Abstract
Inflammatory responses mediated by the transcription factor nuclear factor kappa-light-chain enhancer of activated B cells (NF-κB) play key roles in immunity, autoimmune diseases, and cancer. NF-κB is directly regulated through protein–protein interactions, including those with IκB and 14-3-3 proteins. These two important regulatory proteins have been reported to interact with each other, although little is known about this interaction. We analyzed the inhibitor of nuclear factor kappa B α (IκBα)/14-3-3σ interaction via a peptide/protein-based approach. Structural data were acquired via X-ray crystallography, while binding affinities were measured with fluorescence polarization assays and time-resolved tryptophan fluorescence. A high-resolution crystal structure (1.13 Å) of the uncommon 14-3-3 interaction motif of IκBα (IκBαpS63) in a complex with 14-3-3σ was evaluated. This motif harbors a tryptophan that makes this crystal structure the first one with such a residue visible in the electron density at that position. We used this tryptophan to determine the binding affinity of the unlabeled IκBα peptide to 14-3-3 via tryptophan fluorescence decay measurements.
Introduction
The κB-protein family is well known for their involvement in cell proliferation, cell survival, apoptosis, and regulation of the immune response.1−4 The family is divided into transcription factors, summarized as nuclear factor kappa-light-chain enhancer of activated B cells (NF-κB), and their inhibitors (IκB). NF-κB is sequestered by IκB in the cytosol, and upon activation, IκB is phosphorylated and degraded, while NF-κB is translocated into the nucleus. The transcriptional activity of NF-κB regulates over 500 genes, and many of those are involved in a variety of diseases.5 The most abundant representatives of this protein family are the heterodimer p50/p65 forming NF-κB and its inhibitor of nuclear factor kappa B α (IκBα), which is fundamental for the regulation of NF-κB activity.6
The most prominent regulatory pathway of IκBα is its phosphorylation at Ser32 and Ser36, which induces ubiquitination and proteasomal degradation of IκBα.7,8 Several studies reported IκBα as an essential component for the inhibition of the transcriptional activity of NF-κB,9−11 with the nuclear export of the IκBα/p65 complex depending on a third binding partner, the 14-3-3 proteins.12 Overexpression of 14-3-3 increases the IκBα amount in the cytosol, while mutation of the possible IκBα motifs for 14-3-3 binding leads to an increased nuclear signal for both IκBα and NF-κB (Figure 1A).12
Figure 1.
Phosphorylated IκBα peptide bound to 14-3-3. (A) Schematic representation of the key elements of the NF-κB pathway. In short, upon an inflammatory signal (here represented via TNFα and its receptor TNFR), a signaling cascade (thick dashed arrow) leads to proteasomal degradation of IκBα and translocation of NF-κB into the nucleus. Binding of 14-3-3 to IκBα and NF-κB is required for either nuclear export or cytosolic retention of the IκBα/NF-κB complex (red dashed arrow). (B) IκBα protein consists of an unstructured N-terminus, six helical ankyrin repeats, and the proline (P), glutamic acid (E), serine (S), and threonine (T) rich (PEST) domain. The 14-3-3 is supposed to bind to S63 at the unstructured N-terminal region. (C) Fluorescence polarization (FP) with the IκBαS63 peptide and all human 14-3-3 isoforms. (D) FP with the IκBαpS63 peptide and all human 14-3-3 isoforms.
The 14-3-3 proteins are dimeric scaffold proteins that interact with several hundred different proteins.13 The seven human isoforms (β, γ, ε, ζ, η, σ, and τ) are highly abundant in almost all human cells, and they exist as homo- or heterodimers.14 The 14-3-3 monomer consists of nine helices that form an amphipathic binding groove. By binding one or two interaction partners within the two binding grooves of the 14-3-3 dimer, the activity or cellular localization of the target proteins is altered.15 Interaction partners of 14-3-3 bind to 14-3-3 via distinguished phosphorylated motives in most often unstructured regions.16
Physical interaction of 14-3-3 and IκBα was demonstrated by co-immunoprecipitation, whereby the truncation analysis of IκBα suggested only one active 14-3-3 binding motif.12 This motif, 59-PRGSEPW-67, follows the 14-3-3 binding motif RXpSXP, although phosphorylation of S63 is not yet reported. Co-immunoprecipitation of the IκBα/14-3-3 complex in the presence and absence of phosphatases revealed no significant differences, indicating that the phosphorylation of this residue might not be required for binding.12 However, later studies showed that 14-3-3 binding protects its interaction partners from phosphatases,17−19 wherefore further investigations are needed to confirm the phosphorylation status of S63 of IκBα. Nevertheless, this region of IκBα is most likely unstructured and therefore compatible with 14-3-3 binding. Fortunately, the binding sequence of IκBα contains a Trp residue at position 66. An aromatic amino acid at this position, +3 from the phosphosite, is rare within the known 14-3-3 interactome, which makes this Trp especially interesting. It has been well documented that Trp is an environmentally sensitive fluorophore that is able to reflect even subtle changes in its interaction network by spectral shifts and changes in its quantum yield and fluorescence lifetime.20−22 The specific Trp response depends on a number of intramolecular and/or solvent interactions (e.g., Coulombic interaction, local polarity, H-bonding, Trp mobility, etc.), all of which depend on a detailed protein structure.23,24 Trp emission can be therefore used for monitoring binding-induced conformational transitions and variations of microenvironmental polarity. Generally, increased hydrophobicity leads to blue spectral shift and prolonged fluorescence lifetime of indole emission.25 We, therefore, utilized Trp66 fluorescence for the detection of interaction between IκBα peptides and the 14-3-3 protein.
In this study, the interaction of IκBα and 14-3-3 is characterized by a peptide-based approach. The binding of the IκBα peptide to 14-3-3 was measured by fluorescence polarization (FP) assay and the interface of IκBα and 14-3-3 was structurally characterized by X-ray crystallography. Furthermore, the intrinsic fluorescence of Trp66 of the peptide was used to establish a label-free method for the determination of binding affinities based on fluorescence decay. Our findings expand our knowledge of the IκBα/14-3-3 interaction and further provide a technique for the characterization of binding affinities.
