Abstract
Methylation of RNA and DNA, notably in the forms of N6-methyladenosine (m6A) and 5-methylcytosine (5mC) respectively, plays crucial roles in diverse biological processes. Currently, there is a lack of knowledge regarding the cross-talk between m6A and 5mC regulators. Thus, we systematically performed a pan-cancer genomic analysis by depicting the molecular correlations between m6A and 5mC regulators across ~ 11,000 subjects representing 33 cancer types. For the first time, we identified cross-talk between m6A and 5mC methylation at the multiomic level. Then, we further established m6A/5mC epigenetic module eigengenes by combining hub m6A/5mC regulators and informed a comprehensive epigenetic state. The model reflected status of the tumor-immune-stromal microenvironment and was able to predict patient survival in the majority of cancer types. Our results lay a solid foundation for epigenetic regulation in human cancer and pave a new road for related therapeutic targets.
Keywords: m6A regulators, 5mC regulators, Pan-cancer analyses, Genomic alterations, Tumor microenvironment, Survival
To the Editor,
Nucleotide methylation, notably in the forms of 5-methylcytosine (5mC) in DNA and N6-methyladenosine (m6A) in mRNA, carries important information for gene regulation [1]. Recent research advances highlight the biological importance of m6A methylation as a dynamic and reversible post-transcriptional modification [2]. 5mC DNA methylation, a conserved epigenetic modification along with m6A RNA modification, also plays critical roles in fundamental biological processes [3, 4]. In addition, recent studies have identified 5mC methylation as a modulator of alternative mRNA splicing at the post-transcriptional level [5, 6]. Although Zhou and colleagues [7] established a molecular link between 5mC DNA methylation and m6A mRNA methylation during fruit ripening, the potential cross-talk still remains uncharacterized in human cancers.
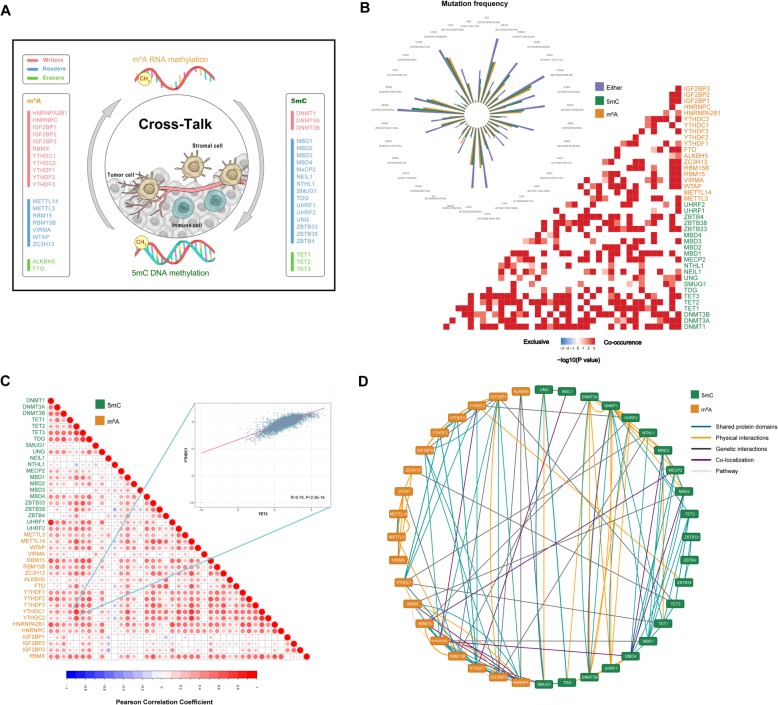
To address this issue, we curated a catalog of 20 and 21 genes that function mainly as regulators of RNA and DNA methylation, respectively (Fig. 1a). The genome-wide omics data comprising of 11,080 human samples across 33 cancer types from the The Cancer Genome Atlas (TCGA) were obtained for analyses (please see Methods and Table S1). First, most of the m6A and 5mC regulators were found to exhibit comparable expression levels across the 33 cancer types (Supplementary Fig. S1). Basing on the Gene Set Cancer Analysis (GSCA) web server [8], we further assessed the gene set differential expression profiles among 14 cancer types with available paired tumor-normal tissue expression data. Across multiple cancer types, the differentially expressed genes (upregulated or downregulated) included both m6A and 5mC regulators (Supplementary Fig. S2). Then, we investigated the mutation frequencies of the m6A and 5mC regulators. Intriguingly, m6A and 5mC regulators exhibited comparable levels of mutation frequency, and significant co-occurrences of genetic alterations were observed between the two regulators (Fig. 1b). Our results showed correlated expression patterns for genes within the same regulator class and even high correlations between the expression of m6A and 5mC regulators (Fig. 1b). Moreover, these m6A and 5mC regulators interacted with one another frequently in protein-protein interaction networks (Fig. 1d).
