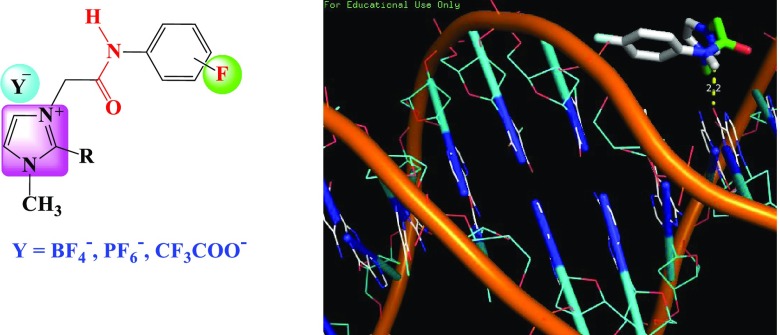
Abstract
Newer imidazolium ionic liquid (IL) halides 4a–f appending variety of fluorinated phenylacetamide side chains were designed and synthesized through quaternization of 1-methyl and/or 1,2-dimethylimidazole with appropriate 2-chloro-N-(fluorinatedphenyl)acetamides. The resulting ILs were converted to their respective ionic liquid analogues carrying fluorinated counteranions (PF6–, BF4–, and/or CF3COO–) 5a–r. All newly synthesized ILs were fully characterized using several spectroscopic experiments such as 1H, 13C, 11B, 19F, 31P NMR, and mass analysis. The synthesized ionic liquids were investigated for their DNA binding and anticancer activities. The obtained DNA binding constants ranged from 1.444 × 105 to 3.518 × 105, indicating a reasonably good binding affinity. The percentage of anticancer activities ranged from 48 to 59 with H-1229 cell line, showing quite good anticancer potential. The modeling studies indicated the interactions of the reported molecules with DNA via hydrogen bonds. These were in agreement with those of DNA binding and anticancer results. Briefly, the designed ionic liquids may be used as good anticancer candidates for treating human cancer.
1. Introduction
The drug discovery is at the cutting edge of the most promising medicinal chemistry. The drug design is at a crossroad, facing growing strategies for the synthesis of new active pharmaceutical ingredients (APIs).1 These strategies faced several challenges in the development of such scaffolds for effective drug delivery. These challenges are further exacerbated when drug compounds resulted from the combination of simple and active moieties with unique and tunable physicochemical and biological properties. Consequently, the development of potent anticancer agents is a major trend in drug discovery efforts in medicinal chemistry.1,2 Ionic liquids (ILs) have been a topic of great interest in organic synthesis owing to their potential pharmaceutical properties and hold an important challenge in medicinal chemistry, especially in the race to synthesize new therapeutic agents or active pharmaceutical ingredients (APIs) tethered such moieties.3 Generally, ILs are synthesized by combining organic cations such as imidazolium, pyridinium, ammonium, guanidinium, and phosphonium4 with a wide variety of anions including halides (Cl–, Br–), hexafluorophosphate (PF6–), tetrafluoroborate (BF4–), trifluoroacetate (CF3COO–), bis(trifluoromethylsulfonyl)amide (NTf2), and dicyanamide (DCA).5 These classes of ILs are well known as tunable molecules with unique physicochemical properties including low flammability, extremely low vapor pressure at room temperature, high ionic conductivity, and high thermal and chemical stabilities.6 By modifying the cations and anions with special functional groups, all these properties may be adjustable with fascinating applications such as antiviral,7 antibacterial,8 antifungal,9 anti-inflammatory,10 and anticancer11 activities.
In addition, several studies have been devoted to the use of ILs as antitumor agents against several human cancer cells such as breast,12 brain,13 colon,14 lung,15 liver,16 osteosarcomas,17 leukemia, and prostate.18 In view of encouraging observations and as a continuation of our interest in the development of novel functionalized bioactive ionic liquids,19−27 we have anticipated the synthesis of novel imidazolium-ionic liquids carrying fluorinated phenylacetamide. The synthesized ILs were tested with DNA binding and screened for their anticancer activities. Additionally, simulation studies were also investigated to determine the anticancer mechanism.
2. Results and Discussion
2.1. Chemistry
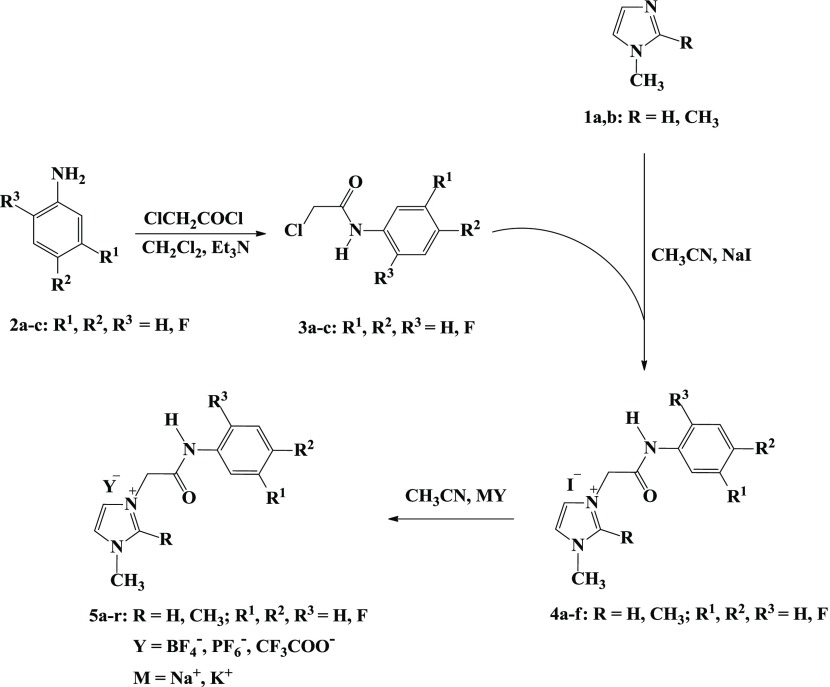
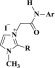
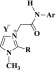
A more holistic approach to design the desired imidazolium ionic liquids 4a–f and 5a–r is described in Scheme 1 and comprises quaternization and metathesis reactions.
Scheme 1. Synthesis of Ionic Liquids Bearing Imidazole Ring and Fluorinated Phenylacetamide Linkages 4a–f and 5a–r.
