Abstract
Size-exclusion chromatography employing aqueous mobile phases with volatile salts at neutral pH combined with electrospray-ionization mass spectrometry (SEC-ESI-MS) is a useful tool to study proteins in their native state. However, whether the applied eluent conditions actually prevent protein–stationary phase interactions, and/or protein denaturation, often is not assessed. In this study, the effects of volatile mobile phase additives on SEC retention and ESI of proteins were thoroughly investigated. Myoglobin was used as the main model protein, and eluents of varying ionic strength and pH were applied. The degree of interaction between protein and stationary phase was evaluated by calculating the SEC distribution coefficient. Protein-ion charge state distributions obtained during offline and online native ESI-MS were used to monitor alterations in protein structure. Interestingly, most of the supposedly mild eluent compositions induced nonideal SEC behavior and/or protein unfolding. SEC experiments revealed that the nature, ionic strength, and pH of the eluent affected protein retention. Protein–stationary phase interactions were effectively avoided using ammonium acetate at ionic strengths above 0.1 M. Direct-infusion ESI-MS showed that the tested volatile eluent salts seem to follow the Hofmeister series: no denaturation was induced using ammonium acetate (kosmotropic), whereas ammonium formate and bicarbonate (both chaotropic) caused structural changes. Using a mobile phase of 0.2 M ammonium acetate (pH 6.9), several proteins (i.e., myoglobin, carbonic anhydrase, and cytochrome c) could be analyzed by SEC-ESI-MS using different column chemistries without compromising their native state. Overall, with SEC-ESI-MS, the effect of nonspecific interactions between protein and stationary phase on the protein structure can be studied, even revealing gradual structural differences along a peak.
Assessment of protein higher-order structures (HOS) is critical, not only when defining the quality of, for example, enzymes and biopharmaceuticals, but also in understanding the function of these complex molecules. Changes in HOS can have a significant impact on the biological activity and physicochemical properties of proteins. As a result, there is an increasing demand to analyze proteins in their native state, preferably under near-physiological conditions.1−6 However, analysis of proteins while preserving their three-dimensional structure remains challenging. This is related to the separation and detection conditions imposed by analytical methodologies, which potentially distort the structural integrity, molecular conformation, and/or activity of proteins.4,7
A number of separation approaches employing aqueous mobile phases in principle allow preservation of the HOS of proteins. Exemplary are capillary electrophoresis (CE),8,9 asymmetrical flow-field flow fractionation (AF4),10−12 and size-exclusion chromatography (SEC).2,13 The latter is regarded a golden standard in many fields of application when it comes to size-based separation of biomacromolecules. Commonly combined with light scattering detection methodologies, SEC can provide insights into the physical size (hydrodynamic volume) of macromolecules, from which for instance the compactness of protein conformation or the extent of oligomerization may be deduced.4,11 Yet, more detailed structural information, e.g., on post-translational modifications, requires hyphenation of SEC with mass spectrometry (MS). Over the years, native electrospray ionization (ESI) has proven to be very useful for obtaining multiply charged ions of intact proteins under near-physiological conditions. ESI is soft and preserves protein conformation and noncovalent interactions in the gas phase14−16 while providing the possibility to attain accurate molecular mass information by high-resolution MS. Coupling of native ESI-MS to a protein separation technique decreases the complexity of protein mass spectra so that more meaningful information can be obtained.
Despite the advantages of hyphenating SEC to MS, there is a potential risk that the applied separation conditions change the protein structure.17 Alterations might be due to, for instance, dilution in the mobile phase or interaction with the stationary phase material. Although SEC is considered a soft separation technique, separation conditions need to be carefully considered in order to ensure reliable conclusions are drawn on protein structures.2,13,18−20 It is, for example, common practice to add organic solvents to the aqueous mobile phase in order to improve the separation performance for protein and biopharmaceutical characterization.18,21−27 However, protein conformation and aggregates may be affected even by small percentages of organic solvent. Ideally, native SEC-ESI-MS provides both optimal protein resolution and preservation of structural integrity.
A number of studies have been reported using SEC-ESI-MS under presumably native conditions for the characterization of small oligomers,28 intact antibodies,19,23,24,29 and protein aggregates.27 However, these studies were mainly focused on the application and did not verify whether the analyzed proteins were actually in their native state under the applied separation conditions. Still, there are several studies using either SEC or ESI-MS that show the importance of the choice of analytical conditions on the conservation of the native state of proteins.30−33
In this study, the impact that (volatile) mobile phases may have on the retention and ionization of proteins during SEC-ESI-MS analysis is investigated. For this, solutions of the most commonly used volatile salts (ammonium acetate, formate, and bicarbonate) at varying ionic strength and pH will be used. Myoglobin is selected as the model protein, as it can reveal potential interactions with the stationary phase during SEC separation34 and denaturation during separation and ionization by monitoring of its charge state distribution and loss of the hemichrome moiety.33,35−38 SEC of myoglobin using UV absorbance detection will be employed to study the protein chromatographic behavior at the different conditions. Subsequently, direct-infusion (DI) ESI-MS and SEC-ESI-MS experiments are performed in order to assess potential protein denaturation during ionization and separation, respectively. For verification, experiments were performed using two different (in terms of column chemistry and dimensions) SEC columns and by including an acidic (carbonic anhydrase; CA) and basic (cytochrome C; CC) protein. The main goal of the study was to reveal sources of protein denaturation during SEC analysis by probing protein unfolding with native ESI-MS. Ultimately, from these studies, suitable conditions for genuine native SEC-ESI-MS of proteins could be derived.
Materials and Methods
Chemicals
For the mobile phase preparation, sodium phosphate dibasic (≥98.5%), sodium phosphate monobasic (≥99.0%), sodium sulfate (≥99.0%), and sodium azide (≥99.5%) from Sigma-Aldrich (Schnelldorf, Germany) were used. For the preparation of the volatile solutions used for DI-ESI-MS and SEC-ESI-MS experiments, ammonium acetate (≥98%), ammonium formate (≥98%), and ammonium bicarbonate (≥99.5%) from Sigma-Aldrich were used. Solutions were adjusted to a final pH of 5.9, 6.9, or 7.5 with ammonium hydroxide (28–30% NH3 in water), acetic acid (≥99%), or formic acid (≥97%) from Sigma-Aldrich. A Milli-Q purification system (Millipore, Bedford, MA, USA) provided ultrapure water. The gel-filtration protein standard (# 1511901) was purchased from Bio-Rad Laboratories (California, United States). It contains thyroglobulin (from bovine; MW, 670 kDa; pI, 4.5), γ-globulin (from bovine; MW, 158 kDa; pI, 7.2), ovalbumin (from chicken; MW, 44 kDa; pI, 4.6), myoglobin (from horse; MW, 17 kDa; pI, 7.2 (major component) and 6.8 (minor component)), and vitamin B12 (MW, 1350 Da). Carbonic anhydrase isozyme II (from bovine erythrocytes; MW, 30 kDa; pI 5.4) and cytochrome C (from equine heart; MW, 13 kDa; pI 10.8) were both purchased from Sigma-Aldrich.
Sample Preparation
For the SEC-UV experiments, 25 mg/mL myoglobin (from horse heart, Sigma-Aldrich) in water was filtered using the Phenomenex (California, United States) syringe filter tool (0.45 μm) and diluted to 1 mg/mL in mobile phase. For the DI-ESI-MS experiments, 0.5 mL of 3.5 mg/mL myoglobin in water was added to 5.0 mL of mobile phase solution yielding a final concentration of 20 μM (0.32 mg/mL). Carbonic anhydrase and cytochrome C (20 μM) were prepared similarly from 15 mg/mL stock solutions. For the SEC-UV-MS experiments, the gel-filtration standard protein mixture was used. The solution was prepared according to the manufacturer’s instructions. Shortly, the proteins were rehydrated with deionized water and gently swirled. The final myoglobin concentration in this mixture was 0.5 mg/mL. For additional measurements, 1 mg/mL solutions of carbonic anhydrase and cytochrome C were prepared by diluting the stock solutions in mobile phase.