Fig. 1.
Cross-talk identified among the m6A and 5mC regulators. a m6A and 5mC regulator genes and a diagram of their potential cross-talk. b Correlations between the expression of m6A and 5mC regulators. The scatter plot shows the strong positive correlation between YTHDC1 and TET2. The Pearson correlation coefficients (R) are shown. c Co-occurrence of genetic alterations in m6A and 5mC regulators. The log2 (odds ratio) is colored as a heat map. The Nightingale rose diagram shows the mutation frequency distribution of m6A and 5mC regulators across different cancer types. d Protein-protein interactions among the m6A and 5mC regulators based on the GeneMANIA database
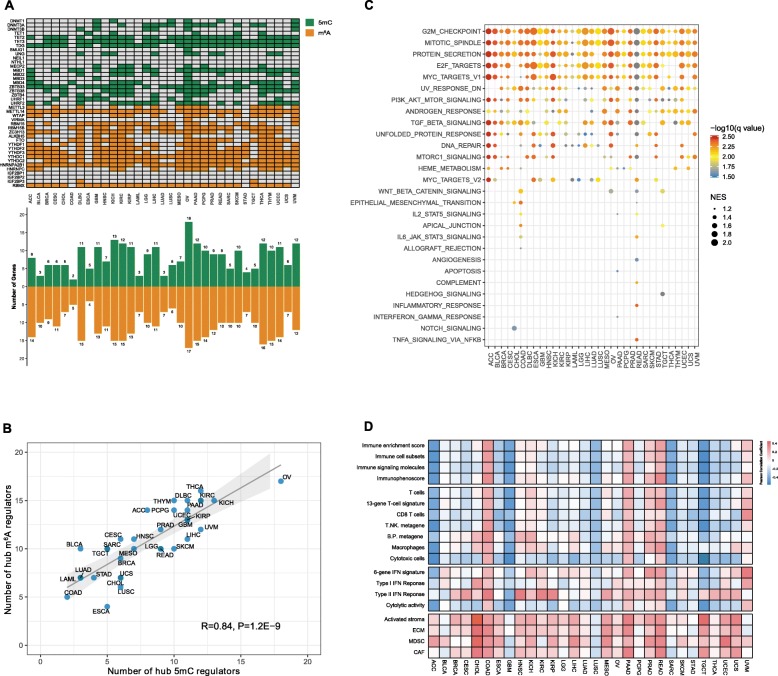
To identify the hub regulators involved in RNA and DNA methylation, we then applied weighted gene coexpression network analysis (WGCNA) to determine the hub genes in m6A and 5mC regulators (Fig. 2a). Strikingly, the number of hub m6A regulators was highly correlated with that of hub 5mC regulators in different cancer types (R = 0.84; Fig. 2b), which may be explained by the cross-talk. We then combined the hub m6A/5mC genes to develop an epigenetic module eigengene (EME), which may reflect both the pre- and post-transcriptional modification statuses. Next, we examined the correlation between EMEs and the activity of hallmark oncogenic pathways (Fig. 2c). Interestingly, our results indicate that high expression of the EME may reflect a highly proliferative and aggressive status in the majority of tumors. In addition, we applied GSCA [8] to analyze the effect (activation or inhibition) of m6A/5mC regulators on cancer-related pathways and confirmed that the m6A and 5mC regulators may be functionally related (Supplementary Fig. S3).
Fig. 2.
Development and characterization of the m6A/5mC epigenetic module eigengenes (EMEs). a Module membership-based hub m6A and 5mC regulators across 33 cancer types. The lower panel shows the number of hub m6A and 5mC regulators in each cancer type. b Correlations between the number of hub m6A regulators and the number of hub 5mC regulators. The Pearson correlation coefficients (R) are shown. c Gene set enrichment analysis (GSEA) results (normalized enrichment scores [NES] and q values) regarding the hallmark oncogenic pathways for EMEhigh versus EMElow subgroups across 33 cancer types. Enrichment score terms with an FDR < 0.05 are shown. d Heatmap showing the Pearson correlation coefficients between the EMEs and immuno-stromal signatures across 30 cancer types. Diffuse large B cell lymphoma (DLBC), acute myeloid leukemia (LAML), and thymoma (THYM) were excluded, as they mainly consist of immune cells
In addition to the tumor compartments, we further investigated the associations between the EME and immuno-stromal signatures representing different statuses of the immune and stromal cells (Table S2) across cancer types. In general, relatively low expression of inflammatory markers and low infiltration of immune cells were observed in the EMEhigh versus EMElow subgroups across cancer types (Fig. 2d). Interestingly, the high enrichment of stromal-related signatures was observed in the EMEhigh subgroups in almost all cancer types, indicating that hub m6A/5mC regulators may generally be involved in stroma activation (Fig. 2d).
