Abstract
The complexity of schizophrenia (SZ) and bipolar disorder (BD) has slowed down progress in understanding their genetic roots. Alternative genomic approaches are needed to bypass these difficulties. We attempted a multimodal approach to follow-up on reported linkage findings in SZ and BD from the Eastern Quebec kindreds in chromosomes 3q21, 4p34, 6p22, 8p21, 8p11, 13q11-q14, 15q13, 16p12, and 18q21. First, in 498 subjects, we measured RNA expression (47 K Illumina chips) in SZ and BD patients that we compared with their non-affected relatives (NARs) to identify, for each chromosomal region, genes showing the most significant differences in expression. Second, we performed SNP genotyping (700 K Illumina chips) and cis-eQTN analysis. Third, we measured DNA methylation on genes with RNA expression differences or eQTNs. We found a significant overexpression of the gene ITGB5 at 3q25 in SZ and BD after multiple testing p value adjustment. SPCS3 gene at 4q34, and FZD3 gene at 8p21, contained significant eQTNs after multiple testing corrections, while ITGB5 provided suggestive results. Methylation in associated genes did not explain the expression differences between patients and NARs. Our multimodal approach involving RNA expression, dense SNP genotyping and eQTN analyses, restricted to chromosomal regions having shown linkage, lowered the multiple testing burden and allowed for a deeper examination of candidate genes in SZ or BD.
Subject terms: Schizophrenia, Genetics research, Bipolar disorder
Introduction
Schizophrenia (SZ) and bipolar disorder (BD) are heritable brain disorders involving impairments in cognition, perception, and motivation that usually manifest late in adolescence or in early adulthood [1, 2]. The pathogenic mechanisms of these brain disorders are unknown, but pathological features involving excessive loss of gray matter have been reported [3], as well as reduced numbers of synaptic structures on neurons [4]. Treatments exist for the psychotic symptoms but no preventive or curative therapies are available [1]. So far, GWAS and family linkage studies have not provided breakthroughs in the illness genomic underpinnings that would be useful to derive therapies, despite genes being undisputedly involved in etiology [5]. New ways of approaching genomic studies and new theories must thus be imagined to decipher the pathophysiology [6, 7].
One major GWAS study has identified 108 loci in the human genome that contain single nucleotide polymorphism (SNP) haplotypes associated with SZ [8]. However, the functional alleles and mechanisms at these loci remain to be elucidated. The identification of genetic cis associations between SNPs and gene expression may help to find functionally relevant pathways that contribute to the etiology of psychiatric disorders [9]. Despite controversies, RNA and protein expression profiles from peripheral blood would present patterns that reflect those found in brain tissue [10–12], and gene expression of peripheral blood lymphocytes may discriminate SZ patients [13–16] as BD patients [17, 18] from controls. Gene expression has also been used in attempts to pinpoint the genes most likely to be the source of genetic signals. For instance, Fromer et al. [19] sequenced RNA from dorsolateral prefrontal cortex in SZ patients and found 693 genes showing differential expression of which the genes FURIN, TSNARE1, CNTN4, CLCN3, and SNAP91 were among the 108 GWAS loci identified previously [8]. Also, Lopez de Lara et al. have successfully combined linkage and expression studies performed in the same set of families [20]. Indeed, they screened 26 BD families with 811 microsatellite markers to identify the linkage regions in which gene expression could identify candidate genes. They found evidence of linkage to lithium-responsive BD on 3p25, 3p14, and 14q11 as well as dysregulated gene expression in these regions suggesting altered synaptic and mitochondrial function [20].
Altered DNA methylation has also been observed in SZ [21] in peripheral leukocytes [22]. For instance, methylation-wide analysis has successfully refined results from GWAS with the top three methylation findings located near genes SDCCAG8, CREB1, and ATXN7 [23]. Analysis of methylation also helped to refine functional/regulatory variations associated with SZ polygenic risk scores and genetic variants in it [24]. With methods combining evaluation of DNA methylation and gene expression levels from whole blood of 260 SZ patients and 250 controls, van Eijk et al. [25] identified 432 CpG sites with differential methylation levels that were also associated with differential gene expression. These findings suggest that inspecting concurrently gene expression and methylation might improve interpretation of findings.
We have already reported significant linkage results in SZ, BD, and both diseases in 12 chromosomal regions from 9 different chromosomes [26–28]. Since then, we have added markers and subjects to the 2005 sample [26] and have reanalyzed the data ending up with very similar updated signals in nine regions from eight different chromosomes (including six of the original regions) as presented in Supplementary Table S1. In all, the latter nine linked regions included some 2051 genes that were all candidates by position.
Herein, our objective was to follow up of these linkage findings and assess the likelihood that some of these 2051 genes be involved in SZ or BD, or in both (CL, common loci). First, as depicted in Supplementary Fig. S1, our multimodal follow-up involved differential gene RNA expression. Second, we selected genes with significant differences in RNA expression for a cis-eQTN (expression Quantitative Trait Nucleotide) analysis to identify SNP that could potentially regulate gene expression. Third, genes showing significant differential RNA expression and/or cis-eQTN analyses were investigated for DNA methylation in relation to gene expression, DNA variations, and phenotypic status. In each region the analysis was restricted to the phenotype yielding the strongest linkage signal. Among the genes in the nine linkage regions, our experimental procedure yielded the best multimodal convergence for ITGB5, SPCS3, and FZD3 located, respectively, on chromosomes 3q21, 4p34, and 8p21.