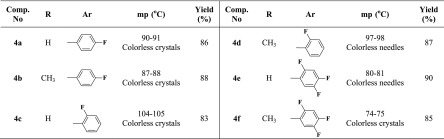
Thus, the quaternization of sp2 nitrogen atom of the substituted imidazoles 1 and/or 2 was carried out through their thermal alkylation by some aromatic acetamide chlorides 3a–c for 2 h to afford the halogenated IL-based imidazolium–amide hybrids 4a–f in 83–90% yields (Table 1).
Table 1. Physical and Analytical Data for the Imidazolium IL Halides 4a–f.
It should be noted that the fluorinated phenyl acetamide precursors 3a–c have been synthesized via base-assisted nucleophilic acylation of the appropriate fluorinated anilines with chloroacetyl chloride using triethylamine as a basic catalyst and dichloromethane as a solvent.
The structures of the resulted imidazolium iodides 4a–f were elucidated based on their spectroscopic data. Their 1H NMR spectra showed clearly the appearance of two distinct singlets at 4.64–5.29 and 10.39–10.77 ppm assigned to the NCH2 and NHCO protons, which confirmed the success of the quaternization reaction. All the remaining protons were recorded in their respective area (see Experimental Section). In addition, the 13C NMR spectra were in agreement with the designed structures. They exhibited new signals at 163.75–164.99 and 50.54–51.56 ppm belonging to the acetamide carbonyl (NHCO) and methylene (NCH2) carbons, respectively.
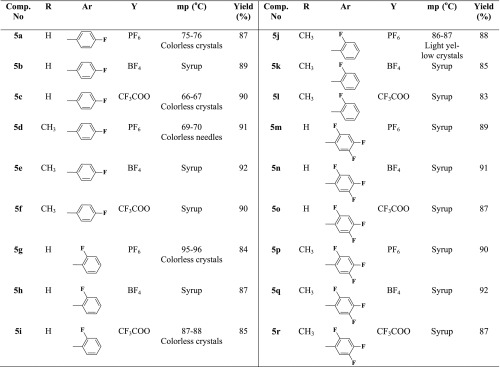
The resulting imidazolium iodides 4a–f underwent a metathetical anion exchange via their treatment with appropriate fluorinated metal salts in acetonitrile furnishing on the elaboration of the desired task-specific imidazolium ionic liquids incorporating specific fluorinated anions (BF4–, PF6–, and CF3COO–) as counteranions 5a–r (Scheme 1).
The structures of the obtained ILs 5a–r were deduced from their spectroscopic data. It is noticeable that no changes were recorded on the signals that appeared in their 1H and 13C NMR spectra. This confirmed that the exchange occurred only on the counteranion. Thus, the presence of PF6– anion was evidenced by the 31P and 19F NMR spectra. The presence of a characteristic septet between −157.43 and −131.04 ppm in 31P spectra, as well as a diagnostic doublet ranging from −69.18 to −69.16 ppm in their 19F NMR spectra, supported the success of the PF6– anion exchange (Table 2).
Table 2. Physical and Analytical Data of the Imidazolium ILs Carrying Fluorinated Counteranions 5a–r.
On the other hand, the incorporation of tetrafluroborate (BF4–) anions on the specific imidazolium ionic liquids was evidenced based on their 31B and 19F NMR spectral data. Their 31B NMR data showed a diagnostic multiplet near −1.32 to −1.29 ppm, but their 19F NMR spectra exhibited two characteristic doublets around −148.23 to −148.14 ppm. However, the structures of the imidazolium ionic liquids, bearing the trifluoroacetate (CF3COO–) as a counter anion, were deduced based on their 19F NMR spectra and showed a distinct singlet around −73.67 to −73.52 ppm attributed to such an anion. Additionally, all the spectra showed variable multiplets ranging from −141.60 to −118.31 ppm belonging to the aromatic fluorine atoms of the substituted phenyl rings (see Experimental Section).
2.2. DNA Binding Study
DNA is among the most significant pharmaceutical targets of anticancer medications.28−30 Thus, studying the interactions of target compounds with DNA is important to get an indication about their anticancer impacts and imaginable mechanisms of action. In general, a compound and DNA formed covalent and noncovalent bonds. Through covalent bonding, a labile compound is switched by a nitrogen atom of the DNA base, such as N7 of guanine, in which the interactions like electrostatic, intercalation, and groove binding are conceivable in non-covalent binding.31 Meanwhile, The change in the wavelength or absorption or both is characteristic of the interactions and intercalative modes including strong stacking interactions between DNA base pairs and aromatic chromophores.32 It is supposed that bathochromism means breakage of the secondary structure of DNA and hyperchromism involves covalent binding. Moreover, considerable red shifts revealed that the compounds were coordinated to DNA via N7 position of guanine.33 On the other hand, no or slight shifts in UV spectra are indicative of an outside groove binding, generally with minor hyperchromicity.34
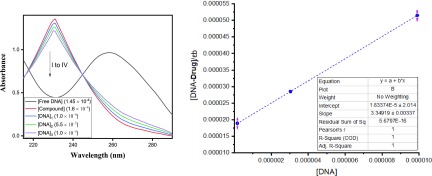
In this study, the absorption bands of the tested ionic liquids 4a–f and 5a–r are recorded in the absence and presence of DNA. The spectra for 4a are shown in Figure 1 as the representative one. The UV–vis spectral data for ILs 4a–f and 5a–r are summarized in Table 3. The DNA spectra for the remaining compounds are provided in the Supporting Information. Obviously, the absorption spectra of the compounds revealed the presence of absorption bands around 200 nm. The trivial shifts of the bands were assigned to 240–320 nm by adding DNA owing to the intraligand π → π* and n → π* transitions.35,36 These minor shifts of the bands pointed to bathochromic shifts of all the compounds, which confirmed their interactions with DNA. Additionally, hypochromaticities were recorded for all the compounds with the addition of DNA in different concentrations (1.45 × 10–4 to 1.0 × 10–5 M). Such changes are obvious evidence of the formation of DNA adducts.37 It is believed that these hypochromic shifts are probably due to the covalent and noncovalent bonds, which were observed in all target compounds.38
Figure 1.
DNA binding study of compound 4a.