Finally, we assessed the prognostic value of the EME in various types of cancers. We found that the EME showed oncogenic features in most cancer types, with overall survival (OS) hazard ratios larger than one (Supplementary Fig. S4a–c). Of these, high expression of the EME was significantly associated with unfavorable OS in cancer types such as KICH, ACC, and LGG (Supplementary Fig. S4a, b). Among HNSC, KIRC, and READ, improved survival was observed in the EMEhigh versus EMElow groups (Supplementary Fig. S4a, c).
In summary, to our best knowledge, this is the first study suggesting potential cross-talk between m6A and 5mC regulators in human cancers. This study provides essential insights into epigenetic regulation in cancer and paves new ways for related therapeutic targets.
Supplementary information
Additional file 1: Figure S1. Gene expression profile of m6A/5mC regulators across 33 cancer types. Pan-cancer normalized RNA-Seq by Expectation-Maximization (RSEM) data were used. For a given m6A/5mC regulators in a given cancer type, the non-scaled median expression level is presented.
Additional file 2: Figure S2. Gene set differential expression profile of m6A/5mC regulators among 14 cancer types with available paired tumor-normal tissue expression data calculated by Gene Set Cancer Analysis (GSCA).
Additional file 3: Figure S3. Heatmap showing percentage of cancers in which a pathway may be activated (red) or inhibited (blue) by the m6A/5mC regulators calculated by Gene Set Cancer Analysis (GSCA). Reverse phase protein array (RPPA) data of 32 cancer types from The Cancer Proteome Atlas (TCPA) are used for the calculation; acute myeloid leukemia (LAML) is not included. A total of 10 cancer related pathways are included (i.e., TSC/mTOR, RTK, RAS/MAPK, PI3K/AKT, Hormone ER, Hormone AR, EMT, DNA Damage Response, Cell Cycle, and Apoptosis pathways), and only m6A/5mC regulators that have function (activate or inhibit) in at least five cancer types are shown by GSCA.
Additional file 4: Figure S4. Clinical relevance of the EMEs across 33 cancer types. a Forest plots showing the hazard ratios (HRs; squares) and 95% confidence intervals (CIs; horizontal ranges) of overall survival (OS) across 33 cancer types. Significant results are indicated by red (unfavorable prognosticators) or blue (favorable prognosticators) squares. b Kaplan-Meier plots showing unfavorable OS in the EMEhigh versus EMElow groups for KICH, ACC, LGG, CESC, SARC, LIHC, BRCA, and LUAD. P values for the two-sided log-rank test are shown. c Kaplan-Meier plots showing improved OS in the EMEhigh versus EMElow groups for HNSC, KIRC, and READ. P values for the two-sided log-rank test are shown.
Additional file 5. Materials and Methods.
Additional file 6: Table S1. Details of the 33 cancer types from the TCGA.
Additional file 7: Table S2. Immuno-stromal signatures used in the current study.
Acknowledgements
We would like to thank the staff members involved in the TCGA Research Network.
Abbreviations
- 5mC
5-Methylcytosine
- ACC
Adrenocortical carcinoma
- BRCA
Breast cancer
- CESC
Cervical squamous cell carcinoma and endocervical adenocarcinoma
- CNV
Copy number variation
- EME
Epigenetic module eigengene
- GSEA
Gene set enrichment analysis
- HNSC
Head and neck squamous carcinoma
- KICH
Kidney chromophobe
- KIRC
Kidney renal clear cell carcinoma
- KIRP
Kidney renal papillary cell carcinoma
- LGG
Brain low-grade glioma
- LIHC
Liver hepatocellular carcinoma
- LUAD
Lung adenocarcinoma
- m6A
Methylation of N6 adenosine
- OS
Overall survival
- PCPG
Pheochromocytoma and paraganglioma
- READ
Rectal adenocarcinoma
- SARC
Sarcoma
- SKCM
Skin cutaneous melanoma
- TCGA
The Cancer Genome Atlas
- THCA
Thyroid cancer
- UCEC
Uterine corpus endometrial carcinoma
- UVM
Uveal melanoma
- WGCNA
Weighted gene coexpression network analysis
Authors’ contributions
YPC, WFL, and ZXW designed the study; ZXW, JYS, YTC, DPC, JWL, RG, PPZ, WFL, and YPC analyzed and interpreted the data; YPC, JYS, DPC, and ZXW wrote and edited the manuscript; and all authors read and approved the final manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (81802707, 81702682) and the Fundamental Research Funds for the Central Universities (19ykpy186). YPC also received support from the Sun Yat-sen University Cancer Center Promotion Program for Talented Youth of the National Natural Science Foundation of China.
