploaded variants is http://www.lovd.nl/DNMT3A
DC Disease causing, M medium, T tolerated, D damaging/deleterious, P pathogenic, LP likely pathogenic, N/A not applicable, ND not determine
agnomAD exomes, gnomAD genomes, Kaviar, 1000 G phase III, ESP
bdbNSFP (MutationTaster, MutationAssessor, FATHMM, FATHMM-MKL, MetaSVM, MetalR, Provean, LRT, SIFT)+CADD phred score
cOvarian serous carcinoma [26]
Fig. 2.
DNMT3A gene NC_000002.12(NM_022552.4) and DNMT3A protein structure. Details of the seven disease-causing variants described in this work (one at PWWD, three at the Zinc Finger and three at the C-5 cytosine methyltransferase domains)
Three out of the seven variants produce aberrant proteins; two would lead to truncated proteins and one a splice site variant predicting an exon skipping. These three changes may lead to mRNA degradation through the nonsense mediated decay (NMD) machinery. The rest of the changes are missense variants, three classified as likely pathogenic and one as VUS.
Discussion
Since the first publication of variants in DNMT3A related to an overgrowth disorder in 2014 [2], 55 patients have been compiled so far in a recent review [24]. There are other five individuals included by genetic laboratories in the ClinVar database with disease-causing variants and TBRS but without additional clinical information of them. Here we describe seven novel patients with DNMT3A variants emphasizing on the neuropsychiatric findings of this entity.
Clinical features of the published patients with TBRS are listed in Supplementary Table 1. The most common clinical features (frequency above 75%) of these patients include macrocephaly, tall stature, and ID. Other less common features (between 25% and 75% of frequency) are horizontal and thick eyebrows, weight above+2 SD/ obesity, narrow palpebral fissures, broad forehead, hypotonia, and scoliosis. Clinical findings such as the facial gestalt and large hands and feet in present patients are shown in Fig. 1. Facial dysmorphic features allowing distinction with SoS are triangular face with pointed chin and macrodolichocephaly [27]. Also, advanced bone age is very common in SoS (≈80–85% of the cases) but not in TBRS. Skeletal abnormalities are not usually seen in TBRS, helping to distinguish from Weaver syndrome, in which patients suffered for clinodactyly, scoliosis, talipes equinovarus, and pectus excavatum/carinatum in a highest proportion [28].
All the cases presented in this report were macrosomic newborns and have macrocephaly and ID in the follow-up. Anthropometry (when available) at birth showed that five had weight >2 SD. Macrocephaly is a common clinical sign but not present in all cases (50%). Almost all reported cases have an impaired neurocognitive development, mainly mild, or moderate ID (~85% [10]) like the patients describe herein and only ~15% were reported with severe ID. Additional neuropsychiatric abnormalities described in our series include profound global developmental delay, schizophrenia, aggressiveness, attention deficit-hyperactivity disorder, impulsivity, stereotypic behavior, cognitive impairment, and sleep disturbances. Abnormalities of the vision were not reported in TBRS cases, but this is well-known to be present in ~75% of individuals with Weaver syndrome [29], therefore this can give a clue for differential diagnosis between these two conditions.
Facial gestalt in TBRS is different from others overgrowth disorders. It includes a long oval face, thick horizontal eyebrows, narrow palpebral fissures, and marked philtrum. This would be useful to distinguish from others well-known disorders such as SoS, Beckwith-Wiedemann (BWS) (MIM 130650), Simpson-Golabi-Behmel (SGBS) (MIM 312870), Weaver (277590), and Fragile X (MIM 300624) syndromes, among others. Nevertheless, TBRS is still difficult to be distinguished from some recently described overgrowth disorders such as the Thauvin-Robinet-Faivre (TROFAS) (MIM 617107), Tenorio (MIM 616260), Rahman (MIM 617537), Kosaki (MIM 616592), and Luscan-Lumish (LLS; MIM 616831), syndromes because only a few patients have been reported so far. Thus, more individuals and a more detailed clinical description are necessary to have a better understanding of their clinical characteristics. Clinical comparison between TBRS and Weaver, Sotos, and Malan syndrome is detailed in Supplementary Table 3.
