Abstract
Background
Malaria is a major public health problem in Cameroon. The study of the genetic diversity within parasite population is essential for understanding the mechanism underlying malaria pathology and to determine parasite clones profile in an infection, for proper malaria control strategies. The objective of this study was to perform a molecular characterization of highly polymorphic genetic markers of Plasmodium falciparum, and to determine allelic distribution with their influencing factors valuable to investigate malaria transmission dynamics in Cameroon.
Methods
A total of 350 P. falciparum clinical isolates were characterized by genotyping block 2 of msp-1, block 3 of msp-2, and region II of glurp gene using nested PCR and DNA sequencing between 2012 and 2013.
Results
A total of 5 different genotypes with fragment sizes ranging from 597 to 817 bp were recorded for GLURP. Overall, 16 MSP-1 genotypes, including K1, MAD20 and RO33 were identified, ranging from 153 to 335 bp. A peculiarity about this study is the RO33 monomorphic pattern revealed among the Pfmsp-1 allelic type. Again, this study identified 27 different Pfmsp-2 genotypes, ranging from 140 to 568 bp in size, including 15 belonging to the 3D7-type and 12 to the FC27 allelic families. The analysis of the MSP-1 and MSP-2 peptides indicates that the region of the alignment corresponding K1 polymorphism had the highest similarity in the MSP1and MSP2 clade followed by MAD20 with 93% to 100% homology. Therefore, population structure of P. falciparum isolates is identical to that of other areas in Africa, suggesting that vaccine developed with K1 and MAD20 of Pfmsp1 allelic variant could be protective for Africa children but these findings requires further genetic and immunological investigations. The multiplicity of infection (MOI) was significantly higher (P < 0.05) for Pfmsp-2 loci (3.82), as compare with Pfmsp-1 (2.51) and heterozygotes ranged from 0.55 for Pfmsp-1 to 0.96 for Pfmsp-2.
Conclusion
High genetic diversity and allelic frequencies in P. falciparum isolates indicate a persisting high level of transmission. This study advocate for an intensification of the malaria control strategies in Cameroon.
Trial registration This study was approved by Cameroon National Ethics Committee. It is a randomized controlled trial retrospectively registered in NIH U.S. National Library of Medicine, ClinicalTrials.gov on the 28/11/2016 at https://clinicaltrials.gov/ct2/show/NCT02974348 with the registration number NCT02974348
Keywords: MSP-1, MSP-2, GLURP protein, Plasmodium, Heterozygote, Infection control
Background
In spite of enhanced control efforts, malaria continues to be a major public health problem in sub-Saharan Africa and south-eastern Asia, and Plasmodium falciparum infection is prevalent in most of the endemic countries. The World Health Organization (WHO) estimated that 219 million malaria cases occurred worldwide of which 78% were in Africa [1]. About 407,000 fatal cases were registered, 92% in Africa and 61% of the global death in children under 5 years of age, with P. falciparum being the major cause of all deaths [1, 2]. Malaria in Cameroon is caused by three human malaria species: P. falciparum, Plasmodium ovale and Plasmodium malariae. The prevalence of P. falciparum is about 99% while P. malariae and P. ovale share the remaining 1%. Anopheles arabiensis, a member of the Anopheles gambiae complex, is a principal malaria vector in Cameroon [3]. Malaria is a leading public health problem in Cameroon and is reported as the first cause of morbidity and mortality accounting for 16% outpatient visits, 20% hospital admissions and 27% inpatient deaths [4].
Despite the current efforts to control malaria in Cameroon, the situation has not improved, mainly due to the increasing vector resistance to insecticides [5], and resistance to almost all anti-malarial drugs, including some resistance to artemisinin-based combination therapy (ACT) [6, 7]. Merozoite surface protein (MSP-1) and merozoite surface protein-2 (MSP-2) are two proteins challenging the human immune system [8] and are important candidates for development of blood stage malaria vaccines [9]. The msp-1 gene is located on chromosome 9 and contains 17 blocks of sequences, of which 7 are variable, flanked by conserved regions [10]. The block 2 msp-1 is particularly polymorphic and 3 distinct allelic families have been described as MAD20, K1 and RO33. The msp-2 gene is located on chromosome 2 composed of five blocks of which the most polymorphic is the central block 3.
The polymorphic central domain of the gene encoding MSP-2 belongs to 2 distinct families; 3D7 and Fc27 [11, 12]. Allelic forms of these antigen genes have been reported from different parts of the world [12–14]. Differences in allelic types as well as the number of repeat sequences in msp-1 and msp-2 can be detected by PCR, followed by dot-blot hybridization [15]. Size polymorphism is used not only for the msp genes but also for other markers genes, including glutamate rich protein (glurp) and circumsporozoite protein (csp) in which variations are detected by sequencing. Genetic diversity of P. falciparum populations determines the intensity of malaria transmission [16, 17], thereby providing a baseline data for any anti-malarial drug efficacy trials and the possibility of implementing control strategies based on modern interventions or vaccines. Merozoite surface proteins 1 and 2 genotyping is widely used in malaria molecular epidemiology studies to assess the allelic diversity and multiplicity of infection as a proxy of transmission level in molecular monitoring of interventions [18]. The Pfmsp1, Pfmsp2 and glurp markers are commonly selected because they are located on different chromosomes, and this reduces the likelihood of a linkage [19, 20]. Genotyping of these genes has been effectively used to trace individual clones over time in cohort studies and to measure duration of infection [21].
These markers are equally essential in distinguishing recrudescence from reinfection of the parasite in an anti-malarial treatment trial [22], since the discriminating power of these markers is dependent on the extent of allelic diversity and on the frequency of each allele within a population [12]. Genetic diversity of P. falciparum has been used to implement specific strategies for control and to evaluate the impact of interventions on changes in malaria epidemiology [5, 23]. However, in Cameroon, there are no data on the multiplicity of infection (MOI) I and limited data on P. falciparum genetic diversity, especially when msp-1, msp-2 and glurp need to be considered together as molecular markers in genotyping studies.
Thus, the aim of this study was to characterize the highly polymorphic genetic markers of P. falciparum field isolates, including the merozoite surface protein 1 (MSP-1), the merozoite surface protein 2 (MSP-2) and glutamate rich protein (GLURP), and to determine the allele distribution and factors influencing the MOI and heterozygosity as indicators of malaria parasite transmission dynamics.
Methods
Study site
This study was carried out in the areas occupied by the Cameroon Development Corporation (CDC). This is an agro-industrial para-state company practicing plantation farming based in the south-west region of Cameroon, specialized in the production of rubber, palm oil, and banana. Cameroon climate which varies with terrain, from equatorial rain forest with mean annual temperature of 24.5 °C in the south coastal regions to 26.5 °C in the semiarid northern regions. Elevation extremes are 0 m at sea level at the Atlantic Ocean in the south west and with highest peak at Mount Fako (Mount Cameroon) at 4095 m above the sea level. The CDC is rural communities in the South West region of the country located at 45 km away from Douala (Fig. 1). The climate is characterized by fairly constant temperatures and two seasons: a short dry season (November–February) and a long rainy season (March–November) with abundant precipitation (2000–10,000 mm). The mean annual rainfall is 2625 mm, relative humidity is constantly high (75–80%), and the temperature varies from 18 °C in August to 24.5 °C in March [3]. The CDC Company has headquarters in Limbe town, with a multicultural population of about 45,000 plantation workers consisting primarily of peasant farmers, with over 60% coming from the north-west and western regions of Cameroon. Limbe has 8 primary and two secondary schools. The community has good access roads, good pipe-borne water and electricity supplies in some localities. CDC has good health system consisting of two reference hospitals, Tiko and cottage hospital which are primary health facilities belonging to the Tiko health District and, 23 satellite clinics located in different CDC estates of the south west region of Cameroon. Apart from the CDC Primary Health Centre and hospitals, there are also private clinics, mission and government health centres and hospitals. This site was selected to conduct both clinical trial studies with artemisinin-based combination therapy and malaria parasite genetic diversity studies.
Fig. 1.
Map of major health facilities of the Cameroon Development Corporation (CDC), South West Region of Cameroon and Geographic map of Cameroon.
Source: https://www.worldatlas.com/webimage/countrys/africa/cm.htm
The study site is harbouring all Plasmodium species found in the country. Malaria transmission in this area is perennial with high intensity and prevalence varying from 7 to 29% [24] with peak periods corresponding to the rainy season. Three Anopheles vectors including An. gambiae sensu lato, Anopheles funestus and Anopheles nili have been identified in malaria transmission in the South West Region. Anopheles gambiae, the most dominant in terms of aggressiveness and activity, accounts for up to 72.7% of transmission, Infection rates of 87 infective bites/person/year [3] and overall entomologic inoculation rate (EIR) estimated recently at 3.93 infective bites/person/night [24]. In addition, houses are grouped into camps which are structured buildings provided by the company to accommodate the workers and their families. Because the patients coming to CDC health facilities are located in various camps and grouped in well-organized communities it makes it easy for the investigators to track them after enrolment at D0, in case they do not return for follow-up at health facilities. This has an impact in reducing the rate of loss for patients follow-up. Besides, this study site is one of the settings carefully selected by the NMCP as sentinel site to carry out epidemiological survey in order to obtain information paramount in guiding the national malaria management policy.
