Abstract
Several published protocols exist for isolating contractile or myofibrillar (MF) proteins from skeletal muscle, however, achieving complete resuspension of the myofibril pellet can be technically challenging. We performed several previously published MF isolation methods with the intent of determining which method was most suitable for MF protein isolation and solubilization. Here, we provide an optimized protocol to isolate sarcoplasmic and solubilized MF protein fractions from mammalian skeletal muscle suitable for several downstream assays.
Keywords: protein isolation, muscle, MF protein, actin, myosin
INTRODUCTION
Several researchers have had an interest in isolating and solubilizing skeletal muscle myofibrillar (MF) proteins to assess MF protein synthesis rates [1-12], or chronic changes in myofibril protein concentrations in response to exercise training [13-21]. However, isolating and solubilizing MF proteins is technically challenging given that skeletal muscle is made up of ~80% MF protein (on a dry mass basis) [22], and contractile proteins possess large molecular masses and are highly interconnected. Different methods for MF protein isolation and quantification have been published. For instance, some studies have used the phenol phase from the guanidinium thiocyanate-phenol-chloroform extraction method (TRIzol) to extrapolate vastus lateralis (VL) MF protein changes both acutely [14] and following long-term resistance training [16]. Likewise, we recently examined how 12 weeks of resistance training affected VL MF and sarcoplasmic protein concentrations in college-aged male subjects [13] by utilizing a protocol from Goldberg’s laboratory [23] (referred to herein as MF method 1). This method isolates sarcoplasmic and MF proteins, and our analyses confirmed that prominent myosin and actin bands were present through SDS-PAGE and Coomassie imaging of the isolated MF protein fraction [13]. However, as noted above, myofibril pellet solubility in the final resuspension solution was relatively poor and required continuous manual disaggregation. Beyond these two aforementioned methods, several tracer studies examining the fractional synthetic rate of VL MF protein have allocated a method similar to Goldberg’s for the separation of MF and sarcoplasmic constituents followed by the solubilization of the MF protein pellet through the addition of 0.3 M NaOH and heating/vortexing [18,24-26]. Several laboratories also use commercially-made RIPA or general cell lysis buffers to process muscle tissue for downstream assays such as Western blotting. However, based on our extensive experience, these buffers typically leave behind a large protein pellet following homogenization and centrifugation, and much of this residual protein pellet likely contains MF proteins.
While numerous methods for MF protein isolation have been reported throughout the literature, no study has thoroughly investigated if these methods yield similar protein concentrations and protein integrity. Comparing and contrasting these methods and determining which method yields optimal results (i.e., completely solubilized myofibrils) are imperative given that several different methods for muscle tissue homogenizing exist. Therefore, the purpose of this study was to compare the five different protein isolation methods with respect to MF protein yield and integrity (Experiment 1). Additionally, we leveraged results from Experiment 1 to develop an optimized and proteomic-validated procedure, which we have coined as the “myofibril isolation and solubilization technique” or “MIST”, that can be allocated to isolate soluble MF and sarcoplasmic protein fractions (Experiment 2).
MATERIALS AND METHODS
Ethical approval for rodent study and gastrocnemius dissections
Due to the relatively large tissue requirement needed for Experiment 1, rat gastrocnemius muscle was analyzed. Gastrocnemius muscles were obtained from three 6-month old and three 18-month old male Fisher rats in a protocol which was previously approved by the Auburn University Animal Care and Use Committee. The sampling of this muscle from younger and older male rats was due to convenience given that we had a plentiful amount of muscle left over from previously published studies [27,28]. It should be noted that the purpose of this study was not to perform age comparisons for outcome variables. Rather, we were primarily interested in determining which method was optimal for isolating and solubilizing MF protein. Readers are encouraged to refer to [27] for further experimental details regarding animal husbandry and tissue procurement.
Ethical approval for obtaining human muscle
Prior to engaging in data collection, the human portion of this study was approved by the Institutional Review Board at Auburn University (Protocol # 18-442 MR 1812) and conformed to the standards set by the latest revision of the Declaration of Helsinki. College-aged males from the local community were recruited to participate in this study. Participants provided both verbal and written informed consent, and completed a medical history form prior to screening.
Muscle biopsies were collected using a 5-gauge needle under local anesthesia as previously described [29]. Immediately following tissue procurement, ~100 mg of tissue was teased of blood and connective tissue, wrapped in pre-labelled foils, flash frozen in liquid nitrogen, and subsequently stored at -80°C until assays described below.
Experiment 1 procedures
On the day of homogenization, muscles were powdered on a liquid nitrogen-cooled ceramic mortar and pestle. Frozen tissue (20–25 mg) from the six rats was weighed from each muscle using an analytical scale with a sensitivity of 0.0001 g (Mettler-Toledo; Columbus, OH, USA) and quickly placed in the first buffer of the five methods listed below. For all methods except the general cell lysis (GCL) method we began protocols with ~20 mg muscle and 200 μl of buffer 1, and resuspended the MF pellet from all methods in 300 μl of the final buffer. The GCL method, however, is a one-step method; thus, ~20 mg muscle was added to 300 μl of GCL buffer described below. The remainder of this section discusses the five protocols used for comparative analyses.