Table 3. UV–Vis Data for Compounds 4a–f and 5a–ra.
| comp. no. | Af | Ab | Δλmax (nm) | % hypochromism | Kb (M–1) |
|---|---|---|---|---|---|
| 4a | 1.311 | 1.123 | 0.188 | 14 | 1.825 × 105 |
| 4b | 2.068 | 2.154 | 0.086 | 4 | 2.124 × 105 |
| 4c | 1.594 | 1.422 | 0.172 | 10.7 | 2.547 × 105 |
| 4d | 1.384 | 1.204 | 0.18 | 13 | 2.568 × 105 |
| 4e | 0.553 | 0.503 | 0.05 | 9 | 1.5245 × 105 |
| 4f | 0.029 | 0.025 | 0.004 | 13.7 | 2.356 × 105 |
| 5a | 1.359 | 1.145 | 0.214 | 15.7 | 2.739 × 105 |
| 5b | 0.13 | 0.113 | 0.017 | 13 | 1.832 × 105 |
| 5c | 0.120 | 0.132 | 0.012 | 1 | 1.506 × 105 |
| 5d | 0.218 | 0.226 | 0.008 | 3.6 | 1.444 × 105 |
| 5e | 0.028 | 0.029 | 0.001 | 3.5 | 5.566 × 104 |
| 5f | 2.350 | 2.261 | 0.089 | 3.7 | 1.693 × 105 |
| 5g | 1.520 | 1.424 | 0.096 | 6 | 2.5 × 105 |
| 5h | 1.745 | 1.549 | 0.196 | 11 | 1.928 × 105 |
| 5i | 0.987 | 0.882 | 0.105 | 10 | 2.716 × 105 |
| 5j | 0.023 | 0.026 | 0.003 | 13 | 2.40 × 105 |
| 5k | 0.997 | 0.945 | 0.052 | 5 | 1.829 × 105 |
| 5l | 1.265 | 1.153 | 0.112 | 8.8 | 1.935 × 105 |
| 5m | 0.449 | 0.469 | 0.02 | 4 | 1.802 × 105 |
| 5n | 0.591 | 0.523 | 0.068 | 11.5 | 4.543 × 104 |
| 5o | 1.126 | 1.074 | 0.052 | 4.6 | 3.518 × 105 |
| 5p | 0.145 | 0.150 | 0.005 | 3 | 2.303 × 105 |
| 5q | 0.141 | 0.151 | 0.01 | 7 | 3.109 × 105 |
| 5r | 1.549 | 1.305 | 0.244 | 15.7 | 3.032 × 105 |
% Hypochromism (H%) = [change in λmax/Af] × 100, where Af and Ab are the absorbances of free and bound compounds, respectively, Kb = binding constants, λf = λmax (free), λb = λmax (bound to DNA).
Furthermore, the values of DNA binding constants for all ILs range from 4.543 × 104 to 3.518 × 105, signifying a good interaction with DNA. The regression analysis was investigated by origin software for DNA binding readings. The resulted correlation coefficient (R2) for each compound is also given in the respective figure and showed that almost all compounds interpolated through the minor groove with Ct-DNA. These results are bolstered by the accessible literature.39 The literature referred that the compounds forming adducts with DNA through minor grooves and hydrophobic interactions as well as hydrogen bonds make major contribution adducts.40,41 All these facts are well supported by the simulation studies.
2.3. Anticancer Study
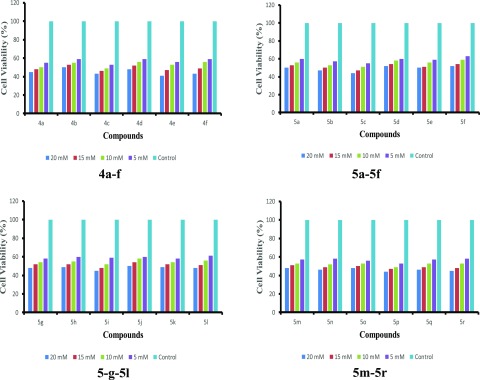
In vitro percentage inhibitions of lung cancer cell line H-1229 were used to assess the anticancer profiles of the tested ILs at 5.0, 10.0, 15.0, and 20.0 mM concentrations. The results are represented in Figure 2.
Figure 2.
Anticancer profiles of the reported molecules with H-1229 cell line.
Initially, the tested molecules 4a–f, 5a–5f, 5-g–5l, and/or 5m–5r were dissolved in DMSO (0.1%), and natural cell by DMSO was used as vehicle control. The resulting mixtures were accumulated until the control cells touched an inactive phase. After 24 h, the cells were counted. The proliferation inhibitions were clearly enhanced with the increasing amount of compounds. The results exhibited a noteworthy inhibition in cancer cell proliferation. The maximum percentage of inhibitions were in the range of 48–59%. The inhibition ranges of 4a–f, 5a–5f, 5-g–5l, and 5m–5r compounds were 50–59, 48–56, 50–55, and 52–56%, respectively. The percentage inhibitions were almost the same in all four series. The variations in the percentage inhibition were observed with the dissociation tendencies of the reported compounds. It was observed that highly dissociated molecules exhibited higher anticancer activities. This is probably because highly dissociate molecules provided a high concentration of positive ionic liquid parts to the DNA, leading to the higher binding with DNA and hence more activity. In the same manner, it was observed that bulky molecules showed low anticancer activities due to the steric effect. Thus, large-size molecules might have restricted their interactions with DNA. Overall, the total percentage inhibitions of 48–59% are quite good, and the reported molecules may be considered as potential anticancer agents for lung cancer.
2.4. Docking Study
It is important to mention here that out of 24 compounds, the basic organic moieties are 6 in 4a–f. It is very clear that DNA docking study is carried out with organic moieties. Therefore, the docking study was carried out with six moieties of 4a–f molecules. The obtained binding energies of the reported imidazolium ILs with DNA are summarized in Table 4 and were in the range from −3.9 to −4.4 kcal/mol. It is noticeable that the binding energies were variable due to the structure variation of the tested imidazolium compounds. Also, these structural differences also caused variation in hydrogen bond formation with DNA. Hydrogen bondings were found responsible for six moieties with DNA binding one hydrogen bond was formed between DNA and compounds 4a, 4d, and 4e, while two hydrogen bonds were formed between DNA and compound 4b. However, three hydrogen bonds were formed between DNA and compound 4c. The hydrophobic interactions were also responsible for DNA interaction with the reported compounds. All the hydrophobic interactions with DNA are shown in Table 4. It was found that the DNA fragments involved with imidazolium were dc9, dc13, dc15, dg10, dg14, dc15, and dt8. The representative model of DNA binding with compound 4a is shown in Figure 3, while the rest are given in the Supporting Information. On the contrary, the docked results clearly showed that all tested compounds reacted with DNA minor grooves. The docking model in Figure 3 clearly indicates DNA binding via DNA minor grooves. The DNA interaction process revealed that the orientation of all compounds was the same and such that their active sites were inside minor grooves. The DNA binding results were in agreement with the docking results.
