Abstract
Free-fatty acid receptor-4 (FFA4), previously termed GPR120, is a G protein–coupled receptor (GPCR) for medium and long-chained fatty acids, agonism of which can regulate a myriad of metabolic, sensory, inflammatory, and proliferatory signals. Two alternative splice isoforms of FFA4 exist that differ by the presence of an additional 16 amino acids in the longer (FFA4-L) transcript, which has been suggested to be an intrinsically β-arrestin–biased GPCR. Although the shorter isoform (FFA4-S) has been studied more extensively, very little is known about mechanisms of regulation or signaling of the longer isoform. Because β-arrestin recruitment is dependent on receptor phosphorylation, in the current study, we used the endogenous agonist docosahexaenoic acid (DHA) to examine the mechanisms of FFA4-L phosphorylation, as well as DHA-dependent β-arrestin recruitment and DHA-dependent extracellular-signal regulated kinase-1/2 (ERK1/2) signaling in human embryonic kidney 293 cells. Our results reveal differences in basal phosphorylation of the two FFA4 isoforms, and we show that DHA-mediated phosphorylation of FFA4-L is primarily regulated by G protein–coupled receptor kinase 6, whereas protein kinase-C can also contribute to agonist-induced and heterologous phosphorylation. Moreover, our data demonstrate that FFA4-L phosphorylation occurs on the distal C terminus and is directly responsible for recruitment and interactions with β-arrestin-2. Finally, using CRISPR/Cas9 genome-edited cells, our data reveal that unlike FFA4-S, the longer isoform is unable to facilitate phosphorylation of ERK1/2 in cells that are devoid of β-arrestin-1/2. Together, these results are the first to demonstrate phosphoregulation of FFA4-L as well as the effects of loss of phosphorylation sites on β-arrestin recruitment and ERK1/2 activation.
SIGNIFICANCE STATEMENT
Free-fatty acid receptor-4 (FFA4) is a cell-surface G protein–coupled receptor for medium and long-chained fatty acids that can be expressed as distinct short (FFA4-S) or long (FFA4-L) isoforms. Although much is known about FFA4-S, the longer isoform remains virtually unstudied. Here, we reveal the mechanisms of docosahexaenoic acid–induced phosphorylation of FFA4-L and subsequent β-arrestin-2 recruitment and extracellular-signal regulated kinase-1/2 activity.
Introduction
Long-chain free-fatty acids (FFAs) are critical dietary nutrients that modulate a variety of cellular processes including mitochondrial β-oxidation and maintenance and structural integrity of the cell membrane. These FFAs have also been shown to play important roles in cell signaling and gene transcription, effects that were historically thought to be mediated by FFA action on nuclear receptors such as peroxisome proliferator-activated receptor-γ or the retinoid-X receptor. However, in the mid-2000s, a family of cell-surface FFA receptors belonging to the rhodopsin-like G protein–coupled receptor (GPCR) superfamily was discovered and noted to modulate many of the effects of FFAs (Alvarez-Curto and Milligan, 2016; Moniri, 2016). Of these, free-fatty acid receptor-1 (FFA1, previously termed GPR40) and free-fatty acid receptor-4 (FFA4, previously termed GPR120) have now been well-described as receptors for long-chain fatty acids, including the omega-3 fatty acids α-linolenic acid and docosahexaenoic acid (DHA), which are enriched in dietary flax seed oil and fish oil, respectively.
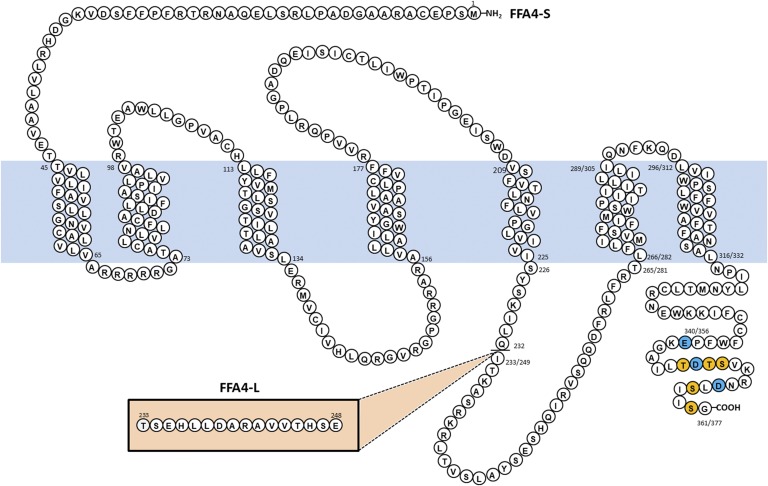
FFA4 is expressed ubiquitously in humans, with dense localization in the lung, gastrointestinal tract, adipose, liver, and immune cells, where it modulates a variety of physiologic functions, including hormone secretion, glucose uptake, and modulation of inflammation, among others (Ulven and Christiansen, 2015; Moniri, 2016; Milligan et al., 2017). The human FFA4 gene can be alternatively spliced yielding a 361 amino acid protein (FFA4-S) or a longer isoform (FFA4-L) that retains exon 3 to encode for an additional 16 amino acids within the third intracellular loop, a receptor domain that influences intracellular signaling and desensitization (Fig. 1) (Moore et al., 2009; Burns and Moniri, 2010). To date, expression of FFA4-L has been detected only within the human colon or colon epithelial cell lines (Galindo et al., 2012; Kim et al., 2015), and the colon expresses the highest FFA4 transcript within the gastrointestinal tract (Hirasawa et al., 2005). Interestingly, FFA4-L is not detectable in rodents, felines, canines, bovines, or nonhuman primates (Moore et al., 2009; Habara et al., 2015; Miyabe et al., 2015; Song et al., 2015; Moniri, 2016), and current evidence suggests that this isoform is a distinct human gene product.
Fig. 1.
Amino acid diagram of human FFA4-S with numbers relative to the initiating Met. The known sites of phosphorylation of FFA4-S are shown in orange while three acidic residues that contribute to the negative charge responsible for β-arrestin recruitment are shown in blue. The inset depicts additional 16 residues of FFA4-L, which changes the numbering scheme after Ile233 (short)/249 (long).
Previous studies have determined that FFA agonist response does not differ greatly at the two receptor isoforms (Moore et al., 2009; Burns and Moniri, 2010; Galindo et al., 2012). Similar to other Gαq/11-coupled GPCRs, agonism of FFA4-S with FFA elicits robust Ca+2-mobilization and downstream activation of PKC or ERK1/2, as well as G-protein independent effects through recruitment of β-arrestin-2 partner proteins, which are capable of scaffolding other signaling molecules (Oh et al., 2010; Watson et al., 2012; Hudson et al., 2013; Burns et al., 2014). On the contrary, agonism of native FFA4-L does not facilitate Ca+2-mobilization but does retain β-arrestin-2–scaffolded outcomes, suggesting that the additional 16 amino acids within the third intracellular loop of FFA4-L counters inherent G-protein coupling and provides intrinsic β-arrestin-bias to this isoform (Watson et al., 2012). Importantly, we have previously shown that agonism with α-linolenic acid and DHA leads to rapid phosphorylation of both isoforms (Burns and Moniri, 2010), demonstrating that differences in signal bias are dependent on more than the additional 16 amino acid region of intracellular loop 3. Indeed, our own studies (Burns et al., 2014), as well as those of others (Butcher et al., 2014), demonstrate that FFA4-S phosphorylation occurs at Ser/Thr residues localized to the C terminus, which work in concert with three nearby acidic residues to create an area of negative charge that serves as the functional β-arrestin phosphosensor (Butcher et al., 2014). Despite these studies, there are no reports related to phosphoregulation of FFA4-L in the literature, and hence, the goal of the current study was to begin to explore such mechanisms, as well as the functional consequences of C-terminal phosphorylation on FFA4-L–mediated β-arrestin recruitment and FFA4-L–mediated ERK1/2 activation. Herein, we have used a transiently expressing human embryonic kidney 293 (HEK293) cell system to investigate phosphorylation of FFA4-L and the impact on β-arrestin recruitment. Our results show that agonist-stimulated homologous phosphorylation of FFA4-L is predominantly mediated by GRK6 as well as protein kinase-C (PKC), whereas the latter also modulates heterologous phosphorylation. We also show that tonic phosphorylation of FFA4-L is significantly lower than that of the shorter isoform and that homologous and heterologous phosphorylation occurs entirely within the receptor C terminus. Our results further reveal that although truncation of the C terminus of FFA4-S fully inhibits β-arrestin recruitment, truncation of the FFA4-L C terminus only significantly reduces this ability. Finally, using CRISPR/Cas9 edited cells, our results show that FFA4-L is incapable of inducing phosphorylation of ERK1/2 in β-arrestin-1/2 deleted cells, whereas this effect is surprisingly significantly enhanced in the absence of Gαq/11.
