Abstract
The structure of a protein complex needs to be controlled appropriately to maximize its functions. Herein, we report the linear polymerization of bacterial alkaline phosphatase (BAP) through the site-specific cross-linking reaction catalyzed by Trametes sp. laccase (TL). We introduced a peptide loop containing a tyrosine (Y-Loop) to BAP, and the Y-Looped BAP was treated with TL. The Y-Looped BAP formed linear polymers, whereas BAP fused with a C-terminal peptide containing a tyrosine (Y-tag) showed an irregular shape after TL treatment. The sterically confined structure of the Y-Loop could be responsible for the formation of linear BAP polymers. TL-catalyzed copolymerization of Y-Looped BAP and a Y-tagged chimeric antibody-binding protein, pG2pA-Y, resulted in the formation of linear bifunctional protein copolymers that could be employed as protein probes in an enzyme-linked immunosorbent assay (ELISA). Copolymers comprising Y-Looped BAP and pG2pA-Y at a molar ratio of 100:1 exhibited the highest signal in the ELISA with 26- and 20-fold higher than a genetically fused chimeric protein, BAP-pG2pA-Y, and its polymeric form, respectively. This result revealed that the morphology of the copolymers was the most critical feature to improve the functionality of the protein polymers as detection probes, not only for immunoassays but also for other diagnostic applications.
Introduction
Conjugation of protein molecules into a large protein complex may expand the functionalities of proteins because the protein units in the complex may cooperatively work together. However, it is challenging to assemble a large number of proteins into a protein complex while retaining the functionality of each protein unit. Some protein units might get deactivated during conjugation if the conjugation occurs at a critical site, and some proteins may not be able to function in the complex because of steric hindrance. Therefore, it is essential to conjugate protein units in a site-specific manner, and the structure of the protein complex needs to be controlled appropriately so that the protein units in the complex can work efficiently.1
Genetic fusion of proteins is a common method to create artificial protein assemblies or protein polymers.2−7 However, the possible number of protein units that can be conjugated is limited because of the difficulty in the expression of fusion proteins, and the tandem fusion strategy requires careful linker designs to organize the different proteins and control their characteristics.1,5,6 Such strategies also decrease the degrees of freedom of the protein dynamics, which may lead to impaired activity and protein misfolding.1,2,8,9 Another approach to assemble proteins into a protein complex is to use post-translational modifications (PTMs).10−12 PTMs enable a higher degree of polymerization than the genetic fusing method. In particular, PTMs using enzymatic reactions are ideal since enzymes can provide high site-selectivity with rapid and mild reaction conditions to retain the activity of protein units.13−16
Horseradish peroxidase (HRP) and laccase have been utilized for making covalently cross-linked protein complexes with a high degree of conjugation, i.e., protein polymers. HRP and laccase are more suitable for making huge protein polymers than other PTM enzymes, such as transglutaminase17,18 and sortase A.19,20 This is because they conduct oxidation reactions on tyrosine residues to yield tyrosyl radicals, and the cross-linking between tyrosyl radicals occurs nonenzymatically without forming tripartite complexes, which are necessary for other enzymes.21−23 In our previous studies, we demonstrated the polymerization of bacterial alkaline phosphatase (BAP), as well as HRP, by a Trametes sp. laccase (TL) reaction. Both BAP and HRP were recombinantly prepared and tethered with peptide tags containing tyrosine residues (Y-tag).16,24 The Y-tagged BAPs and HRPs were site-specifically cross-linked by TL to form polymers. Laccase has an advantage over HRP because it requires only molecular oxygen to catalyze the oxidation of tyrosine residues, while HRP requires hydrogen peroxide, which is potentially harmful to proteins.25
The obtained dendritic protein polymers of BAPs and HRPs were utilized as detection probes in an enzyme-linked immunosorbent assay (ELISA) by copolymerizing with antibody-binding proteins (ABPs), such as protein A (pA) and protein G (pG). Although the polymerization did not affect the activity of enzyme units in the protein polymers, a large number of enzyme units were required to copolymerize with the ABPs to surpass the performance of the genetically fused 1:1 conjugate. In previous work, the ELISA signal of the protein polymers became saturated at high molar ratios of the enzyme to ABPs and even decreased at the highest molar ratio (100:1, enzyme units/ABPs).16,24 These results suggest that some of the enzyme and ABP units are buried inside the dendritic structure of the protein polymers and could not function appropriately. Also, the three-dimensional dendritic protein polymers could not be packed efficiently onto the surface of an ELISA plate, and they occupy a large surface area per protein polymer resulting in fewer enzyme units being immobilized.16,24
One possible strategy to solve this problem is to create linear protein polymers.28 A linear polymer is much simpler and less sterically hindered than three-dimensional (3D) dendritic structures; thus, protein units in a linear protein polymer can work more efficiently. Also, a linear molecular structure is an ideal shape for maximizing the amount of protein polymer immobilized on the surface.
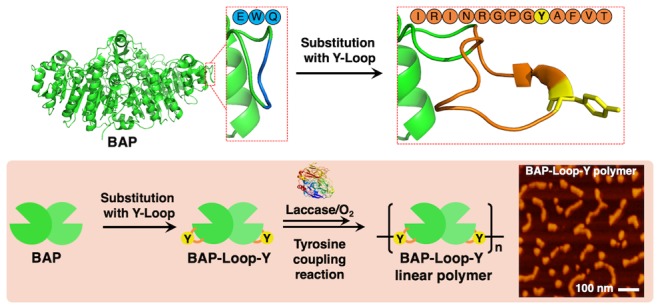
The conventional Y-tag sequences were highly flexible, so the cross-linked dityrosine could again be a substrate for TL, leading to the formation of branched structures.23,26,27,29 We anticipated that by introducing appropriate steric hindrance to the cross-linked dityrosine, the TL could not recognize it and thereby prevent the formation of branched structures, favoring linear polymers. Herein, we report the linear polymerization of engineered BAPs using a tyrosine-containing loop peptide (Y-Loop). We genetically modified the BAP by substituting the loop domain of the BAP structure (219–221 aa) with a 13-mer Y-Loop (IRINRGPGYAFVT) to construct BAP-Loop-Y (Scheme 1A). We designed the Y-Loop by modifying the loop peptide sequence from the previous reports.30−32 We anticipated that the Y-Loop could be recognized by TL to cross-link only once because the cross-linked Y-Loop will be too sterically encumbered to react again with TL (Scheme 1B). We conducted scanning probe microscopy (SPM) analysis to confirm the structural differences between the BAP-Loop-Y polymer and the Y-tagged BAP (BAP-Y) polymer, which were prepared by TL treatment. Furthermore, copolymerization of BAP-Y or BAP-Loop-Y and a single Y-tagged ABP, pG2pA-Y, was performed to form BAP-Y/pG2pA-Y and BAP-Loop-Y/pG2pA-Y copolymers. The ability of these copolymers to function as ELISA probes was also evaluated. Our proof-of-concept study on linear protein polymerization could contribute a new strategy to create highly functional one-dimensional protein complexes in biotechnological applications.
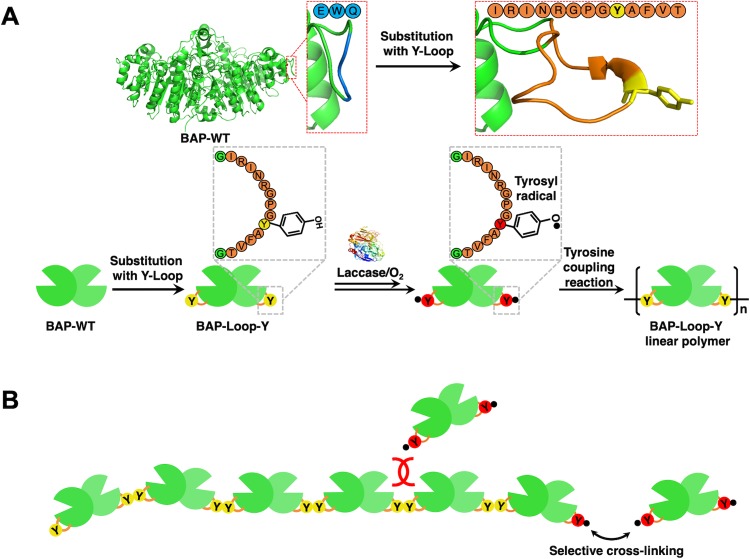
Scheme 1. Polymerization Reaction of a Y-Looped Bacterial (Escherichia coli) Alkaline Phosphatase (BAP), BAP-Loop-Y, Catalyzed by TL.
