Abstract
Influenza vaccines historically have been multivalent, whole virus inactivated products. The first bivalent, intranasal, live attenuated influenza vaccine (LAIV; Ingelvac Provenza), with H1N1 and H3N2 subtypes, has been approved for use in swine. We investigated the LAIV hemagglutinin (HA) sequences in diagnostic cases submitted to the Iowa State University Veterinary Diagnostic Laboratory and potential vaccine virus reassortment with endemic influenza A virus (IAV) in swine. From January 3 to October 11, 2018, IAV HA sequences demonstrating 99.5–99.9% nucleotide homology to the H1 HA or 99.4–100% nucleotide homology to the H3 HA parental strains in the LAIV were detected in 58 of 1,116 (5.2%) porcine respiratory cases (H1 HA A/swine/Minnesota/37866/1999[H1N1; MN99]; H3 HA A/swine/Texas/4199-2/1998[H3N2; TX98]). Nine cases had co-detection of HA genes from LAIV and wild-type IAV in the same specimen. Thirty-five cases had associated epidemiologic information that indicated they were submitted from 11 states representing 31 individual sites and 17 production systems in the United States. Whole genome sequences from 11 cases and another subset of 2 plaque-purified IAV were included in our study. Ten whole genome sequences, including 1 plaque-purified IAV, contained at least one internal gene from endemic IAV detected within the past 3 y. Phylogenetic analysis of whole genome sequences indicated that reassortment occurred between vaccine virus and endemic field strains circulating in U.S. swine. Our data highlight the need and importance of continued IAV surveillance to detect emerging IAV with LAIV genes in the swine population.
Keywords: influenza A virus, live attenuated influenza virus vaccine, reassortment, swine
Introduction
Influenza A virus (IAV), which is one of the most challenging respiratory disease agents in swine worldwide, causes high economic losses (Donovan TS, et al. Influenza isolate selection methodology for timely autogenous vaccine use. Proc 39th Am Assoc Swine Vet Ann Meeting; Mar 2008; San Diego, CA). In addition, IAV in swine is a public health concern given the potential transmission between pigs and humans, as observed from the emergence of the pandemic A(H1N1)pdm09 and recent A(H3N2)v cases detected in humans.9,19,20 Influenza viruses in U.S. swine populations are detected throughout the year with 2 seasonal epidemic peaks; passive surveillance has detected frequent viral reassortment and rapid changes in genetic diversity with as many as 16 co-circulating genetic clades over the past 5 y.21,22,25 Despite the extent of the observed diversity, vaccination remains one of the most commonly utilized methods to help prevent clinical signs and reduce transmission among swine.24,33
In 2017, the first commercial live attenuated influenza vaccine (LAIV), Ingelvac Provenza (Boehringer Ingelheim, St. Joseph, MO) was approved for use in swine as young as 1 d of age.11,12 Ingelvac Provenza is a bivalent H3N2 and H1N1 intranasal LAIV consisting of a hemagglutinin (HA) and neuraminidase (NA) from parental strains A/swine/Texas/4199-2/1998 (TX98) H3N2 and A/swine/Minnesota/37866/1999 (MN99) H1N1, in which the LAIV H1 HA and N1 NA are expressed on the TX98 backbone through reverse genetics. The LAIV is attenuated through truncation of the nonstructural protein (NS1), which impairs the anti-interferon activity of the vaccine virus when administered to pigs.28 Previous studies using LAIVs have consistently demonstrated the benefits of this platform through broad cross-protection against antigenically distinct IAV, the ability to overcome maternally derived antibody, increased levels of mucosal IgA, and decreased transmission of heterologous IAV.10,13,31,32 In addition, LAIVs were shown to be safe when administered to pigs through the intranasal route, which further justified their use in swine.31
Influenza sequences were detected in 2018 at the Iowa State University Veterinary Diagnostic Laboratory (ISU VDL; Ames, IA) in the H1 gamma2-beta–like genetic clade using the swine H1 clade classification tool in the Influenza Research Database (IRD; www.fludb.org)40 or H3 cluster I (C-I) genetic clade using internal reference sequences.35 The unexpected detection of historic IAV phylogenetic clades not currently circulating in swine prompted further investigation that revealed that the HA detected in these diagnostic submissions were genetically similar to the LAIV MN99 or TX98 HA. Our aim was to rule out simultaneous detection of LAIV and wild-type virus in diagnostic submissions through whole genome sequencing (WGS) and plaque-purified isolates and to investigate potential LAIV reassortment with wild-type virus.
Materials and methods
IAV diagnostic submissions
The IAV diagnostic results and related sequences are maintained in a proprietary laboratory information management system (LIMS) developed at the ISU VDL. The LIMS database was queried using a test identifier for accessions, from January to October 11, 2018, that reported an IAV sequence. Accessions with an IAV sequence were downloaded into a spreadsheet with case-level metadata that had been reported on the corresponding submission forms. The sequences were used in additional NCBI GenBank BLASTn and phylogenetic analyses that detected 58 of 1,116 (5.2%) of the submissions from January 3 to October 11, 2018 with IAV HA sequences genetically similar to the H1N1 and H3N2 parental strains of Ingelvac Provenza. The parental LAIV H1 HA and N1 NA gene was from A/swine/Minnesota/37866/1999(H1N1) (MN99; GenBank EU139827, EU139837) and the LAIV H3 strain was A/swine/Texas/4199-2/1998(H3N2) (TX98; GenBank CY095672–CY095679). Twenty-three submitters requested only HA gene sequencing; these cases were included in the phylogenetic analysis but lacked additional case-level information and were excluded from the epidemiologic analysis. Thirty-five submissions included at minimum IAV reverse-transcription real-time PCR (RT-rtPCR) cycle threshold (Ct) value, IAV subtyping RT-rtPCR, state location of the pig site in the United States, RT-rtPCR sample type, and HA sequence. Submissions that were RT-rtPCR positive with a Ct <38 were subtyped and HA gene sequenced. Seven of the 35 identified submissions were enrolled in the USDA IAV surveillance program.37 NS1 gene sequencing was requested on 10 of 35 cases including 8 clinical samples and 2 virus isolates, and 11 of 35 submissions were randomly selected for WGS. Clinical specimens included lung, nasal swab, oral fluid, or virus isolated from clinical samples. No processing was required of oral fluid specimens or virus isolates prior to extraction of nucleic acid. Nasal swabs were processed by dipping the swab in 2 mL of phosphate-buffered saline, and lungs were processed into a 10% homogenate. IAV was isolated from lung tissue of 7 of 35 submissions of which 2 isolates were further plaque-purified yielding 2 and 3 plaques, respectively. A total of 6 clinical samples, 5 isolates, and 5 plaque-purified IAV from 2 isolates were included for WGS and a separate phylogenetic analysis of internal gene segments.