Trizol method
Approximately 20 mg of muscle from each rat were placed in 1.7 ml tubes containing 200 μl of Trizol (Ribozol; Ameresco, Solon, OH, USA; Cat. # N580), manually homogenized using a tight-fitting microtube pestle, and allowed to incubate at room temperature for 10 min. During the muscle crushing procedure, pestles were visually inspected with rigor to ensure no residual tissue remained on the pestle as it was being removed from the 1.7 ml tubes. Notably, residual tissue on the pestle would result in a loss of protein and an underestimation of protein content when normalized to input muscle weights. Chloroform (40 μl; Ameresco) was then added to tubes, and tubes were vigorously shaken for 15 s and incubated at room temperature for 3 min. Tubes were then centrifuged at 12000 g for 15 min at 4°C. The resultant clear supernatant (containing RNA) was completely removed, 60 μl of 100% ethanol (Ameresco) was added to tubes which contained the inter- and organic phases, and tubes were vigorously shaken for 15 s and incubated at room temperature for 3 min. Tubes were centrifuged at 2000 g for 5 min at 4°C, and phenol/ethanol supernatants (containing protein) were transferred to new 1.7 ml tubes. Isopropanol (300 μl; Ameresco) was added to the phenol/ethanol fraction, and tubes were vigorously shaken for 15 s and incubated at room temperature for 10 min. Tubes were centrifuged at 12000 g for 10 min at 4°C, the supernatant was discarded, and the protein pellet was dried in a lyophilizing apparatus for 5 min. The protein pellet was then resuspended in 300 μl of 1% sodium dodecyl sulfate solution (w/v in deionized water; Ameresco) using a tight-fitting micropestle. Tubes were then centrifuged at 10000 g for 10 min at 4°C to remove insoluble material, and the final supernatant was transferred to a new 1.7 ml microtube and stored at -80°C for protein analyses described below. Notably, no putative sarcoplasmic fraction was yielded from this procedure, and 1% SDS was largely ineffective at solubilizing the protein pellet.
General cell lysis method: Approximately 20 mg of muscle from each rat was placed in 1.7 ml tubes containing 300 μl of ice-cold pre-fabricated 1× general cell lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% Triton, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, 1 μg/ml leupeptin; Cell Signaling, Danvers, MA, USA). Samples were homogenized using tight-fitting pestles, insoluble proteins were removed with centrifugation at 500 g for 5 min, and supernatants were transferred to a new 1.7 ml tube and stored at -80°C for protein analyses described below. Again, pestles were visually inspected with rigor to ensure that no residual tissue remained on the pestle as it was being removed from the 1.7 ml tubes. As with the Trizol method, no sarcoplasmic fraction was yielded from this procedure.
MF method 1
Approximately 20 mg of muscle from each rat was placed in 1.7 ml tubes containing 200 μl of ice cold homogenizing buffer (buffer 1: 20 mM Tris-HCl, pH 7.2, 5 mM EGTA, 100 mM KCl, 1% Triton-X100; all chemicals from Ameresco). Samples were homogenized using tight-fitting pestles and centrifuged at 3000 g for 30 min at 4°C. Again, pestles were visually inspected with rigor to ensure that no residual tissue remained on the pestle as it was being removed from the 1.7 ml tubes. Supernatants (sarcoplasmic fraction) were transferred to new 1.7 ml tubes and stored at -80°C until protein analyses described below. As a wash step, resultant pellets (MF fraction) were resuspended in buffer 1, and samples were centrifuged at 3000 g for 10 min at 4°C. Resultant supernatants from this step were discarded, pellets were resuspended in 200 μl of ice cold wash buffer (buffer 2: 20 mM Tris-HCl, pH 7.2, 100 mM KCl, 1 mM DTT; all chemicals from Ameresco), and tubes were centrifuged at 3000 g for 10 min at 4°C; this step was performed twice. Final myofibril pellets were resuspended in 300 μl of ice cold storage buffer (buffer 3: 20 mM Tris-HCl, pH 7.2, 100 mM KCl, 20% glycerol, 1 mM DTT; all chemicals from Ameresco), and stored at -80°C for protein analyses described below.
MF method 2
We sought to improve MF method 1 given that this method yields insoluble MF aggregates when the pellet is resuspended in the final resuspension buffer (buffer 3). Others have previously demonstrated that the addition of the polyamine spermidine increases protein solubility during tissue processing [30]. Notwithstanding, it remained to be determined if modifying MF method 1 through the addition of spermidine in the final resuspension solution (buffer 3) would further increase MF protein solubility and, ultimately, protein yield. Procedures for MF method 2 were identical to MF method 1 except that MF pellets were resuspended in 300 μl of ice cold storage buffer as described above (buffer 3: 20 mM Tris-HCl, pH 7.2, 100 mM KCl, 20% glycerol, 1 mM DTT; all chemicals from Ameresco) with the addition of 6.4 M spermidine (Alfa Aesar, Haverhill, MA USA) to yield a working concentration of 50 mM spermidine in Buffer 3.
MF method 3
Approximately 20 mg of muscle from each rat was placed in 1.7 ml tubes containing 200 μl ice-cold buffer (buffer 1: 25 mM Tris, pH 7.2, 0.5% Triton X-100; all chemicals from Ameresco). Samples were homogenized using tight-fitting pestles and centrifuged at 1500 g for 10 min at 4°C. Again, pestles were visually inspected with rigor to ensure that no residual tissue remained on the pestle as it was being removed from the 1.7 ml tubes. Supernatants (sarcoplasmic fraction) were transferred to new 1.7 ml tubes, and stored at -80°C until protein analyses described below. As a wash step, resultant MF pellets were resuspended in buffer 1, and centrifuged at 1500 g for 10 min at 4°C. Supernatants were discarded, MF pellets were solubilized in 300 μl NaOH (0.3 M; Ameresco), and tubes were heated to 50°C for 30 min with vortex mixing every 10 min. Tubes were centrifuged at 4000 g for 5 min at 4°C, the supernatant containing the MF fraction was collected, placed in new 1.7 ml tubes, and stored at -80°C until protein analyses described below. Notably, while others have reported that insoluble collagen pellets remain after this step [25], we did not observe such pellets.
Protein concentration determination
Protein concentrations of sarcoplasmic isolates (when applicable) as well as putative MF resuspensions were quantified using the bicinchoninic acid (BCA) colorimetric assay (Thermo Scientific, Waltham, MA, USA). First, samples were diluted 5-fold with distilled water prior to assaying. Each diluted sample (20 μl) and bovine serum albumin (BSA) standards ranging from 0.125–2 μg/μl (20 μl) were loaded in triplicate wells on clear polystyrene 96-well microplates. Blank buffers from each isolation method (20 μl) were also loaded in triplicate wells for background subtraction. BCA reagent (200 μl) was added to all wells, and plates were incubated on an orbital plate shaker at 500 rpm for ~60 min at room temperature. The microplate was then read at an absorbance of 590 nm using a spectrophotometer (BioTek Synergy H1; BioTek Instruments, Inc., Winooski, VT USA). Accompanying software was used for analysis where standard curves were generated, buffer-only absorbance values were subtracted from each respective sample, and final protein concentrations were calculated back to the standard curve after being corrected for the 5× dilution factor. For total MF protein yield, final protein concentrations were multiplied by 300 (i.e., the total volume of buffer used to resuspend the MF pellet). These values were then divided by input wet muscle weights to yield μg MF protein per mg wet muscle. Sarcoplasmic fractions from MF method 1 were not quantified given that the steps were identical in obtaining this fraction between MF methods 1 and 2.
