T-cell receptor (TCR) driven MYC translocations characterize a rare but aggressive subtype of T-cell acute lymphoblastic leukemia (T-ALL). In these tumors, the proto-oncogene MYC is juxtaposed to enhancer elements of the TCR α/δ (TRA/TCRD) locus by the translocation, t(8;14)(q24;q11), eventually resulting in its constitutive activation(Erikson, et al 1986). Given that MYC regulates leukemia initiating capacity of malignant T-cells (King, et al 2013), elevated MYC levels might have a severe impact on the clinical behaviour of this rare T-ALL subtype. Indeed, TRA/TRD-MYC positive T-ALLs have been associated with an unfavorable prognosis, rapid disease progression and poor response to conventional therapy (Parolini, et al 2014). Here, we performed a detailed molecular genetic characterization of an extensive series of t(8;14)(q24;q11) positive pediatric T-ALL patients (n=26, Table S1) and evaluated a new therapeutic strategy for the treatment of this poor prognostic subtype of human leukemia.
TRA/TRD-MYC positive T-ALLs were characterized by frequent loss of the T-ALL tumor suppressor genes PTEN (23%), CDKN2A/B (73%) and LEF1 (8%), and often displayed genomic deletions that cause aberrant activation of the STIL-TAL1 or LMO2 oncogenes (30%) (Fig 1A, frequency for a general T-ALL group is reported in brackets (Liu, et al 2017)). Sequence analysis revealed lack of NOTCH1 or FBXW7 mutations, but a high number of loss-of-function mutations targeting PTEN (34%). Therefore, t(8;14)(q24;q11) positive leukemias represent a NOTCH1 independent subtype of T-ALL that often depends on activated PI3K/AKT signaling (PTENmut/del in 12 out of 26 (46%)) (La Starza, et al 2014). In line with this notion, both t(8;14)(q24;q11) positive T-ALL cell lines, KE-37 and MOLT16, lack NOTCH1/FBXW7 mutations and present with genomic loss of PTEN, displaying aberrant pAKT activation in the absence of activated NOTCH1 (Fig S1).
Figure 1. Genetic characterization of TRA/TRD-MYC translocated T-ALLs.
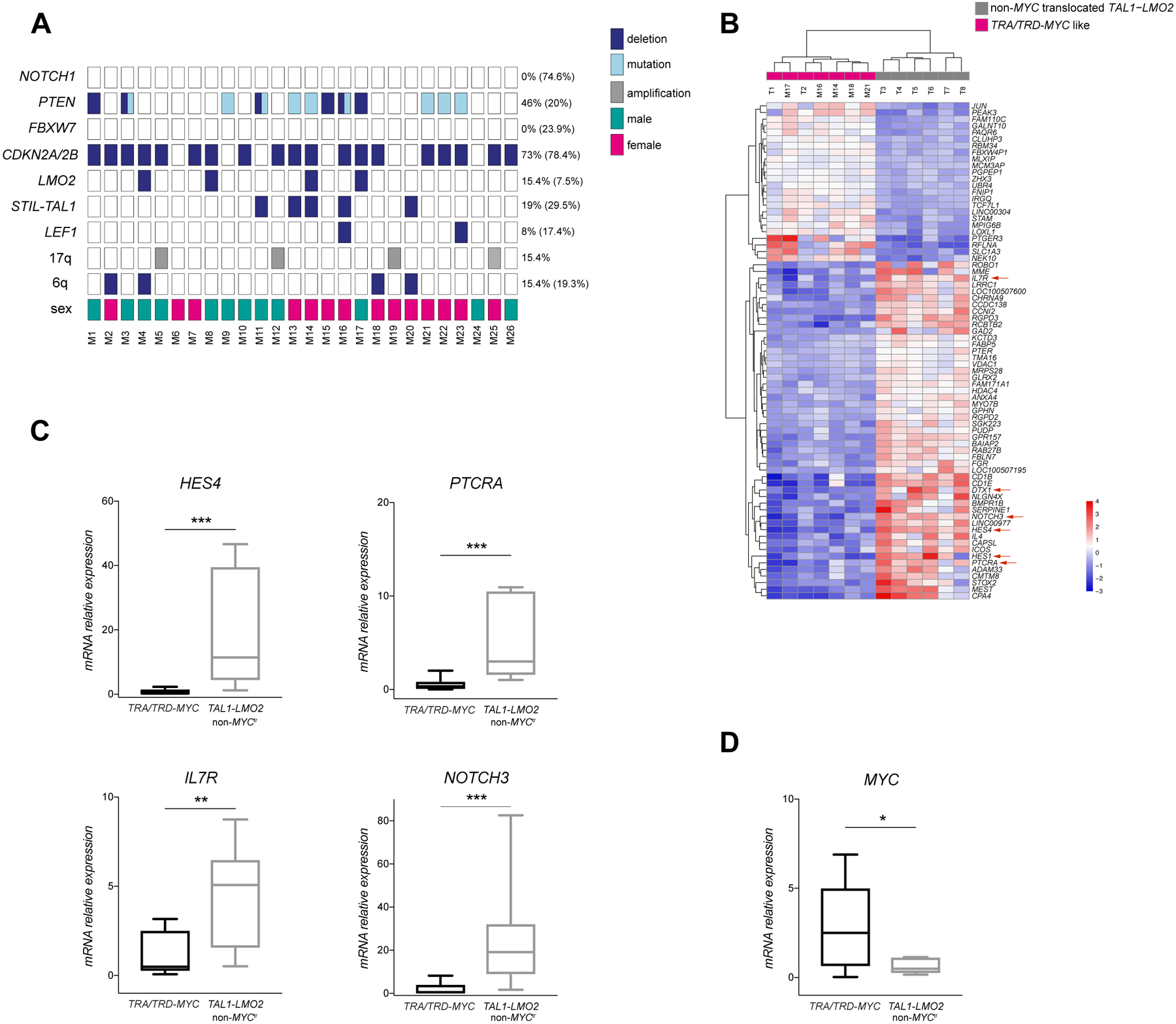