Results
14-3-3 Binding Motif of IκBα
The IκBα protein contains only one 14-3-3 binding site, which follows a typical 14-3-3 binding motif, 57-PRGSEP-66. To investigate the binding affinity of 14-3-3 and IκBα, two peptides were synthesized, representing the unphosphorylated (IκBαS63) and phosphorylated (IκBαpS63) binding motifs (Figure 1B). In both peptides, the centered Ser63 is flanked by six amino acids on each side of the wild-type sequence of IκBα.
For the initial measurements of the binding affinities, the peptides were N-terminally labeled with a fluorescein isothiocyanate (FITC) dye and fluorescence polarization (FP) assays were performed for both peptides with all seven human 14-3-3 isoforms (Figure 1C,D). The unphosphorylated peptide showed no affinity to either of the seven isoforms, whereas the phosphorylated peptide showed binding. No peptide reached sufficient binding for a reliable calculation of dissociation constant (KD), although differences between the isoforms were detectable. The KD values of IκBαpS63 and 14-3-3 are expected to be in the high micromolar to millimolar range, with the best binding to 14-3-3η and the weakest binding to 14-3-3σ (Figure 1D).
This binding behavior was previously reported in the literature for peptides representing 14-3-3 binding motifs.26−28 In those cases, a second binding site was essential for sufficient representation of the bivalent binding to the 14-3-3 dimer. Therefore, we elaborated on a hypothetical presence of additional binding sites with the prediction server 14-3-3-Pred.29 This server analyzes the amino acid sequence of 14-3-3 interaction partners with three different methods, resulting in a score for each method plus an overall score. The IκBα sequence revealed only one site, pT168 (sequence: VLTQSCpT168TPHL; Supporting Information, Table 1), as a likely binding site. This residue is located next to the second ankyrin repeat of the IκBα protein (Supporting Information, Figure S1), and previous truncation assays did not reveal any effect on the 14-3-3 binding.12 Therefore, we continued with the single phosphorylated IκBαpS63 peptide only.
Crystal Structure of IκBα Peptide with 14-3-3σ with Truncated C-Terminus ΔC232-248 (14-3-3σΔC)
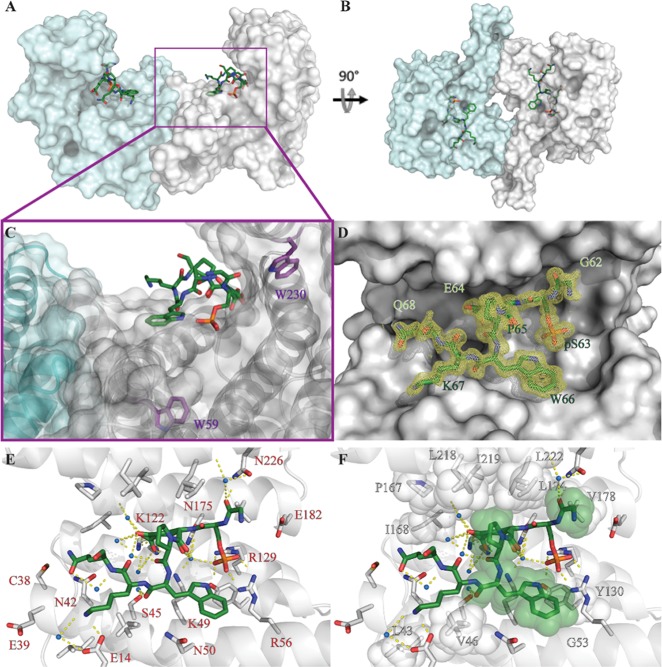
To obtain structural information on the binding interface, co-crystallization trials were performed. The complex of IκBαpS63 with 14-3-3σΔC was grown to the best diffracting crystals among all 14-3-3 isoforms, allowing to solve the structure with a high resolution of 1.13 Å (PDB ID: 6Y1J, Supporting Information, Table S2). The crystals belonged to the space group C2221, with one 14-3-3 monomer in the asymmetric unit. One peptide is binding in each of the two binding grooves of the 14-3-3 dimer (Figure 2A,B). The peptide is binding in the proximity of both tryptophan residues of 14-3-3, Trp59, and Trp230 (Figure 2C). In total, seven of the 13 amino acids of the peptide could be assigned to the electron density map (Figure 2D), whereas pSer63 till Trp66 is the most rigid amino acids with the lowest B factors (Supporting Information, Figure S2). Consistently with other 14-3-3/phosphopeptide structures Arg56, Arg129, and Tyr130 of the protein form polar contacts with the phospho group of IκBαpS63. Except for phosphoserine, the only additional direct polar contact detected by Pymol software appeared to be between Lys67 of the peptide and Glu14 of the protein (Figure 2E). Nevertheless, more polar contacts are mediated via waters, like Gly62 of the peptide with Asn226 and the backbone of Leu222 of the protein, Glu64 of the peptide and the backbone of Pro167 and Lys122 of the protein, and the backbone of Trp66 of the peptide and Ser45 of the protein. Additionally, there are hydrophobic contacts between Gly62 and Val178, Pro65 and Leu218, Ile219, Leu122, and Trp66, and the aliphatic chain of Lys46 and Gly53, Lys67 and Val46 (Figure 2F).
Figure 2.
Crystal structure of the interface of the IκBαpS63 peptide and 14-3-3σΔC. Side (A) and top (B) views of the surface of a 14-3-3 dimer, with IκBαpS63 shown as green sticks in the binding groove of each monomer. (C) Zoom-in view shows the transparent surface and cartoon display of 14-3-3 with Trp59 and Trp230 of 14-3-3σ in violet. (D) IκBαpS63 peptide with eight of 13 amino acids covered by the electron density map (yellow mesh, σ = 1, carve = 1.3 Å). (E) Polar contacts (yellow dashed lines) between IκBαpS63, 14-3-3σΔC, and the water shell (blue spheres). (F) Possible hydrophobic contacts indicated via sphere representation of hydrophobic residue PDB ID: 6Y1J.