SDS-PAGE and Coomassie staining for Experiment 1
SDS-PAGE sample preps from MF protein resuspensions were prepared using: 10 μl resuspensions from each method + 65 μl distilled water + 25 μl 4× Laemmli buffer. Sample preps (5 μl) were then loaded on pre-casted gradient (4%–15%) SDS-polyacrylamide gels (Bio-Rad Laboratories, Hercules, CA USA) and subjected to electrophoresis at 200 V for 40 min using pre-made 1× SDS-PAGE buffer (Ameresco). Gels were rinsed in distilled water for 15 min following electrophoresis, and immersed in Coomassie stain (LabSafe GEL Blue; G-Biosciences; St. Louis, MO, USA) for 60 min. Thereafter, gels were destained in tap water for 60 min, bright field imaged using a gel documentation system (UVP; Upland, CA, USA), and band densities were determined using associated software. Given that the same volume per sample was loaded for SDS-PAGE, resultant myosin and actin band densities were divided by input muscle weights and expressed as arbitrary density units (ADU)/mg wet muscle.
Western blotting
Western blotting for myosin and actin detection was performed using an anti-myosin antibody against all myosin isoforms (DSHB, Iowa City, IA, USA; Cat. # A4.1025) and an anti-actin antibody against actin (α-sarcomeric) (Sigma-Aldrich, Saint Louis, MO, USA; Cat. # A2172), respectively, per the methods of Zergeroglu et al. [31]. Briefly, SDS-PAGE was performed on SDS-PAGE sample preps from MF protein resuspensions, and proteins were transferred to polyvinylidene difluoride membranes at 200 mA for 2 h (Bio-Rad Laboratories). Membranes were then blocked for 60 min at room temperature with 5% nonfat milk powder in Tris-buffered saline with 0.1% Tween-20 (TBST; Ameresco). Membranes were then incubated with myosin and actin antibody solutions (1:5000 dilution of each stock antibody in TBST with 5% BSA) overnight at 4°C. The following day, membranes were washed in TBST and incubated with horseradish peroxidase (HRP)-conjugated anti-rabbit IgG (Cat. # 7074; Cell Signaling; Danvers, MA, USA) or HRP-conjugated anti-mouse IgG (Cat. # 7072; Cell Signaling) in TBST with 5% BSA (1:2000) at room temperature for 60 min. Membrane development was performed using an enhanced chemiluminescent reagent (Luminata Forte HRP substrate; EMD Millipore, Billerica, MA, USA), and digital images were captured using a gel documentation system (UVP, Cambridge, UK).
Experiment 2: development of an optimized procedure from Experiment 1 results
Experiment 1 indicated that the sarcoplasmic isolation from MF method 3 and the MF isolation from MF method 2 yielded the highest fidelity fractions. Thus, in Experiment 2 we sought to determine if an optimized procedure which combined both methods could be applied to human skeletal muscle to isolate high fidelity sarcoplasmic and MF proteins. Briefly, skeletal muscle from six human subjects (~20 mg) were placed in 1.7 ml tubes containing 200 μl ice-cold buffer (buffer 1 from MF method 3: 25 mM Tris, pH 7.2, 0.5% Triton X-100). Tubes were homogenized using tight-fitting pestles and centrifuged at 1500 g for 10 min at 4°C. Supernatants (sarcoplasmic fraction) were collected, transferred to new 1.7 ml tubes, and stored at -80°C until BCA and proteomic assays. As a wash step, the resultant MF pellet was resuspended in buffer 1, and centrifuged at 1500 g for 10 min at 4°C. The supernatant was discarded, and the MF pellet was solubilized in 300 μl of ice cold storage buffer (buffer 3 from MF method 2:20 mM Tris-HCl, pH 7.2, 100 mM KCl, 20% glycerol, 1 mM DTT, 50 μM spermidine). The solubilized MF pellet was then stored at -80°C until BCA and proteomic assays.
Protein quantification of each fraction obtained from human muscle occurred as described above. Shotgun proteomics was also performed at Florida State University (Tallahassee, FL, USA) on the sarcoplasmic and MF protein fractions to identify enriched proteins in each fraction, and then determine if there was significant contamination of these proteins in each opposing fraction. Each protein sample for triplicate technical runs (30 μg for each run) was prepared for LC-MS/MS analysis using EasyPep Mini MS Sample Prep Kit (Thermo Scientific). Protein sample was transferred into a new microcentrifuge tube and the final volume was adjusted to 100 μl with general cell lysis buffer (Cell Signaling). Reduction alkylation solutions (50 μl each) were added to samples, gently mixed, and incubated at 95°C using heat block for 10 min to reduce and alkylate samples, respectively. After incubation, samples were removed and cooled to room temperature. The reconstituted enzyme Trypsin/Lys-C Protease Mix solution (50 μl) was added to the reduced and alkylated samples and incubated with shaking at 37°C for 2 h to digest the protein sample. Digestion stop solution (50 μl) was then added to samples, and peptides were cleaned using a peptide clean-up column according to the kit instructions.