For example, patients reported with TROFAS by Akawi et al. [29], have thick eyebrows similar to those with TBRS, but it is not clear whether this feature is typical of the syndrome or corresponds to a family trait. LLS patients [30] have high forehead but their facial findings are clearly different from those observed in SoS and TBRS syndrome.
DNMT3A encodes for a DNA methyltransferase (MTase) alpha 3, a member of the MTases involved in several human development processes by establishing DNA methylation patterns during development, specifically in histones proteins and CpG sites within the genome, playing an important role in paternal and maternal imprinting [31]. Specifically, DNMT3A has three main functional domains. First, the PWWD domain, which is basically a core of Pro-Trp-Trp-Pro and that is present in proteins functioning as transcriptional factors regulating developmental process. Seven variants were detected in TBRS in this domain in previous reports [10] (Supplementary Table 2). We have detected one individual with a missense variant within this domain. Missense gain-of-function variants within the PWWD domain were recently reported in patients with microcephalic dwarfism and the authors suggested that the substitutions impairs the binding of the H3K36me2 and H3K36me3 histone residues, leading to an abnormal DNA methylation in patient cells [32]. Secondly, the zinc finger FYVE/PHD-type domain that is involved in protein-protein and protein-DNA binding interactions. We identified three variants at this domain, one stop gain, one splicing and one missense variant. Thirdly, the SAM dependent MTase C-5 cytosine methyltransferase domain is essential in these proteins to carry out the methyltransferase activity that predominantly regulates the methylation patterns at CpG sites in many imprinted regions. It is recruited to trimethylated Lys-36 of histone H3 (H3K36me3). Thus, the majority of disease-causing variants associated with TBRS are located within exons that encode for this domain. In our series, we detected three additional variants in this domain, two missense variants located in contiguous codons (p.(Arg736His and p.(Leu737Phe)) and one frameshift variant, which was predicted to be degraded by the NMD mRNA processing machinery. Remarkable, the three described patients with variants in the SAM dependent MTase c5-type –all in exon 19- had neuropsychiatric disorders, including schizophrenia with paranoid behavior, which could be related with defects in this domain of DNMT3A and the phenotypic spectrum of the disease. In fact, genetic analysis of DNMT1, DNMT3A, DNMT3B, and DNMT3L in a cohort of patients with schizophrenia, revealed several SNPs in DNMT1 and DNMT3B that are significantly associated with this neuropsychiatric disorder [18]. Specifically, for DNMT3B it has been found that the presence of the polymorphism rs2424932 increases the risk of developing schizophrenia in males but not in females, and the SNP rs1569686 correlates with early onset schizophrenia and also with a family history of schizophrenia [18]. Schizophrenia is a multifactorial disease with a heterogeneous etiology, including genetic predisposition, environmental aspects, drug abuse, and a comorbidity of other diseases. Hundreds of differentially expressed genes have been found to be altered after microarray expression of brain tissue of patients with schizophrenia compared with control samples, but the specific mechanisms and dysregulation that produce this neuropsychiatric disorder remain unclear. Several biological pathways have been found to be involved in schizophrenia: synaptic [33], mitochondrial [17], immune system [34], GABA-ergic [35], and oligodendrocytic [36].
Metabolic pathways have also been seen to have a major role in schizophrenia due to the reduction in expression involved in the regulation of ornithine and polyamine metabolism, mitochondrial malate shuttle system, the transcarboxylic acid cycle, aspartate and alanine metabolism, and ubiquitin metabolism [17]. Also, DNMT3A participates in several metabolic pathways, including the regulation of the cysteine and methionine metabolism by creating a protein complex with DNTM1 and DNMT3B, which catalyzes the conversion of S-adenosyl-L-methionine to S-adenosyl-L-homocysteine+DNA containing 5-methylcytosine. SAM is an enzyme that catalyzes transmethylation reactions. The function is similar to the C5-cytosine MTase domain of DNMT3A. This enzyme was found to be increased in postmortem brains of patients with schizophrenia [37]. Thus, it is possible that defects in the catalytic function of DNMT3A caused by constitutive variants in the SAM dependent MTase c5-type domain found in families 1, 3, and 4 might produce the alterations of metabolic pathways that could finally lead to neuronal defects and the schizophrenic or schizophrenic-like phenotype. Therefore, metabolite accumulations in the brain prefrontal cortex can be observed and lead to changes in both gene expression and protein function that finally lead to the dysfunction in neurons’ communication in response to stimulus.