Measurement of KD via Mean Tryptophan Fluorescence Lifetime
The crystal structures of IκBαpS63 and 14-3-3σΔC revealed extensive contacts between the Trp66 residue and the binding groove of 14-3-3, with the indole moiety being located in a more hydrophobic environment. This binding mode suggested that the mean fluorescence lifetime τmean of Trp66 might change upon peptide binding and can be therefore used to detect the binding of the peptide without an artificial fluorescence labeling. To separate the fluorescence of the peptide from the Trp fluorescence of 14-3-3ζ, we made 14-3-3ζ fluorescently silent by mutating its two tryptophan residues, Trp59 and Trp228 (equals Trp230 in 14-3-3σ), to phenylalanine (W59F and W228F double mutation of 14-3-3ζ (14-3-3ζnoW)). Then, only the fluorescence of the peptide is detected.
Indeed, time-resolved fluorescence decays clearly indicate a progressive formation of IκBαpS63/14-3-3ζnoW complex by a significant increase of the Trp66 lifetime upon 14-3-3ζnoW addition (Figure 3A). This concentration-dependent increase of τmean ranges from 3.42 ns for the IκBαpS63 alone to 4.30 ns at 763 μM 14-3-3ζnoW (Figure 3A). In contrast, the unphosphorylated IκBαS63 indicated a much weaker interaction with 14-3-3ζnoW (Figure 3B). This finding is consistent with the results of the polarization assay shown in Figure 1. The τmean for the peptide alone was 3.04 ns, while it increased to 3.27 ns at a maximum protein concentration of 640 μM, confirming that the binding sequence needs to be phosphorylated for efficient binding.
Figure 3.
Binding of IκBα peptides to 14-3-3 monitored by Trp66 fluorescence. Panels (A) and (B) represent the peak-normalized fluorescence decays of IκBαpS63 and IκBαp63, respectively, in the presence of different concentrations of 14-3-3ζnoW. (C) τmean of IκBαS63 (opened circles) and IκBαpS63 (closed circles) as a function of 14-3-3ζnoW concentration. Solid lines represent the best fit to the simple two-state single-binding-site model: EC50 = min + (max – min)·[x]/(KD + [x]), where [x] is a concentration of the titrant. (D) FP assay with 14-3-3ζnoW (circles) and 14-3-3ζ (squares) and FITC-labeled IκBαpS63 (closed) and IκBαS63 (open). Lines represent the best fit of peptide binding to 14-3-3ζnoW (solid) or 14-3-3ζ (dashed) to the equation described in (C).
The change in τmean due to different protein concentrations encouraged us to use this change for an estimation of the binding affinity of IκBαpS63 to 14-3-3ζnoW. Therefore, τmean was plotted as a function of the 14-3-3ζnoW concentration, which revealed a binding curve with incomplete saturation (Figure 3C). The data were fitted to a simple 1:1 binding model resulting in a KD value of about 370 ± 50 μM for the IκBαpS63/14-3-3ζnoW complex. It has to be noted that the KD values are slightly underestimated since the fitting model neglected the increased quantum yield of the Trp in the complex. To validate the obtained dissociation constant, FP assays with 14-3-3ζ and 14-3-3ζnoW were repeated with the highest possible protein concentrations and both are labeled as IκBαS63/pS63 peptides. The FP assay also showed incomplete saturation, but the fitting converged to the KD of about 380 ± 1 and 415 ± 30 μM for IκBα with 14-3-3ζ and 14-3-3ζnoW, respectively (Figure 3D). Although the accuracy of such a fitting might be questionable, the similarity of the fitted dissociation constants is promising. Fortunately, due to the comparable molecular weight of the phosphorylated and unphosphorylated peptide, the plateau of all FP curves in Figure 3D, defined by the overall rotational diffusion of the complex, should be very similar. This makes the comparison of KDs more reliable.
Within the uncertainty, the fluorescence decay data agree well with the fluorescence polarization data, whereas both techniques revealed KD values close to 400 μM for the analogous IκBαpS63/14-3-3ζnoW complex. Due to limits of protein solubility, very weak interaction, and thereof inability to reach saturation, the KD for the unphosphorylated complex was not reliably quantifiable.
Discussion
This study focuses on an extension of our knowledge of the IκBα interaction with 14-3-3 and establishing label-free measurements of dissociation constants via fluorescence decay on a difficult system. In the first part, we could validate the 14-3-3 binding motif of IκBα on the basis of an IκBα-peptide–14-3-3 interaction with the traditional FP assay. The biophysical assays showed that phosphorylation of the IκBα peptide is necessary for binding to 14-3-3, although the rather weak binding made a quantitative determination of affinity difficult. Regarding the bivalent binding mode due to the two phospho-accepting pockets of the 14-3-3 dimer, this low affinity is consistent with those of other 14-3-3 interactions. Several studies showed similar binding behaviors for single phosphorylated peptides, and double phosphorylated binding partners reached affinities in the low-micromolar to the high-nanomolar range.26,27 The transition from a peptide motif to a protein domain or even full-length protein–protein interaction is expected to considerably enhance the binding affinity. In the case of 14-3-3 PPIs, only a few studies succeeded in the analysis of the complex formation of full-length domains due to the binding of 14-3-3 to unstructured regions of its interaction partners.28,30−36 In the case of calcium/calmodulin-dependent protein kinase kinase 2 (CaMKK2), binding to 14-3-3 was established via unstructured peptide-like motifs, which could be crystallized in a complex with 14-3-3.33 Complementary small-angle X-ray scattering (SAXS) and binding affinity measurements showed that additional contacts were made with the outside of the 14-3-3 binding groove, while the peptide-like binding conformation in the binding groove was confirmed. Similarly, the crystal structure of the neutral trehalase of Saccharomyces cerevisiae (Nth1)/14-3-3 complex30 revealed that there are extensive interactions between 14-3-3 