Materials and methods
Phenotype definition
A best-estimate lifetime DSM-IV diagnosis was made as outlined in previous reports [29–31]. In brief, all available information across lifetime from different sources (all medical records, family informant interviews, personal structured interview) was reviewed blindly by four research diagnosticians. To test our hypothesis, we also defined a “common locus” (CL) phenotype. The CL phenotype included BD, SZ, and the schizoaffective disorder (SAD). Our SZ definition included SZ plus schizophreniform disorder and schizotypal personality. The BD phenotype included both BD I and BD II, and recurrent major depression.
Gene RNA expression measurement
Sample
We analyzed family members from 29 of the original 48 kindreds in Maziade et al. [28], including 17 kindreds showing linkage signals with one or more of the nine linked chromosomal regions giving preference to the families with the greatest number of subjects. This represented 498 subjects: 156 cases affected by SZ (N = 58), SAD (N = 13), or BD (N = 85) and 342 non-affected relatives (NAR) (see Supplementary 1).
RNA expression in immortalized lymphocytes (IML)
The detailed protocol is reported in Supplementary 1. Briefly, RNA harvested from IML in culture for the 498 subjects was hybridized to HumanHT-12 Expression Beadchip v4 (47 K probes; Illumina; Génome Quebec). IML cultures were monitored to be harvested in exponential growth. RNA quality (RNA integrity number or RIN) was evaluated on a Bioanalyser with RNA 6000 Nano Kit (Agilent) and RIN values close to ten were generally observed.
Signal extraction from probe intensities
Probe intensities were background corrected and quantile normalized across all chips. Normalized intensities were then log2 transformed, and called log2-expression values from now on. A probe was declared expressed in a subject if the p value of the Illumina detection test was <0.05. We retained for analysis all probes that were expressed in 75% or more of the subjects from at least one family where at least four members had been measured, to account for the possibility of family-specific expression. Additional details are provided in Supplementary 1.
SNP genotyping
Whole-genome SNP genotyping was made using DNA extracted from IMLs by affinity column (Midi prep Qiagen) for the 498 subjects using Infinium Human OmniExpress24 Illumina microchips (700 K; Genome Quebec). Some SNPs were completed for the remaining subjects of the cohort using TaqMan kit (Life Technologies) and a real time PCR (LightCycler LC480, Roche) where some 10% blind replicates were added for genotyping quality control. Mendelian inheritance within families was also checked. In the case where more than 5% of non-replication or more than five non-Mendelian inheritance is observed, the marker was genotyped again completely after improving genotyping conditions (salt and DNA contents, reaction volume, annealing temperature).
Expression quantitative trait nucleotides (eQTNs) analysis
For the genes listed in Table 1, we evaluated if proximal SNPs within 50 kb of the gene were associated through the RNA expression of this gene (cis-eQTN) to the phenotype identified through linkage in its corresponding chromosomal region. See statistical analysis below for details.
Table 1.
Most significant mean RNA expression ratio (MER) observed between affected and unaffected-relative subjects for the phenotype specific to each linked region
| Gene | Linked region | Probe ID | Probe_Coord | Affected phenotype | MER | z | P nominal | Bonfb | |
|---|---|---|---|---|---|---|---|---|---|
| # probes | p | ||||||||
| ITGB5 | 3q21 | 2650114 | 125,964,619–125,964,668 | CL | 1.124 | 3.85 | 0.000085 | 201 | 0.017 |
| SPCS3 | 4q34 | 7050026 | 177,487,232–177,487,281 | BD | 1.023 | 2.45 | 0.0143 | 13 | 0.186 |
| ATXN1 | 6p22 | 520601 | 16,407,774–16,407,823 | SZ | 0.978 | −2.89 | 0.00385 | 48 | 0.185 |
| 5080154 | 16,407,623–16,407,672 | SZ | 0.958 | −3.07 | 0.00214 | 0.103 | |||
| GOLGA7 | 8p11 | 2100037 | 41,348 085–41,348,134 | BD | 0.944 | −3.14 | 0.00169 | 57 | 0.096 |