Table 4. Simulation Studies Results of the Reported Compounds with DNA.
| comp. no. | no. of H bonds | bond length (Å) | affinity in (kcal/mol) | hydrophobic interaction |
|---|---|---|---|---|
| 4a | 1 | .263/A/DG′10/O6 & ‘H’ of −CONH- gp. (2.2) | –4.4 | C1::dc15&dc9, C2::dc9, C3::dt8&dc9, C4::dc9, C5::O6, C8::dc15&dg14, C10::dg14&dc15, C12::O, N3::O6 |
| 4b | 2 | .263/A/DG′10/O6 & ‘H’ of −CONH- gp. (2.4) | –4.3 | C2::dg14&O6, C4::dg14, C5::O6, C7::dc15, C8::dc15, C9::dc15, C10::dg14&dc15, C13::O6, N2::dc15, O::dc9 |
| .127/A/DC′9/N4 & ‘O’ of −CONH- gp. (3.4) | ||||
| 4c | 3 | .263/A/DG′10/O6 & ‘H’ of −CONH- gp. (2.3) | –4.4 | C2::dt8&dc9, C3::dc9, C5::O6, C6::dg14, C7::dg14, C8::dc15, C10::dc15, C12::dc15&O6, N2::dc15 |
| .263/A/DG′10/O6 & ‘O’ of −CONH- gp. (3.3) | ||||
| .218/B/DG′14/N7 & ‘O’ of −CONH- (3.4) | ||||
| 4d | 1 | .263/A/DG′10/O6 & ‘H’ of −CONH- gp. (2.5) | –3.9 | C3::dc9, C4::dt8&dc9, C6::O6, C7::dg14, C8::dg14, C11::dc9&O6, C13::O6, N3::dg10&O6 |
| 4e | 1 | .263/A/DG′10/O6 & ‘H’ of −CONH- gp. (2.1) | –4.4 | C1::dc9&dc15, C3::O6, C6::dg14&dc15, C8::dg14&dc15, C9::dc9&dt8, C10::dt8, C12::O6 |
| 4f | 2 | .263/A/DG′10/O6 & ‘H’ of −CONH- gp. (2.6) | –4.3 | C1::dg14&O6, C4::O6, C5::dc15, C6::dc15, C7::dc15, C8::dc15&dg14, C10::dg14, C13::dc9&O6, N2::dc15 |
| .127/A/DC′9/N4 & ‘O’ of −CONH- gp. (3.4) |
Figure 3.
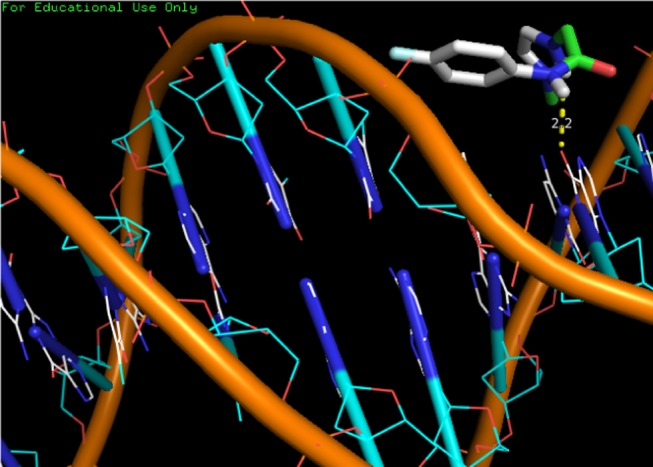
Docking model of compound 4a with DNA.
3. Conclusions
Ionic liquids are gaining importance as anticancer molecules. Therefore, novel imidazolium ionic liquid halides tethering different fluorinated phenylacetamide side chains were prepared by quaternization of 1-methyl and/or 1,2-dimethylimidazole with appropriate 2-chloro-N-(fluorinatedphenyl)acetamides. These were converted to their ionic liquid analogues having fluorinated counteranions (PF6–, BF4–, and/or CF3COO–). All newly synthesized ILs were characterized by various spectroscopic methods. The synthesized compounds were studied for DNA binding and anticancer activities with H-1229 cell line. Besides, the modeling studies were also carried out to investigate the DNA interaction mechanism of these molecules. The results of DNA binding, modeling, and anticancer activities indicated good interactions of these classes of compounds with DNA. Finally, it was concluded that these ionic liquids may be used as excellent anticancer candidates for treating human cancer.
4. Experimental Section
4.1. General Methods
Melting points were measured with a Stuart Scientific SMP1 apparatus and were uncorrected. All synthesized compounds were fully characterized by 1H, 13C, 19F, 31P, and 11B NMR spectroscopy and HRMS. TLC was performed on aluminum plates silica gel (Kieselgel, 0.25 mm, 60 F254, Merck, Germany) and spots were visualized by ultraviolet (UV) light absorption using ethyl acetate/hexane as a developing solvent system. The NMR spectra were recorded with a 400 MHz Bruker NMR spectrometer. Chemical shifts (δ) were expressed in ppm using tetramethylsilane (TMS) as an internal standard. High-resolution mass spectroscopy (HRMS) was carried out using an LC-MS/MS impact II.
4.2. Synthesis and Characterization of 2-Chloro-N-(substituted phenyl)acetamides 3a–c
Chloroacetyl chloride (12 mmol) was added dropwise under stirring to a solution of appropriate fluorinated aniline 2a–c (10 mmol) and triethylamine (12 mmol) in dichloromethane (40 mL) for 2 h at 0 °C. Then, the stirring was continued at room temperature for 6 h. The obtained precipitate was filtered, washed with water, and recrystallized from ethanol to afford the desired phenylacetamides 3a–c.
4.2.1. 2-Chloro-N-(4-fluorophenyl)acetamide (3a)
Colorless crystals, mp: 128–129 °C (lit. mp: 130 °C).421H NMR (400 MHz, DMSO-d6): δH = 4.34 (s, 2H, NCH2), 7.55–7.60 (m, 2H, Ar-H), 7.79 (d, J = 8.0 Hz, 2H, Ar-H), 10.38 (s, 1H, CONH). 13C NMR (100 MHz, DMSO-d6): δC = 43.82 (NCH2), 116.25, 121.97, 124.51, 135.68, 147.26, 158.70 (Ar-C), 164.87 (C=O). 19F NMR (377 MHz, DMSO-d6): δF = −118.57 to −118.50 (m, 1F, Ar-F).