Chromatographic System
A Shimadzu SIL-20AD Prominence Ultra-Fast Liquid Chromatography (UFLC) system (‘s-Hertogenbosch, The Netherlands) equipped with a pump, autosampler, column oven, and UV absorbance detector set at 280 nm was used for all SEC measurements. An injection volume of 20 μL, an eluent flow rate of 0.8 mL/min, a column oven temperature of 25 °C, and an autosampler temperature of 8 °C were used throughout the study. A TOSOH TSKgel G2000SWXL column (Griesheim, Germany; 7.8 mm i.d. × 30 cm, 5 μm particle size, 125 Å pore size; Column 1) was used for the separations. It was preceded by a TSKgel SWXL Type Guard Column (6 mm i.d. × 4 cm, 7 μm particle size, 125 Å pore size). Additional experiments were performed using the Agilent AdvanceBioSEC column (Wilmington, DE, USA; 2.7 μm, 150 × 4.6 mm, 300 Å; Column 2) and AdvanceBioSEC guard column (2.7 μm, 50 × 4.6 mm, 250 Å). The AdvanceBioSEC column was operated with an injection volume of 5 uL and a flow rate of 0.3 mL/min. Both columns were tested according to the manufacturer’s standard method using thyroglobulin (from bovine; MW, 670 kDa; pI, 4.5), albumin from bovine serum (MW, 66 kDa; pI 4.7), myoglobin (from horse; MW, 17 kDa; pI, 7.2 (major component) and 6.8 (minor component)), and uracil as a calibration standard; an example is provided in Figure S1. For the mobile phase eluents, stock solutions of 0.2 M of every ammonium salt were prepared and consecutively diluted to the desired concentration (i.e., 0.01–0.1 M). All eluents were prepared using ultrapure water and filtered over Whatman (Maidstone, United Kingdom) regenerated cellulose membrane filters (0.45 μm). Adjustment of the solution’s pH was performed after filtration. The pH of the mobile phase was checked using a EL 20 pH meter of Mettler Toledo (Ohio, United States) at 25 °C. Equilibration of the column with the respective mobile phase was performed for at least five column volumes prior to protein injection.
Mass Spectrometry
A micrOTOF-Q (Bruker Daltonics, Bremen, Germany) with an ESI source operating in positive ion mode was used. DI-ESI-MS experiments were performed at a flow rate of 180 μL/h using a 0.5 mL gastight syringe (Hamilton, Reno, USA) and a syringe pump (Cole-Parmer, Vernon Hill, USA). For the coupling of the TOSOH TSKgel G2000SWXL SEC column and ESI-MS, a flow splitter (1:50; Agilent Technologies, Waldbronn, Germany) was used in order to ensure a flow of 16 μL/min was directed toward the mass spectrometer, while the residual flow was guided toward the UV absorbance detector. For coupling of the AdvanceBioSEC column with ESI-MS, a homemade 1:10 flow splitter was used. The specific configurations were used to prevent contamination of the source, since sensitivity was not compromised. The ESI settings were as follows: source temperature, 200 °C; capillary voltage, 4.8 kV; dry gas flow, 4 L/min; nebulizer gas, 0.4 bar; ion energy, 5 eV; collision energy, 10 eV; in-source collision-induced dissociation, 0 eV. Ion funnels were set at values of 300 and 400 Vpp, respectively. Mass spectra were acquired in the range of 100 to 5000 m/z. Data analysis was performed using Bruker Compass DataAnalysis Version 5.0 (Bruker Daltonics).
Data Evaluation
SEC distribution coefficients (Kd) were calculated using eq 1, where V0 and Vi are the void volume and intraparticle volume, respectively, as obtained from the most recently established calibration curve, and Vr is the elution volume of the protein of interest.
| 1 |
For the estimation of the fraction of folded myoglobin based on the DI-ESI-MS and SEC-ESI-MS experiments, data evaluation was performed with an in-house developed script in Matlab version 2015b (MathWorks, Massachusetts, United States). Details are in the Supporting Information. Shortly, data is obtained within the 750–2500 m/z range, i.e., covering the myoglobin charge state distribution (CSD). A structurally folded protein with increased compactness of the polypeptide chain has proven to result in lower charge states, and it is considered to be a native-like component.41 Therefore, myoglobin signals below m/z 1700 were regarded as to originate from unfolded protein, whereas signals above m/z 1700 were treated as caused by folded protein (native).33,35,36,39−41 The fraction folded (or native) was calculated taking the sum of the intensity (I) of the [M + 8H]8+, [M + 9H]9+, and [M + 10H]10+ ions from the CSD and dividing it by the total intensity (Itot) of all charge states (excluding the signal of the heme group at m/z 616), as described in eq 2. It is important to note that the obtained charge states might depend on the used instrumentation, as the geometry of the ESI source can have an impact.
| 2 |
To compare between the DI-ESI-MS and SEC-ESI-MS experiments using the two columns, the average charge state (CS) of each of the three proteins was calculated. The CS was calculated based on the intensity of any given charge state (i) multiplied by the net charge of the specific charge state (qi), divided by the sum of the signal intensities.
| 3 |
Results and Discussion
This work aims to evaluate possible structural alterations imposed on proteins during SEC analysis while using eluents of varying composition (nature of salt), pH, and ionic strength. Native ESI-MS was used as tool to detect structural changes by means of unfolding. Mobile phase effects were studied using SEC and ESI-MS individually and in combination (SEC-ESI-MS), providing a comprehensive picture on the influence of the different parameters. As a test protein that is prone to structural changes during analysis, myoglobin was selected, as it readily undergoes conformational changes when stressed.33,35−37 Since diol-modified silica columns are still predominantly used for SEC of proteins,21 the majority of the research has been performed on such a column.
