Key Points
Question
What cellular processes are dysregulated in cerebral organoids generated from stem cells with patient-specific genetic backgrounds in schizophrenia?
Findings
In this case-control study, cerebral organoids from patients with schizophrenia showed differences in the pattern of expression of genes involved in synaptic biology, neurodevelopment, and immune response as well as genes involved in mitochondrial function and modulation of excitation/inhibition balance. Cerebral organoids showed specific deficits in mitochondrial physiology and a diminished response to stimulation and depolarization.
Meaning
Patient-specific 3-dimensional brain organoids may reveal differences in specific biological pathways in schizophrenia.
Abstract
Importance
Three-dimensional cerebral organoids generated from patient-derived induced pluripotent stem cells (iPSCs) may be used to interrogate cellular-molecular underpinnings of schizophrenia.
Objective
To determine transcriptomic profiles and functional characteristics of cerebral organoids from patients with schizophrenia using gene expression studies, complemented with investigations of mitochondrial function through measurement of real-time oxygen consumption rate, and functional studies of neuronal firing with microelectrode arrays.
Design, Setting, and Participants
This case-control study was conducted at Massachusetts General Hospital between 2017 and 2019. Transcriptomic profiling of iPSC-derived cerebral organoids from 8 patients with schizophrenia and 8 healthy control individuals was undertaken to identify cellular pathways that are aberrant in schizophrenia. Induced pluripotent stem cells and cerebral organoids were generated from patients who had been diagnosed as having schizophrenia and from heathy control individuals.
Main Outcomes and Measures
Transcriptomic analysis of iPSC-derived cerebral organoids from patients with schizophrenia show differences in expression of genes involved in synaptic biology and neurodevelopment and are enriched for genes implicated in schizophrenia genome-wide association studies (GWAS).
Results
The study included iPSC lines generated from 11 male and 5 female white participants, with a mean age of 38.8 years. RNA sequencing data from iPSC-derived cerebral organoids in schizophrenia showed differential expression of genes involved in synapses, in nervous system development, and in antigen processing. The differentially expressed genes were enriched for genes implicated in schizophrenia, with 23% of GWAS genes showing differential expression in schizophrenia and control organoids: 10 GWAS genes were upregulated in schizophrenia organoids while 15 GWAS genes were downregulated. Analysis of the gene expression profiles suggested dysregulation of genes involved in mitochondrial function and those involved in modulation of excitatory and inhibitory pathways. Studies of mitochondrial respiration showed lower basal consumption rate, adenosine triphosphate production, proton leak, and nonmitochondrial oxygen consumption in schizophrenia cerebral organoids, without any differences in the extracellular acidification rate. Microelectrode array studies of cerebral organoids showed no differences in baseline electrical activity in schizophrenia but revealed a diminished response to stimulation and depolarization.
Conclusions and Relevance
Investigations of patient-derived cerebral organoids in schizophrenia revealed gene expression patterns suggesting dysregulation of a number of pathways in schizophrenia, delineated differences in mitochondrial function, and showed deficits in response to stimulation and depolarization in schizophrenia.
This study investigates transcriptomic profiles and functional characteristics of cerebral organoids from patients with schizophrenia.
Introduction
Cellular reprogramming methods enable generation of induced pluripotent stem cells (iPSCs) from human somatic cells.1 Induced pluripotent stem cells have the potential to generate any cell or tissue type, including neuronal cells and tissue.2 Induced pluripotent stem cells from patients provide a convenient way to study neuronal cells with disease-specific genetic backgrounds. Most studies to date of iPSC-derived neurons in schizophrenia have used 2-dimensional neuronal cultures.3,4,5,6,7,8 Technical advances provide methods for generation of iPSC-derived 3-dimensional brain organoids,9,10 self-organizing structures that mirror human cortical development and give rise to different cell types and interconnections found in the human brain.11,12,13
In this study, iPSCs of 8 patients with schizophrenia and 8 healthy control individuals were used to grow cerebral organoids. Transcriptomic studies were carried out to identify disease-specific differences in the cerebral organoids. RNA sequencing (RNA-seq) results showed aberrant gene expression in pathways involved in synaptic biology, nervous system development, immune response, and mitochondrial function and indicated an imbalance in excitatory and inhibitory pathways. Mitochondrial function was further studied through measurement of oxygen consumption. Functional studies of the cerebral organoids were undertaken using microelectrode arrays, which showed that schizophrenia organoids had basal electrical firing patterns similar to control organoids, but they had a diminished response to electrical stimulation and to depolarization. These results show the power and utility of interrogating schizophrenia disease biology using 3-dimensional cellular models generated from patient iPSCs.14,15
Methods
Generation of 3-Dimensional Cerebral Organoids
Fibroblasts were collected with approval from McLean Hospital and Massachusetts General Hospital institutional review boards. Written informed consent was obtained from study participants. Demographic and clinical descriptions are shown in eTable 1 in Supplement 1. Induced pluripotent stem cells were reprogrammed through induction with modified mRNA or with transient transfection with retroviruses (eFigure 2A in Supplement 1).16,17,18 The iPSCs were differentiated along the telencephalic lineage to generate cerebral organoids patterned after the dorsal forebrain (eFigure 1A-C in Supplement 1).13 The iPSCs were plated at high density on U-bottom plates to form embryoid bodies (EBs) and maintained in EB formation media for 5 days, with media change every 2 days. Using the STEMdiff Cerebral Organoid Kit (catalog 08570; STEMCELL Technologies) and the STEMdiff Cerebral Organoid Maturation Kit (catalog 08571; STEMCELL Technologies), EBs were resuspended in induction media for 2 days and then embedded in Matrigel on day 7 and maintained on expansion media. On day 10, the media were switched to expansion media, which was changed every 3 to 4 days. On day 30, brain-derived neurotrophic factor was added to the media, with media changed every 3 to 4 days.
Sample Preparation for Immunohistochemistry
For the optimal cutting temperature method, organoids were fixed in paraformaldehyde, 4%, placed in sucrose, 30%, overnight, embedded in molds in Tissue-Plus OCT Compound (Fisher), placed in dry ice until completely frozen, and sectioned at 20-μm thickness on a cryostat. For the paraffin method, samples were fixed in paraformaldehyde, 4%, dehydrated in a series of ethanol and xylene washes, embedded in paraffin, and sectioned at 20-μm thickness on a microtome.
Immunohistochemistry and Image Acquisition
Slides were stained with primary antibodies diluted in phosphate-buffered saline overnight at 4 °C. Secondary antibodies diluted in phosphate-buffered saline were applied for 1 hour at room temperature and put on a coverslip using Vectashield Hardset Mounting Medium. Antibodies are listed in eTable 5 in Supplement 1. Organoids sections were imaged at 20 × objective on the PerkinElmer Opera Phoenix High-Content Screening System.