An externally calibrated Thermo Q Exactive HF (high-resolution electrospray tandem mass spectrometer) was used in conjunction with Dionex UltiMate3000 RSLC Nano System (Thermo Scientific). Sample (5 μl) was aspirated into a 50 μl loop and loaded onto the trap column (Thermo μ-Precolumn 5 mm, with nanoViper tubing 30 μm i.d. × 10 cm). Flow rate was set to 300 NL/min for separation on the analytical column (Acclaim pepmap RSLC 75 μM × 15 cm nanoviper; Thermo Scientific). Mobile phase A was composed of 99.9% H2O (EMD Omni Solvent; Millipore, Austin, TX, USA) and 0.1% formic acid, and mobile phase B was composed of 99.9% acetonitrile and 0.1% formic acid. A 60-minute linear gradient from 3% to 45% B was performed. The LC eluent was directly nanosprayed into the mass-spectrometer. During chromatographic separation, the Q-Exactive HF was operated in a data-dependent mode and under direct control of the Thermo Excalibur 3.1.66 (Thermo Scientific). MS data were acquired using the following parameters: 20 data-dependent collisional-induced-dissociation MS/MS scans per full scan (350 to 1700 m/z) at 60000 resolution. MS2 data were acquired in centroid mode at 15000 resolution. Ions with a single charge or charges more than 7 as well as unassigned charge were excluded. A 15-second dynamic exclusion window was used. All measurements were performed at room temperature, and three technical replicates were performed for each sample. Raw data files were analyzed using the Proteome Discoverer software package (version 2.0, Thermo Scientific) with SequestHT and Mascot search nodes using a human-specific tremble .fasta database (20180308HumanSwissprot.fasta) and the Percolator peptide validator. The resulting .msf files were further analyzed by the proteome validator software Scaffold v4.0 (Portland, OR, USA).
Statistics
One-way analysis of variance (ANOVA) tests were used to examine dependent variables in Experiment 1. If P values reached a threshold for significance (P < 0.05), LSD post hoc tests were used to determine which dependent variable differed between methods. All protein targets yielded from the proteomics data from human muscle were compared between fractions using dependent samples t-tests. All data presented in figures and results are mean ± standard deviation (SD) values.
RESULTS
Protein yields in Experiment 1
BCA protein assays were performed in order to determine protein yields from each method in Experiment 1. Figure 1A demonstrates that similar amounts of rat gastrocnemius were used for each method which is critical given that different amounts of protein input between methods may skew results. MF protein concentrations from MF methods 2 and 3 were significantly greater than all other methods (Fig. 1B). The coefficient of variation (CV) values for triplicates from each assay were as follows: the Trizol method yielded a CV of 1.63%, the GCL method yielded a CV of 1.72%, MF method 1 yielded a CV of 16.52%, MF method 2 yielded a CV of 4.08%, and MF method 3 yielded a CV of 1.20%.
Figure 1.
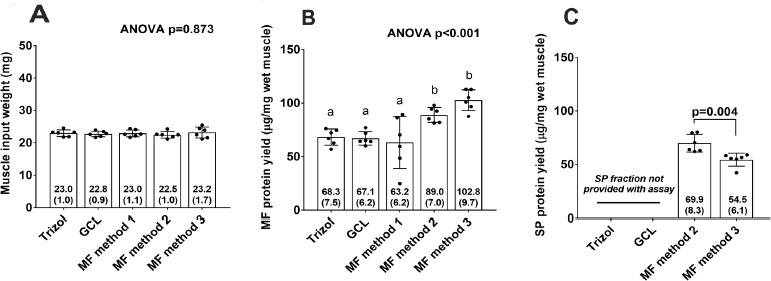
Bicinchoninic acid assay results for rat tissue (Experiment 1). A. Muscle input weights for each method (six rats per method; specifically, three 6-month old and three 18-month old male Fisher rats). B. Total MF protein yield per method. C. SP protein yield per method. In (B), superscript letters that are different indicate significant differences between methods.
Only MF methods 1–3 yielded a sarcoplasmic protein fraction and, given that MF methods 1 and 2 have identical means of isolating the sarcoplasmic fraction, sarcoplasmic protein yield is only presented for MF methods 2 and 3. An independent samples t-test indicated that MF method 2 yielded more sarcoplasmic protein compared to MF method 3 (P = 0.004; Fig. 1C).
Actin and myosin determination by Coomassie staining in Experiment 1
Performing SDS-PAGE and Coomassie staining of the MF fractions from each method enabled us to determine how much relative myosin heavy chain and actin were yielded given that these two proteins make up roughly 70% of the MF protein pool [22]. Coomassie staining results of the MF fractions from all five procedures are presented in Figure 2. The thick bands at 43 kD and 220 kD represent actin and myosin heavy chain, respectively. MF methods 1 and 2 yielded the densest bands at 220 kD (Fig. 2A) and 43 kD (Fig. 2B) compared to the Trizol and GCL methods (P < 0.05) indicating that MF protein yield as well as fidelity was superior using the former two versus the latter two procedures. MF method 3 yielded a smear which is likely due to the addition of 0.3 M NaOH during MF pellet dissolution catalyzing alkaline-mediated hydrolysis. Notably, the Trizol method yielded very little protein overall.
Figure 2.
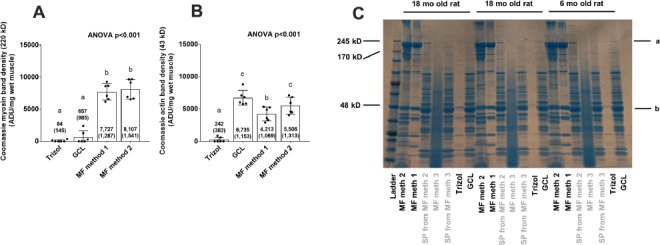
Coomassie staining results for rat tissue (Experiment 1). A. Coomassie band densities at 220 kD for all methods except MF method 3 (note that MF method 3 has band-smearing due to alkaline-mediated hydrolysis). B. Coomassie band densities at 43 kD for all methods except MF method 3. C. representative Coomassie staining gel for 3 rats (2 older and 1 younger); left to right orientation: MF isolate of MF method 2, MF isolate of MF method 1, sarcoplasmic protein, isolate from MF method 1&2, MF isolate of MF method 3, sarcoplasmic protein isolate from MF method 3, putative MF isolate of Trizol method, and putative MF isolate of GCL method. The letters “a” and “b” in (C) indicate myosin (220 kD) and actin (43 kD) bands, respectively with 6 rats per method and specifically three 6-month old male Fisher rats and three 18-month old male Fisher rats. In (A) and (B), superscript letters that are different indicate significant differences between methods.