and Nth1 beyond those involving the 14-3-3 binding grooves. Another example of a biphosphorylated interaction partner of 14-3-3 is procaspase-2.28 In this case, both binding motifs were tested in fluorescence polarization assays, whereas only one site showed μM binding. However, both phosphorylation sites were necessary to establish a complex formation of the full-length proteins, while no complex formation could be observed for either single phosphorylated procaspase 2. These data suggest that an additional binding site is necessary for efficient binding of IκBα to 14-3-3. This can be either via a not yet identified 14-3-3 binding site of IκBα or a third binding partner that is simultaneously binding to IκBα and 14-3-3. A ternary complex formation has been reported among others for the A20/RAF proto-oncogene serine/threonine-protein kinase (C-Raf)/14-3-337 and glycogen synthase kinase-3 β (GSK3beta)/Tau/14-3-3 interactions.38 A high-affinity binding of the second binding site might even mediate binding of the unphosphorylated IκBαS63 motif,12 although the isolated motif needs to be phosphorylated to show activity in binding assays. For the IκBα/14-3-3 interaction, only one active binding motif could be identified via co-immunoprecipitation, and also the sequence analysis of the IκBα protein revealed only one additional possible, but in the structural context unlikely, binding site (pT168; Supporting Information, Table S1 and Figure S1). In this case, it is more likely that the additional binding partner NF-κB is essential to enhance the binding of IκBα to 14-3-3. Binding of 14-3-3 to the NF-κB subunit p65 and to IκBα is needed for sufficient export of the IκBα/NF-κB nuclear export or retention in the cytoplasm.12
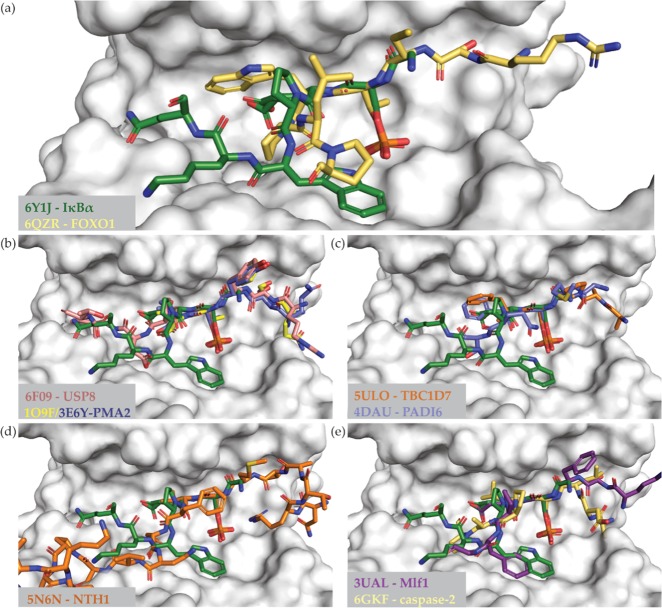
Noteworthy is also the binding mode of the IκBαpS63 peptide as Trp66 at the +3-position of the phosphorylated residue shows a remarkable conformation. Multiple 14-3-3 binding sequences contain aromatic residues; however, even within the constantly growing collection of peptides crystallized in complex with 14-3-3, only a small number contain tryptophan residues. Lately, the first crystal structure was published with a Trp residue, which was actually covered in electron density, located at the +1-position (Figure 4A).39 Two other examples of peptides binding to 14-3-3 and containing a tryptophan resides are the serine/threonine-protein kinase 26 (MST4) derived peptide (PDB ID: 5XY9) and the serine/threonine-protein kinase STK11 (LKB1) derived peptide (PDB ID: 5WXN). The tryptophan residue of MST4 is at the +4-position in relation to the phosphorylated residue, while that of LKB1 is at the −4-position; however, the indol moieties of both tryptophan residues were not visible in the electron density. That makes this study the second one with a Trp residue visible in a 14-3-3 crystal structure.
Figure 4.
Comparison of the IκBαpS63 binding mode with other binding 14-3-3 partners. (a) Only two interaction motifs of 14-3-3 hosting a tryptophan residue within the binding groove. All other displayed binding modes contain at least one aromatic residues at (b) −1-position, (c) +1-position, (d) +2-position, or (e) +3-position in relation to the phosphorylated residue. The 14-3-3 is shown as a white surface and the IκBαpS63 peptide as green sticks. Other binding partners are displayed as colored sticks with the associated PDB ID and the name of the interaction partner. For sequence comparison and Cα distances, see Supporting Information Table S3.
The amphipathic binding groove of 14-3-3 provides a lot of options for aromatic residues to bind (Figure 4). Multiple structures have been published with aromatic residues at the −1-position or +1-position of the phosphorylated residue, where they make contacts with the hydrophobic residues of the binding groove of 14-3-3 (Figure 4B,C).40−43 An aromatic residue at the +2-position is rare and for the structure shown, the interaction of the yeast Nth1 domain with 14-3-3, the phenylalanine does not make contact with 14-3-3 (Figure 4D).30 It is only stable in the context of the full domain, which is not shown for clarity. Actually, there is no other structure published with tryptophan at the +3-position in addition to the IκBαpS63. However, there are at least two structures available with a phenylalanine at that position, whereby for one the side chain was not visible in the electron density map (PDB ID: 6GKF; Figure 4E).44 Nevertheless, the peptide–protein interface of myeloid leukemia factor 1 (Mfl1) and 14-3-3ε comprises a phenylalanine at the +3-position with the same orientation as Trp66 of IκBαpS63 (PDB ID: 3UAL; Figure 4E).45 The conformation of the binding motifs deviates C-terminally from the phospho site more than they do N-terminally, whereas the Mfl1 motif shows the only comparable conformation. Measurements of the distances of the Cα atoms, Trp66 of IκBα, and phenylalanine of Mfl1 are 1.5 Å apart, while in all other structures, the residues at the +3-position are more than 2.6 Å apart (Supporting Information Table S3). This demonstrates that not only the binding groove in general but also the environment of the phosphorylation site can accommodate bulky hydrophobic residues like aromatic amino acids.
The tryptophan, clearly visible in the electron density, offers a nice opportunity to use its environmentally sensitive emission for direct biophysical affinity measurements without a need for a bulky artificial dye that could eventually bias interactions between IκBα peptides and 14-3-3. For KD quantification, we used the FP assay with externally labeled protein, which relies on changed hydrodynamic properties of the complex. In parallel, we applied intrinsic Trp fluorescence directly reflecting interaction-induced variations of the Trp microenvironment. For our system, both methods gave comparable results, proving the importance of the IκBα phosphorylation and yielding KD of the IκBαpS63/14-3-3ζnoW complex formation in the 10–4 M range.