4.2.2. 2-Chloro-N-(2-fluorophenyl)acetamide (3b)
Colorless crystals, mp: 119–121 °C (lit. mp: 120–122 °C).431H NMR (400 MHz, DMSO-d6): δH = 4.41 (s, 2H, NCH2), 7.21–7.30 (m, 3H, Ar-H), 7.82–7.90 (m, 1H, Ar-H), 10.24 (s, 1H, CONH). 13C NMR (100 MHz, DMSO-d6): δC = 43.5 (NCH2), 116.36, 123.24, 125.23, 135.67, 146.32, 155.35 (Ar-C), 165.16 (C=O). 19F NMR (377 MHz, DMSO-d6): δF = −124.74 to −124.67 (m, 1F, Ar-F).
4.2.3. 2-Chloro-N-(2,4,5-trifluorophenyl)acetamide (3c)
Colorless crystals, mp: 116–117 °C (lit. mp: 118–119 °C).261H NMR (400 MHz, DMSO-d6): δH = 4.34 (s, 2H, NCH2), 7.66–7.70 (m, 1H, Ar-H), 7.94–8.02 (m, 1H, Ar-H), 10.30 (s, 1H, CONH). 13C NMR (100 MHz, DMSO-d6): δC = 43.46 (NCH2), 106.65, 112.46, 123.12, 144.90, 147.08, 147.28, 150.67 (Ar-C), 165.85 (C=O). 19F NMR (377 MHz, DMSO-d6): δF = −141.76 to −141.62 (m, 1F, Ar-F), −139.11 to −139.04 (m, 1F, Ar-F), −125.60 to −125.51 (m, 1F, Ar-F).
4.3. General Quaternization Procedure for the Synthesis of Imidazolium IL Halides 4a–f
2-Chloro-N-(4-fluorophenyl)acetamide, 2-chloro-N-(4-fluorophenyl)acetamide, and/or 2-chloro-N-(2,4,5-trifluorophenyl)acetamide 3a–c (10 mmol) were added with stirring to a solution of 1-methyl and/or 1,2-dimethyl imidazole 1a,b (10 mmol) in acetonitrile (30 mL). Then, the mixture was refluxed for 2 h, until the completion of the reaction, as indicated by TLC (silicagel, hexane-ethyl acetate). The excess of the solvent was removed under reduced pressure. The obtained product was filtered and/or extracted by chloroform to yield the desired ionic liquids 4a–f.
NB: Sodium iodide (10 mmol) was added during the synthesis of compounds 4a, 4b, 4e, and 4f.
The characterization of the imidazolium ILs 4a–f is reported in the Supporting Information.
4.4. General Metathesis Procedure for the Synthesis of Imidazolium ILs Carrying Fluorinated Counteranions 5a–r
To a mixture of imidazolium IL halides 4a–f (1 mmol) in acetonitrile (20 mL) was added suitable metal salts: potassium hexafluorophosphate, sodium tetrafluoroborate, and/or sodium trifluoroacetate (1.1 mmol) at room temperature under stirring. The reaction mixture was heated under reflux for 24 h. Then, the reaction mixture was cooled and filtered to eliminate solid metal halide. The acetonitrile was evaporated to afford quantitatively the desired imidazolium ionic liquids 5a–r.
The characterization of the imidazolium ILs 5a–r is reported in the Supporting Information.
4.5. DNA Binding Studies
The DNA binding experiments were carried out in Tris buffer (10–2 M, pH 7.2). Initially, the concentration of DNA was adjusted by recording the absorption spectrum of the CT-DNA solution. This solution showed UV absorbance at 230/260 nm (=1.8), indicating the free nature of DNA, with ε value equal to 6600 M–1 cm–1.44 The different solutions of the tested compounds and DNA were prepared and kept at 4 °C. First, the compounds were dissolved in DMSO (0.1%) and then diluted with Tris buffer till their concentrations became 2.4 × 10–4 M. Then, DNA was added in various concentrations (1.2–1.5 × 10–5 M) to record the respected absorption. Eventually, the binding constants (Kb) were calculated according to the Benessi–Hilderbrand eq 1.45
| 1 |
where absorption coefficients εa, εf, and εb are related to Aobs/[compound], extinction coefficient for the compounds in free as well in fully bound form. The binding constants (Kb) for different compounds were determined from slopes and the intercepts of the plots of
4.6. Anticancer Studies
The antiproliferative activity of the target imidazolium ILs was performed against lung cancer cell line H-1229 using the MTT assay. Thus, the cells were seeded (density of 8000 cells/well) in a 96-well plate and incubated. Around 60–70% confluency, the treatment of the cells with different concentrations of the reported compounds (5.0, 10.0, 15, and 20.0 mM) was carried out, followed by incubation for 24 h. The cells were assayed by adding 15.0 μL of 5.0 mg/mL MTT. Then, the respective media from each well were aspirated at 37 °C of incubation for 4 h. In 100 μL of DMSO, the cells were resuspended and the plate was instantly covered with aluminum foil and shook gently for 15 min. Absorbance was measured at 540 nm46 and the percent inhibition of cellular proliferation was calculated using eq 2.
| 2 |
4.7. Docking Study
The docking readings were conducted using Windows XP operating system on Intel dual CPU (1.86 GHz). Marwin sketch was used in the drawing of the ligand 3D structure, which was transformed to the PDB file format. Ligand preparation was achieved by using AutoDock Tool (ADT) 4.0 version to allocate Gastegier charges, nonpolar hydrogens were merged, and then saved in PDBQT file format.47 X-ray DNA crystal structure (PDB ID: 1bna) was retrieved from the Protein Data Bank (http://www.rcsb.org/pdb). The receptors were saved in the PDB file format using AutoDock Tools (ADT) 4.0, leaving heteroatoms (water). Besides, Gastegier charges were allocated to the receptor and then saved employing previous tools in the PDBQT file format. Preparation of parameter files for grid and docking was performed by means of ADT. Docking was carried out with AutoDock 4.0 (Scripps Research Institute) considering conformationally all the rotatable bonds of the ligand as rotatable and receptor as rigid.41 Grid box size of 60 × 80 × 114 Å3 with 0.375 Å spacing was used that encompassed the whole DNA. Macromolecule docking was done using an empirical-free energy function and Lamarckian genetic algorithm (LGA), with an initial population of 150 randomly placed individuals, a maximum number of 2 500 000 energy assessments, a crossover rate of 0.80, and a mutation rate of 0.02. For each ligand and serum–ligand adduct, at least 50 independent docking runs were performed for the lowest free energy of binding confirmation from the largest cluster and then saved in PDBQT format.