(A) Copy number and targeted mutation screening of 26 TRA/TRD-MYC rearranged T-ALLs. Graphical representation of deletions (dark blue), mutations (light blue) and amplifications (gray) present in a set of T-ALL oncogenes and tumor suppressor genes. Male and female T-ALL cases are represented in green and pink rectangles, respectively. All studied T-ALLs are pediatric cases (age <18 years). Frequency of described aberrations is reported for this cohort and in brackets for a general T-ALL group. (B) Top 75 most differentially expressed genes between TRA/TRD-MYC like T-ALLs (n=7) and non-MYC rearranged TAL1-LMO2 T-ALLs (n=6) based on RNA sequencing. NOTCH1 target genes are indicated by red arrows. M= TRA/TRD-MYC positive T-ALL; T= TAL1-LMO2 T-ALL. Value shown as color scale are mean centered regularized log counts. (C) Validation of NOTCH1 targets expression in an independent set of TRA/TRD-MYC translocated T-ALLs (n=16) and non-MYC translocated TAL1-LMO2 T-ALLs (n=7). HES4, PTCRA, IL7R, NOTCH3 mRNA expression was assessed by qRT-PCR. Mann–Whitney test was performed to compare the different groups (**P<0.01, ***P<0.001). Horizontal lines represent the median for each group. (D) MYC expression in TRA/TRD-MYC translocated T-ALLs (n= 13) and in TAL1-LMO2 T-ALLs (n=7). Mann–Whitney test was performed to compare the different groups (*P<0.05). Horizontal lines represent the median for each group.
Although TRA/TRD-MYC-rearranged T-ALL patients analyzed in this study were treated according to different protocols, the available clinical information confirmed the aggressive nature of this specific genetic subtype of pediatric leukemia. Indeed, most cases (19 out of 22 (86%)) presented with high white blood cell counts at diagnosis (>100×109/L), poor response to glucocorticoid therapy and largely unfavorable outcomes. More specifically, the leukemia was fatal in 13 of 26 (50%) of TRA/TRD -MYC positive T-ALLs due to progressive disease, the development of a secondary malignancy, specific toxicities or infections. Moreover, relapse of leukemia occurred in 8 out of 23 cases (Table S1). Although the prognostic significance of PTEN alterations in T-ALL remains highly debated (Jenkinson, et al 2016, Zuurbier, et al 2012), some studies have suggested that this particular genetic subtype (PTEN loss in the absence of NOTCH1/FBXW7 mutations) identifies a subset of highly aggressive human T-ALLs (Petit, et al 2017).