Future research will further evaluate the binding of IκBα/p65 to 14-3-3 and analyze the contribution of this complex regarding the nuclear export mechanism of IκBα and p65.
Materials and Methods
Peptide Synthesis
Peptides were synthesized via Fmoc-solid-phase peptide synthesis with an automated Intavis MultiPep RSi peptide synthesizer with a Rink amide AM resin (Novabiochem; 0.59 mmol/g loading). After the synthesis was completed, the resin was split, with half of the peptide N-terminally acetylated (1:1:3 acetic anhydride (Ac2O)/pyridine/N-methyl-2-pyrrolidone (NMP)) and the other one isothiocyanate (FITC)-labeled via a β-alanine linker as described above.46 The peptides were cleaved with a 2.5:2.5:2.5:92.5 EDT/TIS/MQ/TFA mixture and purified with a preparative reversed-phase high-performance liquid chromatography (HPLC) system with mass spectrometry (MS) detection (Supporting Information, Figure S3). The purified peptide was lyophilized and stored at −30 °C.
14-3-3 Expression
The 14-3-3 isoforms were transiently expressed in Escherichia coli Nico21(DE3) or BL21(DE3) competent cells via a pPROEX HTb plasmid. The cells were grown to a density of OD600 = 0.8–1, and protein expression was induced with 0.4 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) for 18 h by 18 °C. The proteins were purified following standard procedures for Ni-affinity chromatography. For crystallography, the His6-Tag was removed by TEV cleavage and an additional Ni-NTA column, followed by size-exclusion chromatography ensuring the highest purity. All proteins were dialyzed against 25 mM N-(2-hydroxyethyl)piperazine-N′-ethanesulfonic acid (Hepes) pH 7.5, 100 mM NaCl, 10 mM MgCl2, 0.5 mM tris(2-carboxyethyl)phosphine, and stored at −80 °C. The exact mass of the proteins was confirmed via hybrid quadrupole time-of-flight (QTOF) measurements, whereby the number of peaks in the chromatogram indicated the purity of the protein (Supporting Information, Figure S4).
Crystallography
14-3-3σΔC232-248 and IκBαS63/pS63 complexes were mixed in a ratio of 1:2 in a final protein concentration of 12 mg/mL in crystallography buffer (20 mM Hepes pH 7.5, 2 mM MgCl2, 2 mM β-mercaptoethanol) and incubated overnight at 4 °C. In a hanging drop approach, the peptide/protein complex was mixed in a 1:1 ratio with a precipitation buffer (0.95 mM Hepes pH 7.1, 28% PEG400, 5% glycerol, 0.19 M CaCl2), crystals grew within one week. The crystals could be directly fished and flash-cooled in liquid nitrogen. The data set was measured at the Synchrotron DESY beamline P11, Hamburg, Germany.
For the indexing, integration, and molecular replacement, the CCP4 suite47 was used (iMosflm,48 Aimless,49,50 and MolRep51,52). As a search model for molecular replacement, PDB ID code 4FR3 was used. The refinement was done with phenix53 and the model building in Coot.54
Fluorescence Polarization (FP) Assay
The FP was measured with FITC-labeled peptides at a final concentration of 100 nM in fluorescence polarization buffer (10 mM Hepes, pH 7.4, 150 mM NaCl, 0.1% Tween 20, 1 mg/mL bovine serum albumin (BSA)). The 14-3-3 isoforms were titrated as indicated. All data sets were measured in at least three independent experiments in Corning black round-bottom 384-well plates with a Tecan Infinite F500 plate reader (excitation 485 nm, emission 535 nm).
Time-Resolved Tryptophan Fluorescence Measurements
Measurements of time-resolved Trp emission were performed on an apparatus comprised of a femtosecond Ti:sapphire laser (Chameleon Ultra II; Coherent) with the repetition rate reduced to 4 MHz by a pulse picker (APE). The fluorescence signal was collected using time-correlated single-photon counting detection SPC150 (Becker&Hickl) with a cooled MCP-PMT (R3809U-50; Hamamatsu). Trp fluorescence was excited at 298 nm by a tripled output of the laser and the emission was isolated under the “magic angle” conditions at 355 nm using a combination of a monochromator and a stack of UG1 and BG40 glass filters (Thorlabs) placed in front of the input slit. Fluorescence decays were typically accumulated in 1024 channels with a time resolution of 50 ps/channel until 105 counts in the decay maximum were reached. All experiments were performed at 23 °C in a buffer containing 20 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA), and 5 mM dithiothreitol (DTT). The IκBαS63/pS63 peptide and 14-3-3ζnoW concentrations were 1 and 0–763 μM, respectively. Fluorescence decays were analyzed by the model-independent maximum entropy method55 allowing accurate determination of the mean fluorescence lifetime τmean, calculated as τmean = ∑αi τi2/∑αiτi, where τi and αi are the fluorescence lifetime components and the corresponding amplitudes revealed by the analysis, respectively.
Acknowledgments
This study was supported by the Initial Training Network, funded by the H2020 Marie Curie Actions of the European Commission under Grant Agreement 675179. P.H. acknowledges the EU Operational Program No. CZ.1.05/4.1.00/16.0340 and the institutional support UNCE/SCI/010.
Glossary
Abbreviations
- 14-3-3ζnoW
W59F W228F double mutation of 14-3-3ζ
- 14-3-3σΔC
14-3-3σ with truncated C-terminus ΔC232-248
- A20, TNF α induced protein 3
also known as TNFAIP3
- CaMKK2
calcium/calmodulin-dependent protein kinase kinase 2
- C-Raf
RAF proto-oncogene serine/threonine-protein kinase
- DTT
dithiothreitol
- EDT
1,2-ethanedithiol
- EDTA
ethylenediaminetetraacetic acid
- FITC
fluorescein isothiocyanate
- FP
fluorescence polarization
- GSK3beta
glycogen synthase kinase-3 β
- IκBα
inhibitor of nuclear factor kappa B α
- KD
dissociation constant
- LKB1
serine/threonine-protein kinase STK11
- Mfl1
myeloid leukemia factor 1
- MST4
serine/threonine-protein kinase 26
- NF-κB
nuclear factor kappa-light-chain-enhancer of activated B cells
- Nth1
neutral trehalase of Saccharomyces cerevisiae
- PEST
proline (P), glutamic acid (E), serine (S), and threonine (T) rich sequence
- SAXS
small-angle X-ray scattering
- τmean
mean fluorescence life
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.9b04413.