Sampling and parasite collection
The sample size was estimated using the following formula for sample size calculation as previously described [25]. n = Z2 * p (1 − p)/e2, Z = 1.96, p = prevalence of malaria in Cameroon (29%), e = error rate = 0.05; n = 317 patients, with 10% increment in case of PCR failure, making a total of 350 patients. The P. falciparum isolates were collected from microscopically diagnosed P. falciparum positive patients aged 6 months to 6 years, in clinics and hospitals of CDC Cameroon. Blood sample was taken from each patient using finger prick for thick and thin blood films. Two to three drops of blood were collected on 3 MM Whatman filter paper. A total of 350 blood samples were collected from 23 clinics and 2 hospitals of the CDC Cameroon belonging to 2 different health districts.
Parasitaemia estimation
Blood films were stained with 10% Giemsa at pH7.2 and then examined microscopically for the presence of malaria parasites; 200 fields under 1000× magnification were examined from the thick film before the slide was considered negative. For positive slides, parasitaemia (parasite density) was determined by counting only the asexual stages against 200 white blood cells (WBC) and then multiplied by 25, assuming the average of total WBC count of individuals equal to 8000 cells/μl of blood [26, 27]. The level of parasitaemia was graded as low (< 1000 parasites/μl of blood), moderate (1000–9999 parasites/μl of blood) and severe (≥ 10,000 parasites/μl of blood) [28].
Genomic DNA extraction and genotyping
Genomic DNA was extracted from blood spots collected on filter papers. Briefly, a disc of the filter paper was punched out from the blood spot using a paper puncher and placed in 1.5 ml centrifuge tubes using clean forceps. Genomic DNA was extracted using Qiagen blood and tissue kit (QIAGEN, Germany) according to the manufacturer’s instructions.
PCR amplification and allele detection
DNA was eluted using 50 μl AE (10 mM Tris–Cl; 0.5 mM EDTA; pH 9.0) elution buffer (QIAGEN, DNeasy® Blood & Tissue Kit, Cat. no. 69506, Germany) and kept at − 20 °C until used for PCR. Genomic DNA was amplified by primary and Nested PCR using allelic specific Primers (Table 1) and conditions for PCR amplification were followed as previously described [8, 19] for family specific allele analysis of msp-1 (block 2), msp-2 (block 3) and glurp (region II). In the primary PCR, a 25 μl PCR mixture was used containing 1 μl of DNA template, 0.2 µM of each primer, 1× TBE buffer, 250 μM of dNTPs, nuclease free water, 1 U of Taq polymerase enzyme, 2 mM MgCl2, 10 mM KCl, and 10 mM Tris–HCl, at pH 8.3. All reagents were from TIAGEN (Biotechnology, Inc., Beijing). Cycling conditions for the primary PCR were as follows; starting with three single steps of denaturation at 94 °C for 5 min, annealing at 58 °C for 2 min and extension at 72 °C for 2 min. This was followed by 35 cycles of denaturation at 94 °C for 1 min, annealing at 58 °C for 1 min and extension at 72 °C for 1 min, then a single annealing step at 58 °C for 2 min and final extension at 72 °C for 10 min. P. falciparum genotypes were further analysed by amplification of the two highly polymorphic regions of msp-1 (Block 2) and msp-2 (Block 3) using nested-PCR as previously described [19] with slight modifications for the cycling conditions of the secondary PCR. Briefly, oligonucleotide primers sets (Table 1), were used for detecting the family-specific (K1, MAD20 and RO33 in MSP-1; FC27 and IC in MSP-2). Three microlitres of primary PCR products were used as the DNA templates in the secondary PCR, which had similar concentrations to the primary PCR. The cycling conditions for the secondary PCR were as follows: starting with a single step of denaturation at 95 °C for 10 min followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 58 °C for 30 s and extension at 72 °C for 1 min, and a final extension at 72 °C for 10 min. PCR reaction mixtures were incubated in a thermal cycler Perkin-Elmer Cetus PE 9600 (Norwalk, CT). The secondary PCR products were separated by electrophoresis at 100 V on 1.5% molecular grade agarose gel (Caisson, Utah, USA), stained with ethidium bromide, submerged in 0.5 × TBE (Tris–borate EDTA) buffer and visualized by UV transilluminator (BioDoc-It UVP, Cambridge, UK) at 302 nm on gel documentation system. The number and size of DNA fragments were estimated based on their mobility related to a 100 bp DNA ladder (Vivantis, Selangor Darul Ehsan, Malaysia). DNA fragment sizes were binned into different classes of 20 and 50 bp ranges with each bin assigned as an allele. Alleles in paired samples were considered a match for msp2 and msp1 if within 20 base pairs and for glurp if within 50 base pairs [11, 14].
Table 1.
Sequences of primers for msp1, msp2 and glurp genes of P. falciparum from Cameroon
| Msp-1 (block 2) | |
| Primary PCR |
CHM1-OF: 5′ CTAGAAGCTTTAGAAGATGCAGTATTG-3′ CHM1-OR: 5′ CTTAAATAGTATTCTAATTCAAGTGGATCA-3′ |
| Secondary PCR |
CHM1-KF: 5′ AAATGAAGAAGAAATTACTACAAAAGGTGC-3′ CHM1-KR: 5′ GCTTGCATCAGCTGGAGGGCTTGCACCAGA-3′ CHM1-MF: 5′ AAATGAAGGAACAAGTGGAACAGCTGTTAC-3′ CHM1-MR: 5′ ATCTGAAGGATTTGTACGTCTTGAATTACC-3′ CHM1-RF: 5′TAAAGGATGGAGCAAATACTCAAGTTGTTG-3′ CHM1-RR: 5′ CATTTGAAGGATTTGCAGCACCTGGAGATC-3′ |
| Msp-2 (block 3) | |
| Primary PCR |
M2-OF: 5′ ATGAAGGTAATTAAAACATTGTCTATTATA-3′ M2-OR: 5′ CTTTGTTACCATCGGTACATTCTT’3′ |
| Secondary PCR |
M2-FCF: 5′ ATATTAAGAGTGTAGGTGCARATGCTCCA-3′ M2-FCR: 5′ TTTTATTTGGTGCATTGCCAGAACTTGAAC-3′ M2-ICF: 5′AGAAGTATGGCAGAAAGTAAKCCTYCTACT-3′ M2-ICR: 5′ GATTGTAATTCGGGGGATTCAGTTTGTTCG-3′ |
| Glurp (region II) | |
| Primary PCR |
CHG-OF: 5′ -TGAATTTGAAGATGTTCACACTGAAC-3′ 3′ CHG-OR: 5′ -GTG GAATTGCTTTTTCTTCAACACTAA-3′ |
| Secondary PCR |
CHG-NF: 5′-TGAATTTGA AGA TGT TCA CAC TGA AC-3′ CHG-OR: 5′-GTG GAATTGCTT TTTCTTCAACAC TAA-3′ |
Multiplicity of infection (MOI) and expected heterozygosity (HE)
Multiclonal infections were defined as those having more than one allele in at least one locus out of the loci genotyped. The MOI was determined by calculating the number of different alleles at any one locus detected in the sample [9]; single infections were those with only one allele per locus at all of the genotyped loci. The mean MOI was determined as the quotient of the total number of P. falciparum genotypes detected in MSP-1 or MSP-2 by the number of samples positive for either msp-1 or msp-2 [29].
As a measure for genetic diversity, the expected heterozygosity (HE) which represents the probability of being infected by two parasites with different alleles at a given locus and ranging between 0 and 1, [12] was calculated by using the following formula:
where n is the number of isolates sampled and pi is the allele frequency at a given locus [29].
Statistical analysis
Data was analysed using the SPSS for windows software version 17 of the Statistical Package of Social Science (SPSS) (SPSS Inc, Chicago, IL, USA) [30]. For descriptive analysis, proportion was used to present the distribution of different allelic families while the mean was used to present the MOI. Independent. Independent one way ANOVA method was used to assess the relative size of variance among group means (between group variance) compared to the average variance within groups (within group variance). Thus this method was used to compare the mean MOI according to, gender, parasitaemia and across msp1, msp2 and glurp family specific alleles. The F value calculated from the observed data with the critical value set at an α error level of 0.05 in the F table. A P-value ≤ 0.05 was considered indicative of a statistically significant difference.
msp-1 and msp-2 sequence analysis
Purified PCR products of isolates representing different alleles of msp-1 and msp-2 were sequenced in both directions with the primers of the secondary PCR using the ABI PRISM® BigDyeTM terminator Ready Reaction Cycle Sequencing Kit (Biometra Thermocycler, England) according to the manufacturer’s instruction. The sequences were then analysed using the DNASTAR software package (DNASTAR, Madison, WI). The sequences were used to correct the estimated molecular weight and to confirm the nature and size of the amplified product. To understand the identity of Cameroonian isolates with respect to isolates of other regions, sequence data available in public domains were downloaded for allelic families of msp-1 and msp-2 and aligned using ClustalW method (EMBL-EBI, Hixton, and Cambridge, UK), with query sequences and word match set at 50 and 99% as identity threshold, in a FASTA file database then analysed by MEGA version 5.1 [31]. Sequence differences were identified using BLASTN sequence homology searches. Each allelic family msp-1 (including K1, MAD20 and R033) and msp-2 (including FC27 and 3D7/IC) were analysed separately in order to estimate the average number of nucleotide substitutions for each msp-2 allele, and to examine the mode of evolution of FC27 type repeats. Individual repeat units were assembled and aligned with DNAstar and BLAST searched in GenBank to analyse the distribution of the pairwise proportion of nucleotide differences among individual repeat units using isolates derived from GenBank, namely the isolate from Gabon accession number AY372506 for FC27, the isolate HM568631 from India for 3D7/IC allelic family. As for msp-1 alleles, the isolate from Brazil (accession:JX416338) for the alignment of K1-type allele, while the isolates EU032224 from central sub-Saharan Africa and AY138508 from Iran were used as reference isolate for the sequence analysis of R033 and MAD20, respectively [32].
