Abstract
Gastric cancer (GC) is one of the most common types of cancer and the second leading cause of cancer-associated mortality. Identification of novel biomarkers is critical to prolonging patient survival. MicroRNAs (miRNAs) proved to play diverse roles in the physiological and pathological state in cancers including GC. Herein we aimed at performing a meta-analysis on miRNA profiling studies that used microarray platforms. Relevant studies were retrieved from PubMed and GEO databases. We used the robust rank aggregation to perform the meta-analysis. Moreover, for meta-signature miRNAs target genes, we performed pathway enrichment and GO molecular function enrichment analysis. A total of 19 upregulated miRNAs and seven downregulated miRNAs in GC samples were identified. However, only three upregulated and one downregulated miRNA reached statistical significance after multiple test correction. Here we showed that hsa-miR-21-5p, hsa-miR-93-5p, hsa-miR-25-3p, and hsa-miR-375 are differentially expressed in GC samples.
Key Words: Gastric cancer, microRNA, GO enrichment, biomarkers
Gastric cancer (GC) is the fourth most prevalent malignancy in the world and the second leading cause of cancer- related death worldwide (1). Currently, surgical resection is the most effective treatment for GC, which specially prolongs the survival of patients in early stages. Nonetheless, in advanced condition, GC recurs frequently as hematological and nodal metastases and peritoneal dissemination. The prognosis for individuals with advanced disease consequently remains poor (2). Thus, investigation to identify biomarkers for early detection and precise diagnosis of GC is valuable to prolong survival of patients.
MicroRNAs (miRNAs) are known as a distinct class of small noncoding RNA molecules (18-25 nucleotides in length) regulating the gene expression of, at least, about one third of all protein- coding mRNAs. These highly conserved molecules influence, either directly or indirectly, nearly entire cellular pathways (3). Due to the findings of ENCODE (Encyclopedia of DNA Elements) Project as well as recent emergence of different classes of noncoding RNAs, the potential roles of miRNA expression profiling has achieved a high level of importance specially in cancer research (4, 5).
Over the past few years, altered expression of miRNAs have been correlated with several diseases, particularly cancers (6). Recent findings from integrative and mechanism- based studies have provided essential evidence about the miRNAs roles in normal and disease conditions (7, 8). In particular these studies are helpful in obtaining a deeper comprehension of the underlying molecular mechanisms in GC. There are several lines of evidence that support the implication of miRNA expression deregulation in GC pathogenesis (9). To date a number of microarray, based experiments have explored the miRNA expression profile in GC samples. Unfortunately, the results appear to be inconsistent between these studies. This could be partially explained by applying different technological platforms, limited sample size, different analytical pipelines and ongoing miRNA discovery (10, 11).
In order to minimize these limitations, we aimed at performing a meta-analysis applying the robust rank aggregation method, followed by pathway enrichment to identify deregulated miRNAs and pathways in GC. This included the combination of several independent studies to increment the statistical power and to identify the fundamental miRNAs in carcinogenesis of GCs. The method of leave-one-out cross-validation was utilized to assess robustness of the results. Possibly identification of miRNA meta- signature and pathways regulated by them, could provide promising targets for further biomarker discovery in GC.
Material and methods
Search strategy
GC miRNA expression profiling studies were retrieved from PubMed and GEO database by means of a combination of the keywords ‘gastric cancer’,‘stomach cancer’, ‘microRNAs’, ‘profiling’ and ‘human’. We performed the last search in March 2018. The following criteria had to be met by eligible studies: (i), they were miRNA expression profiling studies in gastric cancer patients; (ii), they used tumor tissue samples and either corresponding noncancerous tissue and adjacent normal tissues for comparison; (iii), the use of miRNA microarray platforms. Meanwhile, studies were not eligible for meta-analysis if they met the following selection criteria: (i) using only gastric cancer cell lines, (ii) investigating effects of Helicobacter pylori on gene expression (iii) profiling circulating miRNA, (iv) using different miRNA profiling technologies or no sufficient data on microarray platform used, (v) review articles and meta-analysis.