Recent studies have suggested a possible relationship between methylation hallmarks in dnmt3a and social behavior in adolescent and adult rats, showing that in rats, which suffered for stressful social experience, dnmt3a levels were elevated compared with the control group. Specifically, methylation of several positions in the histone (H3K4, H3K9, and H3K14ac) was different between these two groups [38].
Previous descriptions of patients with variants in exon 19 of DNMT3A have not reported schizophrenia, although neuropsychiatric symptoms such as depression, anxiety, attention deficit- hyperactivity disorder, and behavior problems have been clearly described [39]. Furthermore, one of the patients described in the present report (patient 3; Fig. 1c) developed a brain tumor with right intraventricular lesions caused by glial proliferation without histological signs of aggressiveness and another patient developed a neuroblastoma requiring surgery at the age of 6 months. A potential link among germline variants in DNMT3A and tumors has to be further explored in order to investigate whether individuals with DNMT3A abnormalities have an increased risk of tumor development.
Patient 4 was clinically diagnosed with Simpson-Golabi-Behmel syndrome, but GPC3 analysis was negative. After that, Beckwith–Wiedemann was also suspected but 11p15.5 MS-MLPA was also negative. Therefore, she was included in whole exome sequencing, which reveals a de novo missense variant in DNMT3A:p.(Arg736Hys). This variant is located within the C-5 cytosine methyltransferase domain, in which most of the disease-causing variants in TBRS were described. In addition to the de novo DNMT3A variant, a second variant was also found in COL11A1 (NM_001854.3:p.(Ile708Thr)), in this patient who was inherited from her father. Variants in COL11A1 are related with Stickler syndrome type II in an autosomal dominant inheritance mode. Thus, the variant in COL11A1 in this patient may have contributed with the phenotypic characteristics. In fact, ophthalmological and hearing defects are commonly related to Stickler syndrome, such as myopia and auditory impairment, and clinical screening of these features have to be considered in the follow-up of the patient and her father. This observation reinforces the notion that about 0.5–1.5% of patients that are screened by WES have more than one disease-causing variant associated with more than one different monogenic disease [40–43].
In patient 6 and his father, aortic root dilatation was detected, which in combination with the tall stature suggested a Marfan syndrome as the first diagnosis. FBN1 variants were discarded, and then other genes were screened, including TGFBR1, being all negative. There is no previous evidence for cardiac anomalies in TBRS, suggesting that this finding is probably not caused by the variant in DNMT3A and maybe other genetic/disruptive defects can be involved.
In summary, we report seven new patients with TBRS with novel variants. Among the clinical features of these patients, schizophrenia, hallucinations, seizures, delayed speech, and language development and an abnormal psychiatric behavior seem to be important findings to be added and to follow-up in adult individuals with this disorder. Therefore, after molecular confirmation of TBRS, neuropsychiatric evaluation is strongly recommended in these patients in order to prevent stigmata during childhood and adolescence. In our cohort, overweight and obesity are less frequent than previous reports and none of the patients suffered for severe ID. None of our patients present pes planus compared with the 20% previously reported. Although one of the patients developed a brain tumor, it is unclear its relationship with constitutive genetic defects in DNMT3A. Due to the high overlap between several OGS, massive paralleled techniques such as customized targeted panels, whole exome sequencing or even whole genome sequencing should be the first molecular approach for these patients.
It is also necessary to carry out further studies focusing on methylation signatures, based on the effect of these genes in chromatin regulation, and therefore, to inquire how the changes in these genes are affecting the methylation signature.
Supplementary information
Acknowledgements
We would like to thank all the families and professionals who participate in the SOGRI consortium registry initiative. This project was supported by ISCII, FEDER funds grant: FIS-PI15/01481.