Ethical approval and consent to participate
This study was approved by the scientific and Ethical Committee of the Cameroon Ministry of Public Health. Field administrative approval was provided by the health administrative authorities of the CDC. A written informed consent for voluntary participation in this study was obtained from parents or guardians of children before their enrolment in this study.
Results
Study profile
Out of 315 DNA samples at baseline, 137 samples for msp-1 and 307 samples for msp-2 were successfully amplified by PCR, among which 107 for msp-1 and 243 for msp-2 were successfully sequenced.
Genetic diversity and haplotype frequency
The number of genotypes observed at each marker is shown in Table 2. Sixteen (16) different msp-1 genotypes were observed, representing K1 (9 genotypes), MAD20 (6 genotypes) and RO33 (1 genotype) allelic families. The msp-1 fragment sizes ranged from 153–335 bp, while the msp-2 fragments ranged from 140 to 568 bp both for the FC27 and IC allelic families (Table 2). The majority (87.5%) of these genotypes especially for the k1 (100%) and MAD20 (97.5%) belonging to the msp1 allelic family occurred at a frequency below 10% (Table 2). However, one genotype of the MAD20 family (167–187 bp) occurred above 10%. The R033 family was found to be monomorphic with an amplified fragment size of 155 bp and occurred at a frequency of 78.1% (107/137) (Tables 2 and 4) of the overall msp1 genotypes.
Table 2.
Base pair range and number of detected genotypes of the respective msp-1 and msp-2 gene families in P. falciparum isolates from 315 malaria patients in Cameroon
| Marker | msp-1 | msp-2 | |||
|---|---|---|---|---|---|
| K1 | MAD20 | R033 | FC27 | 3D7/IC | |
| Number of different genotypes per allele | 9 | 6 | 1 | 12 | 15 |
| Allele range (bp) | 153–335 | 175–205 | 155 | 140–387 | 200–568 |
| Total per locus | 16 | 27 | |||
| Genotypes frequency for allele type occurring at a frequency < 10% | 9 (100%) | 5 (97.5%) | 0 (0%) | 10 (83.3%) | 14 (95.3%) |
| Total per allele | 14 (87.5%) | 24 (88.9%) | |||
| Genotypes frequency for allele type occurring at a frequency > 10% | 0 (0%) | 1 (16.7%) | 1 (100%) | 2 (16.7%) | 1 (6.7%) |
| Total per allele | 2 (12.5%) | 3 (11.1%) | |||
Table 4.
Heterozygosity, and multiplicity of infection (MOI) per msp1 and msp2 allelic families in P. falciparum isolates from 315 malaria patients in Cameroon
| Family specific alleles | Frequency (%) | No. fragments | He | MOI |
|---|---|---|---|---|
| K1 | 80 (58.39) | 197 | 0.65 | 2.46 |
| MAD20 | 40 (29.19) | 40 | 0.50 | 1.00 |
| R033 | 107 (78.10) | 107 | 0.50 | 1.00 |
| Total/msp1 (N = 137) | 227 | 344 | 0.55 | 2.51 |
| 3D7 (IC) | 295 (96.1) | 621 | 0.99 | 2.10 |
| FC 27 | 298 (97.1) | 552 | 0.94 | 1.88 |
| Total/msp2 (N = 307) | 593 | 1173 | 0.96 | 3.82 |
A total of 27 different msp-2 genotypes (size range from 140 to 568 bp) were recorded of which 15 belonged to the 3D7-type and 12 to the FC27 allelic families (Table 2). The majority of these genotypes occurred at a frequency below 10%. However, 1 genotype from the 3D7/IC allelic family (420–440 bp), and 2 from the FC27 family (176–201 bp and 201–226 bp) occurred above 10%. The 3D7/IC family was found to be highly polymorphic (P = 0.01) as compared to FC27.
The glurp diversity showed 5 different glurp genotypes (size range from 597 to 817 bp) with the majority of the allelic families occurring at above 10% (Table 3). However, only two alleles occurred below 10% of which, 4.67% and 9.34% belonged to the 550–600 bp and 652–702 bp allelic group.
Table 3.
Distribution of allelic variants of GLURP RII repeat region of P. falciparum isolates amongst malaria patients in Cameroon
| Genotypes | Allelic size variants (bp) | Frequency (%) | No. fragments | MOI |
|---|---|---|---|---|
| I | 550–600 | 5 (4.5) | 5 | 1.00 |
| II | 652–702 | 18 (16.36) | 19 | 1.10 |
| III | 754–804 | 63 (57.3) | 64 | 1.10 |
| IV | 805–855 | 14 (12.7) | 14 | 1.00 |
| V | 856–906 | 10 (9.1) | 10 | 1.00 |
| Total glurp | 110 | 112 | 1.02 |
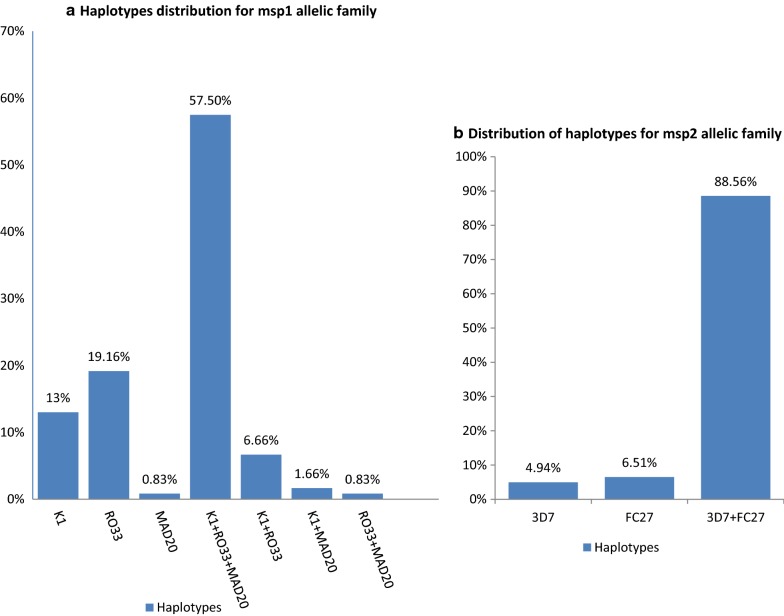
This study revealed that more than half of the msp-1 positive samples harboured all the three types of alleles of the msp-1 gene. Thus the combination of RO33, MAD20 and K1 allelic families was identified with an overall frequency of 57.50% (Fig. 2a). The RO33 allelic family was predominant as it was identified in 78.10% (107/137) of the samples. One-third (33%) of the blood samples positive for msp-1 were identified as monoclonal infections while two-third (66%) exhibited a polyclonal pattern of K1, MAD20 and R033 combination in a set of two or three alleles (Fig. 2a). Among the polyclonal infections, K1/RO33, K1/MAD20 and Ro33/MAD20 constituted 6.66%, 1.6% and 0.83% of the msp-1 positive isolates respectively. The distribution of the identified msp-2 allelic families is illustrated in Fig. 2b. Overall, 11.42% of msp-2 positive isolates were identified as monoclonal infections either for 3D7/IC or FC27 allelic families against 88.56% msp-2 positive isolates (Fig. 2b) displaying a polyclonal pattern of infections. The frequency of the 3D7/IC and FC27 haplotypes combinations was found to be higher than the frequency of samples with only 3D7 or FC27 allelic families (Fig. 2b).
Fig. 2.
a Distribution of msp-1 haplotypes.b Distribution of haplotypes for msp2 allelic family
Multiplicity of infection and expected heterozygosity
Plasmodium falciparum isolates in this study had higher rates of multiple genotypes infection with an overall mean multiplicity of infection of 3.16. The mean MOI for msp-2 loci was the highest (3.82), as compared with msp-1 (2.51) (Table 4). In the children, there was no difference in mean MOI between male and female (P = 0.1) (Table 5). However, the influence of parasite density on genetic diversity showed that children with parasite density ranging from 2000 to 5000 had the lowest MOI (Table 6). Meanwhile those with parasite ranging from 20,000 and above exhibited the highest MOI difference (P = 0.001), as compared with various range of parasite densities. The overall mean multiplicity of infection, 3.16 observed for msp1 and msp2 is significantly higher (P < 0.05) than revealed among glurp genotypes (1.02) (Table 7).
Table 5.
Distribution of MOI by gender amongst Cameroonian children with falciparum malaria
| PCR+ | No. 3D7/IC fragments | No. FC27 fragment | No. FC27 + 3D7 fragment | MOI | P-value | |
|---|---|---|---|---|---|---|
| Female | 157 | 43 | 21 | 175 | 1.52 | 0.08 |
| Male | 150 | 76 | 22 | 202 | 2 | |
| Total | 307 | 119 | 43 | 377 |
F-ratio value = 3.7, df = 1; P = 0.08
Table 6.