Data abstraction
Two investigators (HGH and AZ) independently reviewed all full-text articles and GEO entries. Any disagreement was resolved by team discussion. From the eligible studies, the following items were collected and recorded: first author, region, number of probe, number of samples, GSE or PMID accession number and microarray platform. To have a more comprehensive list of miRNAs, GEO datasets were reanalyzed by the GEO2R tools implemented in the database. The lists of miRNAs with statistically significant expression changes were extracted from the publications. We used the miRBase version 21 (12) to obtain standard name for miRNAs. Non-human miRNAs probe e.g. viral miRNAs probe and also non-miRNA probes were excluded from meta-analysis.
Statistical analysis
Based on statistical test P-values (<0.05 were considered significant), the miRNA records in each study were prioritized. To perform a meta-analysis, we used the robust rank aggregation approach implemented as an R package RobustRankAggreg (13). The method is based on the comparison of real data with a null model which assumes random order of input lists. After computation, in the aggregated list each element has an assigned P-value that indicated how much better it was ranked than expected. To control false positive results, Bonferroni correction was performed. Meanwhile, to assess the robustness of P-values, leave one out (sensitivity analysis) was applied on the robust rank aggregation algorithm.
MiRNA target genes and enrichment analysis
Target genes of miRNAs were obtained from the IMOTA (https://ccb-web.cs.uni-saarland. de/ imota/) (14). This database is an interactive multi-omics-tissue atlas that provides a list of miRNA targets with regards to miRNA and mRNA co-expression data. IMOTA provides three types of target genes regarding evidence level. Strong target (ST), and weak target (WT) are experimentally validated target genes, while predictive targets (PT) have no supporting experimental data. To identify the pathways of miRNA targets, Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology terms were carried out with DAVID web tool (https://david.ncifcrf.gov/) (15).
Results
Study selection and data extraction
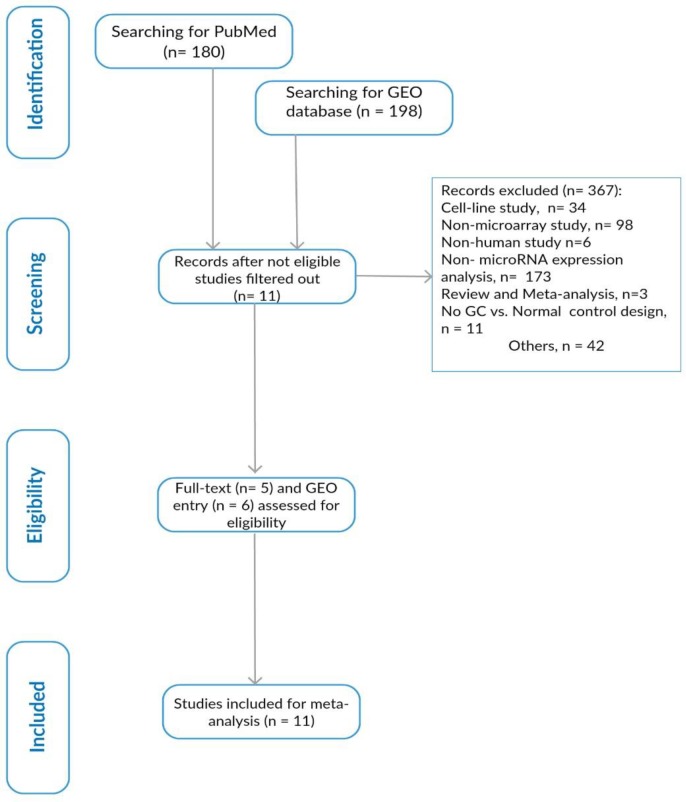
Through the database retrieval initially we found a total of 378 records and 11 studies met the inclusion criteria (Figure 1).
Fig. 1.