The SOGRI Consortium:
Alberto L. Rosa1,2, Aleixandre Blanquer1,2, Alfredo García Alix1,2, Alfredo Santana1,2, Alicia Delicado1,2, Almudena Alonso1,2, Amaya Rodríguez1,2, Amparo Sanchis1,2, Ana Moreno1,2, Ana Patiño García1,2, Ana Vega1,2, Analía Bredani1,2, Andrea Paula Solari1,2, Andrea Villavicencio1,2, Angelina Acosta1,2, Aníbal Nieto1,2, Anna María Cueto González1,2, Antonio Baldellon1,2, Antonio González Meneses1,2, Antonio Martínez Carrascal1,2, Aranzazu Díaz de Bustamante1,2, Arteche Ocasar1,2, Blanca Gener1,2, Blasco González1,2, Boris Groisman1,2, Bradford Coffee1,2, Carlos Alcalde Martín1,2, Carmen Aragón Fernández1,2, Carmen Benito1,2, Carmen González Armengod1,2, Carmen Martín Seisdedos1,2, Carmen Roche1,2, Claudia Arberas1,2, Claudia Perandones1,2, Claudia Toledo Pacheco1,2, Claudio Contessotto1,2, Cristina Olivas1,2, Daniel Armenta1,2, Denise Cavalcanti1,2, Dolores Elorza1,2, Eduardo Castilla1,2, Elena Zamora1,2, Elisa Zambrano1,2, Elisabeth Steichen1,2, Encarna Guillén Navarro1,2, Enrique Caro Cruz1,2, Enrique Galán Gómez1,2, Enriqueta Román1,2, Ernesto Goldschmidt1,2, Esteban Marfil1,2, Esther Gean1,2, Eugenia Antolín1,2, F. Javier Gascón Jiménez1,2, Fco. Javier Martínez Sarries1,2, Feliciano Ramos1,2, Fermina López Grondona1,2, Fernández Córdoba1,2, Fernando Santos1,2, Fernando Vargas1,2, Francisco Martínez1,2, Giovannucci Uzielli1,2, Gloria Gacio1,2, Graciela Mercado1,2, Hamilton Cassinelli1,2, Ieda Orioli1,2, Ignacio Arroyo1,2, Ignacio Díez López1,2, Ignacio Onsurbe Ramírez1,2, Ignacio Pascual Castroviejo1,2, Ignacio Pascual Pascual1,2, Ignacio Vázquez Rio1,2, Inés Bueno1,2, Isabel Espejo Portero1,2, Isabel Lorda Sánchez1,2, Jaime Sánchez del Pozo1,2, Jaume Campistol1,2, Javier Arcas1,2, Javier Fernández1,2, Javier García Planells1,2, Javier López Pisón1,2, Jesús Barreiro1,2, Jesús del Valle Núñez1,2, Joaquín Fernández Toral1,2, Joaquín Ramírez1,2, Jordi Rosell1,2, Jorge Vilaplana1,2, José Carlos Cabral de Almeida1,2, José Ignacio Labarta1,2, José L. Herranz1,2, José Luis Fernández Luna1,2, José Luis Fuster1,2, José M. Díaz1,2, José M. Gairi1,2, José Miguel García Sagredo1,2, Juan A. Piñero1,2, Juan Carlos López Gutiérrez1,2, Juan Manuel Fernández1,2, Juan P. López Siguero1,2, Juan Tovar1,2, Judith Armstrong1,2, Julián Lara1,2, Laura Rodríguez Leandro Soriano1,2, Leila Cardoso1,2, Leonor Arranz1,2, Liliana De Alba1,2, Loreta Cimbalistiene1,2, Loreto Martorell1,2, Luis González Gutiérrez Solana1,2, Luis Pérez Jurado1,2, M Asunción López Ariztegui1,2, M. Antonia Molina1,2, M. Cruz García1,2, M. Ferrer Lozano1,2, M. Jesús Alija Merillas1,2, M. Luisa Martínez-Frías1,2, M. Rocío Jadraque1,2, Mª Asunción García Pérez1,2, Mª Montserrat Rodríguez Pedreira1,2, Mª Nieves Martínez Guardia1,2, Mª Pilar Ribate1,2, Mª Teresa González López1,2, Mª Teresa Moral Pumarega1,2, Mabel Segovia1,2, Macarena Lizama1,2, Manuel J. Lorente1,2, Manuel Pombo1,2, Margarita Martínez1,2, Margarita Tabernero1,2, María Antonia Ramos1,2, María Ballesta1,2, María Belar1,2, María Jesús Lautre1,2, Marta Cruz1,2, Mercedes Artigas1,2, Mercedes Villanueva1,2, Meritxell Torrabías1,2, Miguel del Campo1,2, Miguel Tomás Vila1,2, Miguel Urioste1,2, Mónica Rosello1,2, Nazneen Rahman1,2, Nik Kantaputra1,2, Pablo Prieto Matos1,2, Paloma Dorao1,2, Paula Casano1,2, Paula Lalaguna Mallada1,2, Pedro Olivares1,2, Pilar Tirado1,2, Pricila Bernardi1,2, Rafael Camino León1,2, Ramón Cañete1,2, Ramón