Previous studies have also shown that TRA/TRD-MYC-rearranged T-ALLs cluster with TAL1/LMO2 rearranged mature leukemias based on their gene expression signature (Homminga, et al 2011, La Starza, et al 2014). To further characterize the transcriptional differences between TAL1/LMO2 rearranged T-ALLs with and without MYC translocations, we performed RNA sequencing of 13 TAL1/LMO2 rearranged T-ALLs, including 5 TRA/TRD-MYC positive and 8 TRA/TRD-MYC negative leukemias. Unsupervised clustering of RNA sequencing data revealed the presence of two clusters, including one group that consisted of all 5 TRA/TRD-MYC positive T-ALLs and 2 additional TRA/TRD-MYC negative cases (Fig S2). Notably, copy number profiling and mutational analysis revealed that both of these TRA/TRD-MYC negative T-ALLs also displayed PTEN alterations without NOTCH1 abnormalities, resembling the characteristic genetic landscape of TRA/TRD-MYC T-ALLs. Therefore, we grouped these leukemias together and termed them TRA/TRD-MYC-like T-ALL. Next, differential expression analysis revealed a common transcriptional signature of these TRA/TRD-MYC-like T-ALLs compared to the 6 others non-MYC rearranged TAL1/LMO2 T-ALLs (Fig 1B), with 1856 transcripts differentially expressed between both tumor entities (adj. p-value < 0.05; 852 up and 1004 down in TRA/TRD-MYC like). Interestingly, several canonical NOTCH1 target genes, including NOTCH3, HES1, HES4, PTCRA, IL7R and DTX1, were significantly downregulated in the TRA/TRD-MYC like group, in line with the lack of NOTCH1 or FBXW7 mutations in this genetic subtype (Fig 1B). Differential expression analysis of NOTCH1 target genes was confirmed by qRT-PCR analyses using a larger series of TRA/TRD-MYC rearranged cases and an independent cohort of non-MYC rearranged TAL1/LMO2 T-ALLs (Fig 1C, Fig S3). Nevertheless, and as expected, TRA/TRD-MYC leukemias displayed higher MYC expression as compared to their TAL1/LMO2 rearranged counterparts (Fig 1D).
BET bromodomain inhibitors, such as JQ1, exploit the transcriptional addiction of cancer cells. At low concentrations, it has been shown that JQ1 preferentially targets enhancer elements with the highest levels of H3K27ac(Hnisz, et al 2013). Here, we performed H3K27ac chromatin immunoprecipitation (ChIP) sequencing analysis on the t(8;14)(q24;q11) positive MOLT16 cells and identified the highest levels of H3K27ac in the enhancer elements of the TRA/TRD locus (Fig 2A, Fig S4). Therefore, and given that these strong TRA/TRD locus control regions drive MYC expression in these tumors, we anticipated that BET bromodomain inhibition could serve as a valuable therapeutic strategy for this aggressive T-ALL subtype.
Figure 2. BET bromodomain inhibition in t(8;14)(q24;q11) positive T-ALL.