Results of the 14-3-3pred webserver for IκBα (uniprot ID: P25963) (Table S1); crystal structure of IκBα (Figure S1); B-factor coloring of 14-3-3σΔC and the IκBαpS63 peptide (Figure S2); crystallographic statistics of the X-ray structure of the IκBαpS63/14-3-3σΔC232-248 complex (PDB ID: 6Y1J) (Table S2); quantitative comparison of 14-3-3 binding motifs (Table S3); LC–MS analysis of the in-house synthesized and purified peptides (Figure S3); and QTOF measurements of the purified 14-3-3 isoforms (Figure S4) (PDF)
Accession Codes
Given are the accession codes of UniProt. 14-3-3β: P31946 (1433B_HUMAN) 14-3-3γ: P61981 (1433G_HUMAN) 14-3-3ε: P62258 (1433E_HUMAN) 14-3-3ζ: P63104 (1433Z_HUMAN) 14-3-3η: Q04917 (1433F_HUMAN) 14-3-3σ: P31947 (1433S_HUMAN) 14-3-3τ: P27348 (1433T_HUMAN) IκBα: P25963 (IKBA_HUMAN)
Author Contributions
C.O. supervised the project and provided scientific guidance. T.O. supervised and provided scientific guidance for tryptophan fluorescence experiments. M.W. performed FP and crystallography. D.L.S. performed protein purification of 14-3-3noW and, together with P.H., fluorescence decay experiments. M.W., A.B., and F.C. performed peptide synthesis/purification and protein expression/purification. M.W. wrote the manuscript, followed by its revision by all. These authors contributed equally.
The authors declare no competing financial interest.
Supplementary Material
References
- Hayden M. S.; Ghosh S. NF-κB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev. 2012, 26, 203–234. 10.1101/gad.183434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viatour P.; Merville M.-P.; Bours V.; Chariot A. Phosphorylation of NF-κB and IκB proteins: implications in cancer and inflammation. Trends Biochem. Sci. 2005, 30, 43–52. 10.1016/j.tibs.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Ghosh S.; May M. J.; Kopp E. B. NF-κB AND REL PROTEINS: Evolutionarily Conserved Mediators of Immune Responses. Annu. Rev. Immunol. 1998, 16, 225–260. 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- Baldwin A. S. Jr. The NF-kappa B and I kappa B proteins: new discoveries and insights. Annu. Rev. Immunol. 1996, 14, 649–683. 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- Gupta S. C.; Sundaram C.; Reuter S.; Aggarwal B. B. Inhibiting NF-κB activation by small molecules as a therapeutic strategy. Biochim. Biophys. Acta, Gene Regul. Mech. 2010, 1799, 775–787. 10.1016/j.bbagrm.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulero M. C.; Bigas A.; Espinosa L. IκBα beyond the NF-kB dogma. Oncotarget 2013, 4, 1550–1551. 10.18632/oncotarget.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiDonato J.; Mercurio F.; Roette C.; Wu-Li J.; Suyang H.; Ghosh S.; Karin M. Mapping of the Inducible IκB Phosphorylation Sites That Signal Its Ubiquitination and Degradation. Mol. Cell. Biol. 1996, 16, 1295–1304. 10.1128/MCB.16.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonen H.; Bercovich B.; Orian A.; Carrano A.; Takizawa C.; Yamanaka K.; Pagano M.; Iwai K.; Ciechanover A. Identification of the Ubiquitin Carrier Proteins, E2s, Involved in Signal-induced Conjugation and Subsequent Degradation of IκBα. J. Biol. Chem. 1999, 274, 14823–14830. 10.1074/jbc.274.21.14823. [DOI] [PubMed] [Google Scholar]
- Simeonidis S.; Liang S.; Chen G.; Thanos D. Cloning and functional characterization of mouse IκBε. Proc. Natl. Acad. Sci. U.S.A. 1997, 94, 14372–14377. 10.1073/pnas.94.26.14372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malek S.; Chen Y.; Huxford T.; Ghosh G. IκBβ, but not IκBα, functions as a classical cytoplasmic inhibitor of NF-κB dimers by masking both NF-κB nuclear localization sequences in resting cells. J. Biol. Chem. 2001, 267, 45225–45235. 10.1074/jbc.M105865200. [DOI] [PubMed] [Google Scholar]
- Tran K.; Merika M.; Thanos D. Distinct Functional Properties of IκBα and IκBε. Mol. Cell. Biol. 1997, 17, 5386–5399. 10.1128/MCB.17.9.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera C.; Fernández-Majada V.; Inglés-Esteve J.; Rodilla V.; Bigas A.; Espinosa L. Efficient nuclear export of p65-IκBα complexes requires 14-3-3 proteins. J. Cell Sci. 2016, 129, 2472. 10.1242/jcs.192641. [DOI] [PubMed] [Google Scholar]
- Johnson C.; Tinti M.; Wood N. T.; Campbell D. G.; Toth R.; Dubois F.; Geraghty K. M.; Wong B. H. C.; Brown L. J.; Tyler J.; Gernez A.; Chen S.; Synowsky S.; MacKintosh C. Visualization and Biochemical Analyses of the Emerging Mammalian 14-3-3-Phosphoproteome. Mol. Cell. Proteomics 2011, 10, M110.005751 10.1074/mcp.M110.005751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H.; Subramanian R. R.; Masters S. C. 14-3-3 Proteins: Structure, Function, and Regulation. Annu. Rev. Pharmacol. Toxicol. 2000, 40, 617–647. 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- Aitken A. 14-3-3 proteins: A historic overview. Semin. Cancer Biol. 2006, 16, 162–172. 10.1016/j.semcancer.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Yaffe M. B.; Rittinger K.; Volinia S.; Caron P. R.; Aitken A.; Leffers H.; Gamblin S. J.; Smerdon S. J.; Cantley L. C. The Structural Basis for 14-3-3:Phosphopeptide Binding Specificity. Cell 1997, 91, 961–971. 10.1016/S0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- Palmer D.; Jimmo S. L.; Raymond D. R.; Wilson L. S.; Carter R. L.; Maurice D. H. Protein Kinase A Phosphorylation of Human Phosphodiesterase 3B Promotes 14-3-3 Protein Binding and Inhibits Phosphatase-catalyzed Inactivation. J. Biol. Chem. 2007, 282, 9411–9419. 10.1074/jbc.M606936200. [DOI] [PubMed] [Google Scholar]