Total RNA Sequencing and Analysis
RNA sequencing libraries were constructed by using Illumina RiboZero TruSeq Stranded Total RNA Library Prep Kit (Illumina) and sequenced on the Illumina NovaSeq 6000 platform in the 100-nt, paired-end configuration. Total RNA-seq was performed for organoids from 8 patients with schizophrenia and 8 control individuals. From each sample, we obtained a mean of 60 million reads. For gene expression analyses, trimmed reads with Cutadapt were aligned to the reference genome (hg38 UCSC assembly) using TopHat, version 2.0.14, and Bowtie, version 2.10 (Johns Hopkins University), with default parameters and RefSeq annotation (genome-build GRCh38.p9).19 The distribution of alignments was analyzed using Cufflinks, version 2.2.1 (Trapnell Lab), and fragments per kilobase of exon model per million reads mapped values were quantile normalized. Differential expression testing was performed using Cuffdiff, version 2.2.1 (Trapnell Lab; eAppendix in Supplement 2).20,21 The false discovery rate was 0.05, and sex differences were not considered. Quantitative polymerase chain reaction validation performed on selected genes are presented in eFigure 5 in Supplement 1.
Gene Ontology Analysis and Gene Set Enrichment Analysis
Upregulated and downregulated genes were subjected to gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis using the Functional Enrichment Analysis unit of Hypergeometric Optimization of Motif EnRichment, version 3 (University of California, San Diego), for biological process, cellular component/protein localization, and molecular function.22 Metabolic processes were analyzed using MetaCore+MetaDrug, version 19.1, build 69600 (Clarivate Analytics). The list shown in the figures reached significance (P < .05). Default parameters were used to perform gene set enrichment analysis (GSEA) using the GSEA software for all expressed genes with fragments per kilobase of exon model per million reads mapped values calculated by Cufflinks against following data sets, version 6.2: Hallmark, REACTOME, and KEGG.23
Generation of Protein-Protein Interaction Network
Protein-protein interaction (PPI) networks for differentially expressed genes (DEGs) were generated using Search Tool for the Retrieval of Interacting Genes, version 11.0 (http://www.string-db.org/), and visualized and analyzed in Cytoscape, version 3.7.1 software (Cytoscape Consortium), which represents PPI networks as graphs with nodes illustrating proteins and edges depicting associated interactions.24 Network analysis was done using the Network Analyzer in Cytoscape, and protein nodes of the PPI network with connectivity degree of at least 10 were identified and represented as hub nodes. The following databases were used for network and GO analyses: schizophrenia, http://www.szdb.org/SZDB/score.php; bipolar disorder, https://www.med.unc.edu/pgc/pgc-workgroups/bipolar-disorder/; and autism spectrum disorder, https://gene.sfari.org/database/human-gene/.
Mitochondrial genes were downloaded from the Human MitoCarta2.0 website25 (https://www.broadinstitute.org/scientific-community/science/programs/metabolic-disease-program/publications/mitocarta/mitocarta-in-0). The data from the analyses have been deposited in National Center for Biotechnology Information’s Gene Expression Omnibus26 and are accessible through Gene Expression Omnibus Series accession number GSE133534 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?&acc=GSE133534).
Mitochondrial Respiration: Seahorse Mito Stress Test Assay
The Agilent Seahorse XF Cell Mito Stress Test (103015-100) assay was used to assess mitochondrial respiration in 9-month-old cerebral organoids, measuring the effect of the following perturbations recorded sequentially: 2mM oligomycin, 2mM carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone, 2mM rotenone, and 2mM antimycin and rotenone. For this assay, we used the Seahorse XFe96 Spheroid microplates, which were coated with 10 μg/mL of laminin (Sigma L2020). Organoids cultured for 9 months were cut into 3 sections and plated on the microplates for the experiments. Data were analyzed with Seahorse Wave desktop software, version 2.4 (Agilent).
Microelectrode Array Experiments
Six-month-old organoids were attached to microelectrode array (MEA) 24-well plates that contained 16 electrodes in each well (Med64; Presto) and had been coated with poly-L-ornithine and laminin. Organoids were cultured for 3 more months in MEA plates, media changed twice weekly, and electrical activity recorded when organoids were aged 9 months. After recording spontaneous activity, 10 electrical pulses at 0.8 V were administered and the electrical activity recorded for 1 minute after each stimulation. While 6-month-old organoids showed no electrical activity, 9-month-old organoids showed regular and consistent activity. One millimolar tetrodotoxin was added to validate neuronal electrical activity (eFigure 2C in Supplement 1). Data were analyzed using the MEA Symphony software (Med64).
Results
Generation and Characterization of Cerebral Organoids
The iPSCs were reprogrammed from fibroblasts using standard protocols (eFigure 2A in Supplement 1).16,17 The iPSCs (eTable 1 in Supplement 1) were differentiated to generate cerebral organoids patterned after the dorsal forebrain (eFigure 1 in Supplement 1).13 Organoids grew continuously, and the mean size was 800 μm to 1 mm in diameter after 30 days, 2 mm to 3 mm at 4 months, and 4 mm to 5 mm at 7 months (eFigure 1A in Supplement 1). Marker analysis showed no gross differences between schizophrenia and control organoids (eFigure 1B and C in Supplement 1) and all organoids expressed the following neuronal markers: MAP-2, Ctip2, Satb2, Tbr1, Cult1, GFAP, Iba1, glutamine synthetase, Nkx2.1, Tuj1, Pax-6, Homer1, Bassoon, Tbr2, and Lhx6 (eFigure 1B, 1C, and 4A in Supplement 1).