(A) BAP-Loop-Y were constructed by substituting the loop domain of the BAP structure (219–221 aa) with a 13-mer Y-Loop (IRINRGPGYAFVT) peptide. TL recognizes and oxidizes phenolic moieties of tyrosine residues on the Y-Loop to form reactive tyrosyl radicals, which then react with each other in a nonenzymatic way to form polymers. (B) Short and sterically hindered Y-Loop made the gap between the BAP-Loop-Y units very small and prevented another activated BAP-Loop-Y reacting at that position.
Results and Discussion
Enzymatic Activity of Recombinant BAPs
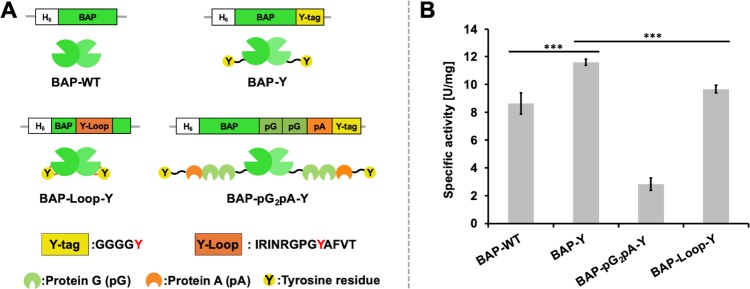
We employed E. coli-expressed BAPs with different structures as model proteins for the protein polymerization reaction. Figure 1A shows the constructs of the BAPs used in this study. We used the wild-type BAP (BAP-WT) as the control. BAP-Y and BAP-pG2pA-Y were tagged with a flexible peptide sequence containing a tyrosine residue (Y-tag) at the C-termini. All of the BAPs were expressed in E. coli with high expression levels (Supporting Information, Figures S1–5 and Table S1). Although the expression level of BAP-Loop-Y was as high as the other BAPs, we observed cleavage of BAP-Loop-Y during expression and purification, probably somewhere around the Y-Loop. The bands of the cleaved BAP-Loop-Y can be seen in Figure S4C at around 20 kDa. The cleaved BAP-Loop-Y eluted at the same elution volume in SEC purification, indicating that the cleaved BAP-Loop-Y retained its tertiary and quaternary structure. Different constructs of BAP showed different specific activities, and the modification of the BAPs with Y-tags, especially BAP-Y, exhibited approximately 34% higher activity than that of the wild type (Figure 1B). However, BAP-pG2pA-Y showed lower activity than the other BAPs; it showed approximately 55% of the specific activity of BAP-WT. This is consistent with the data obtained in our previous report,24 suggesting that genetically fusing pG2pA to BAP is responsible for decreasing the BAP activity. BAP-Loop-Y showed activity comparable to that of BAP-WT. This result also agreed with previous reports.30,31 The Y-Loop peptide was inserted into the BAP structure at a position away from the active site of BAP; therefore, there was no detrimental effect on the ability of the BAPs to recognize their substrates.
Figure 1.
Recombinant bacterial (E. coli) alkaline phosphatases (BAPs) used in this study. (A) Constructs and structural illustration of the recombinant BAPs prepared in this study; BAP-WT, wild-type (unmodified) BAP; BAP-Y, BAP with a peptide tag containing the tyrosine residue (Y-tag) at C-termini; BAP-pG2pA-Y, BAP with a Y-tagged chimeric antibody-binding protein, pG2pA-Y, at C-termini; BAP-Loop-Y, BAP-inserted Y-Loop sequence at 221–223 aa. Three of them were tagged with Y-tags that can be recognized by TL. (B) Specific activities of E. coli-expressed BAPs using p-NPP as the substrate. Error bars denote the standard error of the measurements from three individual samples (***p < 0.001).
TL-Catalyzed Polymerization of BAPs
Next, we evaluated the TL-catalyzed cross-linking reaction of BAPs. We optimized the concentration of TL and the reaction time to perform the polymerization reaction with high efficiency (Supporting Information, Figure S6). Then, we fixed the concentration of TL and reaction time to 2 μM and 2 h, respectively. Figure 2A presents native-PAGE analysis of the TL-treated BAPs. The BAP-Y, BAP-pG2pA-Y, and BAP-Loop-Y formed polymers with high molecular weights that exceeded the upper limit of the separating gel and, therefore, were retained in the stacking gel. A wild-type BAP (BAP-WT) was employed as the control reaction. After the TL treatment, no cross-linked product was observed in the case of BAP-WT. Although there are some tyrosine residues in the structure of BAP-WT, TL did not recognize them as substrates. From these results, TL recognized the tyrosine residues in the Y-tags or Y-Loop, site-selectively. These results agreed with our previous reports.16,24 The presence of Y-tags and Y-Loops, which can be recognized and activated by TL, eliminates the use of phenolic mediators, such as vanillic acid,33,34 ferulic acid,35,36p-coumaric acid,35,37 and caffeic acid,38−40 and are typically used in a laccase-catalyzed cross-linking of proteins.
Figure 2.
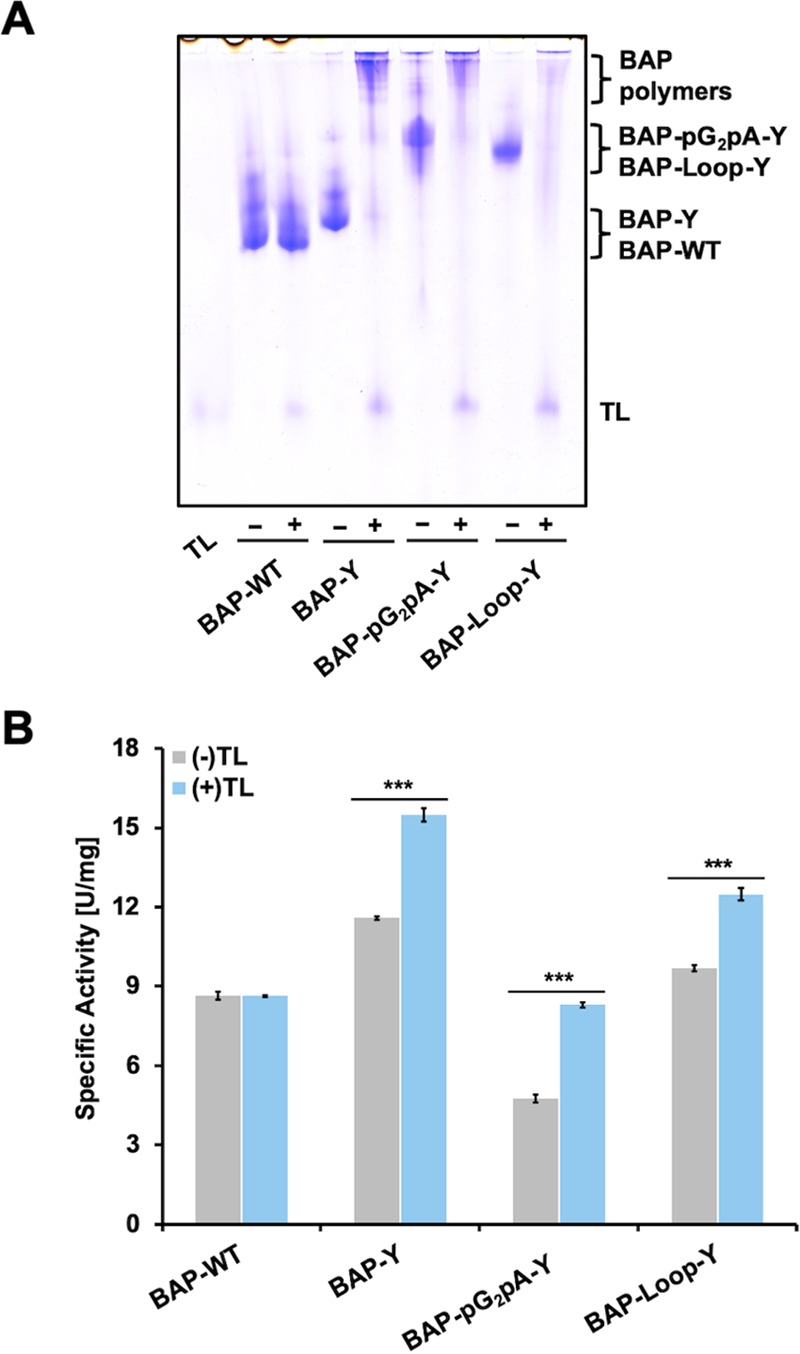
Native-PAGE analysis and relative activity of BAPs before and after TL treatment. (A) Results of native-PAGE analysis of BAPs before and after TL treatment. The concentration of the BAPs and TL were 10 and 2 μM, respectively. The cross-linking reaction of the BAPs was conducted in 10 mM Tris-HCl pH 8.0 at 37 °C for 2 h. (B) Enzymatic activity of the BAPs before and after TL treatment using p-NPP as a substrate. Error bars denote the standard error of the measurements from three individual samples (***p < 0.001).