Gaztañaga1,2, Ramón Velazquez1,2, Ramón Vidal Samahuja1,2, Raquel Pérez Delgado1,2, Raquel Sáez Villaverde1,2, Ricardo Gracia1,2, Richard Scott1,2, Rita Valdez1,2, Rosa Arteaga1,2, Rosa Cedeño1,2, Rosario Cazorla1,2, Rosario Marín Iglesias1,2, Rubén Bronberg1,2, Salvador Climent1,2, Santiago Conde Barreiro1,2, Seema Kapoor1,2, Selma Vázquez Martín1,2, Sixto García Miñaur1,2, Soledad Kleppe1,2, Sonia Santillán1,2, Teresa Calvo1,2, Teresa Vendrell1,2, V. López González1,2, Vanesa López1,2, Vanesa Lotersztein1,2, Vanesa Méndez1,2, Vicente Albiach1,2, Víctor M. Navas López1,2, Virginia Soler1,2, Viviana Cosentino, Yoko Aoki1,2
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Pablo Lapunzina, Phone: +34 91 727 72, Email: pablo.lapunzina@salud.madrid.org.
The SOGRI Consortium:
Alberto L. Rosa, Aleixandre Blanquer, Alfredo García Alix, Alfredo Santana, Alicia Delicado, Almudena Alonso, Amaya Rodríguez, Amparo Sanchis, Ana Moreno, Ana Patiño García, Ana Vega, Analía Bredani, Andrea Paula Solari, Andrea Villavicencio, Angelina Acosta, Aníbal Nieto, Anna María Cueto González, Antonio Baldellon, Antonio González Meneses, Antonio Martínez Carrascal, Aranzazu Díaz de Bustamante, Arteche Ocasar, Blanca Gener, Blasco González, Boris Groisman, Bradford Coffee, Carlos Alcalde Martín, Carmen Aragón Fernández, Carmen Benito, Carmen González Armengod, Carmen Martín Seisdedos, Carmen Roche, Claudia Arberas, Claudia Perandones, Claudia Toledo Pacheco, Claudio Contessotto, Cristina Olivas, Daniel Armenta, Denise Cavalcanti, Dolores Elorza, Eduardo Castilla, Elena Zamora, Elisa Zambrano, Elisabeth Steichen, Encarna Guillén Navarro, Enrique Caro Cruz, Enrique Galán Gómez, Enriqueta Román, Ernesto Goldschmidt, Esteban Marfil, Esther Gean, Eugenia Antolín, F. Javier Gascón Jiménez, Fco. Javier Martínez Sarries, Feliciano Ramos, Fermina López Grondona, Fernández Córdoba, Fernando Santos, Fernando Vargas, Francisco Martínez, Giovannucci Uzielli, Gloria Gacio, Graciela Mercado, Hamilton Cassinelli, Ieda Orioli, Ignacio Arroyo, Ignacio Díez López, Ignacio Onsurbe Ramírez, Ignacio Pascual Castroviejo, Ignacio Pascual Pascual, Ignacio Vázquez Rio, Inés Bueno, Isabel Espejo Portero, Isabel Lorda Sánchez, Jaime Sánchez del Pozo, Jaume Campistol, Javier Arcas, Javier Fernández, Javier García Planells, Javier López Pisón, Jesús Barreiro, Jesús del Valle Núñez, Joaquín Fernández Toral, Joaquín Ramírez, Jordi Rosell, Jorge Vilaplana, José Carlos Cabral de Almeida, José Ignacio Labarta, José L. Herranz, José Luis Fernández Luna, José Luis Fuster, José M. Díaz, José M. Gairi, José Miguel García Sagredo, Juan A. Piñero, Juan Carlos López Gutiérrez, Juan Manuel Fernández, Juan P. López Siguero, Juan Tovar, Judith Armstrong, Julián Lara, Laura Rodríguez Leandro Soriano, Leila Cardoso, Leonor Arranz, Liliana De Alba, Loreta Cimbalistiene, Loreto Martorell, Luis González Gutiérrez Solana, Luis Pérez Jurado, M Asunción López Ariztegui, M. Antonia Molina, M. Cruz García, M. Ferrer Lozano, M. Jesús Alija Merillas, M. Luisa Martínez-Frías, M. Rocío Jadraque, Mª Asunción García Pérez, Mª Montserrat Rodríguez Pedreira, Mª Nieves Martínez Guardia, Mª Pilar Ribate, Mª Teresa González López, Mª Teresa Moral Pumarega, Mabel Segovia, Macarena Lizama, Manuel J. Lorente, Manuel Pombo, Margarita Martínez, Margarita Tabernero, María Antonia Ramos, María Ballesta, María Belar, María Jesús Lautre, Marta Cruz, Mercedes