Acknowledgments
The authors thank the Chemistry Department at Taibah University for providing the facilities in sample analysis.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.9b03468.
Characterization of all newly synthesized compounds as well their respected NMR and mass spectra; DNA binding results and docking model of all newly synthesized compounds (PDF)
Author Contributions
All authors discussed the results and commented on the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Taylor J. B.; Kennewell P. D.; Horwood E. Modern Medicinal Chemistry. Trends Pharmacol. Sci. 1994, 15, 350–351. 10.1016/0165-6147(94)90033-7. [DOI] [Google Scholar]
- Patil S. A. A Role of Medicinal Chemist in the Modern Drug Discovery and Development. Org. Chem.: Curr. Res. 2012, 1, 1–2. 10.4172/2161-0401.1000e110. [DOI] [Google Scholar]
- a Ferraz R.; Branco L. C.; Prudêncio C.; Noronha J. P.; Petrovski Ž. Ionic Liquids as Active Pharmaceutical Ingredients. Chem. Med. Chem. 2011, 6, 975–985. 10.1002/cmdc.201100082. [DOI] [PubMed] [Google Scholar]; b Kellar R.; Nieto N. C.; Koppisch A.; Del Sesto R.. Ionic liquids that sterilize and prevent biofilm formation in skin wound healing devices. U.S. Patent US20180093011A1, Sept 19, 2019.
- Marrucho I. M.; Branco L. C.; Rebelo L. P. N. Ionic Liquids in Pharmaceutical Applications. Annu. Rev. Chem. Biomol. Eng. 2014, 5, 527–546. 10.1146/annurev-chembioeng-060713-040024. [DOI] [PubMed] [Google Scholar]
- Balk A.; Holzgrabe U.; Meinel L. ‘Pro et contra’ ionic liquid drugs-Challenges and opportunities for pharmaceutical translation. Eur. J. Pharm. Biopharm. 2015, 94, 291–304. 10.1016/j.ejpb.2015.05.027. [DOI] [PubMed] [Google Scholar]
- a Egorova K. S.; Gordeev E. G.; Ananikov V. P. Biological activity of ionic liquids and their application in pharmaceutics and medicine. Chem. Rev. 2017, 117, 7132–7189. 10.1021/acs.chemrev.6b00562. [DOI] [PubMed] [Google Scholar]; b Shamshina J. L.; Berton P.; Wang H.; Zhou X.; Gurau G.; Rogers R. D.. Ionic Liquids in Pharmaceutical Industry. In Green Techniques for Organic Synthesis and Medicinal Chemistry, 2nd ed.; Zhang W.; Cue B. W., Eds.; Wiley, 2018. [Google Scholar]; c MacFarlane D. R.; Kar M.; Pringle J. M.. Fundamentals of Ionic Liquids from Chemistry to Application; Wiley-VCH, 2017. [Google Scholar]; d McKee M. A.Ionic liquids for agricultural residue removal. U.S. Patent US20140128626A1, May 8, 2014.; e Qu J.; Luo H.. Corrosion prevention of magnesium surfaces via surface conversion treatments using ionic liquids. U.S. Patent US20150090369A1, April 2, 2015.; f Wu B.; Holbrey J. D.; Reddy R. G.; Rogers R. D.. Ionic liquid temperature sensor. U.S. Patent US6749336B2, June 15, 2004.
- Kumar V.; Malhotra S. V. Synthesis of nucleoside-based antiviral drugs in ionic liquids. Bioorg. Med. Chem. Lett. 2008, 18, 5640–5642. 10.1016/j.bmcl.2008.08.090. [DOI] [PubMed] [Google Scholar]
- Anvari S.; Hajfarajollah H.; Mokhtarani B.; Enayati M.; Sharifi A.; Mirzaei M. Antibacterial and Anti-Adhesive Properties of Ionic Liquids with Various Cationic and Anionic Heads toward Pathogenic Bacteria. J. Mol. Liq. 2016, 221, 685–690. 10.1016/j.molliq.2016.05.093. [DOI] [Google Scholar]
- Ranjan P.; Kitawat B. S.; Sing M. 1-Butylimidazolederived ionic liquids: synthesis, characterisation and evaluation of their antibacterial, antifungal and anticancer activities. RSC Adv. 2014, 4, 53634–53644. 10.1039/C4RA08370A. [DOI] [Google Scholar]
- Azevedo A. M. O.; Costa S. P. F.; Diasa A. F. V.; Marquesa A. H. O.; Pintoa P. C. A. G.; Bicac K.; Ressmannc A. K.; Passosa M. L. C.; Araújoa A. R. T. S.; Reise S.; Saraiva M. L. M. F. S. Antiinflammatory choline based ionic liquids: Insights into their lipophilicity, solubility and toxicity parameters. J. Mol. Liq. 2017, 232, 20–26. 10.1016/j.molliq.2017.02.027. [DOI] [Google Scholar]
- Dias A. R.; Costa-Rodrigues J.; Fernandes M. H.; Ferraz R.; Prudêncio C. Anti-cancer potential of Ionic Liquids. Chem. Med. Chem. 2017, 12, 11–18. 10.1002/cmdc.201600480. [DOI] [PubMed] [Google Scholar]
- Kumar V.; Malhotra S. V. Study on the potential anticancer activity of phosphonium and ammonium-based ionic liquids. Bioorg. Med. Chem. Lett. 2009, 19, 4643. 10.1016/j.bmcl.2009.06.086. [DOI] [PubMed] [Google Scholar]
- Kaushik N. K.; Attri P.; Kaushik N.; Choi E. H. Synthesis and Antiproliferative Activity of Ammonium and Imidazolium Ionic Liquids against T98G Brain Cancer Cells. Molecules 2012, 17, 13727–13739. 10.3390/molecules171213727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frade R. F. M.; Matias A.; Branco L. C.; Afonso C. A. M.; Duarte C. M. M. Effect of Ionic Liquids on Human Colon Carcinoma HT-29 and CaCo-2 Cell Lines. Green Chem. 2007, 9, 873–877. 10.1039/b617526k. [DOI] [Google Scholar]
- Chen H.-L.; Kao H.-F.; Wang J.-Y.; Wei G.-T. Cytotoxicity of Imidazole Ionic Liquids in Human Lung Carcinoma A549 Cell Line. J. Chin. Chem. Soc. 2014, 61, 763–769. 10.1002/jccs.201300632. [DOI] [Google Scholar]