IAV screening RT-rtPCR
IAV extraction and RT-rtPCR were performed at the ISU VDL excluding 23 of 58 submissions for which RT-rtPCR was conducted at an outside laboratory. Viral RNA extraction was performed on lung, nasal swab, or oral fluid specimens (MagMAX pathogen RNA/DNA isolation kit; Kingfisher96 instrument; Thermo Fisher Scientific, Waltham, MA) using the high-volume lysis (HVL) procedure per the manufacturer’s instruction. For the lysis step, 100 μL of sample was added to 240 μL of lysis-binding solution. Internal positive control RNA (VetMAX Xeno internal positive control RNA; Thermo Fisher Scientific) was added to the lysis-binding solution at 20,000 copies per reaction prior to extraction to monitor PCR amplification and detect inhibition. The HVL used 2 washes each of 300 μL and 450 μL of wash solutions I and II, respectively. Nucleic acid was eluted in 90 μL of elution buffer. The HVL extraction was conducted using the Kingfisher program AM1836_DW_HV_v3.
IAV RT-rtPCR was performed on nucleic acid extracts according to the manufacturer’s instructions using PCR reagents with multiple primers and probes targeting different genomic regions (VetMAX Gold SIV detection kit; Thermo Fisher Scientific). One positive extraction control, one positive amplification control, one negative extraction control, and a negative amplification control were included with each extraction and/or PCR run. Each RT-rtPCR reaction included 12.5 μL of 2× multiplex RT-PCR buffer, 1.0 μL of 25× SIV primer–probe mix, 2.5 μL of 10× multiplex RT-PCR enzyme mix, and 1.0 μL of nuclease-free water. A final volume of 25 μL, consisting of 17 μL of master mix and 8 μL of RNA extract, was placed in each well of a 96-well fast PCR plate (Thermo Fisher Scientific). The RT-rtPCR was performed using standard mode on an AB 7500 Fast thermocycler (Thermo Fisher Scientific): 1 cycle at 48°C for 10 min, 1 cycle at 95°C for 10 min, and 40 cycles of 95°C for 15 s, 60°C for 45 s. Amplification curves were analyzed with commercial thermocycler system software. Run data were analyzed using Auto baseline with thresholds of the target and Xeno set according to the kit insert. Samples with Ct <38 were considered positive.
IAV subtyping RT-rtPCR
HA and NA subtyping was performed on RT-rtPCR–positive IAV nucleic acid extracts (VetMAX Gold SIV subtyping kit; Thermo Fisher Scientific). Separate RT-rtPCR reactions were used to detect the presence of H1 or H3 HA or N1 or N2 NA, respectively. Each RT-rtPCR reaction included 12.5 μL of 2× multiplex RT-PCR buffer, 1.0 μL of 25× H1H3 or N1N2 primer–probe mix, 2.5 μL of 10× multiplex RT-PCR enzyme mix, and 1.0 μL of nuclease-free water. Each subtyping plate included the same positive and negative controls used in the swine IAV general RT-rtPCR reaction. Cycling conditions were the same as for the IAV screening RT-rtPCR, and amplification curves were analyzed with commercial thermocycler system software using the Auto baseline with thresholds set as described in the PCR kit insert. Samples with Ct <38 were considered positive.
Sanger sequencing of IAV HA and NS1 genes
Whole gene segments of HA and NS1 were sequenced using Sanger methods for initial sequence analysis. Viral RNA was extracted as described above. Conventional RT-PCR was conducted for each gene segment (primers provided upon request; qScript XLT 1-step RT-PCR kit; Quantabio, Beverly, MA). The sequencing RT-PCR reaction was set up according to the manufacturer’s recommendations, using 200 nM of each primer. One positive extraction control (H1 or H3), one negative extraction control, and one negative amplification control were included with the reaction. The RT-PCR was performed using an ABI 2720 thermocycler (Thermo Fisher Scientific): 1 cycle at 48°C for 20 min, 1 cycle at 94°C for 3 min, 45 cycles of 94°C for 30 s, 55°C for 50 s, and 68°C for 150 s. The final elongation step was 68°C for 7 min. Detection of the RT-PCR product, HA at 1,701 base pairs (bp) and NS1 at 838 bp, was performed on a capillary electrophoresis system (QIAxcel; Qiagen, Germantown, MD), using a DNA screening cartridge and the AM420 method, and purified (ExoSAP-IT PCR cleanup reagent; Affymetrix, Thermo Fisher Scientific) following the manufacturer’s recommendations. Samples were submitted to the Iowa State University DNA facility (Ames, IA) for sequencing. Lasergene SeqMan Pro software v.14.0.0 (DNASTAR, Madison, WI) was used to compile sequences.
Virus isolation and plaque purification
Confluent monolayers of Madin–Darby canine kidney (MDCK) cells were prepared in 25-cm2 flasks (Costar, Corning, NY). Cell culture medium was removed, and monolayers were washed 2 times with IAV post-inoculation medium. Post-inoculation medium included 97.3% Dulbecco modified Eagle medium (DMEM), 1% penicillin–streptomycin (10,000 U/mL penicillin and 10,000 µg/mL streptomycin), 1% 200 mM L-glutamine, 0.1% gentamicin (50 mg/mL), 0.4% amphotericin B (250 µg/mL), and 0.2% tosyl phenylalanyl chloromethyl ketone (TPCK)–trypsin (1 mg/mL). A syringe and needle were used to collect 1 mL of sample that was passed through a 0.22-μm filter (Costar). The flask was incubated with sample inoculum for at least 1 h and subsequently rinsed with 2 mL of post-inoculation medium, twice. Finally, 5 mL of post-inoculation medium was added to the flask and incubated at 37°C and 5% CO2. Cell cultures were evaluated for the appearance of cytopathic effect (CPE) daily. If the flask demonstrated >50% CPE, the flask was frozen at −70°C. The cell culture fluid was tested for HA activity, and HA-positive cell culture fluids were tested with the IAV subtyping RT-rtPCR to confirm the original subtype of the clinical sample. Samples negative for CPE and/or HA units were subjected to a second cell culture passage. Samples were considered negative if CPE and HA units were negative after the second passage on cell culture. Positive and negative inoculation controls were included to validate testing.