Transcriptomic Profiles of Cerebral Organoids
Total RNA-seq was performed on organoids aged 6 months. Principal component analysis revealed a distinct separation between schizophrenia and control organoids (eFigure 4C in Supplement 1). Differentially expressed genes showed a distinct difference in the pattern of gene expression for both coding and noncoding genes between schizophrenia and control organoids (eFigure 3A and B in Supplement 1). Differentially expressed genes were categorized into upregulated and downregulated genes, and the top 25 hits were rank ordered according to significance (P value) (eFigure 3C-E in Supplement 1). The most significant GO processes that were downregulated in schizophrenia were nervous system development, neurogenesis, and generation of neurons, while the most upregulated GO processes were antigen processing and presentation of exogenous peptide antigen via major histocompatibility complex class I, extracellular matrix organization, and cellular response to chemical stimulus (eFigure 3C in Supplement 1). Gene ontology localization analysis showed significant downregulation in the neuron, synapse, and neuronal projections categories in schizophrenia while showing upregulation in categories of the endoplasmic reticulum (ER), ER to Golgi transport, and extracellular regions (eFigure 3D in Supplement 1). Gene ontology molecular process analysis revealed downregulation of cytoskeletal protein binding and upregulation of peptide antigen binding (eFigure 3E in Supplement 1). The upregulation of genes involved in peptide antigen processing and presentation via MHC class I are interesting because the MHC locus has the strongest genetic association for schizophrenia and is suggestive of aberrant synaptic pruning in schizophrenia.27,28,29
Gene Set Enrichment Analysis enables one to determine whether a set of genes can separate 2 biological categories in a statistically significant manner.23 Gene set enrichment analysis was performed with default parameters for all expressed genes with ragments per kilobase of exon model per million reads mapped values calculated by Cufflinks against following the data sets, version 6.2: Hallmark, REACTOME, and KEGG (eFigure 3F-J in Supplement 1). The most significant finding was in the γ-aminobutyric acid system, where a number of pivotal genes were downregulated, including GAD1, GAD2, and GABRB1. Deficits in γ-aminobutyric acid systems are hypothesized to be involved in schizophrenia.30,31,32 The other pathways showed differences in α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor trafficking, unblocking of N-methyl-d-aspartate (NMDA) receptor glutamate binding and activation, potassium channels, and transmission across chemical synapses. These results indicate pivotal roles for synapse biology in schizophrenia.33,34
Analysis of PPI Network
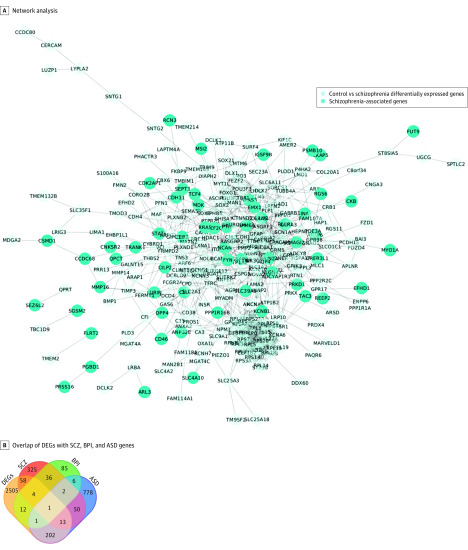
A PPI network was constructed for the DEGs using the Search Tool for the Retrieval of Interacting Genes, a database of known and predicted PPIs.24 The PPI results show that gene expression differences mirror results from schizophrenia GWAS (Table and Figure 1). Seventy-six genes overlapped between DEGs and schizophrenia-associated genes (Figure 1B). The enriched pathway for GO localization analysis was the synapse (eTable 2 in Supplement 1), consistent with enrichment for synaptic proteins in schizophrenia risk genes.35,36,37 The pathway significant for GO process category was nervous system development, while GO molecular function that was significant was phosphatase regulator activity. Twenty-three percent of genes that have genome-wide significance in schizophrenia38 were differentially regulated in schizophrenia and control organoids: 10 GWAS genes were upregulated in schizophrenia organoids while 15 GWAS genes were downregulated (Table).
Table. Schizophrenia GWAS Genes That Were Differentially Expressed in Schizophrenia Cerebral Organoids, Showing the Direction of Change Compared With Control Cerebral Organoids as Well as the Fold Change.
| Gene | Expression in Schizophrenia | log2(fold_change) | P Value |
|---|---|---|---|
| RGS6 | Downregulated | −2.8689 | <.001 |
| TAC3 | Downregulated | −2.39341 | <.001 |
| CNKSR2 | Downregulated | −2.39204 | <.001 |
| GRIN2A | Downregulated | −2.30172 | <.001 |
| CLU | Downregulated | −2.09784 | <.001 |
| MEF2C-AS1 | Downregulated | −1.99342 | .004 |
| GRIA1 | Downregulated | −1.92157 | <.001 |
| PTN | Downregulated | −1.90742 | <.001 |
| MMP16 | Downregulated | −1.23502 | <.001 |
| DOCK4 | Downregulated | −1.07099 | <.001 |
| PRKD1 | Downregulated | −0.9428 | .001 |
| NLGN4X | Downregulated | −0.934536 | .002 |
| TCF4 | Downregulated | −0.892158 | .001 |
| ZEB2 | Downregulated | −0.848958 | .001 |
| PJA2 | Downregulated | −0.701027 | .004 |
| CD46 | Upregulated | 0.723048 | .004 |
| MDK | Upregulated | 0.792983 | .002 |
| RRAS | Upregulated | 0.865922 | .005 |
| FURIN | Upregulated | 1.04331 | <.001 |
| STAT6 | Upregulated | 1.24988 | <.001 |
| PTGIS | Upregulated | 1.5472 | <.001 |
| DPP4 | Upregulated | 1.57011 | <.001 |
| MYO1A | Upregulated | 1.94253 | <.001 |
| TIE1 | Upregulated | 3.07346 | <.001 |
| CD34 | Upregulated | 3.3625 | <.001 |
Abbreviation: GWAS, genome-wide association studies.
Figure 1. Comparison of Differentially Expressed Genes (DEGs) in Schizophrenia (SCZ) Cerebral Organoids With Genes Associated With Neuropsychiatric Disorders.
A, Network analysis of DEGs with SCZ-associated genes. B, Overlap of DEGs with genes associated with SCZ, bipolar disorder (BPI), and autism spectrum disorder (ASD).
There is shared genetic susceptibility between neuropsychiatric disorders, reflected in polygenic overlap and shared neuropathology.39,40,41 The DEGs were compared with databases of autism SFARI genes and bipolar disorder–associated genes.42 Two hundred seventeen genes were shared between DEGs and autism-associated genes while 18 DEGs overlapped with associated genes (Figure 1B; eTable 2 in Supplement 1). Gene ontology localization analyses of DEGs shared with autism genes revealed genes localized to the lumenal side of ER membrane. Gene ontology process analysis showed interferon-γ signaling as the most significant category, while GO molecular function analysis showed the most significant category was peptide antigen binding category, which is associated with inflammation and immune signaling. This suggests that shared biology in schizophrenia and autism may be associated with immune signaling and ER stress, consistent with transcriptomic profiling of cortical tissue in schizophrenia and autism that showed shared dysregulation of immune response genes.43 The GO localization category most significant for overlap of DEGs with bipolar disorder–associated genes was integral component of synaptic membrane, while the most significant GO process was regulation of presynaptic membrane potential and the most significant GO molecular function category was pyruvate carboxylase activity. Previous postmortem results had reported differences in presynaptic proteins in schizophrenia and bipolar disorder.44 An annotated list of all genes in the GO categories are in the eAppendix in Supplement 2.