- Torrecilla J. S.; García J.; Rojo E.; Rodríguez F. Estimation of toxicity of ionic liquids in Leukemia Rat Cell Line and Acetylcholinesterase enzyme by principal component analysis, neural networks and multiple lineal regressions. J. Hazard. Mater. 2009, 164, 182–194. 10.1016/j.jhazmat.2008.08.022. [DOI] [PubMed] [Google Scholar]
- Ferraz R.; Costa-Rodrigues J.; Fernandes M. H.; Santos M. M.; Marrucho I. M.; Rebelo L. P. N.; Prudêncio C.; Noronha J. P.; Petrovski Z.; Branco L. C. Antitumor Activity of Ionic Liquids Based on Ampicillin. Chem. Med. Chem. 2015, 10, 1480–1483. 10.1002/cmdc.201500142. [DOI] [PubMed] [Google Scholar]
- Bubalo M. C.; Radošević K.; Srček V. G.; Das R. N.; Popelier P.; Roy K. Cytotoxicity towards CCO Cells of Imidazolium Ionic Liquids with Functionalized Side Chains: Preliminary QSTR Modeling Using Regression and Classification Based Approaches. Ecotoxicol. Environ. Saf. 2015, 112, 22–28. 10.1016/j.ecoenv.2014.10.029. [DOI] [PubMed] [Google Scholar]
- Rezki N.; Al-Sodies S. A.; Aouad M. R.; Bardaweel S.; Messali M.; El Ashry E. H. An eco-friendly ultrasound-assisted synthesis of novel fluorinated pyridinium salts-based hydrazones and antimicrobial and antitumor screening. Int. J. Mol. Sci. 2016, 17, 766–785. 10.3390/ijms17050766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezki N.; Al-Sodies S. A.; Shreaz S.; Shiekh R. A.; Messali M.; Raja V.; Aouad M. R. Green ultrasound versus conventional synthesis and characterization of specific task pyridinium ionic liquid hydrazones tethering fluorinated counter anions: Novel inhibitors of fungal Ergosterol biosynthesis. Molecules 2017, 22, 1532–1552. 10.3390/molecules22111532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aljuhani A.; El-Sayed W. S.; Sahu P. K.; Rezki N.; Aouad M. R.; Salghi R.; Messali M. Microwave-assisted synthesis of novel imidazolium, pyridinium and pyridazinium-based ionic liquids and/or salts and prediction of physico-chemical properties for their toxicity and antibacterial activity. J. Mol. Liq. 2018, 249, 747–753. 10.1016/j.molliq.2017.11.108. [DOI] [Google Scholar]
- Rezki N.; Messali M.; Al-Sodies S. A.; Naqvi A.; Bardaweel S. K.; Al-blewi F. F.; Aouad M. R.; El Ashry E. H. Design, Synthesis, in-silico molecular docking and evaluation of di-cationic pyridinium ionic liquids as potential anticancer scaffolds. J. Mol. Liq. 2018, 265, 428–441. 10.1016/j.molliq.2018.06.045. [DOI] [Google Scholar]
- Rezki N.; Al-Sodies S. A.; Messali M.; Bardaweel S. K.; Sahu P. K.; Al-blewi F. F.; Sahu P. K.; Aouad M. R. Identification of new pyridinium ionic liquids tagged Schiff base: Design, synthesis, in silico ADMET prediction and biological evaluation. J. Mol. Liq. 2018, 264, 367–374. 10.1016/j.molliq.2018.05.071. [DOI] [Google Scholar]
- Albalawi A. H.; El-Sayed W. S.; Aljuhani A.; Almutairi S. M.; Rezki N.; Aouad M. R.; Messali M. Microwave assisted synthesis of some potential bioactive imidazolium-based room temperature ionic liquids. Molecules 2018, 23, 1727–1741. 10.3390/molecules23071727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Blewi F. F.; Rezki N.; Al-Sodies S. A.; Bardaweel S. K.; Sabbah D. A.; Messali M.; Aouad M. R. Novel cationic amphiphilic fluorinated pyridinium hydrazones: conventional versus green ultrasound assisted synthesis, characterization, molecular docking, and anticancer evaluation. Chem. Cent. J. 2018, 12, 118 10.1186/s13065-018-0489-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aljuhani A.; Aouad M. R.; Rezki N.; Aljaldy O. A.; Al-Sodies S. A.; Messali M.; Ali I. Novel pyridinium based ionic liquids with amide tethers: Microwave assisted synthesis, molecular docking and anticancer studies. J. Mol. Liq. 2019, 285, 790–802. 10.1016/j.molliq.2019.04.143. [DOI] [Google Scholar]
- Al-blewi F. F.; Rezki N.; Bardaweel S. K.; Naqvi A.; Qutb eddn H. A. B.; Al-Sodies S. A.; Messali M.; Aouad M. R. A profile of the in vitro anti-tumor activity against breast and colon cancer cell lines of novel specific ionic liquids based imidazole, benzothiazole conjugates. Int. J. Mol. Sci. 2019, 20, 2865–2885. 10.3390/ijms20122865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wani W. A.; Al-Othman Z.; Ali I.; Saleem K.; Hsieh M. F. Copper (II), nickel (II), and ruthenium (III) complexes of an oxopyrrolidine-based heterocyclic ligand as anticancer agents. J. Coord. Chem. 2014, 67, 2110–2130. 10.1080/00958972.2014.931947. [DOI] [Google Scholar]
- Ali I.; Wani W. A.; Saleem K.; Hsieh M.-F. Anticancer metallodrugs of glutamic acid sulphonamides: in silico, DNA binding, hemolysis and anticancer studies. RSC Adv. 2014, 4, 29629–29641. 10.1039/C4RA02570A. [DOI] [Google Scholar]
- Ali I.; Wani W. A.; Saleem K.; Hseih M.-F. Design and synthesis of thalidomide based dithiocarbamate Cu (II), Ni (II) and Ru (III) complexes as anticancer agents. Polyhedron 2013, 56, 134–143. 10.1016/j.poly.2013.03.056. [DOI] [Google Scholar]