Materials and Methods
Reagents and Chemicals.
Wild-type (WT) FLAG-tagged FFA4-S and FFA4-L constructs were generated as previously described (Burns et al., 2014). Mutant receptors lacking the β-arrestin phosphosensor were truncated after amino acid residue 340 in FFA4-S (FFA4-S-∆340) and after residue 356 in FFA4-L (FFA4-L-∆356) and created via overlapping PCR and cloned into pcDNA3.1, as we described previously (Burns et al., 2014; Cheshmehkani et al., 2017). We have previously demonstrated equivalent whole-cell expression of these constructs upon transient and stable transfection (Burns and Moniri, 2010; Burns et al., 2014; Cheshmehkani et al., 2017). The pcDNA3-β2-adrenergic receptor and pGFP(N1)-topaz-β-arrestin-2-YFP plasmids were kind gifts from Dr. Robert J. Lefkowitz (Duke University Medical Center) and Dr. Michel Bouvier (University of Montreal), respectively. The pcDNA3.1-histamine H1 receptor plasmid was purchased from the Missouri S&T cDNA Resource Center (www.cdna.org). The FFA4-S and -L Renilla luciferase (Rluc) constructs were created by PCR with 5′-HindIII and 3′-EcoRI restriction sites and subcloned into pSF-CMV-Rluc plasmid (Oxford Genetics, Oxfordshire, UK). The corresponding C-terminal truncated Rluc constructs were generated by sequential PCR leading to creation of the FFA4-S-Δ340 and FFA4-L-Δ356 constructs. All plasmid constructs were validated by DNA sequencing. Purified docosahexaenoic acid oil was purchased from Nu-Chek Prep (Elysian, MN). AH7614 (4-Methyl-N-9H-xanthen-9-yl-benzenesulfonamide), phorbol 12-myristate 13-acetate (PMA), H-89, forskolin (FSK), histamine, bisindolylmaleimide II (BIMII), and isoproterenol hydrochloride were obtained from Sigma-Aldrich (St. Louis, MO). TUG-891 was purchased from Tocris-Bio-Techne (Minneapolis, MN). All other chemicals used were obtained at the highest available purity from Thermo Fisher Scientific (Waltham, MA) or Sigma-Aldrich.
Cell Culture and Transfection.
HEK293 cells were obtained from American Type Culture Collection (Manassas, VA) and cultured in 100 mm plates containing Dulbecco’s modified Eagle’s medium, supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin (Life Technologies, Grand Island, NY). Cells were used for 20–25 passages, at which point a new stock was used. Transfections were performed using LipoD293 reagent (Signagen Laboratories, Gaithersburg, MD) and 5 μg of the respective plasmid DNA according to the manufacturer’s directions. After 24 hours, cells were split into appropriate assay plates and all experiments were performed 48 hours after transfection. For experiments with siRNA, HEK293 cells were cotransfected with plasmids encoding for FFA4 along with siRNA specific to each GRK, or scrambled control, exactly as described previously (Burns et al., 2014). CRISPR/Cas9-mediated genome-edited HEK293 cell clones that were null for Gαq and Gα11, or null for β-arrestin-1 and β-arrestin-2, and the scrambled control clone were very kind gifts from Dr. Graeme Milligan (University of Glasgow). Membrane fractions were purified by sequential centrifugation at 45,000g, as described previously (Moniri and Daaka, 2007).
Immunoblotting.
Immunoblotting was performed as described previously (Burns et al., 2014; Cheshmehkani et al., 2017). Briefly, cells were lysed in RIPA buffer (50 mM Tris-HCl, 150 mM NaCl, 5 mM EDTA, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 10 mM NaF, 10 mM Na2HPO4, pH 7.4) plus protease inhibitor cocktail for 20 minutes and cleared for insoluble debris by centrifugation at 15,000g for 15 minutes at 4°C. Protein concentrations were standardized using DC Protein Assay (Bio-Rad, Hercules, CA) and an aliquot of the lysate was denatured in 2× SDS-sample buffer and boiled for 5 minutes. Where blotting was performed for FFA4, samples were incubated at 37°C for 30 minutes. Equivalent concentrations of lysates were resolved by SDS-PAGE, followed by transfer to polyvinylidene fluoride membranes, and immunoblotted with the appropriate antibody. Blots were visualized with horseradish peroxide–conjugated secondary antibody followed by enhanced chemiluminescence. Blots were stripped by incubation in glycine IgG stripping buffer for 30–45 minutes with agitation (25 mM Glycine, 1% SDS, 50°C, pH 2.0) for reprobing. Rabbit anti-phosphoThr202/Tyr204-p44/p42 MAPK (ERK1/2) (cat. no. 9101) and p44/42 MAPK (ERK1/2) (9102) antibodies were purchased from Cell Signaling Technologies (Beverly, MA), mouse anti-actin-horseradish peroxide (sc-47778) and mouse anti-β-arrestin-1/2 antibody (21B1) were purchased from Santa Cruz Biotechnology (Dallas, TX), and rabbit anti-Gαq/11 antibody (06-709) was purchased from EMD Millipore (Burlington, MA).
Whole Cell Phosphorylation Assay.
Assays were performed as we have previously described (Burns and Moniri, 2010; Burns et al., 2014). Briefly, 48 hours after transfection, to reduce constitutive signaling and deprive cells of FFA endogenous to serum, cells were starved in phosphate-free Dulbecco’s modified Eagle’s medium for 75 minutes and incubated with 0.2 mCi H332PO4 (Perkin Elmer, Waltham, MA) for 45 minutes, and then stimulated as described. Unstimulated control conditions replicated the vehicle for each given agent. Following treatment, cells were lysed at 4°C in RIPA/protease inhibitor cocktail for 60 minutes and then diluted with detergent-free RIPA buffer prior to centrifugation at 14,000g for 15 minutes at 4°C. Lysate protein content was assessed using the DC Protein Assay (Bio-Rad) and equivalent protein concentrations (1.0–1.5 mg/ml) were immunoprecipitated overnight at 4°C with anti-FLAG M2 (F3165) antibody (Sigma Aldrich) and protein G-agarose beads (Santa Cruz Biotechnology). Beads were washed three times with RIPA buffer and proteins eluted by addition of 1× SDS-sample buffer with 2.5% β-mercaptoethanol followed by resolution by SDS-PAGE. Gels were then dried and subjected to autoradiography at −80°C to detect 32P incorporation.
Bioluminescence Resonance Energy Transfer.
Bioluminescence resonance energy transfer (BRET) experiments were performed as we have performed previously (Singh and Moniri, 2012). Briefly, HEK293 cells were transfected with appropriate empty-vector or FFA4-Rluc construct described above and pGFP(N1)-topaz-β-arrestin2-YFP in a 1:5 ratio, following determination of the optimal plasmid concentrations via a full donor-saturation curve. All experiments also included an empty vector control condition and an unfused-YFP control condition to account for the effects of plasmid and YFP saturation, respectively. Cells were serum-starved for 12 hours prior to experiments to remove FFAs from the culture media and allow for serum-FFA–mediated signals to normalize. Immediately prior to BRET, cells were washed three times with BRET buffer (140 mm NaCl, 2.7 mM KCl, 1 mM MgCl2, 1 mM CaCl2, 0.37 mM NaH2PO4, 24 mM NaHCO3, 25 mM HEPES, 0.1% Glucose, pH 7.4 [Issad and Jockers, 2006]), collected via centrifugation, and resuspended in BRET buffer at a concentration of 1 × 106 cells/ml. Cells were loaded into a white 96-well plate at 50,000 cells/well, incubated with 5 µM coelenterazine-h substrate (Promega, Madison, WI), in the presence of DHA, TUG-891, or AH7614, as noted in the figure legends. Acceptor emission (535 nm) and donor emission (485 nm) was detected using a Berthold Mithras LB 940 plate reader and the net BRET signal was calculated from the drug-treated Rluc-YFP condition to account for any nonspecific effects of DHA (e.g., cell membrane or metabolism).