Two IAV isolates were randomly selected for further plaque purification. The IAV isolates were 10-fold serially diluted in post-inoculation medium that included DMEM supplemented with 1× pen-strep and TPCK-trypsin (1.5 µg/mL) in a 96-well plate. Prior to inoculation, confluent MDCK cell monolayers in 6-well plates were washed with post-inoculation medium 2 times. Next, 300 μL of each virus dilution were added to a single well, and the plates were incubated for 1 h at 37°C in a 5% CO2 incubator. After 1 h, the virus inoculum was removed from the cells, and 3 mL of overlay medium was added to each well. The overlay medium contained 1× MEM, 1% sea plaque agarose, NaHCO3 (4.4 g/L), TPCK-trypsin (1.5 µg/mL), and neutral red (0.33%).
After the agar was firm (~1 h), the cells were incubated at 37°C with 5% CO2. Plaque formation was checked daily for 2–3 d. When a plaque was clearly visible, it was selected with a pipette tip, and placed into 300 μL of post-inoculation medium and transferred to confluent MDCK cell monolayers in 6-well plates and incubated 1 h at 37°C in a 5% CO2 incubator. Finally, 3 mL of post-inoculation medium was added to each well, and the cells were incubated at 37°C with 5% CO2 until CPE was observed. When the MDCK cells demonstrated 80% CPE, the plates were subjected to 3 freeze–thaw cycles to collect the purified virus.
Whole genome sequencing
Viral RNA was extracted (MagMAX pathogen RNA kit; KingFisher Flex system; Thermo Fisher Scientific). Sequencing libraries were constructed (TruSeq stranded total RNA library preparation kit, catalog 20020596; Illumina, San Diego, CA). Next-generation sequencing was performed (MiSeq platform; Illumina) following standard Illumina protocols at the ISU VDL.6,38,39 Approximately 2,000,000 raw sequencing reads per sample were pre-processed using Trimmomatic v.0.36. Sequencing was quality-checked with FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/).3 Quality-trimmed total reads were mapped against reference sequences downloaded from the NCBI Influenza Sequence Database (ftp://ftp.ncbi.nih.gov/genomes/INFLUENZA/) using BWA-MEM.17 Mapped reads were extracted using SAMtools and used for de novo assembly.18 For each segment, contigs were assembled using ABySS and SPAdes.2,27 The contigs were manually curated in SeqMan Pro to remove extraneous sequences and trim chimeric contigs, thus generating a consensus sequence per segment.
Phylogenetic analysis
IAV HA sequence data from swine collected in the United States between January 2009 and May 2019 were downloaded from the IRD.29 Downloaded data were then sampled randomly (n = 500) and aligned with 35 H1 or H3 LAIV HA from the ISU VDL using default settings in MAFFT v.7.409.14 Twenty-three cases submitted exclusively for HA sequencing without epidemiologic data or state-level information were included in our phylogenetic analysis, but were excluded from clinical descriptions. The best-known maximum likelihood phylogenetic tree was inferred using FastTree v.2.1,23 implementing a general time-reversible (GTR) model of nucleotide substitution with gamma-distributed rate variation among sites. Genetic clade and evolutionary lineage was assigned using octoFLU (https://github.com/flu-crew/octoFLU).5 Figtree v.1.4.4 was used to view and annotate the phylogenetic trees (http://tree.bio.ed.ac.uk/software/figtree/). Genetic diversity between phylogenetic clades was assessed using MEGA version X.15
Six H3 HA amino acid positions (145, 155, 156, 158, 159, and 189; H3 numbering) found in the globular head of the HA gene were identified to be of importance in antigenic evolution in swine.16 These positions were collectively referred to as “antigenic motif” and are known to play a role in defining the antigenic phenotype. The antigenic motif of LAIV H3 HA was compared to the known swine H3 HA motifs currently described.1,4
Results
Case history
Among 1,116 cases with IAV HA sequences detected at the ISU VDL from January 3 to October 11, 2018, 58 cases contained HA sequences that belonged to historic genetic clades that have not been circulating recently in U.S. swine. Of these 58 cases, 21 resulted in H1 HA sequences classified as 1A.2-3–like (γ2-β–like) based on the IAV global and U.S. clade classification reported through the swine H1 clade classification tool in IRD. Thirty-eight cases included H3 HA sequences phylogenetically similar to C-I reference sequences used to classify H3 IAV sequences at the ISU VDL. The unusual detection of historic H1 γ2-β–like HA and H3 C-I HA circulating in swine prompted a BLAST analysis through NCBI GenBank. The 21 H1 γ2-β–like HA BLAST analysis revealed highest hits with the MN99 HA, with 99.5–99.9% nucleotide homology. The 38 H3 C-I HA BLAST detected the highest hits with the TX98 HA, with 99.4–100% nucleotide homology. The H1 MN99 and H3 TX98 are the parental HA antigens used in the LAIV.
LAIV HA were detected by Sanger sequencing through routine laboratory testing: 16 MN99-like H1, 32 TX98-like H3, and 1 dual MN99-like and TX98-like HA genes. Additionally, LAIV HA were detected in the same sample with other endemic strains of IAV in swine: 4 MN99 HA co-detections with H1-δ1 (1 of 4) or H3-2010.1 (3 of 4) HA, and 5 TX98 HA co-detections with H1-α (1 of 5) or H1-γ (4 of 5) HA, suggesting that swine were infected with an endemic IAV and the LAIV at the time of sample collection in these specific cases (Table 1). Thirty-five of the 58 cases had additional epidemiologic data that included state-level information and various clinical data. These 35 cases included: 11 MN99-like H1, 17 TX98-like H3, 2 MN99-like H1 HA co-detections with contemporary H3-2010.1 HA, and 5 TX98-like H3 HA co-detections with contemporary H1-α and H1-γ HA. These 35 cases with evidence of MN99- or TX98-like sequences were submitted from 11 states (IA, IL, KY, MI, MN, MO, MT, NE, OH, SD, and TN), 17 production systems, and 31 different swine sites; 7 of these cases were enrolled in the USDA swine IAV surveillance program (Supplementary Table 1). Nursery pigs 3- to 10-wk-old represented the largest number of cases (22 of 35), followed by grower pigs 11- to 17-wk-old (4 of 35), finisher pigs 18- to 25-wk-old (4 of 35), and 1 case that originated from adult swine (1 of 35). Four cases did not designate a pig age in their submission. The MN99-like and TX98-like HA sequences were detected in multiple sample types including lung (5 of 35), nasal swab (5 of 35), oral fluids (18 of 35), and virus isolates required by the USDA swine IAV surveillance program (7 of 35). The IAV RT-rtPCR at the ISU VDL detected a range in Ct values from 16.0 (lung) to 34.4 (lung; Supplementary Table 1).
Table 1.