The diagram of searching strategy in this study
All studies were published between 2010 and 2016. Moreover, various microarray platforms were used in the included studies. The average number of hsa-miRNA probes was about 843 (ranging from 72 to 2578). A total of 1058 samples consisted of 596 tumors and 462 noncancerous samples were included. The majority of the studies were, however, designed to compare GC tumor tissue to normal adjacent tissue, in some cases (GSE63121, GSE30070, GSE23739 and PMID2776696) GC tumor tissue compared to unrelated normal tissue. Table 1 presents the main characteristics of the included studies.
Table 1.
Characteristics of included studies
| First author | Country | # Probes |
# Samples
(GC vs. normal) |
GSE/PMID | Assay type |
|---|---|---|---|---|---|
| Huang YS ( 16 ) | China | 1918 | 6 (3 vs. 3) | GSE78091 | miRCURY LNA microRNA Array, 7th generation |
| Shin W ( 17 ) | Korea | 833 | 10 (5 vs. 5) | GSE67354 | Homo sapiens miRNA Ca_Hu_MiRNome_v2 |
| Zhang X ( 18 ) | China | 847 | 30 (15 vs. 15 ) | GSE63121 | [miRNA-1_0] Affymetrix miRNA Array |
| Carvalho J ( 19 ) | Portugal | 709 | 47 (37 vs. 10) | GSE33743 | NCode™ Human miRNA microarray probe set V3 |
| Kim CH ( 20 ) | USA | 414 | 124 (90 vs. 34 ) | GSE30070 | Agilent-015868 Human miRNA Microarray |
| Xinhua Li ( 21 ) | China | 72 | 6 (3 vs.3) | 21874264 | miRCURY Array LNA microRNA chip (v.14.0) |
| Tetsuya U ( 22 ) | Japan | 237 | 673 (344 vs. 329) | 20022810 | Ohio State University custom microRNA microarray chip (OSU_CCC version 3.0) |
| Yoshiyuki T ( 23 ) | Japan | 470 | 27 (22 vs. 5) | 20215506 | G4470A Human MiRNA Microarray (Agilent Technologies) |
| Oh H ( 24 ) | Switzerland | 723 | 80 (40 vs. 40) | GSE23739 | Agilent-019118 Human miRNA Microarray 2.0 G4470B |
| Oleg T ( 8 ) | Germany | 476 | 6 (3 vs. 3) | 20726036 | Invitrogen NCode Multi-Species miRNA Microarray |
| Fehmida B ( 25 ) | Saudi Arabia | 2578 | 49 (34 vs. 15) | 27766962 | Affymetrix Genechip miRNA 4.0 |
GC meta-signature miRNA
We identified a meta-signature of nine upregulated miRNAs and seven downregulated miRNAs in GC samples compared to non-cancerous tissue according to robust rank score (Table 2). We found that only hsa-miR-25-3p dysregulation remained statistically significant after multiple test correction (adjusted P-value: 0.02).
Table 2.
List of miRNAs deregulated consistently in GC tissues
| Up-Regulated | Down-Regulated | ||
|---|---|---|---|
| microRNA | score | microRNA | score |
| hsa-miR-25-3p | 1E-05 | hsa-miR-375 | 9.84E-05 |
| hsa-miR-93-5p | 4.65E-05 | hsa-miR-148a-3p | 2.97E-04 |
| hsa-miR-21-5p | 5.17E-4 | hsa-miR-99a-5p | 0.01 |
| hsa-miR-106b-5p | 2.30E-3 | hsa-miR-146a-5p | 0.01 |
| hsa-miR-18a-5p | 5.99E-3 | hsa-miR-29c-3p | 0.02 |
| hsa-miR-19a-3p | 0.03 | hsa-miR-519e-3p | 0.02 |
| hsa-miR-17-5p | 0.03 | hsa-miR-378a-3p | 0.03 |
| hsa-miR-20a-5p | 0.04 | ||
| hsa-miR-92a-3p | 0.04 | ||
The bold interface indicates miRNA that showed significant dysregulation even after multiple test correction.