SEC Elution Behavior of Myoglobin Using Different Mobile Phase Compositions
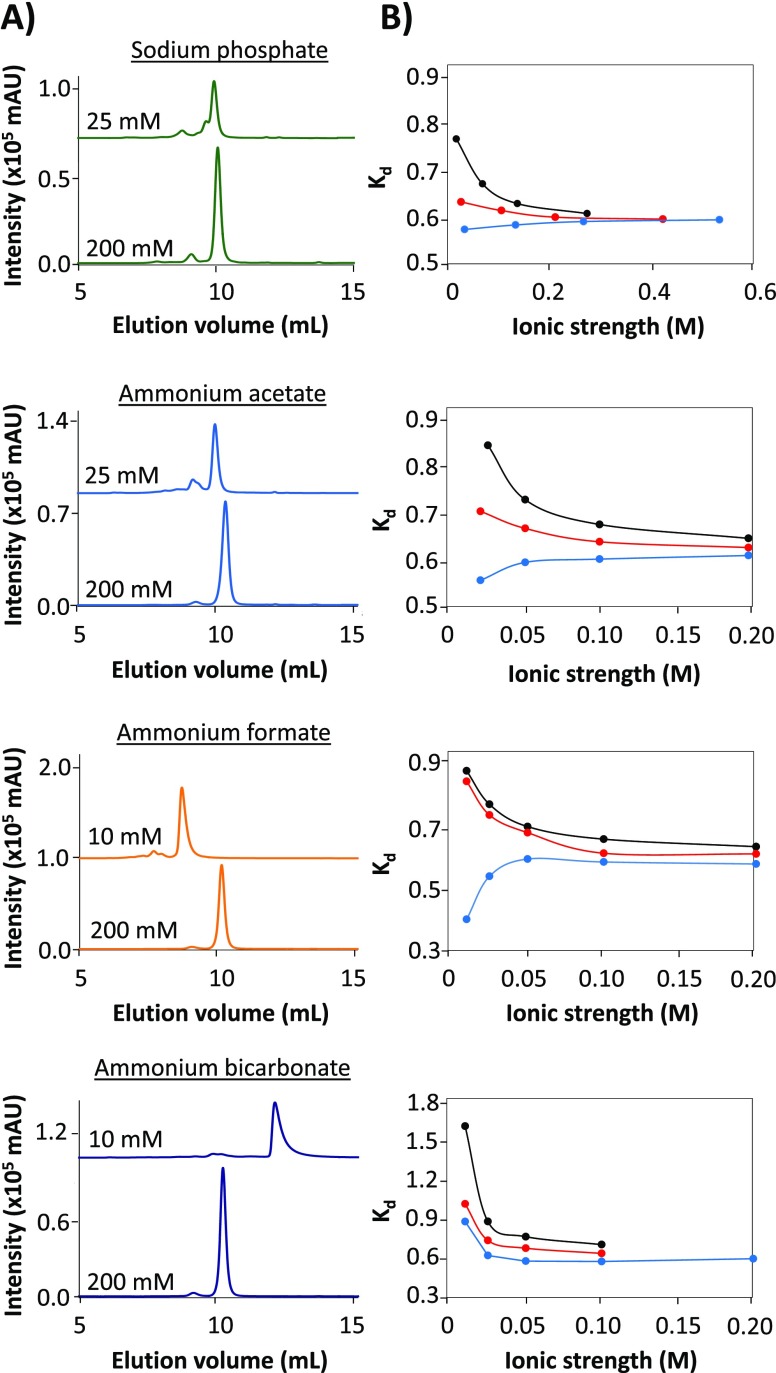
In order to investigate protein elution behavior in aqueous SEC, myoglobin was analyzed using eluents containing four different additives and varying in pH and ionic strength (Figure 1). First, experiments were done using a phosphate-based mobile phase containing sodium sulfate and sodium azide, as this is a standard eluent in aqueous protein SEC. The chromatograms obtained at low and high ionic strength of the eluent (Figure 1A) indicate that the elution volume of the protein depends on the ionic strength. In order to obtain a more comprehensive picture, the distribution coefficient (Kd; eq 1) of myoglobin was plotted against the ionic strength for several eluents of different pH (Figure 1B).34 Monitoring changes of the Kd value under varying pH and ionic-strength conditions can provide insights on the nature of the interactions between the analyte and stationary phase. Focusing on the phosphate eluent, at lower-ionic-strength conditions (<0.1 M), the Kd curves of the examined pH values do not align, which suggests that both size-exclusion and sorption mechanisms are contributing to the separation. Under these nonideal SEC conditions, also a pH dependency was observed. Using an eluent of pH 5.9, which is below the pI of myoglobin (i.e., 7.242), later elution and higher Kd values were obtained. This indicates electrostatic interactions between the positively charged protein and deprotonated silanol groups of the stationary phase. In contrast, at the higher eluent pH of 7.5, earlier elution and lower Kd values were observed, as a result of exclusion of the overall negatively charged protein from the negatively charged stationary phase. Above 0.1 M ionic strength, the Kd lines merge, indicating that the elution volume of myoglobin is not affected by ionic strength nor by pH of the mobile phase, suggesting virtually interaction-free SEC.
Figure 1.
SEC-UV of myoglobin using different eluents varying in nature of salt, ionic strength, and pH. (A) Typical chromatograms obtained with eluents of low and high ionic strength, all at pH 7.5. (B) Plots representing the Kd observed for myoglobin versus the eluent ionic strength using the indicated salt. Eluent pH: 5.9 (black), 6.9 (red), and 7.5 (blue). The connecting lines between the points highlight trends and are not obtained by fitting.
Subsequently, eluents containing the MS-compatible salts ammonium acetate, formate, or bicarbonate were studied. Although the ammonium acetate and formate solutions do not buffer in the examined pH range (5.9–7.5), they are widely used in SEC-MS. The same pH values as for phosphate eluent were tested, whereas the ionic strength of the ammonium salts was selected to be lower to ensure MS compatibility. Myoglobin analyzed using ammonium acetate as eluent showed similar elution behavior as then analyzed using the phosphate eluent. Below 0.1 M ionic strength, protein elution was affected by electrostatic interactions (repulsion or attraction), as revealed from the shift of the Kd values. A different behavior was observed when myoglobin was analyzed with the other two volatile eluents. The high Kd values obtained for myoglobin using ammonium formate indicate that in this situation electrostatic interactions are more pronounced at lower ionic strength as compared to the phosphate and acetate eluent. Ammonium bicarbonate showed the most apparent deviation of the elution behavior of myoglobin. This can be explained by the decomposition of bicarbonate into carbon dioxide at lower pH values.37,38 This instability of the buffer and the corresponding bubble formation were also limiting factors for conducting useful experiments at pH 5.9 and 6.9.
The chromatograms obtained for myoglobin indicate that not only the retention time but also the peak shape were affected by the applied eluent conditions (Figure 1A). With eluents containing phosphate and acetate, the myoglobin peak remained fairly symmetrical when the ionic strength was varied. However, the formate and bicarbonate eluents caused significant broadening and tailing of the myoglobin peak at lower ionic strengths (Figure S2). This can be explained by the nonspecific interactions between the protein and the silica-based stationary phase.23 Moreover, the protein peak area overall was lower at lower ionic strength, which might be caused by a loss of protein due to irreversible adsorption to the column material.18,23 The minimum ionic strength needed to effectively avoid unwanted interactions increased in the order of acetate, formate, and bicarbonate. By comparing the actual chromatograms for myoglobin and the derived Kd plots, it can be concluded that the ammonium acetate eluent performs most similar to the phosphate-based eluent, whereas the mobile phases containing ammonium formate or bicarbonate deviate, especially at lower-ionic-strength conditions. Interestingly, this observation seems to follow the Hofmeister series for anions. Phosphate and acetate are considered as to be kosmotropic anions (i.e., stabilizing protein structures), whereas formate and bicarbonate are more chaotropic anions (i.e., destabilizing protein structures). Although the underlying mechanisms of the Hofmeister effects are not fully understood,43 destabilization of protein structure by the eluent—thereby exposing more amino acid residues—can explain the increased likelihood of interactions with the column material
SEC Elution Behavior of Other Proteins
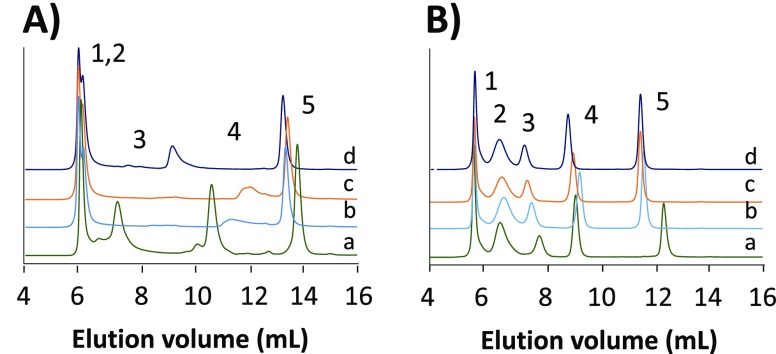
In order to evaluate whether the observations made for myoglobin also hold for other proteins, a mixture of proteins of different pI values and molecular weights was analyzed by SEC-UV. Four different eluents at low (0.01 M) and high (0.2 M) ionic strength and at pH values close to the physiological conditions (pH 6.9, 7.5) were selected (Figure 2). At low ionic strength, electrostatic interactions are not sufficiently suppressed, leading to peak distortion, and certain proteins were not even detected (Figure 2A). Only at high ionic strength, apparent interaction-free SEC separations were achieved for all proteins (Figure 2B). At high ionic strength, the elution behavior of the largest protein (thyroglobulin; MW, 670 kDa) is quite similar for all eluents, but that is most probably due to its elution close to the exclusion limit of the column. For the other proteins (γ-globulin, ovalbumin, and myoglobin) the elution volume depends on the eluent salt used. In the presence of kosmotropic anions, like acetate and phosphate, proteins remain folded, leaving the protein hydrodynamic radius unchanged. In contrast, the chaotropic bicarbonate and formate ions destabilize the protein structure, resulting in a larger hydrodynamic radius and, consequently, shorter elution times. Note that the difference between the potassium eluent and ammonium salts can also be attributed in part to the ability of potassium to more effectively shield the silica gel material.23,44
Figure 2.