Distribution of msp2 allele by parasite density among Cameroonian children with falciparum malaria
| Parasite count | PCR+ | No. 3D7/IC fragments | No. FC27 fragments | No. FC27 + 3D7/IC fragments | Total fragments | MOI |
|---|---|---|---|---|---|---|
| 2000–5000 | 123 | 54 | 39 | 95 | 188 | 1.53 |
| 5001–10,000 | 36 | 6 | 4 | 50 | 60 | 1.67 |
| 10,001–15,000 | 43 | 31 | 24 | 63 | 118 | 2.7 |
| 15,001–20,000 | 36 | 24 | 27 | 35 | 86 | 2.38 |
| 20,001–above | 69 | 64 | 51 | 115 | 230 | 3.3 |
| Total | 307 | 179 | 145 | 358 | 682 | 2.2 |
Table 7.
Distribution of MOI and HE across the three markers in Cameroonian isolates
| Family specific alleles | N | genotypes | No. fragments | HE | MOI | P-value |
|---|---|---|---|---|---|---|
| Msp1 | 137 | 16 | 344 | 0.96 | 2.51 | 0.02 |
| Msp2 | 307 | 27 | 1173 | 0.55 | 3.82 | |
| Glurp | 110 | 5 | 112 | 0.01 | 1.02 |
F-ration value = 7.9; df = 1; P = 0.02
The mean MOI was found to be higher in male patients whose parasitaemia ranged from moderate to high level, than that of male patients with low parasitaemia. The expected heterozygosity (HE) was calculated which used to estimate the fraction of all parasites that would be heterozygous for any of the two loci. Table 4 shows generally high HE values in each allelic family ranging from 0.5 to 0.65 for msp-1 and from 0.94 to 0.96 for msp-2.
Sequence analysis of genetic polymorphism of msp-1 block 2 and msp-2 block3
The msp-1 block 2 and msp-2 block 3 DNA fragments from PCR products with either similar or different molecular sizes in agarose gels were sequenced with the aim of further estimating the genetic diversity of the parasite population. Partial gene sequences were obtained from each end of the PCR product using the same primers for the nested PCR reactions, then DNA sequences assembled and BLAST searched in GenBank to identify identical or similar sequences. Analysis of msp-1 and msp-2 sequence data revealed above 80% identity of study isolates among themselves in general and above 83% with isolates from other countries with a few exceptions (Table 8). Moreover, the RO33 allelic sequences of Cameroonian isolates were all identical and had shown above 99% similarity (Table 8) with sequences reported for isolates of Kenya [33]. In the K1 family, 93 to 99% similarity was observed with K1 allelic sequences reported for isolates of Tanzania [34], Indonesia (accession no. U72950), Vietnam and Brazil [35]. Likewise, 83 to 100% identity was observed in MAD20 family with isolates of Iran [36], Sudan [37], Vietnam [35]. Allelic families of msp-2, FC27 and 3D7/IC showed 97 to 100% identity with isolates of Gambia [38], Brazil (accession no. DQ115969), Tanzania (accession no. AY378316), and Gabon (accession no. AY372518). No significant identity was found with either the Nigerian or the Brazilian isolates with respect to the 3D7/IC allelic family of this study. However 89 to 97% identity were recorded with isolates from Gambia and Thailand [38].
Table 8.
Plasmodium falciparum msp1 and msp2 haplotype sequence diversity amongst malaria patients in Cameroon compared with isolate from different geographical regions
| Family specific allele | Nr of different genotypes | Actual fragment size range (bp) | Degree of identity between alleles of study isolates (%) | Reported identical sequences/accession | Degree of identity (%) | Origin | References |
|---|---|---|---|---|---|---|---|
| K1 | 9 | 153–335 | 90–99 | AF509714 | 90–94 | Brazil | Ferreira et al. [35] |
| AF191061 | 93–98 | Indonesia | Unpublished | ||||
| AF061134 | 93–99 | Tanzania | Jiang et al. [68] | ||||
| AF509651 | 93–99 | Vietnam | Ferreira et al. [35] | ||||
| M77730 | 93–99 | Thailand | Jongwutiwes et al. [69] | ||||
| JX416338 | 99 | Brazil | Unpublished | ||||
| MAD20 | 5 | 175–205 | 83–92 | AY714585 | 92 | Brazil | Scopel et al. [70] |
| AF251345 | 83–93 | China | Unpublished | ||||
| AY138509 | 82 | Iran | Unpublished | ||||
| AF034635 | 83 | Sudan | Cavanagh et al. [37] | ||||
| AF509653 | 91 | Vietnam | Ferreira et al. [35] | ||||
| M77722 | 83 | Thailand | Jongwutiwes et al. [69] | ||||
| EU32224 | 100 | Central Sub Saharan Africa | Noranate et al. [32] | ||||
| R033 | 1 | 155 | 99–100 | AY538507 | 99 | Iran | Unpublished |
| AF462453 | 99 | Kenya | Takala et al. [33] | ||||
| AF191064 | 98 | Indonesia | Unpublished | ||||
| M77737 | 99 | Thailand | Jongwutiwes et al. [69] | ||||
| AY138508 | 98 | Iran | Unpublished | ||||
| IC | 15 | 200–568 | 97 | HM568631 | 97 | India | Unpublished |
| FC27 | 12 | 140–387 | 97 | DQ115973 | 97 | Brazil | Peyerl-Hoffmann et al. [46] |
| DQ338451 | 98 | Iran | Unpublished | ||||
| AF329577 | 97 | Ghana | Unpublished | ||||
| AF329579 | 96 | PNG | Unpublished | ||||
| U91668 | 97 | Gambia | Unpublished | ||||
| AY532386 | 98 | Tanzania | Unpublished | ||||
| AF104696 | 99 | Vietnam | Weisman et al. [71] | ||||
| AY372506 | 100 | Gabon | Mayengue et al. [72] |
Discussion
The aim of this study was to perform a molecular characterization of highly polymorphic markers in P. falciparum clinical isolates. In this first study, conducted after the introduction of artemisinin-based combination therapy (ACT), it was observed that P. falciparum field isolates in Cameroon exhibited a high degree of genetic polymorphism in P. falciparum msp-1 and msp-2 markers. These genes encoding for individual functional proteins expressed on the surface of the merozoite appear to play an essential role in the invasion of the red blood cell [39] and are of interest as potential vaccine candidates and as drug targets for inhibiting blood-stage replication [40]. This study identified a high proportion of multiclonal isolates and MOI and a total of 16 different Pfmsp-1 and 27 different Pfmsp-2 gene types. This figure is not exhaustive since it is obvious that with nested PCR genotyping some of the sub-populations present in mixed infections would not be fully typed by all amplification steps [21]. Moreover, more amplified product was found for the msp2 gene than msp1, this could be explained by non-synonymous substitutions introduced in template DNA that could jeopardize the proper annealing of the primer at its binding site in msp1 gene or could be explained by the fact that natural selection is more efficient when acting on msp-1 than msp-2 [39, 41]. This suggests that MSP-1 as compared with MSP-2 proteins are under strong functional constraints in a complex interaction with the host leading to an increased in host’s immunological response. The two markers including msp-1 (16 genotypes) and msp-2 (27 genotypes) revealed considerably greater parasite diversity than glurp (5 genotypes). Thus, the observed genetic polymorphism in the these two P. falciparum major merozoite surface proteins, Pfmsp1 and Pfmsp2 could be explained by balancing selection occurring as a result of different mechanisms of interaction with the host [42]. This distribution of families of msp-1 and msp-2 and their allelic variations were similar to that reported from other countries with meso- to high endemicity of malaria [43]. Genotyping procedure recommended in anti-malarial drug trials stipulates consecutive analysis of the three markers starting with msp2 or glurp, and then msp1 [23]. Based on allelic profile of each gene obtained in this study, parasitological outcome assessment could be more accurate when both markers msp-1 and msp-2 are included in the genotyping of recurrent parasitaemias in anti-malarial drug trials, consistent with other studies [21]. The results of this study equally raise concern over the use of glurp genotyping in anti-malarial drug trials since this marker showed limited allelic families as compared to msp-1 and msp-2. However, genotyping by SNPs and indels employing NGS could show more allelic families than identified by nPCR [44, 45].