To confirm the activity changes of the BAPs after TL treatment, we measured the enzymatic activity of the BAP monomers and polymers using p-nitrophenyl phosphate (p-NPP) as a substrate (Figure 2B). Interestingly, the relative activity of the BAP polymers was higher than that of their monomers. We found similar results when we polymerized HRP by TL in our previous work.16 Although it is not clear why the activity increased after the polymerization of enzymes, we expect that the small substrate molecules require less time to be recognized by the catalytic sites of the protein as there are many catalytic sites in the polymer, resulting in faster reactions. TL is a more useful enzyme for the preparation of protein polymers than peroxidases, such as HRPs, which need additional hydrogen peroxide (H2O2) to initiate the polymerization reaction. Additional H2O2 may cause changes in enzymatic activity and deactivation of the model proteins (Supporting Information, Figure S7).
The expected schematic polymerization reaction of a BAP by TL is shown in Scheme 1A. TL oxidizes the phenolic moieties of the Y-tags or Y-Loop and generates free tyrosyl radicals, which react with each other to form dityrosine, trityrosine, or an even higher degree of cross-linked tyrosine derivatives.41,42 Although the reactivity of Y-Loop should be lower than that of the Y-tags attached to the C-terminus of the BAPs (because of the steric hindrance), all of the BAP-Loop-Y units were cross-linked by TL treatment, indicating that the reactivity of Y-Loop is sufficient to make polymers. In SDS-PAGE analysis of the TL-treated BAPs (Supporting Information, Figure S8), the cross-linked BAP-Loop-Y showed clearer bands in a lower-molecular-weight region than BAP-Y and BAP-pG2pA-Y. The sharper bands of the cross-linked BAP-Loop-Y suggest that the BAP-Loop-Y monomers were cross-linked with each other in a more uniform way than the other Y-tagged BAPs. Since the cross-linked Y-Loops are more sterically encumbered than the cross-linked Y-tags at the C-terminus, further cross-linking to Y-Loops to yield oligomeric BAPs was inhibited; this should lead to the formation of linear BAP-Loop-Y polymers (Scheme 1B). The multiple bands seen in the cross-linked BAP-Loop-Y is mainly caused by the cleaved BAP-Loop-Y, which is also present in the purified BAP-Loop-Y and can be seen at around 20 kDa. The main product of cross-linked BAP-Loop-Y is its dimer, which appeared at around 140 kDa in SDS-PAGE analysis.
Scanning Probe Microscopy (SPM) of BAP Polymers
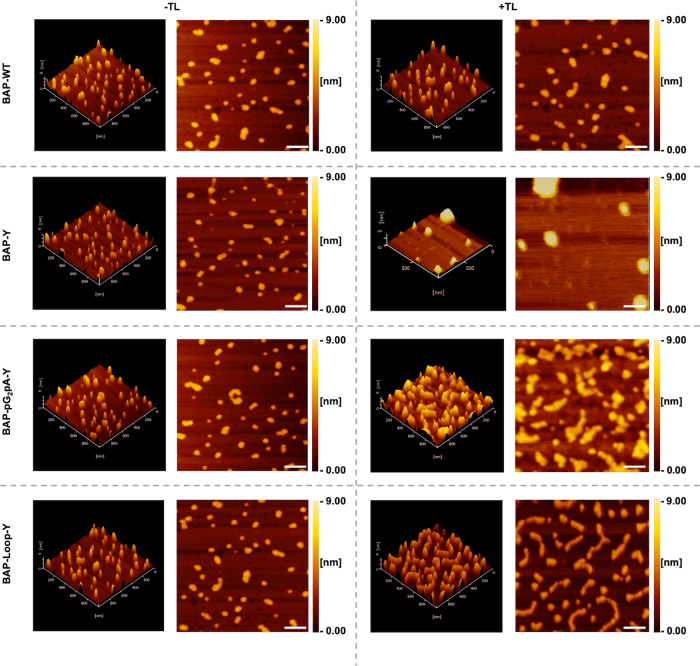
To validate the structural differences of BAP polymers, we analyzed the shape of BAP monomers and polymers using an SPM in dynamic force mode. Figure 3 shows the results of SPM analysis of BAP monomers and polymers. Thanks to the high molecular weight of BAPs (97.6, 99.6, 142.0, and 100.5 kDa for BAP-WT, BAP-Y, BAP-pG2pA-Y, and BAP-Loop-Y, respectively), the monomers of the BAPs were successfully visualized as globular forms with heights ranging from 3.5 to 4.0 nm (Supporting Information, Figure S9–13). However, some aggregates of BAP monomers were also observed. These may be caused by the drying process before SPM analysis. There were no shape differences between the BAP-WT monomer and TL-treated BAP-WT, indicating that BAP-WT was intact after TL treatment. In contrast, BAP-Y formed a huge globular shape, suggesting that the BAP-Y monomers were connected to each other after the TL treatment. As the Y-tag at the C-terminus can cross-link multiple times, the BAP-Y polymer can form highly branched structures, which would be visualized as a huge globular shape in SPM analysis. The BAP-pG2pA-Y formed large irregular structures after TL treatment, but the shapes of the TL-treated BAP-pG2pA-Y were different from those of the BAP-Y polymers. This is probably because of the presence of the chimeric protein, pG2pA, the structure of which is linear, so the molecular structure of the BAP-pG2pA-Y monomer and also its polymers are not globular.
Figure 3.
Results of SPM analysis of the BAPs before and after TL treatment. The BAPs were diluted to a final concentration of 0.2 μM or ∼0.02 mg/mL in 10 mM Tris-HCl pH 8.0. Scale bars: 100 nm.
Interestingly, compared to other BAPs, the BAP-Loop-Y polymers had mostly linear shapes. The longest length of a BAP-Loop-Y polymer observed was approximately 350 nm, and the average length of BAP-Loop-Y polymers was 205 ± 17 nm (Supporting Information, Figure S14). This result strongly suggests that the BAP-Loop-Y units were cross-linked through the designed Y-Loop to yield linear polymers. A short and rigid Y-Loop consisting of 13 amino acid residues could eliminate the formation of branched structures because of the steric hindrance caused by the BAP-Loop-Y structure. Although the polymer length could not be controlled in this system yet, this is the first demonstration of the use of steric hindrance to control the structures of protein polymers, which were made via tyrosyl radical coupling reactions. Because of the better molecular packing on solid interfaces, linear protein polymers have the potential to exhibit better performance than hyperbranched globular protein polymers, especially in applications such as ELISA or western blotting where the reactions and interactions occur on a solid surface.