SEC-UV of the protein test mixture containing thyroglobulin (1), γ-globulin (2), ovalbumin (3), myoglobin (4), and vitamin B12 (5) using eluents consisting of solutions of (a) sodium phosphate, pH 6.9, (b) ammonium acetate, pH 6.9, (c) ammonium formate, pH 7.5, and (d) ammonium bicarbonate, pH 7.5 at concentrations of (A) 0.01 M and (B) 0.2 M.
The SEC-UV experiments reveal a protein elution behavior that depends on both ionic strength and nature of the eluent. The proteins seem to undergo structural changes related to the elution conditions and interactions with the stationary phase, leading to shifts in elution volumes. In order to confirm the conformational changes, native MS experiments were carried out.
Direct-Infusion ESI-MS of Myoglobin
Native ESI-MS in principle is an excellent tool to probe protein structural alterations.14 Direct-infusion (DI)-ESI-MS of myoglobin was performed using different aqueous solvents. The protein was dissolved in ammonium acetate, formate, and bicarbonate solutions of varying concentrations (0.01–0.2 M) and pH (5.9, 6.9, and 7.5), allowing correlations with the obtained SEC data.
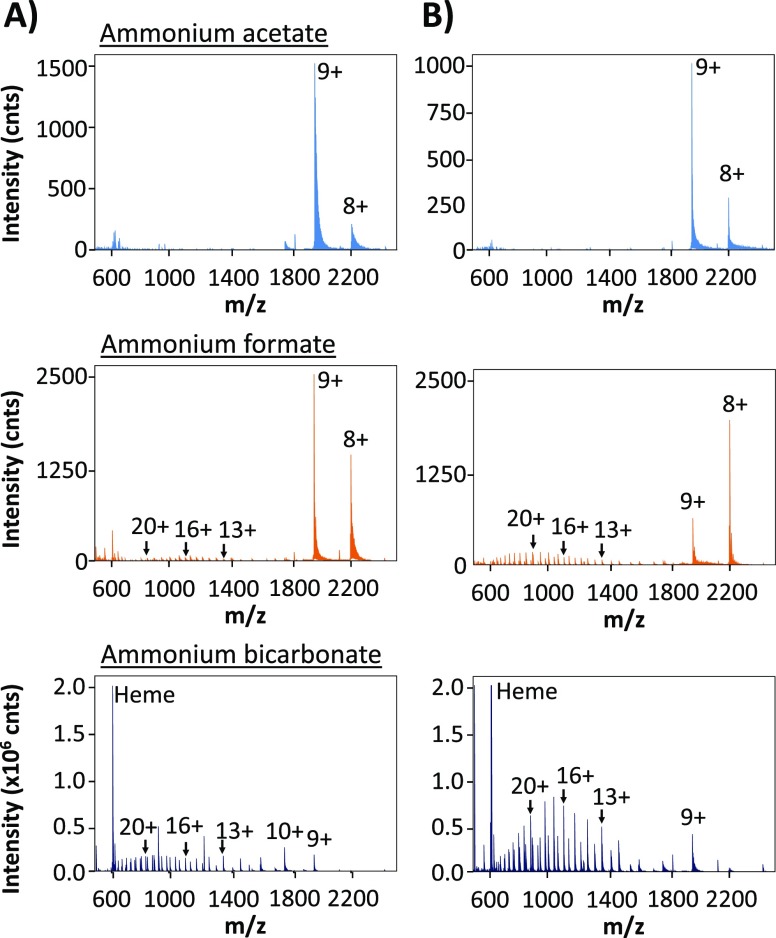
The mass spectra of myoglobin obtained using 0.01 and 0.2 M ionic strength at pH 6.9 are presented in Figure 3. When dissolved in ammonium acetate, myoglobin showed three intense signals between m/z 1900 and 2500, which were assigned to its [M + 10H]10+, [M + 9H]9+, and [M + 8H]8+ charge states. The observed monomodal charge state distribution (CSD) is considered to correspond with the native/nondenatured conformation of myoglobin.33,35,36,39−41 When comparing the mass spectra obtained for myoglobin dissolved in acetate, formate, and bicarbonate, clearly different profiles were observed. For the latter two eluent salts, at higher concentrations, a second CSD between m/z 600–1700 was observed in the respective mass spectra. Additionally, the signal at m/z 616 present in the spectrum obtained with a high concentration of ammonium bicarbonate suggests also loss of the heme group. The loss of the heme group (m/z 616)36 and the appearance of higher charge states indicate that myoglobin was partly denatured when dissolved in 0.2 M ammonium formate or bicarbonate. Deconvolution of these mass spectra indeed yielded the molecular masses of both intact myoglobin and myoglobin without the heme group (data not shown). The deconvoluted mass spectra of myoglobin dissolved in ammonium acetate solutions only showed the intact myoglobin.
Figure 3.
Mass spectra obtained during DI-ESI-MS of myoglobin dissolved in 0.01 M (A) or 0.2 M (B) ammonium acetate, ammonium formate, or ammonium bicarbonate (all pH 6.9).
In order to get a more quantitative insight in the myoglobin structure under the various eluent conditions (nature of salt, ionic strength, and pH), the fraction of native protein was calculated from the obtained mass data (eq 2, Materials and Methods). The summed intensities of the 8+, 9+, and 10+ charge states (representing native myoglobin) were expressed as a fraction of the total intensity of all observed charge states for myoglobin, assuming that the native and denatured forms have equal ionization efficiencies.37 The fraction native was plotted for each tested eluent salt and pH against the ionic strength (Figure 4). What becomes immediately clear is that ammonium acetate does not cause any protein denaturation (Figure 4A), regardless of the pH and ionic strength of the solvent. This is nicely in line with the SEC-UV results obtained when using ammonium acetate at high ionic strength: Kd and peak width and shape were virtually constant for each pH tested. When dissolved in ammonium formate and bicarbonate, the fraction of native myoglobin rapidly decreases with increasing ionic strength (Figures 4B,C). This trend is opposite of what was observed with SEC-UV, where at higher ionic strength, the Kd and peak width and shape improve rather than deteriorate. This difference in behavior then has to be explained by gas phase processes.
Figure 4.
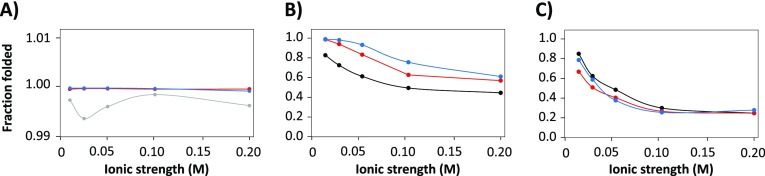
Fraction folded of myoglobin as derived from DI-ESI-MS of myoglobin dissolved in (A) ammonium acetate, (B) ammonium formate, and (C) ammonium bicarbonate of different ionic strength. Eluent pH: 5.9 (black), 6.9 (red), and 7.5 (blue). The connecting lines between the points highlight trends and are not obtained by fitting.