Artigas, Mercedes Villanueva, Meritxell Torrabías, Miguel del Campo, Miguel Tomás Vila, Miguel Urioste, Mónica Rosello, Nazneen Rahman, Nik Kantaputra, Pablo Prieto Matos, Paloma Dorao, Paula Casano, Paula Lalaguna Mallada, Pedro Olivares, Pilar Tirado, Pricila Bernardi, Rafael Camino León, Ramón Cañete, Ramón Gaztañaga, Ramón Velazquez, Ramón Vidal Samahuja, Raquel Pérez Delgado, Raquel Sáez Villaverde, Ricardo Gracia, Richard Scott, Rita Valdez, Rosa Arteaga, Rosa Cedeño, Rosario Cazorla, Rosario Marín Iglesias, Rubén Bronberg, Salvador Climent, Santiago Conde Barreiro, Seema Kapoor, Selma Vázquez Martín, Sixto García Miñaur, Soledad Kleppe, Sonia Santillán, Teresa Calvo, Teresa Vendrell, V. López González, Vanesa López, Vanesa Lotersztein, Vanesa Méndez, Vicente Albiach, Víctor M. Navas López, Virginia Soler, Viviana Cosentino, and Yoko Aoki
Supplementary information
The online version of this article (10.1038/s41431-019-0485-3) contains supplementary material, which is available to authorized users.
References
- 1.Lapunzina P. Risk of tumorigenesis in overgrowth syndromes: a comprehensive review. Am J Med Genet C Semin Med Genet. 2005;137C:53–71. doi: 10.1002/ajmg.c.30064. [DOI] [PubMed] [Google Scholar]
- 2.Tatton-Brown K, Seal S, Ruark E, Harmer J, Ramsay E, Del Vecchio Duarte S, et al. Mutations in the DNA methyltransferase gene DNMT3A cause an overgrowth syndrome with intellectual disability. Nat Genet. 2014;46:385–8. doi: 10.1038/ng.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lemire G, Gauthier J, Soucy JF, Delrue MA. A case of familial transmission of the newly described DNMT3A-overgrowth syndrome. Am J Med Genet Part A. 2017;173:1887–90. doi: 10.1002/ajmg.a.38119. [DOI] [PubMed] [Google Scholar]
- 4.Lyko F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat Rev Genet. 2018;19:81–92. doi: 10.1038/nrg.2017.80. [DOI] [PubMed] [Google Scholar]
- 5.Rinaldi L, Datta D, Serrat J, Morey L, Solanas G, Avgustinova A, et al. Dnmt3a and Dnmt3b associate with enhancers to regulate human epidermal stem cell homeostasis. Cell Stem Cell. 2016;19:491–501. doi: 10.1016/j.stem.2016.06.020. [DOI] [PubMed] [Google Scholar]
- 6.Melberg A, Hetta J, Dahl N, Nennesmo I, Bengtsson M, Wibom R, et al. Autosomal dominant cerebellar ataxia deafness and narcolepsy. J Neurol Sci. 1995;134:119–29. doi: 10.1016/0022-510x(95)00228-0. [DOI] [PubMed] [Google Scholar]
- 7.Winkelmann J, Lin L, Schormair B, Kornum BR, Faraco J, Plazzi G, et al. Mutations in DNMT1 cause autosomal dominant cerebellar ataxia, deafness and narcolepsy. Hum Mol Genet. 2012;21:2205–10. doi: 10.1093/hmg/dds035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klein CJ, Botuyan MV, Wu Y, Ward CJ, Nicholson GA, Hammans S, et al. Mutations in DNMT1 cause hereditary sensory neuropathy with dementia and hearing loss. Nat Genet. 2011;43:595–600. doi: 10.1038/ng.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–57. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 10.Tatton-Brown K, Zachariou A, Loveday C, Renwick A, Mahamdallie S, Aksglaede L, et al. The tatton-brown-rahman syndrome: a clinical study of 55 individuals with de novo constitutive DNMT3A variants. Wellcome Open Res. 2018;3:46. doi: 10.12688/wellcomeopenres.14430.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okamoto N, Toribe Y, Shimojima K, Yamamoto T. Tatton-Brown-Rahman syndrome due to 2p23 microdeletion. Am J Med Genet Part A. 2016;170A:1339–42. doi: 10.1002/ajmg.a.37588. [DOI] [PubMed] [Google Scholar]