TL-Catalyzed Copolymerization of Y-Tagged Proteins
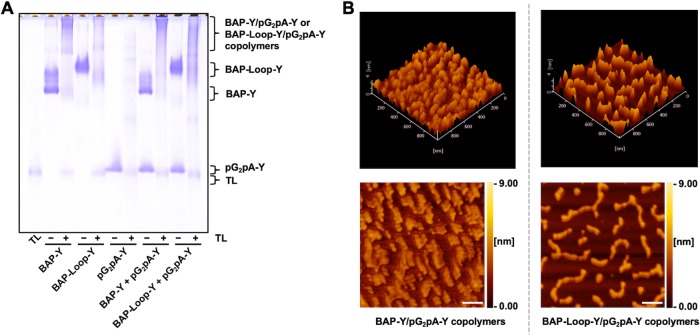
We performed the copolymerization of BAP-Loop-Y or BAP-Y, with a single Y-tagged chimeric antibody-binding protein, pG2pA-Y, to create a bifunctional protein polymer that could be applied as a detection probe in an immunosorbent assay. The copolymer of BAP-Y and pG2pA-Y should form highly branched protein polymers since both proteins possess flexible Y-tags at their C-termini. On the contrary, we expected the BAP-Loop-Y/pG2pA-Y copolymer to form linear structures incorporating pG2pA-Y, mainly at the termini of the polymer. There are also some possibilities for pG2pA-Y to attach to BAP-Loop-Y polymers as side branches. Figure 4A shows native-PAGE analysis of the copolymers that were prepared by the copolymerization reaction of BAP-Y or BAP-Loop-Y with pG2pA-Y catalyzed by TL. After TL treatment, both bands of pG2pA-Y and BAP-Y or BAP-Loop-Y disappeared, and new bands in a high molecular-weight region were produced, indicating the formation of cross-linked copolymers. The co-cross-linking of pG2pA-Y did not affect the activity of the BAPs since no decrease in activity was detected after TL treatment (Supporting Information, Figure S15).
Figure 4.
Copolymerization reaction of BAPs and pG2pA-Y catalyzed by TL. (A) Results of native-PAGE analysis of BAPs/pG2pA copolymers after TL treatment. The concentration of both proteins was 10 μM. TL concentration was 2 μM. The reaction was conducted for 2 h at 37 °C. (B) SPM analysis of BAP-Y/pG2pA-Y and BAP-Loop-Y/pG2pA-Y copolymers performed at a 1:1 molar ratio of BAPs to pG2pA-Y. The copolymers were adsorbed onto mica after a 50 times dilution with 10 mM Tris-HCl pH 8.0. Scale bars: 100 nm.
Figure 4B shows the results of SPM analysis of the BAP-Y/pG2pA-Y and BAP-Loop-Y/pG2pA-Y copolymers. The shape of the BAP-Y/pG2pA-Y copolymer was different from that of the BAP-Y polymers (Figure 3). Incorporation of pG2pA-Y changed the globular structure of the BAP-Y polymers to form smaller globular protein polymers since pG2pA-Y possesses only one Y-tag in a molecule; thus, the attachment of pG2pA-Y to the BAP-Y polymers terminates the polymerization reaction. In contrast, the shape of the BAP-Loop-Y/pG2pA-Y copolymer was as linear as the BAP-Loop-Y polymers. The longest and average length of a BAP-Loop-Y/pG2pA-Y copolymer observed was approximately 500 and 255 ± 35 nm, respectively (Supporting Information, Figure S14). The monomer of BAP-Loop-Y appeared as 47 ± 1 nm of globular shape in SPM analysis (Figure 3). It suggests that the degree of polymerization of BAP-Loop-Y was in the range of 5–10. This suggests that the pG2pA-Y units were incorporated at the termini of BAP-Loop-Y polymers. Although some pG2pA-Y might have been incorporated at the cross-linking site of the BAP-Loop-Y units in the polymers and bound as small branches, they are probably not visible by SPM. There is also a possibility that the pG2pA-Y mostly cross-linked with itself and not with BAP-Loop-Y, which would result in pG2pA-Y oligomers. The presence of the pG2pA unit is essential for the BAP-Loop-Y polymer to bind to IgG. Therefore, the function of the BAP-Loop-Y/pG2pA-Y copolymer as a detection probe in an immunoassay was evaluated next.
Performance of the BAP-Loop-Y/pG2pA-Y Copolymers as Protein Probes in ELISA
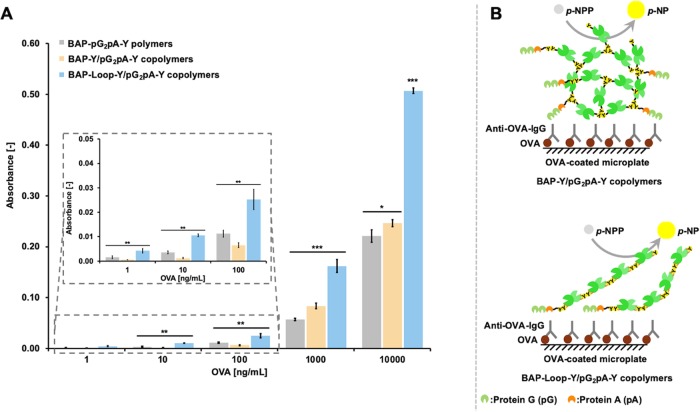
We evaluated the functionality of the BAP-Y/pG2pA-Y and BAP-Loop-Y/pG2pA-Y copolymers in an ovalbumin (OVA)-detecting ELISA (Figure 5). A polymer of genetically fused BAP-pG2pA-Y was used as a control, which corresponds to a copolymer of BAP and pG2pA cross-linked at a 1:1 molar ratio. Both copolymers showed absorbance changes in the ELISA, indicating that the pG2pA-Y and BAP-Y or BAP-Loop-Y were cross-linked to each other by TL treatment. The BAP-Loop-Y/pG2pA-Y copolymer exhibited 2-fold higher absorbance in the ELISA than the BAP-Y/pG2pA-Y copolymer (Figure 5A), despite the specific activity of BAP-Loop-Y being approximately 20% lower than that of BAP-Y (Figure 1).
Figure 5.
Results of the OVA-detecting ELISA using BAP-Y/pG2pA-Y and BAP-Loop-Y/pG2pA-Y copolymers. (A) Results of the OVA-detecting ELISA using BAP-Y/pG2pA-Y and BAP-Loop-Y/pG2pA-Y copolymers prepared at a 1:1 molar ratio of BAPs to pG2pA-Y. The polymer of genetically fused BAP-pG2pA-Y was used as a control. The concentration of the BAP polymers and copolymers was fixed to 0.5 U/mL. The absorption of OVA onto the microplate was conducted at concentrations of 1 ng/mL to 10 μg/mL in Tris-buffered saline (TBS) (pH 7.4). (B) Expected structure of the BAP-Y/pG2pA-Y and BAP-Loop-Y/pG2pA-Y copolymers in the ELISA. The protein G (pG) (green) or protein A (pA) (orange) can bind to the IgG specifically. Error bars denote the standard error of the measurements from three individual samples (*p < 0.05, **p < 0.01, ***p < 0.001).