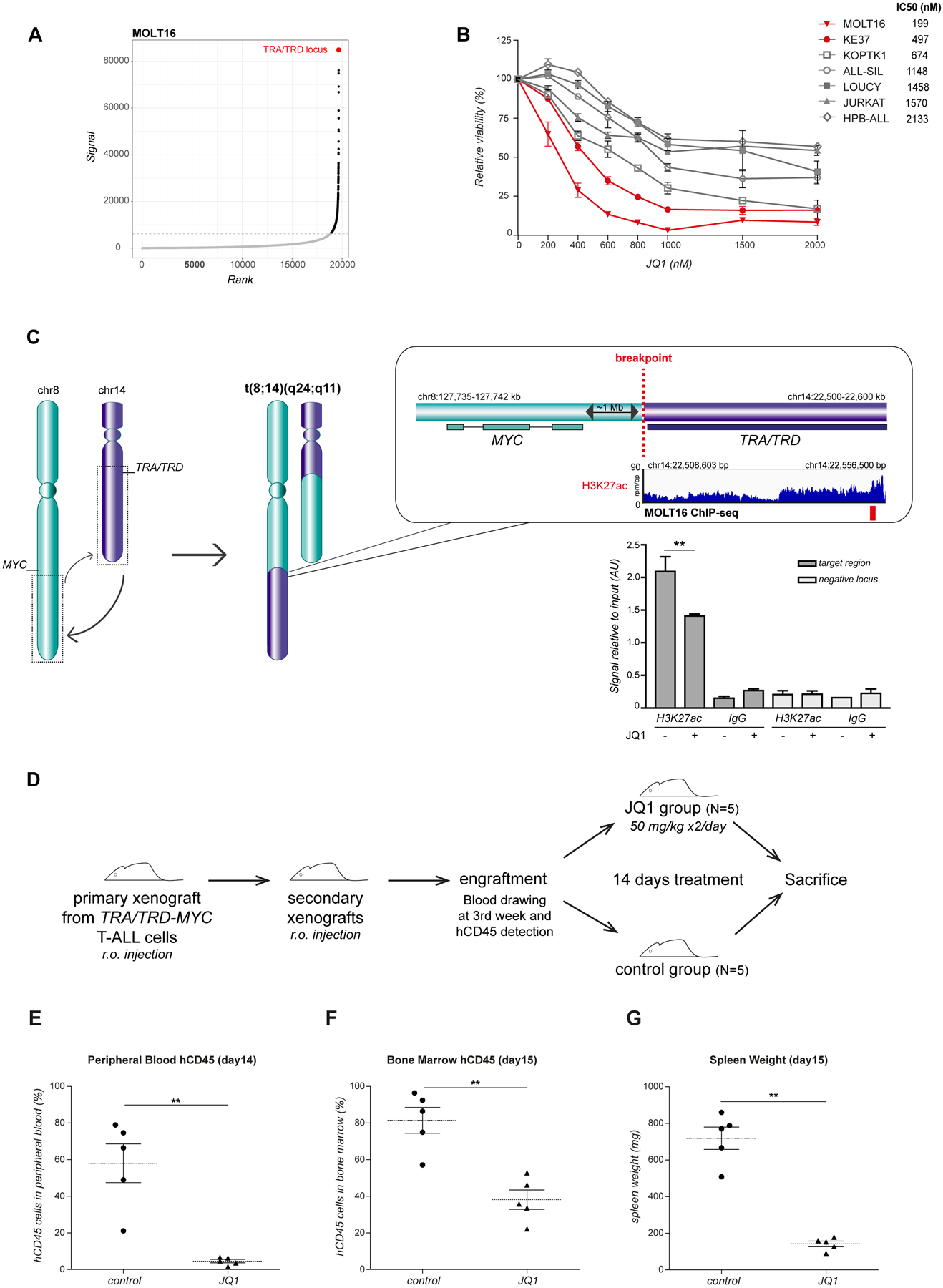
(A) Hockey-stick plot representing the normalized rank and signal of H3K27ac peaks in t(8;14)(q24;q11) positive MOLT16 cells. TRA/TRD enhancer elements (in red) showed the highest level of H3K27ac. (B) Cell viability in a panel of human T-ALL cell lines after 72 hours of JQ1 treatment, relative to control cells treated with dimethylsulfoxide. TRA/TRD-MYC rearranged T-ALL cell lines are represented in red. Average and standard deviation of 3 independent experiments are plotted. IC50 values (nM) are reported for each cell line. (C) Schematic representation of the t(8;14)(q24;q11) translocation, H3K27ac ChIP sequencing tracks at TRA/TRD locus in MOLT16 cell line and H3K27ac levels after JQ1 treatment (7 hours, 2μM) as evaluated by ChIP qPCR analysis. Primers used were designed on putative TRA/TRD enhancer regions (H3K27ac positive targets, red bar). Signal enrichment at target regions is reported in H3K27ac and IgG ChIP vs. relative inputs. Negative regions downstream of the positive target were analyzed as control (chr14:22,626,300–22,626,420). Means were calculated on 4 replicates with standard deviation represented by the error bars (**P<0.01). (D) JQ1 in vivo treatment experimental design. NSG mice were retro-orbital injected with TRA/TRD-MYC translocated cells from T-ALL patient (case 4, see Table S1) to generate primary xenografts. After leukemia engraftment, blasts were isolated from primary models and injected in other NSG mice to obtain a larger cohort of secondary xenografts for treatment. hCD45 positivity was checked from peripheral blood after 3 weeks. Upon engraftment, JQ1 was intraperitoneally administered twice/day for 14 days (50mg/kg bodyweight). Vehicle was administered to the control group following the same schedule. At the end of the experiment, animals were sacrificed and tissues analyzed. (E) Percentage of hCD45 leukemic cells in peripheral blood of NSG mice xenotransplanted with TRA/TRD-MYC T-ALL cells after 14 days of JQ1 treatment vs. DMSO. (F) Percentage of hCD45 leukemic cells in the bone marrow at the end of the experiment (day 15). (G) Xenografts spleen weight (mg) after 14 days of JQ1 treatment vs. DMSO. Mann–Whitney test was used to compare the treatment groups (**P<0.01). Horizontal lines on the graph indicate the median for each group.