Sensitivity analysis
The sensitivity analysis was performed by leave-one-out method. For down regulation scenario no study was found to have significant effect on the obtained P-values. However, omitting six studies (8, 19, 20, 23-25) showed significant influence on the obtained P-value.
GC meta-signature miRNA target genes and enrichment analysis
The target genes for GC meta-signature miRNAs were extracted as for miR-21, 824 targets (ST:89, WT:459, PT:276), for miR-25, 1238 targets (ST:21, WT:387, PT:830), for miR-93, 1968 targets (ST:15, WT:1100, PT:853) and for miR-375, 672 targets (ST:25, WT:415, PT:232). Furthermore, to obtain insights into the biological function of GC-miRNA meta-signature, enrichment analyses were carried out using target genes. Enriched KEGG pathways for GC meta-signature miRNAs target genes were most frequently associated with cell signaling (HIF-1 signaling pathway, FoxO signaling pathway, sphingolipid signaling pathway, PI3K-Akt signaling pathway), cell mobility and differentiation (focal adhesion, signaling pathways regulating pluripotency of stem cells, axon guidance) and tumorigenesis (pathways in cancer, colorectal cancer, prostate cancer, small cell lung cancer, transcriptional misregulation in cancer, viral carcinogenesis) as well as cancer cell metabolism (central carbon metabolism, choline metabolism, proteoglycans) (Figure 2). The most enriched GO molecular processes regulated by the GC meta-signature miRNA target genes include protein binding and DNA binding process (Figure 3).
Discussion
In the present study, we used the robust rank aggregation approach to perform a meta-analysis on 596 GC samples and 462 non tumoral samples from 11 independent profiling experiments. Despite the fact that the preferred approach for gene expression meta-analysis promises analysis of raw expression datasets, these rigorous methods are often not possible due to the raw data unavailability. The technological platforms employed in any specific study and alterations in the number of miRNAs recognized at the moment would make the applicable integration of raw datasets too complicated. Additionally, the relatively small sample size and microarray data noisiness have resulted in an inconsistency of proper conclusions. To overcome such limitations, a meta-analysis based on robust rank aggregation approach was applied. In total, after Bonferroni correction method, used to control the false positive rate and to make the results more reliable, we identified four unique meta-signature miRNAs which were significantly deregulated in GC. Our findings indicated hsa-miR-21-5p, hsa-miR-93-5p, hsa-miR-25-3p and hsa-miR-375 as GC meta-signature microRNAs, of which the first three displayed up-regulation and the last one showed down-regulation. Also, sensitivity analysis revealed that the obtained scores for hsa-miR-21-5p, hsa-miR-93-5p, hsa-miR-25-3p and hsa-miR-375 are robust and credible as sequential leaving of individual study did not affect significant levels dramatically. Moreover, we found extra 16 upregulated and 6 down regulated miRNAs that showed differential expression in primary analysis (P-value < 0.05), however correction for multiple testing turned their statistics to insignificant levels (Table 2).
Due to the significant aberrant expression of miRNAs found in cancer and, moreover, lack of complex transcriptional and translational alterations in comparison with mRNAs and proteins, the application of miRNAs as novel biomarkers for cancer seems to have a substantial potential. The probable usefulness of miRNA-expression profiles, as the biomarkers for cancer diagnosis and prognosis, has been examined in numerous studies, based on tissue-specific deregulation pattern of miRNA expression (26, 27).
hsa-miR-21-5p is the most upregulated miRNA with the highest score in our study. This is a broadly studied miRNA in tissue which is aberrantly expressed in most cancers, highlighting its characteristic potential as a cancer biomarker for detection and prediction (28).