Mitochondrial Function in Cerebral Organoids
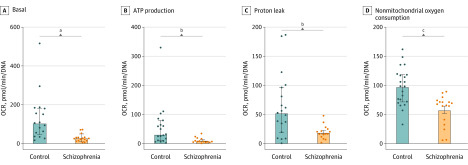
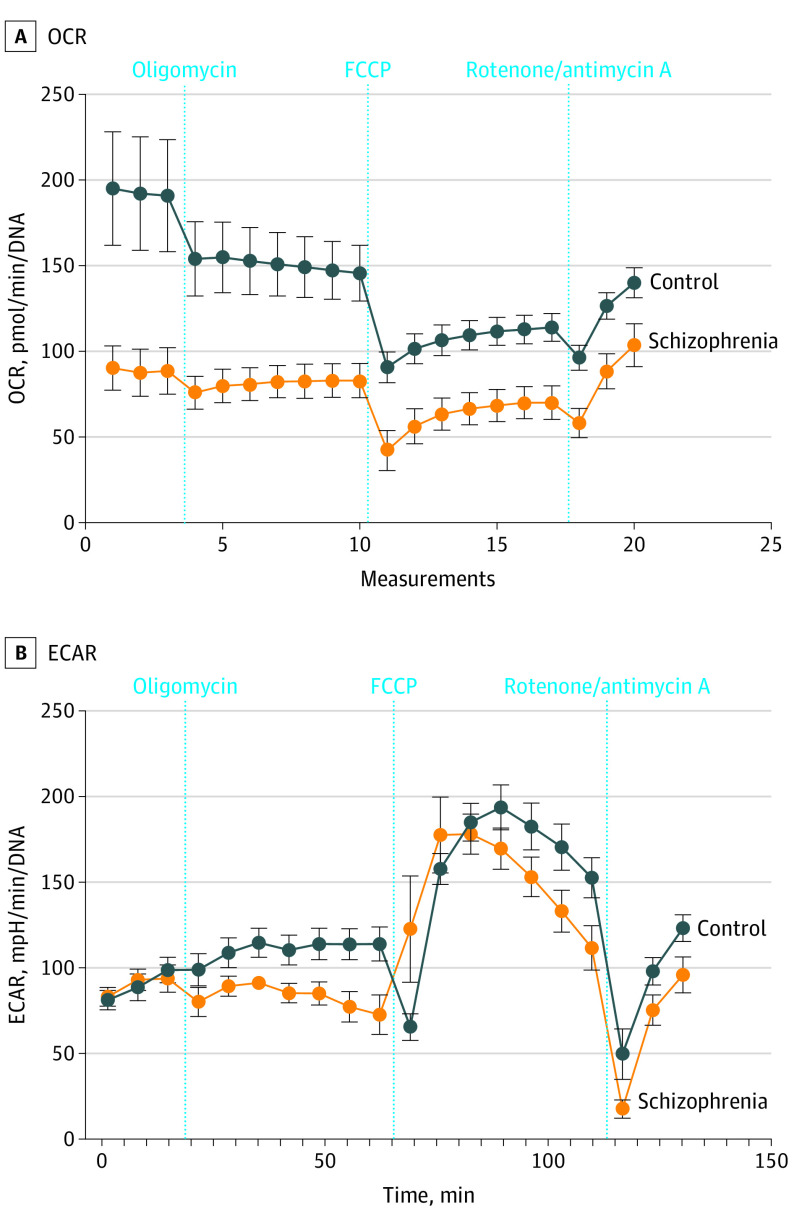
Gene ontology analyses of DEGs revealed downregulation of the phosphatidylserine and hexadecanoylcarnitine pathways (eFigure 7A in Supplement 1). Phosphatidylserine is imported into mitochondria for decarboxylation to form phosphatidylethanolamine via a domain of the ER mitochondria-associated membranes that transiently forms associations between the ER and mitochondria and regulates mitochondrial network morphology.45 Phosphatidylethanolamine is mainly found in white matter in the brain, where it is especially abundant in the inner mitochondrial membrane and required for activity of respiratory chain complexes.46 It has a role in preventing oxidative damage, and this pathway has been explored for potential therapeutic approaches in neurodegenerative disorders.47,48 We compared DEGs with genes in the human MitoCarta, version 2.0, inventory,25 and 39 mitochondrial genes were represented in DEGs (eFigure 7B in Supplement 1 and the eAppendix in Supplement 3). To examine the role of oxidative stress and mitochondrial dysfunction in schizophrenia,49 live-cell metabolic experiments were undertaken with a Seahorse Cell Mito Stress Test using 9-month-old organoids.50 Schizophrenia organoids had significantly lower basal oxygen consumption rate, adenosine triphosphate production, proton leak, and nonmitochondrial oxygen consumption when compared with control organoids (Figure 2A-D). There was significant difference in the oxygen consumption rate between schizophrenia and control organoids in the setting of the different perturbations but not in the extracellular acidification rate (Figure 3). Interestingly, a study in lymphoblastoid cell lines from patients with schizophrenia had previously demonstrated decreased oxygen consumption rates.51
Figure 2. Mitochondrial Function in Cerebral Organoids Through Seahorse Cell Mito Stress Test Assay Conducted With 9-Month-Old Cerebral Organoids.
Eight schizophrenia and 8 control cerebral organoids were studied. Oxygen consumption rate results shown for basal rate, adenosine triphosphate production, proton leak, and nonmitochondrial oxygen consumption rate as well as values for extracellular acidification rate. Data collected from 3 technical replicates in each condition and values are shown as median (IQR). Normality was tested via Kolmogorov-Smirnov, but because all data were not normally distributed, Mann-Whitney U test was performed. A, Basal: 2-tailed, sum of ranks in control column, 396; schizophrenia column, 165; Mann-Whitney U, 29. B, ATP production: 2-tailed, sum of ranks in control column, 409; schizophrenia column, 152; Mann-Whitney U, 47. C, Proton leak: 2-tailed, sum of ranks in control column, 412; schizophrenia column, 183; Mann-Whitney U, 63. D, Nonmitochondrial oxygen consumption rate: sum of ranks in control column, 564; schizophrenia column, 216; Mann-Whitney U, 63.
aP < 01.
bP = .001.
cP = .005.
Figure 3. Oxygen Consumption Rate (OCR) and Extracellular Acidification Rate (ECAR) in Cerebral Organoids.
A. Oxygen consumption rate. B. Extracellular acidification rate. Data shown for schizophrenia and control cerebral organoids in the setting of different perturbations.