Visualization of β-Arrestin Recruitment.
HEK293 cells were transiently cotransfected with the appropriate FFA4 plasmid as in the phosphorylation assays, in the presence of pGFP(N1)-topaz-β-arrestin-2-YFP (2 μg). Cells were seeded onto glass coverslips 24 hours posttransfection, serum-starved overnight to deprive the cells of endogenous serum-containing FFAs, and treated with 100 μM DHA for the noted times. Following agonism, cells were washed in ice-cold PBS and fixed in 4% paraformaldehyde for 15 minutes at 4°C. Once dried, coverslips were mounted onto glass slides and images were obtained using a Leica TCS SP8 confocal laser scanning microscope.
Data Analysis.
Autoradiographic data were quantified by densitometric analysis within the linear range using NIH Image J (Bethesda, MD). The results were imported and graphed using GraphPad Prism (San Diego, CA). Data are expressed as mean ± S.D. Where shown, pEC50 ± S.D. was calculated using the sigmoidal concentration-response algorithm in Prism using pooled data from all experiments (n denoted in figure legends). Where not visible, error bars fall within the symbol size. Statistical analysis was performed in GraphPad Prism or Instat using one-way analysis of variance and post hoc Bonferroni analysis or unpaired, two-tailed Student’s t test, as described in the figure legends. Statistical significance is represented as a descriptive (nonhypothesis testing) P value using a single symbol for P < 0.05, a double symbol for P < 0.01, and a triple symbol for P < 0.001, as noted in the figure legends. Reported P values are accompanied by Cohen’s d value as a measure of the effect size to convey practical significance.
Results
Homologous Phosphorylation of FFA4-L is Mediated by GRK6.
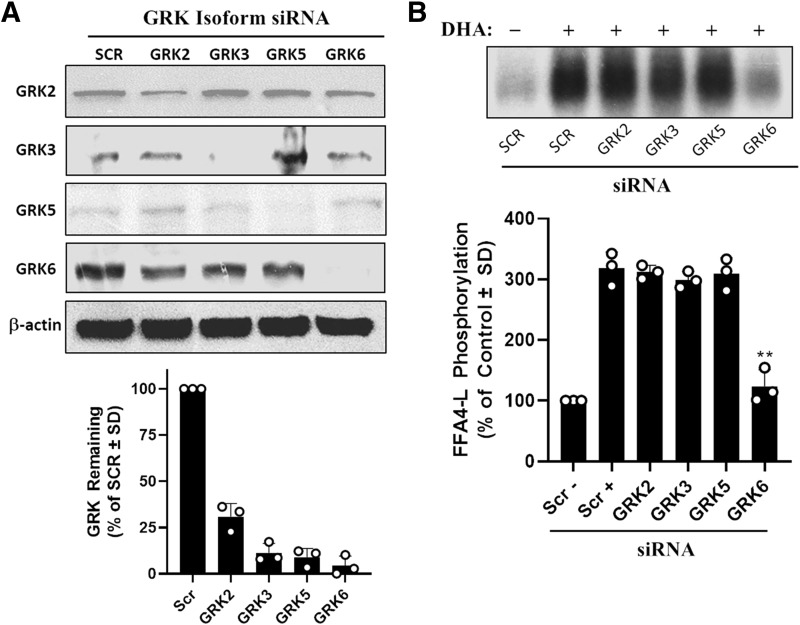
Agonist-induced (i.e., homologous) phosphorylation of GPCRs is mediated via recruitment of G protein–coupled receptor kinases (GRKs) that are activated via liberated Gβγ subunits, Ca+2 signals, or other proteins and protein kinases (Pitcher et al., 1992; Penela et al., 2003; Willets et al., 2003; Pack et al., 2018). Seven distinct GRK isoforms have been identified, with GRKs 2, 3, 5, and 6 being ubiquitously expressed, whereas the others are expressed in a tissue-specific manner (Freedman and Lefkowitz, 1996; Pitcher et al., 1998). The HEK293 cell model was used here to study phosphoregulation of FFA4-L. These cells have previously been shown to express the four ubiquitously expressed GRK isoforms (Ren et al., 2005), and we have also demonstrated that agonism of ectopically expressed FFA4-S or FFA4-L in these cells leads to rapid and transient receptor phosphorylation (Burns and Moniri, 2010). Using isoform-selective siRNA, we achieved greater than 75% knockdown of each specific GRK (Fig. 2A). To ensure that knockdown of a GRK does not alter basal phosphorylation of FFA4-L, we assessed basal phosphorylation in the absence of DHA (i.e., HBSS vehicle only), and results showed no differences in basal FFA4-L phosphorylation in cells expressing each individual siRNA (data not shown). We then assessed agonist-induced phosphorylation of FFA4-L by stimulating cells that coexpressed FFA4-L and GRK isoform-specific siRNA with the endogenous FFA4 agonist DHA. Our results demonstrate that DHA induces robust phosphorylation of FFA4-L in cells that express scrambled-control siRNA as well as siRNA targeting GRKs 2, 3, and 5, but the DHA-induced effect was nearly abolished in cells expressing GRK6-targeted siRNA (P < 0.01; d = 6.6 vs. DHA-stimulated scramble control condition). These results demonstrate that GRK6 is the predominant isoform that mediates agonist-dependent phosphorylation of FFA4-L.
Fig. 2.
Homologous agonist-mediated phosphorylation of FFA4-L. (A) Immunoblotting with isoform-specific GRK antibodies was used to verify siRNA-mediated knockdown of each in HEK293 cells. SCR is the scrambled control siRNA sequence that does not target any known human sequence. β-actin is used as a loading control for each set. Representative immunoblots are shown (upper) and pooled data from three to four independent experiments were quantified (lower) to show the density of each GRK remaining after siRNA transfection. (B) Whole cell phosphorylation assays in cells expressing FFA4-L show that agonism with DHA (100 μM, 5 minutes) induces robust phosphorylation of FFA4-L (305.9% ± 23.7% of scrambled control), which is not affected by siRNA knockdown of GRK2 (317.6% ± 7.9%), GRK3 (300.1% ± 19.1%), or GRK5 (297.7% ± 22.8%), but is significantly decreased upon GRK6 knockdown (134.4% ± 28.4%). A representative autoradiograph (upper) is shown while the graph depicts pooled data from three independent experiments (lower). ** P < 0.01 vs. scrambled, DHA-stimulated control (Scr +).
Heterologous Phosphorylation of FFA4-L is Mediated by PKC and Phosphorylation of FFA4-L Occurs on Its C Terminus.