- 12.Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, et al. DNMT3A mutations in acute myeloid leukemia. New Engl J Med. 2010;363:2424–33. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamashita Y, Yuan J, Suetake I, Suzuki H, Ishikawa Y, Choi YL, et al. Array-based genomic resequencing of human leukemia. Oncogene. 2010;29:3723–31. doi: 10.1038/onc.2010.117. [DOI] [PubMed] [Google Scholar]
- 14.Emperle M, Rajavelu A, Kunert S, Arimondo PB, Reinhardt R, Jurkowska RZ, et al. The DNMT3A R882H mutant displays altered flanking sequence preferences. Nucleic acids Res. 2018;46:3130–9. doi: 10.1093/nar/gky168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hollink I, van den Ouweland AMW, Beverloo HB, Arentsen-Peters S, Zwaan CM, Wagner A. Acute myeloid leukaemia in a case with Tatton-Brown-Rahman syndrome: the peculiar DNMT3A R882 mutation. J Med Genet. 2017;54:805–8. doi: 10.1136/jmedgenet-2017-104574. [DOI] [PubMed] [Google Scholar]
- 16.Shen W, Heeley JM, Carlston CM, Acuna-Hidalgo R, Nillesen WM, Dent KM, et al. The spectrum of DNMT3A variants in Tatton-Brown-Rahman syndrome overlaps with that in hematologic malignancies. Am J Med Genet Part A. 2017;173:3022–8. doi: 10.1002/ajmg.a.38485. [DOI] [PubMed] [Google Scholar]
- 17.Middleton FA, Mirnics K, Pierri JN, Lewis DA, Levitt P. Gene expression profiling reveals alterations of specific metabolic pathways in schizophrenia. J Neurosci. 2002;22:2718–29. doi: 10.1523/JNEUROSCI.22-07-02718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saradalekshmi KR, Neetha NV, Sathyan S, Nair IV, Nair CM, Banerjee M. DNA methyl transferase (DNMT) gene polymorphisms could be a primary event in epigenetic susceptibility to schizophrenia. PloS One. 2014;9:e98182. doi: 10.1371/journal.pone.0098182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhubi A, Veldic M, Puri NV, Kadriu B, Caruncho H, Loza I, et al. An upregulation of DNA-methyltransferase 1 and 3a expressed in telencephalic GABAergic neurons of schizophrenia patients is also detected in peripheral blood lymphocytes. Schizophr Res. 2009;111:115–22. doi: 10.1016/j.schres.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu X, Wu C, Li C, Boerwinkle E. dbNSFPv3.0: a one-stop database of functional predictions and annotations for human nonsynonymous and splice-site SNVs. Hum Mutat. 2016;37:235–41. doi: 10.1002/humu.22932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fickie MR, Lapunzina P, Gentile JK, Tolkoff-Rubin N, Kroshinsky D, Galan E, et al. Adults with Sotos syndrome: review of 21 adults with molecularly confirmed NSD1 alterations, including a detailed case report of the oldest person. Am J Med Genet Part A. 2011;155A:2105–11. doi: 10.1002/ajmg.a.34156. [DOI] [PubMed] [Google Scholar]
- 28.Tatton-Brown K, Murray A, Hanks S, Douglas J, Armstrong R, Banka S, et al. Weaver syndrome and EZH2 mutations: Clarifying the clinical phenotype. Am J Med Genet Part A. 2013;161A:2972–80. doi: 10.1002/ajmg.a.36229. [DOI] [PubMed] [Google Scholar]
- 29.Akawi N, Ben-Salem S, Lahti L, Partanen J, Ali BR, Al-Gazali L. A recessive syndrome of intellectual disability, moderate overgrowth, and renal dysplasia predisposing to Wilms tumor is caused by a mutation in FIBP gene. Am J Med Genet Part A. 2016;170:2111–8. doi: 10.1002/ajmg.a.37741. [DOI] [PubMed] [Google Scholar]