Acetate is a kosmotropic ion and stabilizes the protein structure. The chaotropic ions formate and bicarbonate reduce the protein conformation stability, partly leading to higher charge states of myoglobin during the ESI process.37,38,45−48 An additional explanation for the observed difference between ammonium acetate and formate is related to differences in pKa, being approximately 1 unit lower for formate. As a consequence, acetate may reduce the acidification of the droplets during the final stages of shrinkage more than formate.49 This would also explain the clear pH dependency of unfolding using ammonium formate, with the lower pH values leading to more pronounced protein denaturation (Figure 4B). When using the ammonium bicarbonate solvent, significantly more denatured protein is observed, even at low ionic strength, regardless of the pH (Figure 4C). This can be partly ascribed to the chaotropic nature of bicarbonate in solution, but additional effects may play a role here.16,37,38 For example, the formation of carbon dioxide from bicarbonate during the ESI process can lead to “foaming” in the produced microdroplets, resulting in supercharging of the protein, even from solutions with a pH close to physiological conditions.16,38
Overall, the DI-ESI-MS results show that using a solution of ammonium acetate appears to preserve the myoglobin structure quite efficiently, whereas formate and especially bicarbonate cause much higher fractions of denatured species. Moreover, it shows that structural alterations of myoglobin can occur, even under what are normally considered to be native conditions.
SEC-ESI-MS of Myoglobin
Online SEC-ESI-MS experiments were performed in order to verify whether potential structural alterations induced by the applied SEC conditions can be probed by ESI-MS and subsequently explain the obtained SEC-UV results. The protein test mixture was analyzed using eluents containing ammonium acetate, formate, or bicarbonate at 0.01 and 0.2 M ionic strength with a pH of 6.9 or 7.5. Figure 5 shows the obtained chromatograms and average mass spectra from the myoglobin peak when using the ammonium acetate eluent. The results for ammonium formate and bicarbonate can be found in Figure S3 of the SI.
Figure 5.
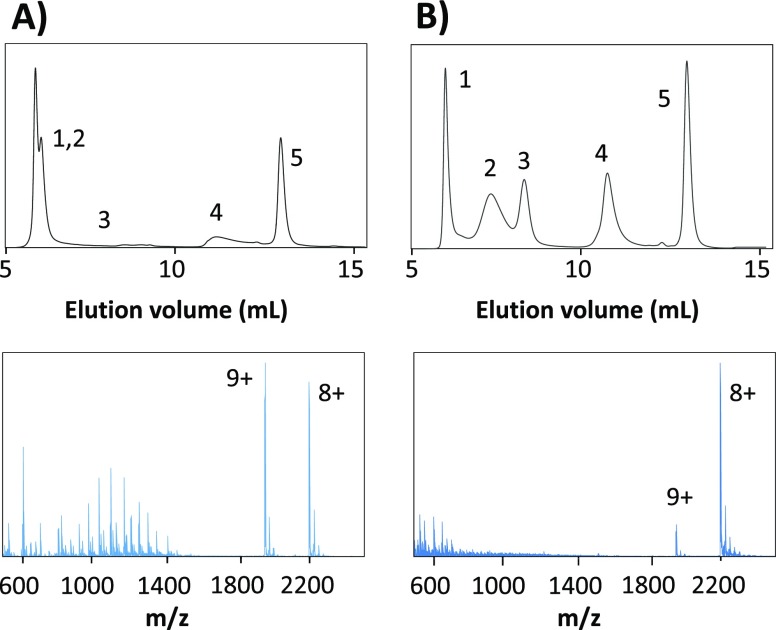
SEC-ESI-MS of the protein test mixture using an eluent of (A) 0.01 M ammonium acetate and (B) 0.2 M ammonium acetate (both pH 6.9). Average mass spectra of the myoglobin peaks are provided. Peaks: thyroglobulin (1), γ-globulin (2), ovalbumin (3), myoglobin (4), and vitamin B12 (5).
As can be expected, the elution behavior of myoglobin during SEC-ESI-MS analysis is similar to that observed using SEC-UV. However, the mass spectra of myoglobin obtained with SEC-ESI-MS clearly differ from the mass spectra obtained with DI-ESI-MS using the same eluent conditions. When using 0.01 M ammonium acetate and formate, only myoglobin in its native form was observed (CSD in m/z 1700–2500 range) with DI-ESI-MS. Remarkably, the mass spectra obtained during SEC-ESI-MS using the same eluent (Figure 5A) indicated a significant presence of denatured myoglobin (CSD between m/z 600 and 1700). For all tested eluent conditions, the fraction native myoglobin was calculated when using SEC-ESI-MS and compared to the fraction values obtained with DI-ESI-MS (Figure 6).
Figure 6.
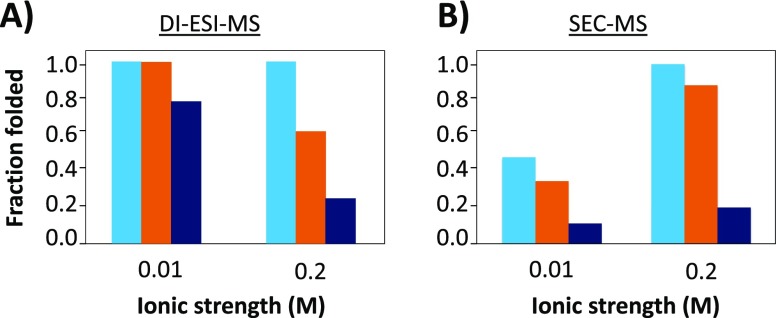
Fraction folded of myoglobin calculated after analysis by (A) DI-ESI-MS and (B) SEC-ESI-MS using different eluents of low (0.01 M) and high (0.2 M) ionic strength. Eluents: ammonium acetate (light blue), ammonium formate (orange), and ammonium bicarbonate (dark blue).
The effect of ionic strength appeared to be opposite for the two analytical setups: whereas when using DI-ESI-MS a increasing ionic strength leads to a higher degree of denaturation, when using SEC-ESI-MS, a decreasing ionic strength leads to more protein denaturation. Notably, for both analytical systems, the measured fraction of folded protein at high ionic strength is similar. Hence, the effect of difference between the setups should be contributed to the lower ionic strength. In order to verify this observation, additional DI-ESI-MS experiments were performed. Initially with DI-ESI-MS, the incubation time of the protein in buffer was only a few minutes. Extending the incubation time to 30 min (i.e., the SEC analysis time), the protein mass spectrum did not change (data not shown), excluding exposure to the buffer as cause of denaturation. Consequently, also in light of the peak tailing of the myoglobin peak and the loss of other proteins during SEC analysis, it seems most plausible that the nonspecific interactions between the protein and the stationary phase at low salt concentration are the cause of this effect. Although extensive equilibration of the column with the respective mobile phases was performed, residual silanols might still give rise to protein denaturation. As is known from many fields, proteins can easily adsorb onto a (silica) surface.18,34,50−53 During this process, the electrostatic interactions, hydrogen-bonding, dipole–dipole van der Waals interactions, and hydrophobic effects between the silica and the protein can result in a conformational change of the latter. This suggests that ESI-MS can be used as a read out of the actual structure of the protein after separation. Consequently, this then would also allow monitoring of the effect of the SEC stationary phase/elution process on protein conformation.