To understand how miR-21 expression is involved in different types of cancer, multiple functional studies have been implanted, predicting and validating a number of miR-21 target genes. Notably, most of these targets are tumor suppressor genes, including phosphatase and tensin homolog (PTEN), tropomyosin 1 (TPM1) and reversion-inducing cysteine-rich protein with Kazal motifs (RECK) (29-31). Interestingly, a large-scale miRNA analysis performed on 540 samples in six particular types of solid tumors revealed that miR-21 is the only miRNA upregulated in all cancer classes (32), such as breast (32), ovarian (32), colorectal (33), prostate (34), pancreatic (35), lung (36) thyroid (37) and glioma (38, 39).
Among the four resultant miRNAs of this study, miR-93 had the highest score for the involvement of pathways in cancer, as followed by miR-21, miR-375 and miR-25, respectively. This is consistent with Kong et al.’s report that miR-93, via upregulation, displays a pivotal role in the cancer development (40). In line with this, our results showed that "pathways in cancer (KEGG PATHWAY: map05200)" is significantly enriched by hsa-miR-21-5p target genes. In addition, the same is true for the other two upregulated miRNAs: hsa-miR-93-5p and hsa-miR-25-3p (41-44). On the other hand, the significant reduced expression of has-miR-375 target genes was attributed to their engagement with several carcinogen signal pathways such as TP53, WNT, MAPK and vascular endothelial growth factor (45).
Moreover, as shown in Figure 2, the GC meta-signature miRNAs target genes were enriched in cell signaling pathways, such as HIF-1, FoxO, sphingolipid and PI3K-Akt Aberrations in such signaling pathways and their contribution to malignancy development are discussed thoroughly in the literature (46-49). From molecular function enrichment analysis, it is indicates that these meta-signature microRNAs regulate cancer cell behavior. Through modulating cell mobility, cell fate determination and cancer cell metabolism modulation, development of malignancy phenotype would be tuned.
Through our analysis of target genes and molecular function GO, we found GC meta-signature miRNAs target genes are involved in protein and DNA binding processes (Figure 2). To date, it becomes clear that miRNA target genes mediate their essential signals by binding to different classes of proteins, such as phosphoserine- containing proteins, and DNA. So, in this way miRNAs control the malignant phenotypes of cancers including GC cells (50). Further, miRNAs could participate in epigenetic cell regulation by binding to methyl- CpG- binding proteins, and modulate their interaction with DNA (50, 51). Additionally, McLean et al. (52) mentioned that not only genetic factors are influenced by these binding processes, but also Helicobacter pylori infection, diet and other GC risk factors can interplay with them. In an interesting manner, all four studied miRNAs showed, almost, the same contribution and score for protein binding process as the first rank in GO enrichment analysis.
In another meta-analysis study, Wang et al. (53) extracted studies using quantitative RT-PCR and microarray, and gathered the miRNA information of different specimen types. Regardless of distinct search strategy, our findings are partly in agreement with their results. However, due to tissue heterogeneity and including studies with various techniques in meta-analysis, they found a wide range of miRNAs associated with GC risk.
Although here we showed that miRNAs might be involved in promoting GC progression by targeting some key genes within the important pathways of cancer regulation, there is still a lot more to know about the interpretation of miRNA impact on GC. Future work should keep continuous concentration on the critical mechanisms by which miRNAs are regulating occurrence, progression and, eventually, metastasis of GC. Studies having the same platform and larger sample size could shed light on our current knowledge of this area.
In conclusion, we strongly suggest that hsa-miR-21-5p, hsa-miR-93-5p, hsa-miR-25-3p and hsa-miR-375 are essential regulatory drivers in the carcinogenic process, which would be an appropriate target for GC diagnosis and therapy.
Acknowledgments
The present article was financially supported by “Research Department of the School of Medicine Shahid Beheshti University of Medical Sciences” (Grant No 11319)
Conflict of interest
The authors have read the journal’s policy on conflicts of interest and disclose no conflicts.