These results shed light on the importance of controlling the molecular structure of protein polymers to maximize their functionality. The detection probes in ELISA are on a surface, so it is important to control the structure of the protein polymer so that they work efficiently.43,44 A surface can be considered as a set of lines, so the ideal structure of protein polymer to maximize the molecular packing on a surface is linear. The copolymerization reaction of BAP-Y and pG2pA-Y resulted in the formation of large, highly branched structures,24 while the BAP-Loop-Y/pG2pA-Y copolymer formed linear structures (Figure 5B). The BAP-Loop-Y/pG2pA-Y copolymer showed higher signals than the BAP-Y/pG2pA-Y copolymers at the conditions of low OVA coating concentrations, where the molecular packing on a surface has less effects. This suggests that the pG2pA-Y units in the linear copolymer work more efficiently than the units in the branched copolymers. The pG2pA-Y units in the BAP-Y/pG2pA-Y copolymers are embedded within the branched structures. On the other hand, the pG2pA-Y units in the BAP-Loop-Y/pG2pA-Y copolymer are most likely to be conjugated at the termini of the linear structure, which is suitable for binding to IgGs. Also, the linear structures of BAP-Loop-Y/pG2pA-Y copolymers provide better molecular packing on the surface of the ELISA plate than the 3-dimensional highly branched BAP-Y/pG2pA-Y copolymer, resulting in a higher signal in the ELISA. The molecular packing on the surface was particularly significant when the OVA coating concentration was high, and there were many antibodies on the ELISA plate surface.45 The BAP-Loop-Y/pG2pA-Y copolymer showed remarkably higher absorbance than the BAP-Y/pG2pA-Y copolymers when there was a high OVA coating concentration.
BAP-Loop-Y/pG2pA-Y Copolymers Prepared with Various Molar Ratios
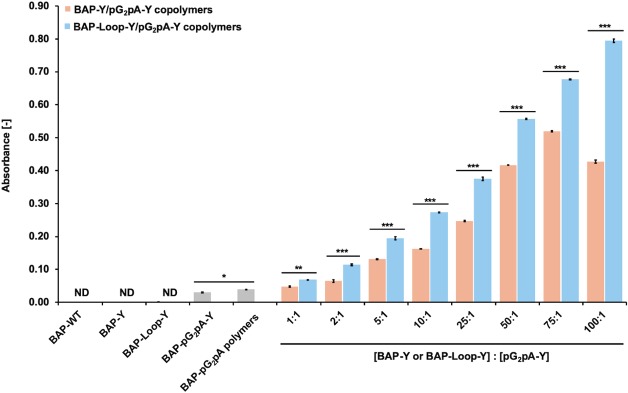
To enhance the activity and functionality of the BAP-Y/pG2pA-Y and BAP-Loop-Y/pG2pA-Y copolymers as detection probes, it is crucial to increase the molar ratio of BAP-Y or BAP-Loop-Y against pG2pA-Y. The copolymers of BAP-Y or BAP-Loop-Y and pG2pA-Y were prepared at various molar ratios (Supporting Information Figure S16). Figure 6 shows the results of OVA-detecting ELISA using the copolymers of BAPs and pG2pA-Y that were prepared at different molar ratios. As the ratio of the BAP against pG2pA-Y increased, the absorbance increased in the ELISA for the BAP-containing copolymers, indicating that a greater number of BAP units were incorporated into the copolymers with pG2pA-Y. Intriguingly, the BAP-Loop-Y/pG2pA-Y copolymer showed an almost linear increase in absorbance in proportion to the ratio of BAP-Loop-Y, while the BAP-Y/pG2pA-Y copolymer reached its peak at a ratio of 75:1; the absorbance decreased at a ratio of 100:1. At a ratio of 100:1, the BAP-Loop-Y/pG2pA-Y copolymer exhibited 26- and 20-fold higher absorbance than BAP-pG2pA-Y and its polymer, respectively. To fully understand the functionality of BAP and pG2pA copolymers, we purified them from the remaining monomers using size exclusion chromatography (SEC). The copolymers with high molecular weights were completely separated from the monomers by SEC (Supporting Information Figure S17A,B). To confirm the functionality and activity of the purified BAP/pG2pA copolymers, we applied them again in an OVA-detecting ELISA (Supporting Information, Figure S17C). The purified copolymers performed better as detection probes than those that were not purified. The BAP-Loop-Y/pG2pA-Y copolymer showed higher signals in ELISA than the BAP-Y/pG2pA-Y copolymers. This result revealed that the morphology of the copolymers was the most critical feature to improve the functionality of the protein polymers as detection probes, not only for immunoassays but also for other diagnostic applications.
Figure 6.
Results of the OVA-detecting ELISA using BAP and pG2pA-Y copolymers prepared at various molar ratios. The concentration of pG2pA-Y was fixed to 0.5 μM. While the concentrations of BAP-Y and BAP-Loop-Y were varied from 0.5–50 μM. The concentration of TL was 2 μM. The reaction was conducted in 10 mM Tris-HCl, pH 8.0, at 37 °C, for 2 h. All protein polymer solutions were diluted to fix the concentration of pG2pA-Y as a binding domain to 1 nM. The concentration of OVA that coated the wells was 1 μg/mL in TBS (pH 7.4). ND: not detected. Error bars denote the standard error of the measurements from three individual samples (*p < 0.05, **p < 0.01, ***p < 0.001).
The use of a large number of enzymes in the copolymerization reaction with binding proteins results in the formation of highly branched structures in which the binding proteins tend to be buried inside the structures and are not able to function correctly. Therefore, in previous studies of protein polymers using Y-tagged proteins, there is often an optimum ratio of enzyme to binding proteins to extract the best functionality as detection probes.16,24,46 Since the molecular structure of the BAP-Loop-Y/pG2pA-Y copolymer is linear and the pG2pA-Y units are mostly conjugated at the terminus of the polymers, the pG2pA-Y could efficiently bind to the antibody on the ELISA plate, while carrying a large number of enzymes. The results here demonstrate the advantages of linear protein polymers over highly branched protein polymers.
Conclusions
We demonstrated a new strategy for controlling the structure of protein polymers by utilizing tyrosine-containing loop peptide, Y-Loop. The Y-Looped BAP, BAP-Loop-Y, formed linear polymers, whereas BAPs fused with a C-terminal Y-tag showed irregular shapes in SPM analysis. The sterically confined structure of the Y-Loop and a closely cross-linked BAP-Loop-Y caused steric hindrance around the cross-linked tyrosine residues at Y-Loops and prevented further cross-linking reactions and branch formations. Furthermore, linear BAP-Loop-Y/pG2pA-Y copolymers that were prepared from the copolymerization reaction of BAP-Loop-Y and pG2pA-Y showed excellent functionality and activity as protein probes in an OVA-detecting ELISA. These results suggested that the linear structure of the BAP-Loop-Y/pG2pA-Y copolymers allowed them to pack better on the anti-OVA IgG surface than the globular BAP-Y/pG2pA-Y copolymers. Interestingly, the absorbance of the BAP-Loop-Y/pG2pA-Y copolymers that were prepared with higher molar ratios of reporter enzyme to IgG-binding protein (BAP-Loop-Y to pG2pA-Y) increased across the whole range tested. Further study on the SEC fractions of the purified BAP-Loop-Y/pG2pA-Y confirmed its excellent functionality and activity as an ELISA probe, which showed a higher signal than the BAP-Y/pG2pA-Y copolymers. It can be concluded that the size of polymers containing the reporter enzyme is not the only factor to improve their performance as a protein probe in an immunoassay. The shape of the protein polymers could be the most important factor in improving the functionality and activity of protein polymers. This newly proposed linear polymerization of the BAP strategy can contribute to the further development of functional protein polymers for specific applications in biotechnology, biomedical sciences, bioimaging, and diagnostics.
Materials and Methods
Materials
TL was provided by Amano Enzyme Inc. (Nagoya, Japan). TL was dissolved in 100 mM sodium phosphate buffer, pH 6.0, and purified from impurities using an Amicon ultrafiltration device (50 kDa MWCO). Dipotassium hydrogen phosphate, sodium chloride, potassium dihydrogen phosphate, para-nitrophenyl phosphate (p-NPP), sodium dodecyl sulfate (SDS), hydrochloric acid, tetramethylethylenediamine (TEMED), and sodium ampicillin were purchased from Wako Pure Chemical Industries, Ltd. (Osaka, Japan). Tryptone and yeast extract were purchased from Difco (Miami). Trizma base and imidazole were purchased from Sigma Aldrich (Tokyo, Japan). Isopropyl-β-D-thiogalactopyranoside (IPTG) was purchased from Takara Bio Inc. (Shiga, Japan). Acrylamide/bis solution 30% was purchased from Nacalai Tesque, Inc. (Kyoto, Japan). All reagents were used as received.