In vitro drug sensitivity screening, using a panel of 7 human T-ALL cell lines, revealed that the TRA/TRD-MYC positive cell lines, MOLT16 and KE-37, showed the highest sensitivity towards JQ1 treatment (MOLT16 IC50= 199nM; KE-37 IC50=497nM) (Fig 2B, Fig S5). In addition, using ChIP qPCR, we confirmed that loss of MYC expression upon JQ1 treatment was accompanied by decreased levels of H3K27ac at the rearranged enhancer region of the TRA/TRD locus (Fig 2C).
Finally, we established patient derived xenograft (PDX) models from t(8;14)(q24;q11) positive primary T-ALLs to study JQ1 drug efficacy in vivo. A primary xenograft was first treated for 14 days with one single administration a day of JQ1 (50mg/kg), revealing a decrease of leukemic blasts in the peripheral blood and a reduction in splenomegaly, albeit limited effect was observed in the bone marrow (Fig S6). Therefore, the therapeutic schedule was reset and a second PDX was treated with JQ1 double dosage (50mg/kg, twice/day) (Fig 2D). Notably, the intense treatment resulted in a marked reduction of human leukemic blasts both in peripheral blood (Fig 2E) and bone marrow (Fig 2F), and produced a significant decrease in splenomegaly (Fig 2G). JQ1 in vivo effect was further confirmed by treating an additional xenograft model established from a different TRA/TRD -MYC translocated T-ALL patient, following the same treatment schedule (Fig S7).
Altogether, our study reveals that TRA/TRD-MYC rearranged T-ALL is an aggressive and NOTCH1-independent high-risk subtype of human leukemia that displays therapeutic sensitivity towards BET bromodomain inhibition.
Supplementary Material
Acknowledgements
We want to thank all members of the Van Vlierberghe laboratory for critical review of the manuscript and their comments. In addition, we would like to thank the Bradner Laboratory for providing in vivo quantities of JQ1.
Funding
This work was supported by the following funding agencies: Fund for Scientific Research Flanders (FWO), the Ghent University Special Research Fund (BOF), Kom op tegen Kanker (Stand up to Cancer; the Flemish cancer society), Kinderkankerfonds (a non-profit childhood cancer foundation under Belgian law), NHMRC Australia APP1057746, National Cancer Institute Outstanding Investigator Award R35 CA197695 and Cancer Center Core Grant P30CA021765. We acknowledge provision of biobanked patient samples by the Tissue Banks at Children’s Cancer Institute (Sydney, Australia), St. Jude Research Hospital (Memphis, USA) and Bloodwise Childhood leukaemia Cell bank.
Footnotes
Additional methodological details are included as Supplemental Methods.