References
- 1.Ang TL, Fock KM. Clinical epidemiology of gastric cancer. Singapore Med J. 2014;55:621–8. doi: 10.11622/smedj.2014174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hohenberger P, Gretschel S. Gastric cancer. Lancet. 2003;362:305–15. doi: 10.1016/s0140-6736(03)13975-x. [DOI] [PubMed] [Google Scholar]
- 3.Di Leva G, Croce CM. miRNA profiling of cancer. Curr Opin Genet Dev. 2013;23:3–11. doi: 10.1016/j.gde.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baffa R, Fassan M, Volinia S, et al. MicroRNA expression profiling of human metastatic cancers identifies cancer gene targets. J Pathol. 2009;219:214–21. doi: 10.1002/path.2586. [DOI] [PubMed] [Google Scholar]
- 6.Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct Target Ther. 2016;1:15004. doi: 10.1038/sigtrans.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Z, Yu X, Wang Y, et al. By downregulating TIAM1 expression, microRNA-329 suppresses gastric cancer invasion and growth. Oncotarget. 2015;6:17559–69. doi: 10.18632/oncotarget.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tchernitsa O, Kasajima A, Schafer R, et al. Systematic evaluation of the miRNA-ome and its downstream effects on mRNA expression identifies gastric cancer progression. J Pathol. 2010;222:310–9. doi: 10.1002/path.2759. [DOI] [PubMed] [Google Scholar]
- 9.Li Z, Lei H, Luo M, et al. DNA methylation downregulated mir-10b acts as a tumor suppressor in gastric cancer. Gastric Cancer. 2015;18:43–54. doi: 10.1007/s10120-014-0340-8. [DOI] [PubMed] [Google Scholar]
- 10.Reid G, Kirschner MB, van Zandwijk N. Circulating microRNAs: Association with disease and potential use as biomarkers. Crit Rev Oncol Hematol. 2011;80:193–208. doi: 10.1016/j.critrevonc.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 11.Tong F, Cao P, Yin Y, et al. MicroRNAs in gastric cancer: from benchtop to bedside. Dig Dis Sci. 2014;59:24–30. doi: 10.1007/s10620-013-2887-3. [DOI] [PubMed] [Google Scholar]
- 12.Griffiths-Jones S, Grocock RJ, van Dongen S, et al. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–4. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kolde R, Laur S, Adler P, et al. Robust rank aggregation for gene list integration and meta-analysis. Bioinformatics. 2012;28:573–80. doi: 10.1093/bioinformatics/btr709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palmieri V, Backes C, Ludwig N, et al. IMOTA: an interactive multi-omics tissue atlas for the analysis of human miRNA-target interactions. Nucleic Acids Res. 2018;46:D770–D5. doi: 10.1093/nar/gkx701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang YS, Jie N, Zou KJ, et al. Expression profile of circular RNAs in human gastric cancer tissues. Mol Med Rep. 2017;16:2469–76. doi: 10.3892/mmr.2017.6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.GAv N. miR-495 regulates multiple epigenetic modifiers in gastric cancer. 2015. Available from: https:// www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE67354.