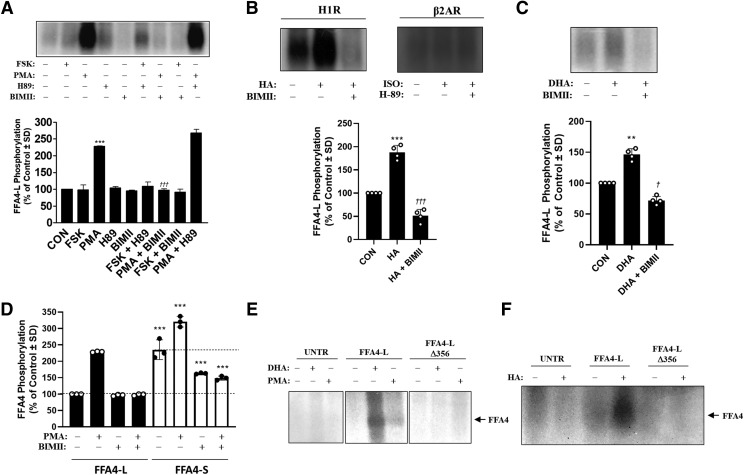
Because GPCRs can also be phosphorylated by second-messenger–linked kinases (e.g., PKC or PKA) that result from heterologous agonism of other GPCRs, we wished to assess heterologous mechanisms of FFA4-L phosphorylation. Given that HEK293 cells express seven distinct isoforms of PKC, which would make siRNA-targeting approaches difficult, we used a pharmacological approach that used well-characterized activators and inhibitors of both PKA and PKC to assess FFA4-L phosphorylation. Stimulation of FFA4-L expressing cells with the PKC activator PMA (1 μM, 20 minutes) facilitated a significant increase in FFA4-L phosphorylation (228% ± 0.8% of control) (P < 0.001; d = 167 vs. unstimulated control), whereas the adenylyl cyclase (and hence downstream-PKA) activator forskolin (FSK) (1 μM, 20 minutes) had no significant effect above basal (Fig. 3A). These results suggest that PKC, but not PKA, modulates phosphorylation of FFA4-L. Indeed, the PKC inhibitor bisindolylmaleimide II (BIMII) fully blocked the PMA-induced phosphorylation of FFA4-L (97.6% ± 2.4% of control) (P < 0.001; d = 52 vs. PMA-stimulated condition), whereas the PKA inhibitor H-89 failed to alter the effects of PMA (267% ± 8% of control) (Fig. 3A). A slight but statistically insignificant increase in phosphorylation (109% ± 12% of control) was seen in the forskolin (FSK) + H-89 condition. To confirm the effects of PKC on FFA4-L phosphorylation, we coexpressed FFA4-L with the histamine H1 receptor (H1R) or the β2-adrenergic receptor (β2AR), which signal via Gαq/11-PKC and Gαs/PKA, respectively, upon agonism. Following agonism of cells expressing FFA4-L/H1R with 10 μM histamine for 10 minute, a robust FFA4-L phosphorylation signal was seen (188% ± 8% of control) (P < 0.001; d = 8.2 vs. unstimulated control) (Fig. 3B), and importantly, this effect was significantly decreased to below control levels by pretreatment with BIMII (51% ± 7% of control) (P < 0.001; d = 9.4 vs. histamine-stimulated condition) (Fig. 3B). Meanwhile, no increase in phosphorylation was seen after agonism of cells expressing FFA4-L/β2AR with 10 μM isoproterenol for 10 minutes (Fig. 3B). Together, these results confirm that PKC, and not PKA, can regulate phosphorylation of FFA4-L. Interestingly, experiments with DHA in the presence of BIMII reveal a BIMII-sensitive effect of DHA-stimulated FFA4-L phosphorylation, suggesting that PKC is also involved in agonist-induced responses (P < 0.01; d = 10.3 vs. control condition; P < 0.05; d = 9.8 vs. BIMII-treated condition) (Fig. 3C). Because we had previously observed and reported that basal phosphorylation of FFA4-S is consistently higher per milligram protein compared with that of FFA4-L, and their expression is similar upon transfection in these cells (Burns and Moniri, 2010), we directly compared the BIMII-sensitivity of this effect. Indeed, our results demonstrate that in the absence of stimulus, basal FFA4-S phosphorylation is twofold higher (212% ± 12%) than basal FFA4-L phosphorylation (P < 0.001; d = 7.5). Although BIMII treatment did not significantly alter basal FFA4-L phosphorylation between experiments, it did do so for basal FFA4-S phosphorylation, decreasing the effect by 23% (Fig. 3D). Interestingly, although PMA-induced effects on FFA4-S were significantly blocked by BIMII, the degree of this blockage was the same as that seen with BIMII treatment in the basal state (ca. 150% of FFA4-L; Fig. 3D), suggesting that inhibition of PKC alters basal FFA4-S phosphorylation more so than FFA4-L.
Fig. 3.
Heterologous phosphorylation of FFA4-L. (A) Whole cell phosphorylation assays in cells stimulated with the downstream PKA activator forskolin (FSK) (1 µM, 20 minutes) or PKA inhibitor H89 (10 µM, 20 minutes) did not demonstrate differences in phosphorylation of FFA4-L vs. control. Significant increases in FFA4-L phosphorylation was seen in cells stimulated with the PKC activator PMA (1 µM, 20 minute) (228% ± 0.8% of control), an effect that was inhibited by the PKC inhibitor BIMII (10 µM, 20 minute) (91% ± 2.4%). A representative autoradiograph (upper) is shown while the graph depicts pooled data from four independent experiments (lower). (B) Stimulation of cells expressing FLAG-FFA4-L/H1R with histamine (10 μM, 10 minutes) significantly increased FFA4-L phosphorylation (187% ± 7.6% of control), an effect that was blocked by BIMII pretreatment (51.28% ± 6.9%) (upper left and lower). Stimulation of cells expressing FLAG-FFA4/β2AR with isoproterenol hydrochloride (1 μM, 10 minutes) had no effect on FFA4-L phosphorylation in the absence or presence of H89 (upper right). Representative autoradiographs are shown while the graph depicts pooled data from two (for β2AR) or four (for H1R) independent experiments. ***P < 0.001 vs. unstimulated control; †††P < 0.001 vs. histamine-stimulated condition. (C) DHA-stimulated FFA4-L phosphorylation (150% ± 3.3% of control) is blocked by BIMII pretreatment (87% ± 3%). A representative autoradiograph is shown while the graph depicts pooled data from four independent experiments. **P < 0.01 vs. unstimulated control; †P < 0.05 vs. the DHA-stimulated condition. (D) Pooled results from three independent experiments show that basal phosphorylation of FFA4-S is significantly higher than that of FFA4-L as are the effects of PMA and BIMII alone and in combination. ***P < 0.001 vs. complimentary group in FFA4-L. (E and F) Homologous and heterologous phosphorylation of FFA4-L are inhibited upon truncation of the C-terminal tail following Pro356. Phosphorylation is induced in WT FFA4-L upon agonism with DHA or treatment with PMA (E) and upon treatment of cells coexpressing H1R/WT-FFA4-L with histamine (F), but these effects are completely absent in cells expressing FFA4-L-Δ356. UNTR represents parallel experiments in untransfected cells.
To determine if homologous and heterologous phosphorylation loci of FFA4-L was limited to the C-terminal tail, we created an FFA4-L mutant that was truncated after Pro-356 (FFA4-L-Δ356) (Fig. 1), which lacks all five phosphorylation sites as well as the three negatively charged acidic residues that are known to comprise the FFA4-S phosphosensor. We have previously reported that the expression of FFA4-S and FFA4-L protein is approximately equivalent in our transfected HEK293 cells, and that expression of FFA4-L and FFA4-L-Δ356 transcript is also equivalent upon transfection (Cheshmehkani et al., 2017). Neither DHA or PMA elicited significant phosphorylation of FFA4-L-Δ356 compared with wild-type FFA4-L (Fig. 3E), and similarly, histamine failed to promote phosphorylation of FFA4-L-Δ356 in cells coexpressing H1R and FFA4-L-Δ356 (Fig. 3F). Taken together, these results demonstrate the GRK6 and PKC phosphorylate FFA4-L at sites that are fully localized to the C terminus.
The C Terminus of FFA4-L Modulates β-Arrestin-2 Interactability.