Preparation of Y-Tagged Proteins
We constructed E. coli expression vectors of the proteins used in this study using the plasmid vectors of pET22b(+)-BAP and pET22b(+)-pG2pA as templates.14,24,47 The pG2pA is a chimeric protein composed of two protein G (pG) domains and a protein A (pA) domain fused in tandem.47 To construct the expression vector of the BAP-Loop-Y, the DNA encoding the EWQ at the position of the loop domain of BAP was replaced with the DNA encoding the Y-Loop sequence (IRINRGPGYFVT). We designed the sequence of Y-Loop based on the loop peptide sequence from the previous reports.30,31 The amino acid sequences of the proteins used in this study are listed in the Supporting Information. All of the proteins were expressed and purified according to a previously reported protocol.14,24
Enzymatic Assay of BAPs
p-NPP was used as a substrate to measure the activity of the BAPs. The BAPs were diluted to an appropriate concentration using 10 mM Tris-HCl (pH 8.0). We pipetted 10 μL of BAP solutions into 990 μL of 1 mM p-NPP in 1 M Tris-HCl (pH 8.0) in a 1 mL quartz cell for UV–vis spectrophotometry. We monitored the absorbance at 410 nm for 3 min at 25 °C. The measurement was replicated three times for three individual samples of each BAP.
TL-Catalyzed Polymerization of Proteins
We conducted the polymerization of the BAPs and the copolymerization of BAP-Loop-Y/pG2pA-Y and BAP-Y/pG2pA-Y by mixing with TL in 10 mM Tris-HCl (pH 8.0) in a total volume of 50 μL. The final concentration of the BAP was 10 μM, and we optimized the concentration of TL and reaction time (Supporting Information). The polymerization reaction was performed by incubating a mixture of the Y-tagged proteins and TL at 37 °C for an optimized length of time. The samples were analyzed by SDS-PAGE and native-PAGE.
Scanning Probe Microscopy (SPM) Imaging of the BAP Polymers
Samples of BAP monomers and polymers were diluted 10–50 times using 10 mM Tris-HCl (pH 8.0). Four microliters of the sample solution were then placed onto mica, which was freshly cleaved (Nilaco Corp., Tokyo, Japan), and incubated for 5–10 min at room temperature. The mica was then rinsed once using ultrapure water and dried under ambient conditions. The SPM images were taken using a Nanocute (Hitachi High-Tech Science Corp., Japan) using an SI-DF3P2 microcantilever (Hitachi)48 in the dynamic force mode.
ELISA Using BAP Polymers
The Y-tagged BAP polymers were used as detection probes in an ELISA. The OVA (albumin from chicken eggs) was diluted to concentrations ranging from 10 μg/mL to 0.1 ng/mL using Tris-buffered saline (TBS), pH 7.4, and pipetted into a flat bottom microplate (96-well, 100 μL/well) and the microplate was incubated overnight at 4 °C. The OVA-coated wells were then washed with 200 μL of TBS containing 0.1% of Tween-20 (TBST) three times. The blocking solution (TBST solution containing 1% bovine serum albumin (BSA)) was added (200 μL/well) and incubated for 1 h at RT. The wells were then washed with 200 μL of TBST three times. The rabbit anti-OVA-IgG (C6534–2 mL; Sigma-Aldrich), used as the primary antibody, was diluted with TBST containing 1% BSA to a concentration of 1 μg/mL and then 100 μL was added to each well, followed by 2 h incubation at RT. The wells were washed with 200 μL of TBST three times. The diluted BAPs, BAP-Loop-Y/pG2pA, and BAP/pG2pA polymers were then added to each well (100 μL/well) followed by 1 h incubation at RT. After washing the wells with 200 μL of TBST three times, 100 μL of p-NPP solution (1 mM in 1 M Tris-HCl (pH 8.0)) was added to each well. A 96-well microplate reader (BioTek, Synergy HTX) was used to monitor the changes of the absorbance at 410 nm for 1 h at 25 °C.
Size Exclusion Chromatography (SEC) of BAP Copolymers
The SEC samples of BAP copolymers were prepared according to the same procedures for TL-catalyzed polymerization of proteins, which are mentioned above. The molar ratio of BAPs to pG2pA-Y was set at 100:1 (50–0.5 μM) with a total volume of 1 mL in 10 mM Tris-HCl, (pH 8.0). The concentration of TL was adjusted to 5 μM to facilitate high copolymerization efficiency between the BAPs and pG2pA-Y in a large volume. The polymerization reaction was conducted at 37 °C for 18 h. The copolymer solution was then injected into the SEC column (HiLoad Superdex 200 pg, GE Healthcare). The SEC mobile phase was 10 mM Tris-HCl (pH 8.0). The fractions containing polymers were pooled and concentrated using an ultrafiltration membrane (30 kDa MWCO) and analyzed using SDS-PAGE and native-PAGE. The enzymatic activity of the polymers was measured, and they were functionally evaluated as probes in ELISA.
Statistical Analysis
Analysis of variance (ANOVA) and Student’s t-test functions in Microsoft Excel (Version 2016 for Macintosh, Microsoft Corp, Redmond, WA) were used to perform statistical analysis. P values of < 0.05 were considered to be statistically significant.
Acknowledgments
D.P. is grateful to the Research and Innovation in Science and Technology Project (RISET-Pro) of the Ministry of Research, Technology, and Higher Education (MoRTHE) of the Republic of Indonesia (World Bank Loan No. 8245-ID) for a Ph.D. scholarship. We thank Amano Enzyme Inc. for providing us with Trametes sp. laccase. We thank James Murray, Ph.D., of Edanz Group (www.edanzediting.com/ac) for editing a draft of this manuscript.
Glossary
Abbreviations
- BAP
bacterial (E. coli) alkaline phosphatase
- BAP-WT
wild-type (unmodified) BAP
- BAP-Y
BAP with a peptide tag containing tyrosine residue (Y-tag) at C-termini
- BAP-pG2pA-Y
BAP with a Y-tagged chimeric antibody-binding protein, pG2pA-Y, at C-termini
- BAP-Loop-Y
BAP-inserted Y-Loop sequence at 221–223 aa
- BSA
bovine serum albumin
- ELISA
enzyme-linked immunosorbent assay
- OVA
ovalbumin
- pG
protein G
- pA
protein A
- pG2pA-Y
a Y-tagged chimeric antibody-binding protein consist of two pG domains and one pA domain
- PTM
post-translational modification
- TBS
Tris-buffered saline
- TBST
TBS containing 0.1% of Tween-20
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.9b04163.
Amino acid sequences of BAPs and pG2pA-Y; purification results of BAPs and pG2pA-Y (Figures S1–S5); SDS-PAGE results of optimization of the polymerization reaction (Figure S6); HRP (Figure S7) and TL-catalyzed BAP polymerization (Figure S8); height analysis of BAP monomers and polymers in SPM analysis (Figure S9–S13); length analysis of BAP-Loop-Y polymers and BAP-Loop-Y/pG2pA-Y copolymers (Figure S14); SDS-PAGE analysis and enzymatic activity of BAPs/pG2pA copolymers (Figure S15); and SDS-PAGE analysis of TL-catalyzed copolymerization of two Y-tagged proteins with various molar ratios (Figure S16); size exclusion chromatography (SEC) of BAPs and pG2pA-Y copolymers (Figure S17) (PDF)
Author Present Address
∥ Department of Applied Chemistry, Graduate School of Engineering, Kyushu University, 744 Motooka, Nishi-ku, Fukuoka 819-0395, Japan (D.P.).
Author Contributions
This manuscript was written through contribution of all authors. All authors have given approval to the final version of the manuscript.