| FZD3 | 8p21 | 1470121 | 28,420,818–28,420,867 | CL | 0.957 | −2.81 | 0.00495 | 128 | 0.634 |
| HS.121655 | 13q11 | 6290523 | 52,124,159–52,124,208 | CL | 0.972 | −3.28 | 0.00104 | 113 | 0.118 |
| SZ | 0.962 | −2.97 | 0.00298 | 99 | 0.295 | ||||
| GOLGA8F (LOC100132565)a | 15q13 | 6370280 | 26,301,632–26,301,663 | BD | 1.038 | 1.86 | 0.0629 | 27 | 1.0 |
| CDR2 | 16p12 | 2140228 | 22,357,429–22,357,478 | BD | 1.008 | 2.64 | 0.00829 | 76 | 0.630 |
| C18orf12 (HS.334493)a | 18q21 | 770465 | 44,033,137–44,033,186 | BD | 0.950 | −2.13 | 0.0332 | 44 | 1.0 |
aPrevious name of the gene used on the chips
bBonferroni corrected for the number of probes tested in the linked region
CpG analysis
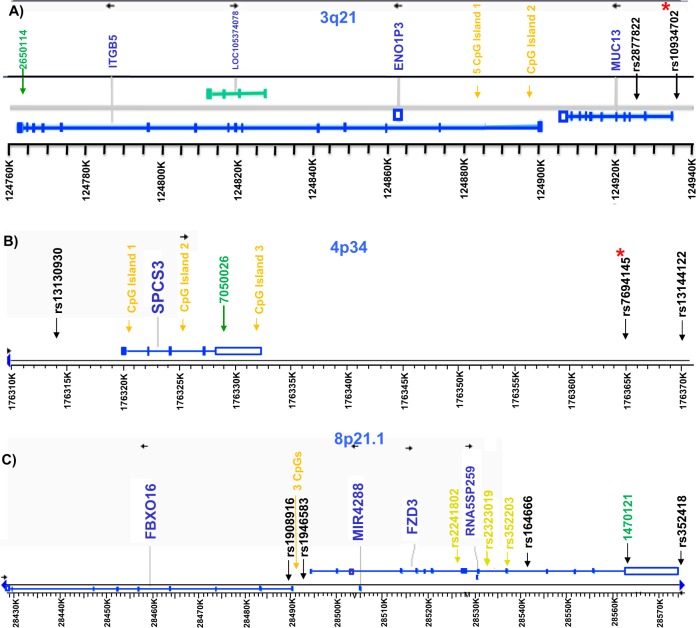
We contrasted DNA methylation in DNA extracted from IMLs between NAR, SZ, and BD subjects according to the level of expression of the selected genes. For the validation run of the chosen CpGs, we selected nine subjects (three NAR, three SZ, and three BD) to evaluate the level of methylation. If DNA methylation was higher than 10% with a good quality call, we selected the first 10 subjects with the lowest, median and highest expression for each NAR, SZ, and BD groups according to adjusted expression values, taking care to avoid overlap between expression categories among phenotypic status. CpG islands were identified [32] and CpG sites selected for analysis. For ITGB5, we analyzed ten CpG sites within a 1.5 Kb region including five CpG islands located at 15 Kb from the 5′ end of ITGB5 (Isl 1), 5 CpGs located in a CpG island located in the first 5 Kb of the gene (Isl 2) and the CpG created by the SNP rs10934702:C > T, NC_000003.11:g.124653868 C > T (see Fig. 1a). For SPCS3, 13 CpG sites located in a CpG island of 721 bp located in the first 1 Kb of the 5′ region of the gene (Isl 1), four CpGs located in intron 3 (Isl 2), four CpGs located in the 3′ untranslated region of the gene (Isl 3), and the CpG created by the SNP rs7694145:A > G, NC_000004.11:g.177285894 A > G (see Fig. 1b) were evaluated in 24 SZ cases, 22 BD cases, and 126 NARs. Finally, for FZD3 three CpG sites located within 3, 22, and 31 nucleotides 5′ of the SNP rs1946583:T > C, NC_000008.10:g.28350753 T > C were analyzed (see Fig. 1c). Initial promising results led us to measure FZD3 methylation in all 58 SZ and 85 BD cases, and in a sample of 199 NARs enriched with subjects having low level of expression. After DNA was treated with bisulfite (EpiTect Fast DNA Bisulfite Kit, Qiagen), 10 ng of cleaned bisulfited DNA was used for PCR followed by DNA pyrosequencing (Pyromark MD, Qiagen). Primers used and sequences analyzed after bisulfite treatment can be found online in Supplementary 2. A biotin was added 5′ of the reverse primers to allow capture of a single DNA strand for pyrosequencing.
Fig. 1.
Genomic structure of the genes highlighted in the results section. a ITGB5, b SPCS3, and c FZD3. CpG islands for which methylation was measured are indicated in dark yellow. Single nucleotide polymorphisms (SNPs) are indicated by their reference sequence number. a, b SNPs marked with an asterisk create a CpG site; c SNPs marked with an asterisk showed association with SZ in a Chinese sample [37] (Color figure online)
Statistical analysis
Comparison of gene RNA expression levels between SZ/BD and NAR (analysis 1)
We compared the RNA expression levels of the 156 CL affected subjects to the 342 NARs, and of the 58 SZ and 85 BD subjects to the 342 NAR (after excluding SAD subjects) in separate linear mixed effects models of the log2-expression values of each probe, age at blood draw and sex included as fixed covariates and the Illumina slide and a polygenic effect as random effects to deal with kinship, in addition to the first 10 principal components of 51 housekeeping probes as fixed covariates to control unwanted variation [33]. Mean expression ratios (MERs) were computed as 2b, where b is the estimated coefficient for the phenotype group tested (CL, SZ, or BD). Wald tests were performed on these coefficients, and a Bonferroni correction was applied by multiplying the p values by the number of probes in the linked region.