Western blotting results for Experiment 1
While actin and myosin heavy chain bands were highly apparent using SDS-PAGE and Coomassie staining, we wanted to confirm their presence in different fractions using antibody-based detection via Western blotting. Notably, we elected not to analyze MF method 3 using this method given that hydrolysis of the MF fraction occurred (Fig. 2), and we elected to analyze only MF method 2 rather than both MF methods 1 and 2 given that the protein quantification results indicated more MF protein was yielded using spermidine addition (i.e., MF method 2).
MF method 2 yielded the greatest amount of myosin heavy chain and actin compared to the GCL and Trizol methods (Fig. 3A). Beyond determining which method yielded the greatest amounts of actin and myosin heavy chain in the MF fraction, we were also interested in determining which method contained the greatest amount of MF protein contamination in the sarcoplasmic fraction. Given that MF methods 1 and 2 have an identical initial procedure to yield sarcoplasmic protein, only the sarcoplasmic fraction from MF method 2 was analyzed. Further, MF method 3 was the only other technique that yielded a sarcoplasmic fraction and was also analyzed. MF methods 2 and 3 yielded sarcoplasmic fractions that contained trace amounts of myosin heavy chain, although the sarcoplasmic fraction of MF method 2 contained significantly more myosin heavy chain than the sarcoplasmic fraction of MF method 3 (887 ± 297 density units vs. 118 ± 45 density units, P < 0.001; Fig. 3B). Both methods contained similar amounts of actin (MF method 2 = 409 ± 85 ADU, MF method 3 = 333 ± 62 ADU, P = 0.103; Fig. 3B).
Figure 3.
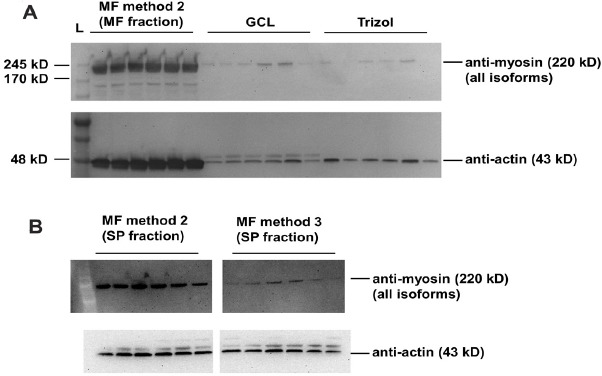
Western blotting results for rat tissue (Experiment 1). A. Myosin and actin immunoblotting for MF protein isolates from MF method 2 as well as the GCL and Trizol methods (MF method 3 was not analyzed due to alkaline-mediated hydrolysis, and MF method 1 was not analyzed because MF method 2 was superior at yielding more MF protein). B. Myosin and actin immunoblotting for sarcoplasmic protein isolates from MF methods 2 and 3 to determine relative MF contamination (six rats per method; specifically, three 6-month old and three 18-month old male Fisher rats).
Experiment 2: an optimized approach for isolating myofibrillar and sarcoplasmic protein fractions from human muscle samples
Results from Figure 1-3 above clearly demonstrate that: MF method 2 was best for yielding intact and solubilized MF protein, and buffer 1 from MF method 3 yielded sarcoplasmic proteins which were least contaminated by myosin heavy chain. Thus, Experiment 2 served to develop an optimized technique combining these two methods, and we elected to employ this method using six human muscle biopsy samples. Muscle input weights and BCA assay results using this optimized protocol are presented in Figure 4A and 4B, and Figure 4C summarizes the compositional results when considering protein concentrations from each fraction.
Figure 4.
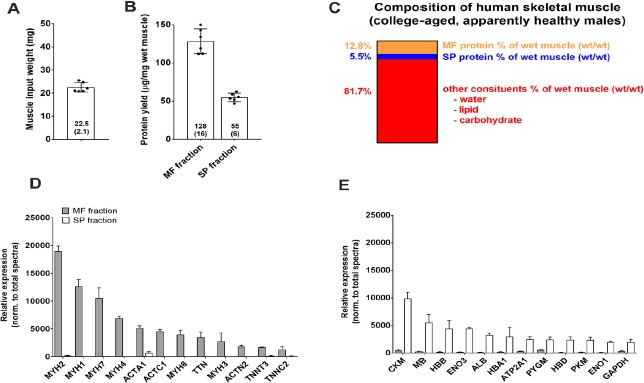
Proteomic validation of optimized technique using human skeletal muscle (Experiment 2). Data are from 6 college-aged males using the MF and SP isolation techniques optimized from rodent experiments (Table 1 procedure). A. Muscle input weights. B. MF and SP protein concentrations. C. Estimated muscle constituents per data in (A) and (B). D. The top 12 enriched proteins (mean ± SD) in the MF fraction (gray bars), showing negligible contamination in the sarcoplasmic fraction (white bars); E. The top 12 enriched proteins (mean ± SD) in the sarcoplasmic fraction (white bars), showing negligible contamination in the MF fraction (gray bars).
We then wanted to use proteomics to validate our optimized approach. Performing proteomics on the sarcoplasmic and MF fraction enabled us to examine the top-enriched proteins in each fraction while also determining if these proteins contaminated the other fraction. The top dozen enriched proteins in the MF fraction from proteomic analysis are presented in Figure 4D. All of these proteins were contractile proteins and included myosin-2 (MYH2), myosin-1 (MYH1), myosin-7 (MYH7), myosin-4 (MYH4), actin, alpha skeletal muscle (ACTS), actin, alpha cardiac muscle (ACTC), myosin-8 (MYH8), titin (TTN), myosin-3 (MYH3), alpha-actinin-2 (ACTN2), troponin T (TNNT), and troponin C (TNNC). Importantly, all of these proteins were significantly more enriched in the MF versus sarcoplasmic protein fraction (P < 0.01 for each target).
The top dozen enriched proteins in the sarcoplasmic fraction from proteomic analysis are presented in Figure 4E. All of these proteins were non-contractile proteins and included creatine kinase, m-type (CKM), myoglobin (MB), hemoglobin subunit-beta (HBB), enolase-3 (ENO3), albumin (ALB), hemoglobin subunit alpha 1 (HBA1), ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 (ATP2A1), glycogen phosphorylase, muscle associated (PYGM), hemoglobin subunit, delta (HBD), pyruvate kinase (PKM), enolase-1 (ENO1), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Importantly, all of these proteins were significantly more enriched in the sarcoplasmic versus MF protein fraction (P < 0.01 for each target).