Functional Studies of Cerebral Organoids with Microelectrode Arrays
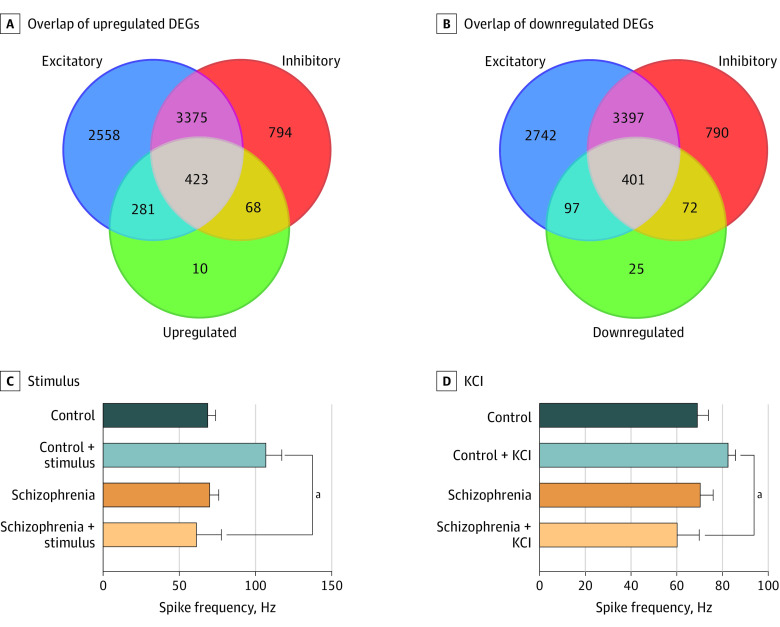
To investigate DEGs involved in synaptic transmission, a PPI network was built using a network from the neural subtype transcriptome52 and schizophrenia-associated genes (eFigure 4B in Supplement 1). The DEGs were divided into upregulated and downregulated genes (Figure 4A and B; eTables 3 and 4 in Supplement 1). Among upregulated DEGs, 281 genes were expressed in excitatory synapses and 68 in inhibitory synapses, while 423 were expressed in both (Figure 4A). Among downregulated DEGs, 97 were expressed in excitatory synapses and 72 were expressed in inhibitory synapses, while 401 were expressed in both (Figure 4B). Among the hub genes in the inhibitory module, 15 genes were downregulated in DEGs, including transcription factors DLX-1, DLX-2, and ARX required for inhibitory neuron formation, as well as GAD1 and ABAT needed for GABA synthesis (eFigure 3F in Supplement 1).53 These data suggest aberrations in the excitatory/inhibitory balance in schizophrenia that would affect synaptic transmission, functional connectivity, and signal processing.54,55,56,57
Figure 4. Functional Characterization of Cerebral Organoids.
A, Overlap of upregulated differentially expressed genes (DEGs) with genes expressed in excitatory and inhibitory neurons. B, Overlap of downregulated DEGs with genes expressed in excitatory and inhibitory neurons. C, Quantification of raster plot data collected before and after stimulus. D, Quantification of raster plot data collected with and without 30mM potassium chloride (KCl). Data collected from 4 control and 4 schizophrenia cerebral organoids, with 2 technical replicates in each condition. Values are shown as mean (SD). The data have a normal distribution confirmed via Kolmogorov-Smirnov. Unpaired t test with Welch correction: control stimulus vs schizophrenia stimulus, R2, 0.7985; 95% CI, −51.40 to −39.72; P < .001. Control KCl vs schizophrenia KCl, R2, 0.8430; 95% CI, −29.83 to −14.67; P < .001.
An MEA system was used to evaluate functional properties in organoids.58 There were no differences in spontaneous firing rate between schizophrenia and control organoids (Figure 4C; eFigure 8A and B in Supplement 1). When neuronal firing was measured with electrical stimulation at 0.8 V, control organoids, but not schizophrenia organoids, showed a significant increase in spike frequency (Figure 4C; eFigure 8A and B in the Supplement). When spike frequency was measured in setting of neuronal depolarization with 30mM potassium chloride, spike frequencies were again significantly increased in control organoids but not in schizophrenia organoids (Figure 4D; eFigure 8A and 8B in Supplement 1). These experiments indicate that schizophrenia cerebral organoids have similar baseline electrical activity as control organoids, but they exhibit a diminished response to electrical stimulation and potassium chloride depolarization.
Discussion
Patient-derived iPSCs provide new opportunities to develop ex vivo cellular models of psychiatric disorders.59,60 Induced pluripotent stem cells can be differentiated to neuronal cells relevant to disease biology.61,62,63,64 While most cellular models have focused on 2-dimensional neuronal cultures, new methodologic advances enable generation of 3-dimensional brain organoids.65,66 In this study, patient-derived iPSCs were used to generate cerebral organoids. Transcriptomic studies of these organoids point to processes that may be aberrant in schizophrenia, including synapse biology, nervous system development, immune response, and modulation of excitatory and inhibitory neurotransmission. While a transcriptomic study with iPSC-derived neural progenitor cells and neurons did not show enrichment at pathway or network level,3 another study showed DEGs were expressed in modules that had genes associated with risk variants for schizophrenia and other neurodevelopmental disorders.4 In a transcriptomic study of iPSC-derived neurons from patients with schizophrenia and 22q11.2 deletion, DEGs converged on pathways associated with CDC45 and PRODH.5
Studies with neuroimaging, postmortem brains, and patient-derived cells have implicated mitochondrial dysfunction in schizophrenia.67 Gene ontology analyses of our transcriptomic data point to differences in phosphatidylserine and hexadecanoylcarnitine pathways, which are hypothesized to play a role in the pathophysiology of schizophrenia.68,69,70 Many human studies have difficulty separating contribution of genetic factors from downstream effects of the disease process or confounding factors from medications and nonprescription drugs. The iPSC-based cellular models allow us to capture contributions from genetic backgrounds since reprogramming results in erasure of epigenetic effects.71 Hence, while information on medication treatment history is included in the eTable 1 in Supplement 1, such prior exposure to medications or to other environmental factors should not affect iPSC-based studies.
The transcriptomic results are consistent with hypotheses about alterations in excitatory/inhibitory balance in schizophrenia.54,55,56,57 Basal activity of schizophrenia organoids were not different compared with control organoids, but they did not show characteristic increase in spike frequency in response to electrical stimulation or depolarization. In rats, the NMDA-receptor antagonist MK-801 results in decreased burst firing, which was ameliorated by antipsychotic drugs.72 The decreased response to electrical stimulation and to depolarization in schizophrenia is interesting in the context of these animal studies.