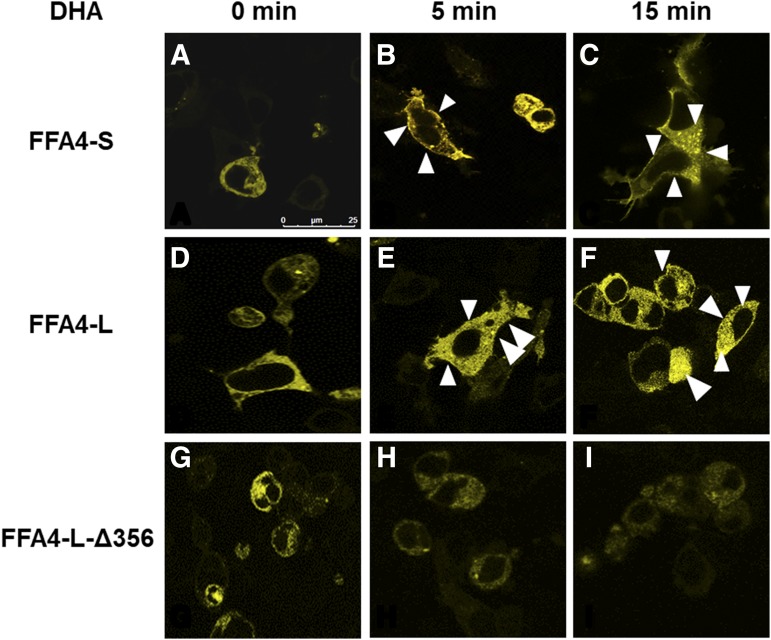
Next, we sought to determine if the C terminus of FFA4-L is responsible for its recruitment of β-arrestin proteins, which are a significant partner protein of FFA4 in general, and for FFA4-L in particular, which has been shown to be fully β-arrestin biased (Watson et al., 2012). To do so, we first visualized DHA-induced β-arrestin-2-YFP recruitment via confocal microscopy. Using FFA4-S as a positive control, DHA-induced rapid cell-surface recruitment of β-arrestin-2-YFP within 5 minutes of addition (Fig. 4, A and B), and this led to significant internalization of FFA4-S as noted by densely localized intracellular fluorescent puncta seen upon 15 minutes of agonism (Fig. 4C). Similar results were seen with FFA4-L (Fig. 4, D–F), yet the cell-surface recruitment of β-arrestin-2-YFP fluorescence as well as internalized fluorescent puncta were visibly decreased using this method in cells expressing FFA4-L-Δ356 (Fig. 4, G–I–I).
Fig. 4.
DHA-mediated FFA4-L-β-arrestin-2 recruitment. Recruitment of β-arrestin-2-YFP in response to agonism with DHA (100 μM; 5 and 15 minutes) was assessed in cells expressing FFA4-S (positive control) (A–C), FFA4-L-WT (D–F), or FFA4-L-Δ356 (G–I). Cell surface and intracellular puncta fluorescence and were visualized with both WT receptors, but not with FFA4-L-Δ356.
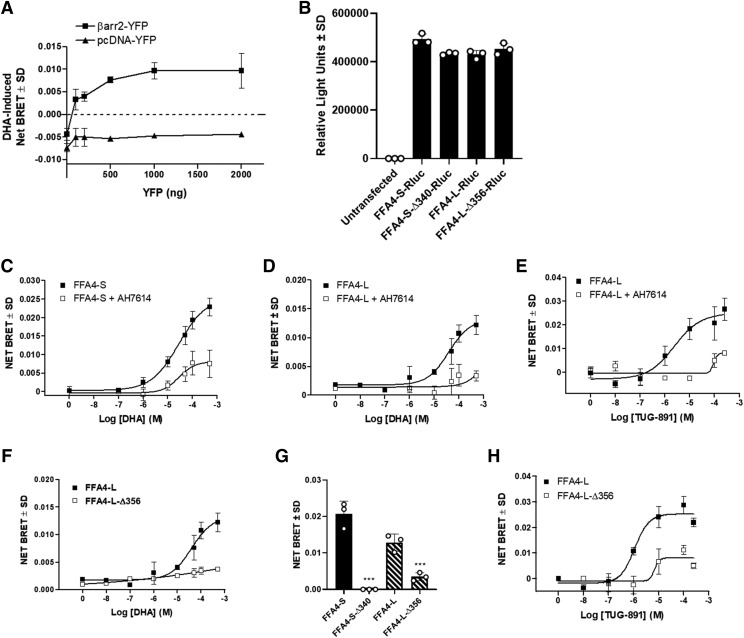
To confirm these results in a more quantitative manner, we assessed DHA-induced BRET between FFA4-Rluc and β-arrestin-2-YFP as a measure of β-arrestin-2 recruitment. BRET acceptor titration experiments showed a saturable DHA-specific response that was completely absent in control experiments where β-arrestin-YFP was replaced by empty-YFP vector (Fig. 5A). Wild-type Rluc-tagged FFA4-S and FFA4-L, and their respective Rluc-tagged truncated mutants exhibited similar amounts of light emission upon expression (Fig. 5B), demonstrating equivalent whole cell expression of each Rluc-tagged protein in this transfected system. Our results demonstrate that DHA produces robust concentration-dependent β-arrestin-2 recruitment signal to both FFA4-S (pEC50 4.6 ± 0.4) and FFA4-L (pEC50 4.4 ± 0.2) (Fig. 5, C and D). The effect of both receptor isoforms was blocked in a noncompetitive manner by the allosteric FFA4 antagonist AH7614 (10 μM) (Watterson et al., 2017) (Fig. 5, C and D). The effect at FFA4-L was also confirmed with the synthetic selective-FFA4 agonist TUG-891 (Fig. 5E) and was similar to that shown previously for FFA4-S (Watterson et al., 2017).
Fig. 5.
DHA-mediated FFA4-L-β-arrestin-2 BRET. (A) DHA-induced BRET acceptor-saturation assay was performed to determine the optimal β-arrestin-2-YFP concentration and to ensure responses were specific to β-arrestin-2-YFP because they were absent in cells expressing empty-YFP vector. (B) Expression and light emission of all FFA4 constructs was compared with ensure equivalent donor emission, and receptor expression. (C and D) DHA produces robust and concentration-dependent β-arrestin-2 recruitment signal to both FFA4-S (C) (pEC50 4.6 ± 0.4) and FFA4-L (D) (pEC50 4.4 ± 0.2), both of which are inhibited noncompetitively by the allosteric FFA4 antagonist AH7614 (10 μM). Similar results were also seen with the synthetic agonist TUG-891 (E). (F–H) Truncation of the FFA4-L C terminus after Pro356 significantly decreases β-arrestin-2 BRET. The DHA (100 μM) induced BRET signal was abolished upon C-terminal truncation of FFA4-S but was markedly reduced upon truncation of FFA4-L (P < 0.01; d = 8.32 vs. FFA4-S-Δ340) (F and G). The FFA4-L-Δ356 signal seen at 10–300 μM DHA is significantly (P = 0.02; d = 3.2) higher than the basal signal in the absence of DHA (F). A similar significantly decreased effect was also seen with TUG-891 at FFA4-L-Δ356 (P < 0.01; d = 6.1 vs. FFA4-L) (H). Each graph represents experiments performed three independent times in triplicate. ***P < 0.001 (FFA4-S vs. FFA4-S-Δ340 and FFA4-S-Δ340 vs. FFA4-L-Δ356).
Next, we assessed the effects of C-terminal truncation on FFA4-L ability to interact with β-arrestin-2 upon agonism. Although our results with human FFA4-S here recapitulate loss of β-arrestin-2 interactability upon C-terminal truncation after Pro340 for the mouse ortholog, shown previously by others (Prihandoko et al., 2016), our results for FFA4-L reveal that C-terminal truncation after the synonymous Pro356 significantly decreases but does not fully abolish β-arrestin-2 recruitment, as was seen for FFA4-S (P < 0.01; d = 8.32 vs. FFA4-S-Δ340) (Fig. 5, F and G). These results were also seen upon agonism with TUG-891, with which a small BRET signal could be detected for FFA4-S-Δ356 only at the highest micromolar concentrations of agonist (P < 0.01; d = 6.1 vs. FFA4-L) (Fig. 5H). Given that FFA4-S and FFA4-L induce similar BRET signals, the noted difference between the truncated variants suggests a significant role for the additional 16 amino acids, even in the absence of the C terminus. Of note, the slopes of the agonist curves for both DHA and TUG-891 at wild-type FFA4-L are near unity (0.7–0.9) as expected; however, as they appear in the presence of AH7614, the slopes appear much steeper at the truncated receptor. This may signify differences in receptor occupancy at FFA4-S-Δ356 that do not emerge with WT FFA4-L and that are required to elicit β-arrestin-2 recruitment, similar to that seen in the presence of the allosteric antagonist.