Summary of porcine cases submitted to the Iowa State University Veterinary Diagnostic Laboratory that contained an influenza A virus (IAV) hemagglutinin (HA) sequence genetically similar to MN99 or TX98 contained in the live attenuated influenza vaccine and detected as a single or mixed sequence detection during January 3 to October 11, 2018.
| HA | Total cases* | Cases with epidemiologic data† |
|---|---|---|
| MN99-like H1 | 16 | 11 |
| TX98-like H3 | 32 | 17 |
| MN99-like H1 + TX98-like H3 | 1 | 0 |
| MN99-like H1 + endemic IAV | 4 | 2 |
| TX98-like H3 + endemic IAV | 5 | 5 |
| Total | 58 | 35 |
Total porcine cases submitted with MN99-like or TX98-like HA sequences.
Subset of total porcine cases with MN99-like or TX98-like HA sequences with clinical epidemiologic metadata.
Most of the porcine cases (18 of 35) did not have associated vaccination information. However, 11 cases from 6 production systems had used the LAIV in the pigs at processing (3- to 5-d-old) through weaning (3-wk-old) prior to submission of diagnostic samples, although the length of time between vaccination and submission of the case to the ISU VDL was unknown. There were 6 cases reporting that no LAIV was administered to the pigs, although prior use of the LAIV in the production system was possible.
Clinical epidemiologic data
Clinical signs of respiratory disease were reported in 17 of 35 of the porcine cases that indicated coughing, with variable reports of respiratory distress, wasting, morbidity, and low mortality (Supplementary Table 2). The remaining cases, 18 of 35, did not report clinical signs, although this does not confirm the absence of respiratory disease at the time of sample collection given that many ISU VDL submissions are received without clinical descriptions. Macroscopic lung lesions including cranioventral consolidation suggestive of influenza were reported in 9 cases from tissues collected at autopsy, although 6 of these submissions had secondary bacterial growth that may have caused similar gross lung lesions. In addition, 4 cases reported pulmonary congestion, hemorrhage, pleuritis, or edema, and 3 cases did not report lesions on the submission form. Nineteen cases that included sequences originating from antemortem samples (nasal swabs and oral fluids) did not include macroscopic lung lesion data on the submission form. Histopathology results from 12 cases described necrotizing bronchiolitis consistent with IAV and 2 cases described bronchopneumonia. Twenty-one cases either had no lesions observed or were unavailable because antemortem sample submissions did not include lung. Various coinfections were detected by rtPCR or RT-rtPCR including porcine reproductive and respiratory syndrome virus (PRRSV), porcine circovirus 2 (PCV-2), and Mycoplasma hyopneumoniae. Routine culture detected secondary bacteria in 12 cases, including single infections or coinfections with Pasteurella multocida, Streptococcus suis, Haemophilus parasuis, and Bordetella bronchiseptica. No significant growth was reported in 3 cases. Bacterial culture was not requested in 20 cases given the submission of antemortem samples or the submitter did not include a request for bacterial culture (Supplementary Table 2).
Hemagglutinin sequencing and phylogenetic analysis
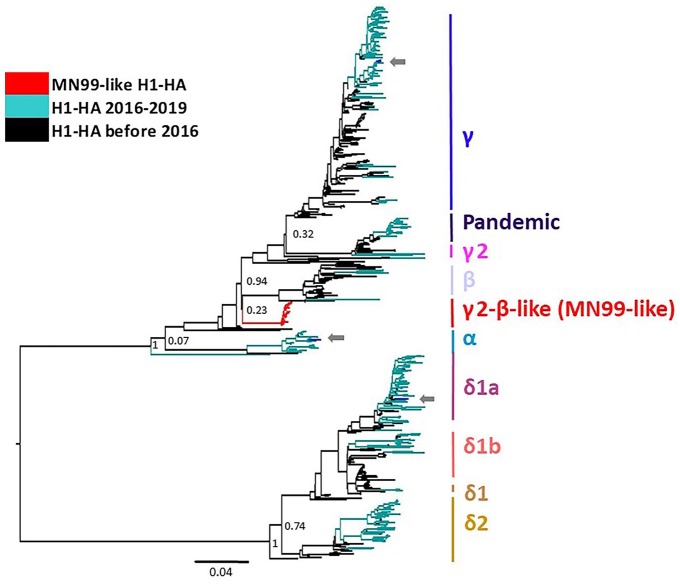
The 21 H1 MN99-like HA were included in an H1 phylogenetic analysis with endemic swine IAV that included an additional 486 randomly sampled H1 HA that represented the genetic diversity of IAV in the United States between 2009 and 2019 (29 H1-α, 37 H1-β, 201 H1-γ, 5 H1-γ2, 137 H1-δ1, 55 H1-δ2, 22 H1pdm09). The H1 MN99-like HA sequences shared a common ancestor with H1-β and H1-γ but formed a statistically supported monophyletic and independent clade (Fig. 1). The genetic diversity of sequences within the MN99-like HA clade averaged 0.3%, and between clade divergence was 8.4% (H1-β) to 29.3% (H1-δ1; Table 2).
Figure 1.
Maximum-likelihood phylogenetic tree of H1 hemagglutinin (HA) genes, including viruses with the MN99-like H1 in the γ2-β–like clade reported in our investigation. Arrows demonstrate co-detection with contemporary influenza A virus (IAV) within their respective phylogenetic clade. MN99-like H1 formed a separate statistically supported monophyletic γ2-β–like clade when compared to contemporary IAV.
Table 2.
Estimates of the within- and between-clade genetic diversity of: A. γ2-β–like H1 phylogenetic clade containing MN99-like H1 hemagglutinin gene sequence labeled as γ2-β–like; and B. cluster I H3 phylogenetic clade containing the TX98-like H3 hemagglutinin gene sequence labeled as C-I.