This study was supported by JSPS KAKENHI Grant Numbers JP16H04581, JP19H00841 (to N.K.), and JP18K14067 (to K.M.).
The authors declare no competing financial interest.
Supplementary Material
References
- Yu K.; Liu C.; Kim B. G.; Lee D. Y. Synthetic Fusion Protein Design and Applications. Biotechnol. Adv. 2015, 33, 155–164. 10.1016/j.biotechadv.2014.11.005. [DOI] [PubMed] [Google Scholar]
- Rizk M.; Antranikian G.; Elleuche S. End-to-End Gene Fusions and Their Impact on the Production of Multifunctional Biomass Degrading Enzymes. Biochem. Biophys. Res. Commun. 2012, 428, 1–5. 10.1016/j.bbrc.2012.09.142. [DOI] [PubMed] [Google Scholar]
- Iturrate L.; Sánchez-Moreno I.; Oroz-Guinea I.; Pérez-Gil J.; García-Junceda E. Preparation and Characterization of a Bifunctional Aldolase/Kinase Enzyme: A More Efficient Biocatalyst for C-C Bond Formation. Chem. - A Eur. J. 2010, 16, 4018–4030. 10.1002/chem.200903096. [DOI] [PubMed] [Google Scholar]
- Krainer F. W.; Darnhofer B.; Birner-Gruenberger R.; Glieder A. Recombinant Production of a Peroxidase-Protein G Fusion Protein in Pichia pastoris. J. Biotechnol. 2016, 219, 24–27. 10.1016/j.jbiotec.2015.12.020. [DOI] [PubMed] [Google Scholar]
- Chen X.; Zaro J. L.; Shen W.-C. Fusion Protein Linkers: Property, Design and Functionality. Adv. Drug Delivery Rev. 2013, 65, 1357–1369. 10.1016/j.addr.2012.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L.; Wang Y.; Tuyishime P.; Gao N.; Li Q.; Zheng P.; Sun J.; Ma Y. Engineering Artificial Fusion Proteins for Enhanced Methanol Bioconversion. ChemBioChem 2018, 19, 2465–2471. 10.1002/cbic.201800424. [DOI] [PubMed] [Google Scholar]
- Wang R.; Xue Y.; Wu X.; Song X.; Peng J. Enhancement of Engineered Trifunctional Enzyme by Optimizing Linker Peptides for Degradation of Agricultural By-Products. Enzyme Microb. Technol. 2010, 47, 194–199. 10.1016/j.enzmictec.2010.07.008. [DOI] [Google Scholar]
- Goeden-Wood N. L.; Conticello V. P.; Muller S. J.; Keasling J. D. Improved Assembly of Multimeric Genes for the Biosynthetic Production of Protein Polymers. Biomacromolecules 2002, 3, 874–879. 10.1021/bm0255342. [DOI] [PubMed] [Google Scholar]
- Kim H. J.; Lee E. J.; Park J. S.; Sim S. J.; Lee J. Reversible and Multi-Cyclic Protein-Protein Interaction in Bacterial Cellulosome-Mimic System Using Rod-Shaped Viral Nanostructure. J. Biotechnol. 2016, 221, 101–106. 10.1016/j.jbiotec.2016.01.033. [DOI] [PubMed] [Google Scholar]
- Foley T. L.; Burkart M. D. Site-Specific Protein Modification: Advances and Applications. Curr. Opin. Chem. Biol. 2007, 11, 12–19. 10.1016/j.cbpa.2006.11.036. [DOI] [PubMed] [Google Scholar]
- Díaz-Rodríguez A.; Davis B. G. Chemical Modification in the Creation of Novel Biocatalysts ′ Guez and Benjamin G Davis. Curr. Opin. Chem. Biol. 2011, 15, 211–219. 10.1016/j.cbpa.2010.12.002. [DOI] [PubMed] [Google Scholar]
- Stephanopoulos N.; Francis M. B. Choosing an Effective Protein Bioconjugation Strategy. Nat. Chem. Biol. 2011, 7, 876–884. 10.1038/nchembio.720. [DOI] [PubMed] [Google Scholar]
- Heck T.; Faccio G.; Richter M.; Thöny-Meyer L. Enzyme-Catalyzed Protein Crosslinking. Appl. Microbiol. Biotechnol. 2013, 97, 461–475. 10.1007/s00253-012-4569-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamihata K.; Goto M.; Kamiya N. Site-Specific Protein Cross-Linking by Peroxidase-Catalyzed Activation of a Tyrosine-Containing Peptide Tag. Bioconjugate Chem. 2011, 22, 74–81. 10.1021/bc1003982. [DOI] [PubMed] [Google Scholar]
- Mattinen M.; Lantto R.; Selinheimo E.; Kruus K.; Buchert J. Oxidation of Peptides and Proteins by Trichoderma Reesei and Agaricus Bisporus Tyrosinases. J. Biotechnol. 2008, 133, 395–402. 10.1016/j.jbiotec.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Permana D.; Minamihata K.; Tatsuke T.; Lee J. M.; Kusakabe T.; Goto M.; Kamiya N. Polymerization of Horseradish Peroxidase by a Laccase-Catalyzed Tyrosine Coupling Reaction. Biotechnol. J. 2019, 14, 1800531 10.1002/biot.201800531. [DOI] [PubMed] [Google Scholar]
- Kamiya N.; Takazawa T.; Tanaka T.; Ueda H.; Nagamune T. Site-Specific Cross-Linking of Functional Proteins by Transglutamination. Enzyme Microb. Technol. 2003, 33, 492–496. 10.1016/S0141-0229(03)00154-6. [DOI] [Google Scholar]
- Lin C.; Ting A. Y. Transglutaminase-Catalyzed Site-Specific Conjugation of Small-Molecule Probes to Proteins in Vitro and on the Surface of Living Cells. J. Am. Chem. Soc. 2006, 128, 4542–4543. 10.1021/ja0604111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck T.; Pham P.-H.; Yerlikaya A.; Thöny-Meyer L.; Richter M. Sortase A Catalyzed Reaction Pathways: A Comparative Study with Six SrtA Variants. Catal. Sci. Technol. 2014, 4, 2946–2956. 10.1039/C4CY00347K. [DOI] [Google Scholar]
- Swartz A. R.; Chen W. SpyTag/SpyCatcher Functionalization of E2 Nanocages with Stimuli-Responsive Z-ELP Affinity Domains for Tunable Monoclonal Antibody Binding and Precipitation Properties. Bioconjug. Chem. 2018, 29, 3113–3120. 10.1021/acs.bioconjchem.8b00458. [DOI] [PubMed] [Google Scholar]
- Zavada S. R.; Battsengel T.; Scott T. F. Radical-Mediated Enzymatic Polymerizations. Int. J. Mol. Sci. 2016, 17, 195 10.3390/ijms17020195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selinheimo E.; Lampila P.; Mattinen M.-L.; Buchert J. Formation of Protein-Oligosaccharide Conjugates by Laccase and Tyrosinase. J. Agric. Food Chem. 2008, 56, 3118–3128. 10.1021/jf0730791. [DOI] [PubMed] [Google Scholar]
- Bailey A. J. The Chemistry of Natural Enzyme-Induced Cross-Links of Proteins. Amino Acids 1991, 1, 293–306. 10.1007/BF00813999. [DOI] [PubMed] [Google Scholar]
- Permana D.; Minamihata K.; Goto M.; Kamiya N. Laccase-Catalyzed Bioconjugation of Tyrosine-Tagged Functional Proteins. J. Biosci. Bioeng. 2018, 126, 559–566. 10.1016/j.jbiosc.2018.05.013. [DOI] [PubMed] [Google Scholar]