To investigate this further, the fraction of folded protein over the peak as obtained with both 0.01 and 0.2 M ammonium acetate was plotted (Figure S4). Using 0.2 M ammonium acetate as an eluent results in a stable folded protein fraction between 0.95 and 1.0 over the whole peak elution window. However, when using the 0.010 M eluent, a clear trend is observed. At the beginning, the fraction folded is approximately 0.5, and it drops to about 0.25 in the tail of the elution window. Indeed, it seems plausible that stronger interaction/adsorption between the protein and column material leads to a higher degree of irreversible denaturation for part of the protein molecules. This process is reduced when using a higher ionic strength,54 as is evident from the preservation of the native protein structure and the lack of any trend throughout the peak profile.
Confirmatory SEC-ESI-MS Experiments
To further study the described effects related to nonspecific interactions, additional proteins were analyzed by SEC-ESI-MS. Both an acidic protein (carbonic anhydrase; CA, pI 5.4) and basic protein (cytochrome C; CC, pI 10.8) were selected for this purpose. They were analyzed using the diol column in combination with ammonium acetate eluents of varying concentrations (0.01, 0.05, and 0.2 M) at pH 6.9. If no on-column interactions take place, these eluents should preserve the protein’s structure in liquid and gas phase. The chromatographic results are shown in Figure S5A. The expected Kd values—based on the calibration curve—were obtained when using the high-ionic-strength (0.2 M) eluent. Lowering the ionic strength makes the previously described protein pI/eluent pH effects become evident. For CA, the elution volume decreases (exclusion), whereas for CC, it increases (electrostatic interactions). The interactions for CC additionally lead to significant peak tailing and at the lowest ionic strength (0.01 M) even result in a full loss of the protein peak. The mass spectra obtained for the two proteins also show a clear trend (Figure S5C). Lowering the ionic strength leads to an increase in average CS (Figure S5E,F). This is in contrast to their CS as obtained with DI-ESI-MS, which remains independent of ionic strength. Hence, the difference between the two experiments must result from stress that the protein experiences when going through the column under low-ionic-strength conditions and is thus effectively shielded at high ionic strength. Denaturation, as observed for myoglobin, was not evident for both proteins. Indeed, exclusion as observed for carbonic anhydrase does not lead to protein–column interaction, and it is reasonable to assume this then does not lead to denaturation. Moreover, for CC, the interactions between the protein and the column were so strong that the protein did not elute and the effect could not be visualized. Apparently, the interactions were limited at 0.05 M and did not lead to full denaturation.
Although diol-based columns are the standard in protein analysis by SEC, improved chemistries have recently been introduced. Therefore, all the three proteins have been analyzed using a SEC column modified with a proprietary hydrophilic chemistry aiming to minimize nonspecific interactions. For CA and CC, a similar trend was found in terms of chromatographic behavior for the proteins; CA experiences exclusion from the pores, whereas CC shows interaction upon ionic-strength decrease (Figure S5B). Interestingly though, especially the electrostatic interaction was reduced to a degree that CC even eluted under the lowest-ionic-strength conditions. The resulting mass spectra show an increase in average CS when the ionic strength is reduced (Figure S5D–F), but the effect is less pronounced compared to when the proteins were analyzed on the diol column. Obviously, myoglobin was also analyzed using this column. Similar to the two other proteins, the chromatographic behavior was improved compared to the diol column. Especially at the lowest ionic strength, the peak position and shape indicate a significant decrease in protein–column interaction (Figure S6A). Interestingly, as a consequence of this, also no protein denaturation but an increase in CS in the native area of the mass spectrum was observed (Figure S6B). So, it seems that even when interactions are limited, SEC-ESI-MS can still give insights into these unwanted effects based on subtle differences in protein elution profile and average CS.
Conclusion
We investigated the influence of SEC conditions on protein structural integrity using native ESI-MS as a selective tool to reveal conformational alterations. The SEC elution and ESI using eluents of several volatile salts of varying ionic strength and pH were examined using the model hemoprotein myoglobin. Results indicate that high-ionic-strength conditions of volatile salts ensure almost interaction-free aqueous SEC under near-physiological pH conditions. Lower ionic strength does not prevent (electrostatic) interactions between the column material and protein, leading to severe adsorption and peak tailing. Variations in the elution behavior of proteins mainly seem to correlate with the kosmotropic/chaotropic nature of the cationic salt additives. The impact of pH and ionic strength in the elution profile are strongly influenced by the physical–chemical properties of the protein as well as the stationary phase material. Therefore, they should be thoroughly assessed on an individual basis. Native ESI-MS revealed critical differences between the ammonium salts regarding their impact on protein denaturation under the examined conditions. Ammonium acetate most effectively preserved the protein structure regardless of ionic strength and pH conditions, whereas formate and especially bicarbonate cause much higher fractions of the denatured species. Coupling of SEC with native ESI-MS enabled monitoring of structural changes during the elution process. Overall, we conclude that the introduction of ESI-MS can reveal the influence of nonspecific interactions between protein and stationary phase on the protein structure. A next, highly relevant, step will be to study large proteins/biopharmaceuticals and protein complexes in order to gain more insights as to whether this platform can be used to monitor structural changes in both liquid and gas phase.
Acknowledgments
I.K.V. and R.L.C.V. acknowledge the HOSAna project, which is funded by The Netherlands Organization for Scientific Research (NWO) in the framework of the Programmatic Technology Area PTA-COAST4 of the Fund New Chemical Innovations (project nr. 053.21.117). The authors want to thank Leo Vleugels, Ynze Mengerink, Jan Jordens, and Harry Philipsen from DSM Materials Science Center for their support and valuable discussions during the project.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.9b04961.
Detailed description of Matlab script used for data analysis, the molecular weight calibration curve obtained on column 1 (Figure S1), values of chromatographic descriptors obtained during SEC-UV of myoglobin (Figure S2), SEC-ESI-MS data (Figure S3), fraction of folded protein measured over full peak width of myoglobin (Figure S4), the chromatograms and mass spectra obtained during SEC-ESI-MS of carbonic anhydrase and cytochrome C using two different columns (Figures S5), and the same data obtained on column 2 for myoglobin (Figure S6) (PDF)
Author Present Address
# MS-Omics, 2950 Vedbæk, Denmark. (D.B.A.M.)
Author Present Address
∇ ChemConnection B.V., Pivot Park, 5349 AB Oss, The Netherlands. (S.K.)
Author Present Address
○ M4I Institute, Faculty Health, Medicine & Life Sciences, University Maastricht, 6229 ER Maastricht, The Netherlands. (M.H.)
The authors declare no competing financial interest.