| Within (%) | Between (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| H1α | H1β | H1γ | H1γ2 | H1δ1 | H1δ2 | H1pdm09 | ||||
| A. γ2-β–like H1 clade | ||||||||||
| H1-α | 7.5 | |||||||||
| H1-β | 4.5 | 15.0 | ||||||||
| H1-γ | 3.5 | 15.9 | 11.4 | |||||||
| H1-γ2 | 7.3 | 16.2 | 12.5 | 11.0 | ||||||
| H1-δ1 | 4.8 | 30.2 | 29.8 | 31.5 | 31.7 | |||||
| H1-δ2 | 4.9 | 30.5 | 29.9 | 30.8 | 30.6 | 12.0 | ||||
| H1pdm09 | 3.2 | 16.5 | 12.0 | 9.8 | 12.1 | 31.0 | 31.2 | |||
| γ2-β–like | 0.3 | 12.6 | 8.4 | 9.5 | 9.9 | 29.3 | 28.8 | 10.5 | ||
| Within (%) | Between (%) | |||||||||
| 2010.1 | 2010.2 | C-IV | C-IVA | C-IVB | C-IVC | C-IVD | C-IVE | C-IVF | ||
| B. Cluster I H3 clade | ||||||||||
| 2010.1 | 1.5 | |||||||||
| 2010.2 | 0.9 | 6.6 | ||||||||
| C-IV | 4.4 | 12.4 | 12.3 | |||||||
| C-IVA | 2.1 | 13.2 | 13.4 | 5.9 | ||||||
| C-IVB | 4.1 | 13.8 | 13.7 | 6.1 | 6.8 | |||||
| C-IVC | NA | 12.6 | 13.0 | 5.6 | 6.4 | 6.5 | ||||
| C-IVD | 1.5 | 12.5 | 12.7 | 5.0 | 6.2 | 6.0 | 5.8 | |||
| C-IVE | 2.0 | 13.3 | 13.5 | 5.5 | 6.3 | 6.7 | 6.3 | 5.5 | ||
| C-IVF | 1.3 | 13.3 | 13.6 | 5.6 | 6.4 | 6.8 | 6.4 | 6.0 | 6.5 | |
| C-I | 0.2 | 11.6 | 11.3 | 8.2 | 9.5 | 10.3 | 9.6 | 9.5 | 9.6 | 9.5 |
NA = a single C-IVC sequence was used in analysis, resulting in no within-clade diversity measure.
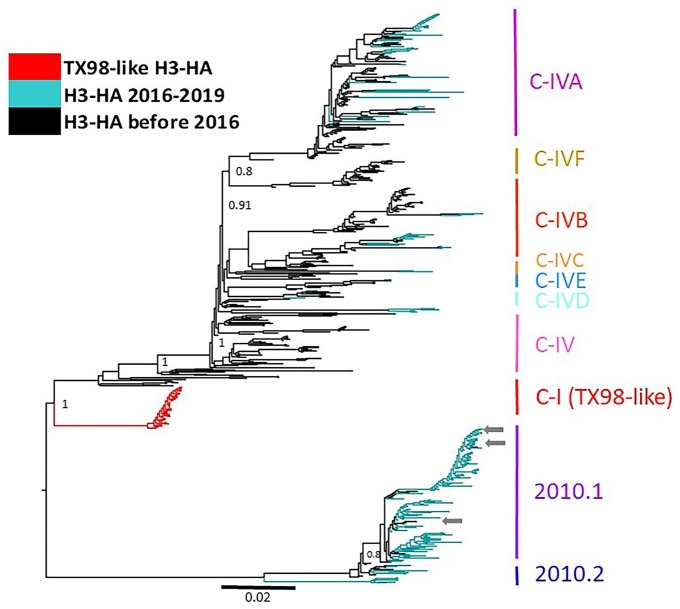
The 38 H3 TX98-like HA were included in an H3 phylogenetic analysis with endemic swine IAV that included an additional 500 randomly sampled H3 HA that represented the genetic diversity of IAV in the United States between 2009 and 2019 (141 2010.1, 6 2010.2, 91 C-IV, 139 C-IVA, 78 C-IVB, 1 C-IVC, 8 C-IVD, 10 C-IVE, 26 C-IVF). The TX98-like HA sequences formed a statistically supported monophyletic and independent clade (Fig. 2). The genetic diversity within the TX98-like HA clade averaged 0.2%, and between clade divergence was 8.2% (H3 C-IV) to 11.6% (H3-2010.1; Table 2). The 38 TX98-like HA detections contained the same antigenic motif, KHKEYS, as the parental TX98 sequence (NCBI accession CY095675).
Figure 2.
Maximum-likelihood phylogenetic tree of the H3 hemagglutinin (HA) genes, including viruses with the TX98-like H3 in the C-I clade reported in our investigation. Arrows demonstrate co-detection with contemporary influenza A virus (IAV) within their respective phylogenetic clade. TX98-like H3 form a separate statistically supported monophyletic C-I clade compared to contemporary IAV.
The HA sequences from 35 cases with epidemiologic data were submitted to GenBank as accessions MN328310–MN328337. Sequences submitted through the USDA surveillance program are identified in GenBank as accessions A02139351, A02156993, A02157797, A02157798, A02157974, A02254795, and A02257614.
Whole genome sequencing
Eleven cases were randomly chosen for WGS, with 5 virus isolates that originated from lung, as well as direct sequencing from 2 lung, 2 oral fluid, and 2 nasal swab samples (Table 3). There were 9 cases from nursery pigs, 1 case from grower swine, and 1 case from adult swine; several of these cases were submitted from swine with clinical respiratory disease and lung lesions consistent with IAV infection. To reduce the possibility of detecting whole genome sequences representing the presence of both LAIV and individual endemic IAV, 2 virus isolates were plaque purified (1P, 7P) and included in the whole genome sequences. The HA sequences generated from WGS were consistent with Sanger sequencing and detected 7 MN99-like H1 HA and 6 TX98-like H3 HA with co-detections in 3 cases with endemic IAV HA including 2 H1-γ with TX98-like HA and 1 H3-2010.1 with a MN99-like HA (Fig. 3). The 2 plaques demonstrated whole genome sequences that were identical to the whole genomes generated from their respective isolates. Clinical epidemiologic data associated with the WGS cases is reported in Table 3 and corresponds to HA numbers 1, 3–7, 9, 10, 13, 17, and 20 in Supplementary Tables 1 and 2.
Table 3.
Clinical epidemiologic data from 11 porcine cases with MN99-like H1 or TX98-like H3 hemagglutinin (HA) and whole genome sequences, including 2 plaque-purified isolates from the Iowa State University Veterinary Diagnostic Laboratory.