DISCUSSION
Several proposed methods exist with the intent of isolating, solubilizing, and quantifying MF proteins. These methods have included the Trizol method [14-16,32], MF method 1 [23], and MF method 3 [18,24-26]. Herein, we sought to determine which of these methods best achieved MF protein isolation and solubilization using the three aforementioned methods along with a modified MF method 1 to improve MF protein solubility (i.e., MF method 2) as well as a general cell lysis buffer method. Additionally, we sought to develop an optimized method for sarcoplasmic and myofibril isolation, and used proteomics to validate this method. A comparative summary outlining the pros and cons of each method from Experiments 1 and 2 is provided in Table 1.
Table 1.
Summary of experimental outcomes.
| Method | Pros | Cons |
|---|---|---|
| GCL |
|
|
| Trizol |
|
|
| MF method 1 |
|
|
| MF method 2 (addition of spermidine to buffer 3 in MF method 1) |
|
|
| MF method 3 |
|
|
| Optimized method (Experiment 2); recommended |
|
|
Our results indicate that MF methods 2 and 3 should be utilized if the goal is to solubilize MF proteins and/or quantify total MF and sarcoplasmic protein concentrations. Additionally, MF method 2 can be used for downstream assays including Coomassie staining, Western blotting and proteomics, whereas MF method 3 is suitable for limited downstream assays (e.g., tracer analysis) given that protein hydrolysis of the MF fraction occurs. The only difference between MF methods 1 and 2 is the addition of spermidine to the myofibril resuspension buffer (or buffer 3 described in the methods). Spermidine is a polyamine, and only one other paper to date has demonstrated that the addition of spermidine to a lysis buffer is capable of increasing protein solubility when processing plant tissues as well as animal Golgi-localized proteins [30]. These authors suggested this effect may be due to spermidine-mediated protein denaturation. Given that the MF protein pellet contains high molecular weight proteins which exist in a complex lattice network in vivo, we contend that spermidine acts to dissociate MF protein aggregates through denaturation. However, our Western blotting and proteomic analyses demonstrated that spermidine does not hydrolyze proteins like what was observed in MF method 3 samples. Therefore, the addition to spermidine to buffer 3 in method 2 (as well as buffer 2 in the optimized method) allows for the complete solubilization of MF proteins without the loss of these proteins through hydrolysis.
While BCA analysis indicated that all methods yielded protein, Coomassie staining and Western blotting demonstrated that the Trizol and GCL methods poorly isolate MF proteins. These results are likely due to much of the MF fraction being poorly soluble in the GCL and 1.0% SDS buffers, respectively. In fact, both methods visually left large protein pellets following the final centrifugation steps, and these pellets were likely MF proteins. Further, it has been previously reported that the Trizol method poorly solubilizes proteins [33,34]. Thus, significant errors in the MF protein concentrations may affect the conclusions of prior studies which have used this method.
Homogenizing muscle with buffer 1 of MF method 3 yielded a sarcoplasmic fraction that contained significantly less myosin relative to homogenizing buffer 1 of MF method 2. This may be the reason why MF method 3 yielded numerically greater MF protein concentrations and significantly less sarcoplasmic protein compared to MF method 2 (Fig. 1B and 1C). Our optimized procedure performed on human muscle samples (Experiment 2), which we have coined to be the “myofibril isolation and solubilization technique” or “MIST” and involved combining MF methods 2 and 3, seemingly optimized the yield and integrity of the sarcoplasmic and MF fractions; this procedure is summarized in Table 2. Proteomics indicated that this procedure yielded lowly-contaminated sarcoplasmic and MF fractions as well as a solubilized MF fraction suitable for downstream assays. Specifically, the top 12 enriched proteins in the MF and sarcoplasmic isolates were contractile and metabolic proteins, respectively, which would be expected to found in this fraction per the commentary of Haun et al. [22].
Table 2.
The “MIST” method from Experiment 2 for myofibrillar and sarcoplasmic protein isolation.
| Step | General notes | Buffer recipe |
|---|---|---|
| 1 | Obtain 10–50 mg of powdered muscle on a LN2-cooled stage, and place in a 1.7 ml microtube (Tube 1) on a pre-tared analytical scale; weigh tissue for standardizing protein results | N/Aa |
| 2 | Place Tube 1 with muscle on ice and add 10 volumes (100–500 μl) of ice cold buffer 1 | Buffer 1: 25 mM Tris, pH 7.2, 0.5% Triton X-100 |
| 3 | Use a tight-fitting microtube pestle to homogenize sample into a slurry, and place Tube 1 back on ice until all samples are homogenized | N/Aa |
| 4 | Centrifuge Tube 1 at 1500 g for 10 min at 4°C | N/Aa |
| 5 | Pipet off supernatant (sarcoplasmic fraction) and place into a new 1.7 ml microtube (Tube 2), but leave ~10 μl of supernatant on top of protein pellet in order to reduce sarcoplasmic contamination with MF protein or loss of MF protein; store Tube 2 at -80°C for downstream assays | N/Aa |
| 6 | Resuspend protein pellet in Tube 1 with 10 volumes (100–500 μl) of ice cold Buffer 1 as a wash step | N/Aa |
| 7 | Centrifuge tubes at 1500 g for 10 min at 4°C | N/Aa |
| 8 | Pipet off supernatant, but leave ~10 μl on top of protein pellet in order to reduce loss of MF protein; allow pellets to dry on ice, or use a fine-tip pipettor (e.g., Western blotting loading tip) to remove the remainder of the wash buffer in order to prevent disrupting the MF pellet | N/Aa |
| 9 | Add 15 volumes of ice cold buffer 2 to pellet in Tube 1, and resuspend using a tight-fitting pestle; remaining/un-suspended protein is putatively collagen | Buffer 2: 20 mM Tris-HCl, pH 7.2, 100 mM KCl, 20% glycerol, 1 mM DTT, 50 mM spermidine |
| 10 | Perform a quick-spin on a desktop centrifuge for 1 min, and transfer supernatant (MF fraction) to a new tube (Tube 3) | N/Aa |
| Notes: |
|
|
aN/A: Not available. Other note: can add 20 volumes (i.e., 400 μl per 20 mg tissue) of buffer 2 to MF pellet to improve solubility.