Genetic diversity for msp-1 and msp-2 allelic families
The present study reported higher numbers of alleles (43 alleles for both MSP-1 and MSP-2) than previously reported to be circulating in the study area and in the central region of Cameroon [11, 24]. A peculiarity of this study is that, RO33 was found to be monomorphic and the most predominant allele type of msp1 compared to the polymorphic K1 and MAD20 allelic families in agreement with previous studies where RO33 was found to be the most predominant allelic family for msp1 locus [19, 35, 43]. However, the results of this study does not corroborate other studies in which RO33 was identified as the least predominant allelic variant type while MAD20 was the most predominant allelic family of the msp1 locus [16, 24]. This discrepancy could be attributed to the difference in the degree of transmission intensity. Another peculiarity of our study is that, the RO33 family of msp-1 did not show any polymorphism, with only 1 variant (155 bp) detected. This result differs from that of Gabon and West Uganda, where the Ro33 family was polymorphic with three and four allelic variants, respectively [8, 46], but was close to that in Senegal [47], and Brazil [48], showing the monomorphic nature of RO33 family of msp-1. The allelic variant K1 was the second highly distributed after RO33. This does not corroborate previous finding in the same region of Cameroon [24], but is consistent with most findings in areas of holoendemic, mesoendemic, and hyperendemic malaria, in which the allelic variant K1 was predominant [19]. The predominance of RO33 and K1 allelic family could be attributed to the balancing selection acting on these two allelic variants. The MAD20 allelic variant was the least predominant among the Pfmsp1 allelic family in agreement with other studies conducted Africa including the Gambia, Nigeria, and Gabon [35]. This result does not corroborate with other studies reporting the predominance of MAD20 allelic variant over K-1 and RO33 allelic variants [13, 19]. The low distribution of MAD20 allelic variant could be partly explained by purifying selection acting on MAD20 allelic type as compared with RO33 and K1 allelic type. This could equally be attributed to a single nucleotide substitution in DNA template that hinders proper annealing with primers designed to amplify the MAD20 allelic type [49]. An association between the distribution of K1 allelic families with severe malaria has been investigated [43], while the RO33 allelic family has been frequently reported in asymptomatic malaria cases [16, 50]. The predominance of RO33 seems to be less harmful for the host since the presence of this allele type is related to reduced risk of clinical malaria [43, 51].
A significant correlation has been established between the genetic diversity of the msp-1 gene of P. falciparum and parasite density. Likewise an association has been observed between msp-1 allele diversity and age group on one hand, and between msp-1 allele diversity and gender among asymptomatic patients on the other hand [52].
The msp-2 allelic families IC/3D7 and FC27 were almost of equal frequencies which is consistent with other findings [36]. In contrast, previous reports showed a significant predominance of FC27 over the 3D7/IC allelic family [16]. The frequencies of individual msp2 genotypes were low with 88.9% occurring at a frequency ≤ 10%. However 2 (16.7%) genotypes, belonging to the FC27 and 1 (6.7%) belonging to the 3D7/IC allelic family were found at frequencies of above 10%. High genetic diversity and low allelic frequencies have been reported previously from other sites including Gabon [8], Uganda [22], Senegal [53], and Burkina Faso [54].
Diversity, expressed as expected heterozygote (He), ranged from 0.55 for msp-1 to 0.96 for msp-2 suggesting that the parasite population in Cameroon, exhibits intermediate to higher heterozygosity reflecting intermediate to high transmission pattern [14] consistent with the findings in Uganda, Congo and Zimbabwe, showing a high heterozygote ranges between 0.78 and 0.8 for msp-2 [16, 55]. The correlation between the genetic variation of P. falciparum and malaria endemicity has been described [16, 46]. In areas with declining endemicity, it is reported that the number and diversity of alleles decrease with decreasing P. falciparum transmission [55]. The present study reveals that the msp-2 gene is highly polymorphic as compared with msp 1 gene. This observation is different in low transmission settings where high diversity has been recorded for msp-1 as compared with the msp-2 gene [13]. Hence, the high allelic diversity together with the low frequency of individual circulating alleles observed in the present study increase the discriminatory power of msp-1 and msp-2 to differentiate between recrudescence and re-infection. Thus, this study reinforce the importance for the genotyping of P. falciparum based on msp-1 and msp-2 in anti-malarial drug efficacy trials, to distinguish between re-infection as recrudescence and emphasize on the importance of implementing msp-1 and msp-2 genotyping in effective malaria management and malaria control strategies in Cameroon and in other endemic areas.
Polyclonal infection expressed as MOI values
In this study, most patients with P. falciparum infections were infected with multiple genetically distinct parasite variants with high level of polyclonal infections observed among msp-2 (88.6%) and msp-1 (33.8%) positive isolates. Such a pattern of parasite structure is typical among P. falciparum populations in areas of high transmission, where more than 10 variants can be routinely detected in an individual [41], and selection among these variants in the host is likely to play an important role in parasite diversity. In contrast, in areas of low transmission, such as in Asia or in Latin America, patients may have infections with as few as a single variant [17]. No data on MOI was available in Cameroon before the introduction of ACT. This renders it difficult to draw a conclusive statement on the impact of ACT on MOI. Nevertheless, in this study, the polyclonal infection expressed as the MOI values were heterogeneous across the different loci, and the mean MOI was highest for msp-2 than msp-1 in accordance with previous studies in neighbouring countries with high intensity of malaria transmission including Congo and Gabon [56]. Higher MOI in Cameroon can be the results of multiple infectious mosquito bites or transmission of genetically diverse sporozoite inoculum from a single mosquito bite. Genetically distinct malaria parasites in natural populations have an extremely high rate of genetic recombination during the sexual stages resulting in multiple strains being transmitted simultaneously [57]. Effective recombination of parasites and mutation occurring in several rounds of DNA replication cycles will likely continue to maintain this genetic diversity. During genetic recombination, novel combinations of alleles can be generated offering beneficial features to the parasite, as driven by positive selection enabling spread alleles through the population. Since the evolutionary selection of malaria occurs both within individual hosts and within populations, determining the number of strain in an infection might be an important indicator of transmission intensity [58]. Effective malaria control measures (ACT, distribution of ITNs) have successfully reduced malaria transmission in many hyperendemic regions of sub-Saharan Africa. After an intervention, the malaria parasite population structure and transmission rate in these regions is expected to become similar to the low transmission rates of the regions of Southeast Asia and South America. The declining malaria transmission, as a result of scaling up interventions, has been shown to affect the genetic diversity pattern and population structure of P. falciparum [18]. Therefore, the high MOI observed in this study reflects a high intensity of malaria transmission in Cameroon, despite several control strategies deployed at the health facilities and in the community. This is in agreement with previous findings that observed an increase of MOI with an increase in malaria endemicity and a low MOI for msp-1 and msp -2 correlated with a low intensity of malaria transmission [17].
Therefore, regular molecular epidemiological surveys need to be performed in order to monitor the genetic diversity of P. falciparum populations in different regions of Cameroon, and then correlate parasite genotypes to the disease phenotypes [52, 59]. Previous studies reported a reduced risk of clinical malaria associated with polyclonal infections and high rate of severe malaria in individual harbouring mono-infections and very common genotypes [60]. This study showed an increase in the mean MOI according to parasite density, but not according to the gender of patients which is consistent with previous studies showing significantly high MOI in patients with moderate to high transmission [57]. In the present study, most of the positive samples were from children aged 0–5 years, and this limited range of age constraint examining the correlation between the MOI and age. However, previous studies showed a pattern of greater MOI in older individuals than younger individuals reflecting more previous exposure to infection [61] while conflicting findings, indicated decreased MOI with age [62]. Thus, determining the MOI in endemic areas is very important since it can be used to predict clinical outcome and target population with higher attention.
Genotyping by gel electrophoresis and direct sequencing
In this study, the polymorphic surface antigens msp2, glurp, and msp1 genes for 304 isolates were successfully amplified by primary and nested PCR and genotyped using agarose gel electrophoresis of which 127 msp-1 and 297 msp-2 gene fragments with single band were selected for further characterization by direct sequencing for identity and MSA purposes, but not to determine the entire parasite population. In this study, the lengths of the repeat units, whose number varies between the different allelic variants, was taken into account to set bin width of 20 bp for msp-1 and msp2 then 50 bp for glurp [14]. However, variations in bin width [22] and the different fragment sizing methods need to be standardized to facilitate the comparison of data for a particular marker between studies. Moreover, genotyping using agarose gel electrophoresis only groups alleles of similar size, but cannot distinguish the presence/absence of SNPs across the gene markers. Length polymorphic markers could introduce bias during the amplification process as this method is known to preferentially amplify shorter fragments [37, 63]. Although, genotyping method using gel electrophoresis may face some confounding factors including the variability in the electrophoretic migration of a given DNA fragment, this genotyping method could be very important where alternative method are not available and is widely used for P. falciparum genotyping [27, 28, 36, 64, 65]. Capillary electrophoresis-PCR (CE-PCR) was not used in this study, which could increase the allele resolution in an agarose gel by determining differences between 2 and 3 bp sizes among bands [14]. Beside, capillary electrophoresis is recognized to have higher resolution power than gel electrophoresis in the ability to distinguish between allelic variants of amplified fragments [20]. However, PCR artefacts are a challenge for this method and MOI determination is often underestimated not only with gel electrophoresis but also with capillary electrophoresis analysis especially when genotyping strategy do not consider separate nPCRs for each allelic family [21]. In contrast to previous studies, a separate nPCR followed by direct sequencing was performed in the present study enabling accurate determination of the fragments sizes for msp1 and msp2 allelic families as well as sequence motifs and nucleotide differences. The development of single-nucleotide polymorphism-based (SNP) genotyping techniques and next-generation sequencing (NGS), might provide highly diverse haplotype markers with sufficient resolution to detect minority population in a mixed infection [44, 45]. Thus, next-generation sequencing technologies and genome-wide characterization is an alternative strategy to accurately analyse polyclonal infections although complete haplotype characterization of multiclonal infections remains a challenge due to PCR artefacts and sequencing errors [66]. Therefore, in order to accurately study the competition and selection between variants in mixed malaria infection, new tools, more sensitive to detect minority populations and quantitative for relative parasite population sizes have being developed including DADA2, PASEC, HaplotypR, SeekDeep among others [45].