- 18.Zhang X, Ni Z, Duan Z, et al. Overexpression of E2F mRNAs associated with gastric cancer progression identified by the transcription factor and miRNA co-regulatory network analysis. PLoS One. 2015;10:e0116979. doi: 10.1371/journal.pone.0116979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carvalho J, van Grieken NC, Pereira PM, et al. Lack of microRNA-101 causes E-cadherin functional deregulation through EZH2 up-regulation in intestinal gastric cancer. J Pathol. 2012;228:31–44. doi: 10.1002/path.4032. [DOI] [PubMed] [Google Scholar]
- 20.Kim CH, Kim HK, Rettig RL, et al. miRNA signature associated with outcome of gastric cancer patients following chemotherapy. BMC Med Genomics. 2011;4:79. doi: 10.1186/1755-8794-4-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li X, Luo F, Li Q, et al. Identification of new aberrantly expressed miRNAs in intestinal-type gastric cancer and its clinical significance. Oncol Rep. 2011;26:1431–9. doi: 10.3892/or.2011.1437. [DOI] [PubMed] [Google Scholar]
- 22.Ueda T, Volinia S, Okumura H, et al. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol. 2010;11:136–46. doi: 10.1016/S1470-2045(09)70343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsukamoto Y, Nakada C, Noguchi T, et al. MicroRNA-375 is downregulated in gastric carcinomas and regulates cell survival by targeting PDK1 and 14-3-3zeta. Cancer Res. 2010;70:2339–49. doi: 10.1158/0008-5472.CAN-09-2777. [DOI] [PubMed] [Google Scholar]
- 24.Oh HK, Tan AL, Das K, et al. Genomic loss of miR-486 regulates tumor progression and the OLFM4 antiapoptotic factor in gastric cancer. Clin Cancer Res. 2011;17:2657–67. doi: 10.1158/1078-0432.CCR-10-3152. [DOI] [PubMed] [Google Scholar]
- 25.Bibi F, Naseer MI, Alvi SA, et al. microRNA analysis of gastric cancer patients from Saudi Arabian population. BMC Genomics. 2016;17:751. doi: 10.1186/s12864-016-3090-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12:847–65. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gigek CO, Chen ES, Calcagno DQ, et al. Epigenetic mechanisms in gastric cancer. Epigenomics. 2012;4:279–94. doi: 10.2217/epi.12.22. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y, Gao X, Wei F, et al. Diagnostic and prognostic value of circulating miR-21 for cancer: a systematic review and meta-analysis. Gene. 2014;533:389–97. doi: 10.1016/j.gene.2013.09.038. [DOI] [PubMed] [Google Scholar]
- 29.Zhang JG, Wang JJ, Zhao F, et al. MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC) Clin Chim Acta. 2010;411:846–52. doi: 10.1016/j.cca.2010.02.074. [DOI] [PubMed] [Google Scholar]
- 30.Gabriely G, Wurdinger T, Kesari S, et al. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol. 2008;28:5369–80. doi: 10.1128/MCB.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu S, Si ML, Wu H, et al. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) J Biol Chem. 2007;282:14328–36. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 32.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X, Xin S, He Z, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor PDCD4 and promotes cell transformation, proliferation, and metastasis in renal cell carcinoma. Cell Physiol Biochem. 2014;33:1631–42. doi: 10.1159/000362946. [DOI] [PubMed] [Google Scholar]
- 34.Folini M, Gandellini P, Longoni N, et al. miR-21: an oncomir on strike in prostate cancer. Mol Cancer. 2010;9:12. doi: 10.1186/1476-4598-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dillhoff M, Liu J, Frankel W, et al. MicroRNA-21 is overexpressed in pancreatic cancer and a potential predictor of survival. J Gastrointest Surg. 2008;12:2171–6. doi: 10.1007/s11605-008-0584-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Markou A, Zavridou M, Lianidou ES. miRNA-21 as a novel therapeutic target in lung cancer. Lung Cancer (Auckl) 2016;7:19–27. doi: 10.2147/LCTT.S60341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sondermann A, Andreghetto FM, Moulatlet AC, et al. MiR-9 and miR-21 as prognostic biomarkers for recurrence in papillary thyroid cancer. Clin Exp Metastasis. 2015;32:521–30. doi: 10.1007/s10585-015-9724-3. [DOI] [PubMed] [Google Scholar]