eQTN analysis (analysis 2)
This analysis involved the same CL, SZ, BD, and NAR samples as the comparison of RNA expression levels above. We used the method of Zhao et al. [34] comprising (1) an outcome model, which is a logistic regression of phenotypic status (either CL, SZ, or BD against NAR) on the log2-expression values of the probe or probes in the gene or genes located near the eQTN, adjusted for age at blood draw, sex and the first 10 principal components of the housekeeping probes, and (2) a transcript model that is a linear regression between the linear predictor of the outcome model and the number of minor alleles of the eQTN. Since the linear predictor of the outcome model captures the association of the gene(s) expression with phenotype, the Wald test of the eQTN coefficient in the transcript model is a test of the eQTN association with phenotype mediated through gene expression. A Bonferroni correction was applied by multiplying the p values by the number of SNPs remaining in each gene after pruning the SNPs on the array so that no r2 exceeds 0.8 in windows of 20 SNPs using Plink (www.cog-genomics.org/plink2).
Comparison of CpGs methylation levels between SZ/BD and NAR (analysis 3)
For FZD3, where we measured methylation in a biased NAR sample, an unbiased estimate of the mean methylation level in NARs was obtained by inverse probability-of-selection weighting. We tested for differences in methylation levels between SZ, BD, and NARs using t-tests, accounting for subject weights in the computation of the standard errors in the denominator of the t statistics.
Ethics and data availability statements
The study was explained to each family member and a signed consent was obtained from the subjects or from a parent for children under 18, as reviewed by the Mental Health and Neuroscience Ethics Review Board of the Centre intégré universitaire de santé et des services sociaux de la Capitale-Nationale (reference numbers MP-13–1996–1, 068–1995).
The RNA expression dataset generated and analysed during the current study is available in the ArrayExpress repository (https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-8018). The SNP genotype dataset generated and analysed during the current study is not publicly available due to absence of study participants consent to publication of their personal information including genotype, but is available from the corresponding author on reasonable request and establishment of a data sharing agreement.
Results
Differentially RNA expressed genes found in several linkage regions
For the phenotype yielding the greatest linkage signal in each chromosomal region (see Supplementary Table S1), Table 1 reports the differentially expressed genes showing the smallest p value of the chromosomal region. Among these genes (see Supplementary Table S2), integrin beta 5 (ITGB5), transcript variant 3, remained significant at the 5% level after adjusting for multiple testing in the 3q21 region (N = 201 probes; p = 0.000085, adjusted p = 0.017), and near significant if adjusted for all probes tested in all regions (N = 686; adjusted p = 0.058). Three other genes, ataxin 1 (ATXN1) at 6p22, golgin A7 (GOLGA7) at 8p11 and the uncharacterized transcript HS.121655 at 13q11 showed suggestive results with adjusted p values below p = 0.15.
eQTNs detected in a few differentially RNA expressed genes
Again for the phenotype yielding the greatest linkage signal in each region, Table 2 presents SNPs associated to the phenotype through the RNA expression of genes with nominally significant MER in analysis 1 using the method of Zhao et al. [34] (analysis 2). Complete results for all SNPs located within the linkage regions and all phenotypes are reported in Supplemental Tables S3–S5. This time, significant or suggestive associations after adjusting for multiple SNPs testing within 50 Kb of each gene were observed for the gene signal peptidase complex subunit 3 (SPCS3) located at 4q32 (adjusted p = 0.027), frizzled class receptor 3 (FZD3) located at 8p21 (adjusted p = 0.052), and ITGB5 at 3q21 (adjusted p = 0.077). We elected to pursue the analysis of these three genes for DNA methylation.
Table 2.
eQTN for genes showing the strongest evidence of differential expression between affected and unaffected-relatives subjects for the phenotype specific to each linked region
| Linked region | Affected phenotype | SNP | Gene expression | ORj | P nominal | #SNPs | Bonfk | Location |
|---|---|---|---|---|---|---|---|---|
| 3q21 | CL | rs2877822:C > Ta | ITGB5 | 0.87 | 0.0370 | 16 | 0.592 | intron 2 MUC13 |
| rs10934702:C > Tb | ITGB5 | 0.81 | 0.0048 | 0.077 | 0.5 kB 5′ MUC13 | |||