Limitations
While iPSC-based ex vivo cellular models provide exciting new avenues to study psychiatric disorders, care needs to be taken to appreciate limitations of these approaches. Schizophrenia manifests fully in late adolescence and early adulthood while cerebral organoids investigated here are younger than 1 year. Gene expression and epigenomic studies indicate that cerebral organoids model fetal cortical development.73 Hence, aspects of the biology related to neurodevelopment may be amenable to investigation in these cellular models. Effects of environmental factors that impinge on the genetic background leading to disease onset are not included in this study. Studies with specific perturbations in conjunction with cellular models can be used to investigate gene-environment interactions that may have relevance to disease biology.74,75 We add a note of caution in the extrapolation of these data. While organoids represent a more physiologically relevant model, RNA-seq of organoids are not as clean compared with RNA-seq with homogeneous cell populations. We present this report as a proof-of-concept study that shows the feasibility of using novel approaches in stem cell–derived cerebral organoids to interrogate the cellular-molecular underpinnings of psychiatric disorders.
Conclusions
Cerebral organoids generated by differentiating iPSCs from schizophrenia patients show aberrant expression of genes involved in neurodevelopment and in synapse biology. Cerebral organoids from schizophrenia patients show similar baseline activity as cerebral organoids from healthy individuals but have a markedly diminished response to electrical stimulation and depolarization.
eFigure 1. Generation and characterization of cerebral organoids from human iPSCs
eFigure 2. iPSC characterization
eFigure 3. Gene ontology (GO) analysis of differentially expressed genes (DEGs) in schizophrenia cerebral organoids
eFigure 4. Cerebral organoid characterization
eFigure 5. RT-PCR validation of DEGs
eFigure 6. Synaptic staining and Western blots of cerebral organoids
eFigure 7. Mitochondrial DEGs in SCZ organoids
eFigure 8. Raster Plots from MEAs
eTable 1. Demographic and treatment information on subjects
eTable 2. GO analysis of DEGs that are associated specifically with SCZ, BPD or ASD.
eTable 3. Excitatory and inhibitory genes: upregulated DEGs
eTable 4. Excitatory and inhibitory genes: downregulated DEGs
eTable 5. Primary antibodies used
eTable 6. KaryoSTAT codes for each line
eAppendix.
eAppendix.
References
- 1.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861-872. doi: 10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]
- 2.Okita K, Yamanaka S. Induced pluripotent stem cells: opportunities and challenges. Philos Trans R Soc Lond B Biol Sci. 2011;366(1575):2198-2207. doi: 10.1098/rstb.2011.0016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffman GE, Hartley BJ, Flaherty E, et al. Transcriptional signatures of schizophrenia in hiPSC-derived NPCs and neurons are concordant with post-mortem adult brains. Nat Commun. 2017;8(1):2225. doi: 10.1038/s41467-017-02330-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roussos P, Guennewig B, Kaczorowski DC, Barry G, Brennand KJ. Activity-dependent changes in gene expression in schizophrenia human-induced pluripotent stem cell neurons. JAMA Psychiatry. 2016;73(11):1180-1188. doi: 10.1001/jamapsychiatry.2016.2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin M, Pedrosa E, Hrabovsky A, et al. Integrative transcriptome network analysis of iPSC-derived neurons from schizophrenia and schizoaffective disorder patients with 22q11.2 deletion. BMC Syst Biol. 2016;10(1):105. doi: 10.1186/s12918-016-0366-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moslem M, Olive J, Falk A. Stem cell models of schizophrenia, what have we learned and what is the potential? Schizophr Res. 2019;210:3-12. doi: 10.1016/j.schres.2018.12.023 [DOI] [PubMed] [Google Scholar]
- 7.Habela CW, Song H, Ming GL. Modeling synaptogenesis in schizophrenia and autism using human iPSC derived neurons. Mol Cell Neurosci. 2016;73:52-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Watmuff B, Berkovitch SS, Huang JH, Iaconelli J, Toffel S, Karmacharya R. Disease signatures for schizophrenia and bipolar disorder using patient-derived induced pluripotent stem cells. Mol Cell Neurosci. 2016;73:96-103. doi: 10.1016/j.mcn.2016.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Renner M, Lancaster MA, Bian S, et al. Self-organized developmental patterning and differentiation in cerebral organoids. EMBO J. 2017;36(10):1316-1329. doi: 10.15252/embj.201694700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelava I, Lancaster MA. Stem cell models of human brain development. Cell Stem Cell. 2016;18(6):736-748. doi: 10.1016/j.stem.2016.05.022 [DOI] [PubMed] [Google Scholar]
- 11.Qian X, Song H, Ming GL. Brain organoids: advances, applications and challenges. Development. 2019;146(8):dev166074. doi: 10.1242/dev.166074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yakoub AM. Cerebral organoids exhibit mature neurons and astrocytes and recapitulate electrophysiological activity of the human brain. Neural Regen Res. 2019;14(5):757-761. doi: 10.4103/1673-5374.249283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jabaudon D, Lancaster M. Exploring landscapes of brain morphogenesis with organoids. Development. 2018;145(22):dev172049. doi: 10.1242/dev.172049 [DOI] [PubMed] [Google Scholar]
- 14.Srikanth P, Lagomarsino VN, Muratore CR, et al. Shared effects of DISC1 disruption and elevated WNT signaling in human cerebral organoids. Transl Psychiatry. 2018;8(1):77. doi: 10.1038/s41398-018-0122-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stachowiak EK, Benson CA, Narla ST, et al. Cerebral organoids reveal early cortical maldevelopment in schizophrenia-computational anatomy and genomics, role of FGFR1. Transl Psychiatry. 2017;7(11):6. doi: 10.1038/s41398-017-0054-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Warren L, Wang J. Feeder-free reprogramming of human fibroblasts with messenger RNA. Curr Protoc Stem Cell Biol. 2013;27:6. [DOI] [PubMed] [Google Scholar]