FFA4-L-ERK1/2 Signaling is Devoid in the Absence of β-Arrestin and is Enhanced in the Absence of Gαq/11.
An elegant previous analysis of mouse FFA4-S signaling in HEK293 cells has demonstrated that the ability of this isoform to signal to ERK1/2 is dependent on canonical Gαq/11 pathways and does not involve β-arrestin scaffolding (Alvarez-Curto et al., 2016). Interestingly, the ERK1/2 signal seen here was amplified by receptor lacking phosphorylation sites, suggesting that recruitment of β-arrestin to phosphorylated FFA4-S modulates ERK1/2 signals based on the more traditional role of arrestins as canonical signal arrestors. Given that agonism of FFA4-L does not stimulate canonical Gαq/11-Ca+2 signaling but is rather solely biased toward β-arrestin-dependent scaffolding (Watson et al., 2012), and because the effects of FFA4-L on ERK1/2 are currently undescribed in the literature, we assessed FFA4-L-ERK1/2 signaling in HEK293 cells as well as in genome-edited HEK293 cells with CRISPR/Cas9-mediated elimination of Gαq/11 or β-arrestin-1/2 (Alvarez-Curto et al., 2016).
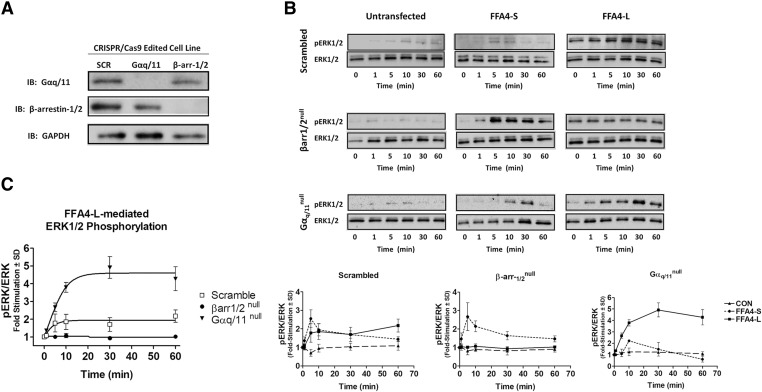
Validation of CRISPR-mediated protein ablation was performed by immunoblotting (Fig. 6A). In the parenteral scramble-edited HEK293 cells, treatment with DHA (100 μM) for the indicated times reveals a rapid but transient phosphorylation of ERK1/2 (pERK1/2) in cells expressing FFA4-S (Fig. 6B), whereas the effect was comparatively more sustained over the time course in cells expressing FFA4-L (Fig. 6B). Notably, the effect of FFA4-S remained similar, whereas the effect of FFA4-L was abolished in β-arrestin-1/2 genome-edited cells (Fig. 6B). Consistent with previous reports of Gαq/11-dependency of FFA4-S to signal to ERK1/2 upon agonism by TUG-891 (Alvarez-Curto et al., 2016; Prihandoko et al., 2016), Gαq/11 ablation caused a decreased pERK signal, particularly at early time points (Fig. 6B), for this isoform. Importantly, the DHA-induced effect was significantly more pronounced at all time points and remained sustained through the 60-minute time course for FFA4-L in Gαq/11 genome-edited cells (Fig. 6B). Taken together, these results may suggest that agonism of FFA4-L with DHA leads to β-arrestin–dependent scaffolds to ERK1/2 that are sustained rather than transient as for the short isoform, which requires Gαq/11 for the ERK1/2 effect. Direct comparison of only the FFA4-L effect in each of the three cell lines (Fig. 6C) demonstrates a clear role for β-arrestin in FFA4-L-mediated ERK1/2 phosphorylation but also reveals that this effect is tonically regulated by presence of Gαq/11 signals because it is significantly amplified in the absence of Gαq/11.
Fig. 6.
β-arrestin and Gαq/11 dependence of DHA-mediated FFA4-L-ERK signaling. (A) CRISPR/Cas9 genome editing of β-arrestin-1/2 and Gαq/11 was validated via immunoblotting (IB) with anti-β-arrestin-1/2 and Gαq/11 antibodies. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) is used as a loading control while SCR denotes scrambled target sequence. (B, upper) DHA-induced (100 μM) phosphorylation of ERK1/2 was detected in parenteral control HEK293 cells (scrambled), or those transiently expressing FFA4-S or FFA4-L, as noted. For FFA4-S, an increased phosphorylation of ERK1/2 was detected in β-arrestin-1/2 genome-edited cells, whereas Gαq/11 ablation caused a decreased pERK signal, particularly at the 1–10 minute times. For FFA4-L, significant ERK1/2 phosphorylation was seen at 5 minutes following agonism and was sustained for the 60-minute duration. This effect was absent in β-arrestin-1/2 genome-edited cells, and significantly heightened in Gαq/11 genome-edited cells. (B, lower) Densitometric quantification of DHA-induced ERK1/2 phosphorylation in scrambled control, β-arrestin-1/2-null, and Gαq/11-null cells expressing either empty-vector (CON), FFA4-S, or FFA4-L. (C) Comparison of the FFA4-L data in scrambled control, β-arrestin-1/2-null and Gαq/11-null cells reveals significant loss of the effect in β-arrestin-1/2-null cells and significant amplification in Gαq/11-null cells. Each graph represents mean ± S.D. of three independent experiments.
Discussion
Herein for the first time, we report on the mechanisms of phosphorylation of FFA4-L as well as the role of the FFA4-L C terminus in β-arrestin recruitment and FFA4-L–dependent signaling to ERK1/2. Our results reveal that as we have reported for the shorter isoform (Burns et al., 2014), DHA-induced FFA4-L phosphorylation is primarily mediated by GRK6 with an additional BIMII-sensitive component, suggesting PKC involvement. Our results also show that PKC is solely responsible for heterologous phosphorylation of FFA4-L. Comparatively, nonagonist mediated basal phosphorylation of FFA4-L is significantly lower than that of the shorter isoform, and in this regard, PKC inhibition did not significantly alter basal FFA4-L phosphorylation, in contrast to that seen with FFA4-S (Burns and Moniri, 2010; Burns et al., 2014). Despite containing four phospholabile Ser/Thr residues in the additional gap of FFA4-L, we have previously shown that these residues are not phosphorylated (Burns and Moniri, 2010), and taken together, these results demonstrate that the 16 additional amino acids in FFA4-L may be responsible for upholding receptor conformations that inhibit basal PKC-mediated phosphorylation seen with the shorter isoform.
Results of our whole-cell phosphorylation assay demonstrate that homologous (i.e., DHA-induced) and heterologous (i.e., PMA or H1R-induced) phosphorylation of FFA4-L is localized to the C-terminal tail as removal of the analogous FFA4-S phosphorylation sites via truncation following Pro356 (FFA4-L-Δ356) abolished our ability to detect 32P incorporation into immunoprecipitated FFA4-L. This translated to an apparent loss of β-arrestin-2-YFP recruitment as visualized by confocal microscopy, where cell-surface and puncta fluorescence were not easily detectable in the FFA4-L-Δ356 mutant. Interestingly, using a more sensitive and quantifiable BRET-based approach, our data show that the absence of the C-terminal phosphorylation sites only significantly decreases, but did not fully abolish FFA4-L-β-arrestin-2 BRET signals seen with DHA as well as TUG-891, unlike that seen with FFA4-S here and as shown by others (Butcher et al., 2014). Based on these data, it is tempting to speculate that this residual β-arrestin responsiveness could be due to the ability of FFA4-L-Δ356 to continue to interact with β-arrestin-2 due to its additional 16 amino acid segment even in the absence of the C-terminal tail, whereas FFA4-S loses all ability to do so upon C-terminal truncation. Another possibility that may explain the difference seen in the β-arrestin recruitment between isoforms is that the sensitivity of the whole-cell phosphorylation assay used in Fig. 3 may fail to capture lower levels of phosphorylation events, for example, a single additional site outside of the C terminus; however, it is unlikely that a single site could provide such negative charge required to create the β-arrestin phosphosensor. Indeed, for FFA4-S, a total of five phosphorylation sites coupled to three acidic residues were required to create two distinct clusters of negative charge required for β-arrestin recruitment (Butcher et al., 2014; Prihandoko et al., 2016).