| 4q34 | BD | rs13130930:C > Tc | SPCS3 | 0.72 | 0.0123 | 17 | 0.209 | 5 kB 5′ |
| rs7694145:A > Gd | SPCS3 | 0.66 | 0.0016 | 0.027 | 35 kB 3′ | |||
| rs13144122:A > Ce | SPCS3 | 0.73 | 0.0128 | 0.218 | 40 kB 3′ | |||
| 6p22 | SZ | rs9396710:T > Cf | ATXN1, HS.408455 | 0.54 | 0.0065 | 94 | 1 | 8 kB 5′ |
| rs493352:T > Cg | ATXN1, HS.408455 | 0.75 | 0.0251 | 1 | intron 3 | |||
| 8p21 | CL | rs1908916:T > Gh | FZD3 | 0.76 | 0.0071 | 13 | 0.092 | 5 kB 5′ |
| rs1946583:T > Ci | FZD3 | 0.76 | 0.0040 | 0.052 | 1 kB 5′ | |||
| 8p11 | BD | na | GOLGA7 | na | ns | 15 | na | |
| 13q11 | CL | na | HS.121655 | na | ns | 3 | na | |
| 13q11 | SZ | na | HS.121655 | na | ns | 3 | na | |
| 15q13 | BD | na | GOLGA8F | na | na | na | na | |
| 16p12 | BD | na | CDR2 | na | ns | 3 | na | |
| 18q21 | BD | na | C18orf12 | na | na | na | na |
aNC_000003.11:g.124645034 C > T
bNC_000003.11:g.124653868 C > T
cNC_000004.11:g.177235586 C > T
dNC_000004.11:g.177285894 A > G
eNC_000004.11:g.177292032 A > C
fNC_000006.11:g.16772207 T > C
gNC_000006.11:g.16744169 T > C
hNC_000008.10:g.28347283 T > G
iNC_000008.10:g.28350753 T > C
jOdds ratio of the phenotype vs. NARs between the homozygous variant and homozygous reference genotypes
kBonferroni adjusted for the number of non-redundant SNPs tested in the gene
No evidence of DNA methylation-phenotype association
Three genes were evaluated for DNA methylation. First, for ITGB5, the level of methylation for the CpGs of ILS1 site (Fig. 1a) was low (0% to 10%) and similar across phenotypes (Table 3A) and their analysis was not pursued. For the ISL2 ITGB5 site located closer to the 5′ end of the gene (Fig. 1a), we found higher levels of methylation with variations ranging from 40% to 100% (Table 3A). However, we detected no obvious DNA methylation differences between CL and NAR subjects. Finally, we observed for rs10934702:C > T, NC_000003.11:g.124653868 C > T around 40–45% and 85–90% methylation in the heterozygotes CT and the homozygotes CC, respectively, with no obvious differences between phenotypes (Table 3A). We concluded that these CpGs were not involved in differential regulation of ITGB5 expression between the CL and NAR phenotypes.
Table 3.
Mean DNA methylation (%) observed for different CpGs island (Isl) located in ITGB5 (A), SPSC3 (B), and FZD3 (C) genes for NAR, BD, and SZ subjects
| (A) ITGB5 | ||||||
|---|---|---|---|---|---|---|
| Island 1 | Island 2 | rs10934702:C > Ta | ||||
| Pheno | N | Mean | N | Mean | N | CpG1 |
| NAR | 3 | 2.9 | 30 | 82.6 | 48 | 42.1 |
| BD | 3 | 2.9 | 30 | 83.0 | 10 | 41.5 |
| SZ | 3 | 2.2 | 30 | 82.7 | 2 | 44.2 |
| min | 0.0 | 15.8 | 4.9 | |||
| max | 28.5 | 98.0 | 91.2 | |||
| (B) SPCS3 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Island 1 | Island 2 | Island 3 | rs7694145:G > Ab,c | |||||||
| Pheno | N | Mean | N | Mean | N | Mean | N GG | N AG | GG | AG |
| NAR | 3 | 3.1 | 10 | 96.0 | 10 | 17.3 | 17 | 109 | 30.8 | 23.3 |
| BD | 3 | 2.5 | 10 | 97.1 | 10 | 16.8 | 1 | 21 | 29.4 | 16.2 |
| SZ | 3 | 3.5 | 10 | 97.0 | 10 | 17.0 | 4 | 20 | 34.2 | 15.7 |
| min | 0.0 | 75.6 | 8.7 | 0.0 | ||||||
| max | 26.5 | 100.0 | 46.6 | 67.7 | ||||||
| (C) FZD3 | ||
|---|---|---|
| Pheno | N | Mean |
| NAR | 199 | 45.2 |
| BD | 85 | 45.8 |
| SZ | 58 | 42.1 |
| min | 2.6 | |
| max | 89.1 | |
aNC_000003.11:g.124653868 C > T
bNC_000004.11:g.177285894 A > G
cOnly heterozygotes AG and homozygotes GG subjects have been analyzed since AA homozygotes are without CpG site
Second, in the case of SPCS3, the first investigated site (Fig. 1b) showed DNA methylation levels of <10% with poor quality calls (Table 3B) and was not pursued. The second SPCS3 CpG site showed very high level of methylation (75–100%) but with no obvious differences between groups, while the third SPCS3 CpG site showed moderate methylation ranging between 10 to 40% that was also similar between groups (Table 3). CpG created by the SNP rs7694145:A > G, NC_000004.11:g.177285894 A > G present variation in methylation from 0 to 70% with a greater methylation in NAR carrying the AG genotype than in BD (p = 0.004; Table 3). The greater methylation observed in NAR corresponded to a lower SPCS3 expression. However, a lower expression of SPCS3 was also observed in AA individuals where no methylation could be observed due to the absence of the CpG site with that genotype (results not shown).
Third, for FZD3, the three CpG sites exhibited a wide range of methylation levels (Table 3C). There was no significant difference in mean methylation level between BD and NARs (p = 0.89) and between SZ and NARs (p = 0.15) using analysis 3.