- Ali I.; Haque A.; Saleem K.; Hsieh M.-F. Curcumin-I Knoevenagel’s condensates and their Schiff’s bases as anticancer agents: Synthesis, pharmacological and simulation studies. Bioorg. Med. Chem. 2013, 21, 3808–3820. 10.1016/j.bmc.2013.04.018. [DOI] [PubMed] [Google Scholar]
- a Shahabadi N.; Kashanian S.; Mahdavi M.; Sourinejad N. DNA interaction and DNA cleavage studies of a new platinum (II) complex containing aliphatic and aromatic dinitrogen ligands. Bioinorg. Chem. Appl. 2011, 2011, 525794 10.1155/2011/525794. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Roviello G. N.; Vicidomini C.; Di Gaetano S.; Capasso D.; Musumeci D.; Roviello V. Solid phase synthesis and RNA-binding activity of an arginine-containing nucleopeptide. RSC Adv. 2016, 6, 14140–14148. 10.1039/C5RA25809J. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Roviello G. N.; Musumeci D.; Moccia M.; Castiglione M.; Sapio R.; Valente M.; Bucci E. M.; Perretta G.; Pedone C. dabPna: Design, Synthesis, And Dna Binding Studies. Nucleosides, Nucleotides Nucleic Acids 2007, 26, 1307–1310. 10.1080/15257770701530640. [DOI] [PubMed] [Google Scholar]
- Erkkila K. E.; Odom D. T.; Barton J. K. Recognition and reaction of metallo intercalators with DNA. Chem. Rev. 1999, 99, 2777–2796. 10.1021/cr9804341. [DOI] [PubMed] [Google Scholar]
- Chen L. M.; Liu J.; Chen J. C.; Tan C. P.; Shi S.; Zheng K.-C.; Ji L. N. Synthesis, characterization, DNA-binding and spectral properties of complexes [Ru (L) 4 (dppz)] 2+(L = Im and MeIm). J. Inorg. Biochem. 2008, 102, 330–341. 10.1016/j.jinorgbio.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Pasternack R. F.; Gibbs E. J.; Villafranca J. J. Interactions of porphyrins with nucleic acids. Biochemistry 1983, 22, 5409–5417. 10.1021/bi00292a024. [DOI] [PubMed] [Google Scholar]
- Zhao P.; Xu L. C.; Huang J. W.; Liu J.; Yu H. C.; Zheng K. C.; Ji L. N. Experimental and DFT studies on DNA binding and photocleavage of two cationic porphyrins: effects of the introduction of a carboxyphenyl into pyridinium porphyrin. Spectrochim. Acta, Part A 2008, 71, 1216–1223. 10.1016/j.saa.2008.03.031. [DOI] [PubMed] [Google Scholar]
- Arjmand F.; Aziz M.; Chauhan M. Synthesis, spectroscopic studies of new water-soluble Co (II) and Cu (II) macrocyclic complexes of 4, 15-bis (2-hydroxyethyl)-2, 4, 6, 13, 15, 17-hexaazatricyclodocosane: their interaction studies with calf thymus DNA and guanosine 5′ monophosphate. J. Inclusion Phenom. Macrocyclic Chem. 2008, 61, 265–278. 10.1007/s10847-008-9417-5. [DOI] [Google Scholar]
- Sun Y.; Zhang H.; Bi S.; Zhou X.; Wang L.; Yan Y. Studies on the arctiin and its interaction with DNA by spectral methods. J. Lumin. 2011, 131, 2299–2306. 10.1016/j.jlumin.2011.04.036. [DOI] [Google Scholar]
- Küng A.; Pieper T.; Wissiack R.; Rosenberg E.; Keppler B. K. Hydrolysis of the tumor-inhibiting ruthenium (III) complexes HIm trans[RuCl4(im)2] and HInd trans-[RuCl4 (ind) 2] investigated by means of HPCE and HPLC-MS. JBIC, J. Biol. Inorg. Chem. 2001, 6, 292–299. 10.1007/s007750000203. [DOI] [PubMed] [Google Scholar]
- Bacac M.; Hotze A. C.; Van Der Schilden K.; Haasnoot J. G.; Pacor S.; Alessio E.; Sava G.; Reedijk J. The hydrolysis of the anti-cancer ruthenium complex NAMI-A affects its DNA binding and antimetastatic activity: an NMR evaluation. J. Inorg. Biochem. 2004, 98, 402–412. 10.1016/j.jinorgbio.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Lipinski C. A.; Lombardo F.; Dominy B. W.; Feeney P. J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Delivery Rev. 2012, 64, 4–17. 10.1016/j.addr.2012.09.019. [DOI] [PubMed] [Google Scholar]
- Kang S.; Zeng H.; Li H.; Wang H. 2-Chloro-N-(4-fluoro-phen-yl)acetamide. Acta Crystallogr., Sect. E: Struct. Rep. Online 2008, 64, o1194 10.1107/S1600536808016152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanja M. W.; Frederik D.; Christoph G.; Herwig H.; Inge L.; Karen R.; Andreas J. S.; Arnold E. S.; Chris A. T.; Stephen G. W.; Heidrun W. Probing the aglycon binding site of a β-glucosidase: a collection of C-1-modified 2,5-dideoxy-2,5-imino-d-mannitol derivatives and their structure-activity relationships as competitive inhibitors. Bioorg. Med. Chem. 2009, 19, 4643. 10.1016/j.bmc.2004.04.037. [DOI] [PubMed] [Google Scholar]
- Wolfe A.; Shimer G. H. Jr.; Meehan T. Polycyclic aromatic hydrocarbons physically intercalate into duplex regions of denatured DNA. Biochemistry 1987, 26, 6392–6396. 10.1021/bi00394a013. [DOI] [PubMed] [Google Scholar]
- Martin A.; Clynes M. Comparison of 5 microplate colorimetric assays for in vitro cytotoxicity testing and cell proliferation assays. Cytotechnology 1993, 11, 49–58. 10.1007/BF00749057. [DOI] [PubMed] [Google Scholar]
- Sanner M. F. Python: A programming language for software integration and development. J. Mol. Graphics Modell. 1999, 17, 57–61. 10.1007/BF00749057. [DOI] [PubMed] [Google Scholar]
- Huey R.; Morris G. M.; Olson A. J.; Goodsell D. S. A semiempirical free energy force field with charge-based desolvation. J. Comput. Chem. 2007, 1145–1152. 10.1002/jcc.20634. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.