- Sun L.; Stoecklin G.; Way S. V.; Hinkovska-Galcheva V.; Guo R.-F.; Anderson P.; Shanley T. P. Tristetraprolin (TTP)-14-3-3 Complex Formation Protects TTP from Dephosphorylation by Protein Phosphatase 2a and Stabilizes Tumor Necrosis Factor-α mRNA. J. Biol. Chem. 2007, 282, 3766–3777. 10.1074/jbc.M607347200. [DOI] [PubMed] [Google Scholar]
- Zhang J.; Chen F.; Li W.; Xiong Q.; Yang M.; Zheng P.; Li C.; Pei J.; Ge F. 14-3-3ζ Interacts with Stat3 and Regulates Its Constitutive Activation in Multiple Myeloma Cells. PLoS One 2012, 7, e29554 10.1371/journal.pone.0029554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obsilova V.; Nedbalkova E.; Silhan J.; Boura E.; Herman P.; Vecer J.; Sulc M.; Teisinger J.; Dyda F.; Obsil T. The 14-3-3 Protein Affects the Conformation of the Regulatory Domain of Human Tyrosine Hydroxylase. Biochemistry 2008, 47, 1768–1777. 10.1021/bi7019468. [DOI] [PubMed] [Google Scholar]
- Herman P.; Lee J. C. Functional Energetic Landscape in the Allosteric Regulation of Muscle Pyruvate Kinase. 2. Fluorescence Study. Biochemistry 2009, 48, 9456–9465. 10.1021/bi900280u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman P.; Vecer J.; Scognamiglio V.; Staiano M.; Rossi M.; D’Auria S. A Recombinant Glutamine-Binding Protein from Escherichia coli: Effect of Ligand-Binding on Protein Conformational Dynamics. Biotechnol. Prog. 2004, 20, 1847–1854. 10.1021/bp049956u. [DOI] [PubMed] [Google Scholar]
- Eftink M. R.Intrinsic Fluorescence of Proteins. In Topics in Fluorescence Spectroscopy; Lakowicz J. R., Ed.; Springer: US, Boston, MA, 2000; Vol. 6, pp 1–15. [Google Scholar]
- Principles of Fluorescence Spectroscopy; Lakowicz J. R., Ed.; Springer: US, Boston, MA, 2006. [Google Scholar]
- Gryczynski I.; Wiczk W.; Johnson M. L.; Lakowicz J. R. Lifetime distributions and anisotropy decays of indole fluorescence in cyclohexane/ethanol mixtures by frequency-domain fluorometry. Biophys. Chem. 1988, 32, 173–185. 10.1016/0301-4622(88)87005-4. [DOI] [PubMed] [Google Scholar]
- Stevers L. M.; de Vries R. M. J. M.; Doveston R.; Milroy L.-G.; Brunsveld L.; Ottmann C. Structural interface between LRRK2 and 14-3-3 protein. Biochem. J. 2017, 7, 1273–1287. 10.1042/BCJ20161078. [DOI] [PubMed] [Google Scholar]
- Stevers L. M.; Lam C. V.; Leysen S. F. R.; Meijer F. A.; van Scheppingen D. S.; de Vries R. M. J. M.; Carlile G. W.; Milroy L. G.; Thomas D. Y.; Brunsveld L.; Ottmann C. Characterization and small-molecule stabilization of the multisite tandem binding between 14-3-3 and the R domain of CFTR. Proc. Natl. Acad. Sci. U.S.A. 2016, 113, E1152–E1161. 10.1073/pnas.1516631113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalabova D.; Smidova A.; Petrvalska O.; Alblova M.; Kosek D.; Man P.; Obsil T.; Obsilova V. Human procaspase-2 phosphorylation at both S139 and S164 is required for 14-3-3 binding. Biochem. Biophys. Res. Commun. 2017, 493, 940–945. 10.1016/j.bbrc.2017.09.116. [DOI] [PubMed] [Google Scholar]
- Madeira F.; Tinti M.; Murugesan G.; Berrett E.; Stafford M.; Toth R.; Cole C.; MacKintosh C.; Barton G. J. 14-3-3-Pred: improved methods to predict 14-3-3-binding phosphopeptides. Bioinformatics 2015, 31, 2276–2283. 10.1093/bioinformatics/btv133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alblova M.; Smidova A.; Docekal V.; Vesely J.; Herman P.; Obsilova V.; Obsil T. Molecular basis of the 14-3-3 protein-dependent activation of yeast neutral trehalase Nth1. Proc. Natl. Acad. Sci. U.S.A. 2017, 114, E9811–E9820. 10.1073/pnas.1714491114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo Y.; Schumacher B.; Landrieu I.; Bartel M.; Smet-Nocca C.; Jang A.; Choi H. S.; Jeon N. L.; Chang K.-A.; Kim H.-S.; Ottmann C.; Suh Y.-H. Involvement of 14-3-3 in tubulin instability and impaired axon development is mediated by Tau. FASEB J. 2015, 29, 4133–4144. 10.1096/fj.14-265009. [DOI] [PubMed] [Google Scholar]
- Kacirova M.; Novacek J.; Man P.; Obsilova V.; Obsil T. Structural Basis for the 14-3-3 Protein-Dependent Inhibition of Phosducin Function. Biophys. J. 2017, 112, 1339–1349. 10.1016/j.bpj.2017.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psenakova K.; Petrvalska O.; Kylarova S.; Lentini Santo D.; Kalabova D.; Herman P.; Obsilova V.; Obsil T. 14-3-3 protein directly interacts with the kinase domain of calcium/calmodulin-dependent protein kinase kinase (CaMKK2). Biochim. Biophys. Acta, Gen. Subj. 2018, 1862, 1612–1625. 10.1016/j.bbagen.2018.04.006. [DOI] [PubMed] [Google Scholar]