Discussion
Our present results build upon several complementary methods to follow-up on our linkage signals previously reported in a large kindred sample. This multimodal procedure has yielded information on association of SNPs with phenotypic status through RNA expression level of genes to approach the disease complexities. Supplementary Table S6 provides an overview of the positive and negative findings. For SPCS3, a greater RNA expression in BD patients than in their NARs appeared independent of the level of methylation at the SNP site. However, very few CpG sites were studied and a more exhaustive analysis using DNA methyl Seq, for example, could highlight functional CpG sites in relation to SZ/BD. We noted that none of the SNPs found associated with FZD3 or SPCS3 RNA expression (cis-eQTNs) would be significantly associated to the CL or BD phenotypes in a simple phenotype—genotype association analysis (results not shown). This highlights the value of the Zhao et al. [34] integrative genomics framework exploiting gene expression to detect disease susceptibility or protective variants when no clear genetic association is detected.
Our experimental approach detected a relatively low number of genes showing a significant difference in RNA expression between SZ or BD patients and their NARs. This contrasted with previous studies analyzing brain or whole blood in smaller or similar sizes of samples of subjects. Four limitations could explain our result. First, gene expression in leukocytes does not correlate perfectly with gene expression in brain cells involved in SZ and BD pathophysiology. We may have missed genes differentially expressed in the brain between patients and NARs but not expressed in leukocytes. Second, we have used cultured IMLs, in which gene expression should be relatively normalized by the similar and controlled culture conditions [35]. In that view, remaining differences between patients and NARs would then come from structural DNA variants and not epigenetic modifications [12]. However, important differences could have been attenuated by the homogeneous culture conditions where immediate environmental factors would have less or no influence to trigger or accentuate genetic effects. Third, we compared patients to NARs, not to unrelated controls. Patients and NARS share a large part of their genome including several gene RNA expression controlling elements, which might have attenuated differences in gene RNA expression. Fourth, our approach targeted solely variants modulating phenotype through gene RNA expression. It would thus have missed variants influencing SZ and BD risk through other routes. For instance, rs1156026:T > C, NC_000013.10:g.43726345 T > C on 13q13–q14, which previously showed an association with SZ in our same sample [36], did not come out in the present analysis because that SNP was not associated to the expression of any transcript present on the chip.
FZD3, a member of a family of genes that encodes a seven-transmembrane domain proteins that are receptors for the wingless type MMTV integration site family of signaling proteins, has already come out as a susceptibility locus for SZ in the Chinese population [37, 38], but only in this population according to a meta-analysis [39]. Congruently with this meta-analysis, when genotyping our sample for two of the SNPs associated with SZ in the Chinese sample (rs352203:C > T, NC_000008.10:g.28394701 C > T and rs2241802:A > G, NC_000008.10:g.28384712 A > G), neither of these SNPs was associated with SZ or to the RNA expression or the methylation of FZD3 in our sample (results not shown). However, FZD3 would be nonetheless implicated in SZ and BD based on molecular genetics and developmental studies of 484 annotated genes on chromosome 8p21 [40]. Knock-out animal models using CRISPER/CAS9 could be used to investigate a possible functional link between SZ/BD and FZD3.
In conclusion, despite the limitations explained above, our multimodal attempt focusing on dense SNP genotyping and RNA expression analyses to previously reported linkage findings, has the advantage of diminishing the multiple statistical testing burdens and allowed us to perform a deeper mining of candidate gene effects related to SZ or BD.
Supplementary information
Acknowledgements
We are grateful to our professional research assistants: Louise Bélanger, Marie-Claude Boisvert, Linda René, Lisette Gagnon, Claudie Poirier, Nicole Leclerc, Julie Lamarche, Pierrette Boutin, Lise St-Germain, Mélanie Mercier, Isabel Moreau (Centre de recherche CERVO), and to the family members, adults and children, who participated in this study. We thank Dr Aurélie Labbe (HEC Montréal) for her statistical advices on gene expression data and Dr Chantal Mérette (Centre de recherche CERVO) for her input on data analysis.
Funding
This work was supported by the Canadian Institutes of Health research (CIHR, grants MOP-74430, MOP-119408 and MOP-114988) and by a Canada Research Chair (# 950–200810) in the genetics of neuropsychiatric disorders of which M. Maziade was the Chair. The data management system was supported by the Canada Foundation for Innovation Leadership Opportunity Fund (grant 27592).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41431-019-0526-y) contains supplementary material, which is available to authorized users.