Availability of data and materials
The datasets used in this study are publicly available. All other relevant data and R and other custom scripts are available upon request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yu-Tong Chen, Jia-Yi Shen, Dong-Ping Chen and Chen-Fei Wu contributed equally to this work.
Contributor Information
Wen-Fei Li, Email: liwf@sysucc.org.cn.
Zi-Xian Wang, Email: wangzx@sysucc.org.cn.
Yu-Pei Chen, Email: chenyup1@sysucc.org.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13045-020-00854-w.
References
- 1.Chen K, Zhao BS, He C. Nucleic acid modifications in regulation of gene expression. Cell Chem Biol. 2016;23(1):74–85. doi: 10.1016/j.chembiol.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu Jinjin, Shen Lujun, Liu Yanli, Ming Hong, Zhu Xinxing, Chu Maoping, Lin Juntang. The m6A methyltransferase METTL3 cooperates with demethylase ALKBH5 to regulate osteogenic differentiation through NF-κB signaling. Molecular and Cellular Biochemistry. 2019;463(1-2):203–210. doi: 10.1007/s11010-019-03641-5. [DOI] [PubMed] [Google Scholar]
- 3.Schubeler D. Function and information content of DNA methylation. Nature. 2015;517(7534):321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- 4.Dor Y, Cedar H. Principles of DNA methylation and their implications for biology and medicine. Lancet. 2018;392(10149):777–786. doi: 10.1016/S0140-6736(18)31268-6. [DOI] [PubMed] [Google Scholar]
- 5.Lev Maor G, Yearim A, Ast G. The alternative role of DNA methylation in splicing regulation. Trends Genet. 2015;31(5):274–280. doi: 10.1016/j.tig.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Yearim A, Gelfman S, Shayevitch R, Melcer S, Glaich O, Mallm JP, et al. HP1 is involved in regulating the global impact of DNA methylation on alternative splicing. Cell Rep. 2015;10(7):1122–1134. doi: 10.1016/j.celrep.2015.01.038. [DOI] [PubMed] [Google Scholar]
- 7.Zhou L, Tian S, Qin G. RNA methylomes reveal the m(6)A-mediated regulation of DNA demethylase gene SlDML2 in tomato fruit ripening. Genome Biol. 2019;20(1):156. doi: 10.1186/s13059-019-1771-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu CJ, Hu FF, Xia MX, Han L, Zhang Q, Guo AY. GSCALite: a web server for gene set cancer analysis. Bioinformatics. 2018;34(21):3771–3772. doi: 10.1093/bioinformatics/bty411. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Gene expression profile of m6A/5mC regulators across 33 cancer types. Pan-cancer normalized RNA-Seq by Expectation-Maximization (RSEM) data were used. For a given m6A/5mC regulators in a given cancer type, the non-scaled median expression level is presented.
Additional file 2: Figure S2. Gene set differential expression profile of m6A/5mC regulators among 14 cancer types with available paired tumor-normal tissue expression data calculated by Gene Set Cancer Analysis (GSCA).
Additional file 3: Figure S3. Heatmap showing percentage of cancers in which a pathway may be activated (red) or inhibited (blue) by the m6A/5mC regulators calculated by Gene Set Cancer Analysis (GSCA). Reverse phase protein array (RPPA) data of 32 cancer types from The Cancer Proteome Atlas (TCPA) are used for the calculation; acute myeloid leukemia (LAML) is not included. A total of 10 cancer related pathways are included (i.e., TSC/mTOR, RTK, RAS/MAPK, PI3K/AKT, Hormone ER, Hormone AR, EMT, DNA Damage Response, Cell Cycle, and Apoptosis pathways), and only m6A/5mC regulators that have function (activate or inhibit) in at least five cancer types are shown by GSCA.
Additional file 4: Figure S4. Clinical relevance of the EMEs across 33 cancer types. a Forest plots showing the hazard ratios (HRs; squares) and 95% confidence intervals (CIs; horizontal ranges) of overall survival (OS) across 33 cancer types. Significant results are indicated by red (unfavorable prognosticators) or blue (favorable prognosticators) squares. b Kaplan-Meier plots showing unfavorable OS in the EMEhigh versus EMElow groups for KICH, ACC, LGG, CESC, SARC, LIHC, BRCA, and LUAD. P values for the two-sided log-rank test are shown. c Kaplan-Meier plots showing improved OS in the EMEhigh versus EMElow groups for HNSC, KIRC, and READ. P values for the two-sided log-rank test are shown.
Additional file 5. Materials and Methods.
Additional file 6: Table S1. Details of the 33 cancer types from the TCGA.
Additional file 7: Table S2. Immuno-stromal signatures used in the current study.
Data Availability Statement
The datasets used in this study are publicly available. All other relevant data and R and other custom scripts are available upon request.