Another strategy could be the combination of sequencing method and efficient computational tool for an effective characterization of allelic variants. A good number of software packages are being developed to analyse genome-wide SNP data of field isolates for the estimation of the presence of multiple genotypes, especially minor allele in multiclonal infections [67]. However, DNA electrophoresis followed by direct sequencing should be used where alternative method are not yet available while waiting for the implementation of newly developed methods for allele genotyping, especially in the surveillance of malaria transmission. This would provide a guideline to policy makers to redefine the malaria control strategies.
Sequence analysis of genetic polymorphism of msp-1 and msp-2
This study showed highly diverse nature of P. falciparum isolates of Cameroon in respect to length and sequence motifs. Sequencing and gene alignment confirmed the identity Pfmsp1 and Pfmsp2 polymorphisms. Thus, when performing the gene alignment, high similarity was observed between the peptides of Pfmsp1 and Pfmsp2 in Cameroon and those of other regions in Africa. However, from all peptides analysed, the region of the alignment corresponding K1 polymorphism had the highest similarity among all the species in the Pfmsp clade included in this study ranging from 93% to 99% homology with previously described polymorphism in isolate from Kenya and Tanzania. The MAD20 peptide sequence polymorphism was the second most conserved with 83% to 100% homology between P. falciparum isolates in Cameroon and those of other regions of Africa as well as with those of other regions of the world. Thus, the development of a vaccine based on K1 and MAD20 allelic variant could likely be effective in providing immune protection against malaria in those regions in Africa, although it is not yet known to what extent the high allelic diversity within the K1-like and MAD20-like allelic types is of immunological significance [42].
However, previous analysis indicated more serological variation among the allelic sequences of the K1-like compared to the MAD20-like type [31], and more effort has been made to incorporate the repeat sequence variation of the K1-like alleles in recombinant antigens towards design of a multivalent vaccine [41, 49] and more than 500 different msp1 block 2 allelic sequences has been described, providing a reference for molecular epidemiological studies and potentially for design of a multi-allelic vaccine [42]. Sequencing and immunological characterization of other allelic variants such as MAD20 for Pfmsp1, alongside with 3D7/IC for Pfmsp 2 should be conducted to obtain more useful information.
Conclusion
In this first study conducted 5 years after the introduction of ACT, the genetic diversity of P. falciparum isolates was investigated among children aged 6 months to 6 years. The present study shows that field isolates of South West Region of Cameroon were found to be mainly polyclonal with high MOI and highly diverse in respect to both msp-1 (block 2) and msp-2 (central repeat region, block 3). These markers appear to be highly polymorphic with low allelic frequencies as compared to glurp. This observation reinforced the value of msp-1 and msp-2 markers of P. falciparum for PCR correction of treatment outcomes in classifying recurrent post-treatment P. falciparum episodes as recrudescence or new-infections in drug clinical trials. This study lays emphasis on the use of both msp-1 and msp-2 genes in monitoring the trend of malaria epidemiology and the use of MOI as an important indicator in the evaluation of malaria control interventions. The high MOI observed in this study is an indication that malaria transmission remains high in Cameroon despite a large distribution of ACT and calls for intensifying control interventions. Besides, the findings reveal that population structure of P. falciparum isolates is identical in Cameroon as shown by presence of common allelic composition and the high level of identity among allelic sequences from Cameroonian isolates and that of other areas in Africa and in the world and that P. falciparum population is a mixture of different strains. A vaccine developed with K1 and MAD20 of Pfmsp1 allelic variant could be protective for Africa children, but this finding will require further genetic and immunological characterization. It is anticipated that clonal selection could not be uniform across the country considering the varied climate range observed in Cameroon moving from the northern region (with Sahel desert) to the south (with equatorial dense forest) corresponding to different malaria epidemiology. Therefore, it will be relevant to carry out studies on the genetic diversity of P. falciparum isolates from other regions of Cameroon with varied malaria epidemiology as well as longitudinal studies involving other malaria transmission related markers such as CSP to further understand the clonal fluctuations associated with transmission intensity.
Acknowledgements
Our profound gratitude to all participants, patients and CDC staff. We acknowledge the technical support of Mrs. Mbanga Christine who set up SPSS database for data entry and for statistical analysis.
Abbreviations
- ACT
Artemisinin-based combination therapy
- CDC
Cameroon Development Corporation
- EDTA
Ethylene diamine tetraacetic acid
- He
Expected heterozygosity
- MOI
Multiplicity of infection
- Pfmsp1
Plasmodium falciparum merozoite surface protein 1
- Pfmsp2
Plasmodium falciparum merozoite surface protein 2
- Glurp
Glutamate rich protein
- CSP
Circumsporozoite protein
- PCR
Polymerase chain reaction
- TBE
Tris-borate EDTA
Authors’ contributions
TNM and X-NZ designed and organized the study. X-NZ, J-HC and RM-S supervised the study. TNM, PF-G, XZ and J-HC carried out the field and laboratory work and preliminary data analysis. TNM drafted and wrote the manuscript. PF-G analysed data and wrote the manuscript. All authors read and approved the final manuscript.
Funding
This study received grant support from the Chinese National Science and Technology Major Project (Grant No. 2012ZX10004-220), China UK Global Health Support Programme (Grant No. GHSP-CS-OP1), and International Collaboration Project from the Ministry of Science and Technology (Grant No. 2010DF33970) as part of their program towards malaria control and malaria elimination. The funders had no role in study design of the trials or in the design of the analysis, data collection, decision to publish, or preparation of the manuscript. Funding was provided by CASTEP (Grant No. China-Africa science and technology program).
Availability of data and materials
All data generated and/or analysed during this study are included in this published article (and its additional files) and are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Parents or guardian were provided a written informed consent before enrollment in this study, in accordance with the principles of the World Health Assembly of 1975 on Ethics in Human experimentation and the Helsinki Declaration. All malaria patients received anti-malarial drugs free of charge. This study was approved by the Ethics Committees of the Cameroon Ministry of Public Health and by the provincial CDC authorities and registered in NIH U.S. National Library of Medicine, ClinicalTrials.gov on the 28/11/2016 at https://clinicaltrials.gov/ct2/show/NCT02974348 with the registration number NCT02974348.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.WHO. Guidelines for the treatment of malaria. 2nd ed. Geneva: World Health Organization; 2010. http://www.who.int/malaria/publications/atoz/9789241547925/en/index.html. [PubMed]
- 2.WHO . World malaria report. Geneva: World Health Organization; 2018. [Google Scholar]
- 3.Wanji S, Tanke T, Atanga SN, Ajonina C, Nicolas T, Fontenille D. Anopheles species of the Mount Cameroon region: biting habit, feeding behaviour and entomological inoculation rates. Trop Med Int Health. 2003;8:643–649. doi: 10.1046/j.1365-3156.2003.01070.x. [DOI] [PubMed] [Google Scholar]
- 4.Moyou-Somo R, Essomba P, Songue E, Tchoubou NN, Ntambo A, Ngo Hiol H, et al. A public private partnership to fight against malaria along the Chad-Cameroon pipeline corridor: I. Baseline data on socio-anthropological aspects, knowledge, attitudes and practices of the population concerning malaria. BMC Public Health. 2013;13:1023. doi: 10.1186/1471-2458-13-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boussougou-Sambe ST, Eyisap WE, Tasse GCT, Mandeng SE, Mbakop LR, Enyong P, et al. Insecticide susceptibility status of Anopheles gambiae (s.l.) in South-West Cameroon four years after long-lasting insecticidal net mass distribution. Parasit Vectors. 2018;11:391. doi: 10.1186/s13071-018-2979-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Metoh Njuabe T, Tahar R, Same-Ekobo A, Foumane Ngane V, Soula G, Basco LK. Molecular epidemiology of malaria in Cameroon. XXIX. Characterization of DHFR and drug resistance markers and efficacy of sulfadoxine–pyrimethamine monotherapy in children in Niete (HEVECAM) Sciences et Médecines d’Afrique. 2010;2:146–152. [Google Scholar]
- 7.Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, et al. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2009;361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aubouy A, Migot-Nabias F, Deloron P. Polymorphism in two merozoite surface proteins of Plasmodium falciparum isolates from Gabon. Malar J. 2003;2:12. doi: 10.1186/1475-2875-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel P, Bharti PK, Bansal D, Raman RK, Mohapatra PK, Sehgal R, et al. Genetic diversity and antibody responses against Plasmodium falciparum vaccine candidate genes from Chhattisgarh, Central India: implication for vaccine development. PLoS ONE. 2017;12:e0182674. doi: 10.1371/journal.pone.0182674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woehlbier U, Epp C, Kauth CW, Lutz R, Long CA, Coulibaly B, et al. Analysis of antibodies directed against merozoite surface protein 1 of the human malaria parasite Plasmodium falciparum. Infect Immun. 2006;74:1313–1322. doi: 10.1128/IAI.74.2.1313-1322.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Basco LK, Tahar R, Escalante A. Molecular epidemiology of malaria in Cameroon. XVIII. Polymorphisms of the Plasmodium falciparum merozoite surface antigen-2 gene in isolates from symptomatic patients. Am J Trop Med Hyg. 2004;70:238–244. doi: 10.4269/ajtmh.2004.70.238. [DOI] [PubMed] [Google Scholar]
- 12.Jamil KF, Supargiyono S, Syafruddin D, Pratama, Silvy S. Genetic diversity of merozoite surface protein-2 in Plasmodium falciparum isolates from Aceh province, Indonesia. In: IOP Conf. Series: Earth and Environmental Science, vol. 125. 2018. p. 012036.