- 38.Zhou X, Zhang J, Jia Q, et al. Reduction of miR-21 induces glioma cell apoptosis via activating caspase 9 and 3. Oncol Rep. 2010;24:195–201. doi: 10.3892/or_00000846. [DOI] [PubMed] [Google Scholar]
- 39.Quintavalle C, Donnarumma E, Iaboni M, et al. Effect of miR-21 and miR-30b/c on TRAIL-induced apoptosis in glioma cells. Oncogene. 2013;32:4001–8. doi: 10.1038/onc.2012.410. [DOI] [PubMed] [Google Scholar]
- 40.Kong YW, Ferland-McCollough D, Jackson TJ, et al. microRNAs in cancer management. Lancet Oncol. 2012;13:e249–58. doi: 10.1016/S1470-2045(12)70073-6. [DOI] [PubMed] [Google Scholar]
- 41.Razumilava N, Bronk SF, Smoot RL, et al. miR-25 targets TNF-related apoptosis inducing ligand (TRAIL) death receptor-4 and promotes apoptosis resistance in cholangiocarcinoma. Hepatology. 2012;55:465–75. doi: 10.1002/hep.24698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lagana A, Russo F, Sismeiro C, et al. Variability in the incidence of miRNAs and genes in fragile sites and the role of repeats and CpG islands in the distribution of genetic material. PLoS One. 2010;5:e11166. doi: 10.1371/journal.pone.0011166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meng X, Joosse SA, Muller V, et al. Diagnostic and prognostic potential of serum miR-7, miR-16, miR-25, miR-93, miR-182, miR-376a and miR-429 in ovarian cancer patients. Br J Cancer. 2015;113:1358–66. doi: 10.1038/bjc.2015.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Du L, Schageman JJ, Subauste MC, et al. miR-93, miR-98, and miR-197 regulate expression of tumor suppressor gene FUS1. Mol Cancer Res. 2009;7:1234–43. doi: 10.1158/1541-7786.MCR-08-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X, Yan Z, Zhang J, et al. Combination of hsa-miR-375 and hsa-miR-142-5p as a predictor for recurrence risk in gastric cancer patients following surgical resection. Ann Oncol. 2011;22:2257–66. doi: 10.1093/annonc/mdq758. [DOI] [PubMed] [Google Scholar]
- 46.Farhan M, Wang H, Gaur U, et al. FOXO Signaling Pathways as Therapeutic Targets in Cancer. Int J Biol Sci. 2017;13:815–27. doi: 10.7150/ijbs.20052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martini M, De Santis MC, Braccini L, et al. PI3K/AKT signaling pathway and cancer: an updated review. Ann Med. 2014;46:372–83. doi: 10.3109/07853890.2014.912836. [DOI] [PubMed] [Google Scholar]
- 48.Maxwell PH, Pugh CW, Ratcliffe PJ. Activation of the HIF pathway in cancer. Curr Opin Genet Dev. 2001;11:293–9. doi: 10.1016/s0959-437x(00)00193-3. [DOI] [PubMed] [Google Scholar]
- 49.Ryland LK, Fox TE, Liu X, et al. Dysregulation of sphingolipid metabolism in cancer. Cancer Biol Ther. 2011;11:138–49. doi: 10.4161/cbt.11.2.14624. [DOI] [PubMed] [Google Scholar]
- 50.Wu WK, Lee CW, Cho CH, et al. MicroRNA dysregulation in gastric cancer: a new player enters the game. Oncogene. 2010;29:5761–71. doi: 10.1038/onc.2010.352. [DOI] [PubMed] [Google Scholar]
- 51.Wada R, Akiyama Y, Hashimoto Y, et al. miR-212 is downregulated and suppresses methyl-CpG-binding protein MeCP2 in human gastric cancer. Int J Cancer. 2010;127:1106–14. doi: 10.1002/ijc.25126. [DOI] [PubMed] [Google Scholar]
- 52.McLean MH, El-Omar EM. Genetics of gastric cancer. Nat Rev Gastroenterol Hepatol. 2014;11:664–74. doi: 10.1038/nrgastro.2014.143. [DOI] [PubMed] [Google Scholar]
- 53.Wang QX, Zhu YQ, Zhang H, et al. Altered MiRNA expression in gastric cancer: a systematic review and meta-analysis. Cell Physiol Biochem. 2015;35:933–44. doi: 10.1159/000369750. [DOI] [PubMed] [Google Scholar]