- 17.Kathuria A, Lopez-Lengowski K, Watmuff B, McPhie D, Cohen BM, Karmacharya R. Synaptic deficits in iPSC-derived cortical interneurons in schizophrenia are mediated by NLGN2 and rescued by N-acetylcysteine. Transl Psychiatry. 2019;9(1):321. doi: 10.1038/s41398-019-0660-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sheridan SD, Theriault KM, Reis SA, et al. Epigenetic characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile X syndrome. PLoS One. 2011;6(10):e26203. doi: 10.1371/journal.pone.0026203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. 2011;17(1):10-12. doi: 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- 20.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trapnell C, Roberts A, Goff L, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562-578. doi: 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heinz S, Benner C, Spann N, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576-589. doi: 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545-15550. doi: 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607-D613. doi: 10.1093/nar/gky1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calvo SE, Clauser KR, Mootha VK. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2016;44(D1):D1251-D1257. doi: 10.1093/nar/gkv1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207-210. doi: 10.1093/nar/30.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sekar A, Bialas AR, de Rivera H, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium . Schizophrenia risk from complex variation of complement component 4. Nature. 2016;530(7589):177-183. doi: 10.1038/nature16549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mokhtari R, Lachman HM. The Major Histocompatibility Complex (MHC) in schizophrenia: a review. J Clin Cell Immunol. 2016;7(6):479. doi: 10.4172/2155-9899.1000479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sellgren CM, Gracias J, Watmuff B, et al. Increased synapse elimination by microglia in schizophrenia patient-derived models of synaptic pruning. Nat Neurosci. 2019;22(3):374-385. doi: 10.1038/s41593-018-0334-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akbarian S, Kim JJ, Potkin SG, et al. Gene expression for glutamic acid decarboxylase is reduced without loss of neurons in prefrontal cortex of schizophrenics. Arch Gen Psychiatry. 1995;52(4):258-266. doi: 10.1001/archpsyc.1995.03950160008002 [DOI] [PubMed] [Google Scholar]
- 31.Hashimoto T, Volk DW, Eggan SM, et al. Gene expression deficits in a subclass of GABA neurons in the prefrontal cortex of subjects with schizophrenia. J Neurosci. 2003;23(15):6315-6326. doi: 10.1523/JNEUROSCI.23-15-06315.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benes FM, Berretta S. GABAergic interneurons: implications for understanding schizophrenia and bipolar disorder. Neuropsychopharmacology. 2001;25(1):1-27. doi: 10.1016/S0893-133X(01)00225-1 [DOI] [PubMed] [Google Scholar]
- 33.Lima Caldeira G, Peça J, Carvalho AL. New insights on synaptic dysfunction in neuropsychiatric disorders. Curr Opin Neurobiol. 2019;57:62-70. doi: 10.1016/j.conb.2019.01.004 [DOI] [PubMed] [Google Scholar]
- 34.Pocklington AJ, O’Donovan M, Owen MJ. The synapse in schizophrenia. Eur J Neurosci. 2014;39(7):1059-1067. doi: 10.1111/ejn.12489 [DOI] [PubMed] [Google Scholar]
- 35.Schijven D, Kofink D, Tragante V, et al. Comprehensive pathway analyses of schizophrenia risk loci point to dysfunctional postsynaptic signaling. Schizophr Res. 2018;199:195-202. doi: 10.1016/j.schres.2018.03.032 [DOI] [PubMed] [Google Scholar]
- 36.Fromer M, Pocklington AJ, Kavanagh DH, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506(7487):179-184. doi: 10.1038/nature12929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Purcell SM, Moran JL, Fromer M, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506(7487):185-190. doi: 10.1038/nature12975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421-427. doi: 10.1038/nature13595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cross-Disorder Group of the Psychiatric Genomics Consortium . Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381(9875):1371-1379. doi: 10.1016/S0140-6736(12)62129-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gandal MJ, Haney JR, Parikshak NN, et al. ; CommonMind Consortium; PsychENCODE Consortium; iPSYCH-BROAD Working Group . Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science. 2018;359(6376):693-697. doi: 10.1126/science.aad6469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hulshoff Pol HE, van Baal GC, Schnack HG, et al. Overlapping and segregating structural brain abnormalities in twins with schizophrenia or bipolar disorder. Arch Gen Psychiatry. 2012;69(4):349-359. doi: 10.1001/archgenpsychiatry.2011.1615 [DOI] [PubMed] [Google Scholar]
- 42.Gandal MJ, Zhang P, Hadjimichael E, et al. ; PsychENCODE Consortium . Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science. 2018;362(6420):eaat8127. doi: 10.1126/science.aat8127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guan J, Cai JJ, Ji G, Sham PC. Commonality in dysregulated expression of gene sets in cortical brains of individuals with autism, schizophrenia, and bipolar disorder. Transl Psychiatry. 2019;9(1):152. doi: 10.1038/s41398-019-0488-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gray LJ, Dean B, Kronsbein HC, Robinson PJ, Scarr E. Region and diagnosis-specific changes in synaptic proteins in schizophrenia and bipolar I disorder. Psychiatry Res. 2010;178(2):374-380. doi: 10.1016/j.psychres.2008.07.012 [DOI] [PubMed] [Google Scholar]
- 45.Vance JE, Tasseva G. Formation and function of phosphatidylserine and phosphatidylethanolamine in mammalian cells. Biochim Biophys Acta. 2013;1831(3):543-554. doi: 10.1016/j.bbalip.2012.08.016 [DOI] [PubMed] [Google Scholar]
- 46.Tasseva G, Bai HD, Davidescu M, Haromy A, Michelakis E, Vance JE. Phosphatidylethanolamine deficiency in Mammalian mitochondria impairs oxidative phosphorylation and alters mitochondrial morphology. J Biol Chem. 2013;288(6):4158-4173. doi: 10.1074/jbc.M112.434183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Longo N, Frigeni M, Pasquali M. Carnitine transport and fatty acid oxidation. Biochim Biophys Acta. 2016;1863(10):2422-2435. doi: 10.1016/j.bbamcr.2016.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nałecz KA, Miecz D, Berezowski V, Cecchelli R. Carnitine: transport and physiological functions in the brain. Mol Aspects Med. 2004;25(5-6):551-567. doi: 10.1016/j.mam.2004.06.001 [DOI] [PubMed] [Google Scholar]
- 49.Hjelm BE, Rollins B, Mamdani F, et al. Evidence of mitochondrial dysfunction within the complex genetic etiology of schizophrenia. Mol Neuropsychiatry. 2015;1(4):201-219. doi: 10.1159/000441252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Divakaruni AS, Paradyse A, Ferrick DA, Murphy AN, Jastroch M. Analysis and interpretation of microplate-based oxygen consumption and pH data. Methods Enzymol. 2014;547:309-354. doi: 10.1016/B978-0-12-801415-8.00016-3 [DOI] [PubMed] [Google Scholar]
- 51.Rosenfeld M, Brenner-Lavie H, Ari SG, Kavushansky A, Ben-Shachar D. Perturbation in mitochondrial network dynamics and in complex I dependent cellular respiration in schizophrenia. Biol Psychiatry. 2011;69(10):980-988. doi: 10.1016/j.biopsych.2011.01.010 [DOI] [PubMed] [Google Scholar]