References
- 1.Sekar A, Bialas AR, de Rivera H, Davis A, Hammond TR, Kamitaki N, et al. Schizophrenia risk from complex variation of complement component 4. Nature. 2016;530:177–83. doi: 10.1038/nature16549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paccalet T, Gilbert E, Berthelot N, Marquet P, Jomphe V, Lussier D, et al. Liability indicators aggregate many years before transition to illness in offspring descending from kindreds affected by schizophrenia or bipolar disorder. Schizophrenia Res. 2016;175:186–92. doi: 10.1016/j.schres.2016.04.038. [DOI] [PubMed] [Google Scholar]
- 3.Cannon TD, Chung Y, He G, Sun D, Jacobson A, van Erp TG, et al. Progressive reduction in cortical thickness as psychosis develops: a multisite longitudinal neuroimaging study of youth at elevated clinical risk. Biol Psychiatry. 2015;77:147–57. doi: 10.1016/j.biopsych.2014.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glausier JR, Lewis DA. Dendritic spine pathology in schizophrenia. Neuroscience. 2013;251:90–107. doi: 10.1016/j.neuroscience.2012.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kendler KS. What psychiatric genetics has taught us about the nature of psychiatric illness and what is left to learn. Mol Psychiatry. 2013;18:1058–66. doi: 10.1038/mp.2013.50. [DOI] [PubMed] [Google Scholar]
- 6.Keshavan MS. Development, disease and degeneration in schizophrenia: a unitary pathophysiological model. J Psychiatry Res. 1999;33:513–21. doi: 10.1016/S0022-3956(99)00033-3. [DOI] [PubMed] [Google Scholar]
- 7.Maziade M, Paccalet T. A protective-compensatory model may reconcile the genetic and the developmental findings in schizophrenia. Schizophrenia Res. 2013;144:9–15. doi: 10.1016/j.schres.2012.12.022. [DOI] [PubMed] [Google Scholar]
- 8.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–7. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim S, Cho H, Lee D, Webster MJ. Association between SNPs and gene expression in multiple regions of the human brain. Transl Psychiatry. 2012;2:e113. doi: 10.1038/tp.2012.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glatt SJ, Everall IP, Kremen WS, Corbeil J, Sasik R, Khanlou N, et al. Comparative gene expression analysis of blood and brain provides concurrent validation of SELENBP1 up-regulation in schizophrenia. PNAS. 2005;102:15533–8. doi: 10.1073/pnas.0507666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sullivan PF, Fan C, Perou CM. Evaluating the comparability of gene expression in blood and brain. Am J Med Genet Part B, Neuropsychiatr Genet. 2006;141B:261–8. doi: 10.1002/ajmg.b.30272. [DOI] [PubMed] [Google Scholar]
- 12.Chagnon YC, Roy MA, Bureau A, Merette C, Maziade M. Differential RNA expression between schizophrenic patients and controls of the dystrobrevin binding protein 1 and neuregulin 1 genes in immortalized lymphocytes. Schizophrenia Res. 2008;100:281–90. doi: 10.1016/j.schres.2007.12.471. [DOI] [PubMed] [Google Scholar]
- 13.Cattane N, Minelli A, Milanesi E, Maj C, Bignotti S, Bortolomasi M, et al. Altered gene expression in schizophrenia: findings from transcriptional signatures in fibroblasts and blood. PLoS One. 2015;10:e0116686. doi: 10.1371/journal.pone.0116686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee J, Goh LK, Chen G, Verma S, Tan CH, Lee TS. Analysis of blood-based gene expression signature in first-episode psychosis. Psychiatry Res. 2012;200:52–4. doi: 10.1016/j.psychres.2012.03.021. [DOI] [PubMed] [Google Scholar]
- 15.Maschietto M, Silva AR, Puga RD, Lima L, Pereira CB, Nakano EY, et al. Gene expression of peripheral blood lymphocytes may discriminate patients with schizophrenia from controls. Psychiatry Res. 2012;200:1018–21. doi: 10.1016/j.psychres.2012.04.030. [DOI] [PubMed] [Google Scholar]
- 16.Takahashi M, Hayashi H, Watanabe Y, Sawamura K, Fukui N, Watanabe J, et al. Diagnostic classification of schizophrenia by neural network analysis of blood-based gene expression signatures. Schizophrenia Res. 2010;119:210–8. doi: 10.1016/j.schres.2009.12.024. [DOI] [PubMed] [Google Scholar]
- 17.Clelland CL, Read LL, Panek LJ, Nadrich RH, Bancroft C, Clelland JD. Utilization of never-medicated bipolar disorder patients towards development and validation of a peripheral biomarker profile. PLoS ONE. 2013;8:e69082. doi: 10.1371/journal.pone.0069082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kato T, Hayashi-Takagi A, Toyota T, Yoshikawa T, Iwamoto K. Gene expression analysis in lymphoblastoid cells as a potential biomarker of bipolar disorder. J Hum Genet. 2011;56:779–83. doi: 10.1038/jhg.2011.101. [DOI] [PubMed] [Google Scholar]
- 19.Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM, et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016;19:1442–53. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopez de Lara C, Jaitovich-Groisman I, Cruceanu C, Mamdani F, Lebel V, Yerko V, et al. Implication of synapse-related genes in bipolar disorder by linkage and gene expression analyses. Int J Neuropsychopharmacol. 2010;13:1397–410. doi: 10.1017/S1461145710000714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kinoshita M, Numata S, Tajima A, Shimodera S, Ono S, Imamura A, et al. DNA methylation signatures of peripheral leukocytes in schizophrenia. Neuromolecular Med. 2013;15:95–101. doi: 10.1007/s12017-012-8198-6. [DOI] [PubMed] [Google Scholar]