- 13.Kang JM, Moon SU, Kim JY, Cho SH, Link K, et al. Genetic polymorphisms of merozoite surface protein-1 in Plasmodium falciparum field isolates from Myanmar. Malar J. 2010;9:131. doi: 10.1186/1475-2875-9-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mwingira F, Nkwengulila G, Schoepflin S, Sumari D, Beck H-P, Snounou G, et al. Plasmodium falciparum msp1, msp2 and glurp allele frequency and diversity in sub-Saharan Africa. Malar J. 2011;10:79. doi: 10.1186/1475-2875-10-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smythe JA, Coppel RL, Day KP, Martin RK, Oduola AMJ, Kemp DJ, Anders RF. Structural diversity in the Plasmodium falciparum merozoite surface antigen2. Proc Natl Acad Sci USA. 1991;88:1751–1755. doi: 10.1073/pnas.88.5.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Babiker HA, Lines J, Hill WG, Walliker D. Population structure of Plasmodium falciparum in villages with different malaria endemicity in east Africa. Am J Trop Med Hyg. 1997;56:141–147. doi: 10.4269/ajtmh.1997.56.141. [DOI] [PubMed] [Google Scholar]
- 17.Atroosh M, Al-Mekhlafi HM, Mahdy MAK, Saif-Ali R, Al-Mekhlaf AM, Surin J. Genetic diversity of Plasmodium falciparum isolates from Pahang, Malaysia based on MSP-1 and MSP-2 genes. Parasit Vectors. 2011;4:233. doi: 10.1186/1756-3305-4-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khaireh BA, Assefa A, Guessod HH, Basco LK, Khaireh MA, Pascual A, et al. Population genetics analysis during the elimination process of Plasmodium falciparum in Djibouti. Malar J. 2013;12:201. doi: 10.1186/1475-2875-12-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Snounou G. Genotyping of Plasmodium spp. nested PCR. Methods Mol Med. 2002;72:103–116. doi: 10.1385/1-59259-271-6:103. [DOI] [PubMed] [Google Scholar]
- 20.Gupta V, Dorsey G, Hubbard AE, Rosenthal PJ, Greenhouse B. Gel versus capillary electrophoresis genotyping for categorizing treatment outcomes in two anti-malarial trials in Uganda. Malar J. 2010;9:19. doi: 10.1186/1475-2875-9-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Messerli C, Hofmann NE, Beck H-P, Felger I. Critical evaluation of molecular monitoring in malaria drug efficacy trials and pitfalls of length-polymorphic markers. Antimicrob Agents Chemother. 2017;61:e01500–e01516. doi: 10.1128/AAC.01500-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cattamanchi A, Kyabayinze D, Hubbard A, Rosenthal PJ, Dorsey G. Distinguishing recrudescence from reinfection in a longitudinal antimalarial drug efficacy study: comparison of results based on genotyping of msp-1, msp-2, and glurp. Am J Trop Med Hyg. 2003;68:133–139. doi: 10.4269/ajtmh.2003.68.133. [DOI] [PubMed] [Google Scholar]
- 23.WHO. Methods and techniques for clinical trials on antimalarial drug efficacy: genotyping to identify parasite populations: informal consultation organized by the Medicines for Malaria Venture and co-sponsored by the World Health Organization. 2008.
- 24.Wanji S, Kengne-Ouafo AJ, Eyong EJ, Kimbi HK, Tendongfor N, Ndamukong-Nyanga JL. Genetic diversity of Plasmodium falciparum merozoite surface protein-1 block 2 in sites of contrasting altitudes and malaria endemicities in the Mount Cameroon region. Am J Trop Med Hyg. 2012;86:764–774. doi: 10.4269/ajtmh.2012.11-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sepúlveda N, Drakeley C. Sample size determination for estimating antibody seroconversion rate under stable malaria transmission intensity. Malar J. 2015;14:141. doi: 10.1186/s12936-015-0661-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.WHO . QA manual for malaria microscopy. 2. Geneva: World Health Organization; 2015. [Google Scholar]
- 27.Sondo P, Derra K, Lefevre T, Diallo-Nakanabo S, Tarnagda Z, Zampa O, et al. Genetically diverse Plasmodium falciparum infections, within-host competition and symptomatic malaria in humans. Sci Rep. 2019;9:127. doi: 10.1038/s41598-018-36493-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Atroosh WM, Al-Mekhlafi HM, Al-Jasari A, Sady H, Al-Delaimy AK, Nasr NA, et al. Genetic variation of pfhrp2 in Plasmodium falciparum isolates from Yemen and the performance of HRP2-based malaria rapid diagnostic test. Parasit Vectors. 2015;8:388. doi: 10.1186/s13071-015-1008-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joshi H, Valecha N, Verma A, Kaul A, Mallick PK, Shalini S, et al. Genetic structure of Plasmodium falciparum field isolates in eastern and north-eastern India. Malar J. 2007;6:60. doi: 10.1186/1475-2875-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nie NH, Bent DH, Hull CH. SPSS: statistical package for the social sciences: update manual. New York: National Opinion Research Center, McGraw-Hill; 2008. p. 343. [Google Scholar]
- 31.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noranate N, Prugnolle F, Jouin H, Tall A, Marrama L, Sokhna C, et al. Population diversity and antibody selective pressure to Plasmodium falciparum MSP1 block2 locus in an African malaria-endemic setting. BMC Microbiol. 2009;9:219. doi: 10.1186/1471-2180-9-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takala SL, Smith DL, Stine OC, Coulibaly D, Thera MA, Doumbo OK, et al. A high-throughput method for quantifying alleles and haplotypes of the malaria vaccine candidate Plasmodium falciparum merozoite surface protein-1 19 kDa. Malar J. 2006;5:31. doi: 10.1186/1475-2875-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang G, Liu R, Daubenberger CA, Pluschke G. Sequence analysis of the MSP 1 gene of Plasmodium falciparum isolates from Hainan, China. Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi. 1999;17:294–297. [PubMed] [Google Scholar]
- 35.Ferreira MU, Ribeiro WL, Tonon AP, Kawamoto F, Rich SM. Sequence diversity and evolution of the malaria vaccine candidate merozoite surface protein-1 (MSP-1) of Plasmodium falciparum. Gene. 2003;304:65–75. doi: 10.1016/S0378-1119(02)01180-0. [DOI] [PubMed] [Google Scholar]
- 36.Zakeri S, Bereczky S, Naimi P, Pedro Gil J, Djadid ND, Farnert A, et al. Multiple genotypes of the merozoite surface proteins 1 and 2 in Plasmodium falciparum infections in a hypoendemic area in Iran. Trop Med Int Health. 2005;10:1060–1064. doi: 10.1111/j.1365-3156.2005.01477.x. [DOI] [PubMed] [Google Scholar]
- 37.Cavanagh DR, Elhassan IM, Roper C, Robinson VJ, Giha H, Holder AA, et al. A longitudinal study of type specific antibody responses to Plasmodium falciparum merozoite surface protein-1 in an area of unstable malaria in Sudan. J Immunol. 1998;161:347–359. [PubMed] [Google Scholar]
- 38.Dobaño C, Khan A, Robinson JV, Taylor RR, McBride JS. Identical alleles of Plasmodium falciparum merozoite surface protein 2 in distant geographic areas and times. Parasitol Int. 1997;46:137–142. doi: 10.1016/S1383-5769(97)00020-2. [DOI] [Google Scholar]
- 39.Escalante AA, Lal AA, Ayala FJ. Genetic polymorphism and natural selection in the malaria parasite Plasmodium falciparum. Genetics. 1998;149:189–202. doi: 10.1093/genetics/149.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tetteh KK, Stewart LB, Ochola LI, Amambua-Ngwa A, Thomas AW, Marsh K, et al. Prospective identification of malaria parasite genes under balancing selection. PLoS ONE. 2009;4:e5568. doi: 10.1371/journal.pone.0005568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tetteh KK, Cavanagh DR, Corran P, Musonda R, McBride JS, Conway DJ. Extensive antigenic polymorphism within the repeat sequence of the Plasmodium falciparum merozoite surface protein 1 block 2 is incorporated in a minimal polyvalent immunogen. Infect Immun. 2005;73:5928–5935. doi: 10.1128/IAI.73.9.5928-5935.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aspeling-Jones H, Conway DJ. An expanded global inventory of allelic variation in the most extremely polymorphic region of Plasmodium falciparum merozoite surface protein 1 provided by short read sequence data. Malar J. 2018;17:345. doi: 10.1186/s12936-018-2475-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kun FJ, Schmidt-Ott RJ, Lehman LG, Lell B, Luckner D, Greve B, et al. Merozoite surface antigen 1 and 2 genotypes and rosetting of Plasmodium falciparum in severe and mild malaria in Lambarene, Gabon. Trans R Soc Trop Med Hyg. 1998;92:110–114. doi: 10.1016/S0035-9203(98)90979-8. [DOI] [PubMed] [Google Scholar]