| WGS | HA clade | IAV co-detection | State | Age | Sample | PCR Ct | LAIV vaccinated | Clinical signs | Histopathology |
|---|---|---|---|---|---|---|---|---|---|
| 1 | C-I | ND | IA | Nursery | Virus isolate | 18.1 | Unknown | Respiratory disease | Necrotizing bronchiolitis |
| 1P | C-I | ND | IA | Nursery | Virus isolate | 18.1 | Unknown | Respiratory disease | Necrotizing bronchiolitis |
| 2 | C-I | ND | IA | Nursery | Lung | 27.1 | Yes | Wasting | Bronchopneumonia |
| 3 | C-I | ND | TN | Nursery | Virus isolate | 16.1 | Unknown | Respiratory disease | Necrotizing bronchiolitis |
| 4 | C-I | H1-γ | MO | Nursery | Nasal swab | 25.1 | Unknown | NR | NA |
| 5 | C-I | H1-γ | MO | Nursery | Oral fluid | 29.4 | No | NR | NA |
| 6 | γ2-β–like | ND | MT | Nursery | Lung | 25.1 | Yes | Respiratory disease | Necrotizing bronchiolitis |
| 7 | γ2-β–like | ND | SD | Nursery | Virus isolate | 20.5 | Unknown | Respiratory disease | Necrotizing bronchiolitis |
| 7P | γ2-β–like | ND | SD | Nursery | Virus isolate | 20.5 | Unknown | Respiratory disease | Necrotizing bronchiolitis |
| 8 | γ2-β–like | ND | NE | Nursery | Oral fluid | 24.9 | Unknown | Respiratory disease | NA |
| 9 | γ2-β–like | ND | IL | Grower | Virus isolate | 19.7 | Unknown | Respiratory disease | Necrotizing bronchiolitis |
| 10 | γ2-β–like | ND | NE | Nursery | Virus isolate | 23.9 | Unknown | Respiratory disease | Necrotizing bronchiolitis |
| 11 | γ2-β–like | H3-2010.1 | OH | Adult | Nasal swab | 32.1 | Unknown | NR | NA |
1P, 7P = plaque-purified isolate; Ct = cycle threshold; IAV = influenza A virus; LAIV = live attenuated influenza vaccine; NA = not applicable; ND = not detected; NR = not reported.
Figure 3.
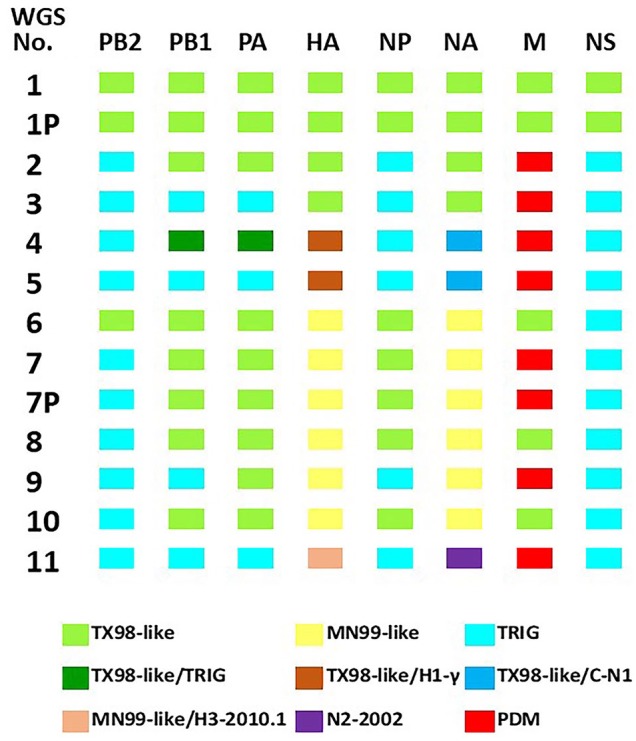
Gene constellations of 11 whole genome sequences and 2 plaques originating from cases that demonstrated MN99-like or TX98-like HA with evidence of mixed infections or reassortment with contemporary influenza A virus (IAV) circulating in swine. C-N1 = classical neuraminidase 1; HA = hemagglutinin; M = matrix; NA = neuraminidase; NP = nucleoprotein; NS = nonstructural; PA = polymerase acidic; PB1 = polymerase basic 1; PB2 = polymerase basic 2; PDM = 2009 H1 pandemic; TRIG = triple reassortant internal gene.
The data generated from WGS demonstrated case 1 and its respective plaque, 1P, with the first 7 gene segments consistent with a TX98-like whole genome contained in the LAIV (Fig. 3). However, the NS1 gene segment had a 185-nt deletion compared to the Provenza NS1 and a 245-nt deletion compared to the TX98 NS1 sequence. In contrast, 10 whole genome sequences (2–11) and plaque 7P demonstrated multiple different genotypes in which LAIV internal gene segments were replaced with contemporary IAV representing the triple reassortant internal gene (TRIG) constellation. There were 9 contemporary TRIG sequences detected in the PB2, 5 TRIG in PB1, 4 TRIG in PA, and 6 TRIG in NP. There were 7 whole genome sequences with a H1pdm09 M replacing the LAIV TX98-like M gene. Only sequence 4 represented co-detections of TX98-like PB1 and PA with TRIG internal genes, and sequences 4, 5, and 11 demonstrated the presence of endemic NA lineages.34 However, all 10 whole genome sequences (2–11) and plaque 7P had replaced the LAIV NS1 gene that contained the characteristic 126 amino acid truncated protein with a contemporary TRIG NS1 gene,26 and Sanger sequencing conducted on 10 additional cases also revealed a contemporary wild-type NS1 gene replacing the LAIV NS1 (data not shown). Phylogenetic trees were constructed to compare each NA and internal gene segments from the ISU VDL cases with published sequence data from IAV in swine (Supplementary Figs. 1–8). For all segments, the TX98 internal gene sequences and the TX98-like detected in WGS formed their own clade that was separate from contemporary TRIG or H1pdm09 IAV genes. Collectively, these data indicate that reassortment had occurred between the LAIV and wild-type IAV circulating in swine.
Discussion
Our results demonstrate the presence of HA sequences similar to the LAIV virus in 58 routine diagnostic swine cases submitted to the ISU VDL. A subset of 35 of these cases had clinical data describing a history of clinical respiratory disease or microscopic lung lesions consistent with IAV infection. Further, WGS analysis of 11 selected accessions demonstrated concurrent detection of an LAIV and endemic IAV HA sequence in the same sample from 3 cases, as well as 10 reassortant viruses that contained at least 1 or more contemporary internal genes from TRIG or pandemic IAV lineages. Although the Provenza vaccine contains attenuated viruses, these data suggest the ability of the LAIV virus to replicate in swine coinfected with wild-type IAV and may result in reassortant strains with a combination of LAIV genes and genes from endemic IAV in swine. It is currently unknown how these reassortant LAIV may influence the ecology of IAV circulating in swine, but they continue to be detected. ISU VDL IAV summary data are publicly available through ISU FLUture (https://influenza.cvm.iastate.edu/).37 To date, 7.5% of 2019 IAV swine sequences were characterized as H1 γ2-β–like or H3 C-I IAV phylogenetic clades (accessed October 16, 2019).