- 22.Nishioka M, Bundo M, Koike S, Takizawa R, Kakiuchi C, Araki T, et al. Comprehensive DNA methylation analysis of peripheral blood cells derived from patients with first-episode schizophrenia. J Hum Genet. 2013;58:91–7. doi: 10.1038/jhg.2012.140. [DOI] [PubMed] [Google Scholar]
- 23.Kumar G, Clark SL, McClay JL, Shabalin AA, Adkins DE, Xie L, et al. Refinement of schizophrenia GWAS loci using methylome-wide association data. Hum Genet. 2015;134:77–87. doi: 10.1007/s00439-014-1494-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hannon E, Dempster E, Viana J, Burrage J, Smith AR, Macdonald R, et al. An integrated genetic-epigenetic analysis of schizophrenia: evidence for co-localization of genetic associations and differential DNA methylation. Genome Biol. 2016;17:176. doi: 10.1186/s13059-016-1041-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Eijk KR, de Jong S, Strengman E, Buizer-Voskamp JE, Kahn RS, Boks MP, et al. Identification of schizophrenia-associated loci by combining DNA methylation and gene expression data from whole blood. Eur J Hum Genet. 2015;23:1106–10. doi: 10.1038/ejhg.2014.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maziade M, Roy MA, Chagnon YC, Cliche D, Fournier JP, Montgrain N, et al. Shared and specific susceptibility loci for schizophrenia and bipolar disorder: a dense genome scan in Eastern Quebec families. Mol Psychiatry. 2005;10:486–99. doi: 10.1038/sj.mp.4001594. [DOI] [PubMed] [Google Scholar]
- 27.Merette C, Roy MA, Bureau A, Fournier A, Emond C, Cliche D, et al. Replication of linkage with bipolar disorder on chromosome 16p in the Eastern Quebec population. Am J Med Genet Part B, Neuropsychiatr Genet. 2008;147B:737–44. doi: 10.1002/ajmg.b.30673. [DOI] [PubMed] [Google Scholar]
- 28.Maziade M, Chagnon YC, Roy MA, Bureau A, Fournier A, Merette C. Chromosome 13q13-q14 locus overlaps mood and psychotic disorders: the relevance for redefining phenotype. Eur J Hum Genet. 2009;17:1034–42. doi: 10.1038/ejhg.2008.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maziade M, Roy MA, Fournier JP, Cliche D, Merette C, Caron C, et al. Reliability of best-estimate diagnosis in genetic linkage studies of major psychoses: results from the Quebec pedigree studies. Am J Psychiatry. 1992;149:1674–86. doi: 10.1176/ajp.149.12.1674. [DOI] [PubMed] [Google Scholar]
- 30.Maziade M, Roy MA, Martinez M, Cliche D, Fournier JP, Garneau Y, et al. Negative, psychoticism, and disorganized dimensions in patients with familial schizophrenia or bipolar disorder: continuity and discontinuity between the major psychoses. Am J Psychiatry. 1995;152:1458–63. doi: 10.1176/ajp.152.10.1458. [DOI] [PubMed] [Google Scholar]
- 31.Roy MA, Lanctot G, Merette C, Cliche D, Fournier JP, Boutin P, et al. Clinical and methodological factors related to reliability of the best-estimate diagnostic procedure. Am J Psychiatry. 1997;154:1726–33. doi: 10.1176/ajp.154.12.1726. [DOI] [PubMed] [Google Scholar]
- 32.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–31. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 33.Gagnon-Bartsch JA, Speed TP. Using control genes to correct for unwanted variation in microarray data. Biostatistics. 2012;13:539–52. doi: 10.1093/biostatistics/kxr034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao SD, Cai TT, Li H. More powerful genetic association testing via a new statistical framework for integrative genomics. Biometrics. 2014;70:881–90. doi: 10.1111/biom.12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karege F, Schwald M, El Kouaissi R. Drug-induced decrease of protein kinase a activity reveals alteration in BDNF expression of bipolar affective disorder. Neuropsychopharmacol. 2004;29:805–12. doi: 10.1038/sj.npp.1300384. [DOI] [PubMed] [Google Scholar]
- 36.Bureau A, Chagnon YC, Croteau J, Fournier A, Roy MA, Paccalet T, et al. Follow-up of a major psychosis linkage site in 13q13-q14 reveals significant association in both case-control and family samples. Biol Psychiatry. 2013;74:444–50. doi: 10.1016/j.biopsych.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kang C, Zhou L, Liu H, Yang J. Association study of the frizzled 3 gene with Chinese Va schizophrenia. Neurosci Lett. 2011;505:196–9. doi: 10.1016/j.neuroscience.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 38.Yang J, Si T, Ling Y, Ruan Y, Han Y, Wang X, et al. Association study of the human FZD3 locus with schizophrenia. Biol Psychiatry. 2003;54:1298–301. doi: 10.1016/S0006-3223(03)00291-9. [DOI] [PubMed] [Google Scholar]
- 39.Pantavou KG, Braliou GG, Kontou PI, Dimou NL, Bagos PG. A meta-analysis of FZD3 gene polymorphisms and their association with schizophrenia. Psychiatr Genet. 2016;26:272–80. doi: 10.1097/YPG.0000000000000155. [DOI] [PubMed] [Google Scholar]
- 40.Tabares-Seisdedos R, Rubenstein JL. Chromosome 8p as a potential hub for developmental neuropsychiatric disorders: implications for schizophrenia, autism and cancer. Mol Psychiatry. 2009;14:563–89. doi: 10.1038/mp.2009.2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.