- 44.Daniels R, Volkman SK, Milner DA, Mahesh N, Neafsey DE, et al. Genetic diversity and protective efficacy of the RTS,S/AS01 malaria vaccine. N Engl J Med. 2015;373:2025–2037. doi: 10.1056/NEJMoa1505819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhong D, Koepfi C, Cui L, Yan G. Molecular approaches to determine the multiplicity of Plasmodium infections. Malar J. 2018;17:172. doi: 10.1186/s12936-018-2322-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peyerl-Hoffmann G, Jelinek T, Kilian A, Kabagambe G, Metzger WG, von Sonnenburg F. Genetic diversity of Plasmodium falciparum and its relationship to parasite density in an area with different malaria endemicities in West Uganda. Trop Med Int Health. 2001;6:607–613. doi: 10.1046/j.1365-3156.2001.00761.x. [DOI] [PubMed] [Google Scholar]
- 47.Zwetyenga J, Rogier C. No influence of age on infection complexity allelic distribution in Plasmodium falciparum infection in Ndiop, a Senegalese village with seasonal, mesoendemic malaria. Am J Trop Med Hyg. 1998;59:726–735. doi: 10.4269/ajtmh.1998.59.726. [DOI] [PubMed] [Google Scholar]
- 48.Sallenave-Sales S, Daubersies P, Mercereau-Puijalon O, Rahimalala L, Contamin H, Druilhe P, et al. Plasmodium falciparum, A comparative analysis of the genetic diversity in malaria-mesoendemic area of Brazil and Madagascar. Parasitol Res. 2000;86:692–698. doi: 10.1007/PL00008554. [DOI] [PubMed] [Google Scholar]
- 49.Tetteh KK, Conway DJ. A polyvalent hybrid protein elicits antibodies against the diverse allelic types of block 2 in Plasmodium falciparum merozoite surface protein 1. Vaccine. 2011;29:7811–7817. doi: 10.1016/j.vaccine.2011.07.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Amodu OK, Adeyemo AA, Ayoola OO, Gbadegesin RA, Orimadegun AE, Akinsola AK, et al. Genetic diversity of the msp-1 locus and symptomatic malaria in south-west Nigeria. Acta Trop. 2005;95:226–232. doi: 10.1016/j.actatropica.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 51.Al-Yaman F, Genton B, Reeder JC, Anders RF, Smith T, Alpers MP. Reduced risk of clinical malaria in children infected with multiple clones of Plasmodium falciparum in a highly endemic area: a prospective community study. Trans R Soc Trop Med Hyg. 1997;91:602–605. doi: 10.1016/S0035-9203(97)90046-8. [DOI] [PubMed] [Google Scholar]
- 52.Chaorattanakawee S, Nuchnoi P, Hananantachai H, Tumkosit U, Saunders D, Naka I, et al. Correction: sequence variation in Plasmodium falciparum merozoite surface protein-2 is associated with virulence causing severe and cerebral malaria. PLoS ONE. 2018;13:e0196694. doi: 10.1371/journal.pone.0196694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Henry M, Bordes J, Diallo I, Pradines B, Ka S, Diatta B, et al. Urban malaria in Dakar, Senegal: chemosusceptibility and genetic diversity of Plasmodium falciparum isolates. Am J Trop Med Hyg. 2006;75:146–151. doi: 10.4269/ajtmh.2006.75.146. [DOI] [PubMed] [Google Scholar]
- 54.Soulama I, Nébié I, Ouédraogo A, Gansane A, Diarra A, Tiono AB, et al. Plasmodium falciparum genotypes diversity in symptomatic malaria of children living in an urban and a rural setting in Burkina Faso. Malar J. 2009;8:135. doi: 10.1186/1475-2875-8-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anderson TJHB, Williams JT, Estrada-Franco JG, Richardson L, Mollinedo R, Bockarie M, et al. Microsatellites reveal a spectrum of population structure in the malaria parasite Plasmodium falciparum. Mol Biol Evol. 2000;17:1467–1482. doi: 10.1093/oxfordjournals.molbev.a026247. [DOI] [PubMed] [Google Scholar]
- 56.Ibara-Okabande R, Koukouikila-Koussounda F, Ndounga M. Reduction of multiplicity of infections but no change in msp2 genetic diversity in P. falciparum isolates from Congolese children after introduction of artemisin combination therapies. Malar J. 2012;11(Suppl 1):P70. doi: 10.1186/1475-2875-11-S1-P70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Talisuna AO, Okello PE, Erhart A, Coosemans M, D’Alessandro U. Intensity of malaria transmission and the spread of Plasmodium falciparum resistant malaria: a review of epidemiologic field evidence. Am J Trop Med Hyg. 2007;77(6 suppl):170–180. [PubMed] [Google Scholar]
- 58.Jiang H, Li N, Gopalan V, Zilversmit MM, Varma S, Nagarajan V, et al. High recombination rates and hotspots in a Plasmodium falciparum genetic cross. Genome Biol. 2011;12:R33. doi: 10.1186/gb-2011-12-4-r33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Somé AF, Bazié T, Zongo I, Yerbanga RS, Nikiéma F, Neya C, et al. Plasmodium falciparum msp1 and msp2 genetic diversity and allele frequencies in parasites isolated from symptomatic malaria patients in Bobo-Dioulasso, Burkina Faso. Parasit Vectors. 2018;11:323. doi: 10.1186/s13071-018-2895-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Durand R, Ariey F, Cojean S, Fontanet A, Ranaivo L, Ranarivelo LA, et al. Analysis of circulating populations of Plasmodium falciparum in mild and severe malaria in two diferent epidemiological patterns in Madagascar. Trop Med Int Health. 2008;13:1392–1399. doi: 10.1111/j.1365-3156.2008.02156.x. [DOI] [PubMed] [Google Scholar]
- 61.Takala SL, Coulibaly D, Thera MA, Dicko A, Smith DL, Guindo AB, et al. Dynamics of polymorphism in a malaria vaccine antigen at a vaccine-testing site in Mali. PLoS Med. 2007;4:e93. doi: 10.1371/journal.pmed.0040093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mayor A, Saute F, Aponte JJ, Almeda J, Gómez-Olivé FX, Dgedge M, et al. Plasmodium falciparum multiple infections in Mozambique, its relation to other malariological indices and to prospective risk of malaria morbidity. Trop Med Int Health. 2003;8:3–11. doi: 10.1046/j.1365-3156.2003.00968.x. [DOI] [PubMed] [Google Scholar]
- 63.Walsh PS, Erlich HA, Higuchi R. Preferential PCR amplification of alleles: mechanisms and solutions. PCR Methods Appl. 1992;1:241–250. doi: 10.1101/gr.1.4.241. [DOI] [PubMed] [Google Scholar]
- 64.Yavo W, Konaté A, Mawili-Mboumba DP, Kassi FK, Mbuyi MLT, Angora EK, et al. Genetic polymorphism of msp1 and msp2 in Plasmodium falciparum isolates from Côte d’Ivoire versus Gabon. J Parasitol Res. 2016;2016:3074803. doi: 10.1155/2016/3074803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mohammed H, Kassa M, Mekete K, Assefa A, Taye G, Commons RJ. Genetic diversity of the msp-1, msp-2, and glurp genes of Plasmodium falciparum isolates in Northwest Ethiopia. Malar J. 2018;17:386. doi: 10.1186/s12936-018-2540-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Acinas SG, Sarma-Rupavtarm R, Klepac-Ceraj V, Polz MF. PCR-induced sequence artifacts and bias: insights from comparison of two 16S rRNA clone libraries constructed from the same sample. Appl Environ Microbiol. 2005;71:8966–8969. doi: 10.1128/AEM.71.12.8966-8969.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhu SJ, Almagro-Garcia J, McVean G. Deconvolution of multiple infections in Plasmodium falciparum from high throughput sequencing data. Bioinformatics. 2018;34:9–15. doi: 10.1093/bioinformatics/btx530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jiang G, Daubenberger C, Huber W, Matile H, Tanner M, Pluschke G. Sequence diversity of the merozoite surface protein 1 of Plasmodium falciparum in clinical isolates from the Kilombero District, Tanzania. Acta Trop. 2000;74:51–61. doi: 10.1016/S0001-706X(99)00045-5. [DOI] [PubMed] [Google Scholar]
- 69.Jongwutiwes S, Tanabe K, Nakazawa S, Yanagi T, Kanbara H. Sequence variation in the tripeptide repeats and T cell epitopes in P190 (MSA-1) of Plasmodium falciparum from field isolates. Mol Biochem Parasitol. 1992;51:81–89. doi: 10.1016/0166-6851(92)90203-V. [DOI] [PubMed] [Google Scholar]
- 70.Scopel KK, et al. Low sensitivity of nested PCR using Plasmodium DNA extracted from stained thick blood smears: an epidemiological retrospective study among subjects with low parasitae-mia in an endemic area of the Brazilian Amazon region. Malar J. 2004;31:8. doi: 10.1186/1475-2875-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weisman S, Wang L, Billman-Jacobe H, Nhan DH, et al. Antibody responses to infections with strains of Plasmodium falciparum expressing diverse forms of merozoite surface protein 2. Infect Immun. 2001;69:959–967. doi: 10.1128/IAI.69.2.959-967.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mayengue PI, Luty AJ, Rogier C, Baragatti M, Kremsner PG, Ntoumi F. The multiplicity of Plasmodium falciparum infections is associated with acquired immunity to asexual blood stage antigens. Micro Infect. 2009;11(1):108–114. doi: 10.1016/j.micinf.2008.10.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated and/or analysed during this study are included in this published article (and its additional files) and are available from the corresponding author on reasonable request.