Twenty-three submissions with LAIV HA lacked clinical and state-level information on the ISU VDL submission form, so were excluded from the epidemiologic analysis given that they may have been submitted from Provenza vaccine field studies. Detection of either the MN99-like or TX98-like HA genes could be expected based on post-vaccination timing of sample collection and may be responsible for the LAIV genotype detected in whole genome sequences 1 and 1P (Fig. 3). In contrast, of the 35 cases submitted with epidemiologic data and sequences similar to MN99 or TX98 HA, 17 included case descriptions with clinical respiratory disease. Eleven of these 17 reported a history of LAIV vaccination, and 6 submissions reported either no LAIV administration in the affected pigs or the information was not provided. However, we cannot differentiate whether sample collection of lung, oral fluid, or nasal swabs occurred shortly after administration of the LAIV, or if these cases represent field scenarios in which LAIV virus persisted longer in the swine population than previously reported, or if the LAIV virus was transmitted from vaccinated to unvaccinated pigs. In addition, cases with clinical respiratory disease may represent concurrent detection of LAIV and an endemic IAV that may have been responsible for the clinical signs at the time of submission.
At least 6 of 35 of the ISU VDL clinical submissions detected MN99-like or TX98-like HA in combination with an endemic H1-α, H1-γ, or H3-2010.1. Importantly, some of these cases included oral fluids, which are by their nature a pooled sample type representing groups of pigs, but pooled samples may increase the opportunity to detect mixed infections. However, mixed IAV subtypes were also detected in nasal swabs that would represent individual pigs if collected properly. Detection of mixed IAV subtypes by PCR in LAIV vaccinated pigs is ambiguous given that the vaccine contains 2 subtypes, H1N1 and H3N2, representing all of the possible HA and NA subtypes circulating in swine. In contrast, detecting multiple HA gene sequences in swine samples may suggest that reassortment may have occurred between LAIV and endemic strains considering the ubiquitous nature of IAV in swine. Interestingly, 10 of 35 cases requested NS1 sequencing through Sanger methods that revealed the presence of contemporary NS1 gene segments without the truncated NS1 from the LAIV virus. However, Sanger sequencing may not discriminate between different IAV if more than one virus is present in a sample given that this method likely detects the target representing the IAV in highest quantity. Collectively, these data suggest the possibility that IAV-infected pigs were simultaneously vaccinated with LAIV, providing an opportunity for either the co-detection of LAIV and wild-type virus or the detection of reassorted viruses. Indeed, LAIV administered to nonclinical but unknowingly infected swine with endemic strains of IAV may provide an opportunity for viral reassortment under appropriate conditions.7,8
Antigenic variability of IAV in swine and other host species occurs through genetic drift and shift.30 Antigenic drift is the result of point mutations in the genome, which is responsible for small genetic changes that accumulate over time. In contrast, antigenic shift is an abrupt, larger change in HA and NA through reassortment that occurs when 2 IAV strains infecting the same cell exchange different genes that are subsequently included in progeny virus.30 The presence of endemic IAV genes, including wild-type NS1, present in ISU VDL submissions with MN99-like and/or TX98-like HA, suggests the occurrence of reassortment between LAIV and wild-type IAV. The results of the whole genome sequence phylogenetic analysis demonstrated that 10 whole genome sequences contained internal genes representing contemporary IAV, and at least 3 whole genome sequences had NA genes consistent with wild-type IAV, indicating that reassortment occurred between LAIV and contemporary IAV. Although it is possible that WGS methods detected 2 exclusive IAV in a sample, whole genome sequences from virus isolates and plaque-purified IAV and a growing number of these detections in swine support evidence of reassortment. Interestingly, plaque-purified IAV demonstrated identical whole genome sequences to the passage 1 virus isolates with 1 plaque (1P), representing a near complete constellation of LAIV gene segments, except that the NS1 contained larger nt deletions compared to the Provenza NS1. In contrast, the additional plaque (7P) had 3 genes (PB2, M, NS1) representing contemporary IAV, which strongly suggests that reassortment occurred between LAIV and wild-type IAV. Although 11 whole genome sequences represent a limited number of cases, 10 whole genome sequences contained an endemic wild-type TRIG NS1, suggesting that reassortant progeny IAV with the non-truncated, contemporary NS1 gene had a selective advantage. Collectively, these data indicate that viral reassortment is possible with LAIV, although the viral phenotype of these reassortant IAV isolates requires further evaluation.
IAV is ubiquitous in swine populations around the world. Endemic infections are common in U.S. breeding herds, and piglets <3-wk-old have been implicated in maintaining endemic IAV in these populations.7,8,36 Vaccination of young piglets may provide protection against infection later in life. However, the ubiquitous nature of IAV infection in swine and particularly the presence of endemic infections in asymptomatic nursing piglets may create an opportunity for viral reassortment in infected swine without clinical signs at the time of vaccination with LAIV. Vaccinating swine free of IAV to avoid potential reassortment presents a challenge. Replication potential and persistence of LAIV virus in swine may vary depending on the method of attenuation and thus influence the potential for reassortment with endemic strains. Our data indicating reassortment emphasizes the need for consistent, herd-level IAV surveillance to detect the presence, magnitude, and frequency of clades that are circulating in production systems to maximize vaccine utility and prevent potential reassortment events and emergence of genetically distinct IAV. However, our investigations also reinforce the need for submitters to the ISU VDL to provide complete descriptions of vaccine history and clinical signs to facilitate higher confidence in interpretation of results.
Supplemental Material
Supplemental material, Supplemental_material for Detection of live attenuated influenza vaccine virus and evidence of reassortment in the U.S. swine population by Aditi Sharma, Michael A. Zeller, Ganwu Li, Karen M. Harmon, Jianqiang Zhang, Hai Hoang, Tavis K. Anderson, Amy L. Vincent and Phillip C. Gauger in Journal of Veterinary Diagnostic Investigation
Acknowledgments
We acknowledge the staff at Iowa State University Veterinary Diagnostic Laboratory for their assistance with testing. We thank Dr. Jennifer Chang at USDA for assistance with clade classification using octoFLU. T.K. Anderson was supported by an appointment to the USDA– Agricultural Research Service (ARS) Research Participation Program administered by the Oak Ridge Institute for Science and Education (ORISE) through an interagency agreement between the U.S. Department of Energy (DOE) and USDA under contract DE-AC05-06OR23100. This research used resources provided by the SCINet project of the USDA-ARS project 0500-00093-001-00-D. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture, DOE, or ORISE. USDA is an equal opportunity provider and employer.
Footnotes
Declaration of conflicting interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The ISU VDL provided funding for additional testing. P.C. Gauger, A. Sharma, and M.A. Zeller were supported by an Iowa State University Presidential Interdisciplinary Research Initiative Award.
ORCID iDs: Tavis K. Anderson  https://orcid.org/0000-0002-3138-5535
https://orcid.org/0000-0002-3138-5535
Phillip C. Gauger  https://orcid.org/0000-0003-2540-8769
https://orcid.org/0000-0003-2540-8769
Supplementary material: Supplementary material for this article is available online.