- 52.Lake BB, Ai R, Kaeser GE, et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 2016;352(6293):1586-1590. doi: 10.1126/science.aaf1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cobos I, Long JE, Thwin MT, Rubenstein JL. Cellular patterns of transcription factor expression in developing cortical interneurons. Cereb Cortex. 2006;16(suppl 1):i82-i88. doi: 10.1093/cercor/bhk003 [DOI] [PubMed] [Google Scholar]
- 54.Gao R, Penzes P. Common mechanisms of excitatory and inhibitory imbalance in schizophrenia and autism spectrum disorders. Curr Mol Med. 2015;15(2):146-167. doi: 10.2174/1566524015666150303003028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Selten M, van Bokhoven H, Nadif Kasri N. Inhibitory control of the excitatory/inhibitory balance in psychiatric disorders. F1000Res. 2018;7:23. doi: 10.12688/f1000research.12155.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kehrer C, Maziashvili N, Dugladze T, Gloveli T. Altered excitatory-inhibitory balance in the NMDA-hypofunction model of schizophrenia. Front Mol Neurosci. 2008;1:6. doi: 10.3389/neuro.02.006.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kimoto S, Zaki MM, Bazmi HH, Lewis DA. Altered markers of cortical γ-aminobutyric acid neuronal activity in schizophrenia: role of the NARP gene. JAMA Psychiatry. 2015;72(8):747-756. doi: 10.1001/jamapsychiatry.2015.0533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Obien ME, Deligkaris K, Bullmann T, Bakkum DJ, Frey U. Revealing neuronal function through microelectrode array recordings. Front Neurosci. 2015;8:423. doi: 10.3389/fnins.2014.00423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wright R, Réthelyi JM, Gage FH. Enhancing induced pluripotent stem cell models of schizophrenia. JAMA Psychiatry. 2014;71(3):334-335. doi: 10.1001/jamapsychiatry.2013.4239 [DOI] [PubMed] [Google Scholar]
- 60.Karmacharya R, Haggarty SJ. Stem cell models of neuropsychiatric disorders. Mol Cell Neurosci. 2016;73:1-2. doi: 10.1016/j.mcn.2016.03.007 [DOI] [PubMed] [Google Scholar]
- 61.Lundin A, Delsing L, Clausen M, et al. Human iPS-derived astroglia from a stable neural precursor state show improved functionality compared with conventional astrocytic models. Stem Cell Reports. 2018;10(3):1030-1045. doi: 10.1016/j.stemcr.2018.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lin M, Lachman HM, Zheng D. Transcriptomics analysis of iPSC-derived neurons and modeling of neuropsychiatric disorders. Mol Cell Neurosci. 2016;73:32-42. doi: 10.1016/j.mcn.2015.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.O’Shea KS, McInnis MG. Neurodevelopmental origins of bipolar disorder: iPSC models. Mol Cell Neurosci. 2016;73:63-83. doi: 10.1016/j.mcn.2015.11.006 [DOI] [PubMed] [Google Scholar]
- 64.Watmuff B, Liu B, Karmacharya R. Stem cell-derived neurons in the development of targeted treatment for schizophrenia and bipolar disorder. Pharmacogenomics. 2017;18(5):471-479. doi: 10.2217/pgs-2016-0187 [DOI] [PubMed] [Google Scholar]
- 65.Chen HI, Song H, Ming GL. Applications of human brain organoids to clinical problems. Dev Dyn. 2019;248(1):53-64. doi: 10.1002/dvdy.24662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Logan S, Arzua T, Canfield SG, et al. Studying human neurological disorders using induced pluripotent stem cells: from 2D monolayer to 3D organoid and blood brain barrier models. Compr Physiol. 2019;9(2):565-611. doi: 10.1002/cphy.c180025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Flippo KH, Strack S. An emerging role for mitochondrial dynamics in schizophrenia. Schizophr Res. 2017;187:26-32. doi: 10.1016/j.schres.2017.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Orologas AG, Buckman TD, Eiduson S. A comparison of platelet monoamine oxidase activity and phosphatidylserine content between chronic paranoid schizophrenics and normal controls. Neurosci Lett. 1986;68(3):293-298. doi: 10.1016/0304-3940(86)90505-7 [DOI] [PubMed] [Google Scholar]
- 69.Wood PL, Holderman NR. Dysfunctional glycosynapses in schizophrenia: disease and regional specificity. Schizophr Res. 2015;166(1-3):235-237. doi: 10.1016/j.schres.2015.05.017 [DOI] [PubMed] [Google Scholar]
- 70.Virmani A, Pinto L, Bauermann O, et al. The Carnitine Palmitoyl Transferase (CPT) system and possible relevance for neuropsychiatric and neurological conditions. Mol Neurobiol. 2015;52(2):826-836. doi: 10.1007/s12035-015-9238-7 [DOI] [PubMed] [Google Scholar]
- 71.Watanabe A, Yamada Y, Yamanaka S. Epigenetic regulation in pluripotent stem cells: a key to breaking the epigenetic barrier. Philos Trans R Soc Lond B Biol Sci. 2013;368(1609):20120292. doi: 10.1098/rstb.2012.0292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Svensson TH. Dysfunctional brain dopamine systems induced by psychotomimetic NMDA-receptor antagonists and the effects of antipsychotic drugs. Brain Res Brain Res Rev. 2000;31(2-3):320-329. doi: 10.1016/S0165-0173(99)00048-X [DOI] [PubMed] [Google Scholar]
- 73.Amiri A, Coppola G, Scuderi S, et al. ; PsychENCODE Consortium . Transcriptome and epigenome landscape of human cortical development modeled in organoids. Science. 2018;362(6420):eaat6720. doi: 10.1126/science.aat6720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang JH, Berkovitch SS, Iaconelli J, et al. Perturbational profiling of metabolites in patient fibroblasts implicates α-aminoadipate as a potential biomarker for bipolar disorder. Mol Neuropsychiatry. 2016;2(2):97-106. doi: 10.1159/000446654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang JH, Park H, Iaconelli J, et al. Unbiased metabolite profiling of schizophrenia fibroblasts under stressful perturbations reveals dysregulation of plasmalogens and phosphatidylcholines. J Proteome Res. 2017;16(2):481-493. doi: 10.1021/acs.jproteome.6b00628 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eFigure 1. Generation and characterization of cerebral organoids from human iPSCs
eFigure 2. iPSC characterization
eFigure 3. Gene ontology (GO) analysis of differentially expressed genes (DEGs) in schizophrenia cerebral organoids
eFigure 4. Cerebral organoid characterization
eFigure 5. RT-PCR validation of DEGs
eFigure 6. Synaptic staining and Western blots of cerebral organoids
eFigure 7. Mitochondrial DEGs in SCZ organoids
eFigure 8. Raster Plots from MEAs
eTable 1. Demographic and treatment information on subjects
eTable 2. GO analysis of DEGs that are associated specifically with SCZ, BPD or ASD.
eTable 3. Excitatory and inhibitory genes: upregulated DEGs
eTable 4. Excitatory and inhibitory genes: downregulated DEGs
eTable 5. Primary antibodies used
eTable 6. KaryoSTAT codes for each line
eAppendix.
eAppendix.