Supplementary Material
References
- Eftekharzadeh B.; Hyman B. T.; Wegmann S. Structural studies on the mechanism of protein aggregation in age related neurodegenerative diseases. Mech. Ageing Dev. 2016, 156, 1–13. 10.1016/j.mad.2016.03.001. [DOI] [PubMed] [Google Scholar]
- Hong P.; Koza S.; Bouvier E. S. A review size-exclusion chromatography for the analysis of protein biotherapeutics and their aggregates. J. Liq. Chromatogr. Relat. Technol. 2012, 35 (20), 2923–2950. 10.1080/10826076.2012.743724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn T. R.; Parker M. J.; Homans S. W.; Radford S. E. Amyloid formation under physiological conditions proceeds via a native-like folding intermediate. Nat. Struct. Mol. Biol. 2006, 13 (3), 195. 10.1038/nsmb1058. [DOI] [PubMed] [Google Scholar]
- Mahler H. C.; Friess W.; Grauschopf U.; Kiese S. Protein aggregation: pathways, induction factors and analysis. J. Pharm. Sci. 2009, 98 (9), 2909–2934. 10.1002/jps.21566. [DOI] [PubMed] [Google Scholar]
- Neupane R.; Källsten M.; Lehmann F.; Bergquist J. Effect of mobile phase composition on the analysis of aggregates of antibody drug conjugates (ADCs) using size exclusion chromatography. Anal. Methods 2018, 10 (9), 938–941. 10.1039/C7AY02696J. [DOI] [Google Scholar]
- Staub A.; Guillarme D.; Schappler J.; Veuthey J.-L.; Rudaz S. Intact protein analysis in the biopharmaceutical field. J. Pharm. Biomed. Anal. 2011, 55 (4), 810–822. 10.1016/j.jpba.2011.01.031. [DOI] [PubMed] [Google Scholar]
- Vetri V.; Foderà V. The route to protein aggregate superstructures: Particulates and amyloid-like spherulites. FEBS Lett. 2015, 589 (19), 2448–2463. 10.1016/j.febslet.2015.07.006. [DOI] [PubMed] [Google Scholar]
- Bertoletti L.; Schappler J.; Colombo R.; Rudaz S.; Haselberg R.; Domínguez-Vega E.; Raimondi S.; Somsen G. W.; De Lorenzi E. Evaluation of capillary electrophoresis-mass spectrometry for the analysis of the conformational heterogeneity of intact proteins using beta 2-microglobulin as model compound. Anal. Chim. Acta 2016, 945, 102–109. 10.1016/j.aca.2016.10.010. [DOI] [PubMed] [Google Scholar]
- Marie A.-L.; Dominguez-Vega E.; Saller F.; Plantier J.-L.; Urbain R.; Borgel D.; Tran N. T.; Somsen G. W.; Taverna M. Characterization of conformers and dimers of antithrombin by capillary electrophoresis-quadrupole-time-of-flight mass spectrometry. Anal. Chim. Acta 2016, 947, 58–65. 10.1016/j.aca.2016.10.016. [DOI] [PubMed] [Google Scholar]
- Fraunhofer W.; Winter G. The use of asymmetrical flow field-flow fractionation in pharmaceutics and biopharmaceutics. Eur. J. Pharm. Biopharm. 2004, 58 (2), 369–383. 10.1016/j.ejpb.2004.03.034. [DOI] [PubMed] [Google Scholar]
- Rambaldi D. C.; Reschiglian P.; Zattoni A. Flow field-flow fractionation: recent trends in protein analysis. Anal. Bioanal. Chem. 2011, 399 (4), 1439–1447. 10.1007/s00216-010-4312-5. [DOI] [PubMed] [Google Scholar]
- Reschiglian P.; Moon M. H. Flow field-flow fractionation: a pre-analytical method for proteomics. J. Proteomics 2008, 71 (3), 265–276. 10.1016/j.jprot.2008.06.002. [DOI] [PubMed] [Google Scholar]
- Fekete S.; Beck A.; Veuthey J.-L.; Guillarme D. Theory and practice of size exclusion chromatography for the analysis of protein aggregates. J. Pharm. Biomed. Anal. 2014, 101, 161–173. 10.1016/j.jpba.2014.04.011. [DOI] [PubMed] [Google Scholar]
- Heck A. J. Native mass spectrometry: a bridge between interactomics and structural biology. Nat. Methods 2008, 5 (11), 927–933. 10.1038/nmeth.1265. [DOI] [PubMed] [Google Scholar]
- Ruotolo B. T.; Robinson C. V. Aspects of native proteins are retained in vacuum. Curr. Opin. Chem. Biol. 2006, 10 (5), 402–408. 10.1016/j.cbpa.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Sterling H. J.; Cassou C. A.; Susa A. C.; Williams E. R. Electrothermal supercharging of proteins in native electrospray ionization. Anal. Chem. 2012, 84 (8), 3795–3801. 10.1021/ac300468a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Engelsman J.; Garidel P.; Smulders R.; Koll H.; Smith B.; Bassarab S.; Seidl A.; Hainzl O.; Jiskoot W. Strategies for the assessment of protein aggregates in pharmaceutical biotech product development. Pharm. Res. 2011, 28 (4), 920–933. 10.1007/s11095-010-0297-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakawa T.; Ejima D.; Li T.; Philo J. S. The critical role of mobile phase composition in size exclusion chromatography of protein pharmaceuticals. J. Pharm. Sci. 2010, 99 (4), 1674–1692. 10.1002/jps.21974. [DOI] [PubMed] [Google Scholar]
- Ehkirch A.; Hernandez-Alba O.; Colas O.; Beck A.; Guillarme D.; Cianférani S. Hyphenation of size exclusion chromatography to native ion mobility mass spectrometry for the analytical characterization of therapeutic antibodies and related products. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2018, 1086, 176–183. 10.1016/j.jchromb.2018.04.010. [DOI] [PubMed] [Google Scholar]
- Garcia M. The effect of the mobile phase additives on sensitivity in the analysis of peptides and proteins by high-performance liquid chromatography–electrospray mass spectrometry. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2005, 825 (2), 111–123. 10.1016/j.jchromb.2005.03.041. [DOI] [PubMed] [Google Scholar]
- Bouvier E. S.; Koza S. M. Advances in size-exclusion separations of proteins and polymers by UHPLC. TrAC, Trends Anal. Chem. 2014, 63, 85–94. 10.1016/j.trac.2014.08.002. [DOI] [Google Scholar]
- Brusotti G.; Calleri E.; Colombo R.; Massolini G.; Rinaldi F.; Temporini C. Advances on size exclusion chromatography and applications on the analysis of protein biopharmaceuticals and protein aggregates: a mini review. Chromatographia 2018, 81 (1), 3–23. 10.1007/s10337-017-3380-5. [DOI] [Google Scholar]
- Goyon A.; D’Atri V.; Colas O.; Fekete S.; Beck A.; Guillarme D. Characterization of 30 therapeutic antibodies and related products by size exclusion chromatography: Feasibility assessment for future mass spectrometry hyphenation. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2017, 1065, 35–43. 10.1016/j.jchromb.2017.09.027. [DOI] [PubMed] [Google Scholar]
- Haberger M.; Leiss M.; Heidenreich A.-K.; Pester O.; Hafenmair G.; Hook M.; Bonnington L.; Wegele H.; Haindl M.; Reusch D.; et al. In Rapid characterization of biotherapeutic proteins by size-exclusion chromatography coupled to native mass spectrometry. MAbs 2016, 8, 331–339. 10.1080/19420862.2015.1122150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kükrer B.; Filipe V.; van Duijn E.; Kasper P. T.; Vreeken R. J.; Heck A. J.; Jiskoot W. Mass spectrometric analysis of intact human monoclonal antibody aggregates fractionated by size-exclusion chromatography. Pharm. Res. 2010, 27 (10), 2197–2204. 10.1007/s11095-010-0224-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar A. C.; Wang L.; Blättler W. A.; Amphlett G.; Lambert J. M.; Zhang W. Analysis of the composition of immunoconjugates using size-exclusion chromatography coupled to mass spectrometry. Rapid Commun. Mass Spectrom. 2005, 19 (13), 1806–1814. 10.1002/rcm.1987. [DOI] [PubMed] [Google Scholar]