References
- Erikson J, Finger L, Sun L, ar-Rushdi A, Nishikura K, Minowada J, Finan J, Emanuel BS, Nowell PC & Croce CM (1986) Deregulation of c-myc by translocation of the alpha-locus of the T-cell receptor in T-cell leukemias. Science, 232, 884–886. [DOI] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA & Young RA (2013) Super-enhancers in the control of cell identity and disease. Cell, 155, 934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homminga I, Pieters R, Langerak AW, de Rooi JJ, Stubbs A, Verstegen M, Vuerhard M, Buijs-Gladdines J, Kooi C, Klous P, van Vlierberghe P, Ferrando AA, Cayuela JM, Verhaaf B, Beverloo HB, Horstmann M, de Haas V, Wiekmeijer AS, Pike-Overzet K, Staal FJ, de Laat W, Soulier J, Sigaux F & Meijerink JP (2011) Integrated transcript and genome analyses reveal NKX2–1 and MEF2C as potential oncogenes in T cell acute lymphoblastic leukemia. Cancer Cell, 19, 484–497. [DOI] [PubMed] [Google Scholar]
- Jenkinson S, Kirkwood AA, Goulden N, Vora A, Linch DC & Gale RE (2016) Impact of PTEN abnormalities on outcome in pediatric patients with T-cell acute lymphoblastic leukemia treated on the MRC UKALL2003 trial. Leukemia, 30, 39–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King B, Trimarchi T, Reavie L, Xu L, Mullenders J, Ntziachristos P, Aranda-Orgilles B, Perez-Garcia A, Shi J, Vakoc C, Sandy P, Shen SS, Ferrando A & Aifantis I (2013) The ubiquitin ligase FBXW7 modulates leukemia-initiating cell activity by regulating MYC stability. Cell, 153, 1552–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Starza R, Borga C, Barba G, Pierini V, Schwab C, Matteucci C, Lema Fernandez AG, Leszl A, Cazzaniga G, Chiaretti S, Basso G, Harrison CJ, Te Kronnie G & Mecucci C (2014) Genetic profile of T-cell acute lymphoblastic leukemias with MYC translocations. Blood, 124, 3577–3582. [DOI] [PubMed] [Google Scholar]
- Liu Y, Easton J, Shao Y, Maciaszek J, Wang Z, Wilkinson MR, McCastlain K, Edmonson M, Pounds SB, Shi L, Zhou X, Ma X, Sioson E, Li Y, Rusch M, Gupta P, Pei D, Cheng C, Smith MA, Auvil JG, Gerhard DS, Relling MV, Winick NJ, Carroll AJ, Heerema NA, Raetz E, Devidas M, Willman CL, Harvey RC, Carroll WL, Dunsmore KP, Winter SS, Wood BL, Sorrentino BP, Downing JR, Loh ML, Hunger SP, Zhang J & Mullighan CG (2017) The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet, 49, 1211–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parolini M, Mecucci C, Matteucci C, Giussani U, Intermesoli T, Tosi M, Rambaldi A & Bassan R (2014) Highly aggressive T-cell acute lymphoblastic leukemia with t(8;14)(q24;q11): extensive genetic characterization and achievement of early molecular remission and long-term survival in an adult patient. Blood Cancer J, 4, e176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit A, Trinquand A, Chevret S, Ballerini P, Cayuela JM, Grardel N, Touzart A, Brethon B, Lapillonne H, Schmitt C, Thouvenin S, Michel G, Preudhomme C, Soulier J, Landman-Parker J, Leverger G, Macintyre E, Baruchel A & Asnafi V (2017) Oncogenetic mutations combined with MRD improve outcome prediction in pediatric T-Cell Acute Lymphoblastic Leukemia. Blood, 131(3):289–300. [DOI] [PubMed] [Google Scholar]
- Zuurbier L, Petricoin EF 3rd, Vuerhard MJ, Calvert V, Kooi C, Buijs-Gladdines JG, Smits WK, Sonneveld E, Veerman AJ, Kamps WA, Horstmann M, Pieters R & Meijerink JP (2012) The significance of PTEN and AKT aberrations in pediatric T-cell acute lymphoblastic leukemia. Haematologica, 97, 1405–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