In conclusion, the intent of this methodological study is to provide guidance for researchers aiming to examine longer-term changes in MF protein concentrations from human and rodent skeletal muscle. While numerous methods on this technique have been published, the standardization of this technique using optimized methods outlined herein can ultimately provide more congruent findings in the literature.
Acknowledgments
We thank Dr. Brooks Mobley (University of Kentucky) for his critical oversight of the original rat project from which muscles were obtained for analysis. We also thank Dr. Rakesh Singh (Translational Science Lab, College of Medicine, Florida State University) for mass spectrometry analysis as well as Dr. Stuart Phillips (McMaster University) for his insights regarding MF method 3. Funding for this project was provided through discretionary laboratory funds from M.D.R. Funding for proteomic analysis was provided through a seed grant to M.D.R. and V.I. from Florida A&M University.
References
- 1.Carrithers JA, Carroll CC, Coker RH, Sullivan DH, Trappe TA. (2007) Concurrent exercise and muscle protein synthesis: implications for exercise countermeasures in space. Aviat Space Environ Med 78: 457-462. PMID: [PubMed] [Google Scholar]
- 2.Kumar V, Selby A, Rankin D, Patel R, Atherton P, et al. (2008) Age-related differences in the dose-response relationship of muscle protein synthesis to resistance exercise in young and old men. J Physiol 587: 211-217. doi: 10.1113/jphysiol.2008.164483. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holm L, van Hall G, Rose AJ, Miller BF, Doessing S, et al. (2009) Contraction intensity and feeding affect collagen and myofibrillar protein synthesis rates differently in human skeletal muscle. Am J Physiol Endocrinol Metab 298: e257-e269 doi: 10.1152/ajpendo.00609.2009. PMID: [DOI] [PubMed] [Google Scholar]
- 4.Kumar V, Atherton PJ, Selby A, Rankin D, Williams J, et al. (2012) Muscle protein synthetic responses to exercise: effects of age, volume, and intensity. J Gerontol A Biol Sci Med Sci 67: 1170-1177. doi: 10.1093/gerona/gls141. PMID: [DOI] [PubMed] [Google Scholar]
- 5.Camera DM, West DWD, Burd NA, Phillips SM, Garnham AP, et al. (2012) Low muscle glycogen concentration does not suppress the anabolic response to resistance exercise. J Appl Physiol (1985) 113: 206-214. doi: 10.1152/japplphysiol.00395.2012. PMID: [DOI] [PubMed] [Google Scholar]
- 6.Witard OC, Jackman SR, Breen L, Smith K, Selby A, et al. (2013) Myofibrillar muscle protein synthesis rates subsequent to a meal in response to increasing doses of whey protein at rest and after resistance exercise. Am J Clin Nutr 99: 86-95. doi: 10.3945/ajcn.112.055517. PMID: [DOI] [PubMed] [Google Scholar]
- 7.Kim PL, Staron RS, Phillips SM. (2005) Fasted-state skeletal muscle protein synthesis after resistance exercise is altered with training. J Physiol 568: 283-290. doi: 10.1113/jphysiol.2005.093708. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Louis M, Poortmans JR, Francaux M, Berré J, Boisseau N, et al. (2003) No effect of creatine supplementation on human myofibrillar and sarcoplasmic protein synthesis after resistance exercise. Am J Physiol Endocrinol Metab 285: doi: 10.1152/ajpendo.00195.2003. PMID: [DOI] [PubMed] [Google Scholar]
- 9.Moore DR, Phillips SM, Babraj JA, Smith K, Rennie MJ. (2004) Myofibrillar and collagen protein synthesis in human skeletal muscle in young men after maximal shortening and lengthening contractions. Am J Physiol Endocrinol Metab 288: doi: 10.1152/ajpendo.00387.2004. PMID: [DOI] [PubMed] [Google Scholar]
- 10.Cuthbertson DJ, Babraj J, Smith K, Wilkes E, Fedele MJ, et al. (2005) Anabolic signaling and protein synthesis in human skeletal muscle after dynamic shortening or lengthening exercise. Am J Physiol Endocrinol Metab 290: doi: 10.1152/ajpendo.00415.2005. PMID: [DOI] [PubMed] [Google Scholar]
- 11.West DWD, Kujbida GW, Moore DR, Atherton P, Burd NA, et al. (2009) Resistance exercise-induced increases in putative anabolic hormones do not enhance muscle protein synthesis or intracellular signalling in young men. J Physiol 587: 5239-5247. doi: 10.1113/jphysiol.2009.177220. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burd NA, Holwerda AM, Selby KC, West DWD, Staples AW, et al. (2010) Resistance exercise volume affects myofibrillar protein synthesis and anabolic signalling molecule phosphorylation in young men. J Physiol 588: 3119-3130. doi: 10.1113/jphysiol.2010.192856. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roberts MD, Romero MA, Mobley CB, Mumford PW, Roberson PA, et al. (2018) Skeletal muscle mitochondrial volume and myozenin-1 protein differences exist between high versus low anabolic responders to resistance training. PeerJ 6:e5338. doi: 10.7717/peerj.5338. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willoughby DS, Nelson MJ. (2002) Myosin heavy-chain mRNA expression after a single session of heavy-resistance exercise. Med Sci Sports Exerc 34: 1262-1269. doi: 10.1097/00005768-200208000-00006. PMID: [DOI] [PubMed] [Google Scholar]
- 15.Willoughby DS, Priest JW, Nelson M. (2002) Expression of the stress proteins, ubiquitin, heat shock protein 72, and myofibrillar protein content after 12 weeks of leg cycling in persons with spinal cord injury. Arch Phys Med Rehabil 83: 649-654. doi: 10.1053/apmr.2002.31184. PMID: [DOI] [PubMed] [Google Scholar]
- 16.Willoughby DS, Rosene J. (2001) Effects of oral creatine and resistance training on myosin heavy chain expression. Med Sci Sports Exerc 33: 1674-1681. doi: 10.1097/00005768-200110000-00010. PMID: [DOI] [PubMed] [Google Scholar]