References
- 1. Abente EJ, et al. The molecular determinants of antibody recognition and antigenic drift in the H3 hemagglutinin of swine influenza A virus. J Virol 2016;90:8266–8280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bankevich A, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012;19:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bolger AM, et al. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bolton MJ, et al. Antigenic evolution of H3N2 influenza A viruses in swine in the United States from 2012 to 2016. Influenza Other Respir Viruses 2019;13:83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chang J, et al. octoFLU: automated classification for the evolutionary origin of influenza A virus gene sequences detected in U.S. swine. Microbiol Resour Announc 2019;8:e00673-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen Q, et al. Metagenomic analysis of the RNA fraction of the fecal virome indicates high diversity in pigs infected by porcine endemic diarrhea virus in the United States. Virol J 2018;15:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Diaz A, et al. Complete genome sequencing of influenza A Viruses within swine farrow-to-wean farms reveals the emergence, persistence, and subsidence of diverse viral genotypes. J Virol 2017;91:e00745-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Diaz A, et al. Association between influenza A virus infection and pig subpopulations in endemically infected breeding herds. PLoS One 2015;10:e0129213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Epperson S, et al. Influenza activity—United States, 2013–14 season and composition of the 2014–15 influenza vaccines. MMWR Morb Mortal Wkly Rep 2014;63:483–490. [PMC free article] [PubMed] [Google Scholar]
- 10. Gauger PC, et al. Live attenuated influenza A virus vaccine protects against A (H1N1) pdm09 heterologous challenge without vaccine associated enhanced respiratory disease. Virology 2014;471:93–104. [DOI] [PubMed] [Google Scholar]
- 11. Genzow M, et al. Live attenuated influenza virus vaccine reduces virus shedding of newborn piglets in the presence of maternal antibody. Influenza Other Respir Viruses 2018;12:353–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kaiser TJ, et al. Influenza A virus shedding reduction observed at 12 weeks post-vaccination when newborn pigs are administered live-attenuated influenza virus vaccine. Influenza Other Respir Viruses 2019;13:274–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kappes MA, et al. Vaccination with NS1-truncated H3N2 swine influenza virus primes T cells and confers cross-protection against an H1N1 heterosubtypic challenge in pigs. Vaccine 2012;30:280–288. [DOI] [PubMed] [Google Scholar]
- 14. Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Bio Evol 2013;30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kumar S, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 2018;35:1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lewis NS, et al. Substitutions near the hemagglutinin receptor-binding site determine the antigenic evolution of influenza A H3N2 viruses in U.S. swine. J Virol 2014;88:4752–4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics 2009;25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nelson MI, et al. Global transmission of influenza viruses from humans to swine. J Gen Virol 2012;93:2195–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nelson MI, et al. Continual reintroduction of human pandemic H1N1 influenza A viruses into swine in the United States, 2009 to 2014. J Virol 2015;89:6218–6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nelson MI, et al. Global migration of influenza A viruses in swine. Nat Commun 2015;6:6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nelson MI, et al. Evolution of novel reassortant A/H3N2 influenza viruses in North American swine and humans, 2009–2011. J Virol 2012;86:8872–8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Price MN, et al. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 2010;5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rajao DS, et al. Pathogenesis and vaccination of influenza a virus in swine. Curr Top Microbiol Immunol 2014;385:307–326. [DOI] [PubMed] [Google Scholar]
- 25. Rajao DS, et al. Reassortment between swine H3N2 and 2009 pandemic H1N1 in the United States resulted in influenza A viruses with diverse genetic constellations with variable virulence in pigs. J Virol 2017;91:e01763-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Richt JA, et al. Vaccination of pigs against swine influenza viruses by using an NS1-truncated modified live-virus vaccine. J Virol 2006;80:11009–11018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Simpson JT, et al. ABySS: a parallel assembler for short read sequence data. Genome Res 2009;19:1117–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Solorzano A, et al. Mutations in the NS1 protein of swine influenza virus impair anti-interferon activity and confer attenuation in pigs. J Virol 2005;79:7535–7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Squires RB, et al. Influenza research database: an integrated bioinformatics resource for influenza research and surveillance. Influenza Other Respir Viruses 2012;6:404–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vincent AL, et al. Swine influenza viruses: a North American perspective. Adv Virus Res 2008;72:127–154. [DOI] [PubMed] [Google Scholar]
- 31. Vincent AL, et al. Efficacy of intranasal administration of a truncated NS1 modified live influenza virus vaccine in swine. Vaccine 2007;25:7999–8009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vincent AL, et al. Live attenuated influenza vaccine provides superior protection from heterologous infection in pigs with maternal antibodies without inducing vaccine-associated enhanced respiratory disease. J Virol 2012;86:10597–10605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vincent AL, et al. Influenza A virus vaccines for swine. Vet Microbiol 2017;206:35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Walia RR, et al. Regional patterns of genetic diversity in swine influenza A viruses in the United States from 2010 to 2016. Influenza Other Respir Viruses 2019;13:262–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Webby RJ, et al. Evolution of swine H3N2 influenza viruses in the United States. J Virol 2000;74:8243–8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. White LA, et al. Influenza A virus in swine breeding herds: combination of vaccination and biosecurity practices can reduce likelihood of endemic piglet reservoir. Prev Vet Med 2017;138:55–69. [DOI] [PubMed] [Google Scholar]
- 37. Zeller MA, et al. ISU FLUture: a veterinary diagnostic laboratory web-based platform to monitor the temporal genetic patterns of influenza A virus in swine. BMC Bioinformatics 2018;19:397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zeller MA, et al. Complete genome sequences of two novel human-like H3N2 influenza A viruses, A/swine/Oklahoma/65980/2017 (H3N2) and A/Swine/Oklahoma/65260/2017 (H3N2), detected in swine in the United States. Microbiol Resour Announc 2018;7:e01203-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang J, et al. High-throughput whole genome sequencing of porcine reproductive and respiratory syndrome virus from cell culture materials and clinical specimens using next-generation sequencing technology. J Vet Diagn Invest 2017;29:41–50. [DOI] [PubMed] [Google Scholar]
- 40. Zhang Y, et al. Influenza Research Database: an integrated bioinformatics resource for influenza virus research. Nucleic Acids Res 2017;45:D466–D474. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Supplemental_material for Detection of live attenuated influenza vaccine virus and evidence of reassortment in the U.S. swine population by Aditi Sharma, Michael A. Zeller, Ganwu Li, Karen M. Harmon, Jianqiang Zhang, Hai Hoang, Tavis K. Anderson, Amy L. Vincent and Phillip C. Gauger in Journal of Veterinary Diagnostic Investigation