Another significant difference our data reveal regarding the two FFA4 isoforms relates to their Gαq/11 and β-arrestin dependence. Agonism of FFA4-S with TUG-891 has been shown to stimulate phosphorylation of ERK1/2 in a manner that was fully blocked by YM-254890, suggesting Gαq/11-PLC-Ca+2-PKC dependence of this effect (Hudson et al., 2013; Alvarez-Curto et al., 2016; Prihandoko et al., 2016). Our results with DHA as the agonist in CRISPR/Cas9-genome edited cells confirm this for the shorter isoform, although we did detect a very small DHA-induced effect seen at 10–30 minute in cells lacking Gαq/11, suggesting that this effect of DHA may be mediated by other G proteins or β-arrestin scaffolding. On the contrary, in β-arrestin-1/2 deleted cells, agonism of FFA4-S with DHA facilitated a significantly higher phosphorylation of ERK1/2. These results would be consistent with the canonical role of arrestins as G-protein signal arrestors, the absence of which removes the ability to halt G protein signals in these cells, leading to the enhanced ERK1/2 phosphorylation seen here. With regard to FFA4-L, agonism with DHA induces a much more sustained phospho-ERK1/2 signal compared with the more transiently natured response seen with FFA4-S. This signal peaked at 5 minutes following agonism and was approximately equal in degree for 60 minutes. Although it may be tempting to correlate this prolonged signal and β-arrestin dependence with FFA4-L internalization, Watson et al. (2012) have previously shown that the rate and degree of internalization of FFA4-S and FFA4-L are very similar, as are their postinternalization fates, suggesting that the results seen here are not likely due to internalized receptor/β-arrestin scaffolds. The noted transient versus sustained ERK1/2 signaling seen between FFA4 isoforms may have important implications given the differing physiologic effects, for example proliferation versus differentiation, induced by the frequency-dependent activation of ERK1/2, which has been demonstrated to affect the fate of a variety of cell types (Buscà and Ballotti, 2000; Ryu et al., 2015; Arkun and Yasemi, 2018). Also, unlike that seen with FFA4-S, our results demonstrate a clear dependence on β-arrestin-1/2 for FFA4-L mediated ERK1/2 phosphorylation because there was no specific DHA-mediated increase in ERK1/2 phosphorylation seen in β-arrestin–edited cells. Unexpectedly, in Gαq/11-edited cells, there was a dramatically increased DHA-mediated effect, suggesting that the Gαq/11/PLC system may tonically regulate the FFA4-L/ERK cascade, whereas removal of this regulation increases the signaling output. However, in the absence of Gαq/11 to sequester them, this effect may also arise due to an increase in free-Gβγ subunits, which may contribute to downstream β-arrestin recruitment and β-arrestin scaffolding. Although our study points to a significant role for β-arrestins in FFA4-L function, there is also a regulatory role of Gαq/11 in the ERK1/2 effect, which may not be altogether unsurprising given the role of G protein heterotrimers in recruitment of GRKs as well as the variety of inputs that are responsible for regulation of MAPKs (Grundmann et al., 2018; Gurevich and Gurevich, 2018). Given this knowledge, our study does not shed light on whether β-arrestin directly mediates ERK1/2 activity because the results seen herein with β-arrestin–edited cells may also be due to loss of β-arrestin scaffolds that modulate other G protein (non-Gαq/11) processes, independent of any β-arrestin–specific scaffolds. Hence, our data do not point to a specific β-arrestin signal that links FFA4-L to ERK1/2 activity, and, as a consequence, we cannot rule out the possibility of β-arrestin–scaffolded non-Gαq/11-G protein or altogether non-G protein process that may be responsible. Further research will need to be performed to determine the precise signaling and scaffolding cascades that facilitate FFA4-L signals. In summary, herein, we report for the first time that the longer FFA4 isoform is phosphoregulated on the C-terminal tail by PKC and GRK6 and that the C terminus is responsible for β-arrestin interactions. Our data also highlights unexpected differences in regard to basal phosphorylation and β-arrestin-2 interactions between the FFA4 isoforms, as well as potentially significant differences in ERK1/2 outcomes. Further work will be required in cells that endogenously express FFA4-L to determine the physiologic significance of these effects.
Abbreviations
- AH7614
4-methyl-N-(9H-xanthen-9-yl)benzenesulfonamide
- β2AR
β2-adrenergic receptor
- BIMII
bisindolylmaleimide II
- BRET
bioluminescence resonance energy transfer
- DHA
docosahexaenoic acid
- ERK1/2
extracellular-signal regulated kinase-1/2
- FFA
free-fatty acid receptor
- FFA4-L
FFA4-long
- FFA4-S
FFA4-short
- GPCR
G protein–coupled receptor
- GRK
G protein–coupled receptor kinase
- HEK293
human embryonic kidney 293
- H1R
H1 receptor
- H-89
N-[2-[[3-(4-Bromophenyl)-2-propenyl]amino]ethyl]-5-isoquinolinesulfonamide dihydrochloride
- MAPK
mitogen activated protein kinase
- PCR
polymerase chain reaction
- PKA
protein kinase A
- PKC
protein kinase C
- PMA
phorbol 12-myristate 13-acetate
- RIPA filter
radioimmunoprecipitation assay buffer
- Rluc
Renilla luciferase
- siRNA
small interfering RNA
- TUG-891
3-(4-{[5-fluoro-2-(4-methylphenyl)phenyl]methoxy}phenyl)propanoic acid
- WT
wild type
Authorship Contributions
Participated in research design: Senatorov, Burns, Moniri.
Conducted experiments: Senatorov, Cheshmehkani, Burns, Singh.
Contributed new reagents or analytical tools: Senatorov, Cheshmehkani, Burns.
Performed data analysis: Senatorov, Moniri.
Wrote or contributed to the writing of the manuscript: Senatorov, Moniri.
Footnotes
This work was supported by the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases Grant [DK098730] to N.H.M.; and a Diabetes Action Research and Educational Foundation grant to N.H.M. R.N.B. was supported by a pre-doctoral fellowship from the American Foundation for Pharmaceutical Education.
Chemical compounds studied in this article: AH7614 (PubChem CID 233085), forskolin (PubChem CID 47936), H-89 (PubChem CID 5702541), PMA (PubChem CID 27924), docosahexaenoic acid (PubChem CID 445580), Bisindolylmaleimide II (PubChem CID 2397), TUG-891 (PubChem CID 57522038) 3-(4-{[5-Fluoro-2-(4-methylphenyl)phenyl]methoxy}phenyl)propanoic acid.