- Færgemand M.; Otte J.; Qvist K. B. Cross-Linking of Whey Proteins by Enzymatic Oxidation. J. Agric. Food Chem. 1998, 46, 1326–1333. 10.1021/jf970743c. [DOI] [Google Scholar]
- Held M. A.; Tan L.; Kamyabi A.; Hare M.; Shpak E.; Kieliszewski M. J. Di-Isodityrosine Is the Intermolecular Cross-Link of Isodityrosine-Rich Extensin Analogs Cross-Linked in Vitro. J. Biol. Chem. 2004, 279, 55474–55482. 10.1074/jbc.M408396200. [DOI] [PubMed] [Google Scholar]
- Minamihata K.; Yamaguchi S.; Nakajima K.; Nagamune T. Tyrosine Coupling Creates a Hyperbranched Multivalent Protein Polymer Using Horseradish Peroxidase via Bipolar Conjugation Points. Bioconjugate Chem. 2016, 27, 1348–1359. 10.1021/acs.bioconjchem.6b00138. [DOI] [PubMed] [Google Scholar]
- Matsunaga R.; Yanaka S.; Nagatoishi S.; Tsumoto K. Hyperthin Nanochains Composed of Self-Polymerizing Protein Shackles. Nat. Commun. 2013, 4, 2211 10.1038/ncomms3211. [DOI] [PubMed] [Google Scholar]
- Minamihata K.; Goto M.; Kamiya N. Control of a Tyrosyl Radical Mediated Protein Cross-Linking Reaction by Electrostatic Interaction. Bioconjugate Chem. 2012, 23, 1600–1609. 10.1021/bc300137s. [DOI] [PubMed] [Google Scholar]
- Mori Y.; Goto M.; Kamiya N. Transglutaminase-Mediated Internal Protein Labeling with a Designed Peptide Loop. Biochem. Biophys. Res. Commun. 2011, 410, 829–833. 10.1016/j.bbrc.2011.06.073. [DOI] [PubMed] [Google Scholar]
- Mori Y.; Wakabayashi R.; Goto M.; Kamiya N. Protein Supramolecular Complex Formation by Site- Specific Avidin–Biotin Interactions. Org. Biomol. Chem. 2013, 11, 914–922. 10.1039/C2OB26625C. [DOI] [PubMed] [Google Scholar]
- Brennan C. A.; Christianson K.; La Fleur M. A.; Mandecki W. A Molecular Sensor System Based on Genetically Engineered Alkaline Phosphatase. Proc. Natl. Acad. Sci. U.S.A. 1995, 92, 5783–5787. 10.1073/pnas.92.13.5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H.; Forssell P.; Partanen R.; Buchert J.; Boer H. Improving Laccase Catalyzed Cross-Linking of Whey Protein Isolate and Their Application as Emulsifiers. J. Agric. Food Chem. 2011, 59, 1406–1414. 10.1021/jf103591p. [DOI] [PubMed] [Google Scholar]
- Cha J. Y.; Kim T. W.; Choi J. H.; Jang K. S.; Khaleda L.; Kim W. Y.; Jeon J. R. Fungal Laccase-Catalyzed Oxidation of Naturally Occurring Phenols for Enhanced Germination and Salt Tolerance of Arabidopsis thaliana: A Green Route for Synthesizing Humic-like Fertilizers. J. Agric. Food Chem. 2017, 65, 1167–1177. 10.1021/acs.jafc.6b04700. [DOI] [PubMed] [Google Scholar]
- Selinheimo E.; Lampila P.; Mattinen M.-L. L.; Buchert J. Formation of Protein - Oligosaccharide Conjugates by Laccase and Tyrosinase. J. Agric. Food Chem. 2008, 56, 3118–3128. 10.1021/jf0730791. [DOI] [PubMed] [Google Scholar]
- Mattinen M. L.; Kruus K.; Buchert J.; Nielsen J. H.; Andersen H. J.; Steffensen C. L. Laccase-Catalyzed Polymerization of Tyrosine-Containing Peptides. FEBS J. 2005, 272, 3640–3650. 10.1111/j.1742-4658.2005.04786.x. [DOI] [PubMed] [Google Scholar]
- Zhuo R.; Yuan P.; Yang Y.; Zhang S.; Ma F.; Zhang X. Induction of Laccase by Metal Ions and Aromatic Compounds in Pleurotus ostreatus HAUCC 162 and Decolorization of Different Synthetic Dyes by the Extracellular Laccase. Biochem. Eng. J. 2017, 117, 62–72. 10.1016/j.bej.2016.09.016. [DOI] [Google Scholar]
- Struch M.; Linke D.; Mokoonlall A.; Hinrichs J.; Berger R. G. Laccase-Catalysed Cross-Linking of a Yoghurt-like Model System Made from Skimmed Milk with Added Food-Grade Mediators. Int. Dairy J. 2015, 49, 89–94. 10.1016/j.idairyj.2015.04.002. [DOI] [Google Scholar]
- Jus S.; Kokol V.; Guebitz G. M. Tyrosinase-Catalysed Coupling of Functional Molecules onto Protein Fibres. Enzyme Microb. Technol. 2008, 42, 535–542. 10.1016/j.enzmictec.2008.02.012. [DOI] [Google Scholar]
- Daniela A.; Gregor H.; Martina T.; Eva B.; Gibson S.; Georg S. N.; Huber D.; Tegl G.; Baumann M.; Sommer E.; et al. Chitosan Hydrogel Formation Using Laccase Activated Phenolics as Cross-Linkers. Carbohydr. Polym. 2017, 157, 814–822. 10.1016/j.carbpol.2016.10.012. [DOI] [PubMed] [Google Scholar]
- Malencik D. A.; Anderson S. R. Dityrosine Formation in Calmodulin: Conditions for Intermolecular Cross-Linking. Biochemistry 1994, 33, 13363–13372. 10.1021/bi00249a024. [DOI] [PubMed] [Google Scholar]
- Matheis G.; Whitaker J. R. Peroxidase-Catalyzed Cross Linking of Proteins. J. Protein Chem. 1984, 3, 35–48. 10.1007/BF01024835. [DOI] [Google Scholar]
- Qu Z.; Xu H.; Xu P.; Chen K.; Mu R.; Fu J.; Gu H. Ultrasensitive ELISA Using Enzyme-Loaded Nanospherical Brushes as Labels. Anal. Chem. 2014, 86, 9367–9371. 10.1021/ac502522b. [DOI] [PubMed] [Google Scholar]
- Huang X.; Chen R.; Xu H.; Lai W.; Xiong Y. Nanospherical Brush as Catalase Container for Enhancing the Detection Sensitivity of Competitive Plasmonic ELISA. Anal. Chem. 2016, 88, 1951–1958. 10.1021/acs.analchem.5b04518. [DOI] [PubMed] [Google Scholar]
- Shin K.; Cho J. H.; Yoon M. Y.; Chung H. Use of Multiple Peptide-Based SERS Probes Binding to Different Epitopes on a Protein Biomarker to Improve Detection Sensitivity. Anal. Chem. 2016, 88, 3465–3470. 10.1021/acs.analchem.5b04873. [DOI] [PubMed] [Google Scholar]
- Jia L.; Minamihata K.; Ichinose H.; Tsumoto K.; Kamiya N. Polymeric SpyCatcher Scaffold Enables Bioconjugation in a Ratio-Controllable Manner. Biotechnol. J. 2017, 12, 1–8. 10.1002/biot.201700195. [DOI] [PubMed] [Google Scholar]
- Minamihata K.; Goto M.; Kamiya N. Site-Specific Conjugation of an Antibody-Binding Protein Catalyzed by Horseradish Peroxidase Creates a Multivalent Protein Conjugate with High Affinity to IgG. Biotechnol. J. 2015, 10, 222–226. 10.1002/biot.201400512. [DOI] [PubMed] [Google Scholar]
- Wakabayashi R.; Yahiro K.; Hayashi K.; Goto M.; Kamiya N. Protein-Grafted Polymers Prepared Through a Site-Specific Conjugation by Microbial Transglutaminase for an Immunosorbent Assay. Biomacromolecules 2017, 18, 422–430. 10.1021/acs.biomac.6b01538. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.