- 17.Haus JM, Carrithers JA, Carroll CC, Tesch PA, Trappe TA. (2007) Contractile and connective tissue protein content of human skeletal muscle: effects of 35 and 90 days of simulated microgravity and exercise countermeasures. Am J Physiol Regul Integr Comp Physiol 293: doi: 10.1152/ajpregu.00292.2007. PMID: [DOI] [PubMed] [Google Scholar]
- 18.Brook MS, Wilkinson DJ, Mitchell WK, Lund JN, Szewczyk NJ, et al. (2015) Skeletal muscle hypertrophy adaptations predominate in the early stages of resistance exercise training, matching deuterium oxide-derived measures of muscle protein synthesis and mechanistic target of rapamycin complex 1 signaling. FASEB J 29: 4485-4496. doi: 10.1096/fj.15-273755. PMID: [DOI] [PubMed] [Google Scholar]
- 19.Carrithers JA, Tesch PA, Trieschmann J, Ekberg A, Trappe TA. (2002) Skeletal muscle protein composition following 5 weeks of ULLS and resistance exercise countermeasures. J Gravit Physiol 9: P155-P156. PMID: [PubMed] [Google Scholar]
- 20.Cribb PJ, Hayes A. (2006) Effects of supplement timing and resistance exercise on skeletal muscle hypertrophy. Med Sci Sports Exerc 38: 1918-1925. doi: 10.1249/01.mss.0000233790.08788.3e. PMID: [DOI] [PubMed] [Google Scholar]
- 21.Woolstenhulme MT, Conlee RK, Drummond MJ, Stites AW, Parcell AC. (2006) Temporal response of desmin and dystrophin proteins to progressive resistance exercise in human skeletal muscle. J Appl Physiol (1985) 100: 1876-1882. doi: 10.1152/japplphysiol.01592.2005. PMID: [DOI] [PubMed] [Google Scholar]
- 22.Haun CT, Vann CG, Roberts BM, Vigotsky AD, Schoenfeld BJ, et al. (2019) A Critical Evaluation of the Biological Construct Skeletal Muscle Hypertrophy: Size Matters but So Does the Measurement. Front Physiol 10: 247. doi: 10.3389/fphys.2019.00247. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohen S, Brault JJ, Gygi SP, Glass DJ, Valenzuela DM, et al. (2009) During muscle atrophy, thick, but not thin, filament components are degraded by MuRF1-dependent ubiquitylation. J Cell Biol 185: 1083-1095. doi: 10.1083/jcb.200901052. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore DR, Tang JE, Burd NA, Rerecich T, Tarnopolsky MA, et al. (2009) Differential stimulation of myofibrillar and sarcoplasmic protein synthesis with protein ingestion at rest and after resistance exercise. J Physiol 587: 897-904. doi: 10.1113/jphysiol.2008.164087. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burd NA, Andrews RJ, West DWD, Little JP, Cochran AJR, et al. (2011) Muscle time under tension during resistance exercise stimulates differential muscle protein sub-fractional synthetic responses in men. J Physiol 590: 351-362. doi: 10.1113/jphysiol.2011.221200. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Damas F, Phillips SM, Libardi CA, Vechin FC, Lixandrão ME, et al. (2016) Resistance training-induced changes in integrated myofibrillar protein synthesis are related to hypertrophy only after attenuation of muscle damage. J Physiol 594: 5209-5222. doi: 10.1113/JP272472. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mobley CB, Mumford PW, Kephart WC, Haun CT, Holland AM, et al. (2017) Aging in Rats Differentially Affects Markers of Transcriptional and Translational Capacity in Soleus and Plantaris Muscle. Front Physiol 8: 518. doi: 10.3389/fphys.2017.00518. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mumford PW, Romero MA, Osburn SC, Roberson PA, Vann CG, et al. (2019) Skeletal muscle LINE-1 retrotransposon activity is upregulated in older versus younger rats. Am J Physiol Regul Integr Comp Physiol 317: doi: 10.1152/ajpregu.00110.2019. PMID: [DOI] [PubMed] [Google Scholar]
- 29.Haun CT, Vann CG, Mobley CB, Roberson PA, Osburn SC, et al. (2018) Effects of Graded Whey Supplementation During Extreme-Volume Resistance Training. Front Nutr 5: 84. doi: 10.3389/fnut.2018.00084. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yasui K, Uegaki M, Shiraki K, Ishimizu T. (2010) Enhanced solubilization of membrane proteins by alkylamines and polyamines. Protein Sci 19:486-493. doi: 10.1002/pro.326. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zergeroglu MA, McKenzie MJ, Shanely RA, Van Gammeren D, DeRuisseau KC, et al. (2003) Mechanical ventilation-induced oxidative stress in the diaphragm. J Appl Physiol (1985) 95: 1116-1124. doi: 10.1152/japplphysiol.00824.2002. PMID: [DOI] [PubMed] [Google Scholar]
- 32.Shelmadine B, Cooke M, Buford T, Hudson G, Redd L, et al. (2009) Effects of 28 days of resistance exercise and consuming a commercially available pre-workout supplement, NO-Shotgun(R), on body composition, muscle strength and mass, markers of satellite cell activation, and clinical safety markers in males. J Int Soc Sports Nutr 6: 16. doi: 10.1186/1550-2783-6-16. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kopec AM, Rivera PD, Lacagnina MJ, Hanamsagar R, Bilbo SD. (2017) Optimized solubilization of TRIzol-precipitated protein permits Western blotting analysis to maximize data available from brain tissue. J Neurosci Methods 280: 64-76. doi: 10.1016/j.jneumeth.2017.02.002. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hummon AB, Lim SR, Difilippantonio MJ, Ried T. (2007) Isolation and solubilization of proteins after TRIzol extraction of RNA and DNA from patient material following prolonged storage. Biotechniques 42: 467-470, 472. doi: 10.2144/000112401. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]