- Woodard J.; Lau H.; Latypov R. F. Nondenaturing size-exclusion chromatography-mass spectrometry to measure stress-induced aggregation in a complex mixture of monoclonal antibodies. Anal. Chem. 2013, 85 (13), 6429–6436. 10.1021/ac401455f. [DOI] [PubMed] [Google Scholar]
- Muneeruddin K.; Thomas J. J.; Salinas P. A.; Kaltashov I. A. Characterization of small protein aggregates and oligomers using size exclusion chromatography with online detection by native electrospray ionization mass spectrometry. Anal. Chem. 2014, 86 (21), 10692–10699. 10.1021/ac502590h. [DOI] [PubMed] [Google Scholar]
- Lakayan D.; Haselberg R.; Gahoual R.; Somsen G. W.; Kool J. Affinity profiling of monoclonal antibody and antibody-drug-conjugate preparations by coupled liquid chromatography-surface plasmon resonance biosensing. Anal. Bioanal. Chem. 2018, 410 (30), 7837–7848. 10.1007/s00216-018-1414-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck A. J.; van den Heuvel R. H. Investigation of intact protein complexes by mass spectrometry. Mass Spectrom. Rev. 2004, 23 (5), 368–389. 10.1002/mas.10081. [DOI] [PubMed] [Google Scholar]
- Lemaire D.; Marie G.; Serani L.; Laprévote O. Stabilization of gas-phase noncovalent macromolecular complexes in electrospray mass spectrometry using aqueous triethylammonium bicarbonate buffer. Anal. Chem. 2001, 73 (8), 1699–1706. 10.1021/ac001276s. [DOI] [PubMed] [Google Scholar]
- Mortensen D. N.; Williams E. R. Electrothermal supercharging of proteins in native MS: effects of protein isoelectric point, buffer, and nanoESI-emitter tip size. Analyst 2016, 141 (19), 5598–5606. 10.1039/C6AN01380E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash H.; Kansara B. T.; Mazumdar S. Effects of salts on the charge-state distribution and the structural basis of the most-intense charge-state of the gaseous protein ions produced by electrospray ionization. Int. J. Mass Spectrom. 2010, 289 (2), 84–91. 10.1016/j.ijms.2009.09.013. [DOI] [Google Scholar]
- Kopaciewicz W.; Regnier F. Nonideal size-exclusion chromatography of proteins: effects of pH at low ionic strength. Anal. Biochem. 1982, 126 (1), 8–16. 10.1016/0003-2697(82)90102-6. [DOI] [PubMed] [Google Scholar]
- Babu K. R.; Douglas D. Methanol-induced conformations of myoglobin at pH 4.0. Biochemistry 2000, 39 (47), 14702–14710. 10.1021/bi001265t. [DOI] [PubMed] [Google Scholar]
- Feng R.; Konishi Y. Stepwise refolding of acid-denatured myoglobin: Evidence from electrospray mass spectrometry. J. Am. Soc. Mass Spectrom. 1993, 4 (8), 638–645. 10.1016/1044-0305(93)85028-V. [DOI] [PubMed] [Google Scholar]
- Cassou C. A.; Williams E. R. Anions in electrothermal supercharging of proteins with electrospray ionization follow a reverse Hofmeister series. Anal. Chem. 2014, 86 (3), 1640–1647. 10.1021/ac403398j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges J. B.; Vahidi S.; Yue X.; Konermann L. Effects of ammonium bicarbonate on the electrospray mass spectra of proteins: evidence for bubble-induced unfolding. Anal. Chem. 2013, 85 (13), 6469–6476. 10.1021/ac401020s. [DOI] [PubMed] [Google Scholar]
- May J. C.; Jurneczko E.; Stow S. M.; Kratochvil I.; Kalkhof S.; McLean J. A. Conformational landscapes of ubiquitin, cytochrome c, and myoglobin: Uniform field ion mobility measurements in helium and nitrogen drift gas. Int. J. Mass Spectrom. 2018, 427, 79–90. 10.1016/j.ijms.2017.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen D. N.; Williams E. R. Investigating protein folding and unfolding in electrospray nanodrops upon rapid mixing using theta-glass emitters. Anal. Chem. 2015, 87 (2), 1281–1287. 10.1021/ac503981c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šamalikova M.; Matečko I.; Müller N.; Grandori R. Interpreting conformational effects in protein nano-ESI-MS spectra. Anal. Bioanal. Chem. 2004, 378 (4), 1112–1123. 10.1007/s00216-003-2339-6. [DOI] [PubMed] [Google Scholar]
- Graf M.; García R. G.; Wätzig H. Protein adsorption in fused-silica and polyacrylamide-coated capillaries. Electrophoresis 2005, 26 (12), 2409–2417. 10.1002/elps.200410360. [DOI] [PubMed] [Google Scholar]
- Kunz W.; Henle J.; Ninham B. W. ‘Zur Lehre von der Wirkung der Salze’(about the science of the effect of salts): Franz Hofmeister’s historical papers. Curr. Opin. Colloid Interface Sci. 2004, 9 (1), 19–37. 10.1016/j.cocis.2004.05.005. [DOI] [Google Scholar]
- Ohta K.; Tanaka K. Ion chromatographic separation of common mono-and divalent cations on an unmodified silica gel column by elution with oxalic acid containing crown ethers. Analyst 1999, 124 (4), 505–510. 10.1039/a808382g. [DOI] [Google Scholar]
- Honarvar E.; Venter A. R. Ammonium bicarbonate addition improves the detection of proteins by desorption electrospray ionization mass spectrometry. J. Am. Soc. Mass Spectrom. 2017, 28 (6), 1109–1117. 10.1007/s13361-017-1628-9. [DOI] [PubMed] [Google Scholar]
- Merenbloom S. I.; Flick T. G.; Daly M. P.; Williams E. R. Effects of select anions from the Hofmeister series on the gas-phase conformations of protein ions measured with traveling-wave ion mobility spectrometry/mass spectrometry. J. Am. Soc. Mass Spectrom. 2011, 22 (11), 1978. 10.1007/s13361-011-0238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okur H. I.; Hladilkova J.; Rembert K. B.; Cho Y.; Heyda J.; Dzubiella J.; Cremer P. S.; Jungwirth P. Beyond the Hofmeister series: Ion-specific effects on proteins and their biological functions. J. Phys. Chem. B 2017, 121 (9), 1997–2014. 10.1021/acs.jpcb.6b10797. [DOI] [PubMed] [Google Scholar]
- Susa A. C.; Mortensen D. N.; Williams E. R. Effects of cations on protein and peptide charging in electrospray ionization from aqueous solutions. J. Am. Soc. Mass Spectrom. 2014, 25 (6), 918–927. 10.1007/s13361-014-0864-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann L. Addressing a common misconception: Ammonium acetate as neutral pH “buffer” for native electrospray mass spectrometry. J. Am. Soc. Mass Spectrom. 2017, 28 (9), 1827–1835. 10.1007/s13361-017-1739-3. [DOI] [PubMed] [Google Scholar]
- Felsovalyi F.; Mangiagalli P.; Bureau C.; Kumar S. K.; Banta S. Reversibility of the adsorption of lysozyme on silica. Langmuir 2011, 27 (19), 11873–11882. 10.1021/la202585r. [DOI] [PubMed] [Google Scholar]
- Mathé C.; Devineau S.; Aude J.-C.; Lagniel G.; Chédin S.; Legros V.; Mathon M.-H.; Renault J.-P.; Pin S.; Boulard Y.; et al. Structural determinants for protein adsorption/non-adsorption to silica surface. PLoS One 2013, 8 (11), e81346. 10.1371/journal.pone.0081346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerz S. T.; Huber P. pH-dependent selective protein adsorption into mesoporous silica. J. Phys. Chem. C 2015, 119 (48), 27072–27079. 10.1021/acs.jpcc.5b09606. [DOI] [Google Scholar]
- Stutz H. Protein attachment onto silica surfaces–a survey of molecular fundamentals, resulting effects and novel preventive strategies in CE. Electrophoresis 2009, 30 (12), 2032–2061. 10.1002/elps.200900015. [DOI] [PubMed] [Google Scholar]
- Rabiller-Baudry M.; Chaufer B. Specific adsorption of phosphate ions on proteins evidenced by capillary electrophoresis and reversed-phase high-performance liquid chromatography. J. Chromatogr., Biomed. Appl. 2001, 753 (1), 67–77. 10.1016/S0378-4347(00)00431-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.