- 30.Luscan A, Laurendeau I, Malan V, Francannet C, Odent S, Giuliano F, et al. Mutations in SETD2 cause a novel overgrowth condition. J Med Genet. 2014;51:512–7. doi: 10.1136/jmedgenet-2014-102402. [DOI] [PubMed] [Google Scholar]
- 31.Soellner L, Begemann M, Mackay DJ, Gronskov K, Tumer Z, Maher ER, et al. Recent advances in imprinting disorders. Clin Genet. 2017;91:3–13. doi: 10.1111/cge.12827. [DOI] [PubMed] [Google Scholar]
- 32.Heyn P, Logan CV, Fluteau A, Challis RC, Auchynnikava T, Martin CA, et al. Gain-of-function DNMT3A mutations cause microcephalic dwarfism and hypermethylation of Polycomb-regulated regions. Nat Genet. 2018;51:96–105. doi: 10.1038/s41588-018-0274-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Faludi G, Mirnics K. Synaptic changes in the brain of subjects with schizophrenia. Int J Dev Neurosci. 2011;29:305–9. doi: 10.1016/j.ijdevneu.2011.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arion D, Unger T, Lewis DA, Levitt P, Mirnics K. Molecular evidence for increased expression of genes related to immune and chaperone function in the prefrontal cortex in schizophrenia. Biol Psychiatry. 2007;62:711–21. doi: 10.1016/j.biopsych.2006.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hashimoto T, Arion D, Unger T, Maldonado-Aviles JG, Morris HM, Volk DW, et al. Alterations in GABA-related transcriptome in the dorsolateral prefrontal cortex of subjects with schizophrenia. Mol psychiatry. 2008;13:147–61. doi: 10.1038/sj.mp.4002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hakak Y, Walker JR, Li C, Wong WH, Davis KL, Buxbaum JD, et al. Genome-wide expression analysis reveals dysregulation of myelination-related genes in chronic schizophrenia. Proc Natl Acad Sci USA. 2001;98:4746–51. doi: 10.1073/pnas.081071198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guidotti A, Ruzicka W, Grayson DR, Veldic M, Pinna G, Davis JM, et al. S-adenosyl methionine and DNA methyltransferase-1 mRNA overexpression in psychosis. Neuroreport. 2007;18:57–60. doi: 10.1097/WNR.0b013e32800fefd7. [DOI] [PubMed] [Google Scholar]
- 38.Karen C, Rajan KE. Social behaviour and epigenetic status in adolescent and adult rats: the contribution of early-life stressful social experience. Cell Mol Neurobiol. 2019;39:371–85. doi: 10.1007/s10571-019-00655-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xin B, Cruz Marino T, Szekely J, Leblanc J, Cechner K, Sency V, et al. Novel DNMT3A germline mutations are associated with inherited Tatton-Brown-Rahman syndrome. Clin Genet. 2017;91:623–8. doi: 10.1111/cge.12878. [DOI] [PubMed] [Google Scholar]
- 40.Yang Y, Muzny DM, Xia F, Niu Z, Person R, Ding Y, et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312:1870–9. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, Ward PA, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. New Engl J Med. 2013;369:1502–11. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tarailo-Graovac M, Shyr C, Ross CJ, Horvath GA, Salvarinova R, Ye XC, et al. Exome sequencing and the management of neurometabolic disorders. New Engl J Med. 2016;374:2246–55. doi: 10.1056/NEJMoa1515792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Posey JE, Harel T, Liu P, Rosenfeld JA, James RA, Coban Akdemir ZH, et al. Resolution of disease phenotypes resulting from multilocus genomic variation. New Engl J Med. 2017;376:21–31. doi: 10.1056/NEJMoa1516767. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.