References
- Alvarez-Curto E, Inoue A, Jenkins L, Raihan SZ, Prihandoko R, Tobin AB, Milligan G.(2016) Targeted elimination of G proteins and arrestins defines their specific contributions to both intensity and duration of G protein-coupled receptor signaling. J Biol Chem 291:27147–27159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Curto E, Milligan G.(2016) Metabolism meets immunity: the role of free fatty acid receptors in the immune system. Biochem Pharmacol 114:3–13. [DOI] [PubMed] [Google Scholar]
- Arkun Y, Yasemi M.(2018) Dynamics and control of the ERK signaling pathway: sensitivity, bistability, and oscillations. PLoS One 13:e0195513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns RN, Moniri NH.(2010) Agonism with the omega-3 fatty acids alpha-linolenic acid and docosahexaenoic acid mediates phosphorylation of both the short and long isoforms of the human GPR120 receptor. Biochem Biophys Res Commun 396:1030–1035. [DOI] [PubMed] [Google Scholar]
- Burns RN, Singh M, Senatorov IS, Moniri NH.(2014) Mechanisms of homologous and heterologous phosphorylation of FFA receptor 4 (GPR120): GRK6 and PKC mediate phosphorylation of Thr347, Ser350, and Ser357 in the C-terminal tail. Biochem Pharmacol 87:650–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscà R, Ballotti R.(2000) Cyclic AMP a key messenger in the regulation of skin pigmentation. Pigment Cell Res 13:60–69. [DOI] [PubMed] [Google Scholar]
- Butcher AJ, Hudson BD, Shimpukade B, Alvarez-Curto E, Prihandoko R, Ulven T, Milligan G, Tobin AB.(2014) Concomitant action of structural elements and receptor phosphorylation determines arrestin-3 interaction with the free fatty acid receptor FFA4. J Biol Chem 289:18451–18465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheshmehkani A, Senatorov IS, Dhuguru J, Ghoneim O, Moniri NH.(2017) Free-fatty acid receptor-4 (FFA4) modulates ROS generation and COX-2 expression via the C-terminal β-arrestin phosphosensor in Raw 264.7 macrophages. Biochem Pharmacol 146:139–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman NJ, Lefkowitz RJ.(1996) Desensitization of G protein-coupled receptors. Recent Prog Horm Res 51:319–351, NaN–353. [PubMed] [Google Scholar]
- Galindo MM, Voigt N, Stein J, van Lengerich J, Raguse JD, Hofmann T, Meyerhof W, Behrens M.(2012) G protein-coupled receptors in human fat taste perception. Chem Senses 37:123–139. [DOI] [PubMed] [Google Scholar]
- Grundmann M, Merten N, Malfacini D, Inoue A, Preis P, Simon K, Rüttiger N, Ziegler N, Benkel T, Schmitt NK, et al. (2018) Lack of beta-arrestin signaling in the absence of active G proteins. Nat Commun 9:341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich VV, Gurevich EV.(2018) Arrestin-mediated signaling: is there a controversy? World J Biol Chem 9:25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habara M, Tamanuki M, Ishikawa S, Takemitsu H, Mori H, Okada Y, et al. (2015) Identification of free fatty acid receptors GPR40/FFAR1 and GPR120/FFAR4 in a domestic cat. Asian J Anim Vet Adv 10:185–190. [Google Scholar]
- Hirasawa A, Tsumaya K, Awaji T, Katsuma S, Adachi T, Yamada M, Sugimoto Y, Miyazaki S, Tsujimoto G.(2005) Free fatty acids regulate gut incretin glucagon-like peptide-1 secretion through GPR120. Nat Med 11:90–94. [DOI] [PubMed] [Google Scholar]
- Hudson BD, Shimpukade B, Mackenzie AE, Butcher AJ, Pediani JD, Christiansen E, Heathcote H, Tobin AB, Ulven T, Milligan G.(2013) The pharmacology of TUG-891, a potent and selective agonist of the free fatty acid receptor 4 (FFA4/GPR120), demonstrates both potential opportunity and possible challenges to therapeutic agonism. Mol Pharmacol 84:710–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issad T, Jockers R.(2006) Bioluminescence resonance energy transfer to monitor protein-protein interactions. Methods Mol Biol 332:195–209. [DOI] [PubMed] [Google Scholar]
- Kim JM, Lee KP, Park SJ, Kang S, Huang J, Lee JM, Sato K, Chung HY, Okajima F, Im DS.(2015) Omega-3 fatty acids induce Ca(2+) mobilization responses in human colon epithelial cell lines endogenously expressing FFA4. Acta Pharmacol Sin 36:813–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan G, Alvarez-Curto E, Hudson BD, Prihandoko R, Tobin AB.(2017) FFA4/GPR120: pharmacology and therapeutic opportunities. Trends Pharmacol Sci 38:809–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyabe M, Gin A, Onozawa E, Daimon M, Yamada H, Oda H, Mori A, Momota Y, Azakami D, Yamamoto I, et al. (2015) Genetic variants of the unsaturated fatty acid receptor GPR120 relating to obesity in dogs. J Vet Med Sci 77:1201–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moniri NH.(2016) Free-fatty acid receptor-4 (GPR120): cellular and molecular function and its role in metabolic disorders. Biochem Pharmacol 110–111:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moniri NH, Daaka Y.(2007) Agonist-stimulated reactive oxygen species formation regulates beta2-adrenergic receptor signal transduction. Biochem Pharmacol 74:64–73. [DOI] [PubMed] [Google Scholar]
- Moore K, Zhang Q, Murgolo N, Hosted T, Duffy R.(2009) Cloning, expression, and pharmacological characterization of the GPR120 free fatty acid receptor from cynomolgus monkey: comparison with human GPR120 splice variants. Comp Biochem Physiol B Biochem Mol Biol 154:419–426. [DOI] [PubMed] [Google Scholar]
- Oh DY, Talukdar S, Bae EJ, Imamura T, Morinaga H, Fan W, Li P, Lu WJ, Watkins SM, Olefsky JM.(2010) GPR120 is an omega-3 fatty acid receptor mediating potent anti-inflammatory and insulin-sensitizing effects. Cell 142:687–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pack TF, Orlen MI, Ray C, Peterson SM, Caron MG.(2018) The dopamine D2 receptor can directly recruit and activate GRK2 without G protein activation. J Biol Chem 293:6161–6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penela P, Ribas C, Mayor F., Jr.(2003) Mechanisms of regulation of the expression and function of G protein-coupled receptor kinases. Cell Signal 15:973–981. [DOI] [PubMed] [Google Scholar]
- Pitcher JA, Freedman NJ, Lefkowitz RJ.(1998) G protein-coupled receptor kinases. Annu Rev Biochem 67:653–692. [DOI] [PubMed] [Google Scholar]
- Pitcher JA, Inglese J, Higgins JB, Arriza JL, Casey PJ, Kim C, Benovic JL, Kwatra MM, Caron MG, Lefkowitz RJ.(1992) Role of β γ subunits of G proteins in targeting the β-adrenergic receptor kinase to membrane-bound receptors. Science 257:1264–1267. [DOI] [PubMed] [Google Scholar]
- Prihandoko R, Alvarez-Curto E, Hudson BD, Butcher AJ, Ulven T, Miller AM, Tobin AB, Milligan G.(2016) Distinct phosphorylation clusters determine the signaling outcome of free fatty acid receptor 4/G protein-coupled receptor 120. Mol Pharmacol 89:505–520. [DOI] [PubMed] [Google Scholar]
- Ren XR, Reiter E, Ahn S, Kim J, Chen W, Lefkowitz RJ.(2005) Different G protein-coupled receptor kinases govern G protein and beta-arrestin-mediated signaling of V2 vasopressin receptor. Proc Natl Acad Sci USA 102:1448–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H, Chung M, Dobrzyński M, Fey D, Blum Y, Lee SS, Peter M, Kholodenko BN, Jeon NL, Pertz O.(2015) Frequency modulation of ERK activation dynamics rewires cell fate [published correction appears in Mol Syst Biol (2016) 12:866]. Mol Syst Biol 11:838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, Moniri NH.(2012) Reactive oxygen species are required for β2 adrenergic receptor-β-arrestin interactions and signaling to ERK1/2. Biochem Pharmacol 84:661–669. [DOI] [PubMed] [Google Scholar]
- Song T, Peng J, Ren J, Wei HK, Peng J.(2015) Cloning and characterization of spliced variants of the porcine G protein coupled receptor 120. BioMed Res Int 2015:813816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulven T, Christiansen E.(2015) Dietary fatty acids and their potential for controlling metabolic diseases through activation of FFA4/GPR120. Annu Rev Nutr 35:239–263. [DOI] [PubMed] [Google Scholar]
- Watson SJ, Brown AJ, Holliday ND.(2012) Differential signaling by splice variants of the human free fatty acid receptor GPR120. Mol Pharmacol 81:631–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson KR, Hansen SVF, Hudson BD, Alvarez-Curto E, Raihan SZ, Azevedo CMG, Martin G, Dunlop J, Yarwood SJ, Ulven T, et al. (2017) Probe-dependent negative allosteric modulators of the long-chain free fatty acid receptor FFA4. Mol Pharmacol 91:630–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willets JM, Challiss RA, Nahorski SR.(2003) Non-visual GRKs: are we seeing the whole picture? Trends Pharmacol Sci 24:626–633. [DOI] [PubMed] [Google Scholar]