- Park E.; Rawson S.; Li K.; Kim B.-W.; Ficarro S. B.; Pino G. G.-D.; Sharif H.; Marto J. A.; Jeon H.; Eck M. J. Architecture of autoinhibited and active BRAF–MEK1–14-3-3 complexes. Nature 2019, 575, 545–550. 10.1038/s41586-019-1660-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluchanko N. N.; Beelen S.; Kulikova A. A.; Weeks S. D.; Antson A. A.; Gusev N. B.; Strelkov S. V. Structural Basis for the Interaction of a Human Small Heat Shock Protein with the 14-3-3 Universal Signaling Regulator. Structure 2017, 25, 305–316. 10.1016/j.str.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tugaeva K. V.; Kalacheva D. I.; Cooley R. B.; Strelkov S. V.; Sluchanko N. N. Concatenation of 14-3-3 with partner phosphoproteins as a tool to study their interaction. Sci. Rep. 2019, 9, 15007 10.1038/s41598-019-50941-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincenz C.; Dixit V. M. 14-3-3 Proteins Associate with A20 in an Isoform-specific Manner and Function Both as Chaperone and Adapter Molecules. J. Biol. Chem. 1996, 271, 20029–20034. 10.1074/jbc.271.33.20029. [DOI] [PubMed] [Google Scholar]
- Agarwal-Mawal A.; Qureshi H. Y.; Cafferty P. W.; Yuan Z.; Han D.; Lin R.; Paudel H. K. 14-3-3 Connects Glycogen Synthase Kinase-3β to Tau within a Brain Microtubule-associated Tau Phosphorylation Complex. J. Biol. Chem. 2003, 278, 12722–12728. 10.1074/jbc.M211491200. [DOI] [PubMed] [Google Scholar]
- Saline M.; Badertscher L.; Wolter M.; Lau R.; Gunnarsson A.; Jacso T.; Norris T.; Ottmann C.; Snijder A. AMPK and AKT protein kinases hierarchically phosphorylate the N-terminus of the FOXO1 transcription factor, modulating interactions with 14-3-3 proteins. J. Biol. Chem. 2019, 294, 13106–13116. 10.1074/jbc.RA119.008649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centorrino F.; Ballone A.; Wolter M.; Ottmann C. Biophysical and structural insight into the USP8/14-3-3 interaction. FEBS Lett. 2018, 592, 1211–1220. 10.1002/1873-3468.13017. [DOI] [PubMed] [Google Scholar]
- Ottmann C.; Weyand M.; Sassa T.; Inoue T.; Kato N.; Wittinghofer A.; Oecking C. A Structural Rationale for Selective Stabilization of Anti-tumor Interactions of 14-3-3 proteins by Cotylenin A. J. Mol. Biol. 2009, 386, 913–919. 10.1016/j.jmb.2009.01.005. [DOI] [PubMed] [Google Scholar]
- Würtele M.; Jelich-Ottmann C.; Wittinghofer A.; Oecking C. Structural view of a fungal toxin acting on a 14-3-3 regulatory complex. EMBO J. 2003, 22, 987–994. 10.1093/emboj/cdg104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose R.; Rose M.; Ottmann C. Identification and structural characterization of two 14-3-3 binding sites in the human peptidylarginine deiminase type VI. J. Struct. Biol. 2012, 180, 65–72. 10.1016/j.jsb.2012.05.010. [DOI] [PubMed] [Google Scholar]
- Smidova A.; Alblova M.; Kalabova D.; Psenakova K.; Rosulek M.; Herman P.; Obsil T.; Obsilova V. 14-3-3 protein masks the nuclear localization sequence of caspase-2. FEBS J. 2018, 285, 4196–4213. 10.1111/febs.14670. [DOI] [PubMed] [Google Scholar]
- Molzan M.; Weyand M.; Rose R.; Ottmann C. Structural insights of the MLF1/14-3-3 interaction. FEBS J. 2012, 279, 563–571. 10.1111/j.1742-4658.2011.08445.x. [DOI] [PubMed] [Google Scholar]
- Kim Y.-W.; Grossmann T. N.; Verdine G. L. Synthesis of all-hydrocarbon stapled α-helical peptides by ring-closing olefin metathesis. Nat. Protoc. 2011, 6, 761–771. 10.1038/nprot.2011.324. [DOI] [PubMed] [Google Scholar]
- Winn M. D.; Ballard C. C.; Cowtan K. D.; Dodson E. J.; Emsley P.; Evans P. R.; Keegan R. M.; Krissinel E. B.; Leslie A. G. W.; McCoy A.; McNicholas S. J.; Murshudov G. N.; Pannu N. S.; Potterton E. A.; Powell H. R.; Read R. J.; Vagin A.; Wilson K. S. Overview of the CCP 4 suite and current developments. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2011, 67, 235–242. 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battye T. G. G.; Kontogiannis L.; Johnson O.; Powell H. R.; Leslie A. G. W. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2011, 67, 271–281. 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P. Scaling and assessment of data quality. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2006, 62, 72–82. 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- Evans P. R.; Murshudov G. N. How good are my data and what is the resolution?. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2013, 69, 1204–1214. 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebedev A. A.; Vagin A. A.; Murshudov G. N. Model preparation in MOLREP and examples of model improvement using X-ray data. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2008, 64, 33–39. 10.1107/S0907444907049839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A.; Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66, 22–25. 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- Adams P. D.; Afonine P. V.; Bunkóczi G.; Chen V. B.; Davis I. W.; Echols N.; Headd J. J.; Hung L.-W.; Kapral G. J.; Grosse-Kunstleve R. W.; McCoy A. J.; Moriarty N. W.; Oeffner R.; Read R. J.; Richardson D. C.; Richardson J. S.; Terwilliger T. C.; Zwart P. H. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66, 213–221. 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P.; Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004, 60, 2126–2132. 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Vecer J.; Herman P. Maximum Entropy Analysis of Analytically Simulated Complex Fluorescence Decays. J. Fluoresc. 2011, 21, 873–881. 10.1007/s10895-009-0589-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.